Abstract
Key message
We found GmNAC06 plays an important role in salt stress responses through the phenotypic, physiological and molecular analyses of OE, VC, and Mutant composite soybean.
Abstract
Salinization affects 20% of all cultivated land worldwide because of the high salinity of irrigation water and the excessive use of water, and this amount is increasing daily. NAC (NAM, ATAF, and CUC) have been found to be involved in salt stress. In this study, a soybean NAC gene, GmNAC06 (Glyma06g21020.1), was cloned and functionally characterized. The results of expression analysis suggested that salt stress could influence the expression level of GmNAC06. The subcellular localization analysis results suggested that GmNAC06 may function as a transcription factor. Under salt stress, the overexpression technology combined with CRISPR-Cas9 system found that GmNAC06 could cause the accumulation of proline and glycine betaine to alleviate or avoid the negative effects of ROS; similarly, it could control the Na+/K+ ratios in hairy roots to maintain ionic homeostasis. The fresh weight of the transgenic hairy roots and the histochemical ROS staining of wild leaves suggested that transgenic hairy roots influence the function of wild leaves under salt stress conditions. Moreover, the expression levels of GmUBC2 and GmHKT1 were higher in the GmNAC06 hairy roots than in the control. Thus, the overexpression of GmNAC06 in hairy roots notably causes an entire composite plant to exhibit salt tolerance. The phenotype of composite soybean plants and transgenic Arabidopsis plants suggest that GmNAC06 plays a role in response to salt stress and could be useful in generating salt tolerant transgenic crops.
Electronic supplementary material
The online version of this article (10.1007/s11103-020-01091-y) contains supplementary material, which is available to authorized users.
Keywords: Soybean, NAC, Hairy roots, CRISPR-Cas9, Salt tolerance
Introduction
Soybean (Glycine max) is the fourth largest crop in the world. It is an important cash crop for food, fuel and feed, and it has been used as a raw material in human health and industrial products. Therefore, the global demand for soybean is increasing gradually. Soybean production is affected by biotic and abiotic stresses, such as inadequate water supplies, high salinity, and low temperatures. Salinity is a major environmental factor limiting the growth and yield of soybean, and it has a negative influence at the cellular, organ and even whole-plant levels. The growth response to salt stress has two phases (Munns and Tester 2008). During the first, rapid phase, plant growth slows because of osmotic responses to the salt outside the roots, so it is more difficult for the roots to obtain water. Due to the decreased water absorption capacity of roots, the water loss from leaves is accelerated; therefore, salinity stress is also considered hyperosmotic stress (Munns 2005). Next, the growth of the shoots is limited. In addition, the stomata close to protect the plant against the ion flow. Finally, the growth of the young leaves slows. During the second, slower phase, a further decrease in plant growth is observed due to the toxic contents of salt inside the plants. Plants, on the basis of adaptive evolution, can be roughly classified into two major types: halophytes (that can withstand high salinity) and glycophytes (that cannot withstand high salinity and eventually die under saline conditions). Soybean belongs to the second category. Therefore, it is of great significance to study the mechanism of salt tolerance in soybean molecular breeding.
The NAC protein family is one of the largest families of TFs in plants; for example, there are 117 NAC genes in Arabidopsis thaliana (Nuruzzaman et al. 2010), 152 in tobacco (Rushton et al., 2008), 151 in rice (Nuruzzaman et al. 2010), 152 in maize (Shiriga et al. 2014), and 152 in soybean (Le et al. 2011). NAC proteins consist of two parts. The NAC domain at the N-terminus is conserved, contains 150–160 amino acids and is divided into five subdomains (A–E) (Ooka et al. 2003). Every subdomain has its own function; subdomain A may be responsible for the formation of a functional dimer, subdomains B and E may be involved in the functional diversity of NAC genes, and subdomains C and D are used to bind to DNA (Puranik et al. 2012). The second part is a highly divergent C-terminus transcription regulatory (TR) region that acts as a transcriptional activator or repressor and has frequent occurrences of simple amino acid repeats and regions rich in serine and threonine, proline and glutamine, or acidic residues (Olsen et al. 2005; Puranik et al. 2012). NAC proteins are plant-specific TFs that are relevant to the development of plants (Shen et al. 2009). NTM1 regulates cell division (Kim et al. 2006). OsNAC05 influences seed development (Sperotto et al. 2009). The Arabidopsis NAC TF ANAC92 has been shown to control senescence (Balazadeh et al. 2010). GmNAC20 is involved in the formation of the lateral root (Hao et al. 2011). In addition, numerous NAC genes are involved in responses to abiotic stresses, such as salinity, drought and cold. OsNAC45 may enhance the salt tolerance of transgenic rice (Zheng et al. 2009). The overexpression of ZmSNAC01 may enhance tolerance to drought in Arabidopsis (Lu et al. 2012). TaNAC02 increases the cold tolerance of Arabidopsis (Mao et al. 2012).
Because of the high salinity of irrigation water and the excessive use of water, more than 800 million hectares of land are affected by salinization globally (Krishnamurthy et al. 2019). Sodium chloride is soluble and widespread, so roots may take up many Na+ and Cl+, ions that negatively affect the metabolic processes and photosynthetic efficiency of plants (Mäser et al. 2002). The root is the first plant organ to experience salt damage in saline soil, so it is important to study the salt tolerance of roots. Although the transgenic soybean has been cultivated at a large scale in the world, its genetic transformation still presents difficulties. Hinchee et al. (1988) obtained transgenic soybean plants using the Agrobacterium tumefaciens-mediated DNA transfer. Subsequently, some laboratories have studied A. tumefaciens-mediated plant transformation, but few bacterial strains are widely used. Through A. tumefaciens-mediated transformation of soybean has been greatly improved over the years (Chen et al. 2018). However, it still presents several unresolved problems such as low transformation efficiency, complex procedure, susceptibility to contamination, and a long time period (Pareddy et al. 2020). However, genetic transformation technology of hairy root can solve these problems. The A. rhizogenes cucumopine strain K599 (pRi2659) was isolated and characterized because of an outbreak of hairy root disease in cucumber in the 1970s (Weller et al. 2004; Mankin et al. 2007). After K599 is injected into the cotyledonary node, it may effectively induce hairy roots at the infection site and form composite soybean plants consisting of wild-type shoots and transgenic roots. There are many advantages to the A. rhizogenes—mediated transformation system; for example, the experimental procedure is simple, the experiment period is short, and it can be used to study root specific genes. In addition, the transformation efficiency is high. Estrada-Navarrete et al. (2006) tested four strains of A. rhizogenes to induce hairy roots on four species of Phaseolus and found that K599 was the most effective, with transformation frequencies up to 90%. Kereszt et al. (2007) reported that they had induced hairy roots in soybean with K599, with transformation frequencies of 25–80%. Therefore, A. rhizogenes could be used as a tool in functional genomics. Curtin et al. (2011) studied targeted mutations induced through zinc-finger nucleases in soybean hairy roots. Cao et al. (2011) reported that the overexpression of TaNHX2 in hairy roots enhanced the salt tolerance of the composite soybean. Wang et al. (2015) found that GmWRKY27 improved salt and drought tolerance in transgenic soybean hairy roots through overexpression and RNAi analysis. Hairy roots were also used to study targeted mutagenesis caused by the CRISPR-Cas9 system (Sun et al. 2015).
To elucidate the role of GmNAC06 in soybean plants under salt stress, we constructed composite plants though overexpression and CRISPR-Cas9 technology. The physiological parameters, molecular experiment and phenotypic analysis suggested that the overexpression of GmNAC06 in hairy roots enhances the salt tolerance of the composite plants. The transgenic Arabidopsis showed a similar result. However, the salt tolerance of the composite plants edited by the CRISPR-Cas9 system decreased. Based on the results, it is suggested that GmNAC06 is an ideal candidate gene for enhancing salt tolerance of soybean.
Materials and methods
Subcellular localization and transactivation activity analysis
The ORF of GmNAC06 with a mutational stop codon was cloned upstream of the green fluorescent protein (GFP) in a 16318hGFP vector with the Quick-Fusion Cloning Kit (biotool.cn, Basel, Switzerland) and a pair of primers (Table S1). The recombinant 35S::GmNAC06:GFP and a control plasmid 35S::GFP (10 μg) were transformed into Arabidopsis leaf protoplasts as described previously (Wang et al. 2017). 158 amino acids in the N-terminal of AtBZR2 (AT1G19350.3) contained a nuclear localization signal (NLS), which were fused with mCherry as nuclear marker (Zhang et al. 2016). After 18 h, the GFP fluorescence was observed under a laser scanning confocal microscope (Olympus FV1000 viewer, Tokyo, Japan).
To perform the transactivation activity in yeast cells, the ORF of GmNAC06 was cloned into pGBKT7 with the Quick-Fusion Cloning Kit and a pair of primers (Table S1). pGBKT7-AtDREB2A was the positive control, while the pGBKT7 empty vector was the negative control (Sakuma et al. 2006). The Yeastmaker™ Yeast Transformation System 2 (Clontech, United States) was used for yeast transformation.
Construction of soybean and Arabidopsis transgenic plants
The TRIzol reagent (Invitrogen, Carlsbad, United States) was used to isolate Total RNA from soybean (Glycine max L. Merr.) cultivar Williams 82. According to the instructions, genomic DNA was removed by DNase I (Thermo Scientific, Waltham, United States). The first-strand complementary DNA (cDNA) synthesis was performed with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, United States). The primers (Table S1) and Pfu DNA polymerase (TransGen Biotech, Beijing, China) were used to get 1074 bp full-length open reading frames (ORF) of GmNAC06. The products were purified and integrated into the blunt vector (pEASY-Blunt Simple Cloning Kit, Beijing, China) for sequencing. GmNAC06 was cloned into pCAMBIA3301 with the Quick-Fusion Cloning Kit and a pair of primers (Table S1). pUC57-GmU6-10-sgRNA and pCambia3301-Cas9 were used to construct pCas9-GmU6-sgRNA recombinant vector as previously described (Sun et al. 2015). After sequence verification, the recombinant plasmid and pCAMBIA3301 were transformed into A. rhizogenes K599 as overexpression (OE), vector control (VC) and Mutant respectively.
The soybean seeds were placed into identical pots containing mixed soil (humus: vermiculite = 2:1) at a depth of 1–2 cm. The seeds were placed in a greenhouse at 28 °C and watered daily. The 6-day-old seedlings with folded cotyledons (Fig. S3a) were infected by A. rhizogenes strain K599 with the OE, VC and Mutant vector around cotyledonary node area using syringe needle (Fig. S3b and c). The soybean was kept in a 12 h light/12 h dark cycle at 28 °C. After infection, seedlings were covered with a transparent lid (Fig. S3d). After the initiation of the hairy root formation from the wounding sites, the infection sites and parts below it were covered by vermiculite to maintain a high humidity (Fig. S3e). Four weeks later, when the hairy roots were approximately 5–10 cm and could support the plant (Fig. S3f and g), the main roots were removed (Fig. S3h). The OE hairy roots were screened by RT-PCR and qRT-PCR (Fig. S4), the VC hairy roots were screened by PCR analysis of the GUS gene (Fig. S5), T7 endonuclease 1 (T7E1) was used to detect the PCR products of the Mutant hairy roots (Fig. S6), and the original roots were removed from the composite plants. The plants with hairy roots were transferred into mixed soil (humus: vermiculite = 2:1) and watered every 3 days (Fig. S3i).
The overexpression vector and pCAMBIA3301 were transformed into A. tumefaciens strain GV3101 as the overexpression (OE) and vector control (VC) and then into wild-type (WT) Arabidopsis Columbia ecotype by the floral dip method (Clough and Bent 1998). The phosphinothricin resistance and PCR were used to screen seeds (Fig. S7a and b). Homozygous T3-generation plants were used for further analyses.
Stress treatments and quantitative reverse transcription PCR (qRT-PCR) analysis
Williams 82 was used to analyze tissue-specific expression. RNA was extracted from roots, cotyledons, stems and leaves of 20-day-old soybean plants. The RNA of flowers was isolated from mature plants. The seeds were germinated in pots containing vermiculite in a greenhouse at 28 °C with a 12 h light/12 h dark cycle and 50% relative humidity. The roots of the 20-day-old seedlings were immersed in Hoagland solutions embodying 250 mM NaCl (salt treatment), 20% polyethylene glycol (PEG) (dehydration treatment) and 100 μM ABA (ABA treatment). The 20-day-old seedlings were placed at 4 °C for the the cold treatment. RNA was isolated from the roots and leaves at 0.5 h, 2 h, 6 h, 12 h and 24 h after treatment.
cDNA samples, Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific, Waltham, United States), the primers (Table S1) and the Eco Real-Time PCR system (Illumina, San Diego, CA, USA) were used to perform qRT-PCR. All experiments were repeated three times. The data were analyzed with the comparative CT method (2−△△CT method) (Livak and Schmittgen 2001). cDNA samples of GmNAC06 from the OE and VC hairy roots were used to examine the expression level of 14 salt stress-related genes by qRT-PCR (Table S1). The expression level of 2 salt stress-related genes in the OE, VC and WT Arabidopsis were examined by qRT-PCR (Shkolnik et al. 2013; Sun et al. 2020). The soybean CYP2 gene and Arabidopsis UBQ3 gene were used as a reference for normalization (Ni et al. 2013).
After 6 weeks, when the hairy roots had recovered enough healthy, the OE, VC and Mutant composite plants were irrigated with 250 mM NaCl solution. Two days later, the leaves and hairy roots were used to analyze the function of GmNAC06. After 2 weeks, thirty OE, thirty VC, thirty Mutant composite plants under normal conditions and same number under salt stress were used to analyze root growth phenotype. After the salt treatment, thirty OE, thirty VC and thirty Mutant composite plants were used to determine the salt damage index (SDI%) for 2 consecutive weeks (Cao et al. 2011).
Seeds of the VC (VC3), OE6, OE9 and OE17 plants of the homozygous T3 generation were surface-sterilized and planted on MS medium without NaCl or with 100 mM NaCl. After 3 days of vernalization at 4 °C, the percentage germination (%) was measured for 1 consecutive week. Each sample contained 36 seedlings, and the experiments were replicated three times. To evaluate the root growth under normal conditions and the 100 mM NaCl treatment, 5-day-old seedlings with roots of nearly equal length were placed vertically in a growth chamber. After 7 days of incubation, the root elongation lengths of 30 seedlings were determined. For the salt tolerance assay, the homozygous T3 generation was germinated soil chambers in a greenhouse at 22 °C with a 16 h light/8 h dark cycle and 70% relative humidity, and the 4-week-old potted Arabidopsis plants were subjected to a 250 mM NaCl treatment. After 2 weeks, the survival rates were scored. Each sample contained 12 seedlings, and the experiments were replicated three times.
Histochemical and physiological analysis
Under salt stress, some plants can generate hydrogen peroxide (H2O2) and superoxide () to mediate numerous physiological and biochemical processes. H2O2 and were visually detected with 3,3′-diaminobenzidine (DAB) (Beijing Biodee Biotechnology, Beijing, China) and nitro blue tetrazolium (NBT) (Beijing Biodee Biotechnology, Beijing, China). The leaves of the OE, VC and Mutant composite plants were infiltrated with 5 mg/ml DAB at pH 3.8 for 20 h and 0.5 mg/ml NBT at pH 7.5 for 20 h in the dark to detect H2O2 and . Then, the leaves were transferred to ethanol (glycerol: 95% ethanol = 3:7) and boiled for fifteen minutes in a water bath. After cooling, the leaves were extracted at room temperature with ethanol (glycerol: 95% ethanol = 3:7) until the leaves were decolorized completely (Kong et al. 2011). The programmed cell death was analyzed by trypan blue (Huang et al. 2011). Trypan blue can color dead cells into blue; however, living cells are not stained. The leaves of the OE, VC and Mutant composite plants were soaked in 0.4% trypan blue solution (MYM Biological Technology Company Limited, Chicago, IL, USA), boiled for two minutes in a water bath, and then incubated for 8 h. The leaves were transferred to 1.25 g/ml chloral hydrate solution (MYM Biological Technology Company Limited, Chicago, IL, USA) for fading for 3 days, and the chloral hydrate solution was changed one time every day.
For measuring H2O2 content (Liu et al. 2010), O2− production rates (Elstner and Heupel 1976), MDA content (Puckette et al. 2007), electrolyte leakage (Zhao et al. 2004), glycine betaine content (Li et al. 2017) and proline content (Pinedo et al. 2015), leaf and hairy root sample were taken from three different plants and experiments were replicated three times. The Na+ and K+ contents were measured by using an inductively coupled plasma optical emission spectrometer (ICP-OES, United States).
The chlorophyll content was measured in the leaves of the OE and VC Arabidopsis plants before the salt stress treatment and 3 days after the salt stress treatment. Each leaf sample came from three different plants. The chlorophyll were extracted by acetone (80%) and measured absorbances at 663 and 645 nm by Beckman DU800 spectrophotometer (Fullerton, CA, United States) (Aono et al. 1993). All experiments were replicated three times.
Statistical analysis
All experiments were replicated independently at least three times, and data are shown as the mean ± SD of three independent experiments. Data were subjected to analysis of variance (ANOVA) using the Statistical Analysis System (SPSS version 22.0) software. The differences between the means were compared using the Tukey's test (P < 0.05).
Results
Characterization of GmNAC06
A comparison of the amino acid sequences of the N-terminal subdomains showed that GmNAC06 proteins are 32.13–55.47% identical to Arabidopsis AtNAC proteins. Subdomains A, C and D are highly conserved, and subdomains B and E are divergent (Fig. S1a). The phylogenetic tree was constructed using MEGA 6. A neighbor-joining evolutionary phylogeny test and 500 bootstrap replicates were selected for the analysis. The result suggested that GmNAC06 clustered with AtNAC2 (Fig. S1b).
Subcellular localization and transactivation activity analysis of GmNAC06
To determine the subcellular localization of GmNAC06, the 35S::GmNAC06:GFP fusion protein (Fig. S2a) and control 35S::GFP were transferred into the Arabidopsis protoplast. AtBZR2 fused to mCherry as nuclear marker. The control 35S::GFP distributed throughout the whole cell, whereas the 35S::GmNAC06:GFP fusion protein was detected only in the nucleus of the Arabidopsis protoplast (Fig. S2b). This result suggests that GmNAC06 may function as a transcription factor.
In order to test for transactivation activity of GmNAC06, the fusion plasmids pGBKT7-GmNAC06, the positive control pGBKT7-AtDREB2A, and the negative control pGBKT7 were separately transformed into yeast strain AH109. The transformed yeast strain grew well in non-selective medium SD/–Trp and, using X-α-Gal, it was observed that both the positive control and the cells harboring pGBKT7-GmNAC06 displayed β-galactosidase activity, whereas the negative control exhibited no β-galactosidase activity (Fig. S2d), suggesting that GmNAC06 possesses transcriptional activation activity in yeast cells.
Tissue-specific and stress-responsive expression of GmNAC06
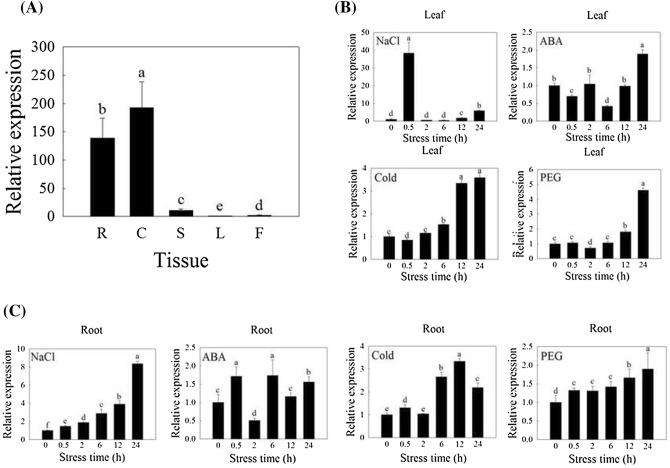
The different soybean organs were collected to analyze the tissue-specific expression of GmNAC06 by qRT-PCR (Fig. 1a). The results showed GmNAC06 was expressed constitutively in soybean. The transcription levels of GmNAC06 in roots were 0.72, 12.97, 138.76 and 59.72 times that of cotyledons, stems, leaves and flowers, respectively.
Fig. 1.
The transcription level of GmNAC06 in various organs and in response to various stresses. a The transcription level of GmNAC06 in different soybean organs was measured by qRT-PCR. b Leaf samples and c root samples under various stresses were collected at different time points to analyze the expression patterns of GmNAC06 by qRT-PCR. The values represent the averages of three independent biological experiments, and the error bars represent standard deviations. Different letters represent significant differences (P < 0.05)
To learn the gene function of GmNAC06, leaf and root samples under various stresses were collected at different time points to analyze the transcription level of GmNAC06 by qRT-PCR (Fig. 1b, c). In the leaves under NaCl stress, the transcriptional level of GmNAC06 at 0.5 h after treatment was 38.4 times that of 0 h. Under the ABA treatment, the transcriptional level of GmNAC06 at 6 h after treatment was 0.4 times that of 0 h. Under the cold and PEG stresses, the transcriptional level of GmNAC06 at 24 h after treatment was 3.6, 4.6 times that of 0 h, respectively. In the roots under NaCl stress, the transcriptional level of GmNAC06 at 24 h after treatment was 8.4 times that of 0 h. Under the ABA treatment, the transcriptional level of GmNAC06 at 2 h after treatment was 0.5 times that of 0 h. Under cold stress, the transcriptional level of GmNAC06 at 12 h after treatment was 3.3 times that of 0 h. Under PEG stresses, the transcriptional level of GmNAC06 at 24 h after treatment was 1.9 times that of 0 h. It suggested that GmNAC06 in the leaf and root responds to salt, exogenous ABA, cold and PEG stresses.
Effect of GmNAC06 overexpression on salt stress tolerance of composite plants
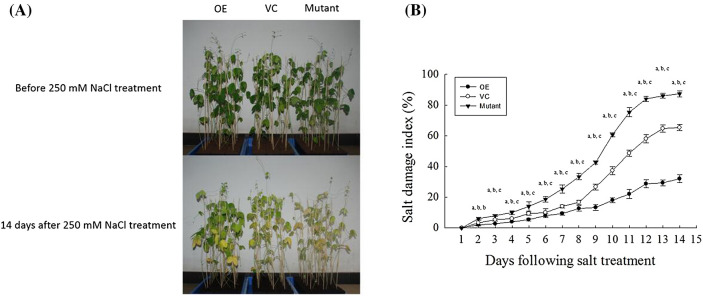
Four weeks after the seedling was infected by A. rhizogenes K599 with the OE, VC and Mutant vectors, the positive hairy roots of the OE, VC and Mutant composite plants were screened, and then the main roots and original roots were removed. After 6 weeks, the composite plants were irrigated with 250 mM NaCl solution three times a week for 2 consecutive weeks (Fig. 2a). After salt treatment, thirty OE individual plants, thirty VC individual plants and thirty Mutant individual plants were used to record the salt damage index (SDI%) for 2 consecutive weeks (Fig. 2b). Fourteen days after the salt stress treatment, the SDI of the VC and Mutant composite plants were 2 times and 2.7 times compared with the OE composite plants, respectively. The OE composite plants displayed salt tolerance, the VC composite plants wilted and died, and 26 Mutant composite plants dead. These results showed that the overexpression of GmNAC06 in hairy roots can enhance the salt tolerance of composite plants.
Fig. 2.
Salt tolerance of composite plants. a The 6-week-old composite plants (OE, VC, Mutant) were irrigated with 250 mM NaCl three times a week for 2 consecutive weeks. The photographs were taken 2 weeks after the salt treatment. b The difference in the SDI (%) among the composite plants with the OE, VC and Mutant hairy roots were recorded every day for 2 consecutive weeks. Different letters represent significant differences (P < 0.05)
Effect of GmNAC06 overexpression on salt stress tolerance of hairy roots
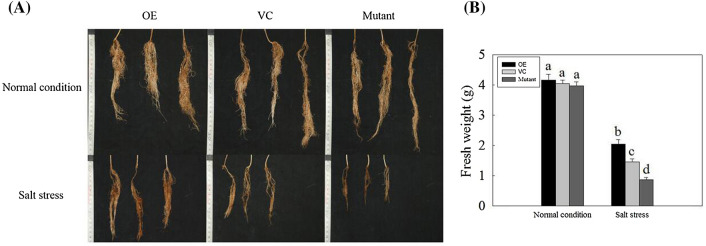
Six-week-old composite plants (OE, VC, Mutant) with positive hairy roots were subjected to salt stress. As a control, composite plants were planted in normal conditions. Two weeks later, photographs of the hairy roots were taken (Fig. 3a). Thirty individual hairy roots of OE, VC and Mutant under normal conditions and salt stress were used to calculate the fresh weight (Fig. 3b). The hairy roots of the OE, VC and Mutant composite plants showed no obvious differences when they were cultivated under normal conditions. Under salt stress, the fresh weight of OE, VC and Mutant hairy roots were 49%, 36% and 22% of that under normal conditions, respectively. These results suggest that GmNAC06 can enhance the salt tolerance of hairy roots.
Fig. 3.
The hairy root growth phenotypes and fresh weights of the composite plants (OE, VC, Mutant) at normal conditions and under salt stress. a To determine the phenotypes of hairy roots, thirty samples were analyzed, and the typical samples were selected. b The fresh weight of the OE, VC, and Mutant hairy roots under normal conditions and salt stress. Different letters represent significant differences (P < 0.05)
Effect of GmNAC06 overexpression on ROS production in composite plants
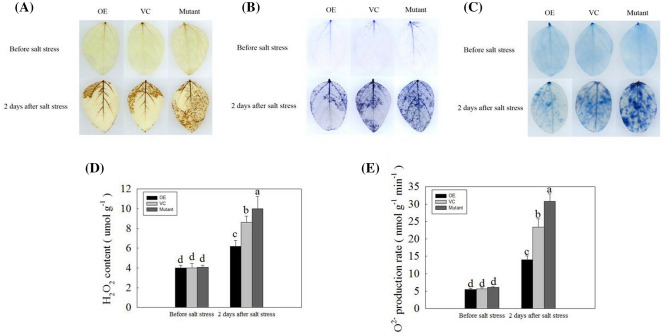
To further investigate the function of GmNAC06 in regulating salt tolerance, the leaves of 6-week-old composite plants (OE, VC, Mutant) before the salt stress treatment and 2 days after the salt stress treatment were stained by DAB and NBT. The leaves of the composite plants had no obvious differences under normal conditions. However, we found lower brown (Fig. 4a) and blue (Fig. 4b) staining intensities in the VC leaves than the Mutant leaves after the salt stress treatment, and the OE leaves had the lowest levels of brown precipitate and blue spots. The H2O2 content (Fig. 4d) and O2− production rate (Fig. 4e) of the Mutant leaves were higher than those of the VC leaves, and both parameters were the lowest in the OE leaves. This result shows that the overexpression of the GmNAC06 gene in composite plants could reduce ROS production during salt stress. The cell death result is consistent with ROS damage (Fig. 4c). These results suggest the overexpression of the GmNAC06 gene could enhance the salt tolerance of composite plants.
Fig. 4.
ROS accumulation and cell death in composite plants before salt stress and 2 days after salt stress. The leaves of the composite plants (OE, VC, Mutant) before salt stress and 2 days after salt stress were stained by a DAB, b NBT, and c trypan blue. The d H2O2 content and e O2− production rate of the composite leaves (OE, VC, Mutant) before salt stress and 2 days after salt stress. Different letters represent significant differences (P < 0.05)
Physiological effects of GmNAC06 overexpression in composite plants
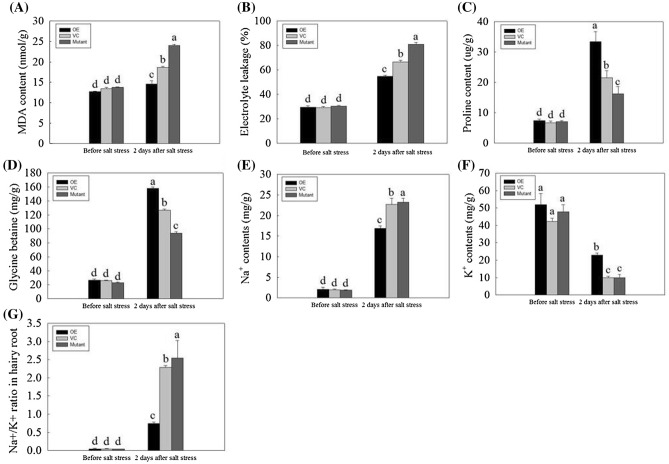
The MDA content is used to suggest lipid peroxidation caused by salt stress (Cao et al. 2007). After salt stress, the MDA content of the hairy roots significantly increased (Fig. 5a). Compared with VC hairy roots, the content of MDA in OE hairy roots decreased by 22%, and that of Mutant hairy roots increased by 29%. Electrolyte leakage is used to suggest the damage caused by salt stress (Zhao et al. 2014). After salt stress, the electrolyte leakage of the hairy roots increased notably. Compared with VC hairy roots, the electrolyte leakage in OE hairy roots decreased by 18%, and that of Mutant hairy roots increased by 22%. (Fig. 5b). Proline and glycine betaine are multifunctional osmolytes, and their accumulation enhances plant stress tolerance, particularly in response to salt stress (Ashraf and Foolad 2007). The proline and glycine betaine accumulation rate among the OE, VC, and Mutant composite plants have no significant difference before salt stress; however, under salt stress, the accumulation of proline and glycine betaine in OE hairy roots increased by 55% and 24% and the accumulation of proline and glycine betaine in Mutant hairy roots decreased by 24% and 26% compared with VC hairy roots (Fig. 5c, d). Maintaining Na+ and K+ homeostasis is very important for enhancing the salt tolerance of composite plants (Mostofa et al. 2015). Before salt stress, the Na+ and K+ contents and the Na+/K+ ratio of the OE, VC, and Mutant hairy roots showed no obvious differences. After salt stress, the Na+ content in OE hairy roots decreased by 26%, and that of Mutant hairy roots increased by 3% compared with VC hairy roots (Fig. 5e). However, the K+ content in OE hairy roots increased by 129%, and that of Mutant hairy roots decreased by 2% compared with VC hairy roots (Fig. 5f). As a result, the Na+/K+ ratio of OE hairy roots decreased by 67%, and that of Mutant hairy roots increased by 12% compared with VC hairy roots (Fig. 5g). We detected the expression of GmNAC06 in the hairy root under normal condition and salt stress (Fig. S6c). Compared with VC hairy roots, the expression level of GmNAC06 of OE hairy roots increased by 496% and 300%, the expression level of GmNAC06 of Mutant hairy roots decreased by 63% and 82% under normal condition and salt stress, respectively. Taken together, the results suggest that the overexpression of GmNAC06 could enhance the salt stress tolerance of composite plants.
Fig. 5.
Physiological changes in composite plants before salt stress and 2 days after salt stress. The a MDA content, b electrolyte leakage, c proline content, d glycine betaine content, e Na+ content, f K+ content and g Na+/K+ ratio of hairy roots in the composite plants before salt stress and 2 days after salt stress. Different letters represent significant differences (P < 0.05)
Expression analysis of salt-responsive genes in the OE and VC hairy roots of composite plants
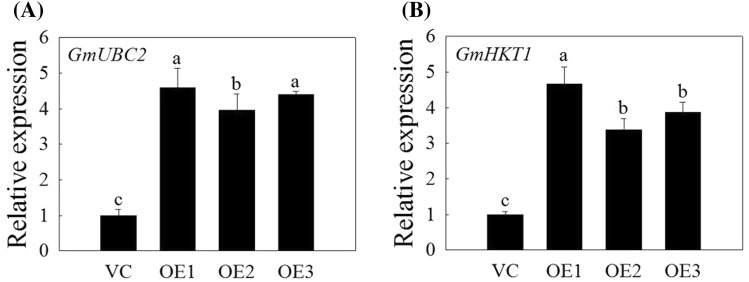
To further understand the function of GmNAC06, the expression levels of the 14 salt-related marker genes were analyzed under normal conditions. The expression levels of GmUBC2 and GmHKT1 in the OE hairy roots were more than three times higher than those in the control (Fig. 6a, b). This suggests that GmNAC06 may enhance the salt tolerance of composite plants by regulating salt stress-related genes.
Fig. 6.
Expression level of salt-related marker genes in the hairy roots of OE and VC composite plants under normal conditions. a The transcription level of GmUBC2. b The transcription level of GmHKT1. Each hairy root sample came from three different plants. All experiments were repeated three times. Different letters represent significant differences (P < 0.05)
Effects of overexpression of GmNAC06 in Arabidopsis
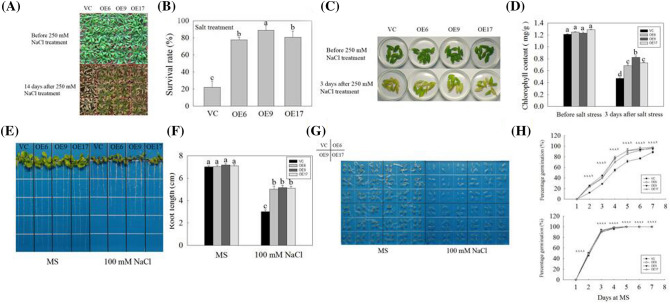
Homozygous T3-generation Arabidopsis were used to further analyze the function of GmNAC06 in salt stress responses. Four-week-old seedlings were treated with 250 mM NaCl solution. Fourteen days after the salt stress treatment, the growth status of the three OE lines was better than that of the VC line (Fig. 7a). The survival rate of the three OE lines was more than three times higher than that of the VC line (Fig. 7b). The leaves of OE and VC Arabidopsis were incubated in 250 mM NaCl solution for 3 days (Fig. 7c). After the salt stress, the green of the VC line leaves faded, but leaves of three OE lines were green. The chlorophyll content was measured in leaves of OE and VC Arabidopsis before salt stress and 3 days after salt stress (Fig. 7d). Before salt stress, the chlorophyll contents of the OE and VC transgenic lines were not obviously different. However, the chlorophyll content of three OE lines was more than 1.5 times higher than that of the VC line under salt stress. Five-day-old seedlings with roots of nearly equal length were placed vertically on MS solid medium with or without NaCl. After 7 days, the root length of the OE line was similar to that of the VC line under normal conditions. However, the root length of the OE line was more than 1.5 times longer than that of the VC line under salt stress (Fig. 7e, f). To determine whether the three OE lines enhanced the salt tolerance of Arabidopsis, seeds from the three OE lines and the VC line were germinated on MS solid medium with or without NaCl. After 3 days of stratifications, the percentage germination was measured for 1 consecutive week. The percentage germination of the OE and VC transgenic lines were not different under normal conditions. Under salt stress conditions, the percentage germination of Arabidopsis was inhibited. However, the percentage germination of the three OE lines was higher than that of the VC line under salt stress (Fig. 7g, h). We detected the expression of marker genes AtUBC2 and AtHKT1;1 under normal conditions. Their expression levels of in the OE line were more than three times higher than those in the VC and WT Arabidopsis (Fig. S8). Therefore, we concluded that overexpression GmNAC06 could enhance the salt tolerance of transgenic Arabidopsis.
Fig. 7.
The phenotype and physiological indexes of transgenic Arabidopsis. a The phenotypes of OE and VC Arabidopsis before salt stress and 14 days after salt stress. b Survival rate after salt stress. The c phenotypes and d chlorophyll content of OE and VC Arabidopsis before salt stress and 3 days after salt stress. e Root growth status, f root length, g emergence performance and h emergence rate of OE and VC Arabidopsis grown for 7 days on MS media with 0 or 100 mM NaCl. Different letters represent significant differences (P < 0.05)
Discussion
Soybean is considered a salt-sensitive glycophyte, and all of its developmental stages are adversely affected by salinity stress (Phang et al. 2008). Salt stress inflicts heavy losses on soybean yield and has a significant negative impact on the quality. Therefore, it is very important to cultivate salt tolerant soybean varieties. Molecular breeding could improve the efficiency of breeding these varieties. NAC proteins constitute one of the largest families of plant-specific transcription factors, and the NAC protein family is present in a wide range of land plants (Olsen et al. 2005). The bioinformatic data provided by a genome-wide transcriptome analysis predict that 20–25% NAC genes function in at least one or more stress responses (Puranik et al. 2012). Increasing evidence has suggested that the NAC family members can enhance salt tolerance in a number of plants, such as Arabidopsis (Mao et al. 2014), wheat (Huang et al. 2015), rice (Hong et al. 2016) and pumpkin (Cao et al. 2017). The soybean NAC family contains 152 members. Due to the limitations of soybean transformation technology, the functions of only a few soybean NAC genes have been studied clearly. An A. rhizogenes—mediated transformation system could save time and improve the transformation efficiency. A tissue-specific expression assay showed GmNAC06 was appropriate for an A. rhizogenes—mediated transformation system (Fig. 1a, b).
Based on the phylogenetic analysis and sequence alignment, the amino acid sequences of the GmNAC06 proteins displayed a high similarity to AtNAC2 (55.47% identity) (Fig. S1a and b). AtNAC2 has been shown to enhance the salt tolerance of groundnut (Patil et al. 2014). The results of qRT-PCR suggested that GmNAC06 might be involved in stress responses, especially in roots under salt stress, and the expression level increased gradually with time (Fig. 1c). The subcellular localization assay suggested that GmNAC06 may function as a transcription factor (Fig. S2b). To study the role of GmNAC06 in salt stress responses, the phenotypes and SDI of OE, VC and Mutant composite plants treated with 250 mM NaCl were observed and recorded (Fig. 2a, b). The results suggested that GmNAC06 likely functions as a positive stress-responsive transcription factor of salt stress in soybean.
In plants, high concentrations of salt result in physiological damage and water stress, while the water deficit leads to the formation of ROS, and cytotoxic oxygen seriously disrupts vital cellular functions by damaging various cellular components such as proteins, lipids, and DNA (Parida and Das 2005). All these effects can lead to reduced plant growth. Under salt stress, the levels of ROS in plant tissues can be dramatically elevated. Therefore, plants maintaining ROS homeostasis depend on the balance between the generation and scavenging of ROS to be less influenced by the oxidative stress (Jin et al. 2017). The results of the histochemical ROS staining (Fig. 4a, b) and measurement of H2O2 and (Fig. 4d, e) showed the OE composite plants could scavenge ROS more effectively than the VC composite plants. The Mutant composite plants accumulated the highest level of ROS because they could not scavenge the ROS in time. The observed occurrence of apoptosis is consistent with the accumulation of ROS (Fig. 4c). Under salt stress, the biomasses of the OE, VC and Mutant hairy roots decreased significantly compared to that of the control (Fig. 3a). However, the OE composite plants showed more developed and faster growing hairy roots compared to the VC composite plants. In contrast, the growth of the Mutant hairy roots suffered serious inhibition (Fig. 3b). This finding suggested GmNAC06 could enhance the salt tolerance of composite plants by maintaining the normal growth of hairy roots under salt stress. The phenotype of the above ground part of the plant was consistent with the underground part. These results suggested that the function of the wild leaves was influenced by the transgenic hairy roots. MDA content is a typical physiological parameter for evaluating stress tolerance in plants (Yu et al. 2016). It can exhibit the damage degree of membranes. The MDA content of the OE, VC and Mutant hairy roots (Fig. 5a) suggested that overexpression of GmNAC06 in hairy roots could alleviate cell membrane injury under salt stress. Electrolyte leakage results (Fig. 5b) were consistent with the MDA content. This suggested that increased membrane damage led to increased solute leakage. Proline in plants plays roles in osmotic adjustment, the protection of cellular macromolecules and the scavenging of hydroxyl radicals (Jin et al. 2010). Glycine betaine has an osmoprotective function and is known to improve salt stress tolerance in most crop plants. Glycine betaine enhances the salt tolerance of plants by accelerating of ROS scavenging systems, protecting membrane integrity and reducing the oxidation of membrane lipids under salt stress (Demiral and Türkan 2006). After salt stress, the proline and glycine betaine content of the OE, VC and Mutant hairy roots suggested that GmNAC06 scavenged ROS by accumulating proline and glycine betaine rapidly to improve the salt tolerance of the hairy roots (Fig. 5c, d). K+ is an important macronutrient for plants. Under salt stress, the capacity for K+ uptake and transport is usually correlated with the salt tolerance of the plant (Ding et al. 2010). However, Na+ could attenuate the intracellular K+ influx by disturbing the ion selectivity of cell membranes (Chen et al. 2017). Two methods of salt detoxification include the transport of redundant Na+ out the cell through the plasma membrane and the storage of Na+ ions in the vacuole (Hasegawa et al. 2000). Plants maintain ionic homeostasis and osmotic equilibrium by preventing Na+ influx and promoting K+ uptake and Na+ extrusion (Zhu 2003). After salt stress, the OE hairy roots maintained a higher K+ content and lower Na+ content and Na+/K+ ratio than the VC hairy roots (Fig. 5e–g). Ensuring a low Na+/K+ ratio in the cytoplasm is very important to improving the salt resistance of plants. All these results suggested the overexpression of GmNAC06 in hairy roots could help maintain Na+ and K+ homeostasis. GmUBC2 had a higher expression level in roots than in stems and leaves. It improved salt tolerance by regulating ion homeostasis, osmolyte synthesis and oxidative stress responses (Zhou et al. 2010). GmHKT1 enhanced salt tolerance by affecting the Na+ and K+ transport and regulating the Na+/K+ homeostasis (Chen et al. 2011). The expression levels of GmUBC2 and GmHKT1 in OE hairy roots were significantly higher than control under normal conditions. This suggests that overexpression of GmNAC06 can improve salt tolerance partly due to the enhanced expression of these genes. Since hairy roots are not heritable, we generated homozygous T3-generation Arabidopsis to investigate the function of GmNAC06. We found that the GmNAC06-overexpression transgenic Arabidopsis increased salt tolerance compared to the control at both the germination and seedling stages. We further characterized the roles of GmNAC06 in salt tolerance.
In summary, we found GmNAC06 plays role in salt stress responses through the phenotypic, physiological and molecular analyses of OE, VC, and Mutant composite soybean. When roots experienced salt stress, overexpression of GmNAC06 induced the accumulation of proline and glycine betaine to scavenge redundant ROS to protect the integrity of cell membranes. At the same time, it maintained ionic homeostasis and osmotic equilibrium by regulating Na+ and K+ transport. These measures ensured that the roots could grow normally under salt stress, which improved the salt tolerance of the leaves. Ultimately, the whole composite plants exhibited strong abilities for salt tolerance. These results suggest GmNAC06 likely functions as a positive regulator of salt tolerance.
Electronic supplementary material
Electronic supplementary material 1 (JPG 96 kb). Figure S1. Sequence comparison and phylogenetic relationship of GmNAC06 and NAC family members from Arabidopsis. (A) Sequence alignment of the N-terminal subdomains in GmNAC06 and Arabidopsis NAC family members. The Arabidopsis Genome Initiative identification numbers of the Arabidopsis members are as follows: AtNAC1(AT3G18400.1), AtNAC2(AT5G39610.1), AtNAC3(AT1G76420.1), AtNAC4(AT3G29035.1), AtNAC5(AT1G32770.1), AtNAC6(AT5G62380.1), AtNAC7(AT3G44350.2) AtNAC8(AT5G18270.1), AtNAC9(AT4G10350.1). (B) The phylogenetic tree was constructed using MEGA 6. A neighbor-joining evolutionary phylogeny test and 500 bootstrap replicates were selected for the analysis.
Electronic supplementary material 2 (JPG 100 kb). Figure S2. Subcellular localization and transactivation activity analysis of GmNAC06. (A) PCR detection of 35S::GmNAC06:GFP. M: DL 2000 maker, 1-8: The amplification of the target fragment by PCR using different liquid bacteria as the template. (B) The vector control (35S::GFP) and fusion protein construct 35S::GmNAC06:GFP were introduced into the Arabidopsis protoplast. Fluorescent materials were observed under Olympus FV1000 viewer confocal laser scanning microscope, 488 nm, argon-ion laser excitation, 507 nm detection GFP; mcherry was 555 nm, argon-ion laser excitation, LP 640 nm IR detection; chloroplast autofluorescence was 488 nm argon-ion laser excitation, SP 630 nm IR detection, pinhole is about 1.0 unit, and the optical section thickness is about 0.5 µm. AtBZR2 fused with mCherry was used as a nuclear marker. Scale bars = 10 μm. (C) CDS and peptide sequence of GmNAC06. (D) Transactivation activity analysis of GmNAC06 in yeast cells. pGBKT7-AtDREB2A and pGBKT7 were used as positive and negative controls, respectively.
Electronic supplementary material 3 (JPG 137 kb). Figure S3. Different stages of the soybean hairy root transformation. The ideal stage for transformation: six-day-old seedlings with unfolded cotyledons. (B) Taking the bacterial mass in the petri dish with the tip of a syringe needle. (C) Stabbing the hypocotyl near the cotyledonary node; the red square suggests which part of the seedling had to be infected for a successful transformation. (D) After infection, the seedlings were kept in a plastic tray covered with a transparent lid. (E) After the initiation of hairy root formation from the infection site, the wounding sites and the following were covered with vermiculite to maintain high humidity. (F) Two weeks after inoculation. (G) Four weeks after inoculation. (H) Four weeks after inoculation, during the removal of the main roots (the part below the red line was removed ). (I) The plants with hairy roots were transferred into mixed soil (humus: vermiculite = 2:1) and watered every three days.
Electronic supplementary material 4 (JPG 38 kb). Figure S4. GmNAC06 overexpression in hairy roots screened by RT-PCR and qRT-PCR. (A) GmNAC06 overexpression in hairy roots screened by RT-PCR. Lanes 1-16: The transcription level of GmNAC06 in the independent hairy root samples screened by RT-PCR; red represents positive hairy roots, and blue represents negative hairy roots. WT: hairy roots induced by K599 (containing no vector) used as wild-type controls. CYP2: internal control. (B) GmNAC06 overexpression in hairy roots screened by qRT-PCR.
Electronic supplementary material 5 (JPG 24 kb). Figure S5. Screened vector control hairy roots. (A) Lanes 1-10: positive hairy roots. WT: PCR products of hairy roots induced by K599 (containing no vector). WT*: PCR products of pCAMBIA3301. M: DL 2000 maker. (B) The transcription level of GmNAC06 in VC hairy roots by RT-PCR. WT: hairy roots induced by K599 (containing no vector) used as wild-type controls. CYP2: internal control.
Electronic supplementary material 6 (JPG 63 kb). Figure S6. CRISPR-Cas9 induced mutations in soybean hairy roots were screened. (A) Detection of mutations using a T7 endonuclease 1 (T7E1) cleavage assay. Lanes 1-15: PCR products amplified from the independent hairy root samples cleaved by T7E1; red represents a mutation, and blue represents no mutation; WT: PCR products of hairy roots induced by K599 (containing no vector) cleaved by T7E1. WT*: PCR products of hairy roots induced by K599 (containing no vector) not cleaved by T7E1. M: DL 2000 maker. (B) Sequence-based detection of mutations by pCas9-GmU6-sgRNA vectors; red represents the protospacer-adjacent motif sequence, and nucleotide substitutions are highlighted in green. +: insertion; -: deletion; S: substitution, X: number of mutations. (C) The expression level of GmNAC06 under normal condition and salt stress was quantified by qRT-PCR.
Electronic supplementary material 7 (JPG 32 kb). Figure S7. PCR, RT-PCR and qRT-PCR analysis of OE and VC transgenic Arabidopsis. (A) Lanes 1-11: The positive lines of OE transgenic Arabidopsis. WT: PCR products of OE vector. M: DL 2000 maker. (B) Lanes 1-10: The positive lines of VC transgenic Arabidopsis. WT: PCR products of wild Arabidopsis. WT*: PCR products of pCAMBIA3301. M: DL 2000 maker. (C) The transcription level of GmNAC06 in OE and VC transgenic Arabidopsis by RT-PCR. UBQ3: internal control. (D) The transcription level of GmNAC06 in OE and VC transgenic Arabidopsis by qRT-PCR.
Electronic supplementary material 8 (JPG 16 kb). Figure S8. Expression level of salt-related marker genes in the OE, VC and WT Arabidopsis under normal conditions. (A) The transcription level of AtUBC2. (B) The transcription level of AtHKT1;1. Different letters represent significant differences (P < 0.05).
Electronic supplementary material 9 (DOCX 34 kb)
Acknowledgements
We thank Jackie Chretien, PhD, from American Journal Experts (https://www.aje.cn/) for editing the English text of a draft of this manuscript.
Author contributions
HZ, ML, and ZH conceived the research. ML and RC performed the experiments, analyzed the data and wrote the manuscript. QJ and XS provided scientific suggestions. HZ and ZH revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFD0101005) and the Agricultural Science and Technology Innovation Program (ASTIP).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ming Li, Email: liming07@caas.cn.
Hui Zhang, Email: zhanghui06@caas.cn.
Zheng Hu, Email: Huzheng@caas.cn.
References
- Aono M, Kubo A, Saji H, Tanaka K, Kondo N. Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant Cell Physiol. 1993;34:129–135. [Google Scholar]
- Ashraf M, Foolad M. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot. 2007;59:206–216. [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62:250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007;143:707–719. doi: 10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hou W, Liu W, Yao W, Wu C, Liu X, Han T, et al. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tissue Organ Cult (PCTOC) 2011;107:541–552. [Google Scholar]
- Cao H, Wang L, Nawaz MA, Niu M, Sun J, Xie J, Bie Z. Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Front Plant Sci. 2017;8:2052. doi: 10.3389/fpls.2017.02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, He H, Yu D, et al. Overexpression of a novel soybean gene modulating Na+ and K+ transport enhances salt tolerance in transgenic tobacco plants. Physiol Plant. 2011;141:1–18. doi: 10.1111/j.1399-3054.2010.01412.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Han Y, Kong X, Kang H, Ren Y, Wang W. Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+/K+ and antioxidant competence. Physiol Plant. 2017;159:161–177. doi: 10.1111/ppl.12492. [DOI] [PubMed] [Google Scholar]
- Chen L, Cai Y, Liu X, Yao W, Guo C, Sun S, Hou W. Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int J Mol Sci. 2018;19:3039. doi: 10.3390/ijms19103039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011;156:466–473. doi: 10.1104/pp.111.172981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiral T, Türkan I. Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ Exp Bot. 2006;56:72–79. [Google Scholar]
- Ding M, Hou P, Shen X, Wang M, Deng S, Sun J, Zhang D. Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol Biol. 2010;73:251–269. doi: 10.1007/s11103-010-9612-9. [DOI] [PubMed] [Google Scholar]
- Elstner EF, Heupel A. Inhibition of nitrite formation from hydroxylammoniumchloride: a simple assay for superoxide dismutase. Anal Biochem. 1976;70:616–620. doi: 10.1016/0003-2697(76)90488-7. [DOI] [PubMed] [Google Scholar]
- Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, Díaz-Camino C, Santana O, Murillo E, et al. Agrobacterium rhizogenes transformation of the Phaseolus spp.: a tool for functional genomics. Mol Plant Microbe Interact. 2006;19:1385–1393. doi: 10.1094/MPMI-19-1385. [DOI] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hinchee MA, Connor-Ward DV, Newell CA, McDonnell RE, Sato SJ, Gasser CS, et al. Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Bio/Technology. 1988;6:915–922. [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XS, Luo T, Fu XZ, Fan QJ, Liu JH. Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliata whose ectopic expression confers dehydration/drought tolerance in transgenic tobacco. J Exp Bot. 2011;62:5191–5206. doi: 10.1093/jxb/err229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Wang Y, Li B, Chang J, Chen M, Li K, He G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015;15:268. doi: 10.1186/s12870-015-0644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Chang Q, Li W, Yin D, Li Z, Wang D, Liu L. Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Tissue Organ Cult (PCTOC) 2010;100:219–227. [Google Scholar]
- Jin C, Li KQ, Xu XY, Zhang HP, Chen HX, Chen YH, Zhang SL. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front Plant Sci. 2017;8:1049. doi: 10.3389/fpls.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Sun L, Zhou Y, Zhang M, Liu Y, Pan J, Li D, et al. ZmMKK4 regulates osmotic stress through reactive oxygen species scavenging in transgenic tobacco. Plant Cell Rep. 2011;30:2097. doi: 10.1007/s00299-011-1116-9. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Gautam RK, Sharma PC, Sharma DK, Singh YP. CSR 53 (Bulk 18)(IC0619320; INGR17029), a rice (Oryza sativa) germplasm with tolerance to salinity stresses up to ECe 10.0 dS/m. Indian J Plant Genet Resour. 2019;32:240–241. [Google Scholar]
- Le DT, Nishiyama RIE, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18:407–417. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hu Z, Jiang QY, Sun XJ, Guo Y, Qi JC, Zhang H. GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean. J Integr Agric. 2017;17:530–538. [Google Scholar]
- Liu ZJ, Guo YK, Bai JG, et al. Exogenous hydrogen peroxide changes antioxidant enzyme activity and protects ultrastructure in leaves of two cucumber ecotypes under osmotic stress. J Plant Growth Regul. 2010;29:171–183. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu M, Ying S, Zhang DF, Shi YS, Song YC, Wang TY, et al. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012;31:1701–1711. doi: 10.1007/s00299-012-1284-2. [DOI] [PubMed] [Google Scholar]
- Mankin SL, Hill DS, Olhoft PM, Toren E, Wenck AR, Nea L, et al. Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell Dev Biol. 2007;43:521–535. [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R, et al. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot. 2012;63:2933–2946. doi: 10.1093/jxb/err462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Chen S, Li A, Zhai C, Jing R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE. 2014;9:e84359. doi: 10.1371/journal.pone.0084359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Mostofa MG, Saegusa D, Fujita M, Tran LSP. Hydrogen sulfide regulates salt tolerance in rice by maintaining Na+/K+ balance, mineral homeostasis and oxidative metabolism under excessive salt stress. Front Plant Sci. 2015;6:1055. doi: 10.3389/fpls.2015.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. Genes and salt tolerance: bringing them together. New Phytol. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82:113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Pareddy D, Chennareddy S, Anthony G, Sardesai N, Mall T, Minnicks T, Shumway N. Improved soybean transformation for efficient and high throughput transgenic production. Transgenic Res. 2020;29:267–281. doi: 10.1007/s11248-020-00198-8. [DOI] [PubMed] [Google Scholar]
- Parida AK, Das AB. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60:324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Patil M, Ramu SV, Jathish P, Sreevathsa R, Reddy PC, Prasad TG, Udayakumar M. Overexpression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol Rep. 2014;8:161–169. [Google Scholar]
- Phang TH, Shao G, Lam HM. Salt tolerance in soybean. J Integr Plant Biol. 2008;50:1196–1212. doi: 10.1111/j.1744-7909.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- Pinedo I, Ledger T, Greve M, Poupin MJ. Burkholderia phytofirmansPsJN induces long-term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front Plant Sci. 2015;6:466. doi: 10.3389/fpls.2015.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckette MC, Weng H, Mahalingam R. Physiological and biochemical responses to acute ozone-induced oxidative stress in Medicago truncatula. Plant Physiol Biochem. 2007;45:70–79. doi: 10.1016/j.plaphy.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Bokowiec MT, Han S, Zhang H, Brannock JF, Chen X, et al. Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008;147:280–295. doi: 10.1104/pp.107.114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Yin Y, Chen F, Xu Y, Dixon RA. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. BioEnergy Res. 2009;2:217–232. [Google Scholar]
- Shiriga K, Sharma R, Kumar K, Yadav SK, Hossain F, Thirunavukkarasu N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene. 2014;2:407–417. doi: 10.1016/j.mgene.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Adler G, Bar-Zvi D, et al. ABI4 downregulates expression of the sodium transporter HKT1; 1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013;73:993–1005. doi: 10.1111/tpj.12091. [DOI] [PubMed] [Google Scholar]
- Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, et al. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta. 2009;230:985–1002. doi: 10.1007/s00425-009-1000-9. [DOI] [PubMed] [Google Scholar]
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhao J, Li X, Li Y, et al. E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol. 2020;227:455–472. doi: 10.1111/nph.16538. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83:224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F. Drought tolerance conferred in soybean (Glycine max L.) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017;58:1764–1776. doi: 10.1093/pcp/pcx111. [DOI] [PubMed] [Google Scholar]
- Weller SA, Stead DE, Young JPW. Acquisition of an Agrobacterium Ri plasmid and pathogenicity by other α-Proteobacteria in cucumber and tomato crops affected by root mat. Appl Environ Microbiol. 2004;70:2779–2785. doi: 10.1128/AEM.70.5.2779-2785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu Y, Wang S, Tao Y, Wang Z, Shu Y, Ma H. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 2016;35:613–627. doi: 10.1007/s00299-015-1907-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang YJ, Yang BJ, Yu XX, Wang D, Zu SH, Lin WH. Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci Rep. 2016;6:1–14. doi: 10.1038/srep31134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zhang F, Guo J, Yang Y, Li B, Zhang L. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004;134:849–857. doi: 10.1104/pp.103.030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ma Q, Jin X, Peng X, Liu J, Deng L, Cheng B. A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 2014;55:1142–1156. doi: 10.1093/pcp/pcu054. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun. 2009;379:985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- Zhou GA, Chang RZ, Qiu LJ, et al. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol Biol. 2010;72:357–367. doi: 10.1007/s11103-009-9575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplementary material 1 (JPG 96 kb). Figure S1. Sequence comparison and phylogenetic relationship of GmNAC06 and NAC family members from Arabidopsis. (A) Sequence alignment of the N-terminal subdomains in GmNAC06 and Arabidopsis NAC family members. The Arabidopsis Genome Initiative identification numbers of the Arabidopsis members are as follows: AtNAC1(AT3G18400.1), AtNAC2(AT5G39610.1), AtNAC3(AT1G76420.1), AtNAC4(AT3G29035.1), AtNAC5(AT1G32770.1), AtNAC6(AT5G62380.1), AtNAC7(AT3G44350.2) AtNAC8(AT5G18270.1), AtNAC9(AT4G10350.1). (B) The phylogenetic tree was constructed using MEGA 6. A neighbor-joining evolutionary phylogeny test and 500 bootstrap replicates were selected for the analysis.
Electronic supplementary material 2 (JPG 100 kb). Figure S2. Subcellular localization and transactivation activity analysis of GmNAC06. (A) PCR detection of 35S::GmNAC06:GFP. M: DL 2000 maker, 1-8: The amplification of the target fragment by PCR using different liquid bacteria as the template. (B) The vector control (35S::GFP) and fusion protein construct 35S::GmNAC06:GFP were introduced into the Arabidopsis protoplast. Fluorescent materials were observed under Olympus FV1000 viewer confocal laser scanning microscope, 488 nm, argon-ion laser excitation, 507 nm detection GFP; mcherry was 555 nm, argon-ion laser excitation, LP 640 nm IR detection; chloroplast autofluorescence was 488 nm argon-ion laser excitation, SP 630 nm IR detection, pinhole is about 1.0 unit, and the optical section thickness is about 0.5 µm. AtBZR2 fused with mCherry was used as a nuclear marker. Scale bars = 10 μm. (C) CDS and peptide sequence of GmNAC06. (D) Transactivation activity analysis of GmNAC06 in yeast cells. pGBKT7-AtDREB2A and pGBKT7 were used as positive and negative controls, respectively.
Electronic supplementary material 3 (JPG 137 kb). Figure S3. Different stages of the soybean hairy root transformation. The ideal stage for transformation: six-day-old seedlings with unfolded cotyledons. (B) Taking the bacterial mass in the petri dish with the tip of a syringe needle. (C) Stabbing the hypocotyl near the cotyledonary node; the red square suggests which part of the seedling had to be infected for a successful transformation. (D) After infection, the seedlings were kept in a plastic tray covered with a transparent lid. (E) After the initiation of hairy root formation from the infection site, the wounding sites and the following were covered with vermiculite to maintain high humidity. (F) Two weeks after inoculation. (G) Four weeks after inoculation. (H) Four weeks after inoculation, during the removal of the main roots (the part below the red line was removed ). (I) The plants with hairy roots were transferred into mixed soil (humus: vermiculite = 2:1) and watered every three days.
Electronic supplementary material 4 (JPG 38 kb). Figure S4. GmNAC06 overexpression in hairy roots screened by RT-PCR and qRT-PCR. (A) GmNAC06 overexpression in hairy roots screened by RT-PCR. Lanes 1-16: The transcription level of GmNAC06 in the independent hairy root samples screened by RT-PCR; red represents positive hairy roots, and blue represents negative hairy roots. WT: hairy roots induced by K599 (containing no vector) used as wild-type controls. CYP2: internal control. (B) GmNAC06 overexpression in hairy roots screened by qRT-PCR.
Electronic supplementary material 5 (JPG 24 kb). Figure S5. Screened vector control hairy roots. (A) Lanes 1-10: positive hairy roots. WT: PCR products of hairy roots induced by K599 (containing no vector). WT*: PCR products of pCAMBIA3301. M: DL 2000 maker. (B) The transcription level of GmNAC06 in VC hairy roots by RT-PCR. WT: hairy roots induced by K599 (containing no vector) used as wild-type controls. CYP2: internal control.
Electronic supplementary material 6 (JPG 63 kb). Figure S6. CRISPR-Cas9 induced mutations in soybean hairy roots were screened. (A) Detection of mutations using a T7 endonuclease 1 (T7E1) cleavage assay. Lanes 1-15: PCR products amplified from the independent hairy root samples cleaved by T7E1; red represents a mutation, and blue represents no mutation; WT: PCR products of hairy roots induced by K599 (containing no vector) cleaved by T7E1. WT*: PCR products of hairy roots induced by K599 (containing no vector) not cleaved by T7E1. M: DL 2000 maker. (B) Sequence-based detection of mutations by pCas9-GmU6-sgRNA vectors; red represents the protospacer-adjacent motif sequence, and nucleotide substitutions are highlighted in green. +: insertion; -: deletion; S: substitution, X: number of mutations. (C) The expression level of GmNAC06 under normal condition and salt stress was quantified by qRT-PCR.
Electronic supplementary material 7 (JPG 32 kb). Figure S7. PCR, RT-PCR and qRT-PCR analysis of OE and VC transgenic Arabidopsis. (A) Lanes 1-11: The positive lines of OE transgenic Arabidopsis. WT: PCR products of OE vector. M: DL 2000 maker. (B) Lanes 1-10: The positive lines of VC transgenic Arabidopsis. WT: PCR products of wild Arabidopsis. WT*: PCR products of pCAMBIA3301. M: DL 2000 maker. (C) The transcription level of GmNAC06 in OE and VC transgenic Arabidopsis by RT-PCR. UBQ3: internal control. (D) The transcription level of GmNAC06 in OE and VC transgenic Arabidopsis by qRT-PCR.
Electronic supplementary material 8 (JPG 16 kb). Figure S8. Expression level of salt-related marker genes in the OE, VC and WT Arabidopsis under normal conditions. (A) The transcription level of AtUBC2. (B) The transcription level of AtHKT1;1. Different letters represent significant differences (P < 0.05).
Electronic supplementary material 9 (DOCX 34 kb)