Abstract
A better understanding of co-evolution between pathogens and hosts holds promise for better prevention and control strategies. This review will explore the interactions between Burkholderia pseudomallei, an environmental and opportunistic pathogen, and the human host immune system. B. pseudomallei causes “Melioidosis,” a rapidly fatal tropical infectious disease predicted to affect 165,000 cases annually worldwide, of which 89,000 are fatal. Genetic heterogeneities were reported in both B. pseudomallei and human host population, some of which may, at least in part, contribute to inter-individual differences in disease susceptibility. Here, we review (i) a multi-host—pathogen characteristic of the interaction; (ii) selection pressures acting on B. pseudomallei and human genomes with the former being driven by bacterial adaptation across ranges of ecological niches while the latter are driven by human encounter of broad ranges of pathogens; (iii) the mechanisms that generate genetic diversity in bacterial and host population particularly in sequences encoding proteins functioning in host—pathogen interaction; (iv) reported genetic and structural variations of proteins or molecules observed in B. pseudomallei—human host interactions and their implications in infection outcomes. Together, these predict bacterial and host evolutionary trajectory which continues to generate genetic diversity in bacterium and operates host immune selection at the molecular level.
Keywords: melioidosis, evolution, burkholderia, host immune system, genetic variants
Introduction
Melioidosis is a serious and often fatal neglected tropical infectious disease caused by Burkholderia pseudomallei, an intracellular bacterial pathogen and also ubiquitous in the environment (Holden et al., 2004). Human hosts can acquire the bacterium after direct environmental exposure either through dermal puncture, ingestion of contaminated food or water supplies, or inhalation of contaminated soil or water aerosols. Following an acquisition, B. pseudomallei can replicate in host non-phagocytic and phagocytic cells as well as spreading intracellularly (Willcocks et al., 2016). The bacterium can either kill the human host rapidly (acute melioidosis) or hide within the host body for a long period (chronic melioidosis). With rare exceptions (Abbink et al., 2001; Ralph et al., 2004; Aziz et al., 2020), B. pseudomallei is not known to transmit from person to person; indicating that B. pseudomallei has not evolved virulence mechanisms through consecutive passage in human hosts. Moreover, B. pseudomallei within-host evolution observed in chronic infections are often linked to attenuated virulence (Price et al., 2013; Viberg et al., 2017; Pearson et al., 2020), which is possibly mediated by host immune evasion. While environment exposure can lead to melioidosis, most patients are elderly or have one or more underlying health risks. The most common health condition is diabetes mellitus which presents in up to half of cases (Kronsteiner et al., 2019). Therefore, it is likely that B. pseudomallei has acquired virulence genes for human infection before being exposed to the human hosts, and that these virulence factors are most effective when the host has underlying health issues. It is possible that a successful human infection is mediated by bacterial redundant virulence mechanisms that were acquired and maintained in the environment. In this review, we will leverage recent pieces of genomic studies to better understand the evolution of virulence mechanisms in B. pseudomallei and host defense system.
A Multi-Host—Pathogen Interaction
Interactions between free-living amoebae and environmental bacteria have been proposed to provide an evolutionary training platform for intracellular pathogen such as Legionella pnemophila (Park et al., 2020), Listeria monocytogenes (Schuppler, 2014), and B. pseudomallei (Inglis et al., 2000, 2003; Noinarin et al., 2016). Many free-living amoebae including the genera Acanthamoeba, Dictyostelium, Naegleria, and Paravahlkampfia share natural habitats with Burkholderia bacteria, including B. pseudomallei (Inglis et al., 2000, 2003; Noinarin et al., 2016). Amoeba-Burkholderia interactions can range from predator-prey to mutualistic relationships. The fate of these interactions is subjected to the species of amoebae host and Burkholderia (genotypes), stages of host, as well as external factors that modify the outcomes. The social amoebae Dictyostelium discoideum life cycle ranges from unicellular amoebae to multicellular slugs where ten of thousands of single-celled D. discoideum aggregate to form a fruiting body. D. discoideum can directly ingest B. pseudomallei and other Burkholderia bacteria as their food; thereby requiring the bacterium to resist host cell phagocytosis, persist inside a unicellular host, and migrate between host cells through the amoeba cytoskeleton. These processes share many similarities with mammalian host infection and likely prime the bacterium for an intracellular lifestyle. The sentinel cells in D. discoideum, which make up approximately 1% of slug cells, can use antimicrobial defense systems similar to those employed by phagocytes in the human innate immune system (Zhang et al., 2016). The sentinel cells are capable of releasing reactive oxygen species to lyse the soil bacterium. Moreover, they can also release an extracellular trap—the reticulated nets of DNA carrying antimicrobial granules—to kill the invading bacteria. The extracellular trap is an ancient host-defense mechanism common among phagocytic cells across vertebrates and invertebrates, thereby providing a training ground for B. pseudomallei infection in mammalian hosts. External factors such as nutrient availability were shown to determine the fate of D. discoideum—Burkholderia relationship. The association is beneficial to both parties when the nutrient is scarce. Under nutrient limited conditions, Burkholderia—associated D. discoideum produced more spores (Brock et al., 2013; DiSalvo et al., 2015) and had an increased uptake of secondary bacteria that can be used as food (Khojandi et al., 2019) which led to a better survival rate than D. discoideum without Burkholderia association. Under this condition, Burkholderia could be detected inside D. discoideum spores, which enable the bacterium to better disseminate. The mechanism underlining the decision to kill or cooperate is unclear.
The species of Burkholderia and genetic variations within the species have been shown to impact the outcomes of amoeba internalization (Haselkorn et al., 2019). It is possible that genetic variations in B. pseudomallei could influence the outcomes of human melioidosis. Heritability scores (Finucane et al., 2015; Hou et al., 2019; Speed et al., 2020) can be used to quantify the proportion of variations in the infection outcomes that can be explained by genetic variations in pathogens and hosts. The technique has been successfully applied to different pathogens (Lees et al., 2017) and human infectious diseases. However, the concept has not been widely adopted for B. pseudomallei – host infection. For B. pseudomallei infection in human hosts with no comorbidity, we estimated that 8% of host mortality could be explained by B. pseudomallei genotypes (h2 = 0.081, SE = 0.050, p = 0.018; see Supplementary Text for method). This new result highlights a moderate proportion of infection outcomes being explained by the bacterial genetics, and suggests that a substantial proportion of the outcomes could be explained by host genetics.
Interactions between pathogens and Homo sapiens have shaped the evolution of modern humans (Karlsson et al., 2014). Currently, less is known about how melioidosis has shaped the human populations, particularly in melioidosis endemic areas. The host immunity could function as a general defense against all invading pathogen mediated by the innate immune system, or a pathogen-specific defense facilitated through an adaptive immune response. For innate immune response, it is possible that interactions between human hosts and other common pathogens may have driven selection on host defense pathways that affect resistance. These may include human interactions with parasites or bacteria that cause malaria (Shimizu et al., 2000; Malaria Genomic Epidemiology Network, 2008; Timmann et al., 2012; Kariuki et al., 2020), tuberculosis (Mahasirimongkol et al., 2012; Thye et al., 2012), or sepsis (Southeast Asia Infectious Disease Clinical Research Network, 2017; Sweeney et al., 2018) in many tropical and subtropical countries, and cholangiocarcinoma (Zou et al., 2014) in Southeast Asia. Using h2, previous works estimate that genetic variations in human host can explain between 32 and 52% of infectious disease susceptibilities, depending on the population studied and the infectious agents (Cooke and Hill, 2001; Jia et al., 2019). Candidate gene approaches have determined host risk- and protective genetic markers for melioidosis infection (West et al., 2012, 2013; Myers et al., 2014; Chaichana et al., 2017; Dickey et al., 2019), many of which were previously identified as gene targets for other common pathogen infections. To cover melioidosis-specific, as well as broad-pathogen host factors, systemic and reproducible genome-wide scans are yet to be conducted.
Natural Selection in Genes Required for Host-Pathogen Interaction
Given an increasing availability of B. pseudomallei and host genomes, a genome-wide scan for signatures of selection, their directions (purifying, balancing, and positive selection) and their magnitudes could be very useful to investigate host-pathogen protein-protein interactions. As both host and B. pseudomallei migrate, detection of directional selection can be challenging after a population bottleneck. A bottleneck leads to reduced levels of genetic variation and a subsequent loss of a selection signal. For a soil microbe like B. pseudomallei, bacterial migration was shown to be infrequent, with major movements tracked and dated (Pearson et al., 2009; Price et al., 2016; Chewapreecha et al., 2017). The bacterial genome evolution is correlated with a strong geographical signal, with the highest diversity observed in Australia and reduced diversity following dissemination out of the Australian origin (Chewapreecha et al., 2017). Nevertheless, frequent recombination observed in B. pseudomallei can introduce new genetic variations, thereby enabling detection of selection signatures in other contemporary populations outside Australia (Chewapreecha et al., 2019). Natural selection on modern humans as a result of pathogen encounter and migration has been reviewed elsewhere (Karlsson et al., 2014; Sironi et al., 2015; Slodkowicz and Goldman, 2020).
For bacteria, evolutionary pressures on orthologous proteins can be quantified using the ratio between substitution rates at non-synonymous (dN) sites, which could have experienced selection, and synonymous (dS) sites which are presumably neutral. The dN/dS ratio is likely to be more than 1 if pressures favor changes in the protein sequence (positive selection). It is likely to be less than 1 if selections suppress changes in the protein sequence (purifying selection). Several studies (Yu et al., 2006; Losada et al., 2010; Nandi et al., 2010; Hayden et al., 2012; Chewapreecha et al., 2019) and this review utilized dN/dS to estimate selection pressures on coding sequences of the pathogenic B. pseudomallei as well as non-pathogenic but closely related species such as B. thailandensis. Although these studies focused on bacterial populations isolated from different melioidosis endemic regions, performed on different sample size and genetic diversity, they largely highlighted three similar patterns (Figure 1). The first common pattern is an elevated level of positive selection in Burkholderia accessory genes (genes that are variably present across different isolates) compared to core genes (genes that are consistently present in all isolates). This observation highlights the role of accessory genomes in mediating B. pseudomallei adaptation, a feature reflected by the open pangenome and large repertoire of accessory genes observed in B. pseudomallei and B. thailandensis (Nandi et al., 2015; Spring-Pearson et al., 2015; Chewapreecha et al., 2017). The second common pattern is the set of genes under purifying selections in B. pseudomallei and B. thailandensis involved in the bacterial replication, transcription and translation machinery; highlighting the conservation of these genes in both species. The third common pattern is that many genes under positive selection in B. pseudomallei and B. thailandensis are required for environmental survival and exchange of genetic materials including secretion systems, response regulator, heat shock proteins, and integration and/or restriction of horizontal gene transfer. However, genes required for host cell invasions such as cell adhesion, fimbriae, intracellular multiplication and macrophage killing are only positively selected in B. pseudomallei but not B. thailandensis. These distinct signals for positive selection in B. pseudomallei and B. thailandensis could mark a different pathogenic potential of the two species.
FIGURE 1.
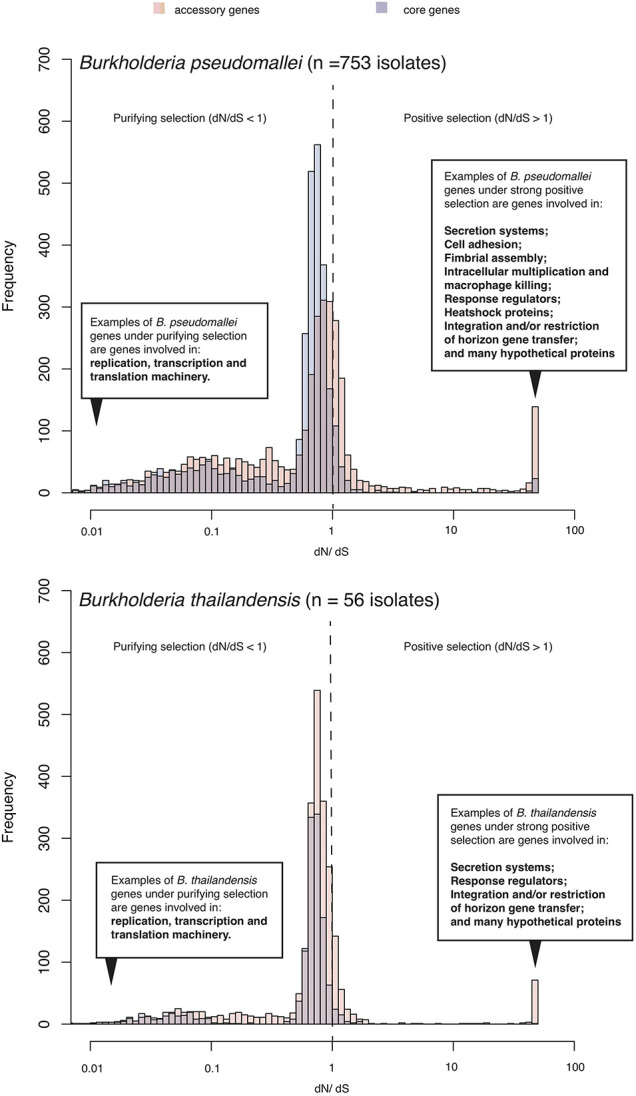
Selection pressure acting on B. pseudomallei population. The histogram summarizes ranges of dN/dS calculated from predicted coding sequences from a collection of diverse B. pseudomallei population from northeast Thailand (Chewapreecha et al., 2019), and B. thailandensis genomes from the public database. B. pseudomallei and B. thailandensis have highly plastic genomes comprising of at least two chromosomes of ∼7–8Mb in size when combined. Using a pan-genome approach, all coding sequences could be categorized as “core” (present in all genomes) or “accessory” (variably present across studied genomes). Accessory genes display an elevated level of dN/dS which is signatures of positive selection or more relaxed purifying selection.
Genes under positive selection could primarily offer B. pseudomallei advantages to survive harsh environmental conditions. Given its tropical and sub-tropical habitats, B. pseudomallei experiences drought and heavy rainfall on a seasonal basis. The bacterium can be found in deeper soil layers during the dry season and moves to the soil surface during the rainy season following water movement (Thomas et al., 1979; Currie and Jacups, 2003; Manivanh et al., 2017), highlighting a spatiotemporal transition. The bacterium is also subjected to heat stress. An in vivo study showed that B. pseudomallei had a slower growth rate and a shift in gene expression toward heat-shock proteins and bacterial motility when exposed to temperature stress (Paksanont et al., 2018). Intriguingly, B. pseudomallei is capable of persisting in a nutrient-free distilled water (Pumpuang et al., 2011). The bacterium was subjected to distilled water since 1994 and this experiment is still ongoing, thereby marking an unusual ability of B. pseudomallei to survive in low nutrient media and high osmotic pressure. Lipopolysaccharide, a known virulence factor, was proposed to facilitate B. pseudomallei survival in water. Statistical analysis suggested that there is a correlation between the presence of LPS and rainfall, in particular, LPS serotype B (Shaw et al., 2019). An experimental study further reported that a gene of inner core LPS biosynthesis cluster (waaE: BPSL2510) is vital for a long-term water incubation (Moore et al., 2008). Moreover, an adhesin BPSL1661 was identified as a hub for co-evolutionary signals in B. pseudomallei population (Chewapreecha et al., 2020). The gene was functionally characterized as essential during nutrient starvation (Chewapreecha et al., 2020), thereby highlighting nutrient limitation as a major evolutionary pressure experienced by this microorganism. The presence of B. pseudomallei is strongly associated with the low-nutrient soil (Limmathurotsakul et al., 2010b; Hantrakun et al., 2016). This led to the hypothesis that B. pseudomallei might have a competitive advantage over benign soil microbes in nutrient-depleted soil but is outcompeted in a nutrient-rich environment. The occurrence of B. pseudomallei in the nutrient-poor agricultural field has linked to crop residue burning, a common practice in the tropics where the burning practice could deplete the soil organic matter (Bot and Benites, 2005). An ability to persist in the hostile environmental conditions allows the bacterium to survive before it could seek shelters in a more protecting reservoir inside the single or multicellular hosts.
Genes that facilitate bacterial transition from the environment to hosts are also under positive selection. A genome-wide association study (GWAS) recently identified the bacterial genetic factors that distinguish between B. pseudomallei isolated from the environment and those causing disease in human (Chewapreecha et al., 2019). Genetic variants associated with cell entry and toxin were found to be more prevalent in disease- than environmental isolates, while variants involved in malfunctional cell adherence were found at higher frequency in environmental- than disease isolates. Together, this highlights significant roles of cell adhesion and cell entry in allowing the bacterium to switch to an intracellular lifestyle, either in an amoeba or in a human host where it causes melioidosis. The same study also quantified the numbers of time the variants were gained or lost from the population phylogenetic tree and highlighted multiple gain-and-loss events for both disease- and environment-associated variants. This genetic evidence suggests a process into which B. pseudomallei can adapt to colonize multiple niches by exploiting existing variations or exchanging disadvantageous for advantageous alleles that promote its survival in a new niche, including the human host.
Mechanisms That Generate Genetic and Molecular Variations
Genetic information in both B. pseudomallei and human host can be passed on vertically, although mistakes during replication process could result in small-scale genetic variations. In B. pseudomallei, larger-scale genetic variations can be introduced by recombination event or horizontal gene transfer (HGT). The former process is akin to sexual reproduction and meiotic crossing-overs in its human hosts. Due to contrasting short- and long- generation time of B. pseudomallei (49 min in the log phase; Ou et al., 2005) and host (22–33 years for modern humans; Ewbank, 2016); the host cannot solely rely on new genetic diversity generated when the new offspring is born. A healthy human possesses a large and dynamic repertoire of B cell- and T cell receptors that recognize the invading pathogen and produce a repertoire of antibody that recognizes a variety of antigenic structure. This molecular diversity is achieved without a change in genetic content. The mechanisms that generate the genetic and molecular diversity of the human immune system have been reviewed elsewhere (Nikolich-Zugich et al., 2004; Robinson, 2015; Dendrou et al., 2018; van den Broek et al., 2018; Adams et al., 2020).
Small-scale genetic variations in B. pseudomallei can be introduced by point mutation which substitutes one nucleotide with another or microindel (an insertion or deletion) that impact 1–50 bp. B. pseudomallei substitution rate was reported to be 1.7–4.9 × 10–7 substitutions per site per year (Viberg et al., 2017; Pearson et al., 2020), which is comparable to those detected in other Burkholderia genera (Lieberman et al., 2014). The rates of indels have not been quantified in B. pseudomallei. However, indels can be detected from isolates obtained from acute and chronic host infections (Hayden et al., 2012; Limmathurotsakul et al., 2014a; Viberg et al., 2017) as well as from the environment (Rachlin et al., 2020), thereby highlighting the role of indels in B. pseudomallei evolution.
Medium-scale genetic variations can be brought in by homologous recombination, with each event contribute to a median recombining size of 5 kb (Nandi et al., 2015) (range 3 bp to 71 kb). A single recombination event can introduce 7.2 times greater nucleotide polymorphisms than a single substitution event (average r/m = 7.2). However, the amounts of SNPs being introduced per recombination event is subjected to the genetic distance between DNA donor and recipient. Sources of imported DNA could be from the same species donor, closely related species such as B. thailandensis and/or other soil microbes. Different B. pseudomallei lineages have been shown to recombine at different rates (Nandi et al., 2015), a characteristic also observed in other recombinogenic bacteria (Chewapreecha et al., 2014; Sanchez-Buso et al., 2014; David et al., 2017). Interestingly, not all DNA donor-recipient pairs are possible, suggesting a structure to the genetic flux within B. pseudomallei population. A lineage-specific restriction-modification system was shown to act as a barrier that restricts gene flow (Nandi et al., 2015). These modification control systems are based on DNA methylation which allows the bacterium to discriminate between correctly methylated “self” DNA, and inappropriately methylated or unmethylated “non-self” DNA. Correctly methylated DNA can be taken up and integrated into the bacterial genome, whereas inappropriately methylated or unmethylated DNA will be degraded. Interestingly, many recombination “hotspots” are focused on bacterial virulence factors (Nandi et al., 2015), highlighting the role of recombination in tuning the bacterial ability to infect hosts.
Large-scale genetic variations can be introduced by HGT or large-scale insertion or deletion. HGT includes the uptake of foreign DNA that can subsequently be integrated into the chromosomes and resulted in the regions termed genomic islands (GIs) (Koonpaew et al., 2000; Holden et al., 2004; Duangsonk et al., 2006). The repetitive genetic composition of B. pseudomallei genomes such as tandem repeats and transposons is believed to aid the integration of horizontally acquired elements into the chromosomes (Holden et al., 2004; U’Ren et al., 2007). Despite several plasmid elements being identified in B. pseudomallei genomes (Holden et al., 2004), four complete plasmids has been characterized so far (NCBI, December 2020) (Nandi et al., 2015). With long-read sequencing technology, the plasmid could be more reliably assembled, thereby allowing more genetic variations to be studied. At least 16 GIs had been identified in the K96243 genome (Holden et al., 2004). Several studies highlight distinct geographical distribution of GIs or their combinations (Ou et al., 2005; Duangsonk et al., 2006; Sim et al., 2008; Tuanyok et al., 2008), leading to a hypothesis that each GI or combination of GIs may confer different fitness under different environments (Sim et al., 2008). GIs carry several virulence factors, many of which are known to interact with hosts, including filamentous hemagglutinin adhesin (fhaB3), and a member of two-partner secretion system (bpaAB) (Sim et al., 2008).
Genetic, Structural or Molecular Variations Observed in Host-Pathogen Interaction
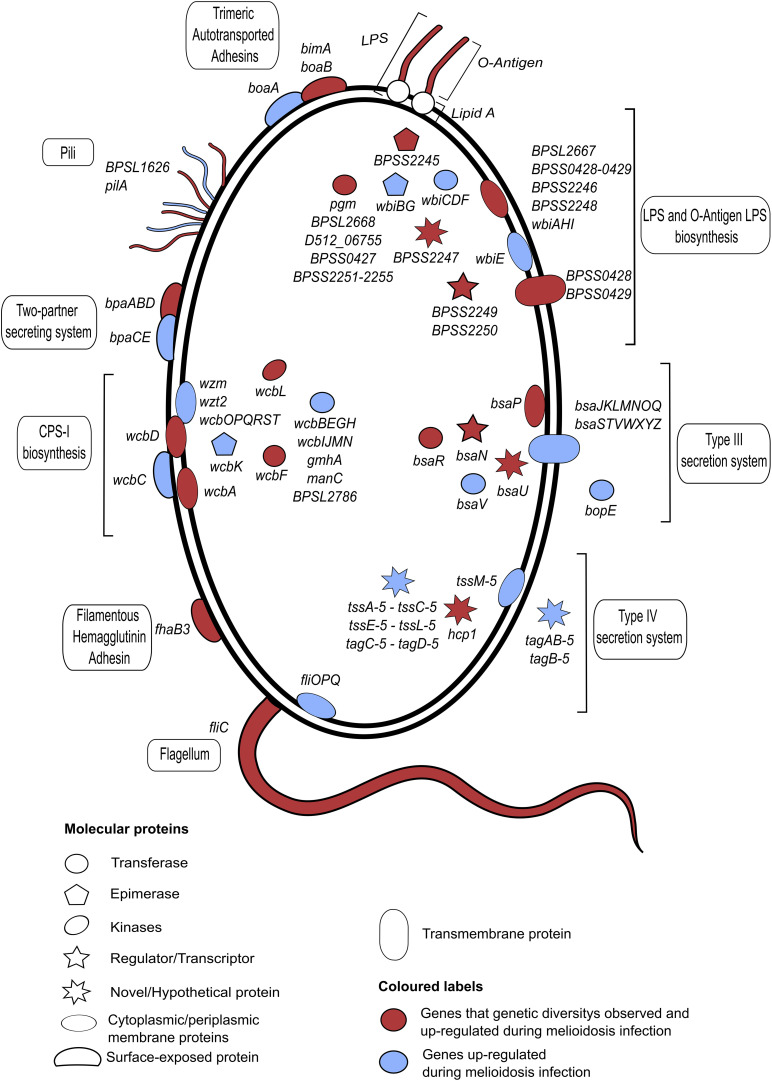
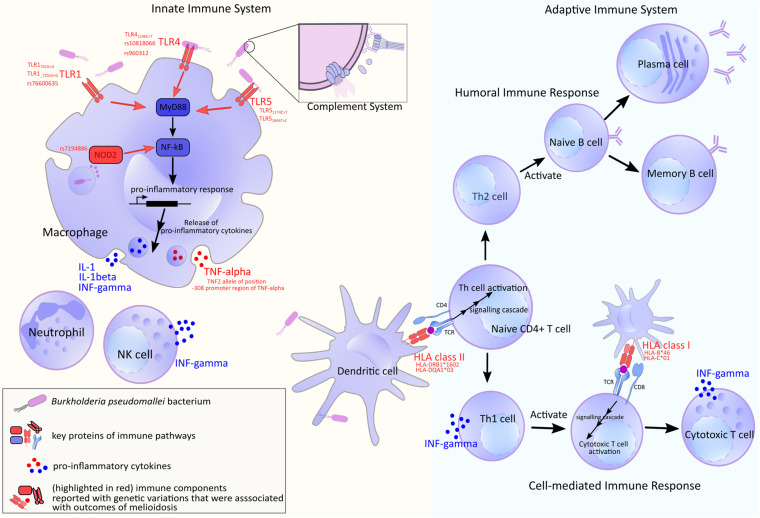
Genetic, structural and molecular variations and their functional impact to B. pseudomallei - host interaction have been the center of melioidosis research over the past few decades. We will summarize genetic and structural diversity in bacterial genes known to mediate the infection (Figure 2 and Supplementary Table 1). The genes discussed here are shown to be essential for infection by a genome-wide saturation mutagenesis (Moule et al., 2015) and also expressed during in vivo infection (Ooi et al., 2013). When host partners are known; variations detected in host proteins, and the host response are also described (Figure 3). It should be noted that there are large overlaps in the host immune system and the examples described here only represent a fraction of the whole machinery.
FIGURE 2.
A summary of B. pseudomallei genes or operons that are expressed during infection and characterized genetic variations. All plotted genes or operons are up-regulated during in vivo infections (Ooi et al., 2013), with a subset that reported genetic variations marked in red. The shape and location of each individual gene indicate the gene function and cellular compartment, respectively. All annotated genes or operons and their functions are described. The bacterium displays a repertoire of antigenic variations, including lipopolysaccharides (LPS), capsular polysaccharides (CPS) and surface proteins. B. pseudomallei LPS is immunologically classified into a number of serotypes A, B, and B2; with each serotype reported to be heterogeneously distributed across distinct geographical locations. Another highly diverse virulent protein is a fimbrial protein which displays a strong geographical distribution between Australia and Asia. Strains from Asia commonly possess a Yersinia-like fimbrial (YLF) gene cluster that believed to be horizontally acquired. B. pseudomallei carries 4 different types of CPS: CPS I, CPS II, CPS III, and CPS IV. A full gene description is provided in Supplementary Table 1.
FIGURE 3.
A summary of human host immune components used in defending against B. pseudomallei. Genes or molecules that show genetic variations are marked in red. Upon infection, the bacterial antigens are recognized by host receptors including Toll-like receptor (TLR) and HLA which also display large genetic diversity. The latter is reported to be varied by ethnic groups (de Bakker et al., 2006; Gourraud et al., 2014). Following an entry into host, B. pseudomallei faces the innate axis of the immune system. Macrophages and neutrophils are recruited to the site early upon the infection; these cells are reported to be essential to the early bacterial containment and clearance (Breitbach et al., 2006; Easton et al., 2007), though excessive recruitment of neutrophils may have a negative outcome that allows B. pseudomallei to propagate intracellularly (Ceballos-Olvera et al., 2011). These innate phagocytes possess pattern recognition receptors (PRRs), such as surface receptors TLR2, TLR4, and TLR5, as well as, cytosolic receptors NOD2; these are reported to be vital for the fight against B. pseudomallei (West et al., 2008; Myers et al., 2014; Weehuizen et al., 2015; Birnie et al., 2019). The signals transduced by these receptors result in mobilization of nuclear factor NF-κB which trigger appropriate immune responses including the synthesis of pro-inflammatory cytokines and initiation of the downstream adaptive immune cascades (Pothlichet and Quintana-Murci, 2013). It is through the antigen-presenting cells and their appropriate antigen-HLA class II complexes that enable the activation of CD4 + T lymphocytes. The naïve CD4 + T cells sit at the central part of the adaptive axis. They can differentiate into Th1 cells which facilitate cell-mediated immune response by CD8 + cytotoxic lymphocytes. A study has shown that strong CD4 + and CD8 + T cell response was elicited during acute melioidosis and the lower cellular response was correlated to fatality (Jenjaroen et al., 2015). On the other hand, naïve CD4 + T cells can also mature to be Th2 cells which initiate class switching of B cells and support humoral immune response.
Genetic Variations in B. pseudomallei Flagella Systems and Host Toll-Like Receptor 5 (TLR5)
Flagella systems of B. pseudomallei enable the motility of the bacterium intracellularly as well as in the environment (Figure 2; DeShazer et al., 1997; French et al., 2011). Flagella filaments are constituted from thousands of flagellin protein monomers, encoded by fliC gene, strung into protofilaments before being braided to form a flagellum (Samatey et al., 2001). Flagellin has been known for its extreme diversity with 113,285 unique nucleotide sequences across the prokaryote phyla (Hu and Reeves, 2020). Flagellin inactivation in B. thailandensis resulted in the reduction of bacterium intracellular motility and cell-to-cell spread (French et al., 2011). Using PCR-restriction fragment length polymorphism analysis, four different alleles of flagellin protein (fliC: BPSL3319) were identified from 100 Malaysian clinical isolates (Tay et al., 2010), and more is expected if investigated with next-generation sequencing. Proteins in the flagella systems were reported to elicit host immune response, highlighting them as melioidosis vaccine candidates (Brett et al., 1994; Chua et al., 2003; Chuaygud et al., 2008; Gregory et al., 2015; Koosakulnirand et al., 2018).
In addition to flagella, a fimbrial gene cluster which is required for cell adherence also displays genetic variation with distinct alleles being predominantly detected in different geographical locations. A yersinia-like fimbrial (YLF) gene cluster, believed to be horizontally acquired was shown to be more prevalence in Southeast Asia and thus was used as a marker for the introduction of B. pseudomallei from this region (Tuanyok et al., 2007; Sarovich et al., 2014; Chewapreecha et al., 2017). A putative type I fimbrial protein BPSL1626 was shown to elicit an immune response and have a potential as a vaccine candidate against melioidosis (Capelli et al., 2018). Whereas, B. thailandensis-like flagellum and chemotaxis (BTFC) gene cluster is believed to be an ancestral sequence in B. pseudomallei and is most common in Australia (Tuanyok et al., 2007). The YLF and BTFC gene clusters are mutually exclusive between the two endemic areas where the latter may implicate in the spread between cell-to-cell by the flagellar protein leading to the formation of multinucleated giant cells (MNGCs) and eventually apoptosis/cell death (French et al., 2011).
It has been established that flagellin is a ligand of TLR5 (Figure 3). The recognition stimulates pro-inflammatory responses including the rise in intracellular calcium ion and upregulation of pro-inflammatory cytokine TNF-α and IL-6 (Hayashi et al., 2001; Chen et al., 2007). TLR5 stop codon polymorphism, TLR51174C>T or rs5744168 was strongly associated with protection against fatality as well as organ failure in a case-control cohort study. When challenged with B. pseudomallei, TLR51174C could mediate the activation of NF-κB, while TLR51174T could not; additionally, TLR51174T saw reduced flagellin-induced cytokines levels (West et al., 2013). Another independent investigation on the same population also found the association between the truncated TLR51174C>T variant and survival from acute melioidosis, as well as, a lower rate of bacteraemia (Chaichana et al., 2017). Furthermore, both studies also found TLR51174C>T variant with a lower level of anti-inflammatory IL-10, which the authors suggested the possibility in which the mortality risk may be modulated by TLR5-driven IL-10 release. Interestingly, a recent study has suggested that the effect of this hypofunctional TLR5 variant may not be restricted to flagellin-driven pathway (Dickey et al., 2019). Another polymorphic variant is TLR51846T>C which was also associated with protection against death and blunted flagellin-driven cytokine response; however, the authors also reported high linkage disequilibrium of the variant with TLR51174C>T (Chantratita et al., 2014), which might reduce the confidence of causal relationship.
Structural Variations in B. pseudomallei LPS and Genetic Variation in Host Toll-Like Receptor 2 and 4 (TLR2 and TLR4)
B. pseudomallei possesses an extensive network of polysaccharides on its outer membrane, namely capsular polysaccharide (CPS) and lipopolysaccharide (LPS) (Figure 2, for a full review on CPS and LPS in Burkholderia spp.; Cloutier et al., 2018). Both are known to play a vital role in virulence of melioidosis and have been used as subunits for vaccine development (Brett and Woods, 1996; Nelson et al., 2004; Wikraiphat et al., 2009; Scott et al., 2014b). Based on a cellular compartment, B. pseudomallei LPS can be divided into (Knirel et al., 1992): lipid A—an endotoxic component embedded in the phospholipid bilayer of the outer membrane; inner and outer core oligopolysaccharide; and O-antigen (Novem et al., 2009). Multiple in vitro studies showed LPS challenge could mount innate and adaptive immune responses, and Nitric Oxide (NO) production, with different LPS serotypes (A, B, and B2) reported to mount different magnitude of responses (Norris et al., 2017). Some genetic data of LPS biosynthesis operons were available. However, many of these were generated from short-read sequencing platforms, which are not ideal to investigate LPS due to the repetitive nature of the LPS locus. Long-read technologies, on the other hand, could overcome this assembly issue, giving complete contigs of the bacterium genome.
To date, 4 serotypes of LPS were previously characterized. Type A LPS is a majority serotype found in both endemic areas, namely Southeast Asia and Australia (Anuntagool et al., 2000, 2006; Tuanyok et al., 2012). Whereas, serotype B is less common in the endemic areas (Australia and Southeast Asia) and is prevalent in both clinical and environmental origins (Tuanyok et al., 2012). However, a recent analysis of clinical B. pseudomallei isolates revealed that Serotype B was highly predominant in India (Shaw et al., 2019). LPS serotype B2 is a variant of LPS serotype B and more commonly found in Australia and Papua New Guinea (Tuanyok et al., 2012). In contrast, LPS R or Rough serotype lacks O-antigen moiety of the LPS structure. It was identified using SDS-PAGE and silver staining techniques with no O-antigen ladder pattern. Type R is relatively rare and found only from the Australian clinical and environmental strains (Anuntagool et al., 2006). Interestingly, it is frequently prevalent in the patients with a relapse history of melioidosis, however, there is no direct association between them (Limmathurotsakul et al., 2014b).
A comparative genomic analysis revealed that there are distinct variations in the core compositions of the O-antigen LPS biosynthesis gene clusters between A, B, and B2 serotypes. However, there are several genes conserved among them. In addition, type A LPS gene operon is also observed in both B. thailandensis and B. mallei, closely related Burkholderia species (Tuanyok et al., 2012). Interestingly, genomic analysis of clinical isolates from Madagascar revealed that there is a 13.5 kb deletion observed in the LPS biosynthesis gene cluster of serotype B, conserving only some genes in the cluster that are essential for the biosynthesis of LPS B2 (Sarovich et al., 2016). The lack of some core genes in the LPS biosynthesis cluster could lead to the reduction of its serological properties. This is supported as some evidence suggested that the strains with LPS type B2 become sensitive to 30% normal human serum whereas the strains with LPS type B remain resistant (Tuanyok et al., 2012).
On the other hand, a single nucleotide insertion of the wbiI gene is observed in the LPS biosynthesis gene cluster of a rough serotype in a patient with more than 16-year chronic lung infection associated with melioidosis. This frame-shift mutation of wbiI gene (an essential gene for the O-antigen synthesis) disrupts the epimerase/dehydratase function of this gene and results in the loss of O-antigen moiety, possibly switching the serotype of B. pseudomallei isolates from type A to type R (Price et al., 2013; Pearson et al., 2020). Pearson and colleagues also revealed the nucleotide insertion of D512_15771 (wbiH) and D512_06755 gene are observed in B. pseudomallei MSHR6686 of the same patient. The mutation in both genes may confer to the reduction in LPS modification and production, assisting in the escape from the host immune response. In addition, a partial deletion of D512_20407, wbiA homolog is also identified in MSHR6686 (Price et al., 2013; Pearson et al., 2020). However, genomic analysis of the rough serotype that is naturally found in the environment or initial infection has not been done to determine the genetic makeup of this serotype.
Not only genetic heterogeneity exists among the serotypes of LPS, further structural diversity is also observed at the O-antigen of LPS serotypes (Stone et al., 2012; Norris et al., 2017). The O-antigen is one of the LPS components with a structure of unbranched disaccharide repeat units. Remarkably, the structural modifications of these sugar chains were observed where substitutions of 2-O-methylated and 4-O-acetylated at talose residues were observed only in about 33% of the LPS serotype A. Whereas the rest bear 2-acetyl substituents at the same residues (Perry et al., 1995). More recently, a structural analysis of O-antigen in the serotype A reported that the modification of the talose residue is more complex than what was previously reported (Heiss et al., 2013). Although multiple gene inactivation studies revealed that wbiA and oacA genes are essential for these modifications at the talose residues (Brett et al., 2003, 2011), it is possible that structural diversities of the LPS O-antigen are further modified at post-transcriptional and post-translational levels, for instance, a length variation of O-antigen chain observed in Escherichia coli O9a which is tightly controlled by the biosynthetic enzymes dynamic (King et al., 2014).
The LPS is common across Gram-negative bacteria and also a well-established pathogen-associated molecular pattern (PAMP) that can trigger the pro-inflammatory innate immune response, such as translocation of NF-κB and TNF-α cytokine release, via its interaction with TLR4, CD-14 and adaptor protein MD-2 (Park and Lee, 2013). Although evidence has been inconclusive, LPS sensing was shown to be through TLR4 in murine models, and TLR4 as well as TLR2 (Wiersinga et al., 2007; West et al., 2008) in human models (Figure 3; Weehuizen et al., 2015). Polymorphisms of TLR4 were reported in humans with TLR41196C>T allelic variant associated with protection against melioidosis when compared to non-hospitalized controls (West et al., 2012). In addition, TLR4 rs10828066 SNP variant was significantly associated to be protective against melioidosis in both adjusted non-hospitalized and hospitalized control groups, whereas TLR4 rs960312 was associated with bacteraemic or lung melioidosis (West et al., 2012).
Genetic Variations in B. pseudomallei CPS and Host B Cell Repertoire
CPS is one of the key virulence determinants in B. pseudomallei (Figure 2). To date, four different types of CPS (CPS I-IV) have been described (Holden et al., 2004). CPS-I biosynthesis cluster is a large 34.5 kb operon harboring on the chromosome one of B. pseudomallei (wcbT to manC). A study in an animal model reported that the CPS-I cluster is required for the full virulence of B. pseudomallei (Sarkar-Tyson et al., 2007). CPS-I exhibits an immunogenic role and the absence of this gene cluster resulted in the attenuation of B. pseudomallei in a mouse model (Atkins et al., 2002; Reckseidler-Zenteno et al., 2005; Parthasarathy et al., 2006). A passive protection was observed when murine intranasal infection models were immunized with anti-CPS monoclonal antibodies (AuCoin et al., 2012), with the later study reported that conjugated CPS could provide the highest degree of protection (Scott et al., 2014a).
The structural basis of CPS-I is highly conserved (Perry et al., 1995). Remarkably, CPS-I is believed to be horizontally transferred between species as the GC content of this CPS-I cluster is about 58% (Reckseidler et al., 2001). In addition, it is possible that the horizontal acquisition of this gene cluster was a key event in the pathogenic evolution of B. pseudomallei compared to B. thailandensis where the cluster is absent (Yu et al., 2006). Nevertheless, several genes involved in the biosynthesis of CPS-I of B. pseudomallei were reported in B. thailandensis with a relatively low sequence identity (75%). In addition, a novel variant of B. thailandensis strains (BTCV) is found to carry a B. pseudomallei like CPS-I (95% sequence identity) gene cluster with an identical organization. Although the origin of the CPS-I gene cluster in B. pseudomallei has yet been identified, several genes involved in the sugar biosynthesis appear to be orthologs to the genes identified in the Yersinia species (Y. pseudotuberculosis H892/87) and some in other gram-negative bacteria (Cuccui et al., 2012). It is possible that the CPSI gene cluster is not entirely acquired from one bacterial species but several organisms. Population studies also detected genetic variations in CPS-I (Challacombe et al., 2014; Chewapreecha et al., 2017). Whether this genetic variation has implications on virulence is a subject of further investigation. Another CPS-I variant could be found in B. pseudomallei isolated from a 16-year chronic melioidosis patient. A single nucleotide insertion is observed within wcbR, an important component involved in the fatty acid synthesis of CPS-I, causing a frameshift mutation in the earlier samples isolated from this patient (Price et al., 2013; Pearson et al., 2020). A CPS-I deletion region, which includes wcbR gene, has shown in the reduction of CPS production in hamster model but not absence entirely (Gutierrez and Warawa, 2016). Price et al. (2013), suggested that this degree of CPS-I-dependent virulence decreases and may consequently be a critical step in the progression for melioidosis to become a chronic-carriage disease.
Unlike CPS-I, CPS-II and CPS-III have been associated with B. pseudomallei persistence in the environment (Reckseidler-Zenteno et al., 2009, 2010), while CPS-IV is less well-characterized. Up to date, no reports suggest the genetic variations of CPS-II, -III, and -IV biosynthesis clusters in B. pseudomallei. The diversity and function of these capsules in the pathogenesis, immunomodulation and environmental adaptation of B. pseudomallei warrant further studies.
We next considered host partners that interact with B. pseudomallei CPS. The CPS has been shown to elicit strong host immune response (Reckseidler et al., 2001; Atkins et al., 2002). CPSs from various B. pseudomallei strains were recognized by the same group of monoclonal antibodies which suggests a limited number of epitopes of this molecule (Zou et al., 2008). The molecule’s monomers can cross-link to B cell receptors and, at an appropriate density, induce a downstream humoral response that is distinct from T cell-dependent pathway (Clarke et al., 2013; Akkaya et al., 2020). Antigen recognition through B-cell receptors is formed through random somatic changes of germline DNA. This results in a repertoire of distinct sequences that enable antigen recognition across wide ranges of pathogens. The B-cell receptors dynamic in melioidosis has not been studied.
Genetic Variations in B. pseudomallei Adhesins and Autotransporters
Filamentous Hemagglutinin Adhesin (FHA) of B. pseudomallei is highly diverse (Figure 2). Early investigation of genomic islands (GIs) from five B. pseudomallei strains identified three different fhaB gene clusters on different GIs of the bacteria. The bacterium could carry multiple fhaB gene clusters, many of with carrying either a combination of cluster I (GI5a/GI5a.1/GI5a.2) and cluster III (GI16/GI16.1), or cluster III alone (Tuanyok et al., 2008). More genetic variations of the FHA loci were reported from the Australian isolates (Chewapreecha et al., 2017). The fhaB3 gene (BPSS2053) has been characterized as an important virulence factor of B. pseudomallei; enabling bacterial binding to the host epithelial cells (Sim et al., 2008), and an anti-macrophage factor (Dowling et al., 2010). It was found in all isolates from Thailand but found in only 83% of Australian strains where the absence of this gene in Australia population correlated with the skin abscess formation and lower mortality rate (Sarovich et al., 2014).
Autotransporter (AT) proteins are one of the largest family of the secretion systems in Gram-negative bacteria, allowing the transportation across the bacterial membrane as well as involving the virulence and immunogenicity of the pathogens (for review Lazar Adler et al., 2011). In B. pseudomallei K96243, a sequence analysis revealed to have at least 11 ATs located in the genome, including putative Trimeric Autotransporter Adhesins (TAAs), bimA (Stevens J.M. et al., 2005) and boaB, two-partner secretion system (TPS), bpaA, bpaB, and bpaD (Campos et al., 2013). Inactivation in some of these genes attenuated and reduced the intracellular survival of B. pseudomallei in the macrophage-like cells (Lazar Adler et al., 2015). ATs are variably present in the genomes. The gene boaB (BPSL1705), which aids bacterial binding to host respiratory cells (Balder et al., 2010), was found to be absent in several strains in Africa, Brazil (Sarovich et al., 2016). When ATs are present, genetic variants can also be observed. Australian-specific and African-specific variants have been reported in bpaA, bpaB, and bpaD (BPSS1434, BPSL2063, and BPSS0088, respectively) (Brown et al., 2004; Tuanyok et al., 2008).
Several experiments demonstrated the role of BimA in actin polymerization and motility (Stevens M.P. et al., 2005; French et al., 2011), promoting a movement of cell-to-cell spread and within the host cells (Lazar Adler et al., 2011). Furthermore, BimA is antigenic (Suwannasaen et al., 2011) and seroactive (Felgner et al., 2009). Two variants of bimA (BPSS1492) were identified: B. pseudomallei bimA (bimABP), and bimA-like B. mallei (bimABM). The latter is an ortholog of bimA from Burkholderia mallei (95% sequence identity) and displays the same domain organization: a single actin monomer binding motif (WH2), a proline-rich domain and a transmembrane anchor domain (Sitthidet et al., 2008). Whereas the bimABP consists of two predicted WH2 domains, a proline-rich domain, a membrane anchor domain and an additional predicted casein kinase II (Stevens M.P. et al., 2005). Of note, the variation was also observed between the predicted proline-rich domain of the bimABM and B. mallei bimA where the former has fewer motifs (Sitthidet et al., 2008). Distinct geographical distribution of bimABP and bimABM have been reported (Sitthidet et al., 2008; Sarovich et al., 2014; Shaw et al., 2019). Benanti et al. (2015) illustrated that both BimAs are functionally similar and associated with the nucleation and elongation of actin filaments with more plaque formation observed in bimABP variant compared to bimABM. A subunit vaccine investigation reported a significant increased survival rate of BALB/c mice immunized with BimABM recombinant, when the mice were challenged with both B. mallei and B. pseudomallei (Whitlock et al., 2010).
Genetic Variations in B. pseudomallei Secretion Systems
Several secretion systems and secreted proteins in B. pseudomallei are vital to combat the host immune defense and environmental stresses. B. pseudomallei has at least three type III secretion systems (T3SSs) (for review, Vander Broek and Stevens, 2017)and six type VI secretion systems (T6SSs).
Among T3SSs, T3SS-3 is better characterized and believed to be an integral part of the full B. pseudomallei virulence in mice and hamster models, as well as associated with the intracellular survival and dissemination. K-mer based sequence analysis has recently demonstrated that variations in the T3SS-3 gene cluster were detected across the global population of B. pseudomallei (Chewapreecha et al., 2017). This included bsaU(BPSS1539), bsaR (BPSS1542), bsaP (BPSS1544), and bsaN (BPSS1546) (Chewapreecha et al., 2017) which are believed to be involved in intracellular escape (Pilatz et al., 2006), predicted chaperone protein (Panina et al., 2005), T3SS-3 secretion regulator (Broek et al., 2015), and T3SS-3 regulator (Chen et al., 2014), respectively. In addition, a partial deletion of genes in the T3SS is also observed in a patient with persisted infection of melioidosis as a result of within-host adaptations (Pearson et al., 2020). For T6SSs, T6SS-5 is better characterized (for review, Lennings et al., 2019)and functionally confirmed to mediate the translocation of effector proteins via contact-dependent manner (Silverman et al., 2012) and MNGC formation (Burtnick et al., 2011; Chen et al., 2011). In addition, an experimental study suggested that T6SS-5 may involve in the intracellular survival of B. pseudomallei in macrophages where the expression level of T6SS-5 is dependent on virAG and bprC regulatory gene (Chen et al., 2011). Genetic variations are also observed in several genes clustered T6SS-5 with distinct geographical distribution (Chewapreecha et al., 2017). Remarkably, a large deletion region on chromosome 2 of an environmental B. pseudomallei A4 and an isogenic strain of K96243 has been demonstrated of which they failed to form a plaque in epithelium cells. Of those, the deleted genes are including genes involved in T3SS-3 and T6SS-5 systems. Interestingly, Saiprom et al. (2020) also identified an absence of multiple T3SS-1 genes of previously described Thai environmental strain RF80.
Genetic Variations in Human Leukocyte Antigen (HLA)
For the rest of the bacterial factors that do not get recognized by any specific innate pattern recognition receptors, these antigens will be processed and presented on HLA of antigen-presenting cells such as dendritic cells before getting recognized by appropriate T cell receptors (TCR) of the adaptive immune axis; this HLA-peptide-TCR interaction kicks start the adaptive immune response. There are three different classes of HLA: class I interacts with TCRs on CD8 + T cells while class II binds to TCRs on CD4 + T cells, and the less well-established class III which is not involved in antigen processing and presentation (Dendrou et al., 2018). Acute melioidosis patients with diabetes mellitus were reported to have lower HLA-DR expression on plasmacytoid dendritic cells than the non-diabetic diseased group (Kronsteiner et al., 2019). In addition, in non-diabetic patients, fatal cases presented with significantly lower expression of HLA-DR on monocytes and plasmacytoid dendritic cells, compared to the survived cases (Kronsteiner et al., 2019). Similar observation was found in murine infection models where B. pseudomallei-infected BALB/c and C57BL/6 mice showed reduced expression of MHC class II on plasmacytoid dendritic cells, however, this was not statistically significant (Williams et al., 2015).
Genetic variations in HLA have been linked to the melioidosis outcomes. A work conducted in an endemic area of Thailand has compared HLA allele frequencies in melioidosis cases and healthy controls. The authors reported a significant increase of DRB1∗1602 frequency in melioidosis patients, compared to healthy controls. Moreover, an increase in HLA-DRB1∗1602 and a decrease in HLA-DQA1∗03 allele frequencies were associated with septicaemic cases of melioidosis (Dharakul et al., 1998). Another study conducted on the same Thai population screened a panel of various HLA class I genotype frequencies in survived and fatal cases of melioidosis. The authors found that HLA-B∗46 and HLA-C∗01 were associated with increased mortality; they were also reported to be in linkage disequilibrium (Dunachie et al., 2017). Interestingly, HLA has been linked with diabetes mellitus, the prominent comorbidity of melioidosis with 12-fold increased risk of developing the disease (Limmathurotsakul et al., 2010a). Several recent publications using GWAS have identified variations of HLA and their corresponding protective or predisposing association with type 2 diabetes (Williams et al., 2011; Scott et al., 2017; Zhao et al., 2017). These prompt further characterization of HLA variants and their relationship with melioidosis progression when modulated by patient diabetic status. However, genetic studies on HLA and melioidosis have suffered from small sample size and availability of reliable HLA typing platform. This is largely impeded by the under-representation of genomic data from the population from melioidosis endemic areas. The availability of data is crucial as this improves imputations and discovery of new causal variants and disease association.
Discussion
The advancement in omic technologies has improved our understanding of co-evolution between B. pseudomallei and different hosts, thereby guiding better control policy, treatment option and vaccine design. Proteins or molecules that participate in host—B. pseudomallei interaction are extremely variable. Their variations have been seen at the genomic, epigenetic, transcriptomic and proteomic levels. Although genome data for human population from melioidois endemic areas is still scarce, genome data for B.pseudomallei has been accumulating (Holden et al., 2004; Hayden et al., 2012; Price et al., 2013, 2016; Sahl et al., 2013, 2016; Daligault et al., 2014; Bugrysheva et al., 2015; Chen et al., 2015; Hsueh et al., 2015; Johnson et al., 2015a,b; McRobb et al., 2015; Nandi et al., 2015; Sidjabat et al., 2015; Song et al., 2015; Spring-Pearson et al., 2015; Viberg et al., 2015; Chapple et al., 2016; Aziz et al., 2017; Chewapreecha et al., 2017, 2019; Podnecky et al., 2017; Viberg et al., 2017; Webb et al., 2019). When combined with spatial and temporal information, this allows further exploration of allelic variants and a shift in allele frequency over space and time. Moreover, a genome-wide saturation mutagenesis can aid prediction of essential genes required under certain conditions (Moule et al., 2015). This can be coupled with transcriptome information to understand variations in the expression patterns (Chieng et al., 2012; Ooi et al., 2013; Price et al., 2018) during the course of infection, and across different host types. A dual host-pathogen transcriptome study has not been conducted for melioidosis but is promising to provide valuable insight into the interaction as well as variations that lead to different infection outcomes.
In this article, we mainly explored variations of B. pseudomallei genes implicated in human host infections at the genetic level. Many of these genes display a strong geographical signal which could either be a result of a founder effect following a migration out of Australia, or an acquisition of new alleles required for local adaptation after an introduction to new geographical location. For each virulence gene, we also noted co-existence of multiple alleles in B. pseudomallei population isolated from the same geographical region. The genetic polymorphism could be maintained by balancing selection where each co-existing allele must be favored under different condition. In the context of virulent genes, these could involve competitions with different soil organisms. Signals for positive selection could be detected in genes that promote bacterial survival under hostile environment, and genes required for cell entry and adaptation to an intracellular lifestyle. The latter can be grouped as virulence genes although they may primarily be used in amoeba hosts rather than mammalian hosts. For multi-host—pathogen interaction, it is thus essential to consider virulence in a broad host and environmental context. The picture is far from complete at the moment, but more incoming omic data is promising to shed light on this complex relationship.
Author Contributions
CCe conceived the study and performed data analyses. CCo and PB performed literature review. AS performed data analyses. All authors wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. CCo was funded by Wellcome International Master Fellowship (221418/Z/20/Z). CCe was funded by Wellcome International Intermediate Fellowship (216457/Z/19/Z) and Sanger International Fellowship and Thailand Research Fund (MRG6280226).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.612568/full#supplementary-material
We estimated the proportion of infection outcome variations (survival vs. mortal outcomes after 28 days) explained by B. pseudomallei nucleotide polymorphisms (SNPs). SNPs were called from Chewapreecha et al. (2019, 2020) using kSNP v3 software. A filtering for minimum allele frequency at 0.01 (MAF = 0.01) resulted in 65,379 SNPs for the analysis. B. pseudomallei population was defined by PopPUNK (Lees et al., 2019). We employed the Genome-wide Complex Trait Analysis (GCTA) (Yang et al., 2011) to calculate h2 using genomic restricted maximum likelihood (GREML) while adjusting for population structure using population clusters defined by PopPUNK.
References
- Abbink F. C., Orendi J. M., de Beaufort A. J. (2001). Mother-to-child transmission of Burkholderia pseudomallei. N. Engl. J. Med. 344 1171–1172. [DOI] [PubMed] [Google Scholar]
- Adams N. M., Grassmann S., Sun J. C. (2020). Clonal expansion of innate and adaptive lymphocytes. Nat. Rev. Immunol. 20 694–707. 10.1038/s41577-020-0307-4 [DOI] [PubMed] [Google Scholar]
- Akkaya M., Kwak K., Pierce S. K. (2020). B cell memory: builing two walls of protection against pathogens. Nat. Rev. Immunol. 20 229–238. 10.1038/s41577-019-0244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuntagool N., Aramsri P., Panichakul T., Wuthiekanun V. R., Kinoshita R., White N. J., et al. (2000). Antigenic heterogeneity of lipopolysaccharide among Burkholderia pseudomallei clinical isolates. Southeast Asian J. Trop. Med. Public Health 31(Suppl. 1) 146–152. [PubMed] [Google Scholar]
- Anuntagool N., Wuthiekanun V., White N. J., Currie B. J., Sermswan R. W., Wongratanacheewin S., et al. (2006). Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am. J. Trop. Med. Hyg. 74 348–352. 10.4269/ajtmh.2006.74.348 [DOI] [PubMed] [Google Scholar]
- Atkins T., Prior R., Mack K., Russell P., Nelson M., Prior J., et al. (2002). Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J. Med. Microbiol. 51 539–553. 10.1099/0022-1317-51-7-539 [DOI] [PubMed] [Google Scholar]
- AuCoin D. P., Reed D. E., Marlenee N. L., Bowen R. A., Thorkildson P., Judy B. M., et al. (2012). Polysaccharide specific monoclonal antibodies provide passive protection against intranasal challenge with Burkholderia pseudomallei. PLoS One 7:e35386. 10.1371/journal.pone.0035386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A., Currie B. J., Mayo M., Sarovich D. S., Price E. P. (2020). Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality. Microb. Genom. 6:e000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A., Sarovich D. S., Harris T. M., Kaestli M., McRobb E., Mayo M., et al. (2017). Suspected cases of intracontinental Burkholderia pseudomallei sequence type homoplasy resolved using whole-genome sequencing. Microb. Genom. 3:e000139. 10.1099/mgen.0.000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balder R., Lipski S., Lazarus J. J., Grose W., Wooten R. M., Hogan R. J., et al. (2010). Identification of Burkholderia mallei and Burkholderia pseudomallei adhesins for human respiratory epithelial cells. BMC Microbiol. 10:250. 10.1186/1471-2180-10-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti E. L., Nguyen C. M., Welch M. D. (2015). Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell 161 348–360. 10.1016/j.cell.2015.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnie E., Weehuizen T. A. F., Lankelma J. M., de Jong H. K., Koh G. C. K. W., van Lieshout M. H. P., et al. (2019). Role of toll-like receptor 5 (TLR5) in experimental melioidosis. Infect. Immun. 87 e409–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot A., Benites J. (2005). The Importance of Soil Organic Matter: key to Drought-Resistant Soil and Sustained Food Production. Rome: Food and Agriculture Organization of the United Nations, 78. [Google Scholar]
- Breitbach K., Klocke S., Tschernig T., van Rooijen N., Baumann U., Steinmetz I. (2006). Role of inducible nitric oxide synthase and NADPH oxidase in early control of Burkholderia pseudomallei infection in mice. Infect. Immun. 74 6300–6309. 10.1128/iai.00966-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett P. J., Burtnick M. N., Heiss C., Azadi P., DeShazer D., Woods D. E., et al. (2011). Burkholderia thailandensis oacA mutants facilitate the expression of Burkholderia mallei-like O polysaccharides. Infect. Immun. 79 961–969. 10.1128/iai.01023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett P. J., Burtnick M. N., Woods D. E. (2003). The wbiA locus is required for the 2-O-acetylation of lipopolysaccharides expressed by Burkholderia pseudomallei and Burkholderia thailandensis. FEMS Microbiol. Lett. 218 323–328. [DOI] [PubMed] [Google Scholar]
- Brett P. J., Mah D. C., Woods D. E. (1994). Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect Immun. 62 1914–1919. 10.1128/iai.62.5.1914-1919.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett P. J., Woods D. E. (1996). Structural and immunological characterization of Burkholderia pseudomallei O-polysaccharide-flagellin protein conjugates. Infect. Immun. 64 2824–2828. 10.1128/iai.64.7.2824-2828.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock D. A., Read S., Bozhchenko A., Queller D. C., Strassmann J. E. (2013). Social amoeba farmers carry defensive symbionts to protect and privatize their crops. Nat. Commun. 4:2385. [DOI] [PubMed] [Google Scholar]
- Broek C. W. V., Chalmers K. J., Stevens M. P., Stevens J. M. (2015). Quantitative proteomic analysis of Burkholderia pseudomallei Bsa type III secretion system effectors using hypersecreting mutants. Mol. Cell. Proteom. 14 905–916. 10.1074/mcp.m114.044875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. F., Logue C. A., Boddey J. A., Scott R., Hirst R. G., Beacham I. R. (2004). Identification of a novel two-partner secretion system from Burkholderia pseudomallei. Mol. Genet. Genom. 272 204–215. 10.1007/s00438-004-1039-z [DOI] [PubMed] [Google Scholar]
- Bugrysheva J. V., Sue D., Hakovirta J., Loparev V. N., Knipe K., Sammons S. A., et al. (2015). Finished Annotated Genome Sequence of Burkholderia pseudomallei Strain Bp1651, a Multidrug-Resistant Clinical Isolate. Genome Announc. 3:e01427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtnick M. N., Brett P. J., Harding S. V., Ngugi S. A., Ribot W. J., Chantratita N., et al. (2011). The Cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79 1512–1525. 10.1128/iai.01218-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos C. G., Byrd M. S., Cotter P. A. (2013). Functional characterization of Burkholderia pseudomallei trimeric autotransporters. Infect. Immun. 81 2788–2799. 10.1128/iai.00526-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli R., Peri C., Villa R., Nithichanon A., Conchillo-Solé O., Yero D., et al. (2018). BPSL1626: reverse and structural vaccinology reveal a novel candidate for vaccine design against Burkholderia pseudomallei. Antibodies (Basel) 7:26. 10.3390/antib7030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Olvera I., Sahoo M., Miller M. A., Del Barrio L., Re F. (2011). Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1β is deleterious. PLoS Pathog. 7:e1002452. 10.1371/journal.ppat.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaichana P., Chantratita N., Brod F., Koosakulnirand S., Jenjaroen K., Chumseng S., et al. (2017). A nonsense mutation in TLR5 is associated with survival and reduced IL-10 and TNF-alpha levels in human melioidosis. PLoS Negl. Trop. Dis. 11:e0005587. 10.1371/journal.pntd.0005587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe J. F., Stubben C. J., Klimko C. P., Welkos S. L., Kern S. J., Bozue J. A., et al. (2014). Interrogation of the Burkholderia pseudomallei genome to address differential virulence among isolates. PLoS One 9:e115951. 10.1371/journal.pone.0115951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantratita N., Tandhavanant S., Myers N. D., Chierakul W., Robertson J. D., Mahavanakul W., et al. (2014). Screen of whole blood responses to flagellin identifies TLR5 variation associated with outcome in melioidosis. Genes Immun. 15 63–71. 10.1038/gene.2013.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple S. N. J., Sarovich D. S., Holden M. T. G., Peacock S. J., Buller N., Golledge C., et al. (2016). Whole-genome sequencing of a quarter-century melioidosis outbreak in temperate Australia uncovers a region of low-prevalence endemicity. Microb. Genom. 2:e000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Schröder I., French C. T., Jaroszewicz A., Yee X. J., Teh B.-E., et al. (2014). Characterization and analysis of the Burkholderia pseudomallei BsaN virulence regulon. BMC Microbiol. 14:206. 10.1186/s12866-014-0206-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wong J., Sun G. W., Liu Y., Tan G.-Y. G., Gan Y.-H. (2011). Regulation of type VI secretion system during Burkholderia pseudomallei infection. Infect. Immun. 79 3064–3073. 10.1128/iai.05148-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L., Chen Y. S., Lin H. H., Chan C. W., Chen S. C., Chen C. H. (2007). Immunostimulatory flagellin from Burkholderia pseudomallei effects on an increase in the intracellular calcium concentration and up-regulation of TNF-alpha by mononuclear cells. Microbiol. Immunol. 51 81–86. 10.1111/j.1348-0421.2007.tb03893.x [DOI] [PubMed] [Google Scholar]
- Chen Y.-S., Lin H.-H., Hsueh P.-T., Liu P.-J., Ni W.-F., Chung W.-C., et al. (2015). Whole-genome sequence of an epidemic strain of Burkholderia pseudomallei vgh07 in Taiwan. Genome Announc. 3:e00345-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Harris S. R., Croucher N. J., Turner C., Marttinen P., Cheng L., et al. (2014). Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46 305–309. 10.1038/ng.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Holden M. T. G., Vehkala M., Välimäki N., Yang Z., Harris S. R., et al. (2017). Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat. Microbiol. 2 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Mather A. E., Harris S. R., Hunt M., Holden M. T. G., Chaichana C., et al. (2019). Genetic variation associated with infection and the environment in the accidental pathogen Burkholderia pseudomallei. Commun. Biol. 2 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C., Pensar J., Chattagul S., Pesonen M., Sangphukieo A., Boonklang P., et al. (2020). Co-evolutionary signals from Burkholderia pseudomallei population genomics highlight its survival strategy in a hostile environment. bioRxiv [Preprint] 10.1101/2020.08.11.245894 [DOI] [Google Scholar]
- Chieng S., Carreto L., Nathan S. (2012). Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genom. 13:328. 10.1186/1471-2164-13-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K. L., Chan Y. Y., Gan Y. H. (2003). Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71 1622–1629. 10.1128/iai.71.4.1622-1629.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuaygud T., Tungpradabkul S., Sirisinha S., Chua K. L., Utaisincharoen P. (2008). A role of Burkholderia pseudomallei flagella as a virulent factor. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1) S140–S144. [DOI] [PubMed] [Google Scholar]
- Clarke E. T., Williams N. A., Findlow J., Borrow R., Heyderman R. S., Finn A. (2013). Polysaccharide-specific memory B cells generated by conjugate vaccines in humans conform to the CD27+IgG+ isotype-switched memory B Cell phenotype and require contact-dependent signals from bystander T cells activated by bacterial proteins to differentiate into plasma cells. J. Immunol. 191 6071–6083. 10.4049/jimmunol.1203254 [DOI] [PubMed] [Google Scholar]
- Cloutier M., Muru K., Ravicoularamin G., Gauthier C. (2018). Polysaccharides from Burkholderia species as targets for vaccine development, immunomodulation and chemical synthesis. Nat. Prod. Rep. 35 1251–1293. 10.1039/c8np00046h [DOI] [PubMed] [Google Scholar]
- Cooke G. S., Hill A. V. (2001). Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2 967–977. [DOI] [PubMed] [Google Scholar]
- Cuccui J., Milne T. S., Harmer N., George A. J., Harding S. V., Dean R. E., et al. (2012). Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect. Immun. 80 1209–1221. 10.1128/iai.05805-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie B. J., Jacups S. P. (2003). Intensity of rainfall and severity of melioidosis, Australia. Emerg. Infect. Dis. J. CDC 9 1538–1542. 10.3201/eid0912.020750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daligault H. E., Davenport K. W., Minogue T. D., Bishop-Lilly K. A., Broomall S. M., Bruce D. C., et al. (2014). Whole-genome assemblies of 56 Burkholderia species. Genome Announc. 2:e01106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S., Sanchez-Buso L., Harris S. R., Marttinen P., Rusniok C., Buchrieser C., et al. (2017). Dynamics and impact of homologous recombination on the evolution of Legionella pneumophila. PLoS Genet. 13:e1006855. 10.1371/journal.pgen.1006855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker P. I. W., McVean G., Sabeti P. C., Miretti M. M., Green T., Marchini J., et al. (2006). A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38 1166–1172. 10.1038/ng1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou C. A., Petersen J., Rossjohn J., Fugger L. (2018). HLA variation and disease. Nat. Rev. Immunol. 18 325–339. [DOI] [PubMed] [Google Scholar]
- DeShazer D., Brett P. J., Carlyon R., Woods D. E. (1997). Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179 2116–2125. 10.1128/jb.179.7.2116-2125.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharakul T., Vejbaesya S., Chaowagul W., Luangtrakool P., Stephens H. A., Songsivilai S. (1998). HLA-DR and -DQ associations with melioidosis. Hum. Immunol. 59 580–586. 10.1016/s0198-8859(98)00052-4 [DOI] [PubMed] [Google Scholar]
- Dickey A. K., Chantratita N., Tandhavanant S., Ducken D., Lovelace-Macon L., Seal S., et al. (2019). Flagellin-independent effects of a Toll-like receptor 5 polymorphism in the inflammatory response to Burkholderia pseudomallei. PLoS Neg. Trop. Dis. 13:e0007354. 10.1371/journal.pntd.0007354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSalvo S., Haselkorn T. S., Bashir U., Jimenez D., Brock D. A., Queller D. C., et al. (2015). Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proc. Natl. Acad. Sci. U.S.A. 112 E5029–E5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling A. J., Wilkinson P. A., Holden M. T. G., Quail M. A., Bentley S. D., Reger J., et al. (2010). Genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS One 5:e15693. 10.1371/journal.pone.0015693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duangsonk K., Gal D., Mayo M., Hart C. A., Currie B. J., Winstanley C. (2006). Use of a variable amplicon typing scheme reveals considerable variation in the accessory genomes of isolates of Burkholderia pseudomallei. J. Clin. Microbiol. 44 1323–1334. 10.1128/jcm.44.4.1323-1334.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunachie S. J., Jenjaroen K., Reynolds C. J., Quigley K. J., Sergeant R., Sumonwiriya M., et al. (2017). Infection with Burkholderia pseudomallei – immune correlates of survival in acute melioidosis. Sci. Rep. 7:12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton A., Haque A., Chu K., Lukaszewski R., Bancroft G. J. A. (2007). Critical role for neutrophils in resistance to experimental infection with Burkholderia pseudomallei. J. Infect. Dis. 195 99–107. 10.1086/509810 [DOI] [PubMed] [Google Scholar]
- Ewbank D. (2016). Measuring selection in human populations using the growth rate per generation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150148. 10.1098/rstb.2015.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner P. L., Kayala M. A., Vigil A., Burk C., Nakajima-Sasaki R., Pablo J., et al. (2009). A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. U.S.A. 106 13499–13504. 10.1073/pnas.0812080106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane H. K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P. R., et al. (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. T., Toesca I. J., Wu T.-H., Teslaa T., Beaty S. M., Wong W., et al. (2011). Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc.Natl. Acad. Sci. U.S.A. 108 12095–12100. 10.1073/pnas.1107183108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourraud P.-A., Khankhanian P., Cereb N., Yang S. Y., Feolo M., Maiers M., et al. (2014). HLA diversity in the 1000 genomes dataset. PLoS One 9:e97282. 10.1371/journal.pone.0097282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory A. E., Judy B. M., Qazi O., Blumentritt C. A., Brown K. A., Shaw A. M., et al. (2015). A gold nanoparticle-linked glycoconjugate vaccine against Burkholderia mallei. Nanomedicine 11 447–456. 10.1016/j.nano.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M. G., Warawa J. M. (2016). Attenuation of a select agent-excluded Burkholderia pseudomallei capsule mutant in hamsters. Acta Trop. 157 68–72. 10.1016/j.actatropica.2015.12.006 [DOI] [PubMed] [Google Scholar]
- Hantrakun V., Rongkard P., Oyuchua M., Amornchai P., Lim C., Wuthiekanun V., et al. (2016). Soil nutrient depletion is associated with the presence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 82 7086–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselkorn T. S., DiSalvo S., Miller J. W., Bashir U., Brock D. A., Queller D. C., et al. (2019). The specificity of Burkholderia symbionts in the social amoeba farming symbiosis: prevalence, species, genetic and phenotypic diversity. Mol. Ecol. 28 847–862. 10.1111/mec.14982 [DOI] [PubMed] [Google Scholar]
- Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410 1099–1103. 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- Hayden H. S., Lim R., Brittnacher M. J., Sims E. H., Ramage E. R., Fong C., et al. (2012). Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One. 7:e36507. 10.1371/journal.pone.0036507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C., Burtnick M. N., Roberts R. A., Black I., Azadi P., Brett P. J. (2013). Revised structures for the predominant O-polysaccharides expressed by Burkholderia pseudomallei and Burkholderia mallei. Carbohydr. Res. 381 6–11. 10.1016/j.carres.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T. G., Titball R. W., Peacock S. J., Cerdeño-Tárraga A. M., Atkins T., Crossman L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou K., Burch K. S., Majumdar A., Shi H., Mancuso N., Wu Y., et al. (2019). Accurate estimation of SNP-heritability from biobank-scale data irrespective of genetic architecture. Nat. Genet. 51 1244–1251. 10.1038/s41588-019-0465-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.-T., Chen Y.-S., Lin H.-H., Liu P.-J., Ni W.-F., Liu M.-C., et al. (2015). Comparison of whole-genome sequences from two colony morphovars of Burkholderia pseudomallei. Genome Announc. 3:e01194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Reeves P. R. (2020). The remarkable dual-level diversity of prokaryotic flagellins. mSystems 5:e00705-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis T. J., Rigby P., Robertson T. A., Dutton N. S., Henderson M., Chang B. J. (2000). Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect. Immun. 68 1681–1686. 10.1128/iai.68.3.1681-1686.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis T. J., Robertson T., Woods D. E., Dutton N., Chang B. J. (2003). Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect. Immun. 71 2280–2282. 10.1128/iai.71.4.2280-2282.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenjaroen K., Chumseng S., Sumonwiriya M., Ariyaprasert P., Chantratita N., Sunyakumthorn P., et al. (2015). T-Cell responses are associated with survival in acute melioidosis patients. PLoS Neg. Trop. Dis. 9:e0004152. 10.1371/journal.pntd.0004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Li Y., Zhang H., Chattopadhyay I., Boeck Jensen A., Blair D. R., et al. (2019). Estimating heritability and genetic correlations from large health datasets in the absence of genetic data. Nat. Commun. 10:5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L., Baker A. L., Chain P. S., Currie B. J., Daligault H. E., Davenport K. W., et al. (2015a). Whole-Genome Sequences of 80 Environmental and Clinical Isolates of Burkholderia pseudomallei. Genome Announc. 3:e01282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L., Bishop-Lilly K. A., Ladner J. T., Daligault H. E., Davenport K. W., Jaissle J., et al. (2015b). Complete genome sequences for 59 burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 3:e00159-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S. N., Marin-Menendez A., Introini V., Ravenhill B. J., Lin Y. C., Macharia A., et al. (2020). Red blood cell tension protects against severe malaria in the Dantu blood group. Nature. 585 579–583. 10.1038/s41586-020-2726-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson E. K., Kwiatkowski D. P., Sabeti P. C. (2014). Natural selection and infectious disease in human populations. Nat. Rev. Genet. 15 379–393. 10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khojandi N., Haselkorn T. S., Eschbach M. N., Naser R. A., DiSalvo S. (2019). Intracellular Burkholderia Symbionts induce extracellular secondary infections; driving diverse host outcomes that vary by genotype and environment. ISME J. 13 2068–2081. 10.1038/s41396-019-0419-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. D., Berry S., Clarke B. R., Morris R. J., Whitfield C. (2014). Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. U.S.A. 111 6407–6412. 10.1073/pnas.1400814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel Y. A., Paramonov N. A., Shashkov A. S., Kochetkov N. K., Yarullin R. G., Farber S. M., et al. (1992). Structure of the polysaccharide chains of Pseudomonas pseudomallei lipopolysaccharides. Carbohydr. Res. 233 185–193. 10.1016/s0008-6215(00)90930-3 [DOI] [PubMed] [Google Scholar]
- Koonpaew S., Ubol M. N., Sirisinha S., White N. J., Chaiyaroj S. C. (2000). Genome fingerprinting by pulsed-field gel electrophoresis of isolates of Burkholderia pseudomallei from patients with melioidosis in Thailand. Acta Trop. 74 187–191. 10.1016/s0001-706x(99)00069-8 [DOI] [PubMed] [Google Scholar]
- Koosakulnirand S., Phokrai P., Jenjaroen K., Roberts R. A., Utaisincharoen P., Dunachie S. J., et al. (2018). Immune response to recombinant Burkholderia pseudomallei FliC. PLoS One 13:e0198906. 10.1371/journal.pone.0198906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronsteiner B., Chaichana P., Sumonwiriya M., Jenjaroen K., Chowdhury F. R., Chumseng S., et al. (2019). Diabetes alters immune response patterns to acute melioidosis in humans. Eur. J. Immunol. 49 1092–1106. 10.1002/eji.201848037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar Adler N., Stevens J., Stevens M., Galyov E. (2011). Autotransporters and their role in the virulence of Burkholderia pseudomallei and Burkholderia mallei. Front. Microbiol. 2:151. 10.3389/fmicb.2011.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar Adler N. R., Stevens M. P., Dean R. E., Saint R. J., Pankhania D., Prior J. L., et al. (2015). Systematic mutagenesis of genes encoding predicted autotransported proteins of Burkholderia pseudomallei identifies factors mediating virulence in mice, net intracellular replication and a novel protein conferring serum resistance. PLoS One 10:e0121271. 10.1371/journal.pone.0121271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J. A., Croucher N. J., Goldblatt D., Nosten F., Parkhill J., Turner C., et al. (2017). Genome-wide identification of lineage and locus specific variation associated with pneumococcal carriage duration. Elife 6:e26255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J. A., Harris S. R., Tonkin-Hill G., Gladstone R. A., Lo S. W., Weiser J. N., et al. (2019). Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29 304–316. 10.1101/gr.241455.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennings J., West T. E., Schwarz S. (2019). The Burkholderia type VI secretion system 5: composition, regulation and role in virulence. Front. Microbiol. 9:3339. 10.3389/fmicb.2018.03339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman T. D., Flett K. B., Yelin I., Martin T. R., McAdam A. J., Priebe G. P., et al. (2014). Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat. Genet. 46 82–87. 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D., Holden M. T. G., Coupland P., Price E. P., Chantratita N., Wuthiekanun V., et al. (2014a). Microevolution of Burkholderia pseudomallei during an acute infection. J. Clin. Microbiol. 52 3418–3421. 10.1128/jcm.01219-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D., Paeyao A., Wongratanacheewin S., Saiprom N., Takpho N., Thaipadungpanit J., et al. (2014b). Role of Burkholderia pseudomallei biofilm formation and lipopolysaccharide in relapse of melioidosis. Clin. Microbiol. Infect. 20 O854–O856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D., Wongratanacheewin S., Teerawattanasook N., Wongsuvan G., Chaisuksant S., Chetchotisakd P., et al. (2010a). Increasing incidence of human melioidosis in Northeast Thailand. Am. J. Trop. Med. Hyg. 82 1113–1117. 10.4269/ajtmh.2010.10-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D., Wuthiekanun V., Chantratita N., Wongsuvan G., Amornchai P., Day N. P. J., et al. (2010b). Burkholderia pseudomallei is spatially distributed in soil in Northeast Thailand. PLoS Neg. Trop. Dis. 4:e694. 10.1371/journal.pntd.0000694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada L., Ronning C. M., DeShazer D., Woods D., Fedorova N., Kim H. S., et al. (2010). Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol. Evol. 2 102–116. 10.1093/gbe/evq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahasirimongkol S., Yanai H., Mushiroda T., Promphittayarat W., Wattanapokayakit S., Phromjai J., et al. (2012). Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J. Hum. Genet. 57 363–367. 10.1038/jhg.2012.35 [DOI] [PubMed] [Google Scholar]
- Malaria Genomic Epidemiology Network (2008). A global network for investigating the genomic epidemiology of malaria. Nature 456 732–737. 10.1038/nature07632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivanh L., Pierret A., Rattanavong S., Kounnavongsa O., Buisson Y., Elliott I., et al. (2017). Burkholderia pseudomallei in a lowland rice paddy: seasonal changes and influence of soil depth and physico-chemical properties. Sci. Rep. 7:3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobb E., Sarovich D. S., Price E. P., Kaestli M., Mayo M., Keim P., et al. (2015). Tracing melioidosis back to the source: using whole-genome sequencing to investigate an outbreak originating from a contaminated domestic water supply. J. Clin. Microbiol. 53 1144–1148. 10.1128/jcm.03453-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. A., Tuanyok A., Woods D. E. (2008). Survival of Burkholderia pseudomallei in water. BMC Res. Notes 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule M. G., Spink N., Willcocks S., Lim J., Guerra-Assunção J. A., Cia F., et al. (2015). Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei. Infect. Immun. 84 701–710. 10.1128/iai.01102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N. D., Chantratita N., Berrington W. R., Chierakul W., Limmathurotsakul D., Wuthiekanun V., et al. (2014). The role of NOD2 in murine and human melioidosis. J. Immunol. 192 300–307. 10.4049/jimmunol.1301436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi T., Holden M. T. G., Didelot X., Mehershahi K., Boddey J. A., Beacham I., et al. (2015). Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 25 129–141. 10.1101/gr.177543.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi T., Ong C., Singh A. P., Boddey J., Atkins T., Sarkar-Tyson M., et al. (2010). A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 6:e1000845. 10.1371/journal.ppat.1000845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Prior J. L., Lever M. S., Jones H. E., Atkins T. P., Titball R. W. (2004). Evaluation of lipopolysaccharide and capsular polysaccharide as subunit vaccines against experimental melioidosis. J. Med. Microbiol. 53(Pt 12) 1177–1182. 10.1099/jmm.0.45766-0 [DOI] [PubMed] [Google Scholar]
- Nikolich-Zugich J., Slifka M. K., Messaoudi I. (2004). The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4 123–132. 10.1038/nri1292 [DOI] [PubMed] [Google Scholar]
- Noinarin P., Chareonsudjai P., Wangsomnuk P., Wongratanacheewin S., Chareonsudjai S. (2016). environmental free-living amoebae isolated from soil in khon kaen, Thailand, Antagonize Burkholderia pseudomallei. PLoS One 11:e0167355. 10.1371/journal.pone.0167355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris M. H., Schweizer H. P., Tuanyok A. (2017). Structural diversity of Burkholderia pseudomallei lipopolysaccharides affects innate immune signaling. PLoS Neg. Trop. Dis. 11:e0005571. 10.1371/journal.pntd.0005571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novem V., Shui G., Wang D., Bendt A. K., Sim S. H., Liu Y., et al. (2009). Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin. Vaccine Immunol. 16 1420–1428. 10.1128/cvi.00472-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi W. F., Ong C., Nandi T., Kreisberg J. F., Chua H. H., Sun G., et al. (2013). The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 9:e1003795. 10.1371/journal.pgen.1003795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou K., Ong C., Koh S. Y., Rodrigues F., Sim S. H., Wong D., et al. (2005). Integrative genomic, transcriptional, and proteomic diversity in natural isolates of the human pathogen Burkholderia pseudomallei. J. Bacteriol. 187 4276–4285. 10.1128/jb.187.12.4276-4285.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paksanont S., Sintiprungrat K., Yimthin T., Pumirat P., Peacock S. J., Chantratita N. (2018). Effect of temperature on Burkholderia pseudomallei growth, proteomic changes, motility and resistance to stress environments. Sci. Rep. 8:9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina E. M., Mattoo S., Griffith N., Kozak N. A., Yuk M. H., Miller J. F. (2005). A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58 267–279. 10.1111/j.1365-2958.2005.04823.x [DOI] [PubMed] [Google Scholar]
- Park B. S., Lee J. O. (2013). Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 45:e66. 10.1038/emm.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. M., Ghosh S., O’Connor T. J. (2020). Combinatorial selection in amoebal hosts drives the evolution of the human pathogen Legionella pneumophila. Nat. Microbiol. 5 599–609. 10.1038/s41564-019-0663-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy N., DeShazer D., England M., Waag D. M. (2006). Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagn. Microbiol. Infect. Dis. 56 329–332. 10.1016/j.diagmicrobio.2006.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T., Giffard P., Beckstrom-Sternberg S., Auerbach R., Hornstra H., Tuanyok A., et al. (2009). Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 7:78. 10.1186/1741-7007-7-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T., Sahl J. W., Hepp C. M., Handady K., Hornstra H., Vazquez A. J., et al. (2020). Pathogen to commensal? Longitudinal within-host population dynamics, evolution, and adaptation during a chronic >16-year Burkholderia pseudomallei infection. PLoS Pathog. 16:e1008298. 10.1371/journal.ppat.1008298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. B., MacLean L. L., Schollaardt T., Bryan L. E., Ho M. (1995). Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect. Immun. 63 3348–3352. 10.1128/iai.63.9.3348-3352.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilatz S., Breitbach K., Hein N., Fehlhaber B., Schulze J., Brenneke B., et al. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74 3576–3586. 10.1128/iai.01262-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky N. L., Rhodes K. A., Mima T., Drew H. R., Chirakul S., Wuthiekanun V., et al. (2017). Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei : deviation from the Norm. mBio 8:e01357-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothlichet J., Quintana-Murci L. (2013). The genetics of innate immunity sensors and human disease. Int. Rev. Immunol. 32 157–208. 10.3109/08830185.2013.777064 [DOI] [PubMed] [Google Scholar]
- Price E. P., Sarovich D. S., Mayo M., Tuanyok A., Drees K. P., Kaestli M., et al. (2013). Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 4:e00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price E. P., Sarovich D. S., Smith E. J., MacHunter B., Harrington G., Theobald V., et al. (2016). Unprecedented melioidosis cases in Northern Australia caused by an asian Burkholderia pseudomallei strain identified by using large-scale comparative genomics. Appl. Environ. Microbiol. 82 954–963. 10.1128/aem.03013-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price E. P., Viberg L. T., Kidd T. J., Bell S. C., Currie B. J., Sarovich D. S. (2018). Transcriptomic analysis of longitudinal Burkholderia pseudomallei infecting the cystic fibrosis lung. Microb. Genom. 4:e000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumpuang A., Chantratita N., Wikraiphat C., Saiprom N., Day N. P., Peacock S. J., et al. (2011). Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans. R. Soc. Trop. Med. Hyg. 105 598–600. 10.1016/j.trstmh.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachlin A., Mayo M., Webb J. R., Kleinecke M., Rigas V., Harrington G., et al. (2020). Whole-genome sequencing of Burkholderia pseudomallei from an urban melioidosis hot spot reveals a fine-scale population structure and localised spatial clustering in the environment. Sci. Rep. 10:5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph A., McBride J., Currie B. J. (2004). Transmission of Burkholderia pseudomallei via breast milk in northern Australia. Pediatr. Infect. Dis. J. 23 1169–1171. [PubMed] [Google Scholar]
- Reckseidler S. L., DeShazer D., Sokol P. A., Woods D. E. (2001). Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69 34–44. 10.1128/iai.69.1.34-44.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reckseidler-Zenteno S. L., DeVinney R., Woods D. E. (2005). The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect. Immun. 73 1106–1115. 10.1128/iai.73.2.1106-1115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reckseidler-Zenteno S. L., Moore R., Woods D. E. (2009). Genetics and function of the capsules of Burkholderia pseudomallei and their potential as therapeutic targets. Mini. Rev. Med. Chem. 9 265–271. 10.2174/138955709787316047 [DOI] [PubMed] [Google Scholar]
- Reckseidler-Zenteno S. L., Viteri D.-F., Moore R., Wong E., Tuanyok A., Woods D. E. (2010). Characterization of the type III capsular polysaccharide produced by Burkholderia pseudomallei. J. Med. Microbiol. 59(Pt 12) 1403–1414. 10.1099/jmm.0.022202-0 [DOI] [PubMed] [Google Scholar]
- Robinson W. H. (2015). Sequencing the functional antibody repertoire–diagnostic and therapeutic discovery. Nat. Rev. Rheumatol. 11 171–182. 10.1038/nrrheum.2014.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Stone J. K., Gelhaus H. C., Warren R. L., Cruttwell C. J., Funnell S. G., et al. (2013). Genome Sequence of Burkholderia pseudomallei NCTC 13392. Genome Announc. 1:e183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Vazquez A. J., Hall C. M., Busch J. D., Tuanyok A., Mayo M., et al. (2016). The effects of signal erosion and core genome reduction on the identification of diagnostic markers. mBio 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiprom N., Sangsri T., Tandhavanant S., Sengyee S., Phunpang R., Preechanukul A., et al. (2020). Genomic loss in environmental and isogenic morphotype isolates of Burkholderia pseudomallei is associated with intracellular survival and plaque-forming efficiency. PLoS Neg. Trop. Dis. 14:e0008590. 10.1371/journal.pntd.0008590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey F. A., Imada K., Nagashima S., Vonderviszt F., Kumasaka T., Yamamoto M., et al. (2001). Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410 331–337. 10.1038/35066504 [DOI] [PubMed] [Google Scholar]
- Sanchez-Buso L., Comas I., Jorques G., Gonzalez-Candelas F. (2014). Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat. Genet. 46 1205–1211. 10.1038/ng.3114 [DOI] [PubMed] [Google Scholar]
- Sarkar-Tyson M., Thwaite J. E., Harding S. V., Smither S. J., Oyston P. C. F., Atkins T. P., et al. (2007). Polysaccharides and virulence of Burkholderia pseudomallei. J. Med. Microbiol. 56(Pt 8) 1005–1010. [DOI] [PubMed] [Google Scholar]
- Sarovich D. S., Garin B., Smet B. D., Kaestli M., Mayo M., Vandamme P., et al. (2016). Phylogenomic analysis reveals an Asian Origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere 1:e00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarovich D. S., Price E. P., Webb J. R., Ward L. M., Voutsinos M. Y., Tuanyok A., et al. (2014). Variable virulence factors in Burkholderia pseudomallei (Melioidosis) associated with human disease. PLoS One 9:e91682. 10.1371/journal.pone.0091682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppler M. (2014). How the interaction of Listeria monocytogenes and Acanthamoeba spp. affects growth and distribution of the food borne pathogen. Appl. Microbiol. Biotechnol. 98 2907–2916. 10.1007/s00253-014-5546-5 [DOI] [PubMed] [Google Scholar]
- Scott A. E., Burtnick M. N., Stokes M. G. M., Whelan A. O., Williamson E. D., Atkins T. P., et al. (2014b). Burkholderia pseudomallei capsular polysaccharide conjugates provide protection against acute melioidosis. Infect. Immun. 82 3206–3213. 10.1128/iai.01847-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. E., Ngugi S. A., Laws T. R., Corser D., Lonsdale C. L., D’Elia R. V., et al. (2014a). Protection against experimental melioidosis following immunisation with a lipopolysaccharide-protein conjugate. J. Immunol. Res. 2014: 392170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. A., Scott L. J., Mägi R., Marullo L., Gaulton K. J., Kaakinen M., et al. (2017). An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 66:2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T., Tellapragada C., Kamath A., Kalwaje Eshwara V., Mukhopadhyay C. (2019). Implications of environmental and pathogen-specific determinants on clinical presentations and disease outcome in melioidosis patients. PLoS Neg. Trop. Dis. 13:e0007312. 10.1371/journal.pntd.0007312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Ao H., Soemantri A., Tiwawech D., Settheetham-Ishida W., Kayame O. W., et al. (2000). Sero- and molecular typing of Duffy blood group in Southeast Asians and Oceanians. Hum. Biol. 72 511–518. [PubMed] [Google Scholar]
- Sidjabat H. E., Cottrell K., Cervin A. (2015). Draft genome sequences of Burkholderia pseudomallei and Staphylococcus aureus, Isolated from a Patient with Chronic Rhinosinusitis. Genome Announc. 3:e01075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J. M., Brunet Y. R., Cascales E., Mougous J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66 453–472. 10.1146/annurev-micro-121809-151619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S. H., Yu Y., Lin C. H., Karuturi R. K. M., Wuthiekanun V., Tuanyok A., et al. (2008). The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoS Pathog. 4:e1000178. 10.1371/journal.ppat.1000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M., Cagliani R., Forni D., Clerici M. (2015). Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16 224–236. 10.1038/nrg3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C., Stevens J. M., Chantratita N., Currie B. J., Peacock S. J., Korbsrisate S., et al. (2008). Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J. Clin. Microbiol. 46 2418–2422. 10.1128/jcm.00368-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slodkowicz G., Goldman N. (2020). Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc. Natl. Acad. Sci. U.S.A. 117 5977–5986. 10.1073/pnas.1916786117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Yu Y., Feng L., He J., Wang T., Zhu H., et al. (2015). Draft genome sequence of Burkholderia pseudomallei strain 350105, isolated in Hainan, China, in 1976. Genome Announc. 3:e01162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southeast Asia Infectious Disease Clinical Research Network (2017). Causes and outcomes of sepsis in southeast Asia: a multinational multicentre cross-sectional study. Lancet Glob. Health 5 e157–e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D., Holmes J., Balding D. J. (2020). Evaluating and improving heritability models using summary statistics. Nat. Genet. 52 458–462. 10.1038/s41588-020-0600-y [DOI] [PubMed] [Google Scholar]
- Spring-Pearson S. M., Stone J. K., Doyle A., Allender C. J., Okinaka R. T., Mayo M., et al. (2015). Pangenome analysis of Burkholderia pseudomallei: genome evolution preserves gene order despite high recombination rates. PLoS One 10:e0140274. 10.1371/journal.pone.0140274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. M., Ulrich R. L., Taylor L. A., Wood M. W., DeShazer D., Stevens M. P., et al. (2005). Actin-Binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA Mutant. J. Bacteriol. 187 7857–7862. 10.1128/jb.187.22.7857-7862.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. P., Stevens J. M., Jeng R. L., Taylor L. A., Wood M. W., Hawes P., et al. (2005). Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56 40–53. 10.1111/j.1365-2958.2004.04528.x [DOI] [PubMed] [Google Scholar]
- Stone J. K., Mayo M., Grasso S. A., Ginther J. L., Warrington S. D., Allender C. J., et al. (2012). Detection of Burkholderia pseudomallei O-antigen serotypes in near-neighbor species. BMC Microbiol. 12:250. 10.1186/1471-2180-12-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannasaen D., Mahawantung J., Chaowagul W., Limmathurotsakul D., Felgner P. L., Davies H., et al. (2011). Human immune responses to Burkholderia pseudomallei characterized by protein microarray analysis. J. Infect. Dis. 203 1002–1011. 10.1093/infdis/jiq142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T. E., Perumal T. M., Henao R., Nichols M., Howrylak J. A., Choi A. M., et al. (2018). A community approach to mortality prediction in sepsis via gene expression analysis. Nat. Commun. 9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S. T., Cheah P. C., Puthucheary S. D. (2010). Sequence polymorphism and PCR-restriction fragment length polymorphism analysis of the flagellin gene of Burkholderia pseudomallei. J. Clin. Microbiol. 48 1465–1467. 10.1128/jcm.01131-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. D., Forbes-Faulkner J., Parker M. (1979). Isolation of Pseudomonas pseudomallei from clay layers at defined depths. Am. J. Epidemiol. 110 515–521. 10.1093/oxfordjournals.aje.a112832 [DOI] [PubMed] [Google Scholar]
- Thye T., Owusu-Dabo E., Vannberg F. O., van Crevel R., Curtis J., Sahiratmadja E., et al. (2012). Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat. Genet. 44 257–259. 10.1038/ng.1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmann C., Thye T., Vens M., Evans J., May J., Ehmen C., et al. (2012). Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 489 443–446. 10.1038/nature11334 [DOI] [PubMed] [Google Scholar]
- Tuanyok A., Auerbach R. K., Brettin T. S., Bruce D. C., Munk A. C., Detter J. C., et al. (2007). A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J. Bacteriol. 189 9044–9049. 10.1128/jb.01264-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanyok A., Leadem B. R., Auerbach R. K., Beckstrom-Sternberg S. M., Beckstrom-Sternberg J. S., Mayo M., et al. (2008). Genomic islands from five strains of Burkholderia pseudomallei. BMC Genom. 9:566. 10.1186/1471-2164-9-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanyok A., Stone J. K., Mayo M., Kaestli M., Gruendike J., Georgia S., et al. (2012). The Genetic and molecular basis of o-antigenic diversity in Burkholderia pseudomallei Lipopolysaccharide. PLoS Neg. Trop. Dis. 6:e1453. 10.1371/journal.pntd.0001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U’Ren J. M., Schupp J. M., Pearson T., Hornstra H., Friedman C. L. C., Smith K. L., et al. (2007). Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiol. 7:23. 10.1186/1471-2180-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek T., Borghans J. A. M., van Wijk F. (2018). The full spectrum of human naive T cells. Nat. Rev. Immunol. 18 363–373. 10.1038/s41577-018-0001-y [DOI] [PubMed] [Google Scholar]
- Vander Broek C. W., Stevens J. M. (2017). Type III secretion in the melioidosis pathogen Burkholderia pseudomallei. Front. Cell. Infect. Microbiol. 7:255. 10.3389/fcimb.2017.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viberg L. T., Price E. P., Kidd T. J., Bell S. C., Currie B. J., Sarovich D. S. (2015). Whole-Genome Sequences of five Burkholderia pseudomallei isolates from Australian cystic fibrosis patients. Genome Announc. 3:e00254-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viberg L. T., Sarovich D. S., Kidd T. J., Geake J. B., Bell S. C., Currie B. J., et al. (2017). Within-host evolution of Burkholderia pseudomallei during Chronic Infection of Seven Australasian cystic fibrosis patients. mBio 8:e00356-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J. R., Rachlin A., Rigas V., Sarovich D. S., Price E. P., Kaestli M., et al. (2019). Tracing the environmental footprint of the Burkholderia pseudomallei lipopolysaccharide genotypes in the tropical “Top End” of the Northern Territory, Australia. PLoS Neg. Trop. Dis. 13:e0007369. 10.1371/journal.pntd.0007369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weehuizen T. A., Prior J. L., van der Vaart T. W., Ngugi S. A., Nepogodiev S. A., Field R. A., et al. (2015). Differential toll-like receptor-signalling of Burkholderia pseudomallei lipopolysaccharide in murine and human models. PLoS One 10:e0145397. 10.1371/journal.pone.0145397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West T. E., Chantratita N., Chierakul W., Limmathurotsakul D., Wuthiekanun V., Myers N. D., et al. (2013). Impaired TLR5 functionality is associated with survival in melioidosis. J. Immunol. 190 3373–3379. 10.4049/jimmunol.1202974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West T. E., Chierakul W., Chantratita N., Limmathurotsakul D., Wuthiekanun V., Emond M. J., et al. (2012). Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun. 13 38–46. 10.1038/gene.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West T. E., Ernst R. K., Jansson-Hutson M. J., Skerrett S. J. (2008). Activation of Toll-like receptors by Burkholderia pseudomallei. BMC Immunol. 9:46. 10.1186/1471-2172-9-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock G. C., Deeraksa A., Qazi O., Judy B. M., Taylor K., Propst K. L., et al. (2010). Protective response to subunit vaccination against intranasal Burkholderia mallei and B. pseudomallei challenge. Proc. Vaccinol. 2 73–77. 10.1016/j.provac.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W. J., Wieland C. W., Dessing M. C., Chantratita N., Cheng A. C., Limmathurotsakul D., et al. (2007). Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (Melioidosis). PLoS Med. 4:e248. 10.1371/journal.pmed.0040248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikraiphat C., Charoensap J., Utaisincharoen P., Wongratanacheewin S., Taweechaisupapong S., Woods D. E., et al. (2009). Comparative in vivo and in vitro analyses of putative virulence factors of Burkholderia pseudomallei using lipopolysaccharide, capsule and flagellin mutants. FEMS Immunol. Med. Microbiol. 56 253–259. 10.1111/j.1574-695x.2009.00574.x [DOI] [PubMed] [Google Scholar]
- Willcocks S. J., Denman C. C., Atkins H. S., Wren B. W. (2016). Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr. Opin. Microbiol. 29 94–103. 10.1016/j.mib.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Williams N. L., Morris J. L., Rush C. M., Ketheesan N. (2015). Plasmacytoid dendritic cell bactericidal activity against Burkholderia pseudomallei. Microb. Infect. 17 311–316. 10.1016/j.micinf.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Williams R. C., Muller Y. L., Hanson R. L., Knowler W. C., Mason C. C., Bian L., et al. (2011). HLA-DRB1 reduces the risk of type 2 diabetes mellitus by increased insulin secretion. Diabetologia 54 1684–1692. 10.1007/s00125-011-2122-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Kim H. S., Chua H. H., Lin C. H., Sim S. H., Lin D., et al. (2006). Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 6:46. 10.1186/1471-2180-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhuchenko O., Kuspa A., Soldati T. (2016). Social amoebae trap and kill bacteria by casting DNA nets. Nat. Commun. 7:10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Rasheed A., Tikkanen E., Lee J.-J., Butterworth A. S., Howson J. M. M., et al. (2017). Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 49 1450–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N., Tsai S., Feng S. H., Newsome T., Kim H. Y., Li B., et al. (2008). Relationship between antigenicity and pathogenicity for Burkholderia pseudomallei and Burkholderia mallei revealed by a large panel of mouse MAbs. Hybridoma (Larchmt) 27 231–240. [DOI] [PubMed] [Google Scholar]
- Zou S., Li J., Zhou H., Frech C., Jiang X., Chu J. S., et al. (2014). Mutational landscape of intrahepatic cholangiocarcinoma. Nat. Commun. 5:5696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We estimated the proportion of infection outcome variations (survival vs. mortal outcomes after 28 days) explained by B. pseudomallei nucleotide polymorphisms (SNPs). SNPs were called from Chewapreecha et al. (2019, 2020) using kSNP v3 software. A filtering for minimum allele frequency at 0.01 (MAF = 0.01) resulted in 65,379 SNPs for the analysis. B. pseudomallei population was defined by PopPUNK (Lees et al., 2019). We employed the Genome-wide Complex Trait Analysis (GCTA) (Yang et al., 2011) to calculate h2 using genomic restricted maximum likelihood (GREML) while adjusting for population structure using population clusters defined by PopPUNK.