Abstract
We have previously shown that withdrawal of folic acid led to metabolic reprogramming and a less aggressive phenotype in a mouse cell model of triple-negative breast cancer (TNBC). Herein, we evaluate the effects of folic acid withdrawal on transcriptomic profiles in these cells. Murine cell lines were originally derived from a pool of spontaneous mammary tumors grown in MMTV-Wnt1 transgenic mice. Based on their differential molecular characteristics and metastatic potential, these cell lines were previously characterized as non-metastatic epithelial (E-Wnt), non-metastatic mesenchymal (M-Wnt) and metastatic mesenchymal (metM-Wntliver) cells. Using custom two-color 180K Agilent microarrays, we have determined gene expression profiles for three biological replicates of each subtype kept on standard medium (2.2 μM folic acid) or folic acid-free medium for 72 hours. The analyses revealed that more genes were differentially expressed upon folic acid withdrawal in M-Wnt cells (1884 genes; Benjamini-Hochberg-adjusted P-value <0.05) compared to E-Wnt and metM-Wntliver cells (108 and 222 genes, respectively). Pathway analysis has identified that type I interferon signaling was strongly affected by folic acid withdrawal, with interferon-responsive genes consistently being upregulated upon folic acid withdrawal in M-Wnt cells. Of note, repressed interferon signaling has been established as one of the characteristics of aggressive human TNBC, and hence reactivation of this pathway may be a promising therapeutic approach. Overall, while our study indicates that the response to folic acid withdrawal varies by molecular subtype and cellular phenotype, it also underscores the necessity to further investigate one-carbon metabolism as a potential therapeutic means in the treatment of advanced TNBC.
Keywords: folic acid, triple-negative breast cancer, epithelial-to-mesenchymal transition, interferon signaling, metastasis, transcriptomics
1. INTRODUCTION
Cancer metastasis is the most common cause of cancer-related death [1–3]. Metastasis is a complex multi-step process, with cells undergoing metabolic reprograming to support their energetic, biosynthetic, and redox needs at various stages of metastasis [4–6]. During invasion, metabolic adaptation also enables cancer cells to survive in nutrient-limited environments [5, 6]. Epithelial-to-mesenchymal transition (EMT), a process where epithelial cancer cells acquire mesenchymal features, plays an important role in cancer cell invasion and metastasis [6, 7]. EMT is regulated by several transcription factors, which control, among others, numerous metabolic genes [5, 6].
One of the key metabolic branches in the cell is the network of folate-dependent biochemical pathways. Folate (vitamin B9) functions as a coenzyme in numerous reactions of one-carbon transfer, which are involved in nucleotide biosynthesis, amino acid metabolism, redox homeostasis, and epigenetic regulation [8–10]. Since humans cannot synthesize folate, folate metabolism is regulated to a significant extent by dietary intake of the vitamin. Low folate intake or status has been associated with an increased risk of certain diseases, most notably neural tube defects [10–12], but also particular forms of cancer [13, 14]. In 1996, the U.S. Food and Drug Administration (FDA) approved mandatory fortification of several types of grain foods in the US with folic acid, a synthetic form of the vitamin, to prevent neural tube defects [15].
Replenishing the intracellular folate pool by the intake of exogenous folates, such as folic acid, is especially important for rapidly dividing cells, including cancer cells. Ample preclinical evidence links cellular replication to folate availability [16–23]. Such a link raised the concern that excessive consumption of dietary folate or folic acid may promote the progression of neoplastic lesions and increase the risk of malignancies and cancer-related death [11, 24–26]. In support of such concern, animal studies have demonstrated that high intake of folic acid promotes the progression of established tumors [27–31]. A similar conclusion has been drawn based on the observed association between serum folate levels and proliferation of cancer cells in men with prostate cancer [32]. Of note, while additional studies support this conclusion [33], other studies have shown inverse associations between dietary folate and tumor development [34]. Nevertheless, the possibility that excessive intake of folates might promote development of malignancies should not be ignored [13, 35, 36].
The role of folate in cancer metastasis is even less clear and the literature on this subject is limited and somewhat controversial. Low extracellular folate is associated with altered expression of genes involved in cell adhesion, migration and invasion [37–39] indicating that the vitamin might play a role in the mechanism of metastasis. Of note, the positive association between folic acid supplementation and the migratory ability of cancer cells, as well as the inhibitory effects of antifolates on cell migration, have been demonstrated [40–42]. In further support of the role of folate metabolism in metastasis, metastasizing melanomas have been shown to have increased dependence on NADPH-generating folate pathways [43]. Also, antifolate pemetrexed has been approved by the FDA for the treatment of advanced or metastatic non-small cell lung cancer [44–46]. Several studies, however, suggested that folate supplementation would inhibit rather than promote metastasis. For example, a recent report has demonstrated that in cultured cells folate deficiency causes the elevation of reactive oxygen species, which leads to the suppression of proliferation, but at the same time enhances metastatic potential [47]. In agreement with these findings, folate deficiency promoted metastasis in a xenograft mouse model [47]. In another study, folate deprivation enhanced invasiveness of colon cancer cells through the activation of sonic hedgehog and NF-κB signaling [48]. Inhibitory effects of folic acid on metastasis and proliferation have been also linked to the activation of the folate receptor-ERK1/2-TSLC1 signaling pathway [49]. Contrary to these studies, we have previously reported that folate deprivation inhibits migration and invasion of cultured cancer cells and decreases tumor growth and metastasis in a mouse model [28, 42, 50].
In a recent study, we have shown that folic acid restriction led to metabolic reprogramming and a less aggressive phenotype in cultured murine triple-negative breast cancer (TNBC) cells [50]. Specifically, cells kept on folic acid-free medium displayed reduced migration and had weaker invasion potential [50]. Interestingly, effects of folic acid withdrawal were more profound in more tumorigenic mesenchymal cells than in epithelial cells [50]. To obtain insight into potential mechanisms related to folate restriction, in the present study we have evaluated transcriptomic profiles in three mouse TNBC cell lines grown in medium with or without folic acid.
2. MATERIALS AND METHODS
2.1. Cell lines
Epithelial (E-Wnt) and mesenchymal (M-Wnt) cells were each derived from the same pool of mammary tumors from mouse mammary tumor virus (MMTV)-Wnt-1 transgenic mice [50, 51]. Spontaneous mammary tumors were excised, dissected and mechanically dissociated, and viable cells were plated [51]. Based on morphology, specific clones with epithelial and mesenchymal properties were selected, further characterized and eventually denoted as E-Wnt and M-Wnt cells, respectively [51]. Metastatic metM-Wntliver cells were derived from a liver metastasis following orthotopic transplant of M-Wnt cells in severe combined immunodeficient (SCID) mice [50, 52]. All cell lines were tested for species verification, karyotyping, and genomic instability and were authenticated by the Molecular Cytogenetics Core facility at the University of Texas MD Anderson Cancer Center (Houston, TX).
2.2. Cell culture experiments
E-Wnt, M-Wnt and metM-Wntliver cells were cultured in 10-cm plates for 72 hours using either standard RPMI 1640 medium (containing 2.2 μM folic acid) with 10% fetal bovine serum (FBS) or RPMI 1640 medium without folic acid with dialyzed FBS. Media were purchased from Invitrogen, Carlsbad, CA and FBS came from Atlanta Biologicals (Flowery Branch, GA).
2.3. RNA isolation and microarrays
Total RNA was extracted from three samples (biological replicates) per cellular subtype using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Complementary RNA (cRNA) was generated using a Low Input Quick Amp labeling kit (Agilent, Santa Clara, CA) with NIH3T3 cells as reference RNA. Gene expression microarrays were processed by the Lineberger Comprehensive Cancer Center Genomics Core (University of North Carolina, Chapel Hill, NC) as previously described [51, 52]. A total of 18 samples were hybridized to custom two-color 180K Agilent microarrays (BARCODE25503) and scanned using an Agilent Technologies Scanner G2505C with Feature Extraction software (Agilent, Santa Clara, CA). Microarray data were submitted to the Gene Expression Omnibus (GEO) repository with number GSE148738.
2.4. Microarray data analysis
Gene expression data were processed and analyzed using the limma package (3.38.3) [53] in R (version 3.5.0). Background correction, within-array normalization using global loess normalization, and between-array quantile normalization were performed before further processing of the data. Positive (n=4254) and negative (n=594) control probes were filtered from the dataset. Probes with a low intensity (< the average signal of 75% of the negative control probes) were excluded from further analyses. The ‘avereps’ function was used to condense the dataset by taking the average of replicate probes, resulting in a final dataset consisting of 78,555 probes used for downstream analyses. The probes were annotated based on the annotation file available through GEO (platform GPL23453). Differentially expressed probes were identified across the cellular subtypes and between folic acid withdrawal and control conditions within each subtype [54]. P-values were corrected for multiple testing by using the Benjamini-Hochberg (BH) approach [55]. BH-adjusted P-values <0.05 were considered statistically significant. Pathway analyses and upstream regulatory analyses were conducted using Ingenuity Pathway Analysis (IPA) software (Qiagen Bioinformatics, Aarhus, Denmark). Additional gene ontology was determined using Enrichr [56, 57]. Integrative pathway analyses based on gene expression data as well as our previously reported metabolite data [50] were performed using the joined pathway module in MetaboAnalyst [58]. For each of the cellular subtypes, the top 100 probes (based on lowest P-values) were analyzed together with the metabolites that were altered upon folic acid withdrawal (n=12 for E-Wnt, n=29 for M-Wnt and n=25 for metM-Wntliver) [50]. Hypergeometric testing was applied for the pathway overrepresentation analyses (with corresponding natural log P-values) and pathway topology analyses (based on degree centrality) were quantified using pathway impact values.
3. RESULTS
3.1. Effects of folic acid withdrawal on transcriptomic profiles in mouse TNBC cell lines
We have previously demonstrated that folic acid withdrawal suppresses proliferation and motility, while promoting metabolic reprogramming, in E-Wnt, M-Wnt and metM-Wntliver cells [50]. Herein, we have determined the transcriptional response of these cells following withdrawal of folic acid. Based on the top 500 probes with most variable gene expression, the epithelial cells (E-Wnt) showed expression profiles distinct from the mesenchymal cells (M-Wnt and metM-Wntliver). Additionally, the M-Wnt cells clustered according to the presence or absence of folic acid in the culture medium (Figure 1A).
Figure 1:
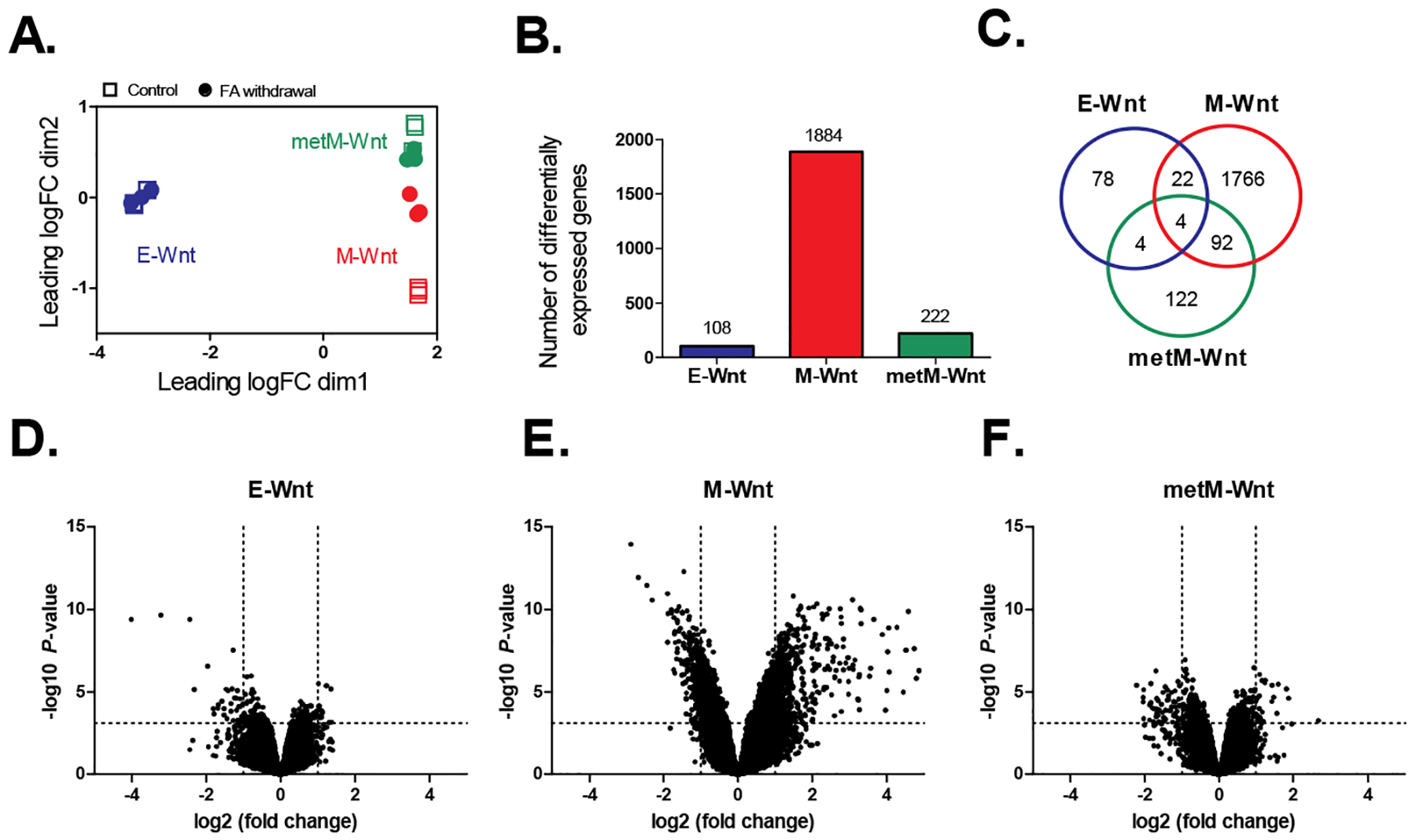
Analysis of transcriptomic data for three mouse TNBC cell lines grown in folic acid-competent (control) or folic acid-free media (withdrawal). A) Multidimensional scaling plot showing the distances between gene expression profiles for the individual samples according to the cellular subtype and treatment based on the top-500 probes. Squares represent control samples (grown in folic acid-competent medium); circles represent samples collected upon folic acid withdrawal. B) Number of differentially expressed genes upon folic acid withdrawal relative to control. BH-adjusted P-values < 0.05. C) Venn diagram showing the overlap between the differentially expressed genes identified for the different subtypes. D-F) Volcano plots showing the (-log10) P-values versus the differences (log2 fold change) in gene expression upon folic acid withdrawal in E-Wnt, M-Wnt and metM-Wntliver cells, respectively. Dashed lines represent the BH-adjusted P-value of 0.05 (horizontal) and fold changes of −2 / 2 (vertical). n=3 biological replicates per group. FA; folic acid.
Upon folic acid withdrawal, differential expression was found for 108, 1884 and 222 genes in the E-Wnt, M-Wnt and metM-Wntliver cells, respectively (BH-adjusted P-value <0.05) (Figure 1B). For four genes (Egr2, Oasl1, Rsad2 and Khdrbs3), differential expression upon folic acid withdrawal was found in all cellular subtypes (Figure 1C). The direction of gene expression changes, however, differed. Expression of Egr2 and Khdrbs3 was upregulated after folic acid withdrawal in M-Wnt and metM-Wntliver cells and downregulated in E-Wnt cells. For Oasl1 and Rsad2, expression was downregulated in the E-Wnt and metM-Wntliver cells and upregulated in the M-Wnt cells. Further exploration of the Venn diagram showed that the majority of the differentially expressed genes was unique for one of the respective cellular subtypes, with some additional overlap (n=92 overlapping genes) found for the M-Wnt and metM-Wntliver cells. These overlapping genes were enriched for the type I interferon signaling pathway (Gene Ontology Biological Process 2018: GO 0060337, q-value 2.8E-09). Based on Volcano plots, it was confirmed that the most pronounced effects on gene expression profiles (in terms of both fold changes and statistical significance) were found for M-Wnt cells as compared to the two other subtypes (Figures 1D–F).
3.2. Expression of interferon signaling genes was differentially regulated upon folic acid withdrawal
Ingenuity Pathway Analysis revealed that the affected pathways upon folic acid withdrawal (compared to control conditions) in M-Wnt cells were predominantly related to (innate) immune responses, and inflammation (Figure 2A). Lipopolysaccharide and various interferon-related factors, such as Irf7, Ifna2, Ifnl1, Irf3 were identified as potential upstream regulators responsible for the observed differences in gene expression upon folic acid withdrawal in the M-Wnt cells (Table 1). All of these pathways and upstream regulators were predicted to be activated, indicating activation of type I interferon signaling, in response to folic acid withdrawal in the MWnt cells.
Figure 2:
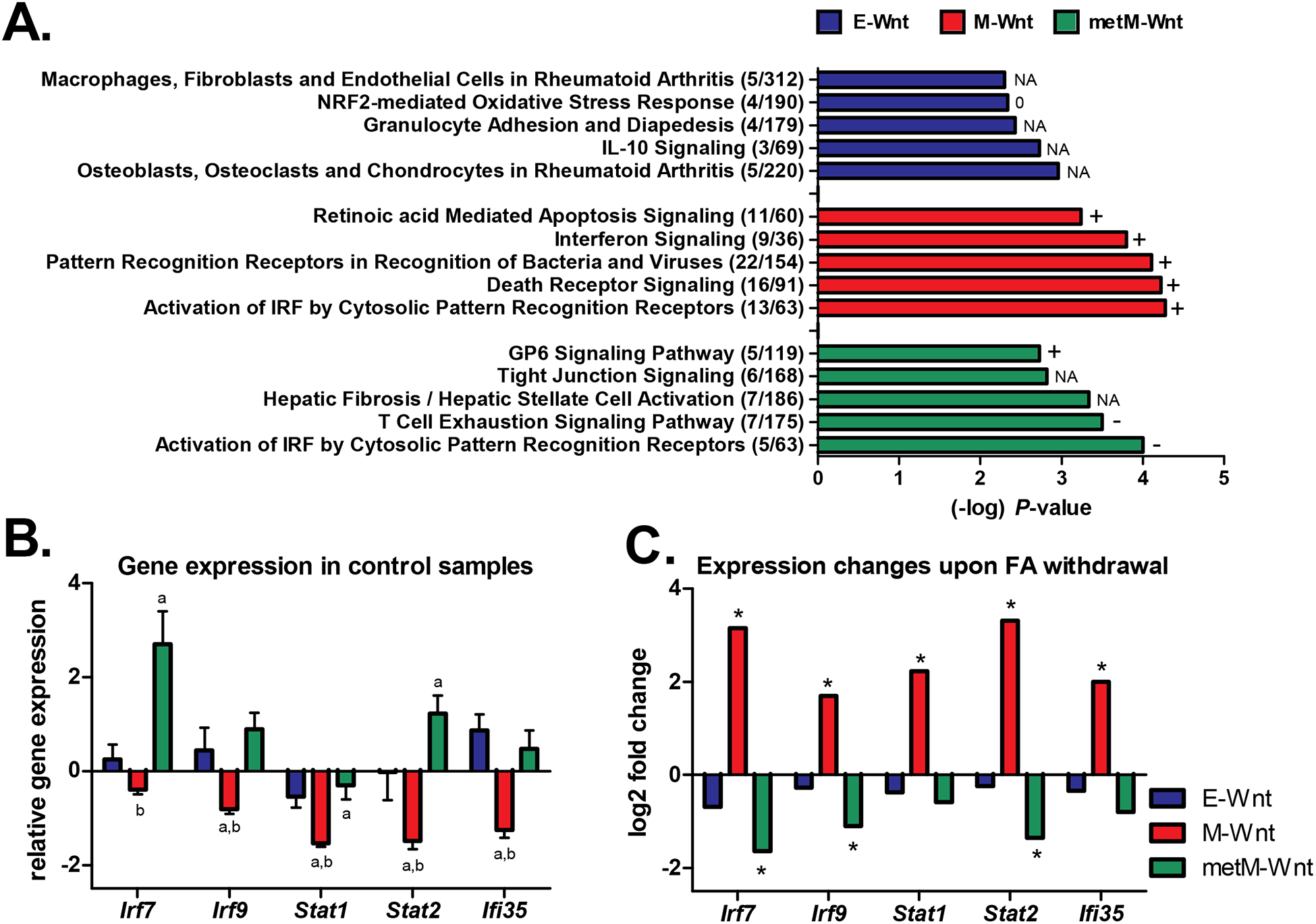
Pathway analyses. A) The top-5 canonical pathways (identified through Ingenuity Pathway Analysis) that were differentially regulated upon folic acid withdrawal in the E-Wnt, M-Wnt or metM-Wntliver cells. The number of differentially expressed genes / the total number of genes in the pathway are shown. A P-value <0.001 was applied as threshold for differential expression. The predicted activation state of the pathways is reflected by + (activation, positive z-score), − (inactivation, negative z-score), 0 (z-score = 0), or NA (activity pattern cannot be assessed). B) Gene expression (mean ± SD) in the control condition (folic acid-competent medium) relative to the common reference NIH3T3 cells according to the microarray data. In case of multiple probes on the array the expression values were averaged. a = BH-adjusted P<0.05 compared to E-Wnt; b = BH-adjusted P<0.05 compared to metM-Wntliver. C) Log2 fold changes in gene expression upon withdrawal of folic acid from the medium (withdrawal versus control conditions). *BH-adjusted P-value <0.05 for comparison folic acid withdrawal versus control condition. Irf7 and Irf9 interferon regulatory factors; Stat1 and Stat2 signal transducer and activator of transcription; Ifi35 interferon-induced protein 35.
Table 1:
The top-10 potential upstream regulators identified through IPA Upstream Regulator Analysis
| Top-10 regulators | Description | Number of target genes in dataset | Predicted activation state | Activation z-score | P-value of overlap |
|---|---|---|---|---|---|
| E-Wnt cells | |||||
| lipopolysaccharide | microbial component | 27 | Inhibited | −3.11 | 2.55E-10 |
| bicuculline | chemical - endogenous | 7 | Inhibited | −2.57 | 1.09E-09 |
| dalfampridine | chemical drug | 7 | Inhibited | −2.65 | 1.81E-09 |
| GnRH-A | chemical reagent | 7 | Inhibited | −2.61 | 7.54E-09 |
| Fosll | transcription regulator | 7 | Activated | 2.18 | 1.01E-08 |
| Igf1 | growth factor | 13 | Inhibited | −3.07 | 1.72E-08 |
| Ifng | cytokine | 21 | Inhibited | −2.01 | 1.75E-08 |
| Pdgfbb | complex | 11 | Inhibited | −3.24 | 2.88E-08 |
| beta-estradiol | chemical - endogenous | 24 | Inhibited | −2.63 | 7.08E-08 |
| Creb1 | transcription regulator | 13 | Inhibited | −2.90 | 1.24E-07 |
| M-Wnt cells | |||||
| lipopolysaccharide | microbial component | 192 | Activated | 6.62 | 1.23E-13 |
| Mapk1 | kinase | 60 | Inhibited | −2.28 | 1.60E-13 |
| Irf7 | transcription regulator | 39 | Activated | 5.83 | 2.27E-13 |
| Ifna2 | cytokine | 46 | Activated | 6.12 | 1.57E-12 |
| Ifnl1 | cytokine | 26 | Activated | 5.01 | 1.73E-12 |
| Tp53* | transcription regulator | 187 | Activated | 4.35 | 1.85E-12 |
| Pntp1 | enzyme | 20 | Inhibited | −4.35 | 2.20E-12 |
| Trim24 | transcription regulator | 28 | Inhibited | −4.29 | 2.74E-12 |
| Irf3 | transcription regulator | 41 | Activated | 5.34 | 2.76E-12 |
| Interferon alpha | group | 66 | Activated | 5.51 | 6.18E-12 |
| metM-Wntllver cells | |||||
| Stat3 | transcription regulator | 25 | Activated | 3.00 | 2.85E-12 |
| Prl | cytokine | 18 | Inhibited | −2.52 | 5.52E-12 |
| Irf7 | transcription regulator | 14 | Inhibited | −3.66 | 4.94E-11 |
| Ifnb1 | cytokine | 16 | Inhibited | −2.77 | 8.31E-10 |
| dexamethasone | chemical drug | 43 | Activated | 2.11 | 8.55E-10 |
| Ifnl1 | cytokine | 10 | Inhibited | −3.10 | 8.87E-10 |
| Ackr2 | G-protein coupled receptor | 8 | Activated | 2.83 | 1.04E-09 |
| Mapk1 | kinase | 17 | Activated | 3.80 | 1.71E-09 |
| Stat6 | transcription regulator | 18 | Activated | 3.13 | 3.59E-09 |
| Ifna2 | cytokine | 14 | Inhibited | −3.18 | 4.50E-09 |
The top-10 potential upstream regulators based on P-value of overlap in regulators with a z-score of <−2 or >2 were considered. Predicted regulators that were flagged for potential bias were excluded.
Trp53 is the mouse variant of the common (human) TP53 output obtained through Ingenuity Pathway Analysis.
For the E-Wnt and metM-Wntliver cells, pathway analyses also indicated that expression of genes involved in immune and inflammatory responses as well as cell death regulation was affected by folic acid withdrawal (Figure 2A). For the immune-, and specifically interferon-related pathways, the activation patterns upon folic acid withdrawal were largely opposite to the effects observed for M-Wnt cells. Further exploration of the transcriptomic data for several individual genes (Irf7, Irf9, Stat1, Stat2) involved in these pathways demonstrated that M-Wnt cells already had lower expression of these genes under control conditions as compared to E-Wnt and metM-Wntliver cells (Figure 2B). Expression of these genes, however, was consistently upregulated upon folic acid withdrawal in M-Wnt cells, whereas it was unchanged or downregulated in E-Wnt and metM-Wntliver cells, respectively (Figure 2C). To further explore the observation that expression of interferon-related genes was repressed in mesenchymal cells and might be restored by folic acid withdrawal, we examined a previously established interferon-responsive gene signature [59]. The gene signature showed that expression of the majority of the interferon-responsive genes was suppressed in M-Wnt cells compared to E-Wnt and metMWntliver cells. M-Wnt cells cultured in folic acid-free medium showed profound recovery of interferon-responsive gene expression, with log2 fold changes up to 4.8 (Figure 3).
Figure 3:
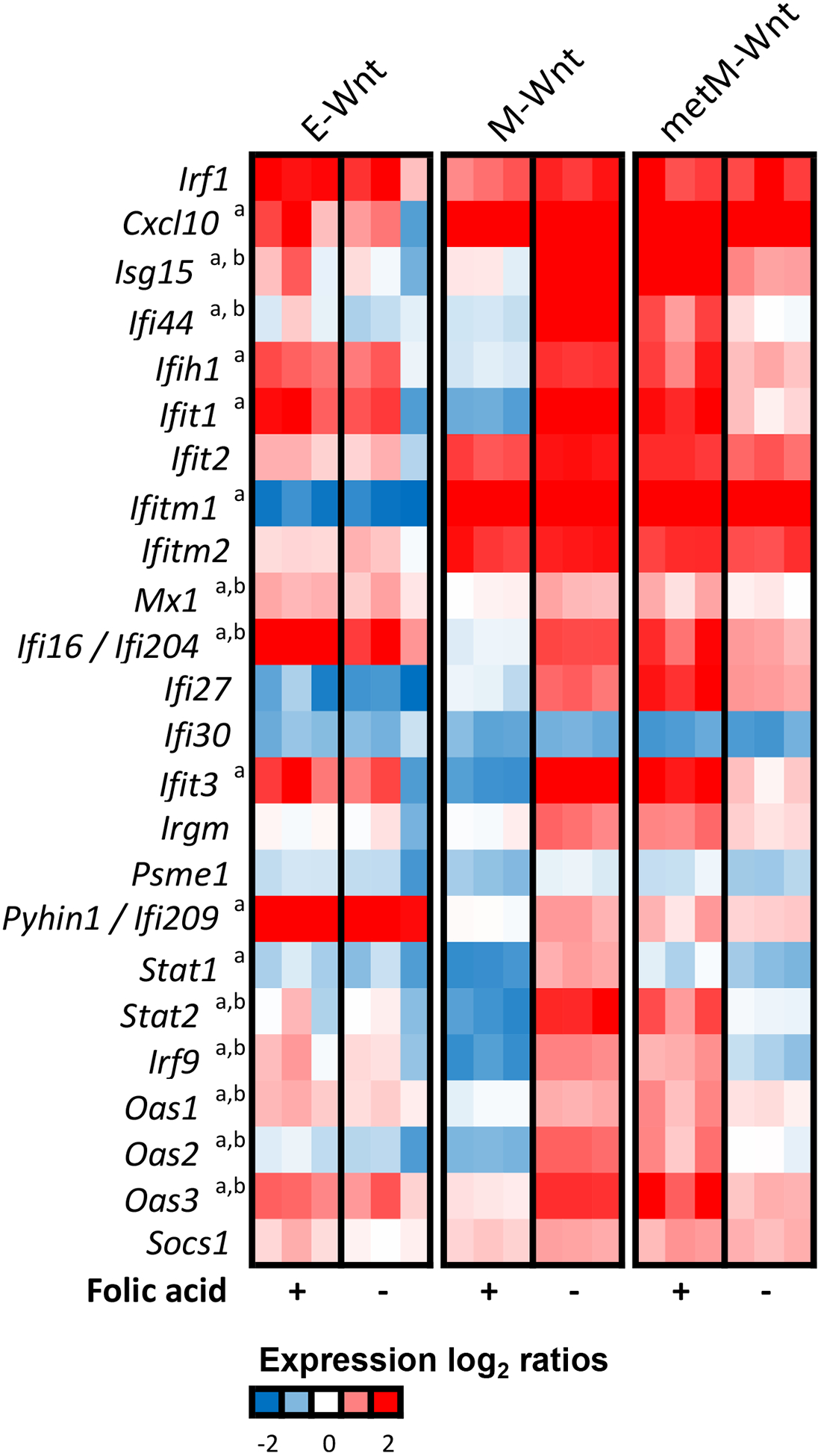
Expression profiles of interferon-responsive genes. Heatmap shows the expression (relative to the common reference NIH3T3 cells) of interferon-responsive genes upon control conditions (+) or folic acid withdrawal (−). A previously reported set of interferon-responsive genes was considered [59]. Colors indicate the expression log2 ratios ranging from blue (−2, low expression) to red (2, high expression) in the current experiment. In case of multiple probes per gene, the average expression value was taken. a Indicates that the expression change upon folic acid withdrawal in M-Wnt cells was statistically significant, b indicates that the expression change upon folic acid withdrawal in metM-Wntliver cells was statistically significant (both BH-adjusted P-value <0.05).
3.3. Folic acid withdrawal did not substantially affect expression of markers for epithelialto-mesenchymal transition in mouse TNBC cell lines
EMT has long been viewed as an important step in the acquisition of invasive and metastatic traits in a variety of cancer types [60]. In the present study, we observed that under control conditions (folic acid-competent medium), the epithelial marker E-cadherin (Cdh1) was strongly expressed in epithelial (E-Wnt) cells (Figure 4A), which is in line with previously reported findings in these cell lines [51]. The mesenchymal markers vimentin (Vim) and fibronectin (Fn1) showed higher expression in the mesenchymal (M-Wnt and metM-Wntliver) cells as compared to the epithelial cells. Also transforming growth factor beta 1 (Tgfb1) and the zinc-finger E-box-binding homeobox factors (Zeb1 and Zeb2) as well as the basic helix-loop-helix factors (Twist1 and Twist2), involved in induction of EMT, were predominantly expressed in the mesenchymal cells. The subtype-specific patterns were less pronounced for the mesenchymal marker N-cadherin (Cdh2), which showed low expression in all cells (relative to the reference NIH3T3 cells). Withdrawal of folic acid from the medium did not result in substantial expression changes for this panel of EMT-related genes (BH-adjusted P-value > 0.05 and log2 fold changes <1) (Figure 4B).
Figure 4:
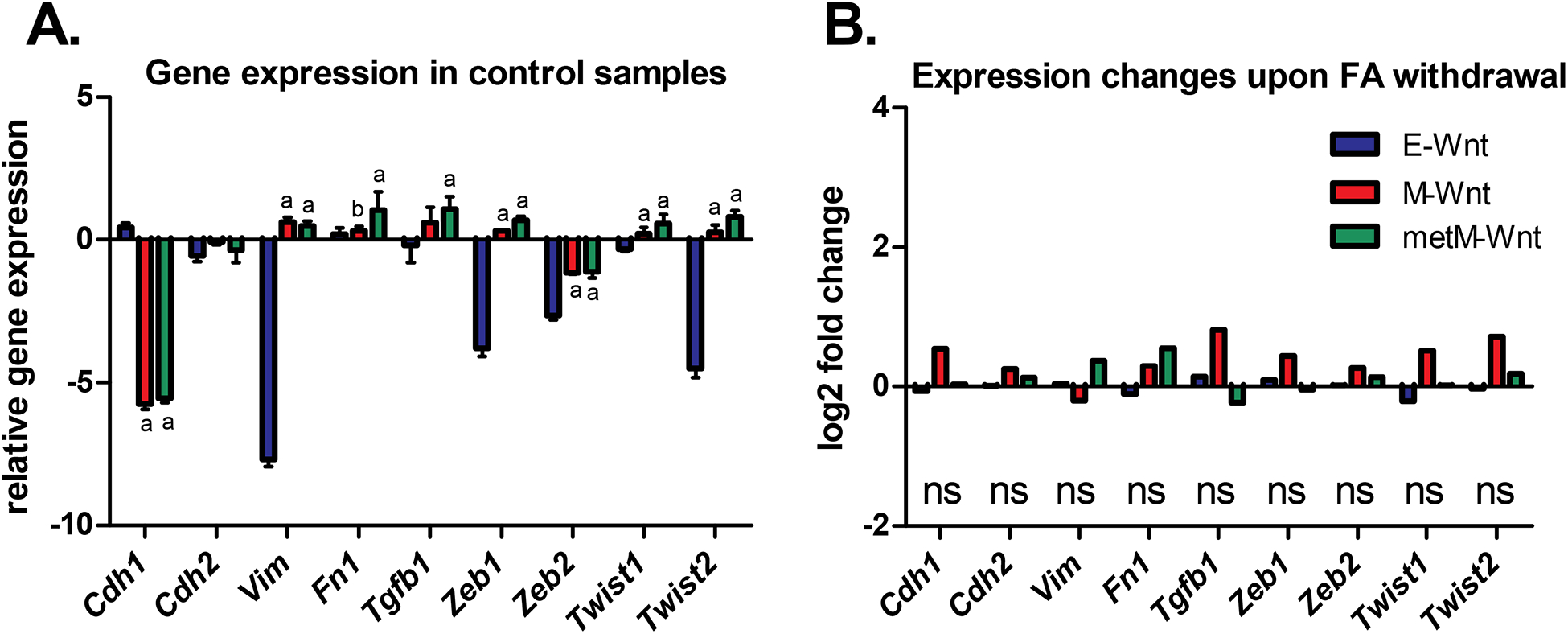
Expression of EMT-related genes in E-Wnt, M-Wnt and metM-Wntliver cells. A) Gene expression (mean ± SD) in the control condition (folic acid-competent medium) relative to the common reference (NIH3T3 cells) according to the microarray data. In case of multiple probes on the array the expression values were averaged. a = BH-adjusted P<0.05 compared to E-Wnt; b = BH-adjusted P<0.05 compared to metM-Wntliver. B) Log2 fold changes in gene expression upon withdrawal of folic acid from the medium (withdrawal versus control conditions; n=3 biological replicates per cellular subtype). Cdh1 E-cadherin; Cdh2 N-cadherin; Vim Vimentin; Fn1 Fibronectin; Tgfb1 transforming growth factor beta 1; Zeb1 and Zeb2 zinc-finger E-box-binding homeobox factors; Twist1 and Twist2 twist family bHLH transcription factors; ns not significant when comparing folic acid withdrawal versus control conditions.
3.4. Integrative pathway analyses
We have also performed an explorative joint pathway analysis [58], which allowed the integration of transcriptomic data with our previous data on metabolite profiling [50]. This analysis indicated that diverse metabolic pathways were affected by folic acid withdrawal (Figure 5). Based on a false-discovery rate (FDR) P-value <0.05, 1, 37 and 19 pathways were identified for E-Wnt, M-Wnt, and metM-Wntliver cells, respectively. The only pathway identified in E-Wnt cells, pantothenate and CoA biosynthesis, was also among the identified pathways in the M-Wnt and metM-Wntliver cells (Supplementary Table 1). All pathways identified in metMWntliver cells were also found for M-Wnt cells, with the exception of pyruvate metabolism and amino sugar and nucleotide sugar metabolism (Supplementary Table 1). Interestingly, pathways uniquely found in M-Wnt cells and not matched with metabolites, were relevant to host response to pathogens, with all identified pathways having links to interferon signaling (Supplementary Table 1). It should be noted that in the present study the application of this approach is rather exploratory due to the limited number of metabolites (n=41) available from our previously published study [50].
Figure 5:
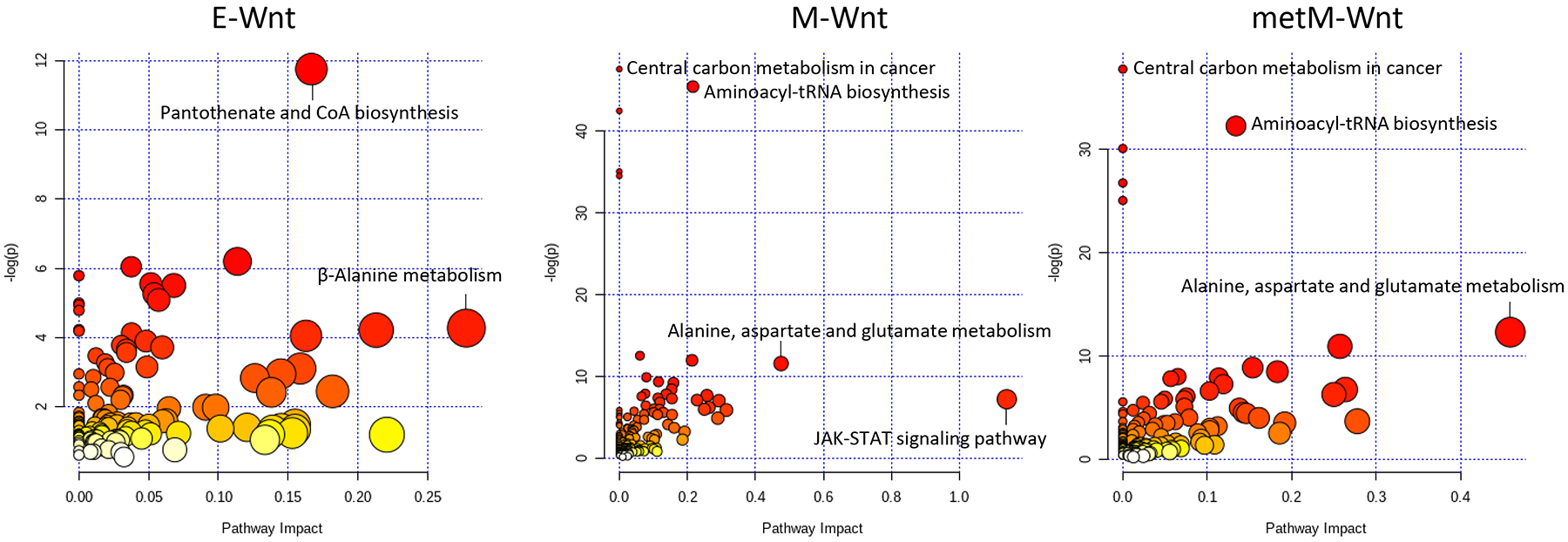
Integrative pathway analyses for transcriptomic and metabolite data using MetaboAnalyst [58]. The top 100 probes showing most pronounced effects of folic acid withdrawal (based on lowest P-values) were analyzed together with the metabolites that were previously reported to be affected by folic acid withdrawal in the E-Wnt (n=12), M-Wnt (n=29) or metM-Wntliver (n=25) cells. Node colors represent -log (ln) P-values for hypergeometric testing as presented on the Y-axis, whereas the X-axis reflects the impact values (with corresponding node sizes) from pathway topology analyses. To clearly illustrate identified pathways for each of the cellular subtypes, scales of the different panels vary.
4. DISCUSSION AND CONCLUSIONS
Folate is known to play an important role in human health and disease due to its participation in diverse metabolic pathways as well as in the regulation of methylation reactions. Pleiotropic effects of folate in cancer cells have been linked to migratory capacity and invasiveness; though effects may differ by cancer type, stage and molecular subtype [50, 61, 62]. We have previously shown that folic acid withdrawal resulted in metabolic and bioenergetic changes indicative of a less proliferative as well as less migratory phenotype in mouse TNBC cell lines [50]. Here we report that transcriptomic profiles of mouse TNBC cell lines possessing a mesenchymal phenotype were more responsive to withdrawal of folic acid than TNBC cells with an epithelial phenotype. These findings are in agreement with our previous work showing that effects on metabolite levels in response to folic acid withdrawal were generally stronger in mesenchymal than epithelial TNBC cells [50]. The transcriptomic effects were also less pronounced in cells derived from liver metastases compared to the parental mesenchymal cell line.
Mesenchymal cells with characteristic traits acquired during EMT are considered crucial for malignant progression of many cancer types including TNBC [63, 64]. We studied the effects of folic acid withdrawal across EMT states in mouse TNBC cell lines that share the same genetic background, but differ in terms of molecular characteristics and metastatic potential in addition to cellular phenotype [51, 52]. We observed that expression of a panel of genes directly involved in EMT did not substantially change upon folic acid withdrawal in any of the TNBC subtypes. Rather, we identified interferon signaling as one of the top pathways affected by folic acid withdrawal in M-Wnt cells. Of note, three out of four genes (Egr2, Oasl1 and Rsad2) affected by folic acid withdrawal in all three studied cell lines are relevant to interferon pathways and immunity. Interestingly, it has been reported that human TNBC cells are characterized by repressed interferon signaling; the phenomenon specific for TNBC cells with mesenchymal, in contrast to epithelial, morphology [59]. Treatment of mesenchymal TNBC cells with interferon-β repressed mesenchymal properties and inhibited migration [59]. Remarkably, the reported change in the expression pattern of interferon signaling pathway genes in response to the sustained interferon-β exposure [59] was almost identical to such a pattern in M-Wnt cells in response to folic acid withdrawal in our study. Furthermore, exploratory joint pathway analysis, based on our transcriptomic and metabolite data, identified several pathways linked to interferon signaling in M-Wnt cells only. The top pathway from this analysis was JAK-STAT signaling, which has a role in both breast cancer etiology and interferon response [65, 66]. Another pathway in the list, NOD-like receptor signaling, has been linked to the STAT-JAK pathway as well as to interferon signaling [67]. Additional pathways identified for M-Wnt cells were host-pathogen interactions and thus also directly relevant to interferon signaling.
The putative link between folate metabolism and interferon signaling as well as other indicators of (innate) immune responses have been described before in cell culture as well as in animal and human studies. High-dose folic acid supplementation or high circulating levels of folic acid in healthy young individuals as well as in obese post-menopausal women were associated with reduced numbers and cytotoxicity of natural killer cells and altered expression profiles of cytokines [68, 69]. Exposure of human colonic cell lines to increasing doses of folic acid resulted in altered expression of inflammatory mediators, with effects differing for non-malignant and malignant cell lines [70]. The relationship between high folic acid intake and reduced natural killer cell cytotoxicity was also confirmed in murine studies and was, at least partially, attributed to reduced interleukin (IL)-10 production with excess folic acid [71]. Our observation that folic acid withdrawal might reactivate interferon signaling in mesenchymal TNBC cells is of particular interest from a clinical perspective since such reactivation has been considered a promising therapeutic target [59, 72, 73]. TNBC cells characterized by active INF / STAT signaling are viewed as ‘hot’ tumors that show prognostic benefits over TNBC subtypes with immune-repressed traits (‘cold’ tumors) [72, 74, 75]. Transformation of TNBC from ‘cold’ to ‘hot’ tumors, may therefore provide a window of opportunity for targeted and immune-based therapies [72].
Towards this end, our data suggest that folate restriction might contribute to such therapeutically beneficial transformation. Therefore, traditional chemotherapeutics targeting folate metabolism could be a possible (additional) treatment option for specific TNBC subtypes. Indeed, methotrexate and 5-fluorouracil in combination with cyclophosphamide have been commonly used for the treatment of TNBC [76, 77]. The new generation antifolate pemetrexed has been in clinical trials for the treatment of advanced breast cancer as well [78]. In general, the drug has shown strong activity with acceptable toxicities for treatment of metastatic breast cancer patients [79]. Yet, resistance to antifolates is a common phenomenon making them largely ineffective in cancer treatment. Multiple mechanisms underlie the development of antifolate resistance, including activation of the drug efflux processes [80]. For tumors relaying on this mechanism, folate restriction removes the need to accumulate drugs in the cell and could directly disable important biosynthetic pathways by depriving cancer cells of important metabolites rather than by inhibiting corresponding enzymes. Moreover, antifolates might not be active in cells with sufficient levels of reduced folate to outcompete the action of drugs, as occurs under conditions of excess folate [11]. It should be expected that the response to folate restriction will be cell-type specific and will be modulated depending on the gene signatures for a specific cell type. For example, thymidylate synthase expression predicts poor response to pemetrexed in patients with advanced breast cancer [81]. Likewise, our study supports differential responses to folic acid restriction varying by EMT status, gene expression profiles and metastatic potential.
These findings may thus provide a rationale to consider folate deprivation as a therapeutic option for TNBC, potentially in combination with other treatment strategies. Obviously, validation of our findings in human cell lines and cancer models is required and ultimately clinical studies are warranted. For example, safe limits of folate restriction as well as its efficacy towards specific cancer subtypes needs to be established; this calls for adaptation of personalized treatment strategies based on molecular subtype and folate status [75]. Importantly, our study indicates a subtype-specific response to folic acid restriction. Perhaps the regulation of interferon signaling in TNBC could be cell type-specific as well. In support of this possibility, it has been shown that the interferon regulatory factor Irf7 was suppressed in tumor cells from a bone metastasis in the 4T1.2 mouse model [82], while this gene was highly expressed in metMWntliver cells in our study. Interestingly, restoration of Irf7 expression or administration of interferon-α1 led to reduced metastasis rates to the spine and femur in the 4T1.2 mouse model, whereas the extent of metastasis to the lung was not reduced [82]. This finding suggests additional, metastasis site-dependent, specificity of the interferon effect.
Overall, our findings suggest that folic acid withdrawal affects gene expression in a mouse cell model of TNBC, with most pronounced effects in cells with a mesenchymal phenotype. These cells revealed repressed expression of genes representing the interferon signaling signature, and this effect was reversed upon folic acid withdrawal. Our research provides novel leads for further studies focusing on folate deprivation as a potential strategy to improve treatment responses, particularly in mesenchymal subtypes of TNBC.
Supplementary Material
HIGHLIGHTS.
Folic acid withdrawal alters gene expression in mouse mammary cancer cell lines
Changes are most pronounced in non-metastatic mesenchymal cells
Interferon signaling pathway genes are upregulated by folic acid withdrawal
Reactivation of the type I interferon pathway may have clinical utility
ACKNOWLEDGMENTS
This study was supported by a grant from the National Cancer Institute (R35 CA197627) to SDH. SAK is supported by the National Institutes of Health grant DK117854. DEK is supported by a Veni grant (016.Veni.188.082) of the Dutch Research Council. Funding agencies had no involvement in study design, collection, analysis or interpretation of data, writing of the report or decision to submit the manuscript. The authors thank Dr. David Horita for carefully reading the manuscript.
ABBREVIATIONS
- BH
Benjamini-Hochberg
- cRNA
complementary RNA
- EMT
epithelial-to-mesenchymal transition
- E-Wnt
epithelial cells
- FA
folic acid
- FBS
fetal bovine serum
- FDA
U.S. Food and Drug Administration
- FDR
false discovery rate
- GEO
gene expression omnibus
- GO
gene ontology
- INF
interferon
- IPA
Ingenuity Pathway Analysis
- ln
natural logarithm
- metM-Wntliver
metastatic mesenchymal cells from liver metastases
- MMTV
mouse mammary tumor virus
- M-Wnt
mesenchymal cells
- NIH3T3
mouse fibroblast cells
- SCID
severe combined immunodeficient
- STAT
signal transducer and activator of transcription
- TNBC
triple-negative breast cancer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF COMPETING INTEREST
The authors declare no conflict of interest
REFERENCES
- [1].Chambers AF, Groom AC, MacDonald IC, Dissemination and growth of cancer cells in metastatic sites, Nat Rev Cancer, 2 (2002) 563–572. [DOI] [PubMed] [Google Scholar]
- [2].Sethi N, Kang Y, Unravelling the complexity of metastasis - molecular understanding and targeted therapies, Nat Rev Cancer, 11 (2011) 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sahai E, Illuminating the metastatic process, Nat Rev Cancer, 7 (2007) 737–749. [DOI] [PubMed] [Google Scholar]
- [4].Thomson TM, Balcells C, Cascante M, Metabolic Plasticity and Epithelial-Mesenchymal Transition, J Clin Med, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lehuede C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM, Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis, Cancer Res, 76 (2016) 5201–5208. [DOI] [PubMed] [Google Scholar]
- [6].Sciacovelli M, Frezza C, Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer, FEBS J, 284 (2017) 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].De Craene B, Berx G, Regulatory networks defining EMT during cancer initiation and progression, Nat Rev Cancer, 13 (2013) 97–110. [DOI] [PubMed] [Google Scholar]
- [8].Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD, Quantitative flux analysis reveals folate-dependent NADPH production, Nature, 510 (2014) 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tibbetts AS, Appling DR, Compartmentalization of Mammalian folate-mediated one-carbon metabolism, Annu Rev Nutr, 30 (2010) 57–81. [DOI] [PubMed] [Google Scholar]
- [10].Ducker GS, Rabinowitz JD, One-Carbon Metabolism in Health and Disease, Cell Metab, 25 (2017) 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Strickland KC, Krupenko NI, Krupenko SA, Molecular mechanisms underlying the potentially adverse effects of folate, Clin Chem Lab Med, 51 (2013) 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stover PJ, Physiology of folate and vitamin B12 in health and disease, Nutr Rev, 62 (2004) S3–12; discussion S13. [DOI] [PubMed] [Google Scholar]
- [13].Kim YI, Folate and cancer: a tale of Dr. Jekyll and Mr. Hyde?, The American journal of clinical nutrition, 107 (2018) 139–142. [DOI] [PubMed] [Google Scholar]
- [14].Williams EA, Folate, colorectal cancer and the involvement of DNA methylation, Proceedings of the Nutrition Society, 71 (2012) 592–597. [DOI] [PubMed] [Google Scholar]
- [15].Honein MA, Paulozzi LJ, Mathews TJ, Erickson JD, Wong L-YC, Impact of Folic Acid Fortification of the US Food Supply on the Occurrence of Neural Tube Defects, JAMA, 285 (2001) 2981–2986. [DOI] [PubMed] [Google Scholar]
- [16].Koury MJ, Horne DW, Apoptosis mediates and thymidine prevents erythroblast destruction in folate deficiency anemia, Proc Natl Acad Sci U S A, 91 (1994) 4067–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Courtemanche C, Elson-Schwab I, Mashiyama ST, Kerry N, Ames BN, Folate deficiency inhibits the proliferation of primary human CD8+ T lymphocytes in vitro, J Immunol, 173 (2004) 3186–3192. [DOI] [PubMed] [Google Scholar]
- [18].Craciunescu CN, Brown EC, Mar MH, Albright CD, Nadeau MR, Zeisel SH, Folic acid deficiency during late gestation decreases progenitor cell proliferation and increases apoptosis in fetal mouse brain, J Nutr, 134 (2004) 162–166. [DOI] [PubMed] [Google Scholar]
- [19].Li D, Rozen R, Maternal folate deficiency affects proliferation, but not apoptosis, in embryonic mouse heart, J Nutr, 136 (2006) 1774–1778. [DOI] [PubMed] [Google Scholar]
- [20].Chen Y, Wang Z, Xie Y, Guo X, Tang X, Wang S, Yang S, Chen K, Niu Y, Ji W, Folic acid deficiency inhibits neural rosette formation and neuronal differentiation from rhesus monkey embryonic stem cells, J Neurosci Res, 90 (2012) 1382–1391. [DOI] [PubMed] [Google Scholar]
- [21].Moussa C, Ross N, Jolette P, MacFarlane AJ, Altered folate metabolism modifies cell proliferation and progesterone secretion in human placental choriocarcinoma JEG-3 cells, Br J Nutr, 114 (2015) 844–852. [DOI] [PubMed] [Google Scholar]
- [22].Lamm N, Maoz K, Bester AC, Im MM, Shewach DS, Karni R, Kerem B, Folate levels modulate oncogene-induced replication stress and tumorigenicity, EMBO Mol Med, 7 (2015) 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hwang SY, Kang YJ, Sung B, Jang JY, Hwang NL, Oh HJ, Ahn YR, Kim HJ, Shin JH, Yoo MA, Kim CM, Chung HY, Kim ND, Folic acid is necessary for proliferation and differentiation of C2C12 myoblasts, J Cell Physiol, 233 (2018) 736–747. [DOI] [PubMed] [Google Scholar]
- [24].Ebbing M, Bonaa KH, Nygard O, Arnesen E, Ueland PM, Nordrehaug JE, Rasmussen K, Njolstad I, Refsum H, Nilsen DW, Tverdal A, Meyer K, Vollset SE, Cancer incidence and mortality after treatment with folic acid and vitamin B12, JAMA, 302 (2009) 2119–2126. [DOI] [PubMed] [Google Scholar]
- [25].Crider KS, Bailey LB, Berry RJ, Folic acid food fortification-its history, effect, concerns, and future directions, Nutrients, 3 (2011) 370–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mason JB, Folate, cancer risk, and the Greek god, Proteus: a tale of two chameleons, Nutr Rev, 67 (2009) 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hansen MF, Jensen SO, Fuchtbauer EM, Martensen PM, High folic acid diet enhances tumour growth in PyMT-induced breast cancer, Br J Cancer, 116 (2017) 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Oleinik NV, Helke KL, Kistner-Griffin E, Krupenko NI, Krupenko SA, Rho GTPases RhoA and Rac1 mediate effects of dietary folate on metastatic potential of A549 cancer cells through the control of cofilin phosphorylation, J Biol Chem, 289 (2014) 26383–26394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bistulfi G, Foster BA, Karasik E, Gillard B, Miecznikowski J, Dhiman VK, Smiraglia DJ, Dietary folate deficiency blocks prostate cancer progression in the TRAMP model, Cancer Prev Res (Phila), 4 (2011) 1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Deghan Manshadi S, Ishiguro L, Sohn KJ, Medline A, Renlund R, Croxford R, Kim YI, Folic acid supplementation promotes mammary tumor progression in a rat model, PLoS One, 9 (2014) e84635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lindzon GM, Medline A, Sohn KJ, Depeint F, Croxford R, Kim YI, Effect of folic acid supplementation on the progression of colorectal aberrant crypt foci, Carcinogenesis, 30 (2009) 1536–1543. [DOI] [PubMed] [Google Scholar]
- [32].Tomaszewski JJ, Cummings JL, Parwani AV, Dhir R, Mason JB, Nelson JB, Bacich DJ, O’Keefe DS, Increased cancer cell proliferation in prostate cancer patients with high levels of serum folate, Prostate, 71 (2011) 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tu H, Dinney CP, Ye Y, Grossman HB, Lerner SP, Wu X, Is folic acid safe for non-muscle-invasive bladder cancer patients? An evidence-based cohort study, Am J Clin Nutr, 107 (2018) 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kuo CS, Lin CY, Wu MY, Lu CL, Huang RF, Relationship between folate status and tumour progression in patients with hepatocellular carcinoma, Br J Nutr, 100 (2008) 596–602. [DOI] [PubMed] [Google Scholar]
- [35].Miller JW, Ulrich CM, Folic acid and cancer--where are we today?, Lancet, 381 (2013) 974–976. [DOI] [PubMed] [Google Scholar]
- [36].Smith AD, Refsum H, Selhub J, Rosenberg IH, Decision on folic acid fortification in Europe must consider both risks and benefits, BMJ, 352 (2016) i734. [DOI] [PubMed] [Google Scholar]
- [37].Jhaveri MS, Wagner C, Trepel JB, Impact of extracellular folate levels on global gene expression, Mol Pharmacol, 60 (2001) 1288–1295. [DOI] [PubMed] [Google Scholar]
- [38].Crott JW, Choi SW, Ordovas JM, Ditelberg JS, Mason JB, Effects of dietary folate and aging on gene expression in the colonic mucosa of rats: implications for carcinogenesis, Carcinogenesis, 25 (2004) 69–76. [DOI] [PubMed] [Google Scholar]
- [39].Crott JW, Liu Z, Keyes MK, Choi SW, Jang H, Moyer MP, Mason JB, Moderate folate depletion modulates the expression of selected genes involved in cell cycle, intracellular signaling and folate uptake in human colonic epithelial cell lines, J Nutr Biochem, 19 (2008) 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Terzis AJ, Fiskerstrand T, Refsum H, Ueland PM, Arnold H, Bjerkvig R, Proliferation, migration and invasion of human glioma cells exposed to antifolate drugs, Int J Cancer, 54 (1993) 112–118. [DOI] [PubMed] [Google Scholar]
- [41].Siu MK, Kong DS, Chan HY, Wong ES, Ip PP, Jiang L, Ngan HY, Le XF, Cheung AN, Paradoxical impact of two folate receptors, FRalpha and RFC, in ovarian cancer: effect on cell proliferation, invasion and clinical outcome, PLoS One, 7 (2012) e47201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oleinik NV, Krupenko NI, Krupenko SA, ALDH1L1 inhibits cell motility via dephosphorylation of cofilin by PP1 and PP2A, Oncogene, 29 (2010) 6233–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ, Oxidative stress inhibits distant metastasis by human melanoma cells, Nature, 527 (2015) 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kazandjian D, Blumenthal GM, Chen HY, He K, Patel M, Justice R, Keegan P, Pazdur R, FDA approval summary: crizotinib for the treatment of metastatic non-small cell lung cancer with anaplastic lymphoma kinase rearrangements, Oncologist, 19 (2014) e5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Arbour KC, Riely GJ, Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review, JAMA, 322 (2019) 764–774. [DOI] [PubMed] [Google Scholar]
- [46].Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ, Powell SF, Cheng SY, Bischoff HG, Peled N, Grossi F, Jennens RR, Reck M, Hui R, Garon EB, Boyer M, Rubio-Viqueira B, Novello S, Kurata T, Gray JE, Vida J, Wei Z, Yang J, Raftopoulos H, Pietanza MC, Garassino MC, K.−. Investigators, Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer, N Engl J Med, 378 (2018) 2078–2092. [DOI] [PubMed] [Google Scholar]
- [47].Su YH, Huang WC, Huang TH, Huang YJ, Sue YK, Huynh TT, Hsiao M, Liu TZ, Wu AT, Lin CM, Folate deficient tumor microenvironment promotes epithelial-to mesenchymal transition and cancer stem-like phenotypes, Oncotarget, 7 (2016) 33246–33256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang TP, Hsu SH, Feng HC, Huang RF, Folate deprivation enhances invasiveness of human colon cancer cells mediated by activation of sonic hedgehog signaling through promoter hypomethylation and cross action with transcription nuclear factor-kappa B pathway, Carcinogenesis, 33 (2012) 1158–1168. [DOI] [PubMed] [Google Scholar]
- [49].Liu Z, Jin X, Pi W, Liu S, Folic acid inhibits nasopharyngeal cancer cell proliferation and invasion via activation of FRalpha/ERK1/2/TSLC1 pathway, Biosci Rep, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ashkavand Z, O’Flanagan C, Hennig M, Du X, Hursting SD, Krupenko SA, Metabolic Reprogramming by Folate Restriction Leads to a Less Aggressive Cancer Phenotype, Mol Cancer Res, 15 (2017) 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dunlap SM, Chiao LJ, Nogueira L, Usary J, Perou CM, Varticovski L, Hursting SD, Dietary energy balance modulates epithelial-to-mesenchymal transition and tumor progression in murine claudin-low and basal-like mammary tumor models, Cancer prevention research (Philadelphia, Pa.), 5 (2012) 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].O’Flanagan CH, Rossi EL, McDonell SB, Chen X, Tsai YH, Parker JS, Usary J, Perou CM, Hursting SD, Metabolic reprogramming underlies metastatic potential in an obesity-responsive murine model of metastatic triple negative breast cancer, NPJ breast cancer, 3 (2017) 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, limma powers differential expression analyses for RNA-sequencing and microarray studies, Nucleic Acids Research, 43 (2015) e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK, Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression, The annals of applied statistics, 10 (2016) 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing, Journal of the Royal Statistical Society. Series B (Methodological), 57 (1995) 289–300. [Google Scholar]
- [56].Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool, BMC bioinformatics, 14 (2013) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update, Nucleic Acids Res, 44 (2016) W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chong J, Wishart DS, Xia J, Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis, Curr Protoc Bioinformatics, 68 (2019) e86. [DOI] [PubMed] [Google Scholar]
- [59].Doherty MR, Cheon H, Junk DJ, Vinayak S, Varadan V, Telli ML, Ford JM, Stark GR, Jackson MW, Interferon-beta represses cancer stem cell properties in triple-negative breast cancer, Proceedings of the National Academy of Sciences of the United States of America, 114 (2017) 13792–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Brabletz T, Kalluri R, Nieto MA, Weinberg RA, EMT in cancer, Nature reviews. Cancer, 18 (2018) 128–134. [DOI] [PubMed] [Google Scholar]
- [61].Chen WJ, Huang RS, Low-folate stress reprograms cancer stem cell-like potentials and bioenergetics metabolism through activation of mTOR signaling pathway to promote in vitro invasion and in vivo tumorigenicity of lung cancers, The Journal of nutritional biochemistry, 53 (2018) 28–38. [DOI] [PubMed] [Google Scholar]
- [62].Feng HC, Lin JY, Hsu SH, Lan WY, Kuo CS, Tian YF, Sun DP, Huang RS, Low folate metabolic stress reprograms DNA methylation-activated sonic hedgehog signaling to mediate cancer stem cell-like signatures and invasive tumour stage-specific malignancy of human colorectal cancers, International journal of cancer, 141 (2017) 2537–2550. [DOI] [PubMed] [Google Scholar]
- [63].Dongre A, Weinberg RA, New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer, Nature reviews. Molecular cell biology, 20 (2019) 69–84. [DOI] [PubMed] [Google Scholar]
- [64].Fedele M, Cerchia L, Chiappetta G, The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas, Cancers, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Koromilas AE, Sexl V, The tumor suppressor function of STAT1 in breast cancer, Jakstat, 2 (2013) e23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Majoros A, Platanitis E, Kernbauer-Holzl E, Rosebrock F, Muller M, Decker T, Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses, Frontiers in immunology, 8 (2017) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Motta V, Soares F, Sun T, Philpott DJ, NOD-like receptors: versatile cytosolic sentinels, Physiological reviews, 95 (2015) 149–178. [DOI] [PubMed] [Google Scholar]
- [68].Paniz C, Bertinato JF, Lucena MR, De Carli E, Amorim P, Gomes GW, Palchetti CZ, Figueiredo MS, Pfeiffer CM, Fazili Z, Green R, Guerra-Shinohara EM, A Daily Dose of 5 mg Folic Acid for 90 Days Is Associated with Increased Serum Unmetabolized Folic Acid and Reduced Natural Killer Cell Cytotoxicity in Healthy Brazilian Adults, The Journal of nutrition, 147 (2017) 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Troen AM, Mitchell B, Sorensen B, Wener MH, Johnston A, Wood B, Selhub J, McTiernan A, Yasui Y, Oral E, Potter JD, Ulrich CM, Unmetabolized folic acid in plasma is associated with reduced natural killer cell cytotoxicity among postmenopausal women, The Journal of nutrition, 136 (2006) 189–194. [DOI] [PubMed] [Google Scholar]
- [70].Niemann B, Nemitz A, Werner J, Mai HD, Steinberg P, Lampen A, Ehlers A, Folic acid modulates cancer-associated micro RNAs and inflammatory mediators in neoplastic and non-neoplastic colonic cells in a different way, Molecular nutrition & food research, 61 (2017). [DOI] [PubMed] [Google Scholar]
- [71].Sawaengsri H, Wang J, Reginaldo C, Steluti J, Wu D, Meydani SN, Selhub J, Paul L, High folic acid intake reduces natural killer cell cytotoxicity in aged mice, The Journal of nutritional biochemistry, 30 (2016) 102–107. [DOI] [PubMed] [Google Scholar]
- [72].Doherty MR, Jackson MW, The Critical, Clinical Role of Interferon-Beta in Regulating Cancer Stem Cell Properties in Triple-Negative Breast Cancer, DNA and cell biology, 37 (2018) 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yang X, Zhang X, Fu ML, Weichselbaum RR, Gajewski TF, Guo Y, Fu Y-X, Targeting the tumor microenvironment with interferon-β bridges innate and adaptive immune responses, Cancer Cell, 25 (2014) 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SAW, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH, Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer, Clin Cancer Res, 21 (2015) 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue MZ, Ruan M, Wang H, Zhao J, Li Q, Wang P, Shi L, Yang WT, Huang W, Hu X, Yu KD, Huang S, Bertucci F, Jiang YZ, Shao ZM, Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer, Clin Cancer Res, 25 (2019) 5002–5014. [DOI] [PubMed] [Google Scholar]
- [76].Colleoni M, Cole BF, Viale G, Regan MM, Price KN, Maiorano E, Mastropasqua MG, Crivellari D, Gelber RD, Goldhirsch A, Coates AS, Gusterson BA, Classical cyclophosphamide, methotrexate, and fluorouracil chemotherapy is more effective in triple-negative, node-negative breast cancer: results from two randomized trials of adjuvant chemoendocrine therapy for node-negative breast cancer, J Clin Oncol, 28 (2010) 2966–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gluz O, Nitz UA, Harbeck N, Ting E, Kates R, Herr A, Lindemann W, Jackisch C, Berdel WE, Kirchner H, Metzner B, Werner F, Schutt G, Frick M, Poremba C, Diallo-Danebrock R, Mohrmann S, West German Study G, Triple-negative high-risk breast cancer derives particular benefit from dose intensification of adjuvant chemotherapy: results of WSG AM-01 trial, Ann Oncol, 19 (2008) 861–870. [DOI] [PubMed] [Google Scholar]
- [78].Gomez HL, Santillana SL, Vallejos CS, Velarde R, Sanchez J, Wang X, Bauer NL, Hockett RD, Chen VJ, Niyikiza C, Hanauske AR, A phase II trial of pemetrexed in advanced breast cancer: clinical response and association with molecular target expression, Clin Cancer Res, 12 (2006) 832–838. [DOI] [PubMed] [Google Scholar]
- [79].Zhou LY, Shi YH, Jia YS, Tong ZS, Potential role of pemetrexed in metastatic breast cancer patients pre-treated with anthracycline or taxane, Chronic Dis Transl Med, 1 (2015) 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Assaraf YG, Molecular basis of antifolate resistance, Cancer Metastasis Rev, 26 (2007) 153–181. [DOI] [PubMed] [Google Scholar]
- [81].Shan F, Liu YL, Wang Q, Shi YL, Thymidylate synthase predicts poor response to pemetrexed chemotherapy in patients with advanced breast cancer, Oncol Lett, 16 (2018) 3274–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bidwell BN, Slaney CY, Withana NP, Forster S, Cao Y, Loi S, Andrews D, Mikeska T, Mangan NE, Samarajiwa SA, de Weerd NA, Gould J, Argani P, Moller A, Smyth MJ, Anderson RL, Hertzog PJ, Parker BS, Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape, Nature medicine, 18 (2012) 1224–1231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


