Abstract
Telomere encompasses a (TTAGGG)n tandem repeats, and its dysfunction has emerged as the epicenter of driving carcinogenesis by promoting genetic instability. Indeed, they play an essential role in stabilizing chromosomes and therefore protecting them from end-to-end fusion and DNA degradation. Telomere length homeostasis is regulated by several key players including shelterin complex genes, telomerase, and various other regulators. Targeting these regulatory players can be a good approach to combat cancer as telomere length is increasingly correlated with cancer initiation and progression. In this review, we have aimed to describe the telomere length regulator's role in prognostic significance and important drug targets in breast cancer. Moreover, we also assessed alteration in telomeric function by various telomere length regulators and compares this to the regulatory mechanisms that can be associated with clinical biomarkers in cancer. Using publicly available software we summarized mutational and CpG island prediction analysis of the TERT gene breast cancer patient database. Studies have reported that the TERT gene has prognostic significance in breast cancer progression however mechanistic approaches are not defined yet. Interestingly, we reported using the UCSC Xena web-based tool, we confirmed a positive correlation of shelterin complex genes TERF1 and TERF2 in recurrent free survival, indicating the critical role of these genes in breast cancer prognosis. Moreover, the epigenetic landscape of DNA damage repair genes in different breast cancer subtypes also being analyzed using the UCSC Xena database. Together, these datasets provide a comprehensive resource for shelterin complex gene profiles and define epigenetic landscapes of DNA damage repair genes which reveals the key role of shelterin complex genes in breast cancer with the potential to identify novel and actionable targets for treatment.
Keywords: Telomere, DNA damage, hTERT, Shelterin complex
Highlights
-
•
Assessed alteration in telomeric function by various telomere length regulators and compares its involvement in regulatory mechanisms that can be associated with clinical biomarkers in cancer.
-
•
Prognostic value of each Shelterin complex genes were determined in breast cancer using the Kaplan–Meier plotter database.
-
•
Summarizes telomere length regulators role in prognostic significance and important drug targets.
-
•
Identified positive correlation of TERF1 and TERF2 in recurrent free survival, indicating the critical role of these genes in breast cancer prognosis.
1. Introduction
Human biology revolves around one term i.e., Deoxyribonucleic acid (DNA), and underpinning the role of DNA, in today's era of science, therefore, it becomes obvious that its protection is critical for our survival. However, the discovery and function of telomeres were hypothesized even way before the DNA was identified as genetic material. While conducting radiation experiment on Drosophila chromosomes, Herman Muller in 1938, found that X rays could induce chromosome rearrangements but not terminal deletions, which led him to conclude special function of the terminal region of chromosomes and later he coined the term “telomere” for this region (derived from Greek words telos = end and meros = part) [1]. Around the same time (1940) Barbara McClintock while working with Zea Mays (where dicentric chromosomes were produced with high frequency) reported that the absence of special end structures of chromosomes led to chromosome instability a detrimental factor for cell survival, congregated on the same conclusion drawn by Muller [2]. Both scientists were awarded Nobel prizes in physiology or Medicine (Hermann Muller in 1946 and Barbara McClintock in 1983) but not for a discovery made in the telomere biology field [3], however, these pioneering studies led the strong foundation for future telomere research [1,2]. Hayflick and Paul Moorhead described that normal human cells have finite life span when cultured in vitro (later this phenomenon is known as Hayflick limit or replicative senescence) which was contrary to earlier belief where it has been demonstrated that all cells have infinite life span [4]. Telomere shortening has been assumed to act as a “mitotic clock” that limit the cell cycle number [5] and critically short telomere eventually triggered cellular senescence [6]. More interestingly, biological aging can be correlated by telomere length as aged persons had shorter telomeres than younger ones [7,8]. Initially, telomere related discoveries were conducted in ciliated protozoa Tetrahymena thermophila, which contain abundant short linear chromosome and therefore provided a unique model system where the sequencing of telomeric DNA was conducted, leading to the discovery of the molecular basis of telomere [9]. Later it has been found that telomeric DNA is the repetitive sequence of TTAGGG in humans [10,11] as well as all vertebrates [12] and variation in tandem repeats i.e (TTAGGG)n, led to variation in telomere length from chromosome to chromosome. In humans, this variation ranges from 3 to 20 kilobase pairs. DNA polymerase needs a labile primer to initiate unidirectional 5′–3′ synthesis to copied full DNA at the 3′ end during DNA replication. Therefore, DNA polymerase unable to synthesize full DNA at the end (3′ end) during DNA replication, and for this reason, this has been termed as an end-replication problem [13,14]. To understand how cancerous cell bypasses this end-replication problem, Greider and Blackburn identified that the presence of telomerase enzyme ensures the cell to bypass telomeric shortening problem [15].
The significance of telomere was demonstrated in many ways, which include coverage for the coding DNA at the end, therefore avoiding loss of genetic information in linear DNA. Moreover, telomere also acts as a cap to protect chromosomes from being recognized as Double-strand breaks (DSBs) avoiding its degradation [16]. The telomerase enzyme is a large multisubunit ribonucleoprotein complex, which adds telomeric DNA repeats at the end of chromosomes. In human, telomerase activity is driven by a complex of two major components: human telomerase reverse transcriptase (hTERT) encoded by the TERT gene which acts as a functional catalytic protein having reverse transcriptase capacity, and the RNA component (also known as human telomerase RNA component: hTERC or hTR) which acts as a template for reverse transcription [17,18]. Both these components are located at chromosomal region 5p15.33 and 3q26 respectively in humans [19]. This enzyme has been demonstrated that it’s components are largely conserved across the species from mammals to vertebrates, therefore demonstrating its critical evolutionary role in biology [20]. Compelling evidence has also accumulated that human telomerase promoter mutations are also strongly correlated with poor disease outcomes [[21], [22], [23]]. Additionally, regulation of telomere length and telomerase enzyme expression also controlled through transcriptional, post-transcriptional, and epigenetic modifications, and therefore gaining insight into these mechanisms will not only provide novel biomarkers and but also help in early detection of disease, identifying novel prognostic markers and therapeutics [24]. Epigenetic marks like trimethylation of histones of certain amino acids also regulate the telomere length, along with some others such as Telomeric repeat-containing RNA (TERRA) [25,26]. Recently, studies related to protein structural resolution of telomere such as a recent cryo-electron microscopy study [27], could be the key finding for targeting human telomerase by small-molecules in days to come.
2. Role of telomere length dynamics in cancer progression and development
Cancer is an umbrella term for a large group of diseases that can be characterized by many capabilities leading to many complications in the human body. These capabilities are best described in one of the highly cited review articles entitled “hallmarks of cancer” by Hanahan and Weinberg in 2000 and 2011 [28]. These hallmarks provide a solid foundation for understanding the biology of cancer which is relevant to today's era as well. A highly complementary hallmark, by which cancer cells can acquire the capability for nourishing proliferative signaling to escape growth suppression and adept for indefinite cell division and growth (immortality) [28]. During cell division, there is a reduction of 50–200 bp of the nucleotide sequence, which is reduced until an acute edge is reached and normal cells undergo replicative senescence [29]. However, cancer cell escapes this replicative senescence crisis and maintain stable but usually shortened telomere lengths by dictating steady level telomerase expression for continued cell growth, which allow them to divide indefinitely [30,31]. Interestingly, in human somatic cells, TERT expression is silenced, however, it's expressed in ~90% of human tumors [32,33] demonstrating the critical role of this enzyme in maintaining cell proliferating capacity of cells, an important hallmark of cancer cells. Due to the critical role of telomerase in cancers various mechanism has been elucidated for hTERT activation, which includes genetic (promoter mutations), amplification, pre-mRNA alternative splicing, epigenetic modification (promoter methylation and miRNAs) and telomere position effect [34,35]. The telomere is also been explored for therapeutic benefit as short telomeres can deliver deleterious effects against tumor cells as they have proliferative characteristics [36]. Telomerase is a good target for anti-cancer drugs as it expresses more in tumor cells than any other marker. Oligonucleotide based drug like Imetelstat (GRN163) is already popular and efficient and small synthetic drug BIBR1532 is one of the most potent inhibitors of human telomerase [37]. BRACO19 and telomestatin are G-quadruplex stabilizing ligands that inhibit the telomerase to bind to the telomere DNA [38]. An anti-telomere drug such as T-oligo is a well-developed drug used to inhibit telomere from various DNA damage responses (DDR) [39]. The development of anti-cancer drugs requires a proper understanding of processes and mechanisms which regulate telomere and its length. Thus, targeting telomerase enzyme and its access to telomeric end thereby regulating telomere homeostasis may be considered a promising approach for cancer therapeutics [[40], [41], [42]].
3. Canonical regulation of telomere length by telomerase enzyme: TERT
Telomerase enzyme was first reported in the early 1980s, however, its genes for RNA component (TER or TERC) was cloned in 1995 [17] and its core protein component (also called TERT) was cloned in 1997 [43,44]. Cryo-EM structure of telomerase revealed that TERT protein contains four domains: Telomerase essential N-terminal (TEN) domain, TERT RNA binding domain (TRBD), Reverse transcriptase domain (RT), and a C-terminal extension (CTE) domain [27]. In most organisms, the telomerase enzyme is a predominant factor for telomere elongation. Initial discoveries involved the origin of the “protein counting model” in S. cerevisiae, which stated that telomere bound proteins inhibit the telomerase access to the telomere end. According to this model, the longer the telomere, have the abundance of telomeric binding proteins, (which play an inhibitory role for telomerase recruitment) hence more the repressive effect on the telomerase on the other hand short telomeres have a less strong repressive effect and hence telomerase can elongate them. This model further gets acceptance in the mammalian system as knockdown of telomere binding proteins resulted in elongated telomeres [45,46]. However, this model was unable to explain how telomere-bound proteins can halt telomerase access from long distances. Later “replication fork” model was better able to explain this problem which stated that telomerase must be deposited at the end of the telomere to elongate it by traveling across the telomere length through the replication fork [47]. Telomere bound proteins and probably nucleosome exerts a negative effect on telomerase movement on replication fork thereby dissociating it and lowering the probability of telomerase reaching the end, where it has to perform its catalytic function [47]. Therefore, short telomeres have a high probability of telomerase to be associated with the telomere and perform its catalytic function to elongate the telomere. Accumulating evidence suggests that G-strand overhang availability is also a major factor for promoting telomerase activity which is positively regulated by POT1-TPP1 heterodimer by inhibiting ATR kinase [48]. Hence, it has been revealed that loss of telomere end protection, telomere length increases aberrantly due to the unregulated access of telomerase to the telomere [45,49,50].
Interestingly, among two important components of telomerase enzyme i.e. TERC and TERT, all human tissues express TERC, while most human adults do not express TERT [51]. TERT gene was silenced in a tissue specific manner during early human fetal development while constitutively expressing telomerase [52]. Apart from its canonical role in telomere length regulation, compelling evidence has been accumulated the essential role of TERT, in cellular immortalization and malignant transformation by stabilizing telomere length and erasing the senescence barrier [53,54]. TERT expression/telomerase reactivation remains a ubiquitous process, which can be detectable in more than 90% of human primary cancers [55], constituting it's one of the important major hallmarks of cancer [56], however its precise mechanism of activation is still unclear [53]. Here we listed some of the mechanisms of TERT regulation in cancers.
3.1. TERT gene mutations
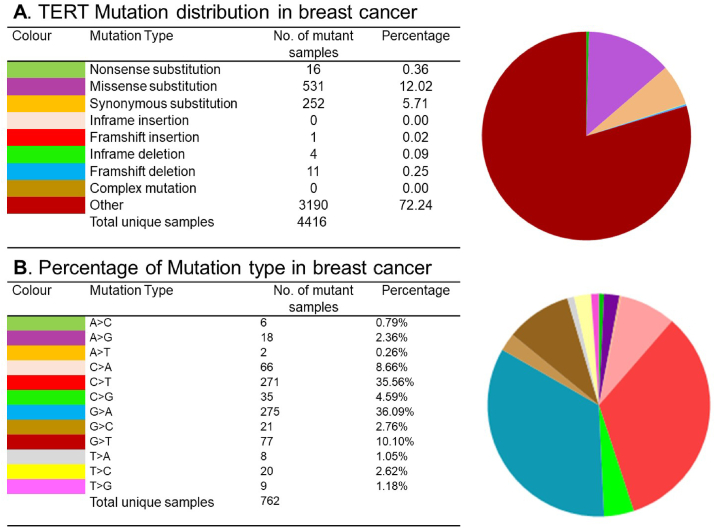
The TERT gene (42 kb long) comprises 16 exons and 15 introns located on the short arm of chromosome 5 (5p.15:33) [18]. TERT promoter is rich with various transcription factor binding motifs such as Sp1 and oncogene MYC. The most prominent form of mutation of the TERT promoter is C > T mutation at −146 or −124 upstream from the start codon (ATG) which is termed as C228T and C250T respectively. Both these mutations are associated with the generation of ETS (E-twenty-six) transcription factors binding motifs (CCGGAA) [57,58]. Family members of ETS transcription factors such as GA binding proteins GABPA and GABPB1 are specifically recruited are main drivers for the mutated promoter activation by promoting TERT transcription and telomerase activation [59]. This mutant promoter displays active chromatin marks such as H3K4me2/3 while wild type promoter remains epigenetically silenced suggested by the H3K27me3 [60]. Studies suggested that TERT promoter mutations are negatively correlated with telomere length [61] leading to genetic instability. To explore whether TERT promoter mutations can be correlated with prevalence and prognostic significance in human breast cancer, we analyzed the COSMIC database (https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=TERT) [62]. Analyzed results described information which includes nonsense, missense, synonymous, substitution, deletion frame, and insertion frameshift mutations. We found that TERT mutations are largely missense and non-sense in Breast cancer (BC) samples (Fig. 1A). BC had 36.09% G > A and 35.56% C > T mutation in TERT coding strand (Fig. 1B).
Fig. 1.
COSMIC database analysis of TERT mutation distributions and its types A. TERT mutations showed in the pie chart illustrate mostly missense and non-sense in BC. B. TERT coding strands largely included G > A (36.09%) and C > T (35.56%) mutations. TERT, Human telomerase reverse transcriptase; BC, breast cancer. TERT, Human telomerase reverse transcriptase; BC, breast cancer.
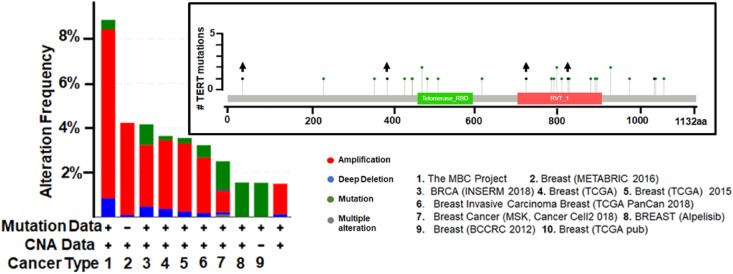
We also analyzed compare the frequency of TERT gene alterations in 9344 samples using the cBioPortal online tool (https://www.cbioportal.org) [63,64]. The summary graph of TERT gene alterations (shown in different colors) in individual breast cancer (total 9 studies) is shown in Fig. 2. Obtained results indicate more than 8% TERT mutation in BC patients. The distribution of the hTERT mutations is shown in the inset (Fig. 2).
Fig. 2.
TERT mutation analysis and visualization using cBioportal database. Accumulated alterations of TERT in BC are summarized in the graph in individual studies deposited in the database. Furthermore, the distribution of these alterations are shown in the inset, black arrows showing nonsense mutations while others showing missense mutation. TERT, telomerase reverse transcriptase; BC, breast cancer.
3.2. TERT amplification and TERT rearrangements
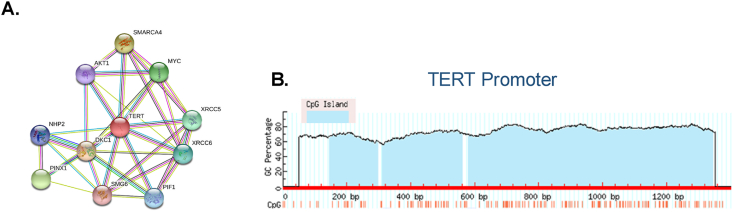
TERT is also activated by several genomic rearrangements that affect the TERT locus. It brings enhancers in close contact with the coding part of the gene. Studies reveal that these rearrangements involve direct overlapping between super-enhancers and the locus of the gene which results in significant chromatin remodeling and subsequent activation of TERT transcription [65,66]. MYCN amplification and ATRX mutation are exclusive during the process of TERT rearrangement observed mainly in high-grade neuroblastoma [66]. Genetic instability is one of the important hallmarks of cancer as it favors cancer cells to accumulate a large number of mutations. Gene amplification refers to the differential increase in a specific portion of the gene and can be considered as an important event in oncogenesis which imparts cancer cells to achieve overexpression of some oncogenes [67]. Recent reports have documented that TERT gene amplification was observed in 4% of tumors in the systematic analysis of the TERT gene [68]. Our STRING interaction analysis (https://string-db.org/) revealed that TERT protein involved in many molecular and biological process and has high-confidence interactions with Dyskerin pseudouridine synthase 1 (DKC1), MYC oncoprotein, XRCC5 (X-Ray Repair Cross Complementing 5), XRCC6 (X-Ray Repair Cross Complementing 6), ATP-dependent DNA Helicase PIF1 (PIF1), telomerase-binding protein EST1A (SMG6), TERF1-interacting telomerase inhibitor 1 (PINX1), NHP2 ribonucleoprotein (NHP2) and SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4) and serine-threonine protein kinase (AKT1) (Fig. 3A).
Fig. 3.
Bioinformatics analysis of CpG island prediction on TERT gene promoter and interaction network using the STRING database. (A) TERT, and its strongly interacting protein interaction network derived from the STRING database. In a biological network, a node is any biological molecule and an edge indicates the interaction between the two nodes. (B) CpG island presence near the promoter of a gene is strongly associated with methylation mark recruitment. Our bioinformatics analysis provided the information that there is a heavy abundance of CpG island near the TERT gene promoter. As the TERT promoter contains a site for various transcription factors, these CpG islands may play a role in the alteration and regulation of TERT transcription. The location of the CpG islands is colored in light blue. TERT, Human Telomerase reverse transcriptase. TERT, human telomerase reverse transcriptase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Epigenetic regulation of TERT promoter
CpG sites are usually the hotspots for the DNA methylation to occur, the process mediated by DNA methyltransferase enzyme which catalyzes the addition of methyl groups (-CH3) at the 5th carbon of the cytosine base. Using CpG island prediction software (https://www.urogene.org/methprimer/) [69], we found that the TERT promoter is rich in CpG island (Fig. 3B). Due to this frequent presence of CpG islands on gene promoters, DNA methylation unarguably plays an important part in gene expression [70]. The TERT gene contains around 52 CpG sites within its promoter and this region is identified as TERT hypermethylated oncological region (THOR). Hypermethylation of THOR led to TERT up-regulation which is an important epigenetic mechanism for malignant transformation [71] Hypomethylation is also observed to initiate the binding of transcriptional activators such as c-MYC on the contrary hypermethylation led to inhibit the binding of repressor proteins like CCCCTC- binding factor (CTCF) and Wilms tumor 1 (WT1) to the promoter region of the TERT [34,71,72]. All these findings conclude that dynamic changes in methylation patterns are an important regulator of TERT transcription.
4. Role of shelterin complex in telomere length regulation
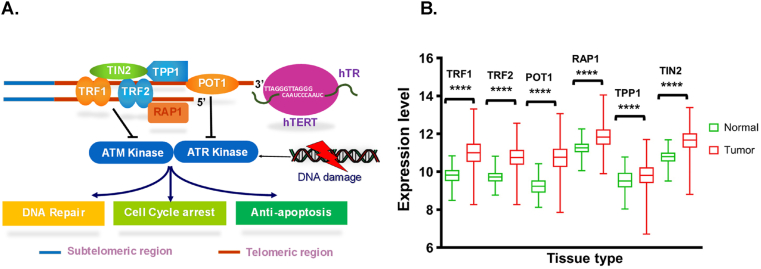
Telomere protects the chromosomal ends to be identified as Double-Stranded Breaks (DSBs) but the telomere itself needs to be protected from DDR [73,74]. So, to protect the protector of telomeric DNA, there lies the most important complex associated with telomere which is known as shelterin complex. It consists of (Fig. 4A) six major proteins- Telomeric repeat-binding factor 1 (TERF1), TERF2, Telomeric repeat-binding factor 2 (TERF2), TRF1 -interacting nuclear protein −2 (TIN-2), Protection of telomeres 1 (POT1), Repressor and Activator Protein 1 (Rap-1) and TPP1 or Adrenocortical dysplasia protein homolog (ACD). Shelterin complex is also known as telosome due to its specific role in protecting telomere.
Fig. 4.
Gene expression levels of shelterin complex genes in breast cancer (TCGA TARGET GTEx RNAseq data) and their link to DDR A. Cartoon describing that shelterin complex consists of majorly six proteins – TERF1, TERF2, Rap1, TIN2, TPP1, and POT1. TERF1 and TERF2 are homodimers binding to the double-stranded telomeric DNA through the Myb domain. Rap1 is an interactive partner of TERF2. TPP1 and POT1 bind to the ssDNA telomere and protect the 3′ overhang sequence of the telomere. TIN2 acts as a bridge between TERF1/TERF2 and TPP1/POT1 and stabilizes the shelterin complex. TERF2 and POT1 block ATM and ATR kinase respectively through which shelterin complex sequesters telomere from DDR, mainly initiated by activation of ATM and ATR kinases resulting in DNA repair mechanisms. B. Shelterin complex genes mRNA expression levels of shelterin complex genes were compared between “normal” (GTEx) and tumor samples (TCGA). The middle line in the boxes represents the median value. Graph-pad Prism software was used to calculate statistical differences by two-tailed Student's t-test. ****P < 0.00001.
TERF1, telomeric repeat-binding factor 1; TERF2, telomeric repeat-binding factor 2; Rap1/TERF2IP, telomeric-repeating binding factor 2-interacting factor; POT1, protection of telomeres protein 1; TPP1/ACD; Adrenocortical dysplasia protein; TIN2/TINF2, TERF1-interacting nuclear factor 2; DDR, DNA damage response, ATM, Ataxia telangiectasia mutated kinase; ATR, Ataxia telangiectasia, and Rad3.
4.1. TERF-1
TERF-1 is a homodimer protein binding directly to the duplex region of the telomere. Both TERF1 and TERF2 contain a TERF homology (TRFH) domain which helps in homodimerization and a Myb domain which is highly specific to the telomeric DNA [75]. There are many theories reported in the past as to how TERF-1 regulates the telomere length. Studies have reported that in the presence of TERF-1, telomere length is decreased [49,50]. With the help of the chromatin immunoprecipitation (ChIP) technique, Loayza and de Lange reported that TERF-1 regulates telomere length via POT1, either directly blocks the telomerase binding or it stabilizes the t-loop structure of telomere, which means telomerase cannot bind to 3’ end [49]. This means TERF-1 does not interact with telomerase directly. Tankyrase-1 overexpression is the most effective method to remove TERF-1 from the telomere [76]. Tankyrase-1 is an enzyme of the Poly (ADP-Ribose) polymerase (PARP) superfamily. PARPs generate the ADP-ribose polymers on the glutamic acid residues of the protein acceptor, which in this case is TERF-1, which results in inhibition of TERF-1 binding to the telomere [77]. Overexpression of the tankyrase enzyme in the nucleus of human cells has shown to remove the TERF-1 from telomeres [76].
4.2. TERF-2
As TERF-1 and TERF-2 are almost similar to each other, TERF-2 is also a homodimer protein and binds to the ds telomeric DNA. It is demonstrated that it helps in the formation of a secondary structure known as the t-loop in which 3′ ss overhang invades into the double-stranded DNA sequence [78]. This remodeling of telomere into t-loop help remove threats of non-homologous end joining (NHEJ) and ataxia telangiectasia mutated kinase (ATM) signaling [79,80]. It also acts as a negative regulator of the telomere length like TERF-1 as t-loop formation discourages telomerase binding to the 3’ end [50]. TERF-2 has also a role in the protection of telomere from DNA damage signaling by inhibiting the autophosphorylation property of ATM kinase [81].
4.3. TIN-2
TIN2 acts as a linchpin of the shelterin complex. It acts as the bridge between telomeric dsDNA and single-stranded DNA (ssDNA) by connecting TERF1 and TERF-2 to TPP1/POT1 heterodimer, thus stabilizing the shelterin complex [73]. As previously reported that tankyrase-1 PARsylates the TERF-1 and inhibit its binding to the telomere, TIN-2 is reported to prevent this action of tankyrase-1 on TERF-1, thereby acting as a negative regulator of telomere length in this case [45,77]. TIN2 mutation that leads to dyskeratosis congenital were shown to decrease the telomere length without any interference with the telomere protection which gives us the clear idea that TIN2 mutations somehow has inhibited the activity of telomerase [82].
4.4. POT1 and TPP1 - homologs of TEBPα and TEBPβ of oxytricha nova
POT1 and TPP1 together bind to the 3’ ss overhang as a stable heterodimer). TEBPα and TEBPβ were the first discovered proteins bound on the single-stranded telomeric DNA in the ciliate Oxytricha nova [83]. It is reported that TPP1 and POT1 are homologs of TEBPα and TEBPβ [[84], [85], [86]]. TPP1 is a very diverse protein in the sense that it interacts with ss DNA binding protein POT1 to form a heterodimer as well as to TIN2 to connect the ds DNA binding proteins (TERF1 & TERF2) to POT1 [45,87]. This heterodimer also prevents ataxia telangiectasia and Rad3 related (ATR) dependent DDR to act on telomere [88]. Both these events overall increase the stability of the shelterin complex as well as a telomere.
N terminal OB domain of TPP1 contains TPP1 glutamate (E) and leucine (L)-rich (TEL) patch which is also reported to regulate the telomere length by interacting with the telomerase-N-terminal (TEN) domain and as a processivity factor for telomerase during telomerase extension by improving template translocation and decreasing primer dissociation [89,90].
4.5. TERF2-Rap1
In mammals, Rap1 is a highly conserved protein present in the shelterin complex [73]. It is recruited by TERF-2 and they form a heterodimer that is essential in protecting telomere from homology-directed repair (HDR) pathways (SLX4 and PARP1) [91]. The absence of this heterodimer is reported to result in telomere shortening and telomere-free chromosome fusion by homology-directed repair (HDR) pathway [91].
5. Differential mRNA expression analysis of shelterin complex genes and its correlation with prognostic potential in breast cancer
We analyzed the mRNA expression of shelterin complex genes in tumor and normal cells, by using the University of California, Santa Cruz (UCSC) XENA platform (https://xenabrowser.net/) [92]. We selected the TCGA TARGET GTEX study which enables us to compare the expression using the TCGA database of tumor tissue and GTEx database for normal tissue samples in breast cancer patients. Taking this cohort study, we filtered down the data to breast cancer and checked the mRNA expression of shelterin complex genes. RSEM expected count dataset was used for the analysis and values were downloaded as log2 (expected_count +1). The Box plot was designed using GraphPad Prism software (8.4.2) and P < 0.05, were considered as significant. Analyzed data suggested that all the shelterin complex genes upregulated in breast cancer (Fig. 4B) compare to normal samples. To the best of our knowledge, no study reported the role of shelterin complex genes in breast cancer, however, mutation/or altered expression in shelterin components at telomeres have been described in various types of cancers [26]. To identify whether these genes might play a role in breast cancer prognosis or survival we used Kaplan–Meier plotter analysis (https://kmplot.com) and assessed the prognostic significance of the shelterin complex genes in all BC patients [93]. The relationship between survival and gene expression level of individual shelterin complex genes were demonstrated in Fig. 5.
Fig. 5.
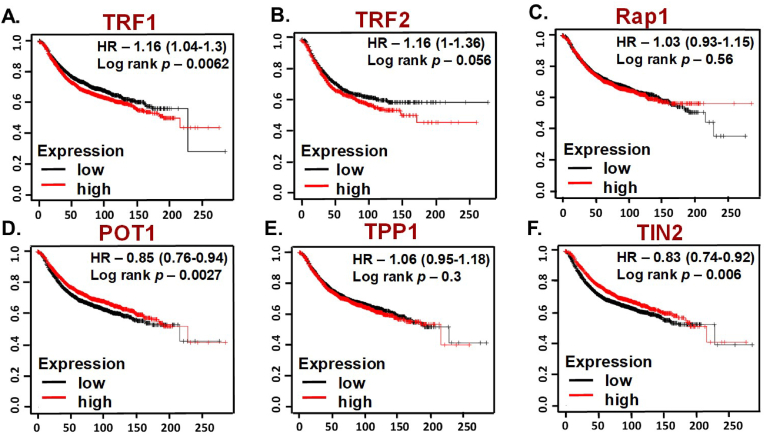
Survival analysis of shelterin complex genes in Breast cancer patients using Kaplan-Meier plotter. Survival curves of (A) TERF1 (Affymetrix ID:203448_s_at); (B) TERF2 (Affymetrix IDs: 1555185_x_at); (C) Rap1 (Affymetrix IDs:201174_s_at); (D) POT1 (Affymetrix IDs:204353_s_at); (E) TPP1 (Affymetrix IDs:204617_s_at); (F) TIN2 (Affymetrix IDs:220052_s_at). Abbreviations: TERF1, telomeric repeat-binding factor 1; TERF2.
Among the six sheltering complex genes we analyzed, high TERF1 and TERF2 expressions were significantly associated with a poor prognosis (TERF1, HR 1.16 [95% CI: 1.04–1.3], p = 0.0062; TERF2, HR 1.16 [95% CI: 1.1–1.36], p = 0.056; Fig. 5A and B (Table 1).
Table 1.
Datasets of the shelterin complex genes in breast cancer (Kaplan-Meier plotter).
| Gene | Affymetrix ID | No. of cases | Cut off value | HR | 95% CI | P-Value |
|---|---|---|---|---|---|---|
| TERF-1 | 203448_s_at | 3951 | 700 | 1.16 | 1.04–1.3 | 0.0062 |
| TERF-2 | 1555185_x_at | 1764 | 44 | 1.16 | 1–1.36 | 0.056 |
| Rap1/TERF2IP | 201174_s_at | 3951 | 1339 | 1.03 | 0.93–1.15 | 0.56 |
| POT1 | 204353_s_at | 3951 | 422 | 0.85 | 0.76–0.94 | 0.0027 |
| ACD/TPP1 | 204617_s_at | 3951 | 414 | 1.06 | 0.95–1.18 | 0.30 |
| TIN2 | 220052_s_at | 3951 | 585 | 0.83 | 0.74–0.92 | 0.0006 |
Abbreviations: HR, hazard ratio; CI, confidence level; TERF1, telomeric repeat-binding factor 1; TERF2, telomeric repeat-binding factor 2; Rap1/TERF2IP, telomeric-repeating binding factor 2-interacting factor; POT1, protection of telomeres protein 1; TPP1/ACD; Adrenocortical dysplasia protein; TIN2/TINF2, TERF1-interacting nuclear factor 2.
However, POT1 and TIN2 mRNA high expression were correlated with significantly better relapse-free survival (RFS) for all BC patients, POT1 (HR 0.85 [95% CI: 0.76–0.94], p = 0.0027; TIN2, (HR 0.83 [95% CI: 0.74–0.92], p = 0.0006 (Fig. 5D and F) and Table 1 In addition, the mRNA expression levels of TPP1, and RAP1 were not correlated with RFS (Fig. 5C and E). Our analysis identifies TERF1 and TERF2 may have prognostic potential in breast cancer. This is correlated with a recent study that shows that TERF-1 inhibition using combinatorial therapy can reduce stemness in glioblastoma xenograft models [94].
In conclusion, our data analysis showed that overexpression of TERF1 and TERF2 mRNA is correlated to a poor prognosis for all BC patients and may be a useful tool for prognosis. The possible responsiveness to these shelterin complex genes expressed by breast cancer cells can be the consequence of activation of DNA damage and suppression of DDR pathways. DNA damage causes genetic instability, and the accumulation of mutation can be increased during DNA damage. It causes the activation of DDR which repairs the DNA by recognizing it as a DSB and its result can be apoptosis or senescence [95]. Activation of two major kinase – ATR and ATM kinases initiate the process of DDR. It is followed by the DNA damage foci formation characterized by the presence of phosphorylated histone H2A.X and at last cell cycle is arrested when cell cycle checkpoints are alarmed such as p21 and CDK inhibitor [96]. Telomeric DNA is sequestered by the shelterin complex and prevents it from being recognized by DNA damage signaling [97]. Association of DDR proteins such as ATM, 53BP1, and MRE11 with uncapped telomere is observed resulting in cell-cycle arrest or apoptosis [98]. TERF2 and POT1 are also reported to block ATM and ATR kinase respectively [74]. ATM and ATR are also reported to help in the assembly of telomerase components which is essential in telomerase function [99].
6. The aberrant epigenetic landscape of DNA repair genes in the basal subtype of breast cancer
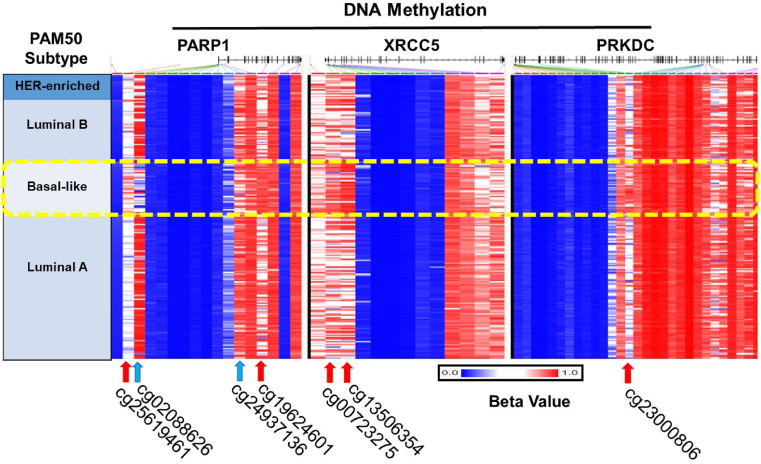
Deficiency in the DNA repair pathway is previously reported to result in tumor development [100]. Therefore, it is possible that tumor functions by inactivating the effectiveness of the DNA repair system by epigenetic gene inactivation which directly affects the DNA repair genes. To investigate the epigenetic landscape of DNA repair genes in breast cancer subtypes, we used the “TCGA Breast Cancer (BRCA)” study in the UCSC XENA browser. Interestingly, we found out that some of the DNA repair genes showed a contrasting pattern in only basal-like breast cancer subtype. Some of the CpG islands of PARP1, XRCC5 (coding gene for protein Ku80), and PRKDC (coding gene for protein DNA-PK) were either hypomethylated or hypermethylated only in basal-like breast cancer subtype (Fig. 6). DNA-PK, in association with Ku70/Ku80 heterodimer protein, functions in recombination and DNA double-stranded break repair. Epigenetic inactivation or activation of these DNA repair genes can be exploited using targeted therapies.
Fig. 6.
Methylation status of DNA damage repair genes correlated with TERF1 in breast cancer. UCSC Xena browser was used to analyze the DNA methylation of PARP1, XRCC5 and PRKDC correlated with TERF1 in breast cancer subtypes. DNA methylation of these genes was found to be distinctly different in the case of basal-like subtype in comparison to otherwise pattern. Red and blue arrows define hypermethylation and hypomethylation consequently. TERF1, telomeric repeat-binding factor 1; PARP1, Poly [ADP-ribose] polymerase 1; XRCC5, Ku80; PRKDC, Protein Kinase DNA Activated Catalytic subunit. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
7. Advancements in clinical therapeutics of anti-cancer drugs- clinical implications of drugs targeting telomere length regulatory factors
How the ends of chromosomes (telomeres) are maintained for the perpetual proliferation of cancer cells cancer cell immortality is still an area of extensive research. Induction of telomerase enzyme is one of the most prominent phenomena occurring in 85–90% of all malignant tumors and thus telomerase inhibitors are an attractive tool for cancer therapeutics [33,101]. However, in the rest 10–15% of cancers — which include some cancers which do have a poor prognosis as well as poor outcome — the telomere elongation is achieved through a mechanism known as alternative lengthening of telomeres (ALT) [102,103]. ALT to adversely impact the outcome of patients having soft tissue sarcoma, mesenchymal, and neuroepithelial tumor [103]. Interestingly, maintenance of the telomere length mechanism and its targeting became more complex as there has been evidence that both the mechanism can coexist within a single tumor [104]. This dynamic nature of tumor in maintaining telomere length suggested that telomerase targeted therapies may require accurate telomere lengthening mechanism characterization before treatment to get a maximum therapeutic benefit.
Altogether, the concept of hindering the telomere length regulating mechanism in cancer cells drug targeting telomere length mechanism is an emerging target for cancer therapeutics. Currently, there are a variety of antitelomerase drugs like small molecule drugs (BIBR1532), antisense oligonucleotides (ASO- GRN163L), ribozyme, and dominant-negative molecules like DN-TERT are in the trial/developmental phase. Here we are summarizing some telomerase targeting drug molecules which are currently in clinics or the developmental phase.
7.1. Imetelstat
Imetelstat is an FDA approved anti-telomerase drug that is a potent competitive inhibitor of the enzymatic activity of TERT. It is a lipid-based conjugate of oligonucleotide GRN163, thus named GRN163L, lipid components helping in an increase in the bioavailability [105]. Gathering clues from a pilot study for myelofibrosis [106], a recent clinical study in intermediate-2 (int-2), or high-risk myelofibrosis patients who are relapsed or refractory to Janus kinase inhibitor (JAKi) demonstrated favorable clinical activity with an acceptable safety profile [107].
Telomerase inhibition by imetelstat has been shown in mouse xenograft models and cell culture systems like breast [108,109], lungs [109,110], and bladder [111]. In addition to these hematological malignancies like multiple myeloma and lymphoma cancer cells are also being studied extensively for its antitelomerase activity [112].
7.2. MST-312
Epigallocatechin gallate (EGCG) chemical derivative known as MST-312 is reported to possess telomerase inhibitory (TI) activity and can be seen as a prototype of anticancer drugs [113,114]. MST-312 has a lower effective dose requirement and drug resistance and chemically more stable than its parent i.e., EGCG [114]. The mechanism of action of this drug is associated with the downregulation of several genes such as TNF-α, c-Myc, IL-6, and Bcl-2 and related pathways of these genes.
MST-312 has been shown to decrease telomerase activity and promoting telomere dysfunction leading to growth arrest in breast cancer cells [115]. According to reports, MST-312 is believed to express that its effects differ based on exposure time. Short term treatment of 90 days led to telomere attrition and senescence [114]. Studies reported that U-266 cells (myeloma cells), when treated with MST-312, can induce acute growth arrest and apoptosis indicating that this drug may have the potential to treat multiple myeloma (MM) which is described to be an incurable disease [116]. Long term exposure (130 days) of MST-312 resulted in TERT overexpression in breast cancer cells [117]. Similar results were also reported in another study where MST-312 can induce resistance by the selection of long telomere cells in vitro [113]. Therefore, the drawback of this drug is resistance induction due to long term treatment according to the limited available knowledge must be acknowledged.
7.3. BIBR1532
BIBR (2-[E]-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid) is a small and non-nucleosidic anti-telomerase drug that is widely used in kinetic, biochemical, and in vivo studies of telomerase function due to its selective nature [118]. It has been shown that BIBR1532 binds to the novel FVYL motif of the thumb domain of telomerase and through its binding, it prevents telomerase complex assembly formation [119]. Owing to this property, BIBR1532 is reported to be a therapeutic agent that suppresses telomerase activity in breast cancer [120] and glioblastoma [121], acute lymphoblastic leukemia, and Kras mutant NSCLC [122]. Moreover, in combination with doxorubicin, BIBR1532 is shown encouraging results to treat acute lymphoblastic leukemia (ALL) [123].
7.4. G- Quadruplex stabilizing ligands
The human genome contains a guanine rich sequence that is capable of forming a four-stranded quadruplex structure which is known as G-quadruplex [124,125]. The presence of these G-quadruplex structures on G-rich telomeric DNA at the chromosomal ends restricts the telomerase access to the telomere and therefore hinders its activity [38]. Small molecules called G4 ligands can facilitate the production of G-quadruplex structures [126,127]. Telomere shortening is induced by G-4 ligands which stabilize G-quadruplex on telomeric DNA [[128], [129], [130]]. A study revealed that ligands such as BRACO-19 and RHPS4 may require monovalent cations like central K+ to stabilize the G-quadruplex structure [131].
7.5. BRACO-19
BRACO-19 is an important ligand that is proved to stabilize and promote the formation of a G-quadruplex structure on telomere [132]. It is evident by the previous studies that BRACO-19 inhibits the catalytic property of telomerase, induce telomere shortening, and enable telomerase positive tumor cells to senescence [133]. Using the glioblastoma cancer model, it was recently shown that BRACO carries that cytoplasmic transport of telomerase and consequent reduction in telomerase activity. Additionally, the release of telomere-binding proteins was also seen in this study [134]. Therefore, it is obvious that these dysfunctional telomeres activate the DDR and undergo senescence. Recently, with the help of molecular dynamics simulations with the latest AMBER force field, detailed structural information was gathered about the BRACO-19 [135].
7.6. Telomestatin
It is one of the most popular and well-explored drugs and has been reported as a potent telomerase inhibitor [136]. It is a G-quadruplex stabilizer isolated from Streptomyces anulatus 3533-SV4. In a study conducted by Tahara and others, it was found that telomestatin not only affects telomerase but also dissociates TERF2 protein from telomere and induces loss of 3’ single-stranded overhang [137]. Telomere shortening and the apoptotic effect is observed in many cancer types and thus ultimately results in the induction of anti-cancer phenotypes [[138], [139], [140]].
7.7. T-oligo
T-oligo is an oligonucleotide resembling the 3’ overhang of the telomere. T-oligo is reported to activate DDR (p53, transcription factor E2F1, ATM) and anti-cancer responses (apoptosis, senescence, and differentiation) in malignant cells [39,141], T-oligo is also reported to inhibit growth and induce apoptosis in prostate, ovarian, and breast cancer cells, melanoma and lymphoma [39,[141], [142], [143]].
8. Conclusion
Telomere length as a clinical marker is getting enough attention today in the scientific world. There is a need to explore this field to find new ways to diagnose cancer and treat it. Players of telomere length modulation are specific and interconnecting in function and there is a need to find a common link between them to understand tumor progression. Understanding telomere length homeostasis as well as identifying the key players affecting this homeostasis in cancer can help to unravel new therapeutic opportunities.
Authors’ contributions
RM and VKB wrote and edit the manuscript; RM, VKB, DK and SK contributed to writing and provided experimental results and information. The author(s) read and approved the final manuscript.
Declaration of competing interest
None.
Acknowledgment
The work was supported in part by Start-up Research Grant (SRG/2019/001049), from the Science and Engineering Research Board (SERB), New Delhi, India, and Enhanced Seed grants EF/2018-19/QE04-11 (to R.M.) from Manipal University Jaipur, Rajasthan, India is gratefully acknowledged.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Muller H.J., Herskowitz I.H. Concerning the healing of chromosome ends produced by breakage in Drosophila melanogaster. Am. Nat. 1954;88:177–208. doi: 10.1086/281830. [DOI] [Google Scholar]
- 2.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. http://www.ncbi.nlm.nih.gov/pubmed/17247004%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1209127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zakian V.A. The ends have arrived. Cell. 2009;139:1038–1040. doi: 10.1016/j.cell.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 5.Olovnikov A.M. Telomeres, telomerase, and aging: origin of the theory. Exp. Gerontol. 1996;31:443–448. doi: 10.1016/0531-5565(96)00005-8. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn E.H., Greider C.W., Szostak J.W. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 7.Hastie N.D., Dempster M., Dunlop M.G., Thompson A.M., Green D.K., Allshire R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 8.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 9.Blackburn E.H., Gall J.G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 10.Nimmo E.R., Cranston G., Allshire R.C. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U. S. A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyne J., Ratliff R.L., Moyzis R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olovnikov A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 14.Watson J.D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 15.Greider C.W., Blackburn E.H. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 16.Grandin N., Charbonneau M. Protection against chromosome degradation at the telomeres. Biochimie. 2008;90:41–59. doi: 10.1016/j.biochi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Feng J., Funk W.D., Wang S.S., Weinrich S.L., Avilion A.A., Chiu C.P., Adams R.R., Chang E., Allsopp R.C., Yu J., Le S., West M.D., Harley C.B., Andrews W.H., Greider C.W., Villeponteau B. The RNA component of human telomerase. Science (80-. ) 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 18.Cong Y.S., Wen J., Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 19.Jafri M.A., Ansari S.A., Alqahtani M.H., Shay J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8 doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrumpfová P.P., Fajkus J. Composition and function of telomerase-A polymerase associated with the origin of eukaryotes. Biomolecules. 2020;10 doi: 10.3390/biom10101425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinothkumar V., Arunkumar G., Revathidevi S., Arun K., Manikandan M., Rao A.K.D.M., Rajkumar K.S., Ajay C., Rajaraman R., Ramani R., Murugan A.K., Munirajan A.K. TERT promoter hot spot mutations are frequent in Indian cervical and oral squamous cell carcinomas. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016;37:7907–7913. doi: 10.1007/s13277-015-4694-2. [DOI] [PubMed] [Google Scholar]
- 22.Qu Y., Dang S., Wu K., Shao Y., Yang Q., Ji M., Shi B., Hou P. TERT promoter mutations predict worse survival in laryngeal cancer patients. Int. J. Canc. 2014;135:1008–1010. doi: 10.1002/ijc.28728. [DOI] [PubMed] [Google Scholar]
- 23.Patel B., Taiwo R., Kim A.H., Dunn G.P. TERT, a promoter of CNS malignancies. Neuro-Oncology Adv. 2020;2 doi: 10.1093/noajnl/vdaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A.K., Sharma S., Sengupta S., Saha D., Kumar P., Hussain T., Srivastava V., Roy S.D., Shay J.W., Chowdhury S. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montero J.J., López De Silanes I., Granã O., Blasco M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cacchione S., Biroccio A., Rizzo A. Emerging roles of telomeric chromatin alterations in cancer. J. Exp. Clin. Canc. Res. 2019;38:1–12. doi: 10.1186/s13046-019-1030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen T.H.D., Tam J., Wu R.A., Greber B.J., Toso D., Nogales E., Collins K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018;557:190–195. doi: 10.1038/s41586-018-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Muraki K., Nyhan K., Han L., Murnane J.P. Mechanisms of telomere loss and their consequences for chromosome instability. Front. Oncol. 2012;2:135. doi: 10.3389/fonc.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shay J.W., Wright W.E. Role of telomeres and telomerase in cancer. Semin. Canc. Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shay J.W., Wright W.E. Telomerase and Telomerase. 2012;21:349–353. doi: 10.1016/j.semcancer.2011.10.001.Role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L.C., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Telomeres are specialized structures at the ends of eukaryotic chromosomes that ap-pear to function in chromosome protection, positioning, and replication (1, 2). In ver-tebrates, telomeres consist of hundreds to thousands of tandem repeats of the se-quen. Science (80-. ) 1994;266:2011–2015. http://science.sciencemag.org/ [Google Scholar]
- 33.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Canc. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 34.Leão R., Apolónio J.D., Lee D., Figueiredo A., Tabori U., Castelo-Branco P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J. Biomed. Sci. 2018;25:1–12. doi: 10.1186/s12929-018-0422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J., Zhao Y., Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2010;1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Mar V., Zhou W., Harrington L., Robinson M.O. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascolo E., Wenz C., Lingner J., Hauel N., Priepke H., Kauffmann I., Garin-chesa P., Rettig W.J., Damm K., Schnapp A. Mechanism of human telomerase inhibition by BIBR1532 , a synthetic. Non-nucleosidic Drug Candidate *. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- 38.Liu W., Wang S., Dotsenko I.A., Samoshin V.V., Xue L. Arylsulfanyl groups - suitable side chains for 5-substituted 1,10-phenanthroline and nickel complexes as G4 ligands and telomerase inhibitors. J. Inorg. Biochem. 2017;173:12–20. doi: 10.1016/j.jinorgbio.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Yaar M., Eller M.S., Panova I., Kubera J., Wee L.H., Cowan K.H., Gilchrest B.A. Telomeric DNA induces apoptosis and senescence of human breast carcinoma cells. Breast Cancer Res. 2007;9:1–13. doi: 10.1186/bcr1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Tang W.-J., Shi J.B., Liu M.M., Liu X.-H. Therapeutic strategies for targeting telomerase in cancer. Med. Res. Rev. 2020;40:532–585. doi: 10.1002/med.21626. [DOI] [PubMed] [Google Scholar]
- 41.Bajaj S., Kumar M.S., Peters G.J., Mayur Y.C. Targeting telomerase for its advent in cancer therapeutics. Med. Res. Rev. 2020 doi: 10.1002/med.21674. [DOI] [PubMed] [Google Scholar]
- 42.Ivancich M., Schrank Z., Wojdyla L., Leviskas B., Kuckovic A., Sanjali A., Puri N. Treating cancer by targeting telomeres and telomerase. Antioxidants. 2017;6 doi: 10.3390/antiox6010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura T.M., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R. Telomerase Catalytic Subunit Homologs from Fission Yeast and Human. Science. 2012:955. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 44.Harrington L., Mcphail T., Mar V., Zhou W., Oulton R., A.E.S.T. Program. Bass M.B., Arruda I., Robinson M. Mammaliaian Telornerase-Associated Protein. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 45.Ye J.Z., De Lange T. 2004. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex; p. 36. [DOI] [PubMed] [Google Scholar]
- 46.Takai K.K., Hooper S., Blackwood S., Gandhi R., De Lange T. In Vivo Stoichiometry of Shelterin Components * □. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greider C.W. Regulating telomere length from the inside out : the replication fork model. Genes & Development. 2016:1483–1491. doi: 10.1101/gad.280578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kibe T., Zimmermann M., de Lange T. TPP1 blocks an ATR-mediated resection mechanism at telomeres. Mol. Cell. 2016;61:236–246. doi: 10.1016/j.molcel.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loayza D., De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;vol. 75:1013–1018. doi: 10.1038/nature01720.1. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia C.K., Wright W.E., Shay J.W. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W., Shay J.W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Yuan X., Larsson C., Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38:6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cong Y.-S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/mmbr.66.3.407-425.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Tergaonkar V. Telomerase reactivation in cancers: mechanisms that govern transcriptional activation of the wild-type vs. mutant TERT promoters. Transcription. 2016;7:44–49. doi: 10.1080/21541264.2016.1160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Low K.C., Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013;38:426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Horn S. 2013. TERT Promoter Mutations in Familial; p. 959. [DOI] [PubMed] [Google Scholar]
- 58.Huang F.W., Hodis E., Xu M.J., V Kryukov G., Garraway L.A. HejishuHS Public Access. 2015;339:957–959. doi: 10.1126/science.1229259. [DOI] [Google Scholar]
- 59.Bell R.J.A., Rube H.T., Kreig A., Mancini A., Fouse S.D., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M., John K., Wrensch M.R., Chang S.M., Walsh K.M., Myong S., Song S., Costello J.F. the mutant TERT promoter in cancer. 2015;vol. 348:1036–1039. doi: 10.1126/science.aab0015. The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stern J.L., Theodorescu D., Vogelstein B., Papadopoulos N., Cech T.R. 2010. Mutation of the TERT promoter , switch to active chromatin , and monoallelic TERT expression in multiple cancers; pp. 2219–2224. GENES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu T., Wang N., Cao J., Sofiadis A., Dinets A., Zedenius J., Larsson C., Xu D. 2013. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas; pp. 1–7. [DOI] [PubMed] [Google Scholar]
- 62.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S.C., Kok C.Y., Noble K., Ponting L., Ramshaw C.C., Rye C.E., Speedy H.E., Stefancsik R., Thompson S.L., Wang S., Ward S., Campbell P.J., Forbes S.A. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2018;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Canc. Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peifer M., Hertwig F., Roels F., Dreidax D., Gartlgruber M., Menon R., Roncaioli J.L., Sand F., Heuckmann J.M., Ikram F., Schmidt R., Nu P., Gloeckner C., Bosco G., Leuschner I., Schweiger M.R., Savelyeva L., Watkins S.C., Bu R., Sullivan R.J.O., Westermann F., Thomas R.K., Fischer M. in high-risk neuroblastoma. 2015 doi: 10.1038/nature14980. [DOI] [Google Scholar]
- 66.Valentijn L.J., Koster J., Zwijnenburg D.A., Hasselt N.E., Van Sluis P., Volckmann R., Van Noesel M.M., George R.E., Tytgat G.A.M., Molenaar J.J., Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Publ. Gr. 2015;47:1411–1415. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 67.Tumors H., Zhang A., Zheng C., Lindvall C., Hou M., Ekedahl J., Lewensohn R., Yan Z., Yang X., Henriksson M., Blennow E., Nordenskjo M., Zetterberg A., Bjo M., Gruber A., Xu D. Advances in Brief Frequent Amplification of the Telomerase Reverse Transcriptase Gene in. 2000:6230–6235. [PubMed] [Google Scholar]
- 68.Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q., Lichtenberg T., Hu J., Zhang J., Zheng S., Verhaak R.G.W. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li L.-C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 70.Deaton M., Bird A. 2011. CpG islands and the regulation of transcription; pp. 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee D.D., Leão R., Komosa M., Gallo M., Zhang C.H., Lipman T., Remke M., Heidari A., Nunes N.M., Apolónio J.D., Price A.J., De Mello R.A., Dias J.S., Huntsman D., Hermanns T., Wild P.J., Vanner R., Zadeh G., Karamchandani J., Das S., Taylor M.D., Hawkins C.E., Wasserman J.D., Figueiredo A., Hamilton R.J., Minden M.D., Wani K., Diplas B., Yan H., Aldape K., Akbari M.R., Danesh A., Pugh T.J. DNA hypermethylation within TERT promoter upregulates. TERT expression in cancer. 2019;129:223–229. doi: 10.1172/JCI121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kyo S., Takakura M., Fujiwara T., Inoue M. 2008. Understanding and Exploiting hTERT Promoter Regulation for Diagnosis and Treatment of Human Cancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Lange T., Shelterin The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 74.W. Palm, T. De Lange, How shelterin protects mammalian telomeres, (n.d.). 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed]
- 75.Tobiszewski A., Wityk P., Tomiczek B., Czub J. Molecular Recognition in Complexes of TRF Proteins with Telomeric DNA. PLoS ONE. 2014;vol. 9 doi: 10.1371/journal.pone.0089460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.S. Smith, T. De Lange, Tankyrase promotes telomere elongation in Human Cells, (n.d.) 1299–1302. [DOI] [PubMed]
- 77.Hsiao S.J., Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 78.Stansel R.M., De Lange T., Grif J.D. 2001. T-loop Assembly in Vitro Involves Binding of TRF2 Near the 3 ¢ Telomeric Overhang. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rai R., Zheng H., He H., Luo Y., Multani A., Carpenter P.B., Chang S. The fusion of dysfunctional telomeres. EMBO J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P., Paoletti J., Gilson E., Giraud-Panis M.-J. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 81.Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H.J., De Lange T. The telomeric protein TRF2 binds the ATM Kinase and Can Inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frescas D., De Lange T. 2014. A TIN2 dyskeratosis congenita mutation causes telomerase-independent telomere shortening in mice; pp. 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gottschling D.E., Zakian V.A. Telomere Proteins : Specific Recognition Protection of the Natural Termini of Oxytricha Macronuclear DNA. 1966;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 84.Baumann P., Cech T.R. 2014. Pot1 , the Putative Telomere End-Binding Protein in Fission Yeast and Humans. 1171. [DOI] [PubMed] [Google Scholar]
- 85.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., Connor M.S.O., Songyang Z. TPP1 is a homologue of ciliate TEBP- b and interacts with POT1 to recruit telomerase. Nature. 2007;vol. 445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 86.Horvath M.P., Schweiker V.L., Bevilacqua J.M., Ruggles J.A., Schultz S.C. Crystal Structure of the Oxytricha nova Telomere End Binding Protein Complexed with Single Strand. DNA. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 87.Corriveau M., Mullins M.R., Baus D., Harris M.E., Taylor D.J. Coordinated Interactions of Multiple POT1-TPP1 Proteins with Telomere. DNA * □. 2013;288:16361–16370. doi: 10.1074/jbc.M113.471896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo X., Deng Y., Lin Y., Cosme-blanco W., Chan S., He H., Yuan G., Brown E.J., Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. The EMBO Journal. 2007:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nandakumar J., Bell C.F., Weidenfeld I., Zaug A.J., Leinwand L.A., Cech T.R. Vol. 492. 2013. HejishuHS Public Access; pp. 285–289. The. [DOI] [Google Scholar]
- 90.Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M. The POT1 – TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;vol. 445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 91.Rai R., Chen Y., Lei M., Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., Zhu J., Haussler D. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. BioRxiv. 2019:326470. doi: 10.1101/326470. [DOI] [Google Scholar]
- 93.Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Canc. Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 94.Bejarano L., Bosso G., Louzame J., Serrano R., Gomez Casero E., Martínez Torrecuadrada J., Martínez S., Blanco‐Aparicio C., Pastor J., Blasco M.A. Multiple cancer pathways regulate telomere protection. EMBO Mol. Med. 2019;11:1–21. doi: 10.15252/emmm.201910292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Attikum H., Gasser S.M. Crosstalk between histone modifications during the DNA damage response. Trends in Cell Biology. 2009;2:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 96.Reaper P.M., Clay-farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G. 2003. A DNA damage checkpoint Response in Telomere-Initiated Senescence; p. 426. [DOI] [PubMed] [Google Scholar]
- 97.D'Adda Di Fagagna F., Teo S.H., Jackson S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 98.Takai H., Smogorzewska A., De Lange T. DNA Damage Foci at Dysfunctional Telomeres. 2003;13:1549–1556. doi: 10.1016/S. [DOI] [PubMed] [Google Scholar]
- 99.Tong A.S., Stern J.L., Sfeir A., Kartawinata M., de Lange T., Zhu X.D., Bryan T.M. ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 2015;13:1633–1646. doi: 10.1016/j.celrep.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turner N., Tutt A., Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat. Rev. Canc. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 101.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 102.Lawlor R.T., Veronese N., Pea A., Nottegar A., Smith L., Pilati C., Demurtas J., Fassan M., Cheng L., Luchini C. Alternative lengthening of telomeres (ALT) influences survival in soft tissue sarcomas: a systematic review with meta-analysis. BMC Canc. 2019;19:232. doi: 10.1186/s12885-019-5424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres : Nat. Publ. Gr. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 104.Gocha A.R.S., Nuovo G., Iwenofu O.H., Groden J. Human sarcomas are mosaic for telomerase-dependent and telomerase-independent telomere maintenance mechanisms: implications for telomere-based therapies. Am. J. Pathol. 2013;182:41–48. doi: 10.1016/j.ajpath.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herbert B., Gellert G.C., Hochreiter A., Pongracz K., Wright W.E., Zielinska D., Chin A.C., Harley C.B., Shay J.W., Gryaznov S.M. 2005. Lipid modification of GRN163 , an N3 0 - P5 0 thio -phosphoramidate oligonucleotide , enhances the potency of telomerase inhibition; pp. 5262–5268. [DOI] [PubMed] [Google Scholar]
- 106.Tefferi A., Lasho T.L., Begna K.H., Patnaik M.M., Zblewski D.L., Finke C.M., Laborde R.R., Wassie E., Schimek L., Hanson C.A., Gangat N., Wang X., Pardanani A. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N. Engl. J. Med. 2015;373:908–919. doi: 10.1056/NEJMoa1310523. [DOI] [PubMed] [Google Scholar]
- 107.Mascarenhas J., Komrokji R.S., Cavo M., Martino B., Niederwieser D., Reiter A., Scott B.L., Baer M.R., Hoffman R., Odenike O., Bussolari J., Zhu E., Huang F., Rose E., Sherman L., Dougherty S., Feller F.M., Kiladjian J.-J. Imetelstat is effective treatment for patients with intermediate-2 or high-risk myelofibrosis who have relapsed on or are refractory to Janus kinase inhibitor therapy: results of a phase 2 randomized study of two dose levels. Blood. 2018;132:685. doi: 10.1182/blood-2018-99-115163. [DOI] [Google Scholar]
- 108.Gomez-Millan J., Goldblatt E.M., Gryaznov S.M., Mendonca M.S., Herbert B.-S. Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2007;67:897–905. doi: 10.1016/j.ijrobp.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 109.Gellert G.C., Dikmen Z.G., Wright W.E., Gryaznov S., Shay J.W. 2006. Effects of a novel telomerase inhibitor , GRN163L , in human breast cancer; pp. 73–81. [DOI] [PubMed] [Google Scholar]
- 110.Teo S., Jackson S.P. GENES; 2004. Functional links between telomeres and proteins of the DNA-damage response; pp. 1781–1799. [DOI] [PubMed] [Google Scholar]
- 111.Dikmen Z.G., Wright W.E., Shay J.W., Gryaznov S.M. Telomerase targeted oligonucleotide thio -phosphoramidates in T24-luc bladder. Cancer Cells (Cold Spring Harbor) 2008;452:444–452. doi: 10.1002/jcb.21635. [DOI] [PubMed] [Google Scholar]
- 112.Koi Y., Tsutani Y., Nishiyama Y., Kanda M., Shiroma Y., Yamamoto Y., Sasada S., Akita T., Masumoto N., Kadoya T., Takahashi R., Tanaka J., Okada M., Tahara H. Diagnostic performance of peripheral leukocyte telomere G‐tail length for detecting breast cancer. Canc. Sci. 2020:1–6. doi: 10.1111/cas.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morais S., Gabriel T.R., Arcanjo S., Guimarães G., De Oliveira D.M., Silva F.P. vols. 1–8. 2019. (Long ‐ term in vitro treatment with telomerase inhibitor MST ‐ 312 induces resistance by selecting long telomeres cells). [DOI] [PubMed] [Google Scholar]
- 114.Seimiya H., Oh-hara T., Suzuki T., Naasani I., Shimazaki T., Tsuchiya K., Tsuruo T., Naasani I., Biophys B., Commun R. Vol. 1. 2002. pp. 657–665. (Telomere shortening and growth Inhibition of Human cancer cells by novel Synthetic Telomerase Inhibitors MST-312). [PubMed] [Google Scholar]
- 115.Gurung R.L., Lim S.N., Hande M.P., Kah G., Low M. MST-312 Alters Telomere Dynamics. Gene Expression Profiles and Growth in Human Breast Cancer Cells. 2015;117597:283–298. doi: 10.1159/000381346. [DOI] [PubMed] [Google Scholar]
- 116.Ameri Z., Ghiasi S., Farsinejad A., Hassanshahi G. Telomerase inhibitor MST-312 induces apoptosis of multiple myeloma cells and down-regulation of anti-apoptotic , proliferative and in fl ammatory genes. Life Sci. 2019;228:66–71. doi: 10.1016/j.lfs.2019.04.060. [DOI] [PubMed] [Google Scholar]
- 117.Morais K.S., Guimarãesb A.F.R., Ramos D.A.R., Silva F.P., De Oliveira D.M. Long-term exposure to MST-312 leads to telomerase reverse transcriptase overexpression in MCF-7 breast cancer cells, Anticancer. Drugs. 2017;28:750–756. doi: 10.1097/CAD.0000000000000508. [DOI] [PubMed] [Google Scholar]
- 118.Damm K., Hemmann U., Garin-chesa P., Hauel N., Kauffmann I., Priepke H., Niestroj C., Daiber C., Enenkel B., Guilliard B., Lauritsch I., Mu E., Pascolo E., Sauter G., Pantic M., Martens U.M., Wenz C., Lingner J., Kraut N., Rettig W.J., Schnapp A. 2001. A Highly selective telomerase inhibitor limiting Human Cancer Cell Proliferation; p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bryan C., Rice C., Hoffman H., Harkisheimer M., Sweeney M., Skordalakes E. Structural basis of telomerase inhibition by the highly specific BIBR1532. Structure. 2015;23:1934–1942. doi: 10.1016/j.str.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abbaszadeh Z. 2018. Investigation of the effect of telomerase inhibitor BIBR1532 on breast cancer and breast cancer stem cells; pp. 1–11. [DOI] [PubMed] [Google Scholar]
- 121.Lavanya C., Venkataswamy M.M., Sibin M.K., Bharath M.M.S., Chetan G.K. Down regulation of human telomerase reverse transcriptase ( hTERT ) expression by BIBR1532 in human glioblastoma. Cytotechnology. 2018;70:1143–1154. doi: 10.1007/s10616-018-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu W., Yin Y., Wang J., Shi B., Zhang L., Qian D., Li C., Zhang H., Wang S., Zhu J., Gao L., Zhang Q., Jia B., Hao L., Wang C., Zhang B. Kras mutations increase telomerase activity and targeting telomerase is a promising therapeutic strategy for Kras-mutant NSCLC. Oncotarget. 2017;8:179–190. doi: 10.18632/oncotarget.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bashash D., Zareii M., Safaroghli-azar A., Davood M., Ghaffari S.H. 2017. Inhibition of telomerase using BIBR1532 enhances doxorubicin-induced apoptosis in pre-B acute lymphoblastic leukemia cells; p. 8454. [DOI] [PubMed] [Google Scholar]
- 124.Kaushik M., Kaushik S., Bansal A., Saxena S., Kukreti S. 2011. Structural Diversity and Specific Recognition of Four Stranded G-Quadruplex DNA; pp. 744–769. [DOI] [PubMed] [Google Scholar]
- 125.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human. Genome. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang S., Singh M., Ling M., Li D., Christison K.M., Xue G.P.L.L. CHEMISTRY Synthesis of nucleobase-neomycin conjugates and evaluation of their DNA binding , cytotoxicities , and antibacterial properties. Med. Chem. Res. 2018 doi: 10.1007/s00044-018-2169-x. [DOI] [Google Scholar]
- 127.Chitranshi P., Xue L. Bioorganic & medicinal chemistry letters utilizing G-quadruplex formation to target 8-oxoguanine in telomeric sequences. Bioorg. Med. Chem. Lett. 2011;21:6357–6361. doi: 10.1016/j.bmcl.2011.08.110. [DOI] [PubMed] [Google Scholar]
- 128.Han H., Hurley L.H. Vol. 21. 2000. pp. 136–142. (G-quadruplex Dna : a potential target for anti-cancer drug design). [DOI] [PubMed] [Google Scholar]
- 129.Gowan S.M., Heald R., Stevens M.F.G., Kelland L.R. Potent inhibition of telomerase by small-molecule pentacyclic acridines Capable of Interacting with. G-Quadruplexes. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 130.Riou J.F., Guittat L., Mailliet P., Laoui A., Renou E., Petitgenet O., Mégnin-Chanet F., Hélène C., Mergny J.L. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Z., Liu J. Journal of Molecular Graphics and Modelling Effects of the central potassium ions on the G-quadruplex and stabilizer binding. J. Mol. Graph. Model. 2017;72:168–177. doi: 10.1016/j.jmgm.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 132.Read M., Harrison R.J., Romagnoli B., Tanious F.A., Gowan S.H., Reszka A.P., Wilson W.D., Kelland L.R., Neidle S. 2001. Structure-based design of selective and potent G quadruplex-mediated Telomerase Inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Incles C.M., Schultes C.M., Kempski H., Koehler H., Kelland L.R., Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. 2004;3:1201–1207. [PubMed] [Google Scholar]
- 134.Zhou G., Liu X., Li Y., Xu S., Ma C., Wu X., Cheng Y., Yu Z., Zhao G., Chen Y. Telomere targeting with a novel G-quadruplex-interactive ligand BRACO-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget. 2016;7:14925–14939. doi: 10.18632/oncotarget.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Machireddy B., Sullivan H. 2019. Binding of BRACO19 to a Telomeric G-Quadruplex DNA Probed by All-Atom Molecular Dynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim M., Vankayalapati H., Shin-ya K., Wierzba K., Hurley L.H. Telomestatin , a potent telomerase inhibitor that interacts quite specifically with the human Telomeric Intramolecular. G-Quadruplex. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 137.Tahara H., Seimiya H., Yamada H., Tsuruo T., Ide T. 2006. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3 0 telomeric overhang in cancer cells. 1955–1966. [DOI] [PubMed] [Google Scholar]
- 138.Shammas M.A., Reis R.J.S., Li C., Koley H., Hurley L.H., Anderson K.C., Munshi N.C. Telomerase Inhibition and Cell Growth Arrest after Telomestatin Treatment in Multiple. Myeloma. 2004;vol. 10:770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 139.Nakajima A., Tauchi T., Sashida G., Sumi M., Abe K., Yamamoto K., Ohyashiki J.H., Ohyashiki K. Telomerase inhibition enhances apoptosis in human acute leukemia cells: possibility of antitelomerase therapy. Leukemia. 2003;17:560–567. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 140.Tauchi T., Shin-Ya K., Sashida G., Sumi M., Nakajima A., Shimamoto T., Ohyashiki J.H., Ohyashiki K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 141.N. Puri, M.S. Eller, H.R. Byers, S. Dykstra, J. Kubera, B.A. Gilchrest, Telomere-based DNA damage responses : a new approach to melanoma, (n.d.) 1373–1381. 10.1096/fj.04-1774com. [DOI] [PubMed]
- 142.Sarkar S., V Faller D. T-Oligos Inhibit Growth and Induce Apoptosis in Human Ovarian Cancer Cells. Oligonucleotides. 2011:21. doi: 10.1089/oli.2010.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gnanasekar M., Thirugnanam S., Zheng G., Chen A., Ramaswamy K. T-Oligo induces apoptosis in advanced prostate cancer cells. Oligonucleotides. 2009;19:287–291. doi: 10.1089/oli.2009.0179. [DOI] [PubMed] [Google Scholar]