Abstract
This study compared the transcriptional changes in Nicotiana benthamiana plants treated with homologous sequences derived from Pepper golden mosaic virus (PepGMV) and heterologous sequences that derived from another begomovirus, Tomato chino La Paz virus (ToChLPV) prior to infection by PepGMV. The results of microarray analyses identified upregulated genes associated with RNAi such as DCL2, DCL4, AGO3, AGO7, AGO10, NRPD2B (Pol IV), DRB3, CMT3, RDR6. The components that participate in different RNAi pathways were identified, including methylation induced by both constructs, as well as the code of these genes in Arabidopsis thaliana and its counterpart in N. benthamiana through different genome assembly. The expression of these genes was validated by quantitative reverse transcription polymerase chain reaction (RT-qPCR), where DCL3, DCL4, AGO1-1, AGO2, RDR6 and PPR1 showed increased expression during plant protection with the heterologous construct compared to those protected with the homologous construct. The results of this study confirmed the activation of the gene silencing mechanism at the transcriptional level with both constructs and established the possibility of their use as a protection system for both homologous and heterologous sequences.
Keywords: Tomato chino La Paz virus (ToChLPV), Gene silencing, Microarray, dsRNA
Introduction
Begomoviruses (family Geminiviridae) are plant pathogenic viruses that are widely disseminated by whiteflies (De Barro et al. 2011). These viruses are commonly found in mixed infections with other members of this genus, favoring their plasticity, emergence of new viruses of pathological importance, and even widen their host range (Navas-Castillo et al. 2011; García-Arenal and Zerbini 2019). Mixed infections are common in nature as it was observed for Pepper golden mosaic virus (PepGMV) with Pepper huasteco yellow vein virus (PHYVV), which can even exhibit a triple infection with Tomato yellow leaf curl virus (TYLCV) (Méndez-Lozano et al. 2003; Morales-Aguilar et al. 2019), or also TYLCV co-infecting with Tomato chino La Paz virus (ToChLPV) (Cárdenas-Conejo et al. 2010). One of the conserved mechanisms that plants use against viral infections is RNA interference (RNAi) (Lin et al. 2011). Some research has focused on evaluating the efficiency of RNAi as a protection system when the viral sequence has high homology to disease-causing begomoviruses. For example, Chellappan et al. (2004) used the sequence of the Rep (AC1) gene, which encodes the replication associated protein of the African cassava mosaic virus (ACMV) in Manihot esculenta plants in challenge experiments with East African cassava mosaic Cameroon virus (EACMCV) and Sri Lankan cassava mosaic virus (SLCMV). These authors reported strong protection through an activating gene silencing mechanism. On the other hand, a sequence obtained from non-coding conserved regions of five different TYLCVs was used to design a construct capable of activating the gene silencing mechanism against three of these viruses and generate resistant Solanum lycopersicum and Nicotiana benthamiana plants (Abhary et al. 2006). The efficacy of this strategy was also demonstrated using other intron-hairpin constructs (ihp-RNA) from the CP (AV1) gene, which encodes the capsid protein, and Rep from the Tomato yellow leaf curl Sardinia virus (Sicily strain; TYLCSV-[Sic]). However, in viral challenge assays, the plants only showed resistance against viruses with homologous sequences and not against those with heterologous ones (Gharsallah-Chouchane et al. 2008). The gene silencing mechanism has been activated using fragments from DNA-A or DNA-B from begomoviruses. However, the best results were obtained using DNA-A fragments (Taha et al. 2016). In contrast to previous reports, a previous study observed extremely low efficiency of RNAi in the complex of the begomoviruses Cotton leaf curl Multan virus (CLCuMV) and Cotton leaf curl Multan β-satellite (CLCuMB) (Mubin et al. 2011). Based on their results, they argued that high efficiency is achieved when the target sequence is highly similar to the constructed sequence, which would allow resistance to be achieved to begomovirus complexes. In addition, in a previous report, the efficiency of these constructs was evaluated against infection by PepGMV, observing decreases in viral loads from 95.6 to 99.5% for the heterologous and homologous constructs, respectively (Medina-Hernández et al. 2013). Although the silencing mechanism was activated in these studies, as evidenced by the observed decreases in symptoms and viral load, the participating components and how they interact when homologous and heterologous sequences are used remains unclear.
RNA interference (RNAi) is gene regulation mechanism conserved in eukaryotic organisms that also plays important roles in the control of basic cellular processes in plants, such as gene expression, heterochromatin formation, and adaptive antiviral defense (Herr and Baulcombe 2004). RNAi has been used as a method to study antiviral defense in plants. It is activated in the presence of double-stranded RNA molecules (dsRNAs) or small interfering RNAs (siRNAs) and acts at different levels, such as in the degradation of homologous RNAs in a sequence-specific manner (homologous sequence) or DNA methylation and chromatin modification (Voinnet 2001; Tenllado et al. 2004; Matzke and Mosher 2014). With respect to the mechanism of RNAi, it is known that DICER cuts dsRNAs that may then generate siRNAs of different sizes, including primary and secondary siRNAs (Wang et al. 2011). Dicer-like 2 (DCL2) acts directly in viral control by participating in the generation of sRNAs and activating the production of a secondary series of siRNAs called secondary or transitive siRNAs. DCL2, DCL3 and DCL4 are known to process endogenous siRNAs in plants, which are categorized into secondary siRNAs. In addition, DCL4 also participates in the defense against viruses by inducing the production of primary siRNAs (Yoshikawa et al. 2005; Axtell et al. 2006; Borges and Martienssen 2015). These siRNAs then bind to the argonaute (AGO) protein to form the RNA-induced silencing complex RISC (Pumplin and Voinnet 2013).
However, the studies have shown that transgenes with heterologous sequences can also initiate the RNAi process (Lin et al. 2011; Mubin et al. 2011). Thus, to achieve effective control strategies against mixed begomoviral infections, it is necessary to understand the various interactions in a heterologous RNAi system. This study analyzed a differential gene expression using microarrays and quantitative real time reverse transcription polymerase chain reaction (RT-qPCR) to evaluate the key components that interact during RNAi activation in N. benthamiana plants, which can be over- or under-expressed using homologous and heterologous dsRNA constructs to PepGMV. The results obtained will allow implementing effective infection control strategies by begomoviruses with homologous and heterologous sequences.
Materials and methods
Plants growth and constructs
Nicotiana benthamiana plants were grown at 28 °C, with a photoperiod of 16 h light and 8 h dark. Plants at the 3–4-leaf stage were used in four treatments. To evaluate the key components that interact during RNA interference activation when heterologous constructs derived from ToChLPV and homologous to PepGMV are used, because viral loads decreased in (95.6 and 99.5%, respectively), as previously described by Medina-Hernández et al. (2013). The constructs comprised inverted repeat sequence fragments of these pathogens and were denominated as homologous CIRP (construct of the intergenic region of PepGMV) and heterologous CIRT (construct of the intergenic region of ToChLPV) and transformed into Agrobacterium tumefaciens GV2260. These constructs express an intron-hairpin comprising 146 nt of the 5′ end of the AV1 gene; the entire 326 nt intergenic region (IR) and 714 nt of the 5′ end of the AC1 gene; the AC1-IR-AV1 total segment comprising 1186 nt with sequence identity of 50% and six regions longer than 22 nt of both viruses, which displays the highest identity from 82 to 100%. CIRP and CIRT contained inverted repeat sequences of the AC1-IR-AV1 region of each virus, including an intron as a spacer with an XbaI restriction digestion to confirm the presence of the sense and antisense arms and orientation of the inverted repeat sequences. Four treatments were performed: (1) CIRP, agro-infiltrated two leaves of N. benthamiana with CIRP and inoculated with PepGMV four days after; (2) CIRT, agro-infiltrated two leaves of N. benthamiana with CIRT and inoculated with PepGMV four days after; (3) positive control, agro-infiltrated two leaves of N. benthamiana with the empty vector (pH7GWIWG2 II) and inoculated with PepGMV four days after; and (4) negative control, agro-infiltrated two leaves of N. benthamiana with the empty vector but not inoculated with PepGMV.
Microarray and data analysis of Nicotiana benthamiana plants protected with homologous and heterologous constructs
Arabidopsis thaliana V.3.0.3 microarrays were used to analyze and compare global gene expression profiles of N. benthamiana experimental plants. Three leaves from each plant were collected at 10 days after inoculation (dpi), immediately frozen in liquid nitrogen and stored at -80 °C until used for RNA extraction. Total RNA was extracted using TRIzol (Invitrogen, CA, USA), and 10 μg of RNA was used for complementary DNA (cDNA) synthesis, incorporating fluorophores dUTP-Alexa Fluor 555 or dITP-Alexa Fluor 647. Incorporation of the fluorophore was analyzed by measuring absorbance at 555 nm for Alexa555 and 650 nm for Alexa647.
An equal quantity of labeled cDNA from the positive control, CIRP or CIRT groups was hybridized with labeled cDNA from controls at 42 °C for 14 h using UniHyb hybridization solution (Arrayit Corporation, Silicon Valley CA, USA). For each spot, the Alexa555 and Alexa647 signal density and background mean values were calculated with Array-Pro Analyzer (Houston, TX, USA). Processing and quantification of the images obtained from the microarray hybridization were performed in an Array-Pro Analyzer 4000 with the corresponding software (Packard BioChips, IL, USA). To analyze the microarray data and identify genes with significant differential expression between sample classes, the genArise package developed by the Computational Unit of the Institute of Cellular Physiology (UNAM, CDMX, MX) was used.
For gene ontology (GO) analysis, the IDs obtained from the microarray data were loaded into the Arabidopsis Information Resource (TAIR) database to convert them to FASTA format. Then, each sequence was searched against the non-redundant (NR) protein database with the Blast2GO tool (Conesa et al. 2005) using the parameters 20 hits per sequence, expected value < 1 × 103. When the names of all the genes were identified, evidence codes (EC) were obtained, which showed the reliability index values of the GO annotation, and each gene was mapped in the GO database. A one-tailed Fisher’s exact test was used to remove double IDs, and a false discovery rate (FDR) cutoff of 0.01 was used to verify a significant overrepresentation of GO annotations. Enrichment of a GO term was considered significant at p < 0.05. After mapping, each gene was annotated, and all of the enzymes were labeled using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Then, the data were labeled with respect to InterPRO protein families (Conesa and Götz 2008).
The TAIR (Arabidopsis.org) annotation data were also obtained from the different databases with available genome and transcriptome assemblies for N. benthamiana, including the draft genomes Niben0.4.4 (NbS000) and Niben1.0.1 generated by the Boyce Thompson Institute for Plant Research (Bombarely et al. 2012), Nbv3K, Nbv6.1 (Nakasugi et al. 2013), and the re-annotated NbD (Kourelis et al. 2018).
Differential gene expression analysis
First, upregulated genes associated with defense response, stress, and systemic acquired resistance were selected. Second, the overexpressed (upregulated) genes obtained in the microarray analysis with a Z score ≤ 1.50–3.0 were selected. To analyze the differential expression of the genes involved in the RNAi activation response by homologous and heterologous sequences to PepGMV, a search was performed for key genes involved in RNA silencing pathways, such as AGO, DCL-type proteins and RNA-dependent RNA polymerases (RDRs) using the tools described above (see the GO analysis section) with the TAIR database. To determine the effects of differential gene expression and interpret the results in the context of a protein–protein interaction network, an analysis was performed using the STRING version 9.1 database, which is a valuable tool for searching protein/protein interactions. The STRING network results were restricted to those genes that are known to be associated with RNAi (Franceschini et al. 2012; Szklarczyk et al. 2016).
RT-qPCR analysis of differential gene expression
For RT-qPCR analysis, RNA from experimental plants was treated with DNase I (1 U/μL; Thermo Fisher Scientific, Inc. MA, USA) to remove genomic DNA. Then, first strand cDNA was synthesized from 5 μg of total RNA with Super Script III Reverse Transcriptase (Thermo Fisher Scientific, Inc. MA, USA) according to the manufacturer’s instructions. Actin (ACT) was used as an internal control for normalization due to its similar expression level in all viral infections and its efficiency in PCR amplification. According to Liu et al. (2012) and Wu et al. (2014), genes associated with RNAi were selected for gene expression analysis, including DCL1, DCL2, DCL3, DCL4, AGO1, AGO2, RDR6, and pentatricopeptide repeat (PPR) proteins using the primers shown in Table 1. This study analyzed the primers previously described for AGO1 with available sequences in the NCBI database for AGO1a, AGO1b, AGO1-1 and AGO1-2 from N. benthamiana, observing alignment with AGO1a and AGO1-1 but not AGO1b or AGO1-2. RT-qPCR was performed in triplicate in a 96-well thermocycler (CFX-96, Bio-Rad, CA, USA) using SsoFast EvaGreen Supermix (Bio-Rad) in a final volume of 20 μL per reaction. The PCR conditions were as follows: one cycle at 95 °C/30 s followed by 39 cycles of 95 °C/0.05 s and 60 °C/0.05 s. After amplification, melting curve analyses were performed to verify the products amplified by their specific melting temperatures (Tm) from 65 to 95 °C with increments of 0.5 °C/0.05 s. A sample without DNA was used as control in each analysis. The amplification efficiency (E), E = 10 (−1/slope) × 100% was determined in accordance with the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines. The RT-qPCR data were normalized with respect to the CT reference gene (Table 1). Relative expression was determined with the 2-∆∆CT method, where ∆∆CT = ([CT1Target-C1TReference] - [CT0Target-CT0Reference]) (Livak and Schmittgen 2001), using the results from three independent biological replicates for each treatment (n = 3). One-way analysis of variance (ANOVA) and statistical significance was determined using IBM SPSS Statistics (version 22.0, IBM Corp. NY, USA) software.
Table 1.
Primer sequences used for quantitative gene expression analysis
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| DCL2F | CGGGATCCCCGGGATTTATTCGTAAT | |
| DCL2R | CCCTCGAGAATGACAAAGCCGCTACT | Wu et al. (2014) |
| DCL3F | ACTTGTTGAATGCGGTGAAG | |
| DCL3R | CCCCTGTCGTTCTAGCTCAT | Wu et al. (2014) |
| DCL4F | CGTCCGTGCCCAGAAATCT | |
| DCL4R | AATGCAATTGCCGCTTTGA | Wu et al. (2014) |
| AGO1F | GCTCTAGAAGATCTGTACAAGACTTGGC | |
| AGO1R | CGAATTCTTATTGGCAAACAACCTAGT | Wu et al. (2014) |
| AGO2F | CATTTGAACCTCCTTTCTATCGAC | |
| AGO2R | CATACCTCTAGAAGTGAGGATCAC | Liu et al. (2012) |
| RDR6F | TTCAGGAATGTCTTCTTCGAGCG | |
| RDR6R | AGTGATCTAGCAACCCAATGAG | Liu et al. (2012) |
| PPR1 | ATGAGGGTCCATTTGAGTGAC | |
| PPR1 | AGGCTGATGTTGGAATCTGG | Liu et al. (2012) |
| ACT | TCCTGATGGGCAAGTGATTAC | |
| ACT | TTGTATGTGGTCTCGTGGATTC | Liu et al. (2012) |
This table shows primer sequences used for quantitative reverse transcription polymerase chain reaction (RT-qPCR) gene expression analysis of treatments of Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) and challenged with PepGMV
Results
Microarray based gene expression analysis of plants previously protected
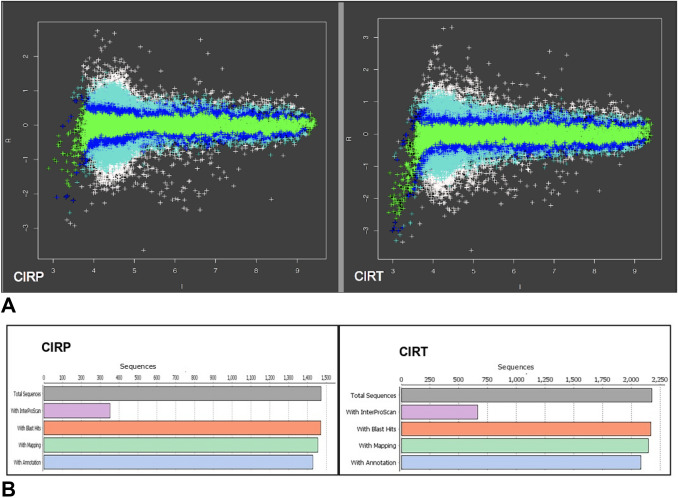
A total of 27,342 and 27,218 genes were obtained in the samples of plants protected with homologous CIRP and heterologous CIRT constructs. Figure 1a shows the microarray results where the data and colors are associated with the differential gene expression, and the color of each point depends on the absolute value of -3 to 3 Z-score associated with the differential expression of down-or upregulated genes. Functional characterization of the differentially expressed genes was performed using TAIR and the GO program Blast2GO, with 1473 upregulated genes were identified in plants with CIRP and 2169 identified in plants with CIRT (Fig. 1b).
Fig. 1.
a Graphic results of the evaluation of Z-scores from the microarrays analysis from Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) and challenged with PepGMV, where ‘up” and “down” values are indicated by green, blue, cyan and white. b Results of the analysis of annotated sequences
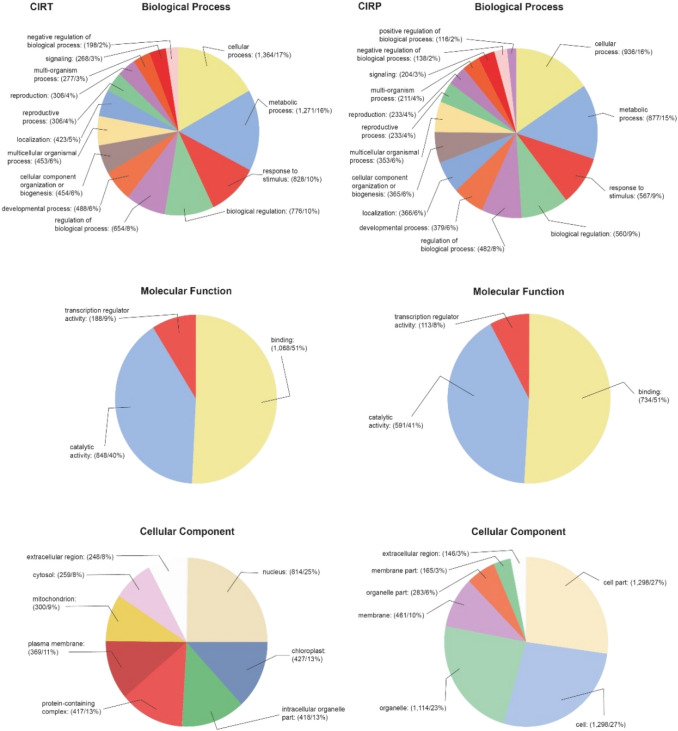
Each identified gene was assigned to one of the three primary GO categories (Fig. 2): (1) biological process (BP); (2) molecular function (MF); and (3) cellular components (CC). BP (GO level 2) was represented by transcripts associated with cellular processes, such as differentiation, organization, growth, cycle, cell–cell signaling (16 and 17% in plants with CIRP and CIRT, respectively), metabolic processes of protein, lipid, carbohydrate, nucleobase-containing compound, DNA (15 and 16% in plants with CIRP and CIRT, respectively) and primary and secondary metabolites (generation of precursor metabolites and energy), as well as response to stimuli (biotic, abiotic, endogenous) or defense (9 and 10% in plants with CIRP and CIRT, respectively). In plants protected by CIRP, these transcripts were associated with response to stress; abiotic, biotic, endogenous and external stimuli mediated through mitogen-activated protein kinase (MAPK) cascades; gene expression regulation and epigenetics (histones methylation and maintenance of DNA methylation). In CIRT, these transcripts were associated with (MAPK cascades) oxide reduction and transcription regulation, response to abscisic acid, acquired systemic resistance, signaling pathways mediated by salicylic acid, hypersensitive response, even positive regulation transcription dependent on DNA, and protein ubiquitination. MF was represented by transcripts associated with binding of proteins, DNA, RNA, nucleotide, lipid, chromatin, signaling receptor, DNA-binding transcription factor activity (51% in plants with both constructs), catalytic activity (41 and 40% in plants with CIRP and CIRT, respectively) and transcription regulator activity (8 and 9% in plants with CIRP and CIRT, respectively). CC was represented in different percentages for each construct. For example, in plants protected by CIRP, transcripts were associated with cell part (27%), cell (27%), organelles (23%), membrane (10%), organelle part (6%), and membrane part (3%). Differences were also observed with respect to the extracellular region (3 and 8% in plants with CIRP and CIRT, respectively), for plants protected by CIRT transcripts associated with nucleus (25%), chloroplast (13%), intracellular organelle (13%), protein-containing complex (13%), plasma membrane (11%), mitochondrion (9%), and cytosol (8%) (Fig. 2).
Fig. 2.
Gene ontology annotation obtained by microarray analysis from Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) and challenged with PepGMV. Transcripts were grouped into functional Blast2GO groups: biological process, molecular function and cellular component categories with Z-scores 2 and 1.5. The percentages of differentially expressed genes during infection in each category are shown within the parentheses
Upregulated genes associated with defense response, stress, systemic acquired resistance and hypersensitive response in plants protected with CIRP and CIRT are shown in Table 2. The data were obtained from different databases with available genome and transcriptome assemblies and have the names of the proteins and biological functions derived from TAIR. The genes were also associated with IDs that correspond to the assembly version and code that identify genome and transcriptome assemblies (Bombarely et al. 2012; Nakasugi et al. 2013; Kourelis et al. 2018).
Table 2.
List of upregulated genes in the transcriptome of protected Nicotiana benthamiana plants, associated with defense response, stress, and systemic acquired resistance
| Gene model | Protein name | GO term | Nicotiana benthamiana | GO or product | CIRP 25 | CIRT 26 |
|---|---|---|---|---|---|---|
| AT2G13360 | AGT | Response to stress |
AGT1 NbD006757 NbS00019758g0008.1 Niben101Scf00797g18009.1 |
Serine-glyoxylate aminotransferase | 1.5 | 2 |
| AT2G14170 | ALDH6B2 | Oxidation–reduction |
mmsA NbD000775 ALDH6B2 NbS00024307g0010.1 |
Response to oxidative stress | 2 | 2 |
| AT2G19560 | EER5 | Response to stress |
Niben101Scf01633g00029.1 Niben101Scf07229g00011.1 EER5 NbS00028917g0001.1 |
Response to stress | 1.5 | 1.5 |
| AT1G60940 | SNRK2.10 | Response to stress |
Niben101Scf01993g08016.1 Niben101Scf01970g 08001.1 SRK2B NbS00005219g0010.1 |
Response to stress | 1.5 | 1.5 |
| AT2G24850 | TAT3 | Response to stress | Nf | 1.5 | 1.5 | |
| AT2G30360 | CIPK11 | Response to stress |
WNK4 NbD010417 CIPK11 NbS00028941g0005.1 |
Response to stress | 2 | 1.5 |
| AT2G38730 | RCA | Systemic acquired resistance |
Niben101Scf04098g00017.1 NbS00029897g0007.1 |
Systemic acquired resistance | 1.5 | 2 |
| AT3G11840 | PUB24 | Hypersensitive response |
PUB24 NbD001850 NbS00032295g0001.1 |
Defense response | 2 | 1.5 |
| AT3G11930 | AT3G11930 | Response to stress |
Niben101Scf09610g00017.1 NbS00029709g0002.1 |
Response to stress | 1.5 | 2 |
| AT3G23250 | MYB15 | Defense response |
MYB4-like NbD008259 Niben101Scf01013g01003.1 NbS00011517g0003.1 |
Defense response | 2 | 2 |
| AT3G32920 | CSNSA | Defense response | Nf | 2 | 1.5 | |
| AT3G58620 | TTL4 | Response to stress | Nf | Nf | 2 | 2 |
| AT4G08500 |
MAPK MEKK1 |
Response to stress |
STK10 Niben101Scf03906g03013.1 MEKK1 NbS00011174g0009.1 |
Response to stress | 1.5 | 1.5 |
| AT4G19530 | AT4G19530 | Defense response | Nf | Nf | 2 | 2 |
| AT4G19840 | ATPP2-A1 | Hypersensitive response |
Niben101Scf03867g02041.1 Niben101Scf02174g03009.1 ATPP2-A1 NbS00041802g0007.1 |
Protein phloem protein 2-LIKE A1 | 2 | 1.5 |
| AT4G29100 | AT4G29100 | Defense response |
bHLH68-like NbD002325 NbS00049384g0003.1 |
Defense response | 2 | 2 |
| AT4G30480 | AT4G30480 | Hsp90 TPR1 |
TTC1 Niben101Scf03964g04005.1 NbS00020723g0011.1 |
TTC1 | 1.5 | 1.5 |
| AT4G31550 | WRKY11 | Response to stress |
WRK22 NbD013410 Niben101Scf01721g08007.1 NbS00042305g0004.1 |
Response to stress | 2 | 1.5 |
| AT5G09420 | ATTOC64-V | Hypersensitive response |
OM64 NbD010896 Niben101Scf01370g00007.1 NbS00018282g0024.1 |
Hypersensitive response | 1.5 | 2 |
| AT5G37930 | AT5G37930 | Ubiquitin-protein | NbS00011136g0002.1 | 2 | 2 | |
| AT5G40010 | AATP1 | Systemic acquired resistance |
AATP1 NbD010669.1 NbS00033104g0001.1 |
AATP1 | 1.5 | 1.5 |
| AT5G40990 | GLIP1 | Systemic acquired resistance | Nf | Nf | 1.5 | 1.5 |
| AT5G42020 | BIP2 | Response to stress |
BIP5 Niben101Scf04126g01011.1 NbS00020855g0001.1 |
BIP5 | 2 | 2 |
| AT5G45800 | MEE62 | Systemic acquired resistance |
Probable LRR NbS00022774g0009.1 NbS00016395g0015.1 |
LRR | 2 | 2 |
| AT5G50720 | HVA22E | Response to stress |
HVA22-like protein Niben101Scf03886g04003.1 NbS00031130g0003.1 |
Response to stress | 1.5 | 1.5 |
| AT5G55990 | CBL2 | Hypersensitive response |
CBL2 NbS00029252g0013.1 |
CBL2 | 1.5 | 1.5 |
The following table shows a list of upregulated genes in the transcriptome of Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) constructs and challenged with PepGMV that are associated with defense response, stress, and systemic acquired resistance. Annotation data obtained from different databases genome and transcriptome assemblies available for N. benthamiana Niben0.4.4 (NbS000) and Niben1.0.1 (Bombarely et al. 2012). Nbv3K, Nbv6.1 (Nakasugi et al. 2013). NbD (Kourelis et al. 2018). Nf not found
Differential expression of genes associated with RNAi and methylation in plants protected by CIRP and CIRT
Regarding the genes associated with RNAi (Table 3), upregulated genes with high identity with genes annotated from N. benthamiana transcriptomes (Bombarely et al. 2012; Nakasugi et al. 2013) were observed in the plants protected with homologous CIRP and heterologous CIRT constructs; as DCL2, DCL4, AGO3, AGO5, AGO6, AGO7, AGO10, nuclear RNA polymerase D2B (NRPD2B/POL IV), NRPD1A, Dawdle (DDL), defective in meristem silencing 3 (DMS3), dsRNA-binding protein 3 (DRB3), DRB4, DRB5, domains rearranged methylase 3 (DRM3), RDR6, chromo-methylase 3 (CMT3), PPR, SU(VAR)3–9 homolog 1 (SUVH1), SUVH5 and HUA enhancer 1 (HEN1). These upregulated genes, which were activated by both constructs, participate in interwoven pathways of gene regulation and plant defense that generate and use different types of sRNAs (siRNA, miRNA, ta-siRNA, nat-siRNA and hc-siRNA) to participate in viral defense, developmental regulation, stress response and transcriptional silencing (Eamens et al. 2008; Nakasugi et al. 2013; Borges and Martienssen 2015). In plants protected with the homologous CIRP construct upregulated genes included AGO4a because AGO9 from A. thaliana is homologous to AGO4 of N. benthamiana AGO4b (Nakasugi et al. 2013), as well as DRM2, variant in methylation 1 (VIM1) and SUVH2. In contrast, in plants protected with the heterologous CIRT construct, upregulated genes included RDR2 and SUVH4. In addition, genes upregulated by CIRP but not CIRT and vice versa were associated with the synthesis of different siRNAs and methylation (Eamens et al. 2008).
Table 3.
List of transcripts associated with RNAi obtained of protected Nicotiana benthamiana plants
| TAIR locus | Protein | Description | Locus in Nicotiana benthamiana | CIRP up or down regulation | CIRT up or down regulation |
|---|---|---|---|---|---|
| AT1G01040 | DCL1 (DICER-LIKE 1) | Dicer is an RNA helicase involved in miRNA and siRNA processing and virus-induced gene silencing |
DCL1 Nbv3K605750463 Nbv6.1trP38701 Niben101Scf03304g01025.1 NbS00015538g0006.1 |
-0.33 | -0.41 |
| AT3G03300 | DCL2 (DICER-LIKE 2) | Functions in the antiviral silencing response. Involved in the production of ta-siRNAs |
DCL2 Nbv3K725833766 Nbv6.1trP4604 Niben101Scf08272g00021.1 Niben101Scf06666g01011.1 |
0.70 | 0.53 |
| AT3G43920 | DCL3 (DICER-LIKE 3) | Required for endogenous RDR2-dependent siRNA (but not miRNA) formation |
DCL3 Nbv3K585704110 Nbv6.1trP10636 |
-0.43 | -0.53 |
| AT5G20320 | DCL4 (DICER-LIKE 4) | Catalyzes processing of ta-siRNA precursors in a distinct sRNA biogenesis pathway. Involved in the production of 21-nt primary siRNAs from both inverted repeat constructs and endogenous sequences as well as the RDR6-dependent 21-nt secondary siRNAs involved in long-range cell-to-cell signaling. Binds DRB4, a dsRNA-binding protein |
DCL4 Nbv3K725837175 Nbv3K625768999 Nbv6.1trP61740 |
0.829 | 0.93 |
| AT1G48410 | AGO1 (ARGONAUTE 1) | Encodes an RNA slicer that selectively recruits microRNAs and siRNAs |
AGO1a Nbv3K705826800 Nbv6.trA73469 Niben101Scf12841g03019.1 AGO1b Niben101Scf08137g02022.1 Niben101Scf05146g06007.1 |
-1.1 | -1.06 |
| AT1G31280 | AGO2 (ARGONAUTE 2) | Encodes an argonaute protein that binds viral siRNAs and is involved in antiviral defense response |
Nbv3K585706870 Nbv3K785652117 Nbv3K705830082 |
-1.05 | -1.7 |
| AT1G31290 | AGO3 (ARGONAUTE 3) | Functions in the defense response to viruses, gene silencing by RNA the regulation of DNA-templated translations and transcription |
AGO2 Nbv3K585706870 |
0.82 | 1.193 |
| AT2G27040 | AGO4 (ARGONAUTE 4) | Protein involved in siRNA-mediated gene silencing. Loss-of-function mutations reduce site-specific CpNpG and CpHpH methylation and increase susceptibility to bacterial pathogens |
AGO4a Nbv3K585737054 AtAGO9—AGO4b Nbv3K5745626388 Niben101Scf05519g01007.1 |
0.18 | -0.123 |
| AT2G27880 | AGO5 (ARGONAUTE 5) | Required for antiviral RNA silencing. confers resistance to Potato virus X |
AGO5 Nbv3K585731374 Nbv6.1trP59647 Niben101Scf04371g04008.1 |
2.13 | 1.05 |
| AT2G32940 | AGO6 (ARGONAUTE 6) | Encodes a nuclear-localized 879-amino-acid protein that contains conserved PAZ and PIWI domains and is important for the accumulation of specific heterochromatin-related siRNAs and for DNA methylation and TGS |
AGO6 Nbv3K705827462 Nbv6.1trP35996 |
0.49 | 0.693 |
| AT1G69440 | AGO7 (ARGONAUTE 7) | Required for the accumulation of TAS3 ta-siRNAs but not miR171, miR173, miR390 or mi391 |
AGO7 Nbv3K585720936 Nbv6.1trP11465 Niben101Scf20224g00004.1 Niben101Scf00272g02007.1 NbD002754.1 |
0.063 | 0.59 |
| AT5G43810 | AGO10 (ARGONAUTE 10) | Function in the defense response to viruses, gene silencing by RNA and miRNA metabolic process |
AGO10 Nbv3K585734208 Nbv6.1trP33879 Niben101Scf01240g11006.1 NbD010099.1 |
0.961 | 1.954 |
| AT3G18090 | Pol IV NRPD2B | Encodes a subunit of RNA polymerase IV (RNA polymerase D). NRPD2b is closely associated with NRPD2a but has lower levels of transcription and does not affect endogenous siRNA when mutated |
NRPD2b NbS00016929g0017.1 |
0.69 | 1.74 |
| AT1G63020 | Pol IV NRPD1A SDE4 | Encodes one of the two alternative largest subunits of putative plant-specific RNA polymerase IV (RNA polymerase D). Required for PTGS |
NRPD1A Nbv3K585708997 |
0.84 | 1.17 |
| AT4G35800 | RNA Pol II (NRPB1) | Encodes the unique largest subunit of nuclear DNA-dependent RNA polymerase II |
NRPB1 NbC23346514g0001.1 |
-0.81 | -0.74 |
| AT3G20550 | DDL (DAWDLE) | Participates in the production of miRNAs involved in miRNA gene silencing | Nf | 0.614 | 1.05 |
| AT3G49250 | DMS3 (DEFECTIVE IN MERISTEM SILENCING 3) | Can potentially link nucleic acids in facilitating RNA1-mediated epigenetic modification involving secondary siRNA and spreading of DNA methylation |
DMS3 NbS00047856g0005.1 Niben101Scf08455g00014.1 |
1.48 | 1.26 |
| AT1G09700 | DRB1 (DSRNA-BINDING PROTEIN 1) | Encodes a nuclear dsRNA-binding protein. Involved in mRNA cleavage |
DRB1 Nbv3K605753726 DRB1501 NbD009575.1 Niben101Scf01181g02012.1 |
-2.44 | -2.036 |
| AT2G28380 | DRB2 (DSRNA-BINDING PROTEIN 2) | Encodes a cytoplasmic dsRNA-binding protein. A maternally expressed imprinted gene. DRB2 and DRB4 have antagonistic impacts on polymerase IV-dependent siRNA levels |
DRB2 Nbv3K585718488 Niben101Scf02669g05009.1 NbD013708 |
-0.46 | -0.58 |
| AT3G26932 | DRB3 (DSRNA-BINDING PROTEIN 3) | dsRNA-binding protein 3; methylation-mediated antiviral defense |
DRB3 Nbv3K725608215 |
1.24 | 2.12 |
| AT3G62800 | DRB4 (DSRNA-BINDING PROTEIN 4) | Encodes a nuclear dsRNA-binding protein that specifically interacts with DCL4. May regulate DCL4 function and thereby affect miRNA biogenesis and also has an impact on polymerase IV-dependent siRNA levels |
DRB4 Nbv3K725839976 Niben101Scf05841g01021.1 |
0.63 | 1.45 |
| AT5G41070 | DRB5 (DSRNA-BINDING PROTEIN 5) | Encodes a dsRNA-binding protein |
DRB5 Nbv3K585684100 |
1.44 | 1.46 |
| AT5G15380 | DRM1 (DOMAINS REARRANGED METHYLASE 1) | Encodes a MTase involved in de novo DNA methylation and the maintenance of asymmetric methylation of DNA sequences |
DRM1 NbS00047903g0006.1 |
-0.88 | -1.56 |
| AT5G14620 | DRM2 (DOMAINS REARRANGED METHYLASE 2) | Functions in DNA methylation, the defense response to fungus, gene silencing, and histone H3K9 methylation |
DRM2 NbS00009663g0003.1 |
3.11 | -1.37 |
| AT3G17310 | DRM3 (DOMAINS REARRANGED METHYLASE3) | Required for normal maintenance of non-CG DNA methylation, establishment of RNA-directed DNA methylation triggered by repeat sequences and accumulation of repeat-associated sRNAs |
DRM3 Nbv3K745624408 DRM2a NbD010724.1 Niben101Scf01334g06016.1 |
0.27 | 1.26 |
| AT4G11130 | RDR2 (RNA-DEPENDENT POLYMERASE 2) | Encodes an RNA-dependent RNA polymerase required for endogenous siRNA (but not miRNA) formation |
RDR2 Nbv3K625766705 Niben101Scf04296g00019.1 |
-1.55 | 0.569 |
| AT3G49500 | RDR6 (RNA-DEPENDENT POLYMERASE 6) SDE1 | Encodes an RNA-dependent RNA polymerase. Involved in the biogenesis of ta-siRNAs and other siRNAs. Required for PTGS and natural virus resistance |
RDR6 Nbv3K585707928 Niben101Scf12609g01010.1 |
1.54 | 1.61 |
| AT1G69770 | CMT3 (CHROMO-METHYLASE 3) | Encodes a chromomethylase involved in methylating cytosine residues at non-CG sites. Involved in gene silencing |
CMT3 Nbv3K625768297 Niben101Scf02006g04014.1 |
0.50 | 0.75 |
| AT1G02420 | PPR (PENTATRICOPEPTIDE REPEAT) | Function unknown |
PPR NbS00000911g0002.1 Niben101Scf00151g05009.1 NbD001629.1 |
1.06 | 0.156 |
| AT1G57820 | VIM1 (VARIANT IN METHYLATION 1) | Functions in DNA methylation of cytosine, DNA methylation of cytosine within a CG sequence, and the maintenance of DNA methylation |
VIM1 NbS00027916g0013.1 Niben101Scf01582g09001.1 ORTH2 NbD012430.1 |
1.29 | -0.55 |
| AT5G04940 | SUVH1 (SU(VAR)3–9 HOMOLOG 1) | Involved in epigenetic control of gene expression and acts as a histone MTase |
SUVH1 NbS00008825g0019.1 |
0.35 | 0.19 |
| AT2G33290 | SUVH2 (SU(VAR)3–9 HOMOLOG 2) | Involved in epigenetic control of gene expression and acts as a histone MTase |
SUVH2 NbC23204936g0001.1 |
0.095 | -0.23 |
| AT1G73100 | SUVH3 SU(VAR)3–9 HOMOLOG 3) | Involved in epigenetic control of gene expression and acts as a histone MTase |
SUVH3 NbS00008825g0007.1 |
-0.091 | -0.23 |
| AT5G13960 | SUVH4 SU(VAR)3–9 HOMOLOG 4) | Encodes an H3K9-specific MTase involved in the maintenance of DNA methylation |
SUVH4 NbS00002211g0017.1 NbD040130.1 |
-0.17 | 0.13 |
| AT2G35160 | SUVH5 SU(VAR)3–9 HOMOLOG 5) | Exhibits histone MTase activity in vitro and contributes to the maintenance of H3-mK9 (methylation of histone H3 at Lys 9) and CMT3-mediated non-CG methylation in vivo |
SUVH5 NbS00025360g0007.1 Niben101Scf00614g00025.1 NbD005519.1 |
1.83 | 1.89 |
| AT2G22740 | SUVH6 | MTase involved in histone methylation |
SUVH6 NbS00018104g0005.1 NbD001814.1 |
-0.5 | -0.29 |
| AT4G20910 | HEN1 HUA ENHANCER 1 | Involved in de novo methylation, methylates the terminal nucleotide of 24-nt sRNAs |
HEN1 Nbv3K645785075 NbD011168.1 |
0.48 | 0.71 |
| AT1G05950 | AT1G05950 | Methylation DNA repair protein |
RECA NbD009068.1 Niben101Scf01116g01005.1 Unnamed protein NbS00029630g0016.1 NbS00029754g0004.1 NbS00043925g0028.1 Niben101Scf02044g06013.1 |
3.86 | 2.44 |
| AT1G65660 | SMP1 | Gene silencing pre-mRNA-splicing factor SLU7-like |
slu7 NbD028145 SMP1 NbS00031927g0008.1 |
2.78 | 2 |
| AT2G04660 | APC2 | Gene silencing |
APC2 Niben101Scf10322g01013.1 NbS00038059g0007.1 |
2 | 2 |
| AT2G19930 | RDR3 (RNA-DEPENDENT POLYMERASE 3) | Gene silencing by RNA |
RDR3 NbD002592 Niben101Scf00257g00018.1 Niben101Scf04189g00002.1 RDR5 NbS00054825g0009.1 |
1.5 | 1.5 |
| AT3G11440 | MYB65 MYB DOMAIN PROTEIN 65 | DNA binding Contains a binding site for miRNA159 |
MYB16 NbD007160 MYB65 NbS00013240g0015.1 |
2 | 2 |
| AT3G12210 | AT3G12210 | DNA binding protein |
Niben101Scf12783g01008.1 NbS00028950g0007.1 |
2 | 2 |
| AT3G13682 | LDL2 LSD1-LIKE2 | Involved in H3K4 methylation |
LDL2 Niben101Scf01976g00017.1 NbS00024949g0008.1 |
1.5 | 1.5 |
| AT3G26932 | DRB3 DSRNA- BINDING PROTEIN3 | Gene silencing |
DRB1 Niben101Scf09603g04002.1 DRB3 Nbv3K725608215 |
2 | 2 |
| AT3G57300 | INO80 ATINO80 | Gene silencing DNA repair Genome stability maintenance |
INO80 NbD012748 Niben101Scf01634g02020.1 NbS00008749g0009.1 |
1.5 | 1.5 |
| AT4G16420 | ADA2B | Gene silencing Chromatin remodeling |
ADA2B Niben101Scf03129g01013.1 ADA2B NbS00013870g0004.1 |
2 | 2 |
| AT4G29830 | VIP3 VERNALIZATION INDEPENDENCE 3 | Involved in histone H3-K36 methylation and histone H3-K4 methylation |
Niben101Scf02790g03009.1 VIP3 NbS00040297g0001.1 |
1.5 | 2 |
| AT5G23570 | SGS3 SUPPRESSOR OF GENE SILENCING 3 | Required for posttranscriptional gene silencing and natural virus resistance |
SGS3 NbE44071533 Niben101Scf01326g11010.1 Niben101Scf05278g04003.1 Nbv3K785651293 NbS00001969g0002.1 |
2 | 2 |
| AT5G55920 | OLI2 OLIGOCELLULA 2 | Methylation |
OLI2 NbS00015992g0001.1 |
1.5 | 1.5 |
This table shows a list of transcripts obtained by microarray analysis of Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) and challenged with PepGMV. The numbers in the up- or down regulation columns indicate fold changes in the expression of the transcripts associated with RNAi. Annotation data obtained from different databases with genome and transcriptome assemblies available for N. benthamiana Niben0.4.4 (NbS000) and Niben1.0.1 (Bombarely et al. 2012). Nbv3K, Nbv6.1 (Nakasugi et al. 2013). NbD (Kourelis et al. 2018). Nf not found
Downregulated genes observed in both plants protected with homologous CIRP and heterologous CIRT constructs included DCL1, DCL3, AGO1, AGO2, RNA Pol II (NRPB1), DRB1, DRB2, DRM1, SUVH3 and SUVH6. Genes that were only downregulated in the plants protected with the homologous CIRP construct included RDR2 and SUVH4, whereas those only observed in the plants protected with the heterologous CIRT construct included AGO4a, DRM2, VIM1 and SUVH2.
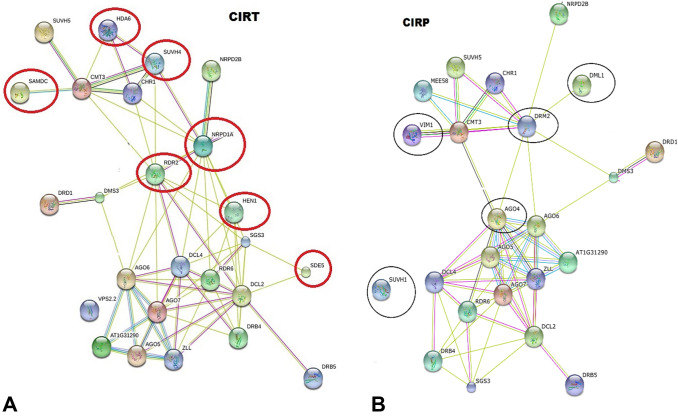
In addition, the protein network analysis using STRING allowed the molecular interactions to be visualized among the differentially expressed genes identified from the microarray. The results of this analysis identified groups of proteins involved in RNAi-activated pathways by CIRT (Fig. 3a) and CIRP (Fig. 3b) constructs, such as the major proteins involved in biogenesis of different small RNAs (sRNAs), including secondary siRNAs and other core components that participate in initiation and maintenance of RNA-directed DNA methylation (RdDM) (Eamens et al. 2008; Nakasugi et al. 2013; Borges and Martienssen 2015).
Fig. 3.
STRING network of functional protein associations with gene silencing. a High connectivity is observed among the proteins associated with gene silencing and methylation in CIRT (construct of the intergenic region of ToChLPV), and b subset of proteins involved in gene silencing and methylation in CIRP (construct of the intergenic region of PepGMV). The colors of lines connecting the proteins indicate an interaction
RT-qPCR gene expression analysis
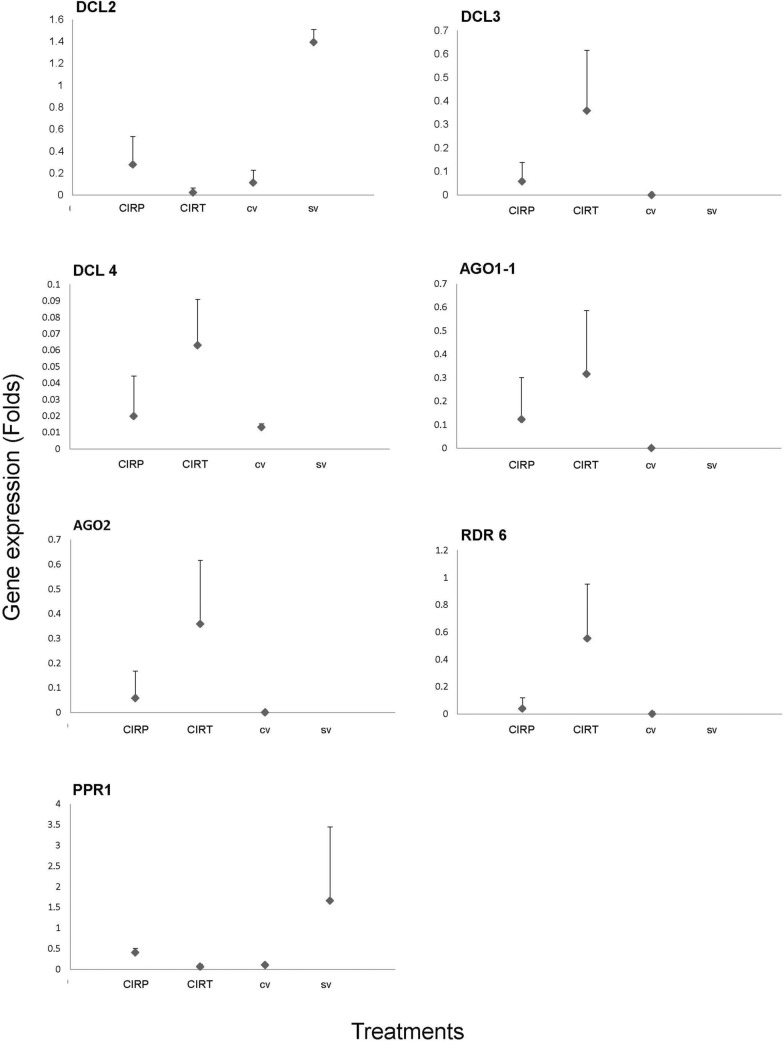
To validate the reference genes, the relative expression level of target genes was analyzed, and the Actin (ACT) gene was used for data normalization. Figure 4 shows the RT-qPCR results for seven RNAi-associated genes, previously identified as being differentially expressed by microarray analyses of N. benthamiana plants protected with homologous CIRP and heterologous CIRT constructs and inoculated with PepGMV. These genes include DCL2, DCL3, DCL4, AGO1-1, AGO2, RDR6, and PPR1, all of which exhibited significant differences in expression (p < 0.05).
Fig. 4.
RT-qPCR expression analysis of DCL2, DCL4, AGO1, AGO2, RDR6 and PPR1 in Nicotiana benthamiana plants protected with CIRP (homologous construct of the intergenic region of PepGMV) or CIRT (heterologous construct of the intergenic region of ToChLPV) and infected with PepGMV. Means indicate significant difference (p < 0.05) among treatments CIRP, CIRT, cv (positive control) and sv (negative control)
The qPCR analysis showed that genes RDR6, AGO2, DCL3 and AGO1-1 had higher expression levels 55, 38, 35, and 31%, respectively when the plants were protected with heterologous CIRT constructs, whilst the same genes in plants protected with homologous CIRP constructs represented 8, 10, 8, and 17, respectively. Expression levels at the controls were close to zero. DCL4 had higher expression level 6% when the plants were protected with heterologous CIRT constructs than when the plants were protected with homologous CIRP constructs 2%, and in the control with virus, it had an expression level of 1.3%. However, PPR1 and DCL2 showed a similar expression pattern when both constructs were used for protection and similar in the controls. PPR1 and DCL2 had an expression level of 6 and 2%, respectively, when the plants were protected with heterologous CIRT constructs and when they were protected with homologous CIRP constructs, representing 40 and 27%, respectively. Whilst the control with virus cv PPR1 and DCL2 had an expression level of 10 and 11%, respectively, and in the controls without virus PPR1 and DCL2 had higher expression levels that represented 160 and 130%, respectively.
Discussion
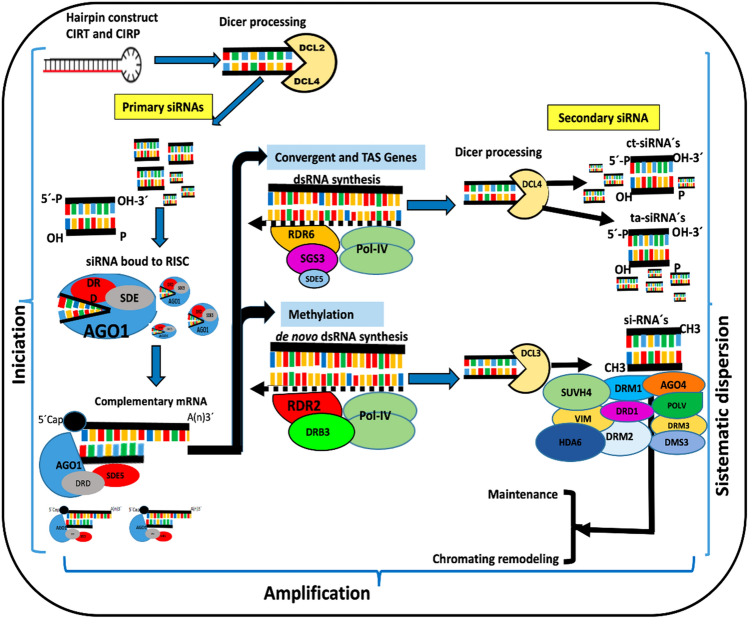
The results of this study indicate that homologous CIRP (construct of the intergenic region of PepGMV) and heterologous CIRT (construct of the intergenic region of ToChLPV) against infection of the PepGMV activate key components of the RNAi gene silencing mechanism. Global gene expression results demonstrated that the overexpression of some key RNAi and methylation-associated components could have been activated by heterologous dsRNA obtained from ToChLPV. Upregulated genes activated by both constructs participated in interwoven pathways of gene regulation and plant defense that generate and use different types of sRNAs (siRNA, miRNA, ta-siRNA, nat-siRNA and hc-siRNA) to participate in viral defense, development regulation, stress response and activate transcriptional silencing (Eamens et al. 2008; Nakasugi et al. 2013). The results showed that the mechanisms activated in response to infection by PepGMV in N. benthamiana recognized the dsRNAs generated from CIRP and CIRT and caused the overexpression of DICER as DCL2, DCL3 and DCL4 (Fig. 5). DICER has been reported to cleave dsRNAs, and these dsRNAs may then generate siRNAs of different sizes, including primary and secondary siRNAs (Wang et al. 2011). Small siRNAs are assembled in the multiprotein complex RISC (Li et al. 2017), and the systemic movement of siRNAs among cells can occur through the phloem (Qin et al. 2017; Zhang et al. 2019). DCL2 acts directly in viral control in a hierarchical manner, participates in the generation of sRNAs, and activates the production of a second series of siRNAs called secondary or transitive siRNAs. In addition, DCL2, DCL3 and DCL4 are known to participate in the processing of endogenous siRNAs in plants, which are categorized as secondary siRNAs, while DCL4 also participates in the defense against viruses by inducing the production of primary siRNAs (Yoshikawa et al. 2005; Axtell et al. 2006; Borges and Martienssen 2015). In this study, microarray and RT-qPCR analyses confirmed that DCL2 and DCL4 were activated by the homologous CIRP and heterologous CIRT constructs. RDR6 is involved in the pathway that triggers the biogenesis of secondary RNAi in cell-to-cell signaling, and plants with mutated RDR6 have shown to be more susceptible to begomoviral infections than wild-type plants (Qu et al. 2005). A transcript associated with RDR6 is SDE5, which has RNA helicase activity and functions with RDR6 in the generation of dsRNA from specific dsRNA to produce trans-acting small-interfering RNA (ta-siRNA), which participates in RNA interference (Hernandez-Pinzon et al. 2007). Both CIRP and CIRT constructs activated SGS3, another transcript that directly interacts with RDR6 in the ta-siRNA production pathway (Xie et al. 2012; Li et al. 2017). HEN1, which is a crucial factor in biogenesis of siRNAs and miRNAs that methylates and protects against uridylation (Movahedi et al. 2018), was also activated by homologous CIRP and heterologous CIRT constructs. In addition, NRPD1A can maintain RNA silencing and transitivity through DNA methylation, participating in sRNA production and amplification together with NRPD2B/Pol IV (Eamens et al. 2008).
Fig. 5.
Schematic representation of the proposed model for RNAi-mediated protection of Nicotiana benthamiana against PepGMV infection with CIRP (homologous construct of the intergenic region of PepGMV) and CIRT (heterologous construct of the intergenic region of ToChLPV) based on the results of microarrays and RT-qPCR analysis from the this study and those of several other studies on this topic
Genome and transcriptome microarray data from A. thaliana vs different genome and transcriptome assembly versions of N. benthamiana were compared, resulting in the identification of genes homologous to transcripts associated with those involved in defense response, stress, systemic acquired resistance; even some genes that participate in MAPK cascades involved in signaling multiple defense responses as the defense gene activation- were identified (Ichimura et al. 2002; Meng and Zhang, 2013) (Table 2), and RNAi (Table 3). The results indicated that AGO3 from A. thaliana is similar to AGO2 of N. benthamiana, AGO1 is homologous to AGO1a or AGO1-1 from N. benthamiana and that AGO9 from A. thaliana is homologous to AGO4b from N. benthamiana (Bombarely et al. 2012; Nakasugi et al. 2013; Kourelis et al. 2018). All of those components were activated by the heterologous CIRT construct, suggesting that CIRT can activate transcriptional silencing.
The protein network analysis using STRING allowed visualizing molecular interactions among the differentially expressed genes induced by the CIRT (Fig. 3a) and CIRP (Fig. 3b) constructs. In both cases, the network of proteins was involved in pathways associated with RNAi, including proteins involved in biogenesis of different small RNAs, secondary siRNAs, other core components that participate in the initialization and maintenance of RNA-directed DNA methylation (RdDM) in N. benthamiana and other dicots (Eamens et al. 2008; Nakasugi et al. 2013; Borges and Martienssen 2015). Furthermore, both constructs could activate PTGS and TGS.
The RT-qPCR results (Fig. 4) for genes associated with RNAi showed that PPR1 was expressed in protected plants with homologous CIRP and heterologous CIRT constructs and in the controls, with higher expression levels in sv. PPR1 is known to participle in growth and developmental stages in plant structure and plant genomes have up to five hundred PPR genes per genome whereas non-plant genomes encode only two to six PPR proteins (Klepikova et al. 2016). High expression levels in sv might be normal in plants, and they were affected by exogenous material in the other treatments. PPR1 belongs to the clade of genes that encode PPR proteins, which have been documented to play important roles in antiviral defense in plants (Wu et al. 2016). In addition, PPR1 participates in the reaction cascade by which gene silencing is propagated via the SGS3/RDR6/DCL4 pathway, and their transcription depends on trans-acting siRNAs (tasiRNAs) (Li et al. 2017). An alternate RNAi route involves small RNAs called ct-siRNAs derived from transcription encoders, which have been shown to function as a regulatory system to maintain controlled protection systems and prevent cells from being overloaded by the overexpression of foreign genes (Zhang et al. 2015). This route solely focuses on the defense against the transcription of invasive DNA (Alvarez et al. 2010), and these small RNAs as well as ta-siRNAs depend on interactions with RDR6, SGS3, DCL2, DCL4 and AGO1 (Li and Wang 2018). The microarray and RT-qPCR analysis results were congruent, indicating that these transcripts were expressed in the assayed plants, when heterologous constructs derived from ToChLPV and homologous to PepGMV are used, because viral loads decreased in (95.6 and 99.5%, respectively), as previously described by Medina-Hernández et al. (2013). DCL2 has been reported to be involved in processing endogenous siRNAs in plants as well in the defense against viruses but it also has specific roles in regulation of genome expression and maintenance (Vazquez and Hohn 2013), which may explain its high expression levels observed in negative control (sv) plants.
The high expression levels of AGO1-1 and AGO2 provide evidence for the activation of RNAi in plants protected with homologous CIRP and heterologous CIRT constructs and infected with PepGMV. Furthermore, several studies have documented the participation of NbAGO1 and NbAGO2 in defense mechanisms against viruses (Ghoshal and Sanfaçon 2014; Kontra et al. 2016; Odokonyero et al. 2017; Paudel et al. 2018).
The results of this study indicate that plants protected with homologous and heterologous constructs against PepGMV activate key components of the RNAi gene silencing mechanism (RNAi). Despite the sequence identity of DNA-A of PepGMV with ToChLPV was 52% and that of the fragments used was approximately 50% with six regions longer than 22 nt of both viruses, with highest identity. This result contributes to understanding cross protection using RNAi, and might be efficient for controlling these pathogens, even in mixed infections (Robinson et al. 2014; Yousaf et al. 2015; Chen et al. 2016; Rasool et al. 2016). Evaluating heterologous dsRNAs is important due to the occurrence of mixed infections in nature and may result in the development of strategies that focus on diseases with symptoms that are difficult to diagnose and control. However, to our knowledge the majority of published studies have not compared transcripts between homologous and heterologous sequences to the challenged virus. Such comparisons may increase our understanding of virus-virus interactions and provide information to confront unknown etiologies. The results in this study were consistent with those that have demonstrated that each type of dsRNA produces a different type of small RNA that functions in diverse pathways involved in gene silencing or DNA methylation activation (Matzke and Mosher 2014). The increased expression of some key factors involved in the TGS and PTGS responses play a crucial role in silencing and epigenetic regulation (Ding 2010; Saze et al. 2012). In this study, different specialized pathways were activated and involved in the production and function of different classes of sRNAs that are required to trigger and propagate the PTGS and TGS responses.
Acknowledgements
This research was financed by the Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) through projects SEP2007-84777 and SEP2015-253828 “Analysis of the regulatory function of sRNAs in a gene silencing system as a mechanism of protection against mixed begomoviral infections” granted to R.J. Holguín-Peña. The authors are grateful to Gerardo Rafael Hernández García for his support in figure editing, the Laboratory of Phytopathology, CIBNOR, México, and Diana Fischer for editorial services.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Diana Medina Hernández, Mayela Vargas Salinas, Guadalupe Fabiola Arcos Ortega and Irasema Elizabeth Luis Villaseñor. The first draft of the manuscript was written by Diana Medina Hernández, Mayela Vargas Salinas, Guadalupe Fabiola Arcos Ortega and all authors commented on previous versions of the manuscript. Review and editing: Mayela Vargas Salinas and Guadalupe Fabiola Arcos Ortega. Funding acquisition: Ramón Jaime Holguín Peña. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mayela Vargas-Salinas, Email: msalinas@pg.cibnor.mx.
Diana Medina-Hernández, Email: dmedina@cibnor.mx.
Guadalupe Fabiola Arcos-Ortega, Email: farcos04@cibnor.mx.
Irasema Elizabeth Luis-Villaseñor, Email: irasemaluis@uas.edu.mx.
Ramón Jaime Holguín-Peña, Email: jholguin04@cibnor.mx.
References
- Abhary MK, Anfoka GH, Nakhla MK, Maxwell DP. Post-transcriptional gene silencing in controlling viruses of the Tomato yellow leaf curl virus complex. Arch Virol. 2006;151:2349–2363. doi: 10.1007/s00705-006-0819-7. [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol. 2010;11:563–576. doi: 10.1111/j.1364-3703.2010.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, et al. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant-Microbe Interact. 2012;25:1523–1530. doi: 10.1094/MPMI-06-12-0148-TA. [DOI] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas-Conejo Y, Arguello-Astorga G, Poghosyan A, et al. First report of Tomato yellow leaf curl virus co-infecting Pepper with Tomato chino La Paz virus in Baja California Sur. Mexico Plant Dis. 2010;94:1266. doi: 10.1094/PDIS-06-10-0444. [DOI] [PubMed] [Google Scholar]
- Chellappan P, Masona MV, Vanitharani R, et al. Broad spectrum resistance to ssDNA viruses associated with transgene-induced gene silencing in cassava. Plant Mol Biol. 2004;56:601–611. doi: 10.1007/s11103-004-0147-9. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin C, Tsai W, et al. Resistance to viral yellow leaf curl in tomato through RNAi targeting two Begomovirus species strains. J Plant Biochem Biotechnol. 2016;25:199–207. doi: 10.1007/s13562-015-0325-7. [DOI] [Google Scholar]
- Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- De Barro PJ, Liu S-S, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Annu Rev Entomol. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Eamens A, Vaistij FE, Jones L. NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis. Plant J. 2008;55:596–606. doi: 10.1111/j.1365-313X.2008.03525.x. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9. 1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arenal F, Zerbini FM. Life on the edge: geminiviruses at the interface between crops and wild plant hosts. Annu Rev Virol. 2019;6(1):411–433. doi: 10.1146/annurev-virology-092818-015536. [DOI] [PubMed] [Google Scholar]
- Gharsallah-Chouchane S, Gorsane F, Nakhla MK, et al. Evaluation of two gene-silencing constructs for resistance to tomato yellow leaf curl viruses in Nicotiana benthamiana plants. Acta Virol. 2008;52:143–149. [PubMed] [Google Scholar]
- Ghoshal B, Sanfaçon H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires argonaute 1. Virology. 2014;456–457:188–197. doi: 10.1016/j.virol.2014.03.026. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Yelina NE, Schwach F, et al. SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 2007;50:140–148. doi: 10.1111/j.1365-313X.2007.03043.x. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Baulcombe DC (2004) RNA silencing pathways in plants. In: Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Laboratory Press, pp 363–370 [DOI] [PubMed]
- Ichimura K, Shinozaki K, Tena G, et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, et al. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- Kontra L, Csorba T, Tavazza M, et al. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016;12:e1005935. doi: 10.1371/journal.ppat.1005935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, Kaschani F, Grosse-Holz FM et al (2018) Re-annotated Nicotiana benthamiana gene models for enhanced proteomics and reverse genetics. bioRxiv 373506
- Li F, Wang A. RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 2018;14:e1007228. doi: 10.1371/journal.ppat.1007228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zhou X. SGS3 cooperates with RDR6 in triggering geminivirus-induced gene silencing and in suppressing geminivirus infection in Nicotiana benthamiana. Viruses. 2017;9:247. doi: 10.3390/v9090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Ku HM, Tsai WS, et al. Resistance to a DNA and a RNA virus in transgenic plants by using a single chimeric transgene construct. Transgenic Res. 2011;20:261–270. doi: 10.1007/s11248-010-9412-7. [DOI] [PubMed] [Google Scholar]
- Liu D, Shi L, Han C, et al. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One. 2012;7:e46451. doi: 10.1371/journal.pone.0046451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Medina-Hernández D, Rivera-Bustamante R, Tenllado F, Holguín-Peña RJ. Effects and effectiveness of two RNAi constructs for resistance to Pepper golden mosaic virus in Nicotiana benthamiana plants. Viruses. 2013;5:2931–2945. doi: 10.3390/v5122931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Lozano J, Torres-Pacheco I, Fauquet CM, Rivera-Bustamante RF. Interactions between geminiviruses in a naturally occurring mixture: Pepper huasteco virus and Pepper golden mosaic virus. Phytopathology. 2003;93:270–277. doi: 10.1094/PHYTO.2003.93.3.270. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–266. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- Morales-Aguilar JJ, Rodríguez-Negrete EA, Camacho-Beltrán E, et al. Identification of Tomato yellow leaf curl virus, Pepper huasteco yellow vein virus and Pepper golden mosaic virus associated with pepper diseases in northern Mexico. Can J Plant Pathol. 2019;41:544–550. doi: 10.1080/07060661.2019.1591509. [DOI] [Google Scholar]
- Movahedi A, Zhang J, Sun W, et al. Plant small RNAs: definition, classification and response against stresses. Biologia (Bratisl) 2018;73:285–294. doi: 10.2478/s11756-018-0034-5. [DOI] [Google Scholar]
- Mubin M, Hussain M, Briddon RW, Mansoor S. Selection of target sequences as well as sequence identity determine the outcome of RNAi approach for resistance against cotton leaf curl geminivirus complex. Virol J. 2011;8:122. doi: 10.1186/1743-422X-8-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasugi K, Crowhurst RN, Bally J, et al. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One. 2013;8:e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S. Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol. 2011;49:219–248. doi: 10.1146/annurev-phyto-072910-095235. [DOI] [PubMed] [Google Scholar]
- Odokonyero D, Mendoza MR, Moffett P, Scholthof HB. Tobacco rattle virus (TRV)-mediated silencing of Nicotiana benthamiana Argonautes (NbAGOs) reveals new antiviral candidates and dominant effects of TRV-NbAGO1. Phytopathology. 2017;107:977–987. doi: 10.1094/PHYTO-02-17-0049-R. [DOI] [PubMed] [Google Scholar]
- Paudel DB, Ghoshal B, Jossey S, et al. Expression and antiviral function of Argonaute 2 in Nicotiana benthamiana plants infected with two isolates of Tomato ringspot virus with varying degrees of virulence. Virology. 2018;524:127–139. doi: 10.1016/j.virol.2018.08.016. [DOI] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- Qin C, Li B, Fan Y, et al. Roles of dicer-like proteins 2 and 4 in intra- and intercellular antiviral silencing. Plant Physiol. 2017;174:1067. doi: 10.1104/pp.17.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Hou G, et al. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol. 2005;79:15209. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool G, Yousaf S, Amin I, et al. Transient expression of synthetic coat protein gene of Cotton leaf curl Burewala virus in tobacco (Nicotiana benthamiana) J Agric Res. 2016;54:21–34. [Google Scholar]
- Robinson KE, Worrall EA, Mitter N. Double stranded RNA expression and its topical application for non-transgenic resistance to plant viruses. J plant Biochem Biotechnol. 2014;23:231–237. doi: 10.1007/s13562-014-0260-z. [DOI] [Google Scholar]
- Saze H, Tsugane K, Kanno T, Nishimura T. DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 2012;53:766–784. doi: 10.1093/pcp/pcs008. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha O, Farouk I, Abdallah A, Abdallah NA. Use of posttranscription gene silencing in squash to induce resistance against the Egyptian isolate of the Squash leaf curl virus. Int J Genomics. 2016;2016:1–9. doi: 10.1155/2016/6053147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado F, Llave C, Díaz-Ruíz JR. RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res. 2004;102:85–96. doi: 10.1016/j.virusres.2004.01.019. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Hohn T. Biogenesis and biological activity of secondary siRNAs in plants. Scientifica (Cairo) 2013;2013:783253. doi: 10.1155/2013/783253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/S0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- Wang X-B, Jovel J, Udomporn P, et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WQ, Fan HY, Jiang N, et al. Infection of Beet necrotic yellow vein virus with RNA4-encoded P31 specifically up-regulates pathogenesis-related protein 10 in Nicotiana benthamiana. Virol J. 2014;11:118. doi: 10.1186/1743-422X-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Li X, Guo S, Wong S-M. Analyses of RNA-Seq and sRNA-Seq data reveal a complex network of anti-viral defense in TCV-infected Arabidopsis thaliana. Sci Rep. 2016;6:36007. doi: 10.1038/srep36007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Ren G, Costa-Nunes P, et al. A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res. 2012;40:4422–4431. doi: 10.1093/nar/gks034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf S, Rasool G, Amin I, et al. Evaluation of the resistance against begomoviruses imparted by the single-stranded DNA binding protein VirE2. Pak J Agri Sci. 2015;52:887–893. [Google Scholar]
- Zhang X, Zhu Y, Liu X, et al. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;80(348):120–123. doi: 10.1126/science.aaa2618. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lai T, Zhang P, et al. Mini review: revisiting mobile RNA silencing in plants. Plant Sci. 2019;278:113–117. doi: 10.1016/j.plantsci.2018.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]