Summary
Glycolysis is a fundamental metabolic pathway for glucose catabolism across biology, and glycolytic enzymes are among the most abundant proteins in cells. Their expression at such levels provides a particular challenge. Here we demonstrate that the glycolytic mRNAs are localized to granules in yeast and human cells. Detailed live cell and smFISH studies in yeast show that the mRNAs are actively translated in granules, and this translation appears critical for the localization. Furthermore, this arrangement is likely to facilitate the higher level organization and control of the glycolytic pathway. Indeed, the degree of fermentation required by cells is intrinsically connected to the extent of mRNA localization to granules. On this basis, we term these granules, core fermentation (CoFe) granules; they appear to represent translation factories, allowing high-level coordinated enzyme synthesis for a critical metabolic pathway.
Subject areas: molecular biology, cell biology
Graphical abstract
Highlights
-
•
Glycolytic mRNAs colocalize and are translated in RNA granules
-
•
The glycolytic mRNA granules are important when cells are fermenting
-
•
Glycolytic protein translation is likely coordinated in these translation factories
-
•
Therefore, “CoFe granules” are core fermentation factories for glycolytic proteins
Molecular biology; cell biology
Introduction
The glycolytic pathway lies at the core of metabolic activity as a virtually ubiquitous biochemical pathway across living cells. The pathway serves both to supply energy and maintain levels of biochemical intermediates (Bar-Even et al., 2012). Multiple genes express a variety of isoforms for many glycolytic enzymes providing abundant scope for adaptable regulation (Masters et al., 1987; Oparina et al., 2013; Postmus et al., 2012; Warmoes and Locasale, 2014). The pathway was gradually pieced together by a succession of influential biochemists including Meyerhof, Embden, and Parnas (Bar-Even et al., 2012; Barnett, 2005; Schurr and Gozal, 2015). After these major biochemical breakthroughs, interest in central metabolism waned over a period where it was often perceived to perform mundane “housekeeping” functions (Bar-Even et al., 2012; Ray, 2010). More recently, the pathway and its regulation have received renewed interest for various reasons, including connections to cancer and cellular proliferation (Diaz-Ruiz et al., 2011; Gill et al., 2016), moonlighting activities of the glycolytic enzymes (Castello et al., 2015; Kim and Dang, 2005), and increased interest in central metabolism as a focus for metabolic engineering in a synthetic biology era (Lim and Jung, 2017).
In many aerobic cells, the pyruvate produced by glycolysis is transported to and oxidized in the mitochondria via respiration (Gray et al., 2014). However, under anaerobic conditions and in various aerobic cells, such as yeast, lymphocytes, and cancer cells, glucose is fermented through to ethanol or lactic acid. Hence, in these cells glycolysis serves as the major source of ATP and intermediates (Lunt and Vander Heiden, 2011). Indeed, the reduction of pyruvate to ethanol or lactic acid can be viewed as an extension of glycolysis.
Given the critical nature of the glycolytic pathway in energy production and cellular metabolism, it is unsurprising to find that the pathway is regulated by a myriad of different mechanisms. These include direct regulation of the enzymes via substrate and product concentration (Wegner et al., 2015), allosteric enzyme regulation by small molecules (Shen et al., 2016), and post-translational covalent modifications (Shenton and Grant, 2003; Tripodi et al., 2015). Aside from controls of enzymatic activity, other regulatory mechanisms act at the level of gene transcription (Chambers et al., 1995; Yeung et al., 2008), mRNA processing/stability (Krieger and Ernst, 1994; Lunghi et al., 2015), protein stability (Benanti et al., 2007; Lu et al., 2014; Riera et al., 2003), translation (Daran-Lapujade et al., 2007; Man and Pilpel, 2007), and protein localization (Jin et al., 2017). Although clearly the glycolytic enzymes and the mRNAs that encode them can be regulated, they are often viewed as providing “housekeeping” functions. Indeed, in yeast, many of the glycolytic mRNAs are among the most abundant, heavily translated mRNAs in the cell. This raises obvious questions, such as how is this level of gene expression attained both at the transcriptional and post-transcriptional levels? Furthermore, how is this scale of gene expression coordinated across the pathway such that appropriate levels of enzyme are produced to generate a metabolic flux that is pertinent to the cellular conditions?
A number of recent observations have supplemented the understanding of glycolysis and the role of glycolytic enzymes in cells. For instance, it has become evident that a number of glycolytic enzymes “moonlight” as RNA binding proteins (Castello et al., 2015). Indeed, it has been suggested that many of the glycolytic proteins bind to glycolytic mRNAs to orchestrate control of the pathway (Matia-Gonzalez et al., 2015). In addition, the localization of two glycolytic mRNAs in yeast, PDC1 and ENO2, has been identified as important in their translation control and in the formation of mRNA processing bodies or P-bodies (PBs) after glucose starvation (Lui et al., 2014).
mRNA localization has been commonly considered as a means to generate localized sources of protein, with specific examples involved in cellular polarization identified across many biological systems—ASH1 mRNA in yeast (Long et al., 1997), Bicoid in Drosophila oocytes (Berleth et al., 1988), and Vg1 in Xenopus oocytes (Melton, 1987). In these cases, translationally repressed mRNAs are localized in a transit process involving motor proteins and cytoskeletal elements (Besse and Ephrussi, 2008). Another situation where translationally repressed mRNAs become localized is under stress conditions, where non-translated mRNAs can enter either PBs or stress granules (SGs) to play roles in mRNA degradation and/or storage (Hoyle and Ashe, 2008; Hubstenberger et al., 2017; Jain and Parker, 2013). More global assessments of mRNA localization suggest that the phenomena is widespread: large numbers of mRNA species are localized in Drosophila, neuronal cells, and yeast (Gadir et al., 2011; Lecuyer et al., 2007; Miyashiro et al., 1994; Pizzinga and Ashe, 2014; Zipor et al., 2009; Zivraj et al., 2010). Even so, mRNA localization is rarely thought to play a role in core housekeeping functions such as central metabolism.
In this study, we show that glycolytic mRNAs in yeast and human cells are specifically localized to granules. In yeast, we define the core fermentation (CoFe) granule, a core glycolytic mRNA granule where glycolytic mRNAs are colocalized and translated. Translation is a prerequisite for CoFe granules, and individual mRNA translation is required for localization. Finally, we show that the presence of mRNA granules correlates with the degree of glycolytic function required by the cell. We suggest that the localization of these mRNAs provides a means to generate the scale of protein expression required for such a critical pathway and permits rapid coordinated regulation or complex formation.
Results
Glycolytic mRNAs localize to granules under active growth conditions
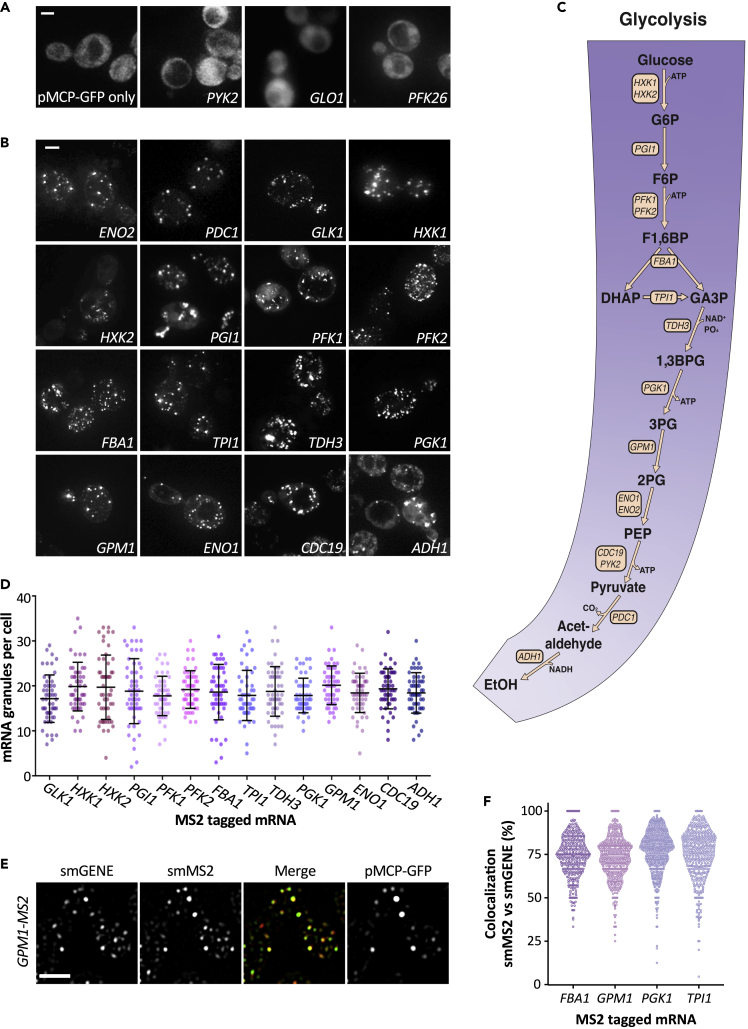
Previous work from our laboratory has highlighted that the glycolytic mRNAs, PDC1 and ENO2, encoding pyruvate decarboxylase and enolase, respectively, are translated in cytoplasmic granules (Lui et al., 2014) (Figure 1A). To evaluate whether glycolytic mRNAs in general are localized to these sites, we have utilized the m-TAG system, where elements of the MS2 bacteriophage are used to tether GFP to an mRNA to study its localization in live cells (Haim-Vilmovsky et al., 2011). Accordingly, MS2 stem loops were directly inserted into the 3’UTR sequences of the glycolytic genes at their endogenous genomic loci. The localization of the resulting mRNAs was then followed via coexpression of the MS2 coat protein-GFP fusion (MS2-CP-GFP). It should be noted that MS2-CP-GFP expression alone generates diffuse fluorescence throughout the cell (Figure 1A). In contrast, when MS2 stem loops are integrated into glycolytic mRNA 3’UTRs, the vast majority of the resulting mRNAs are observed in granules (Figure 1B). This includes mRNAs that encode enzymes at every step of the glycolytic pathway (Figure 1C). Notably, not all MS2-tagged mRNAs localize to granules of this kind (Lui et al., 2014; Simpson et al., 2014); for instance, two non-glycolytic mRNAs, GLO1 and PFK26, where the gene products are involved in the control of glycolysis, and the glycolytic mRNA, PYK2, are not observed in granules (Figure 1A). Previously PDC1 and ENO2 mRNAs were shown to localize to ~20 granules per cell (Lui et al., 2014), and here we show that 14 of the 15 tested glycolytic mRNAs localize to a similar number of granules (Figure 1D).
Figure 1.
MS2-tagged glycolytic mRNAs are localized to granules in S. cerevisiae
(A) and (B) z-stacked images of strains expressing MS2-tagged mRNAs as labeled and the MS2 coat protein GFP fusion. Scale bar: 2 μm.
(C) Diagram of glucose fermentation to ethanol depicting the glycolytic mRNAs investigated in this study.
(D) A dotplot showing the variation in the number of granules per cell for each of the MS2-tagged strains above. n = 50. The mean ± SD are indicated for each strain.
(E) z-stacked images of smFISH performed on strains expressing MS2-tagged mRNAs and the MS2 coat protein GFP fusion. smFISH was performed for the canonical GPM1 gene (smGENE) or the MS2 stem loop sequence (smMS2). Scale bar: 3 μm.
(F) Beeswarm plot showing the proportion of smMS2 foci that colocalize with smGENE foci for a subset of strains expressing MS2-tagged glycolytic mRNAs. Each dot represents a single cell. n > 300.
Recent commentaries have highlighted the potential for an accumulation of fragments of mRNA carrying MS2 stem loops that can impact upon the interpretation of experiments using MS2 tethering systems (Garcia and Parker, 2015, 2016; Haimovich et al., 2016; Heinrich et al., 2017). It is possible that such fragments would accumulate at sites of mRNA degradation. To assess whether this is the case for the granules observed here, a range of approaches were taken. Firstly, it should be noted that all of the experiments presented are conducted on cells actively growing in nutrient replete media. Under these conditions in our experiments, PBs are largely absent (Lui et al., 2014), so the high-level accumulation of mRNA fragments at sites of mRNA decay seems unlikely. Secondly, although most of the glycolytic mRNAs tested are present in granules, the MS2 stem loops have a highly variable impact on the steady state level of the mRNAs: some mRNAs are stabilized, others destabilized, and some remain unchanged (Figure S1A). This profile is not consistent with MS2 fragments explaining the observed localization. Thirdly, the major mRNA species observed on Northern blots under active growth conditions for either the non-tagged or MS2-tagged ENO2 and PDC1 mRNAs were full-length mRNAs (Figure S1B). In contrast, in stressed cells where PBs are present, such as shortly after glucose depletion, degradation fragments for the MS2-tagged mRNAs comprise a high proportion of the total mRNA (Figure S1B). Fourthly, a subset of granule localizing glycolytic mRNAs has been tagged with a version of the MS2 system (termed the “version 6” system) where MS2 fragments do not accumulate (Tutucci et al., 2018). This new MS2 system reveals an identical granular pattern of glycolytic mRNA localization to the original MS2 system (Figure S1C). Finally, a single-molecule fluorescent in situ hybridization (smFISH) strategy where probes were targeted to either the MS2 stem loops or the body of the mRNA revealed greater than 75% signal overlap between these two probes (Figures 1E and 1F). This result suggests that the MS2 region of the mRNA reports the localization of full-length mRNAs. In addition, significant overlap is seen between the MS2-CP-GFP protein signal and either the MS2 RNA probe signal or mRNA body probe signal suggesting that the GFP signal also reports full-length mRNAs (Figure 1F). This overlap is observed despite the fact that the MS2-CP-GFP signal is only seen for those granules that exceed a specific intensity threshold, because, as reported previously, (Pizzinga et al., 2019) the MS2 live cell system predominantly detects multi-mRNA granules. Overall, the combination of different validatory analyses used show that glycolytic mRNAs evaluated using the m-TAG system localize to multi-mRNA granules.
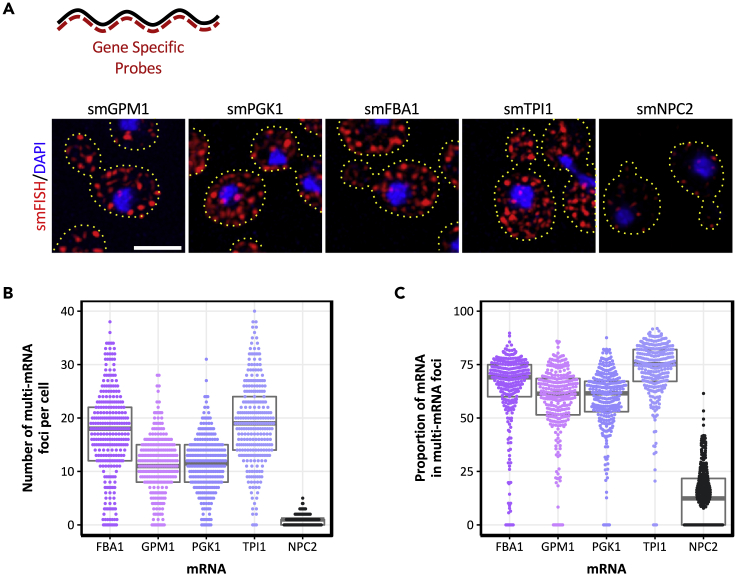
However, insertion of MS2 stem loops could still alter some aspect of an MS2-tagged mRNAs fate. Therefore, in order to provide an independent assessment of the glycolytic mRNA localization, endogenous unmodified mRNAs were evaluated using smFISH. smFISH strategies commonly use ~30–50 fluorescently labeled oligonucleotides that are hybridized to mRNAs in fixed cells (Pizzinga et al., 2019; Tsanov et al., 2016). Because many glycolytic genes are present in yeast as multiple paralogues with very high levels of sequence identity, the use of smFISH to unambiguously study the localization of individual mRNA species is problematic. Therefore, sets of smFISH probes were designed to study the localization of mRNAs encoded by glycolytic genes that either lack paralogues or harbor substantial sequence differences to their paralogues. As a result, four different glycolytic mRNAs, GPM1, FBA1, TPI1, and PGK1, were analyzed and shown to localize specifically to granules (Figure 2A). In terms of the number of granules per cell, the smFISH data for endogenous mRNAs are entirely complementary to the live cell MS2-tagged mRNAs (cf. Figures 2B and 1D). One of the key advantages of the smFISH versus the m-TAG experiments is that for the smFISH data, single mRNA foci are visible. This allows an estimate of mRNA copy number per cell and the proportion of single mRNAs present in multi-mRNA granules (Pizzinga et al., 2019). From the data, it is clear that ~70% of the glycolytic mRNA molecules are present in large granules (Figure 2C). The results correlate well with live cell m-TAG data, where a similar fraction of the PDC1 and ENO2 mRNAs were previously found in multi-mRNA granules (Lui et al., 2014). For the non-glycolytic NPC2 mRNA, which is not localized to large multi-mRNA granules (Pizzinga et al., 2019), a speckled pattern is observed with a reduced, homogeneous fluorescent intensity for each speckle relative to the glycolytic mRNAs. The quantitation of fluorescent intensity profiles shows that the NPC2 mRNA is rarely localized to multi-mRNA granules, and even where NPC2 multi-mRNA granules can be identified, the proportion of mRNA present is very low (Figures 2A–2C). Overall, these data confirm that glycolytic mRNAs are housed in large cytoplasmic bodies or granules and, combined with our concurrent studies on translation factor mRNAs (Pizzinga et al., 2019), indicate that the localization observed with the MS2 system in actively growing cells can be representative of and meaningful to the localization observed for an untagged mRNA.
Figure 2.
smFISH analysis reveals that endogenous glycolytic mRNAs are present in multi-mRNA granules
(A) Upper diagram depicts the smFISH strategy. Multiple probes complementary to an mRNA (black) are tagged with a specific fluorophore (red). Lower panels show z-stacked smFISH images performed for the indicated endogenous glycolytic mRNAs. Both mRNAs (red) and nuclei (blue) are shown. Dotted lines represent the extent of the cell from brightfield micrographs. Scale bar: 3 μm.
(B) Beeswarm plot showing the number of multi-mRNA (>2.5mRNAs) granules per cell for a number of endogenous mRNAs. The gray box and line represent the interquartile range and the median, respectively. Each dot represents a single cell. n > 300.
(C) Beeswarm plot showing the proportion of mRNA that resides within multi-mRNA granules (>2.5mRNAs) per cell. Gray box and line represent the interquartile range and the median, respectively. n > 300.
Glycolytic mRNAs colocalize to the same RNA granules
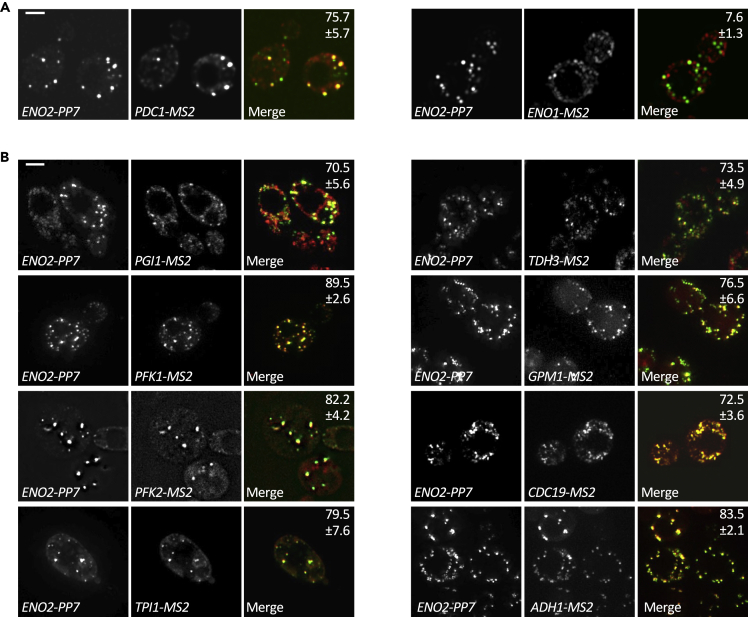
Our previous assessment of the ENO2 and PDC1 mRNAs suggested that these two mRNAs colocalize to the same set of granules (Lui et al., 2014) (Figure 3A) but are distinct from the translation factor mRNA granules we have recently described as factories for the production and inheritance of the translation machinery (Pizzinga et al., 2019). On this basis, we speculated that the colocalization of glycolytic mRNAs might generally allow a concerted production and/or regulation of the pathway of the glycolytic enzymes. More recent work highlights the potential for cotranslational assembly of components of the glycolytic pathway (Shiber et al., 2018), which again hints that actively translating glycolytic mRNAs might colocalize. In both the live cell and fixed cell mRNA localization experiments presented here, both the pattern and number of mRNA granules in a cell is remarkably similar across the different glycolytic mRNAs (cf. Figures 1C and 2A; cf Figures 1D and 2B). This similarity is consistent with a model where many of the glycolytic mRNAs colocalize to the same site.
Figure 3.
Glycolytic mRNAs colocalize to granules in actively growing cells
(A) and (B) z-stacked images show the localization of various MS2-tagged mRNAs (via coexpression of the MS2-CP-mCh fusion) relative to the ENO2-PP7 mRNA (visualized using coexpression of the PP7-CP-GFP fusion). The percentage of observable tagged mRNA colocalizing with the PP7-tagged mRNA is indicated ±SD. Scale bars: 2 μm.
In order to directly assess glycolytic mRNA colocalization, we made use of a PP7/MS2 system, which allows the simultaneous visualization of two mRNAs in the same live cell (Hocine et al., 2013; Lui et al., 2014; Pizzinga et al., 2019). A series of yeast strains were generated carrying PP7-tagged ENO2 mRNA and another MS2-tagged glycolytic mRNA. By coexpressing the MS2 and PP7 coat proteins fused to mCherry and GFP respectively, the localization of each MS2-tagged mRNA was compared directly with that of ENO2 mRNA in the same living cell. As previously shown (Lui et al., 2014), we observed a strong colocalization of PDC1 with ENO2 using this system (Figure 3A). Equally, for many of the glycolytic enzymes tested, a high degree of colocalization with the ENO2 mRNA pattern was observed (Figure 3B). Interestingly, however, despite the high sequence homology between ENO2 and ENO1 mRNAs, these mRNAs localized to discrete foci (Figure 3A).
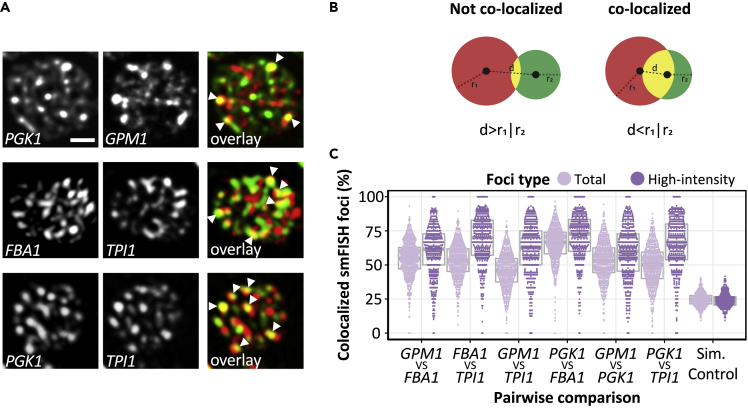
In order to corroborate the colocalization observed using the PP7/ MS2 systems, we assessed pairwise colocalization of endogenous unmodified glycolytic mRNAs using smFISH (Figure 4A). Because, in contrast to the MS2 system where only multi-mRNA granules can be followed, the smFISH technique detects all of the probed mRNA present in a cell, the signal is very congested. This congestion is particularly apparent for glycolytic mRNAs, which are typically estimated to be present at >100 copies per cell (Lahtvee et al., 2017). So, in order to objectively measure pairwise colocalization, a computational strategy was developed. In short, the distance between the centroid of a granule for one mRNA species was measured relative to the centroid of the nearest neighboring granule for the other mRNA species. A spot was deemed to colocalize if its centroid was within the sphere of a spot in the opposite channel (Figure 4B). Using this method, a significant proportion (~50%–60%) of the glycolytic mRNA granules were deemed to overlap with one another (Figure 4C). Because each mRNA is present in ~20 multi-mRNA foci per cell (Figure 2B) with a similar number of single mRNA foci, we were concerned that high levels of colocalization could simply stem from the proportion of cytosolic space occupied by the mRNA foci. Therefore, to control for this, we established a Monte Carlo simulation model (Fletcher et al., 2010), where the position of real foci were randomized within the cell accounting for vacuole space and cross-compared with randomized simulated foci for a second mRNA. From this analysis it is clear that relative to the simulated model, the various tested glycolytic mRNAs display significant colocalization (Figure 4C). Although these smFISH results mirror the colocalization observed using the MS2 and PP7 stem loop systems (Figure 3), the scale of colocalization appears lower. One possible reason for the lower colocalization reflects the extra sensitivity of the smFISH technique in detecting single mRNA foci, which may not colocalize to the same extent as multi-mRNA granules. To explore this, we considered only the most intense foci in the smFISH data, which likely represent multi-mRNA containing foci. This analysis revealed a positive shift in the level of colocalization by ~10%–20% (Figure 4C). Importantly, simulated controls using these brighter, larger spots did not display this shift in colocalization (Figure 4C).
Figure 4.
smFISH confirms that glycolytic mRNAs colocalize in granules
(A) z-stacked images from smFISH colocalization studies using the designated probes. Scale bar: 1 μm.
(B and C) (B) Diagram detailing the colocalization method used to generate data in panel. (C) The centroid of both spots must be within the radius of either channel in order for spots to be deemed colocalized (d < r1|r2). Spots that are touching are not always colocalized, if the distance between centroids is greater than the radius of both channels (d > r1|r2). (C) Beeswarm plot showing the proportion of colocalized smFISH foci considering total foci or only high-intensity foci, as indicated. Colocalization was assessed in a pairwise manner using smFISH foci identified via Fish-quant (see Methods). Simulated colocalization was assessed by sub-sampling foci properties across a number of pairwise comparisons (see Methods). Gray box and line represent the interquartile range and the median, respectively. Each data point represents a single cell, n > 300.
Overall, the smFISH results combined with the live cell studies reveal that in optimally growing yeast there is a high-degree of colocalization to multi-mRNA granules for the majority of glycolytic mRNAs tested. Not every localized glycolytic mRNA colocalizes, for instance the ENO1 mRNA does not, but the vast majority do. These colocalized mRNAs encode enzymes that catalyze most of the reactions that are required for glucose fermentation to ethanol; therefore, we have termed the granules “core fermentation” mRNA granules or CoFe granules.
Glycolytic mRNA granules do not reside on the ER or mitochondria
A number of studies have described the role of organelles in the localization of specific mRNAs in yeast (Fundakowski et al., 2012: Gadir et al., 2011). Given data showing that mRNAs encoding non-organellar proteins can also be enriched with organelles (Jan et al., 2014), it is plausible that the localization of glycolytic mRNAs to multi-mRNA granules could be occurring on specific organelles such as the ER or mitochondria. Although the profile of the glycolytic mRNAs does not necessarily match the known localization pattern for these organelles, we could not rule out that the CoFe granules are somehow anchored or stabilized by interactions with these organelles. In order to test this idea, the localization of the glycolytic mRNAs was assessed in strains carrying the fluorescent tagged organelle markers Sec63p (ER) and Cox4p (mitochondria) (Figure S2). For both markers little, if any, evidence of colocalization was observed. Instead, the glycolytic mRNA granules appear largely separated from either the ER or the mitochondria, suggesting that their structure and localization are not reliant upon either of these organelles.
mRNA translation both occurs in and is required for localization to CoFe granules
Previously, we have shown that in contrast to most mRNA containing granules, which carry translationally repressed mRNAs, the granules housing the glycolytic mRNAs PDC1 and ENO2 are sites where these mRNAs are translated (Lui et al., 2014). A variety of experiments supported this hypothesis, including data from FRAP assays where newly synthesized unbleached protein accumulated in the granules (Lui et al., 2014). In addition, treatment of cells with cycloheximide, which freezes ribosomes on mRNA, caused an increase in the number of mRNA granules. Furthermore, the quantification of ribosome-associated mRNA relative to granule-associated mRNA showed that even though 95% of these mRNAs are translated, at least 70% are present in granules. Finally, under polysome run-off conditions a rapid coalescence of the mRNA granules to form PBs was observed, suggesting that prior to the stress the mRNAs were in a distinct granule, being actively translated.
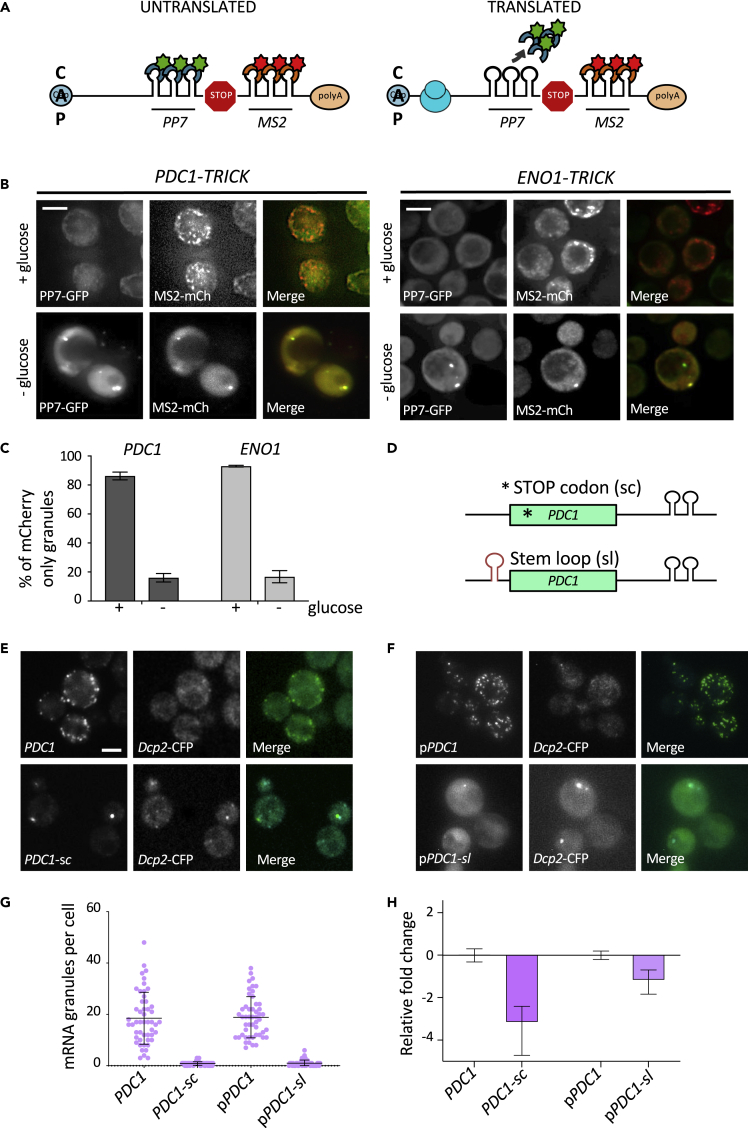
To extend this analysis further, a technique called TRICK (translating RNA imaging by coat protein knockoff) was used, which allows visualization of translation in live cells (Halstead et al., 2015; Pizzinga et al., 2019). This technique relies upon observations that a PP7 coat protein fusion bound to PP7 stem loops upstream of the STOP codon is displaced under active translation conditions, whereas the MS2 coat protein fusion tethered downstream of the STOP codon remains associated (Halstead et al., 2015) (Figure 5A). For PDC1 mRNA under active growth conditions, most granules observed only carry the MS2-CP-mCherry (MS2-CP-mCh) fusion protein (Figures 5B and 5C). In contrast, after a 10-min glucose depletion to elicit a robust and global inhibition of protein synthesis (Ashe et al., 2000), both MS2-CP-mCh and PP7-CP-GFP colocalize to granules (Figures 5B and 5C). This result supports our previous work showing that under active growth conditions the glycolytic mRNAs such as PDC1 and ENO1 are translated in granules, and combined with the colocalization studies, suggest that the glycolytic mRNA granules serve as factories for glycolytic enzyme production.
Figure 5.
mRNAs translation in CoFe granules is required for localization
(A) Schematic of TRICK reporter system. Ribosomes on translated RNAs “knock off” the PP7-CP-GFP fusion, whereas on untranslated RNAs the coat protein remains bound.
(B) z-stacked images of TRICK-tagged mRNAs coexpressing the MS2-CP-mCh fusion and the PP7-CP-GFP fusion, in + and – glucose. Scale bars: 3 μm.
(C) Quantification of MS2-CP-mCh-only granules as a percentage of total granules observed in TRICK-tagged mRNAs in + and – glucose conditions. Error bars are ±SD.
(D) Schematic of PDC1 premature stop codon (sc) and stem loop (sl) insertion.
(E) z-stacked images of cells expressing Dcp2p-CFP- and PDC1-MS2-tagged mRNA. PDC1-MS2 (sc) has a premature stop codon in the ORF.
(F) z-stacked images of strains expressing Dcp2p-CFP with pPDC1-MS2 or pPDC1-MS2 (sl). pPDC1-MS2 (sl) has a stem loop upstream of the ORF.
(G) Scatterplot of mRNA granules per cell in PDC1-MS2-tagged mRNA with or without a premature stop codon and in strains bearing pPDC1-MS2 with or without the stem loop. Error bars are ±SD. Scale bars: 2 μm.
(H) Relative fold change of (1) PDC1 MS2-tagged mRNA relative to untagged PDC1 mRNA, (2) PDC1-MS2 mRNA in strains harboring a premature stop codon (sc) relative to a strain without, (3) PDC1-MS2 mRNA on a plasmid (pPDC1-MS2 mRNA) relative to genomic PDC1-MS2 mRNA, and finally (4) pPDC1-MS2 mRNA in strain with a stem loop upstream of the ORF relative to a pPDC1-MS2 mRNA without a stem loop. Error bars are ±SD.
Another key question is whether translation of a molecule of mRNA is a requirement for entry into the granule. In order to address this question, we selected the PDC1 mRNA and sought to limit its translation, then assess the impact on localization. More specifically, we adopted two different strategies toward reducing PDC1 mRNA translation. In the first approach a STOP codon was inserted immediately downstream of the translation START codon (PDC1-sc) (Figure 5D). We reasoned that this would severely reduce the number of ribosomes associated with this mRNA and significantly increase the pool of non-translated PDC1 mRNA. As a second strategy, a stem loop was inserted into the PDC1 mRNA 5’UTR, upstream of the START codon (PDC1-sl) (Figure 5D). Introduction of this well-characterized stem loop (ΔG value of −41 kcal/mol) has previously been shown to reduce translation of specific mRNAs by limiting scanning of the 43S preinitiation complex through to the AUG Start codon (Palam et al., 2011; Pizzinga et al., 2019; Vattem and Wek, 2004). In strains carrying these altered PDC1 mRNAs, mRNA localization was followed relative to the non-modified mRNA using the MS2 system.
Introduction of either the STOP codon or the stem loop structure into the PDC1 mRNA dramatically reduced the number of PDC1-MS2 mRNA granules: decreasing from ~20 granules per cell to less than 5 (Figures 5E–5G). Coincident with this effect on the number of mRNA granules in the cell, both strategies used to limit PDC1 mRNA translation also resulted in reduced mRNA levels (Figure 5H). Insertion of the STOP codon caused an ~8-fold reduction in PDC1-MS2 mRNA, whereas stem loop insertion reduced mRNA levels ~2-fold. The reduction of mRNA caused by the introduction of the STOP codon is consistent with premature STOP codons leading to nonsense mediated mRNA decay (Hagan et al., 1995). The impact of the stem loop on PDC1 mRNA levels is not as pronounced as the STOP codon insertion, and it is a little surprising that this insertion leads to mRNA destabilization, as this same stem loop has been inserted into a number of mRNAs without impacting upon overall mRNA levels (Palam et al., 2011; Pizzinga et al., 2019; Vattem and Wek, 2004). This suggests that the context of a stem loop in the 5’UTR of an mRNA is important in determining to what extent the insertion impacts upon the fate of the mRNA. These results highlight the intimate connection between the translation of an mRNA and its stability and add to many observations showing that a reduction in translation can lead to mRNA destabilization (Roy and Jacobson, 2013).
A surprising observation was made when the localization of the PB marker Dcp2p was assessed in cells bearing either the PDC1-sc or PDC1-sl mRNAs. In both strains, Dcp2p was constitutively present in PBs even in unstressed cells, and the PDC1-sc and PDC1-sl mRNAs colocalized with these bodies (Figures 5E and 5F). This result is especially intriguing as generally PBs are barely visible unless cells are stressed in some way (Lui et al., 2014). Yet in these unstressed cells, just a single point mutation to introduce an STOP codon into one mRNA species is sufficient to induce PB formation. This result is also interesting with regard to the controversy surrounding MS2 tagging. The specific introduction of a mutation that inhibits translation changes the mRNA localization pattern dramatically and causes PB formation. Therefore, if the RNA granules observed for non-mutated glycolytic mRNAs during exponential growth (Figure 1C) were due to the accumulation of RNA fragments carrying the MS2 stem loops, we would expect a similar colocalization with PB markers. However, we do not observe such colocalization with PB markers in unstressed cells (Lui et al., 2014; Pizzinga et al., 2019).
Overall, these results highlight that in keeping with many observations over the years it is difficult to alter the translation of an mRNA without affecting its stability (Mugridge et al., 2018; Roy and Jacobson, 2013). However, the results do suggest that as well as translation occurring in granules, inhibiting translation of individual glycolytic mRNAs changes the fate of those mRNAs so that instead of entering the granules for translation, alternative mRNA fates become apparent, such as relocalization to PBs.
Glycolytic mRNA localization varies according to the level of fermentation
In order to understand the potential physiological role the CoFe granules play, experiments were undertaken where yeast were grown on a range of carbon sources selected based upon the pathways required for carbon source metabolism. For example, although yeast cells ferment glucose to ethanol even under aerobic conditions, for other carbon sources the degree of fermentation varies. Yeast cells grown on ethanol as the sole carbon source derive their energy from respiration and only require the glycolytic enzymes for gluconeogenesis. Raffinose is catabolized initially via the action of the secreted enzymes invertase (Suc2p) and α-galactosidase (Mel1p). These enzymes yield the monosaccharides glucose, fructose and galactose, which are readily available for fermentation via the glycolytic enzymes (Barnett, 1976). Equally, yeast that are pre-adapted to galactose express enzymes of the Leloir pathway, allowing fermentation of this sugar via entry into the glycolytic pathway (Timson, 2007).
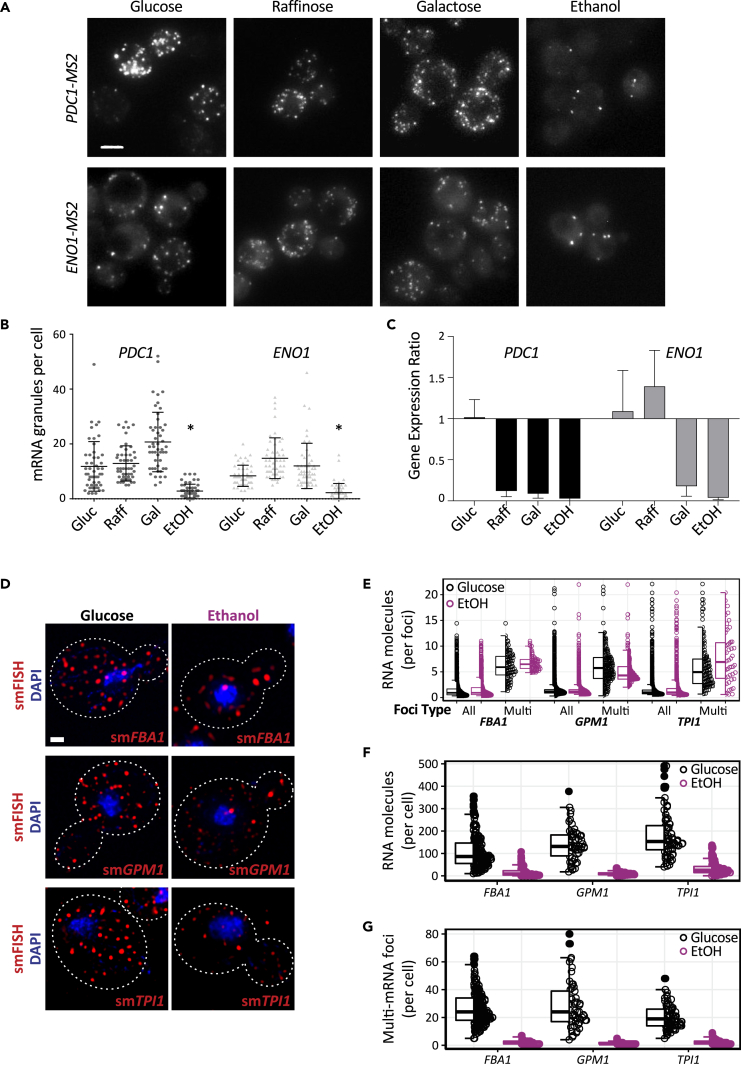
Microscopic analysis revealed that yeast cells grown on glucose, raffinose, or galactose harbored approximately 10–20 granules of either PDC1 mRNA or ENO1 mRNA per cell, whereas cells grown on ethanol harbored significantly fewer granules (Figures 6A and 6B). In terms of the levels of the PDC1 and ENO1 mRNAs, these also vary with carbon source, consistent with glucose representing the preferred yeast carbon source. For both mRNAs, glucose grown cells harbor significantly more glycolytic mRNA than cells grown on most other carbon sources (Figure 6C).
Figure 6.
CoFe granule number varies with quality of carbon source
(A) z-stacked images of PDC1-MS2 or ENO1-MS2 mRNA in strains coexpressing the MS2-CP-GFP fusion, grown in SC media with either 2% glucose, 2% raffinose, 2% galactose, or 3% ethanol. Scale bar: 2 μm.
(B) Quantification of mRNA granules per cell for PDC1 and ENO1 mRNA in strains grown in the different carbon sources (Gluc = Glucose, Raff = Raffinose, Gal = Galactose, EtOH = Ethanol). n = 50. Error bars are ±SD. ∗p < 0.005 relative to other columns from a 1-way ANOVA and the Tukey's HSD test.
(C) Graph representing the gene expression ratio of PDC1 or ENO1 in strains (yMK1586 and yMK2468) grown in the different carbon sources. The gene expression ratio was calculated according to the Pfaffl method. This approach considers the PCR efficiency for the different genes and is expressed as the change in target gene levels (PDC1 and EN O1) between glucose conditions and the different carbon sources over the change in reference gene levels (ACT1) between glucose conditions and the different carbon sources (Pfaffl, 2001) (Gluc = Glucose, Raff = Raffinose, Gal = Galactose, EtOH = Ethanol). Error bars are ±SD.
(D) z-stacked images of FBA1, GPM1, and TPI1 mRNAs probed using smFISH in DAPI-stained wild-type strains. Strains were grown in SC media with either 2% glucose or 3% ethanol. Scale bar: 1 μm.
(E) Graph representing the number of mRNA molecules within each smFISH foci in cells grown in either glucose (black) or ethanol (EtOH; magenta). Foci were separated into either all foci or multi mRNA foci dependent upon the number of mRNAs predicted to reside within that foci (see Methods). Box and line represent the interquartile range and the median, respectively
(F) Quantification of total mRNA molecules per cell in cells grown in either glucose (black) or ethanol (EtOH; magenta), as measured by smFISH. Box and line represent the interquartile range and the median, respectively
(G) Quantification of the number of multi-mRNA containing foci per cell in cells grown in either glucose (black) or ethanol (EtOH; magenta). Box and line represent the interquartile range and the median, respectively
Similar observations were made when the localization of the FBA1, GPM1, and TPI1 mRNAs was investigated for cells grown on either glucose or ethanol using smFISH (Figure 6D). For yeast grown on either carbon source multi-mRNA granules were observed (Figures 6D and 6E), and, consistent with qRT-PCR results above, the number of mRNA molecules per cell was reduced for the ethanol grown cells (Figure 6F). However, despite this, the number of mRNA molecules per multi-RNA granule was similar for yeast grown on either carbon source (Figure 6E), suggesting some form of regulated recruitment to the RNA granule. Perhaps the most striking difference in the localization of the mRNAs was the number of multi-mRNA granules, with ethanol leading to dramatically reduced numbers of RNA granules (Figure 6G), consistent with MS2 experiments performed previously (Figure 6A).
Therefore, these results show that both the level of glycolytic mRNAs and prevalence of CoFe granules vary depending on the carbon sources. In particular, the presence of CoFe granules appears to correlate with a requirement for glycolytic flux to utilize the provided carbon source, with growth on a respiratory carbon source causing a dramatic reduction in the number of granules. Overall, the data are consistent with a view that the localization of glycolytic mRNAs to CoFe granules represents a strategy allowing high-level coordinated production of glycolytic enzymes in translation factories.
Glycolytic mRNA granules are also evident in human cells
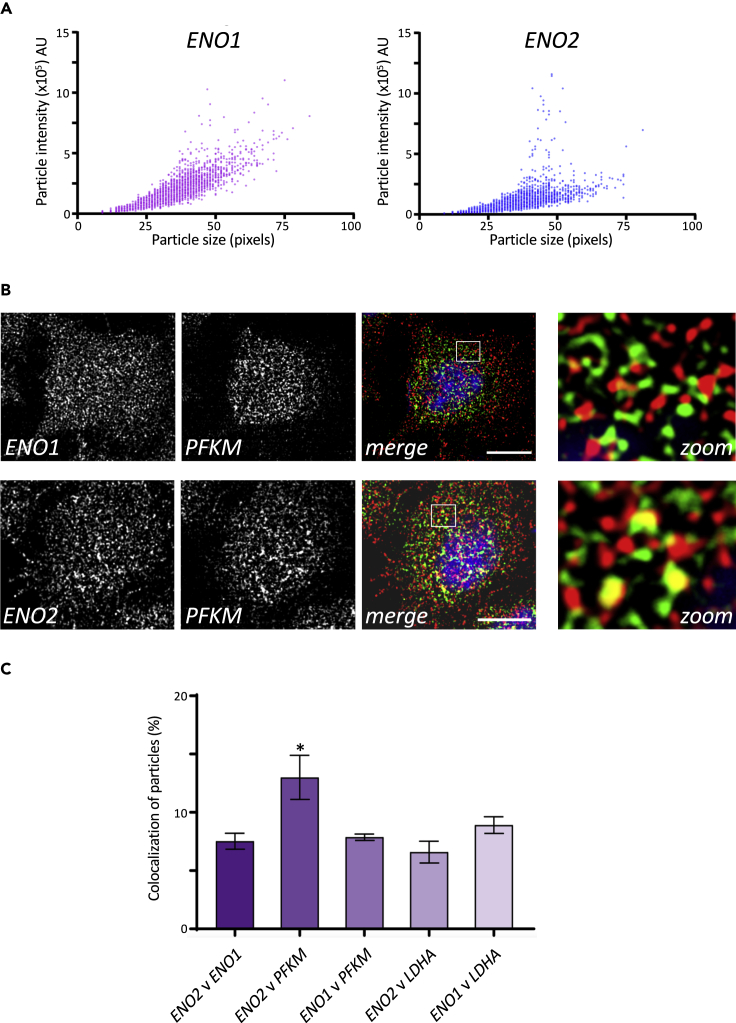
In order to assess whether a similar organization of glycolytic mRNAs might exist in higher eukaryotic cells, smFISH analysis was conducted for four different glycolytic mRNAs in HeLa cells: two enolase mRNAs (ENO1 and ENO2), a lactate dehydrogenase mRNA (LDHA), and a phosphofructokinase mRNA (PFKM). For all four of the selected mRNAs, variation in mRNA signal was observed both in terms of particle size and intensity (Figure 7A, data not shown). In particular, large intense mRNA foci were observed for the glycolytic mRNAs (Figure 7) that were not present for other highly expressed mRNAs such as ACTB mRNA (Figure S3). This suggests that granules harboring multiple mRNAs can also be a feature for glycolytic mRNAs in higher eukaryotic cells. Similar observations were made in other cell lines such as HFF-1 cells and SH-SY5Y cells (data not shown). This opens up the possibility that glycolytic mRNAs might be coordinately localized in higher cells. Therefore, a multichannel smFISH approach was taken. Here, evidence for a specific colocalization of the ENO2 and PFKM mRNAs was obtained (Figures 7B and 7C). Although the degree of colocalization is not as comprehensive as observed in yeast, these data do show that in actively growing human tissue culture cells, glycolytic mRNAs can be localized to granules and that these granules can contain more than one type of glycolytic mRNA.
Figure 7.
Human glycolytic mRNAs are present in granules and can colocalize
(A) Scatterplot of particles detected in z-stacked smFISH images of HeLa cells using probes to the mRNAs indicated. Images from three biological replicates were analyzed using the ImageJ ComDet plugin, which generates readouts of particle size (measured in pixels where each pixel = 45 × 45 nm) and the fluorescent intensity of particles.
(B) Single z-slice smFISH images of HeLa cells using probes to the mRNAs indicated. Scale bar: 10 μm. Insets magnified x2.
(C) Histogram showing the percentage of colocalized mRNA particles calculated using ComDet analysis of z-slices from three biological replicates. The significance across the various combinations was calculated using one-way ANOVA and the Tukey's HSD test. ENO2 versus PFKM (p value <0.05 shown by asterisk) is significantly different to ENO2 versus ENO1, ENO1 versus PFKM, or ENO2 versus LDHA. Error bars are ±SD.
Discussion
mRNA localization serves critical functions in the expression of proteins at specific loci within cells and in the response to stress in terms of PB and SG formation (Pizzinga and Ashe, 2014). In this study, we suggest that mRNA localization to granules can co-ordinate whole pathways of metabolism. We use a combination of live cell experiments and smFISH to show that glycolytic mRNAs localize to granules in yeast and human cells. In stark contrast to mRNAs localizing to PBs, SGs, or transport granules, in yeast the glycolytic mRNAs are translated in CoFe granules, and their translation is a requirement for localization.
Recent evidence suggests that liquid-liquid phase separation (LLPS) within cells produces membraneless compartments or biological condensates where enzymatic reactions and processes can occur. For instance, in the nucleolus, rRNA is produced via numerous highly complex reactions (Brangwynne et al., 2011), whereas in the centrosome microtubule nucleation occurs (Zwicker et al., 2014). The CoFe granules described here conform to many of the properties of phase-separated condensates: they are dynamic, can be observed to fuse, and are disrupted by low concentrations of 1,6-hexanediol (Lui et al., 2014). Therefore, our data suggest that translated glycolytic mRNAs are present in such biological condensates where molecular processes are not only maintained but might actually be enhanced (Kojima and Takayama, 2018).
Enhanced translation of mRNA is therefore one possible explanation as to why the glycolytic mRNAs would be localized within granules. Previous observations from our lab have shown that up to 95% of the glycolytic mRNAs are translated (Lui et al., 2014). In addition, the glycolytic mRNAs are among the most abundant in the cell and so may require rather specific mechanisms to maintain their high rates of translation. Equally LLPS has previously been associated with altered efficiency of a host of biological processes and enzymes (Zhou et al., 2008), so translation may prove to represent another example of such a process, especially where the coordinated generation of high volumes of glycolytic enzyme may be important.
Another possible rationale for localized mRNA translation is to aid the formation of multi-protein complexes. Many of the glycolytic enzymes are present in multimeric complexes. For example, almost all of the glycolytic enzymes function as multimers: in yeast the phosphofructokinase enzyme is present as an octamer (Schwock et al., 2004), phosphoglycerate mutase and pyruvate kinase are tetramers (Jurica et al., 1998; Rigden et al., 1998), and enolase is dimeric (Sims et al., 2006). Co-translation of individual mRNAs at the same site within cells could therefore aid the formation and productive folding pathways for these complexes. We have not formally shown that different mRNA species are cotranslated at these sites. However, in previous work we have shown that for two different glycolytic mRNA species over 90% of these mRNAs are associated with heavy polysomes, whereas 60%–70% of these mRNAs are associated with the mRNA granules (Lui et al., 2014). The fact that most glycolytic mRNAs, including those that are coassociated in granules, are being actively translated, is suggestive that cotranslation at these granules is also occurring. A range of precedents exist for the cotranslational production of complexes across various biological systems (Halbach et al., 2009; Kamenova et al., 2019; Shiber et al., 2018; Wells et al., 2015), whereas a systematic analysis in Schizosaccharomyces pombe suggests that cotranslational production of protein complexes is widespread, with a substantial fraction of proteins copurifying with mRNAs that encode interacting proteins (Duncan and Mata, 2011). Notably, in recent work characterizing the propensity for cotranslational folding in several different protein complexes in Saccharomyces cerevisiae, the PFK1 and PFK2 phosphofructokinase mRNAs were identified as key examples where the translated products are cotranslationally assembled or folded (Shiber et al., 2018).
It has also been shown that, as well as forming multimeric single enzyme complexes, various different glycolytic enzymes can be compartmentalized into much larger complexes (Masters, 1991). A variety of observations suggest that the physical compartmentalization of glycolysis is advantageous. For instance, in protozoan organisms such as Trypanosoma and Leishmania, a specific membrane-bound organelle called the glycosome has evolved, which is thought to provide these pathogens a scope for regulating metabolic activity (Haanstra et al., 2016). Furthermore, in human cells, such as skeletal muscle cells, neurons, and erythrocytes, glycolytic enzymes can be organized as complexes coordinated either on membranes or the cytoskeleton (Knull and Walsh, 1992; Puchulu-Campanella et al., 2013). Moreover, a glycolytic metabolon has also been described in yeast (Masters, 1991), and it is thought to be stabilized by various weak interactions with actin (Araiza-Olivera et al., 2013). These multi-enzyme complexes are likely to promote both the channeling of metabolites from one enzyme to the next, as well as the reduction of potentially toxic intermediates. More recent work in yeast has shown that although glycolytic enzymes are broadly cytosolic under non-stress conditions, they can coalesce into “G-bodies” in response to hypoxic stress (Jin et al., 2017). Overall, therefore, the coproduction of the glycolytic enzymes at the same site by virtue of coordinated mRNA localization could promote the cotranslational formation of some of these higher order complexes of enzymes. Although, it should be noted that our own data suggest that under active growth conditions fluorescent-protein tagged forms of the glycolytic enzymes are generally found throughout the cytosol (Lui et al., 2014).
Another point worth reflecting upon when considering the role of the CoFe granules is that several glycolytic enzymes have extra-glycolytic or “moonlighting” functions outside of their role in glycolysis. For example, many of the glycolytic enzymes have been identified as RNA-binding proteins that appear to interact with their own mRNA (Castello et al., 2015; Matia-Gonzalez et al., 2015). In addition, yeast enolase is important for both the mitochondrial import of tRNALysCUU (Entelis et al., 2006) and for vacuole fusion (Decker and Wickner, 2006), whereas yeast fructose-1,6-bisphosphate aldolase is important for vacuolar H+-ATPase function (Lu et al., 2007). Many further moonlighting functions of glycolytic enzymes, including nuclear functions in transcription, DNA replication/repair, and histone modification have been described (Boukouris et al., 2016). One possible explanation for the presence of CoFe granules could be that they serve as a focus for the coordinated high-level production of the glycolytic machinery en masse, whereas individual translated glycolytic mRNAs outside of factories could provide the capacity for moonlighting protein activities.
One intriguing observation made during the course of our studies is that although the yeast ENO1 mRNA is observed to localize to granules, it does not appear to colocalize with the ENO2 mRNA in CoFe granules. Several possible non-mutually exclusive explanations could account for this. Firstly, the expression levels of the enolase isoforms vary greatly depending upon the fermentation/ respiration status; Eno2p represents the predominant polypeptide in fermenting cells, whereas the distribution is much more even in stationary phase or respiring cells (Entian et al., 1987). So the discrete localization of the two mRNAs could contribute to these expression differences. Secondly, since the enolase enzyme is dimeric, and both Eno2p and Eno1p homodimers, as well as the heterodimer have been described (Holland et al., 1982), the discrete localization of the ENO1 and ENO2 mRNAs could serve to regulate the relative proportion of these different complexes via cotranslational homodimer formation. The rationale for requiring homodimers seems unlikely to reside in their enzymatic function, as the various forms display very similar enzyme kinetics (Holland et al., 1982). Therefore, an alternative possibility relates to the enolase moonlighting functions, with Eno2p playing a more active role than Eno1p in the targeting of the nuclear-encoded tRNALysCUU isoacceptor to mitochondria (Entelis et al., 2006).
Overall, glycolysis is perhaps the most fundamental of all biological pathways. The enzymes of the pathway are regulated at almost every level, and paralogues have evolved distinct functions. The pathology of many disease conditions is intimately connected to the glycolytic pathway. For instance, aerobic glycolysis serves as a hallmark of many malignant cancers and the surrounding stroma, which can serve as a negative prognostic indicator due to increased resistance to therapy (Lee and Yoon, 2015; Ngo et al., 2015). The identification and characterization of factories for the production of glycolytic proteins can only serve to increase understanding of the functions, regulation, and possibility for genetic adjustment of this key metabolic pathway.
Limitations of the study
Although we have interpreted our data to mean that cotranslation of different colocalized mRNAs is occurring in translation factories, due to technical limitations we have not directly shown the translation of two or more mRNAs at the same site. However, in this study and previous work, we have shown translation of single mRNA species occurs at these sites. We have shown that multiple mRNAs colocalize to the sites and that for any single glycolytic mRNA, over 90% of the mRNA is engaged with heavy polysomes. We have interpreted these results to mean that cotranslation of different mRNAs is indeed occurring at the same site.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Mark Ashe (mark.p.ashe@manchester.ac.uk).
Materials availability
Strains and plasmids generated as part of this study are available upon request.
Data and code availability
Code for bespoke yeast cell smFISH analysis is available at the GitHub repository github.com/CPBS/MoralesPolanco.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
We thank L. Berchowitz, J. Gerst, J. Chao, and R. Singer for reagents; P. March and S. Marsden for microscopy advice; and E. Linney for comments on the manuscript. FMP was supported by a CONICYT Becas Chile studentship (72140307). CB and MP were supported by Wellcome Trust (WT) PhD studentships (210002/Z/17/Z and 099732/Z/12/Z). JL, CG, and GF were supported by a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (BB/K005979/1). JC and KG were supported by a BBSRC project grant (BB/P018270/1). The Bioimaging microscopes used in this study were funded by grants from BBSRC, WT, and the University of Manchester Strategic Fund.
Author contributions
All authors generated reagents, performed experiments, evaluated results, and generated figures. FMP, CB, JL, JC, and MPA conceived the study and designed experiments, whereas MPA wrote the manuscript. All authors contributed to the discussion and evaluation of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 19, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2021.102069.
Supplemental information
References
- Araiza-Olivera D., Chiquete-Felix N., Rosas-Lemus M., Sampedro J.G., Pena A., Mujica A., Uribe-Carvajal S. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013;280:3887–3905. doi: 10.1111/febs.12387. [DOI] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A., Flamholz A., Noor E., Milo R. Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat. Chem. Biol. 2012;8:509–517. doi: 10.1038/nchembio.971. [DOI] [PubMed] [Google Scholar]
- Barnett J.A. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 1976;32:125–234. doi: 10.1016/s0065-2318(08)60337-6. [DOI] [PubMed] [Google Scholar]
- Barnett J.A. Glucose catabolism in yeast and muscle. Comprehensive Biochemistry. 2005;44:1–132. [Google Scholar]
- Benanti J.A., Cheung S.K., Brady M.C., Toczyski D.P. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 2007;9:1184–1191. doi: 10.1038/ncb1639. [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F., Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Boukouris A.E., Zervopoulos S.D., Michelakis E.D. Metabolic enzymes moonlighting in the Nucleus: metabolic regulation of gene transcription. Trends Biochem. Sci. 2016;41:712–730. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Hentze M.W., Preiss T. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 2015;26:746–757. doi: 10.1016/j.tem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A., Packham E.A., Graham I.R. Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae) Curr. Genet. 1995;29:1–9. doi: 10.1007/BF00313187. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P., Rossell S., van Gulik W.M., Luttik M.A., de Groot M.J., Slijper M., Heck A.J., Daran J.M., de Winde J.H., Westerhoff H.V. The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc. Natl. Acad. Sci. U S A. 2007;104:15753–15758. doi: 10.1073/pnas.0707476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker B.L., Wickner W.T. Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J. Biol. Chem. 2006;281:14523–14528. doi: 10.1074/jbc.M600911200. [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R., Rigoulet M., Devin A. The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Duncan C.D., Mata J. Widespread cotranslational formation of protein complexes. PLoS Genet. 2011;7:e1002398. doi: 10.1371/journal.pgen.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis N., Brandina I., Kamenski P., Krasheninnikov I.A., Martin R.P., Tarassov I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K.D., Meurer B., Kohler H., Mann K.H., Mecke D. Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1987;923:214–221. doi: 10.1016/0304-4165(87)90006-7. [DOI] [PubMed] [Google Scholar]
- Fletcher P.A., Scriven D.R., Schulson M.N., Moore E.D. Multi-image colocalization and its statistical significance. Biophys. J. 2010;99:1996–2005. doi: 10.1016/j.bpj.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fundakowski J., Hermesh O., Jansen R.P. Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic. 2012;13:1642–1652. doi: 10.1111/tra.12011. [DOI] [PubMed] [Google Scholar]
- Gadir N., Haim-Vilmovsky L., Kraut-Cohen J., Gerst J.E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.F., Parker R. MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA. 2015;21:1393–1395. doi: 10.1261/rna.051797.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.F., Parker R. Ubiquitous accumulation of 3' mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays. RNA. 2016;22:657–659. doi: 10.1261/rna.056325.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill K.S., Fernandes P., O'Donovan T.R., McKenna S.L., Doddakula K.K., Power D.G., Soden D.M., Forde P.F. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim. Biophys. Acta. 2016;1866:87–105. doi: 10.1016/j.bbcan.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Gray L.R., Tompkins S.C., Taylor E.B. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014;71:2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanstra J.R., Gonzalez-Marcano E.B., Gualdron-Lopez M., Michels P.A. Biogenesis, maintenance and dynamics of glycosomes in trypanosomatid parasites. Biochim. Biophys. Acta. 2016;1863:1038–1048. doi: 10.1016/j.bbamcr.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Hagan K.W., Ruiz-Echevarria M.J., Quan Y., Peltz S.W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim-Vilmovsky L., Gadir N., Herbst R.H., Gerst J.E. A genomic integration method for the simultaneous visualization of endogenous mRNAs and their translation products in living yeast. RNA. 2011;17:2249–2255. doi: 10.1261/rna.029637.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G., Zabezhinsky D., Haas B., Slobodin B., Purushothaman P., Fan L., Levin J.Z., Nusbaum C., Gerst J.E. Use of the MS2 aptamer and coat protein for RNA localization in yeast: a response to "MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system". RNA. 2016;22:660–666. doi: 10.1261/rna.055095.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A., Zhang H., Wengi A., Jablonska Z., Gruber I.M., Halbeisen R.E., Dehe P.M., Kemmeren P., Holstege F., Geli V. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009;28:2959–2970. doi: 10.1038/emboj.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead J.M., Lionnet T., Wilbertz J.H., Wippich F., Ephrussi A., Singer R.H., Chao J.A. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Sidler C.L., Azzalin C.M., Weis K. Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA. 2017;23:134–141. doi: 10.1261/rna.057786.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S., Raymond P., Zenklusen D., Chao J.A., Singer R.H. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods. 2013;10:119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M.J., Holland J.P., McAllister L. Structure and expression of yeast glycolytic genes. Basic Life Sci. 1982;19:291–303. doi: 10.1007/978-1-4684-4142-0_23. [DOI] [PubMed] [Google Scholar]
- Hoyle N.P., Ashe M.P. Subcellular localization of mRNA and factors involved in translation initiation. Biochem. Soc. Trans. 2008;36:648–652. doi: 10.1042/BST0360648. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157 e145. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., Weissman J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Parker R. The discovery and analysis of P Bodies. Adv. Exp. Med. Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- Jin M., Fuller G.G., Han T., Yao Y., Alessi A.F., Freeberg M.A., Roach N.P., Moresco J.J., Karnovsky A., Baba M. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 2017;20:895–908. doi: 10.1016/j.celrep.2017.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Mesecar A., Heath P.J., Shi W., Nowak T., Stoddard B.L. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- Kamenova I., Mukherjee P., Conic S., Mueller F., El-Saafin F., Bardot P., Garnier J.M., Dembele D., Capponi S., Timmers H.T.M. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019;10:1740. doi: 10.1038/s41467-019-09749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Dang C.V. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Knull H.R., Walsh J.L. Association of glycolytic enzymes with the cytoskeleton. Curr. Top. Cell. Regul. 1992;33:15–30. doi: 10.1016/b978-0-12-152833-1.50007-1. [DOI] [PubMed] [Google Scholar]
- Kojima T., Takayama S. Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition. ACS Appl. Mater. Interfaces. 2018;10:32782–32791. doi: 10.1021/acsami.8b07573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger K., Ernst J.F. Iron regulation of triosephosphate isomerase transcript stability in the yeast Saccharomyces cerevisiae. Microbiology. 1994;140:1079–1084. doi: 10.1099/13500872-140-5-1079. [DOI] [PubMed] [Google Scholar]
- Lahtvee P.J., Sanchez B.J., Smialowska A., Kasvandik S., Elsemman I.E., Gatto F., Nielsen J. Absolute quantification of protein and mRNA abundances demonstrate variability in gene-specific translation efficiency in yeast. Cell Syst. 2017;4:495–504.e95. doi: 10.1016/j.cels.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee M., Yoon J.H. Metabolic interplay between glycolysis and mitochondrial oxidation: the reverse Warburg effect and its therapeutic implication. World J. Biol. Chem. 2015;6:148–161. doi: 10.4331/wjbc.v6.i3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.H., Jung G.Y. A simple method to control glycolytic flux for the design of an optimal cell factory. Biotechnol. Biofuels. 2017;10:160. doi: 10.1186/s13068-017-0847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Singer R.H., Meng X., Gonzalez I., Nasmyth K., Jansen R.P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Lu M., Ammar D., Ives H., Albrecht F., Gluck S.L. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 2007;282:24495–24503. doi: 10.1074/jbc.M702598200. [DOI] [PubMed] [Google Scholar]
- Lu W., Zhang Y., McDonald D.O., Jing H., Carroll B., Robertson N., Zhang Q., Griffin H., Sanderson S., Lakey J.H. Dual proteolytic pathways govern glycolysis and immune competence. Cell. 2014;159:1578–1590. doi: 10.1016/j.cell.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J., Castelli L.M., Pizzinga M., Simpson C.E., Hoyle N.P., Bailey K.L., Campbell S.G., Ashe M.P. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 2014;9:944–954. doi: 10.1016/j.celrep.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunghi M., Galizi R., Magini A., Carruthers V.B., Di Cristina M. Expression of the glycolytic enzymes enolase and lactate dehydrogenase during the early phase of Toxoplasma differentiation is regulated by an intron retention mechanism. Mol. Microbiol. 2015;96:1159–1175. doi: 10.1111/mmi.12999. [DOI] [PubMed] [Google Scholar]
- Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Man O., Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat. Genet. 2007;39:415–421. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- Masters C. Cellular differentiation and the microcompartmentation of glycolysis. Mech. Ageing Dev. 1991;61:11–22. doi: 10.1016/0047-6374(91)90003-i. [DOI] [PubMed] [Google Scholar]
- Masters C.J., Reid S., Don M. Glycolysis--new concepts in an old pathway. Mol. Cell. Biochem. 1987;76:3–14. doi: 10.1007/BF00219393. [DOI] [PubMed] [Google Scholar]
- Matia-Gonzalez A.M., Laing E.E., Gerber A.P. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 2015;22:1027–1033. doi: 10.1038/nsmb.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D.A. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Miyashiro K., Dichter M., Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc. Natl. Acad. Sci. U S A. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge J.S., Coller J., Gross J.D. Structural and molecular mechanisms for the control of eukaryotic 5'-3' mRNA decay. Nat. Struct. Mol. Biol. 2018;25:1077–1085. doi: 10.1038/s41594-018-0164-z. [DOI] [PubMed] [Google Scholar]
- Ngo H., Tortorella S.M., Ververis K., Karagiannis T.C. The Warburg effect: molecular aspects and therapeutic possibilities. Mol. Biol. Rep. 2015;42:825–834. doi: 10.1007/s11033-014-3764-7. [DOI] [PubMed] [Google Scholar]
- Oparina N.Y., Snezhkina A.V., Sadritdinova A.F., Veselovskii V.A., Dmitriev A.A., Senchenko V.N., Mel'nikova N.V., Speranskaya A.S., Darii M.V., Stepanov O.A. Differential expression of genes that encode glycolysis enzymes in kidney and lung cancer in humans. Genetika. 2013;49:814–823. doi: 10.7868/s0016675813050111. [DOI] [PubMed] [Google Scholar]
- Palam L.R., Baird T.D., Wek R.C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzinga M., Ashe M.P. Yeast mRNA localization: protein asymmetry, organelle localization and response to stress. Biochem. Soc. Trans. 2014;42:1256–1260. doi: 10.1042/BST20140086. [DOI] [PubMed] [Google Scholar]
- Pizzinga M., Bates C., Lui J., Forte G., Morales-Polanco F., Linney E., Knotkova B., Wilson B., Solari C.A., Berchowitz L.E. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol. 2019;218:1564–1581. doi: 10.1083/jcb.201704019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postmus J., Aardema R., de Koning L.J., de Koster C.G., Brul S., Smits G.J. Isoenzyme expression changes in response to high temperature determine the metabolic regulation of increased glycolytic flux in yeast. FEMS Yeast Res. 2012;12:571–581. doi: 10.1111/j.1567-1364.2012.00807.x. [DOI] [PubMed] [Google Scholar]
- Puchulu-Campanella E., Chu H., Anstee D.J., Galan J.A., Tao W.A., Low P.S. Identification of the components of a glycolytic enzyme metabolon on the human red blood cell membrane. J. Biol. Chem. 2013;288:848–858. doi: 10.1074/jbc.M112.428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray L.B. Metabolism. Metabolism is not boring. Introduction. Science. 2010;330:1337. doi: 10.1126/science.330.6009.1337. [DOI] [PubMed] [Google Scholar]
- Riera L., Obach M., Navarro-Sabate A., Duran J., Perales J.C., Vinals F., Rosa J.L., Ventura F., Bartrons R. Regulation of ubiquitous 6-phosphofructo-2-kinase by the ubiquitin-proteasome proteolytic pathway during myogenic C2C12 cell differentiation. FEBS Lett. 2003;550:23–29. doi: 10.1016/s0014-5793(03)00808-1. [DOI] [PubMed] [Google Scholar]
- Rigden D.J., Alexeev D., Phillips S.E., Fothergill-Gilmore L.A. The 2.3 A X-ray crystal structure of S. cerevisiae phosphoglycerate mutase. J. Mol. Biol. 1998;276:449–459. doi: 10.1006/jmbi.1997.1554. [DOI] [PubMed] [Google Scholar]
- Roy B., Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr A., Gozal E. Glycolysis at 75: is it time to tweak the first elucidated metabolic pathway in history? Front. Neurosci. 2015;9:170. doi: 10.3389/fnins.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwock J., Kirchberger J., Edelmann A., Kriegel T.M., Kopperschlager G. Interaction of 6-phosphofructokinase with cytosolic proteins of Saccharomyces cerevisiae. Yeast. 2004;21:483–494. doi: 10.1002/yea.1114. [DOI] [PubMed] [Google Scholar]
- Shen Q., Wang G., Li S., Liu X., Lu S., Chen Z., Song K., Yan J., Geng L., Huang Z. ASD v3.0: unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 2016;44:D527–D535. doi: 10.1093/nar/gkv902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D., Grant C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiber A., Doring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., Bukau B. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018;561:268–272. doi: 10.1038/s41586-018-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.E., Lui J., Kershaw C.J., Sims P.F., Ashe M.P. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 2014;127:1254–1262. doi: 10.1242/jcs.139055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims P.A., Menefee A.L., Larsen T.M., Mansoorabadi S.O., Reed G.H. Structure and catalytic properties of an engineered heterodimer of enolase composed of one active and one inactive subunit. J. Mol. Biol. 2006;355:422–431. doi: 10.1016/j.jmb.2005.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson D.J. Galactose metabolism in Saccharomyces cerevisiae. Dyn. Biochem. Process. Biotechnol. Mol. Biol. 2007;1:63–73. [Google Scholar]
- Tripodi F., Nicastro R., Reghellin V., Coccetti P. Post-translational modifications on yeast carbon metabolism: regulatory mechanisms beyond transcriptional control. Biochim. Biophys. Acta. 2015;1850:620–627. doi: 10.1016/j.bbagen.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Tsanov N., Samacoits A., Chouaib R., Traboulsi A.M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E., Vera M., Biswas J., Garcia J., Parker R., Singer R.H. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods. 2018;15:81–89. doi: 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmoes M.O., Locasale J.W. Heterogeneity of glycolysis in cancers and therapeutic opportunities. Biochem. Pharmacol. 2014;92:12–21. doi: 10.1016/j.bcp.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A., Meiser J., Weindl D., Hiller K. How metabolites modulate metabolic flux. Curr. Opin. Biotechnol. 2015;34:16–22. doi: 10.1016/j.copbio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Wells J.N., Bergendahl L.T., Marsh J.A. Co-translational assembly of protein complexes. Biochem. Soc. Trans. 2015;43:1221–1226. doi: 10.1042/BST20150159. [DOI] [PubMed] [Google Scholar]
- Yeung S.J., Pan J., Lee M.H. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipor G., Haim-Vilmovsky L., Gelin-Licht R., Gadir N., Brocard C., Gerst J.E. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 2009;106:19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj K.H., Tung Y.C., Piper M., Gumy L., Fawcett J.W., Yeo G.S., Holt C.E. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D., Decker M., Jaensch S., Hyman A.A., Julicher F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. U S A. 2014;111:E2636–E2645. doi: 10.1073/pnas.1404855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code for bespoke yeast cell smFISH analysis is available at the GitHub repository github.com/CPBS/MoralesPolanco.