Abstract
Hippo Tumor Suppressor Pathway is the main pathway for cell growth that regulates tissue enlargement and organ size by limiting cell growth. This pathway is activated in response to cell cycle arrest signals (cell polarity, transduction, and DNA damage) and limited by growth factors or mitogens associated with EGF and LPA. The major pathway consists of the central kinase of Ste20 MAPK (Saccharomyces cerevisiae), Hpo (Drosophila melanogaster) or MST kinases (mammalian) that activates the mammalian AGC kinase dmWts or LATS effector (MST and LATS). YAP in the nucleus work as a cofactor for a wide range of transcription factors involved in proliferation (TEA domain family, TEAD1-4), stem cells (Oct4 mononuclear factor and SMAD-related TGFβ effector), differentiation (RUNX1), and Cell cycle/apoptosis control (p53, p63, and p73 family members). This is due to the diverse roles of YAP and may limit tumor progression and establishment. TEAD also coordinates various signal transduction pathways such as Hippo, WNT, TGFβ and EGFR, and effects on lack of regulation of TEAD cancerous genes, such as KRAS, BRAF, LKB1, NF2 and MYC, which play essential roles in tumor progression, metastasis, cancer metabolism, immunity, and drug resistance. However, RAS signaling is a pivotal factor in the inactivation of Hippo, which controls EGFR-RAS-RAF-MEK-ERK-mediated interaction of Hippo signaling. Thus, the loss of the Hippo pathway may have significant consequences on the targets of RAS-RAF mutations in cancer.
Keywords: Cancer, Hippo pathway, Metastasis, Signaling, Tumor suppressor
Introduction
The most common oncogenic pathway in different types of tumor is the RAS transduction pathway and also the active mutations responsible for the direct growth and survival signals.1,92,93 Mutation in all RAS isoforms (K-RAS, H-RAS, and N-RAS) enhances the active form of the protein. In addition, the normal signal for wild-type RAS is membrane tyrosine kinase receptors that can be activated by mutation, gene replication, or high levels of receptor-ligand (EGF).2 Mitogen and other chemicals (such as lysophosphatidic acid and sphingosine-1-phosphate) increase growth and can also send a signal directly to RAS via G-protein-coupled receptors. This has led to the activation of heterotrimeric G-proteins and Gαi,3 that are capable of targeting RAS activity by activating G-protein-independent, β-arrestin, and SRC.4 Also, activation or mutation of G-protein-coupled receptors in cancer can increase cell proliferation activity and survival pathways.92 Undoubtedly, wild-type RAS increases by changing endogenous control mechanisms. It is important that by increasing the expression of GTPase exchange factors or by decreasing the expression of GTPase activating proteins (hydrolysis of GTP to GDP), RAS changes, eventually resulting in the elimination of neurofibromatosis protein 1.5,92 Indeed, after activation, RAS signaling is mediated through interaction with RAS-binding domains (RAF kinases, A-RAF, B-RAF, or C-RAF (RAF-1), PI3K, Tiam1, and RGS12/14) or through the domain RAS association (RA), such as RalGDS, Afadin-6, Rin1, phospholipase C epsilon, and RASSF proteins (RASSF1-10) transmitted to downstream effectors.6,7 Notably, many downstream effectors are oncogenes or tumor suppressor genes that are mutated or silenced in cancers independently of RAS (B-RAF, PI3K, RalGDS, and RASSF1).92 In fact, mitogen-activated protein kinase (MAPK), PI3K-AKT, and RASSF1-MST (Hippo) are downstream pathways that activated in these cases. These pathways also mediate the interaction between EGFR-RAS signaling and the Hippo tumor suppressor pathway. Thus, the mitogen-activated protein kinase cascade comprises several related parallel pathways that activate a set of extracellular regulated kinase 1/2, JNK1/2, p38α/β/γ/δ or extracellular regulated kinase 5.8 Although growth and inflammation can activate different amounts of these events, however, RAS is highly correlated with JNK activation as well as p38 with extracellular regulated kinase 1/2 activation. Activation of RAF kinases by RAS stimulates MEK1/2 activation, which in turn MEK1/2 activates kinases of extracellular regulated kinase 1/2 and increases proliferation by stimulating cell entry into the cell cycle.9 Remarkably, this process supported by a number of scaffolding proteins and places the cascade forming units adjacent to the cell, which also enhances the control of the output signal. Beside, Scaffold proteins may be essential to increase signal diffusion. These proteins are also capable of directing the mechanical position of a signal in a cell and stimulating the output domain of the signal to create the proper cellular response. Many scaffold proteins disrupt cancer cells and cause inappropriate levels of RAS signaling (RAS-RAF scaffolds, CNK, RAF-MEK scaffolds KSR and IQGAP).10,11 In principle, MP1 supports MEK-ERK, and the RASSF1 scaffold also restricts MEK activation (Fig. 1).12,13
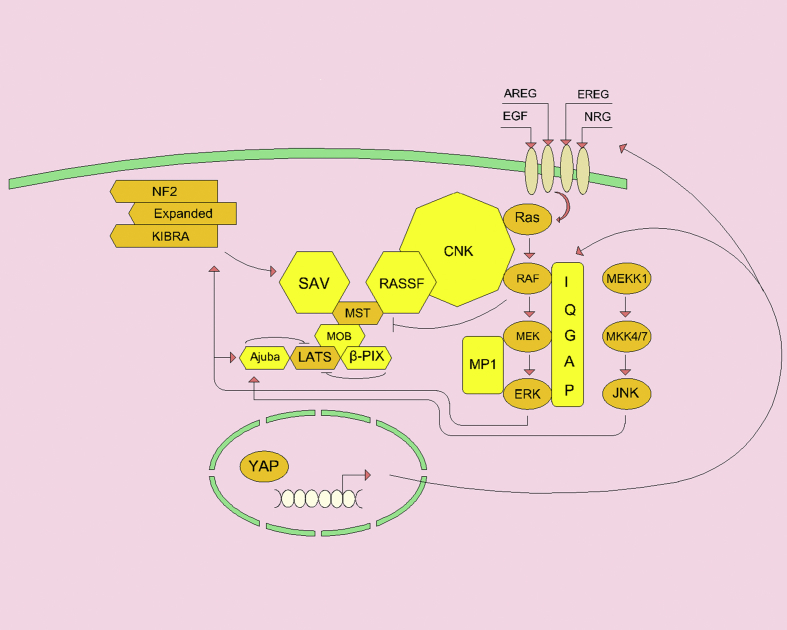
Figure 1.
MOB, RASSF, SAV scaffolding proteins play an essential role in activating the Hippo pathway. β-PIX and Ajuba scaffolds inhibit LATS's ability to phosphorylate YAP and trigger a positive response to JNK or ERK phosphorylation. ERK can phosphorylate KIBRA (upstream activator of SAV) and increase growth. RAS-MAPK scaffolds enhance MAPK signaling and help to suppress Hippo signaling. Activation of YAP transcription increases IQGAP, EGFR, and growth factors.
Certainly, RAS is a potent oncogene and the occurrence of additional mutations (N-RAS and B-RAF mutations in melanoma) in its pathway causes the development of a neoplasm.14 However, mutations (K-RAS, B-RAF, or PI3K) can occur in normal human or mouse tissues without tumorigenesis, that indicate mutations in the RAS pathway limit tumor suppressor pathways.15, 16, 17, 18 Importantly, p53 responds to RAS activation by stimulating the cell cycle, that in most cancers p53 is mutated; accordingly, although germline mutations or loss of TP53 induce tumorigenesis independently of RAS, however, somatic changes are a late event in tumor progression that is often related to metastatic expansion.19, 20, 21 This fact indicates that family members associated with TP53 (p63 and p73) or other tumor suppressor pathways play an important role in the early stages of tumorigenesis, which will certainly help to identify the RAS pathway of cancer.
Hippo pathway
Hippo Tumor Suppressor Pathway
A major pathway for cell growth is the Hippo tumor suppressor pathway that regulates tissue and organ growth by limiting cell growth.22 This pathway is also activated in response to cell cycle arrest signals (cellular polarity, transduction, and DNA damage) and blocked by growth factors or mitogens associated with EGF and LPA.23,24 It should be noted that the main pathway consists of the central kinase Ste 20 MAPK (Saccharomyces cerevisiae), Hpo (Drosophila melanogaster) or MST kinases (mammalian) that activate the mammalian AGC kinase dmWts or LATS effector (MST and LATS).25 The major output of LATS-mediated phosphorylation pathway is the co-factor yes-associated protein (YAP) (dmYki) that participates in various activities (biological responses associated with different transcription factors).26 The number of inputs of cell polarity regulators (Crumbs, Fat, Par 6/aPkc) and FERM domains upstream signal inducer proteins are very limited, as well as direct binding modulators regulate Hippo kinase activity including scaffolds Sav1 (hWW45, dmSalvador), RASSF1-6 (dmRASSF) and MOB1 (dmMats), which actually these regulators support LATS dimerization.27, 28, 29 Importantly, developmental studies indicate that this pathway limits the proliferation of cancer cells in model organisms and human tumors.23 However, genetic alterations in the components of the path increase the risk of tumors that somatic mutations play a central role.30, 31, 32 In fact, activation of the pathway may depend on the type of tumor selected, due to the diverse roles of YAP (proliferation, differentiation, apoptosis and cell cycle arrest) that may limit tumor progression and establishment.33, 34, 35 Studies show that in cancer, Hippo-pathway signaling is deactivated epigenetically by YAP proliferation, deletion (MST2), or gene silencing (MST, LATS, and RASSF1).26,36, 37, 38, 39, 40 Other evidence suggests that in breast cancer, bladder and brain tumors, methylation-associated gene silencing of the RAS and Hippo pathway scaffold (RASSF1) increase by activation of YAP.41 Other studies have shown that the loss of Hippo activator correlates with clinical outcomes in mice.42, 43, 44 However, RAS signaling is an essential factor in Hippo deactivation, with reports suggesting thatEGFR-RAS-RAF-MEK-ERK-mediated interaction controls Hippo signaling.45, 46, 47, 48 Accordingly, loss of the Hippo pathway may have important consequences on the targets of RAS-RAF mutations in cancer.48 That is why we intend to discuss the details of this route.
LATS and MST
The mammalian pathway consists of two central kinases: 1- MST, STe20-like kinases 1 and 2 (MST1 and MST2), homologs of Saccharomyces cerevisiae Ste 20 kinase. 2- LATS; Tumor Suppressors LATS1 and LATS2, homologs of the mitotic yeast Dbf2 kinase.49 Activation of MST1/2 occurs by homo-dimerization or hetero-dimerization and trans-phosphorylation of the remaining T183/T180 in the ATP-binding pocket. This activation (MST1/2) is essential for the coordination of ATP in hydrolysis.25 Dimerization of MST kinases also occurs via the leucine zipper motif at the C-terminal,22 which found as the SARAH domain in the SAV, RASSF1-6, and Hippo kinases. The domains of MST1 and MST2 kinases are located at the N-terminal and are released by apoptosis via caspase cleavage, indicating that the C-terminal sequences restrict kinase activity. Thus, between the kinase domain and the SARAH domains is the auto-inhibitory α-helix that limits the activity of the monomer kinase.25
The SAV and RASSF proteins can bind to the monomeric MST and eliminate the kinase activity of LATS or eliminate its inhibitory activity through dimerization of the RASSF-MST heterodimer.22 RASSF1-MST:MST-RASSF1 complex cause the auto-transphosphorylation of T183/T180.28,50 Evidence suggests that RASSF1, RASSF2, and RASSF5 (NORE1/RAPL) maintain MST kinases in the inhibitory state until induces dimerization, whereas RASSF3, RASSF4, and RASSF6 resemble dRASSF and suppress its activity.51,52 RASSF1 and RASSF5, however, may affect dimerization capacity or activation of MST.53 Interestingly, wild-type RAS activation may play an inhibitory role in activating the Hippo pathway.37 It is important to note that RAF is a RAS effector that binds directly to the MST1/2 SARAH domain and prevents dimerization and association with RASSF1, which limits the activity of the Hippo pathway in mice and Drosophila melanogaster.50,54
The evidence suggests that RAF induces hypertrophy in the heart by stimulating YAP activity, which leads to inhibition of RAF-mediated MST kinase. SAV1 is also involved in inhibiting tissue growth during development and RASSF1A in response to injury.55,43 It appears that the regulation between RAF and MST is reciprocal since activation of MST kinase enhances RAF communication and limits RAF's ability to activate MEK (Fig. 2).13 A mechanism that helps the inhibitor bind to RAF or MEK molecules enhances B-RAF/RAF-1 dimerization, which inhibits MST and facilitates YAP transcription by inactivating RAF-MST.56
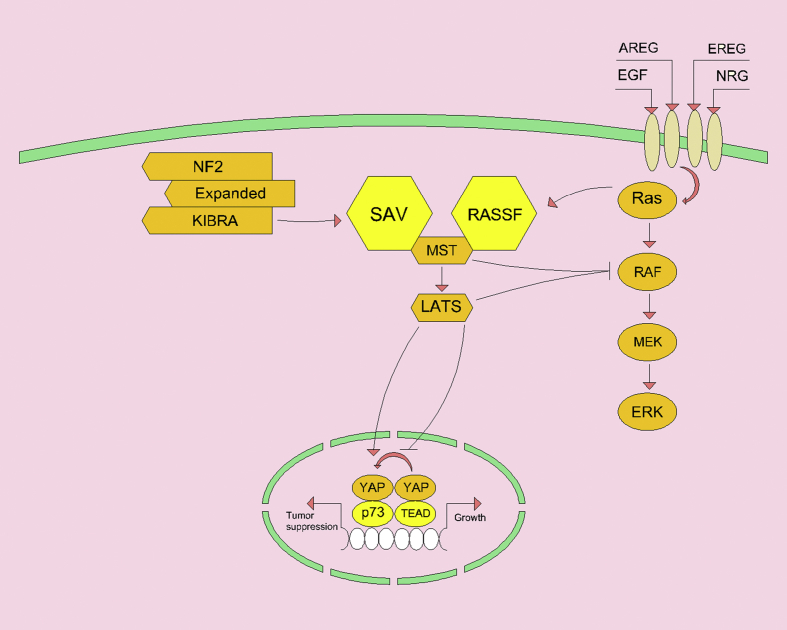
Figure 2.
The MST activation center is the Hippo path. This pathway is stimulated through signals from the NF2/Expanded/KIBRA complex to SA or through the interaction of RAS with RASSF. Activation of MST results in activation of LATS kinase and phosphorylation of YAP, which prevents association with oncogenic TEAD growth-promoting transcription factors. In addition to increasing cytoplasmic levels, YAP phosphorylation decreases the transcription factor association under tumor suppression and differentiation conditions. Increased MST activity inhibits MEK activation and also, LATS inhibits RAF activity through S259 phosphorylation and silencing of RAF.
Upon activation, MST kinase phosphorylates Scaffold MOB1 and causes deformation and ultimately LATS kinase activation.57 Responsible for directing the LATS-MOB1 heterodimer to the spindle apparatus is the RAS-GTPases involved in cell division and mitosis. LATS activation also induces transcriptional activator phosphorylation and YAP. Since MST and LATS have significant effects on the living organism, they are only epistatically associated with YAP.58, 59, 60, 61, 62, 63, 64 One of the conserved proteins in the Hippo pathway is the Ajuba Scaffold protein, which interacts with LATS kinases and SAV, limiting centromere signaling and decreasing inhibition of YAP phosphorylation.65 EFGR-RAS signaling activates MAPKs (ERK and JNK), both of which MAPKs phosphorylate Ajuba proteins, increase LATS binding, and YAP transcriptional activation.66,67 Mammalian MST kinases are involved in JNK activation (with the help of MKK7 and MEKK1); therefore, MST cannot activate JNK in cells lacking MKK7.68, 69, 70 Also, MST kinases respond to actin stress disorder and stimulatory activity of JNK, indicating that there is a physiological pathway downstream of the GTPase RHO-like RAS.71 Recent studies suggest that this pathway regulates Ajuba by JNK (Lim protein domain), increased actin f, increased MST1/2, decreased activation of the Hippo pathway and YAP transduction from the nucleus.72 JNK also inhibits YAP activity, whereby, YAP directly phosphorylated by JNK and leads to apoptosis by p73.73,74
The tumor suppressor of neurofibromatosis 2 is the false Expanded/KIBRA/NF2 (dmMerlin) complex located in the Hippo cassette upstream and contributes to activation of SAV1 and MST.75 Also, one of the MAPK signaling events is KIBRA phosphorylation by ERK, which is essential for cell proliferation. It is worth noting that KIBRA itself can play an important role in collagen-induced ERK signaling,76 indicating another level of regulatory activity between pathways. Another Hippo pathway scaffold is ARFGEF7 (or beta-Pix) that not required for GEF activity.77 ARFGEF7 also stimulates LATS in combination with YAP and TAZ, limiting the output of the Hippo pathway and YAP/TAZ transcription. In general, this scaffold mainly binds to RAS-like GTPases (Rac 1 and Cdc 42) that are active in cancer under different conditions of RAS, RHO, RAC, or CD42.78
In general, increased KRAS activity in RAF-MST, ERK-KIBRA, or ERK-Ajuba and JNK-Ajuba can have inhibitory effects on YAP and TAZ transcriptional activity (Fig. 1). Importantly, Hippo pathway scaffolding proteins and KRAS pathway scaffolds (IQGAP) increase RAS-RAF-MEK-ERK signaling (Fig. 1), which is required to suppress the Hippo pathway and some similar inhibitory events.79 Upstream receptor-driven activation of RAS results in inhibition of the output of the Hippo pathway. In mammals, activators of the ERBB family of EGF, EGF, epiregulin (EREG), amphiregulin (AREG), hairpin-binding EGF (hbEGF) and neuregulin 1 and 2 (NRG1 and NRG2) associated receptors lead to inactivation of the Hippo pathways (Fig. 3).42,54,80 LPA can inhibit the Hippo pathway by activating G heterodimer proteins (this activation mediated by RHO/Rock and directly suppressed by LATS). Also, the effects of LPA on RAS, PKA on RAS/RAF-1, and RHO/Rock on MST can be ambiguous factors that are not considered (Fig. 3).81, 82, 83, 84 All ligands that regulated by EGF were deleted and Gα activation by LPA occurs in the presence of RAS-independent Hippo pathway inhibition signal.81 Accordingly, EGF ligands are different or LPA plays an independent role could be important. Despite specific mechanisms, there are several distinct levels by which RAS-activated cancers are inhibited, including direct inhibition of RAF-MST and suppression of LATS through Ajuba phosphorylation by ERK.
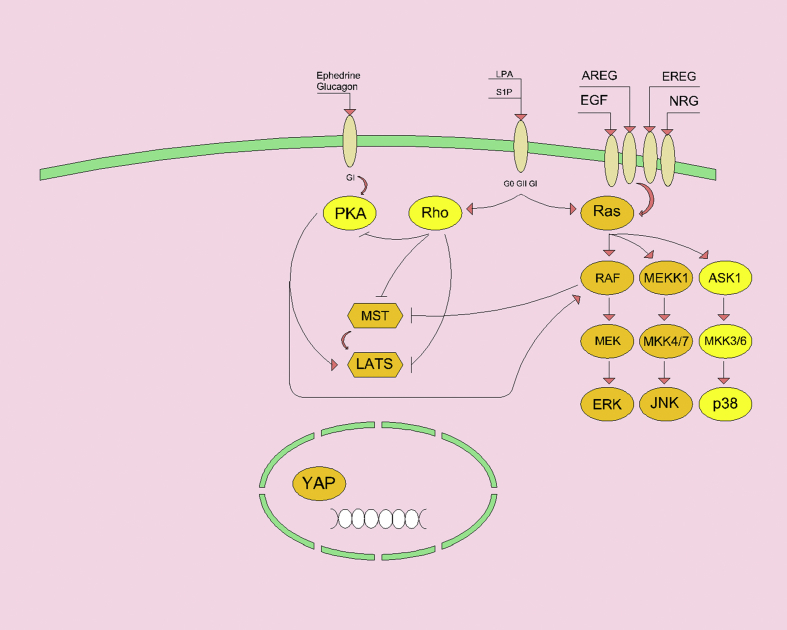
Figure 3.
Activation of ERBB family receptors by growth factors can activate RAS MAPK signaling. The RAF effector deactivates MST1 and MST2 by preventing dimerization. RAS can also be activated by G-protein coupled receptors. Activation of G heterodimer proteins (Gα, G11, and Gq) by GPCR could stimulate RHO activity. RHO activity inhibits MST and LATS by regulating actin dynamics. In addition, RHO inhibits cAMP-dependent protein kinase A. In contrast, GPCR activation activates protein kinase A and suppresses LATS, thereby facilitating YAP nuclear localization. It should be noted that protein kinase A has different effects across a range of substrates (for example, RAF is associated with MST repression and LATS also facilitates YAP activity).
YAP signaling and regulation
YAP is a regulatory protein found in the cytoplasm (in cellular junctions) as well as in the nucleus as a cofactor for a range of transcription factors involved in proliferation (TEA domain family, TEAD1-4), stem cells (Oct 4 mononuclear factor and TGFβ effector related SMAD), differentiation (RUNX1), and cell cycle/apoptosis control (p53, p63 and p73 family members).85 Mostly, proliferative activity occurs in growing tissues during development as well as in cancer cells that have transcription factors containing TEA domain (TEAD1-4). Responsible for nuclear loci and transcriptional activity of SMADs in response to TGFβ, is Yap and its homolog WWTR1 (TAZ).85 YAP and TAZ influence differentiation by regulating RUNX and PPARγ (differentiating into osteoclasts and adipocytes, respectively).85 Also, YAP participates in tumor suppression by p73, which suggests that YAP transcriptional activity synchronized with the switch between proliferation, cell cycle arrest, and differentiation. Indeed, studies have shown that direct phosphorylation by LATS is a pre-p73 communicative signal, whereas the same signal disrupts TEAD and ultimately results in rapid alteration of tumor proliferation and suppression.24,35,86 However, with activation of Hippo, YAP binds to p73, which accumulates in the cytoplasm through association with the catenin complex. Free cytoplasmic YAP is degraded by the ubiquitin ligase βTrCP-SCF complex, which in the presence of RAS, decreases the ligase targeting compounds and thereby increases YAP stability.87 Thus, active RAS can increase YAP levels independently of Hippo, which required for RAS-induced tumorigenesis.
Disruption of the Hippo pathway activity disrupted tumor suppressor activity in p73, which supports tumor growth through TEAD transcription. Evidence suggests that in mice models, the activation of TEAD causes tumorigenesis, with several somatic alterations leading to YAP-TEAD activity in common cancers.36
Studies show that the RAS-RAF-MEK-ERK cascade interacts with the Hippo tumor suppressor pathway to facilitate tumorigenesis. Further studies show the interrelationship between RAS signaling and the Hippo pathway through the feedback loop. Consequently, inactivation of Hippo signaling resulted in increased YAP/TEAD transcription of target genes and increased RAS activation. Importantly, YAP is one of the most important activators of transcriptional stimulation of EGF and AREG ligands. In fact, MAPK signaling reflects a response that further inhibits Hippo activity. With enhancing survival signals (AKT) in response to stimulation of the EGFR family and increasing level of insulin-like growth factor (IGF), the PI3K-AKT signaling increased.37,88 High levels of YAP are maintained by E6 viral oncoprotein through proteasome inhibition.89 Most importantly, deactivation of the NF2 tumor suppressor (dmMerlin) stimulates neurofibromatosis (which is associated with RAS activation by inactivating RAS-GAP). Loss of NF2 is associated with decreased activation of the Hippo and YAP pathways, which in general, K-RAS, H-RAS, and N-RAS transcription factors increase mRNA replication and K-RAS pre-oncogenic activity.90 Certainly, YAP activation directly enhances transcription of the EFGR, EFGR, and RAS family receptor ligands, increases mRNA levels in the TEAD-dependent manner, and generates a positive feedback loop to stabilize Hippo-suppression and enhances MAPK activity (Fig. 1). The most important method of activating the RAS pathway in tumors is enhancement of EGFR, via gene amplification or transcription.91
Transcriptional enhanced associate domain (TEAD)
Since its inception, TEAD has been best studied in the Hippo-YAP/TAZ signaling pathway and in tumorigenesis. There has been extensive evidence on TEAD activity that is limited to its function in the Hippo-YAP/TAZ pathway.94 Recent studies suggest that the determinants of TEAD activity in vitro and in vivo are nucleocytoplasmic silencing, post-translational modifications, and interactions between oncogenic signaling pathways.94 TEAD produced as an important drug candidate for the treatment of human diseases (cancer, cardiovascular disease, and neurological disorders).94 The oncogenic role of TEADs in tumorigenesis and development of TEAD anticancer drugs discussed below.
TEAD family transcription factors
TEAD transcription factor (all four transcription factors) are conserved in most human body tissues,95,96 and are widely expressed as each family member has several names: TEAD1 (TEF1/NTEF), TEAD2 (TEF-4/ETF), TEAD3 (TEF-5/ETFR-1) and TEAD4 (TEF-3/ETFR-2/FR-19). In animal model experiments, despite the high homology and pattern of expression between TEAD1-4, each TEAD plays different roles in specific tissues (heart, neurodevelopment, tropoderm characterization, and fetal growth)97, 98, 99, 100, 101, 102 Their most important role in cell biology is the regulation of cell proliferation and contact inhibition.103,104 TEADs are very similar in domain architectures (Fig. 4). The N-terminal of TEAD contains a TEA/ATTS DNA-binding domain containing 68 conserved amino acids that bind to the MCAT element (5′-CATTCCA/T-3) and serve as a GT-IIC motif (5-ACATTCCAC-3) Simian virus 40 (SV40) amplifiers are defined.105, 106, 107 These sequences, are an 8xGTIIC luciferase plasmid reporter synthetic TEAC luciferase containing 8 GT-IIC motifs used to measure YAP/TAZ and TEAD activity.108 Other transcription factors of TEAD, are more attached to DNA (barely showing transcriptional activity themselves) and are found in the chromatin segment.109,110 Thus, TEAD activity predominantly linked to the C terminal, in which all TEADs share their trans-activation domain to recruit YAP/TAZ transcription coactivators, VGLL1-4 corepressor, NuRD chromatin remodeling factors, and mediators.111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122 TEAD, in addition to interacts with final transcription factors of the Hippo-YAP/TAZ pathway, also interacts with other transcription factors (SMAD, TCF, OCT4, AP-1, and MRTF).123, 124, 125, 126, 127 TEAD transcription involves established genes that are involved in cell growth, proliferation, and tissue homeostasis. In recent studies, in addition to classical target TEAD genes (CTGF and CYT61), there has been focused on WNT5A/B, DKK1, TGFB2, BMP4, AREG, EGFR, PD-L1, MYC, LATS2, SLC38A1/SLC7A5 amino acid carriers and glucose transporter GLUT3 as the TEAD direct target genes.112,128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144 These signaling inputs illustrate protein-protein interactions and the role of TEAD in direct control of WNT, TGFB, RTK, mTOR and Hippo signaling that is involved in tumorigenesis, cancer immunity, stem cell amplification, metabolism, and development.
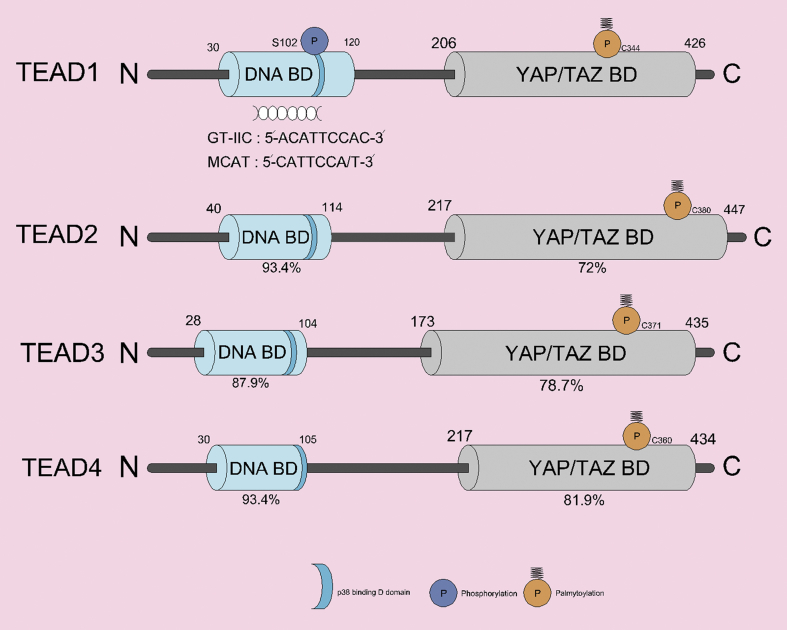
Figure 4.
Domain structure of human TEADs. The N-terminal DNA binding domain and the C-terminal YAP/TAZ binding domain of TEAD1-4 have high similarity in four different patterns. It shows the similarity of each TEAD domain to the percentage (%) of TEAD1. TEAD post-translational modifications include palmitoylation and PKC-mediated phosphorylation (occurring in YAP/TAZ-BD and DNA-BD, respectively). Palmitoylation is essential for TEAD functions. TEAD cytoplasmic translocation occurs through a protein-protein interaction with p38 MAPK, which binds to the p38-binding motif in the DNA binding domain (all TEADs).
TEAD in cancer
Expression and role of TEAD in human cancers and metastasis
Many studies have highlighted the importance of TEAD in cancer progression, with overexpression of TEAD and its high activity in several stages of cancer progression. TEAD may be involved in the reduction of some cancers (breast and kidney or bladder tumors), but the high-level expression of TEAD is associated with poor clinical outcomes as a prognostic marker for various tumors (prostate cancers, colon cancers, gastric cancers, breast cancers, embryonic cell tumors, squamous cell carcinoma of the head and neck, kidney cell carcinoma, and medulloblastomas).145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157 In addition, loss of function of TEAD1 (Y421H) causes Sveinsson's chorioretinal atrophy, resulting in genetic disorder and impaired TEAD1-YAP interaction.158,159 According to a recent meta-analysis, nuclear YAP and TAZ expression are associated with overall survival and disease-free survival of various cancers, indicating the prognostic role of TEADs and YAP/TAZ expression in patients with various malignancies.160,161 TEAD has also been shown to be an important drug target in YAP-induced tumorigenesis. In hepatocellular carcinoma, TEAD negative domain can regulate YAP-induced hepatomegaly reversely. In addition, VGLL4-mimicking peptide (bind to YBD) and verteporfin (YAP-binding small chemical) have therapeutic effects against YAP-induced tumorigenesis through impaired TEAD-YAP interaction.162, 163, 164,190
Most cancer-related death are due to metastasis (the spread of cancer cells from the primary tumor site to the secondary organs). In metastasis, cancer cells must prevent anoikis (apoptosis induced by detachment) and enter the blood vessels and should get out of it (intravasation and extravasation), thereby demonstrating complete metastatic colonization and drug resistance. These are critical features of cancer stem cells that after interacting with platelets, metastatic cancer cells induce TEAD activation through the RHOA-MYPT1-PP1-YAP pathway. Platelet-activated TEAD transcription program in cancer cells results in resistance to anoikis and stimulation of cell viability and metastasis.165 In this way, YAP enhances the resistance of anoikis to the separation of cancer cells.166 Also, exposure of cancer cells to the cut through ROCK-LIMK-cofilin signaling activates TEAD and stimulates cancer cell and metastasis.167 On the other hand, activation of the target TEAD gene, CTGF, and metastatic colonization of breast cancer occurs through leukemia inhibitory factor receptor suppression.168 In colon cancer, RAR via the Hippo pathway stimulates TEAD activation and induces EMT, invasion, and metastasis.169 However, overexpression of TEAD and its nuclear localization in colorectal cancer cells via Hippo-independent mechanism induce EMT and metastasis.148 Activation of TEAD results in induction of osteoclast differentiation by ROR1-HER3 and through Hippo-YAP pathway cause cancer cells metastasis.170 In addition, TEAD activated by MRTF stimulates breast cancer cell metastasis to the lung.123 In breast and melanoma cancer cells, induced cell adhesion to ECM by SRC tyrosine kinase activates TEAD and stimulates tumor growth and enhances metastasis.171 TGFβ-induced TEAD transcriptional activity is required to induce metastatic phenotypes in breast cancer cells. The target gene for TEAD is ARHGAP29, which upregulates actin and metastasis by regulating actin in cancer cells.172 Also, TEADs are involved in metastatic diffusion of cancer cells. For metastatic colonization, tumor cell expansion through TEAD activation by L1CAM-ILK-YAP signaling is important.173 However, these studies indicate that activation of TEAD enhances metastatic tumor formation and inhibition of TEAD can prevent tumor cell survival and proliferation.
EGFR-RAS-RAF-MAPK pathway
TEADs are intermediates of the EGFR-RAS-RAF-MAPK pathway, one of the most important uncontrolled molecular pathways in cancers. TEAD plays a vital role in tumor progression and drug resistance downstream of EGFR, KRAS, or BRAF overactive mutations. In NSCLC patients, TEAD activity is associated with EGFR mutation. EGFR mutations in lung cancer tissues and cell lines indicate that increased YAP expression results in increased TEAD activity.174 EGFR through T790M mutation induces drug resistance to TKI, but inhibition of TEAD decreases the viability of TKI-resistant lung adenocarcinoma cells.175,176 TEAD also contributes to the safety of NSCLC cells by direct transcription of PD-L1 and in turn, causes CD8 + T cell senescence.137,177 In addition, TEAD enhances EGFR expression through direct binding to the EGFR promoter and induces tumorigenesis and drug resistance in esophageal cancer.133 One of the most common proteins in human cancer is the KRAS oncogene located downstream of EGFR. The KRAS activation mutation in Pancreatic Ductal Adenocarcinoma is quite clear. BRAF (unlike EGFR and KRAS) is not considered an appropriate therapeutic target. This is due to downstream effects in the treatment of Pancreatic Ductal Adenocarcinoma. Activation of TEAD-induced target genes (COX2 and MMP7), promotes pancreatic Ductal Adenocarcinoma by KRASG12D in vitro and in vivo. Celebrex (COX2 inhibitor) and marimastat (clinical MMP inhibitor) are used as potential drug agents for the treatment of pancreatic Ductal Adenocarcinoma.178 Besides, several TEAD target genes have been associated with poor prognosis for Pancreatic Ductal Adenocarcinoma.179 TEAD2 cooperates with the E2F transcription factor to stimulate cell-cycle gene expression, which enables the activation of pancreatic Ductal Adenocarcinoma by oncogenic KRAS through a KRAS-independent tumor.180 Approximately, in half of the patients with metastatic melanoma, the BRAFV600E mutation occurs that the most common mutation is the BRAFV600E.181 Although vemurafenib (PLX4032) and dabrafenib have developed for the treatment of BRAFV600-mutant metastatic melanoma, the patients eventually show resistance.182 Numerous studies have shown that TEAD activity contributes to BRAF inhibitor-resistance in melanoma cells. TEAD activity in drug resistant-melanoma cells results in cancer resistance and invasion.183,184 BRAF inhibitor-resistant cells can reduce the immune system by activating TEAD. TEAD-mediated direct transcription of PD-L1 is responsible for PD-1-dependent CD8 + T cell senescence, which enables immune responses by BRAF inhibitor-resistant cells.135,136 In addition, IL-6 and TEAD-induced CSF1-3 cytokines in PDAC mutant cells in KRAS can induce myeloid-derived suppressor cells (MDSCs) and induce immunosuppressive activity.185 Overall, TEAD transcriptional output plays an important role in the pathogenesis of the EGFR-RAS-RAF-MAPK pathway and induces tumor progression through mutation.
Conclusion
Given all the evidence, it is clear that there is a need for an understanding of how the Hippo pathway deactivates in cancer. Although we observe RAS-MAPK suppression of the Hippo pathway at the signaling level, the fact is that the Hippo pathway enhances RAS-MAPK activity. When Hippo eliminate, EGFR-RAS-MAPK is activated and, conversely, when EGFR-RAS-MAPK inhibited, the Hippo signaling pathway is activated.30, 31, 32 Therefore, in the early stages of tumorigenesis, especially in the absence of mutation, the RAS pathway is active. Genetic polymorphisms weaken the Hippo pathway. This type of weakening has been observed in cancers such as soft-tissue sarcomas, lung cancer, and breast cancer. In addition, somatic changes (these changes are very rare) have been observed in cancers such as melanoma.186,187 Recent advances indicate that in endocrine pancreatic β cells, RASSF1 responds to KRAS and prevents MAPK activation.188 In these cells, the Hippo pathway is activated. This has also been observed in colorectal cancer, where activation of K-RAS causes the inhibition of the Hippo tumor by RASSF1, but not in the case of loss of RASSF1 and MST kinases. Expression of RASSF1 disappears in colorectal cancer. Not only mutations, genomic deletion of ch3p21 and epigenetic silencing of RASSF1A have also been linked to the incidence of lung, breast, and ovarian cancers.37,40,189 This condition not only occurs frequently but is a clinical prognosis to tumor onset. Thus, activation of YAP in the pancreas and gastrointestinal tumors may be the result of the loss of the RASSF1-mediated Hippo pathway.45, 46, 47 In this regard, RASSF1 methylation also associated with pre-cancerous colon dysplasia.43 This suggests that inactivation of RASSF1 is likely associated with a reduction of the Hippo pathway and activation of YAP in chronic diseases.41 Overall, this condition suggests that epigenetic inactivation of MST and LAST may be a key sign of Hippo pathway loss and that EGFR-RAS-RAF-MEK genetic disorders should be considered in susceptible patients.
Conflict of Interests
The authors declare no conflict of interests.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Mohammad Reza Zinatizadeh, Email: zinati3333@gmail.com.
Habibollah Mahmoodzadeh, Email: dr94habib@yahoo.com.
References
- 1.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(13):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronte G., Silvestris N., Castiglia M. New findings on primary and acquired resistance to anti-EGFR therapy in metastatic colorectal cancer: do all roads lead to RAS? Oncotarget. 2015;6(28):24780–24796. doi: 10.18632/oncotarget.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kranenburg O., Moolenaar W.H. Ras-MAP kinase signaling by lysophosphatidic acid and other G protein-coupled receptor agonists. Oncogene. 2001;20(13):1540–1546. doi: 10.1038/sj.onc.1204187. [DOI] [PubMed] [Google Scholar]
- 4.Magalhaes A.C., Dunn H., Ferguson S.S. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br J Pharmacol. 2012;165(6):1717–1736. doi: 10.1111/j.1476-5381.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaijmakers J.H., Bos J.L. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284(17):10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.J., Katan M. PLCϵ and the RASSF family in tumour suppression and other functions. Adv Biol Res. 2013;53(3):258–279. doi: 10.1016/j.jbior.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Raman M., Chen W., Cobb M.H. Differential regulation and properties of MAPKs. Oncogene. 2007;26(22):3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 9.Brown M.D., Sacks D.B. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21(4):462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claperon A., Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007;26(22):3143–3158. doi: 10.1038/sj.onc.1210408. [DOI] [PubMed] [Google Scholar]
- 11.White C.D., Brown M.D., Sacks D.B. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583(12):1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teis D., Taub N., Kurzbauer R. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol. 2006;175(6):861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romano D., Nguyen L.K., Matallanas D. Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat Cell Biol. 2014;16(7):673–684. doi: 10.1038/ncb2986. [DOI] [PubMed] [Google Scholar]
- 14.Dhomen N., Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17(1):31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Hafner C., Toll A., Fernandez-Casado A. Multiple oncogenic mutations and clonal relationship in spatially distinct benign human epidermal tumors. Proc Natl Acad Sci USA. 2010;107(48):20780–20785. doi: 10.1073/pnas.1008365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaloglou C., Vredeveld L.C., Soengas M.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 17.Junttila M.R., Karnezis A.N., Garcia D. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldser D.M., Kostova K.K., Winslow M.M. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan E.H., Morton J.P., Timpson P. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene. 2014;33(25):3325–3333. doi: 10.1038/onc.2013.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton J.P., Timpson P., Karim S.A. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107(1):246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren M., Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2) doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irvine K.D., Harvey K.F. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb Perspect Biol. 2015;7(6) doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163(4):811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oren M., Aylon Y. Springer; 2013. The Hippo Signaling Pathway and Cancer. [Google Scholar]
- 25.Rawat S.J., Chernoff J. Regulation of mammalian Ste20 (mst) kinases. Trends Biochem Sci. 2015;40(3):149–156. doi: 10.1016/j.tibs.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey K., Tapon N. The Salvador-Warts-Hippo pathway – an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7(3):182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 27.Couzens A.L., Knight J.D., Kean M.J. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013;6(302) doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton G., Yee K.S., Scrace S., O'Neill E. ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol. 2009;19(23):2020–2025. doi: 10.1016/j.cub.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avruch J., Xavier R., Bardeesy N. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284(17):11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayram S. Association between RASSF1A Ala133Ser polymorphism and cancer susceptibility: a meta-analysis involving 8,892 subjects. Asian Pac J Cancer Prev APJCP. 2014;15(8):3691–3698. doi: 10.7314/apjcp.2014.15.8.3691. [DOI] [PubMed] [Google Scholar]
- 31.Wu C., Xu B., Yuan P. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70(23):9721–9729. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- 32.Yuan H., Liu H., Liu Z. Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. Int J Cancer. 2015;137(3):638–645. doi: 10.1002/ijc.29429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong J.H., Hwang E.S., McManus M.T. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 34.van der Weyden L., Papaspyropoulos A., Poulogiannis G. Loss of RASSF1A synergizes with deregulated RUNX2 signaling in tumorigenesis. Cancer Res. 2012;72(15):3817–3827. doi: 10.1158/0008-5472.CAN-11-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matallanas D., Romano D., Yee K. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27(6):962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 37.Matallanas D., Romano D., Al-Mulla F. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell. 2011;44(6):893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi Y., Miyoshi Y., Takahata C. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11(4):1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- 39.Seidel C., Schagdarsurengin U., Blumke K. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46(10):865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- 40.Grawenda A.M., O'Neill E. Clinical utility of RASSF1A methylation in human malignancies. Br J Canc. 2015;113(3):372–381. doi: 10.1038/bjc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlahov N., Scrace S., Soto M.S. Alternate RASSF1 transcripts control SRC activity, E-cadherin contacts, and YAP-mediated invasion. Curr Biol. 2015;25(23):3019–3034. doi: 10.1016/j.cub.2015.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregorieff A., Liu Y., Inanlou M.R., Khomchuk Y., Wrana J.L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 43.Gordon M., El-Kalla M., Zhao Y. The tumor suppressor gene, RASSF1A, is essential for protection against inflammation -induced injury. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halder G., Johnson R.L. Hippo signaling: growth control and beyond. Development. 2011;138(1):9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W., Nandakumar N., Shi Y. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7(324) doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao D.D., Xue W., Krall E.B. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158(1):171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapoor A., Yao W., Ying H. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin L., Sabnis A.J., Chan E. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47(3):250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avruch J., Zhou D., Fitamant J. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev Biol. 2012;23(7):770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Neill E., Rushworth L., Baccarini M., Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306(5705):2267–2270. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 51.Volodko N., Gordon M., Salla M., Ghazaleh H.A., Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 2014;588(16):2671–2684. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 52.Polesello C., Huelsmann S., Brown N.H., Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16(24):2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avruch J., Praskova M., Ortiz-Vega S., Liu M., Zhang X.F. Nore1 and RASSF1 regulation of cell proliferation and of the MST1/2 kinases. Methods Enzymol. 2006;407:290–310. doi: 10.1016/S0076-6879(05)07025-4. [DOI] [PubMed] [Google Scholar]
- 54.Yu L., Daniels J.P., Wu H., Wolf M.J. Cardiac hypertrophy induced by active Raf depends on Yorkie-mediated transcription. Sci Signal. 2015;8(362) doi: 10.1126/scisignal.2005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tapon N., Harvey K.F., Bell D.W. Salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110(4):467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 56.Rushworth L.K., Hindley A.D., O'Neill E., Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26(6):2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hergovich A., Hemmings B.A. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors. 2009;35(4):338–345. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- 58.Muller-Taubenberger A., Kastner P.M., Schleicher M., Bolourani P., Weeks G. Regulation of a LATS-homolog by Ras GTPases is important for the control of cell division. BMC Cell Biol. 2014;15 doi: 10.1186/1471-2121-15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anguera M.C., Liu M., Avruch J., Lee J.T. Characterization of two Mst1-deficient mouse models. Dev Dynam. 2008;237(11):3424–3434. doi: 10.1002/dvdy.21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oh S., Lee D., Kim T. Crucial role for Mst1 and Mst2 kinases in early embryonic development of the mouse. Mol Cell Biol. 2009;29(23):6309–6320. doi: 10.1128/MCB.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du X., Dong Y., Shi H. Mst1 and mst2 are essential regulators of trophoblast differentiation and placenta morphogenesis. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0090701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.St John M.A., Tao W., Fei X. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21(2):182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 63.McPherson J.P., Tamblyn L., Elia A. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23(18):3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morin-Kensicki E.M., Boone B.N., Howell M. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26(1):77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das Thakur M., Feng Y., Jagannathan R. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20(7):657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun G., Irvine K.D. Ajuba family proteins link JNK to Hippo signaling. Sci Signal. 2013;6(292) doi: 10.1126/scisignal.2004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reddy B.V., Irvine K.D. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell. 2013;24(5):459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graves J.D., Gotoh Y., Draves K.E. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17(8):2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graves J.D., Draves K.E., Gotoh Y., Krebs E.G., Clark E.A. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J Biol Chem. 2001;276(18):14909–14915. doi: 10.1074/jbc.M010905200. [DOI] [PubMed] [Google Scholar]
- 70.Ura S., Nishina H., Gotoh Y., Katada T. Activation of the c-Jun N-terminal kinase pathway by MST1 is essential and sufficient for the induction of chromatin condensation during apoptosis. Mol Cell Biol. 2007;27(15):5514–5522. doi: 10.1128/MCB.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Densham R.M., O'Neill E., Munro J. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol Cell Biol. 2009;29(24):6380–6390. doi: 10.1128/MCB.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Enomoto M., Kizawa D., Ohsawa S., Igaki T. JNK signaling is converted from anti- to pro-tumor pathway by Ras-mediated switch of Warts activity. Dev Biol. 2015;403(2):162–171. doi: 10.1016/j.ydbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Danovi S.A., Rossi M., Gudmundsdottir K. Yes-associated protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ. 2008;15(1):217–219. doi: 10.1038/sj.cdd.4402226. [DOI] [PubMed] [Google Scholar]
- 74.Tomlinson V., Gudmundsdottir K., Luong P. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010;1(2) doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Genevet A., Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436(2):213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 76.Yang S., Ji M., Zhang L. Phosphorylation of KIBRA by the extracellular signal-regulated kinase (ERK)-ribosomal S6 kinase (RSK) cascade modulates cell proliferation and migration. Cell Signal. 2014;26(2):343–351. doi: 10.1016/j.cellsig.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hilton H.N., Stanford P.M., Harris J. KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim Biophys Acta. 2008;1783(3):383–393. doi: 10.1016/j.bbamcr.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 78.Heidary Arash E., Song K.M., Song S., Shiban A., Attisano L. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014;33(24):2997–3011. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anakk S., Bhosale M., Schmidt V.A. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 2013;5(4):1060–1069. doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., Ji J.Y., Yu M. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11(12):1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu F.X., Zhao B., Panupinthu N. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim M., Kim M., Lee S. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013;32(11):1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu F.X., Zhang Y., Park H.W. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27(11):1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dumaz N., Marais R. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signaling pathways. Based on the anniversary prize of the Gesellschaft Fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272(14):3491–3504. doi: 10.1111/j.1742-4658.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- 85.Mauviel A., Nallet-Staub F., Varelas X. Integrating developmental signals: a Hippo in the (path) way. Oncogene. 2012;31(14):1743–1756. doi: 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- 86.Dong J., Feldmann G., Huang J. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130(6):1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hong X., Nguyen H.T., Chen Q. Opposing activities of the Ras and Hippo pathways converge on regulation of YAP protein turnover. EMBO J. 2014;33(21):2447–2457. doi: 10.15252/embj.201489385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xin M., Kim Y., Sutherland L.B. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4(196) doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He C., Mao D., Hua G. The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol Med. 2015;7(11):1426–1449. doi: 10.15252/emmm.201404976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia-Rendueles M.E., Ricarte-Filho J.C., Untch B.R. NF2 loss promotes oncogenic RAS-induced thyroid cancers via YAP-dependent transactivation of RAS proteins and sensitizes them to MEK inhibition. Cancer Discov. 2015;5(11):1178–1193. doi: 10.1158/2159-8290.CD-15-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harris T.J., McCormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7(5):251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 92.Zinatizadeh M.R., Momeni S.A., Zarandi P.K. The Role and Function of Ras-association domain family in Cancer: A Review. Genes Dis. 2019;6(4):378–384. doi: 10.1016/j.gendis.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azmi Asfar. Elsevier; 2016. Conquering RAS: From Biology to Cancer Therapy. [Google Scholar]
- 94.Huh H.D., Kim D.H., Jeong H.S., Park H.W. Regulation of TEAD transcription factors in cancer biology. Cells. 2019;8(6) doi: 10.3390/cells8060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yasunami M., Suzuki K., Ohkubo H. A novel family of TEA domain-containing transcription factors with distinct spatiotemporal expression patterns. Biochem Biophys Res Commun. 1996;228(2):365–370. doi: 10.1006/bbrc.1996.1667. [DOI] [PubMed] [Google Scholar]
- 96.Lin K.C., Park H.W., Guan K.L. Regulation of the hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42(11):862–872. doi: 10.1016/j.tibs.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Z., Friedrich G.A., Soriano P. Transcriptional enhancer factor-1 disruption by a retroviral gene trap leads to heart-defects and embryonic lethality in mice. Genes Dev. 1994;8(19):2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 98.Kaneko K.J., Kohn M.J., Liu C., DePamphilis M.L. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45(9):577–587. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sawada A., Kiyonari H., Ukita K. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28(10):3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yagi R., Kohn M.J., Karavanova I. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134(21):3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 101.Nishioka N., Yamamoto S., Kiyonari H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125(3–4):270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 102.Nishioka N., Inoue K., Adachi K. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16(3):398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 103.Ota M., Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135(24):4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 104.Zhao B., Wei X., Li W. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jacquemin P., Hwang J.J., Martial J.A., Dolle P., Davidson I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem. 1996;271(36):21775–21785. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- 106.Jiang S.W., Desai D., Khan S., Eberhardt N.L. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19(8):507–514. doi: 10.1089/10445490050128430. [DOI] [PubMed] [Google Scholar]
- 107.Anbanandam A., Albarado D.C., Nguyen C.T. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci USA. 2006;103(46):17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dupont S., Morsut L., Aragona M. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 109.Li X., Wang W., Wang J. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol. 2015;11(1) doi: 10.15252/msb.20145504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao J.H., Davidson I., Matthes H., Garnier J.M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor tef-1. Cell. 1991;65(4):551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H., Liu C.Y., Zha Z.Y. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284(20):13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao B., Ye X., Yu J. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vassilev A., Kaneko K.J., Shu H., Zhao Y., DePamphilis M.L. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15(10):1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mahoney W.M., Jr., Hong J.H., Yaffe M.B., Farrance I.K. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388(pt1):217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen H.H., Maeda T., Mullett S.J., Stewart A.F. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis. 2004;39(4):273–279. doi: 10.1002/gene.20055. [DOI] [PubMed] [Google Scholar]
- 116.Koontz L.M., Liu-Chittenden Y., Yin F. The hippo Eector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25(4):388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen H.H., Mullett S.J., Stewart A.F. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279(29):30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 118.Pobbati A.V., Chan S.W., Lee I., Song H., Hong W. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure. 2012;20(7):1135–1140. doi: 10.1016/j.str.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 119.Honda M., Hidaka K., Fukada S.I. Vestigial-like 2 contributes to normal muscle fiber type distribution in mice. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-07149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Figeac N., Mohamed A.D., Sun C. VGLL3 operates via TEAD1, TEAD3 and TEAD4 to influence myogenesis in skeletal muscle. J Cell Sci. 2019;132(13) doi: 10.1242/jcs.225946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim M., Kim T., Johnson R.L., Lim D.S. Transcriptional Co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11(2):270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 122.Galli G.G., Carrara M., Yuan W.C. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol Cell. 2015;60(2):328–337. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jiao S., Li C., Hao Q. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beyer T.A., Weiss A., Khomchuk Y. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5(6):1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 125.Liu X., Li H., Rajurkar M. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016;14(5):1169–1180. doi: 10.1016/j.celrep.2015.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zanconato F., Forcato M., Battilana G. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17(9):1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim T., Hwang D., Lee D. MRTF potentiates TEAD-YAP transcriptional activity causing metastasis. EMBO J. 2017;36(4):520–535. doi: 10.15252/embj.201695137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Park H.W., Kim Y.C., Yu B. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162(4):780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seo E., Basu-Roy U., Gunaratne P.H. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the Osteo-adipo lineage. Cell Rep. 2013;3(6):2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee D.H., Park J.O., Kim T.S. LATS-YAP/TAZ controls lineage specification by regulating TGF beta signaling and Hnf4 alpha expression during liver development. Nat Commun. 2016;7 doi: 10.1038/ncomms11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lai D., Yang X. BMP4 is a novel transcriptional target and mediator of mammary cell migration downstream of the Hippo pathway component TAZ. Cell Signal. 2013;25(8):1720–1728. doi: 10.1016/j.cellsig.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 132.Zhang J., Ji J.Y., Yu M. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11(12):1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Song S., Honjo S., Jin J. The hippo coactivator YAP1 mediates EGFR overexpression and confers chemoresistance in esophageal cancer. Clin Cancer Res. 2015;21(11):2580–2590. doi: 10.1158/1078-0432.CCR-14-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Feng J., Yang H., Zhang Y. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36(42):5829–5839. doi: 10.1038/onc.2017.188. [DOI] [PubMed] [Google Scholar]
- 135.Kim M.H., Kim C.G., Kim S.K. YAP-induced PD-L1 expression drives immune evasion in BRAFi-resistant melanoma. Cancer Immunol Res. 2018;6(3):255–266. doi: 10.1158/2326-6066.CIR-17-0320. [DOI] [PubMed] [Google Scholar]
- 136.Van Rensburg H.J., Azad T., Ling M. The hippo pathway component TAZ promotes immune evasion in human cancer through PD-L1. Cancer Res. 2018;78(6):1457–1470. doi: 10.1158/0008-5472.CAN-17-3139. [DOI] [PubMed] [Google Scholar]
- 137.Miao J.B., Hsu P.C., Yang Y.L. YAP regulates PD-L1 expression in human NSCLC cells. Oncotarget. 2017;8(70):114576–114587. doi: 10.18632/oncotarget.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Neto-Silva R.M., de Beco S., Johnston L.A. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19(4):507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rajbhandari P., Lopez G., Capdevila C. Cross-cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 2018;8(5):582–599. doi: 10.1158/2159-8290.CD-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moroishi T., Park H.W., Qin B. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 2015;29(12):1271–1284. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Park Y.Y., Sohn B.H., Johnson R.L. Yes-associated protein 1 and transcriptional coactivator with PDZ-binding motif activate the mammalian target of rapamycin complex 1 pathway by regulating amino acid transporters in hepatocellular carcinoma. Hepatology. 2016;63(1):159–172. doi: 10.1002/hep.28223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hansen C.G., Ng Y.L., Lam W.L., Plouffe S.W., Guan K.L. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 2015;25(12):1299–1313. doi: 10.1038/cr.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang W., Xiao Z.D., Li X. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17(4):490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Holden J.K., Cunningham C.N. Targeting the hippo pathway and cancer through the TEAD family of transcription factors. Cancers. 2018;10(3) doi: 10.3390/cancers10030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pobbati A.V., Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14(5):390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Knight J.F., Shepherd C.J., Rizzo S. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Canc. 2008;99(11):1849–1858. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liang K., Zhou G., Zhang Q., Li J., Zhang C. Expression of hippo pathway in colorectal cancer. Saudi J Gastroenterol. 2014;20(3):188–194. doi: 10.4103/1319-3767.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu Y., Wang G., Yang Y. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene. 2016;35(21):2789–2800. doi: 10.1038/onc.2015.342. [DOI] [PubMed] [Google Scholar]
- 149.Zhou Y., Huang T., Zhang J. TEAD1/4 exerts oncogenic role and is negatively regulated by miR-4269 in gastric tumorigenesis. Oncogene. 2017;36(47):6518–6530. doi: 10.1038/onc.2017.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou G.X., Li X.Y., Zhang Q. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev APJCP. 2013;14(9):5199–5205. doi: 10.7314/apjcp.2013.14.9.5199. [DOI] [PubMed] [Google Scholar]
- 151.Han W., Jung E.M., Cho J. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47(6):490–499. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 152.Wang C.Y., Nie Z., Zhou Z. The interplay between TEAD4 and KLF5 promotes breast cancer partially through inhibiting the transcription of p27(Kip1) Oncotarget. 2015;6(19):17685–17697. doi: 10.18632/oncotarget.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Korkola J.E., Houldsworth J., Chadalavada R.S. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo dierentiation of human male germ cell tumors. Cancer Res. 2006;66(2):820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 154.Skotheim R.I., Autio R., Lind G.E. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol. 2006;28(5–6):315–326. doi: 10.1155/2006/219786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang W., Li J., Wu Y. TEAD4 overexpression promotes epithelial-mesenchymal transition and associates with aggressiveness and adverse prognosis in head neck squamous cell carcinoma. Cancer Cell Int. 2018;18 doi: 10.1186/s12935-018-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schutte U., Bisht S., Heukamp L.C. Hippo signaling mediates proliferation, invasiveness, and metastatic potential of clear cell renal cell carcinoma. Transl Oncol. 2014;7(2):309–321. doi: 10.1016/j.tranon.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fernandez-L A., Northcott P.A., Dalton J. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23(23):2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bokhovchuk F., Mesrouze Y., Izaac A. Molecular and structural characterization of a TEAD mutation at the origin of Sveinsson's chorioretinal atrophy. FEBS J. 2019;286(12):2381–2398. doi: 10.1111/febs.14817. [DOI] [PubMed] [Google Scholar]
- 159.Fossdal R., Jonasson F., Kristjansdottir G.T. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13(9):975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 160.Sun Z., Xu R., Li X. Prognostic Value of yes-associated protein 1 (YAP1) in various cancers: a meta-analysis. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Feng J., Ren P., Gou J., Li Z. Prognostic significance of TAZ expression in various cancers: a meta-analysis. OncoTargets Ther. 2016;9:5235–5244. doi: 10.2147/OTT.S109540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jiao S., Wang H., Shi Z. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25(2):166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 163.Park H.W., Guan K.L. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci. 2013;34(10):581–589. doi: 10.1016/j.tips.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Haemmerle M., Taylor M.L., Gutschner T. Platelets reduce anoikis and promote metastasis by activating YAP1 signaling. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-00411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao B., Li L., Wang L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26(1):54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee H.J., Diaz M.F., Price K.M. Fluid shear stress activates YAP1 to promote cancer cell motility. Nat Commun. 2017;8 doi: 10.1038/ncomms14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen D., Sun Y., Wei Y. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18(10):1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guo P.D., Lu X.X., Gan W.J. RAR gamma downregulation contributes to colorectal tumorigenesis and metastasis by derepressing the hippo-Yap pathway. Cancer Res. 2016;76(13):3813–3825. doi: 10.1158/0008-5472.CAN-15-2882. [DOI] [PubMed] [Google Scholar]
- 169.Li C., Wang S., Xing Z. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–119. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lamar J.M., Xiao Y., Norton E. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J Biol Chem. 2019;294(7):2302–2317. doi: 10.1074/jbc.RA118.004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Qiao Y., Chen J., Lim Y.B. YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep. 2017;19(8):1495–1502. doi: 10.1016/j.celrep.2017.04.075. [DOI] [PubMed] [Google Scholar]
- 172.Er E.E., Valiente M., Ganesh K. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat Cell Biol. 2018;20(8):966–978. doi: 10.1038/s41556-018-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lee T.F., Tseng Y.C., Chang W.C. YAP1 is essential for tumor growth and is a potential therapeutic target for EGFR-dependent lung adenocarcinomas. Oncotarget. 2017;8(52):89539–89551. doi: 10.18632/oncotarget.19647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee T.F., Tseng Y.C., Nguyen P.A. Enhanced YAP expression leads to EGFR TKI resistance in lung adenocarcinomas. Sci Rep. 2018;8(1):271. doi: 10.1038/s41598-017-18527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu W., Wei Y., Wu S. Up-regulation of the Hippo pathway effector TAZ renders lung adenocarcinoma cells harboring EGFR-T790M mutation resistant to gefitinib. Cell Biosci. 2015;5 doi: 10.1186/2045-3701-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yamauchi T., Moroishi T. Hippo pathway in mammalian adaptive immune system. Cells. 2019;8(5) doi: 10.3390/cells8050398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang W., Nandakumar N., Shi Y. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7(324) doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rozengurt E., Sinnett-Smith J., Eibl G. Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival. Signal Transduct Targeted Ther. 2018;3 doi: 10.1038/s41392-017-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kapoor A., YaoW, Ying H. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davies H., Bignell G.R., Cox C. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 181.Hauschild A., Grob J.J., Demidov L.V. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 182.Fisher M.L., Grun D., Adhikary G., Xu W., Eckert R.L. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget. 2017;8(66):110257–110272. doi: 10.18632/oncotarget.22628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 183.Kim M.H., Kim J., Hong H. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 2016;35(5):462–478. doi: 10.15252/embj.201592081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Murakami S., Shahbazian D., Surana R. Yes-associated protein mediates immune reprogramming in pancreatic ductal adenocarcinoma. Oncogene. 2017;36(9):1232–1244. doi: 10.1038/onc.2016.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Feng X., Degese M.S., Iglesias-Bartolome R. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through atrio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25(6):831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yu F.X., Luo J., Mo J.S. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25(6):822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Garcia-Ocana A., Stewart A.F. “RAS”ling beta cells to proliferate for diabetes: why do we need MEN? J Clin Investig. 2014;124(9):3698–3700. doi: 10.1172/JCI77764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chamberlain C.E., Scheel D.W., McGlynn K. Menin determines K-RAS proliferative outputs in endocrine cells. J Clin Investig. 2014;124(9):4093–4101. doi: 10.1172/JCI69004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kok K., Naylor S.L., Buys C.H. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 190.Liu-Chittenden Y., Huang B., Shim J.S. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]