Abstract
Alternative polyadenylation (APA) is a molecular process that generates diversity at the 3′ end of RNA polymerase II transcripts from over 60% of human genes. APA is derived from the existence of multiple polyadenylation signals (PAS) within the same transcript, and results in the differential inclusion of sequence information at the 3′ end. While APA can occur between two PASs allowing for generation of transcripts with distinct coding potential from a single gene, most APA occurs within the untranslated region (3′UTR) and changes the length and content of these non-coding sequences. APA within the 3′UTR can have tremendous impact on its regulatory potential of the mRNA through a variety of mechanisms, and indeed this layer of gene expression regulation has profound impact on processes vital to cell growth and development. Recent studies have particularly highlighted the importance of APA dysregulation in cancer onset and progression. Here, we review the current knowledge of APA and its impacts on mRNA stability, translation, localization and protein localization. We also discuss the implications of APA dysregulation in cancer research and therapy.
Keywords: 3′ untranslated region, Alternative polyadenylation, Cancer, Gene regulation, Polyadenylation signals
Introduction
It is well-established that mRNA conveys genetic information from DNA to the protein level.1 Newly synthesized precursor mRNA (pre-mRNA) undergoes multiple type of RNA processing events to lead to production of mature mRNA, which is ultimately translocated to the ribosome to act as a protein translation template. While alternative splicing has been known to generate diversity within transcripts, the process of transcriptional termination was long thought to be a constitutive event. Termination is an obligatory RNA processing occurrence and includes a cleavage step in which sequence is enzymatically removed from the 3′ end of the transcript, followed by the addition of a series of adenosine monophosphate units to form a poly(A) tail that plays a role in the transcript's stability, nuclear export, and translation. Work over the past two decades have enabled the realization that the majority of human pre-mRNAs contain multiple alternative PAS sequences. This suggests that a single gene is capable of utilizing multiple cleavage and polyadenylation signals, giving rise to two or more mRNA transcripts with distinct 3′ ends.2 Therefore APA is critical for controlling mRNA stability, localization, translation, protein coding and protein localization, and emerging evidence suggests that APA also plays key roles in the regulation of gene expression and gene function.3, 4, 5 Similar to splicing, APA is regulated by cis-regulatory elements within the pre-mRNA that are recognized by trans-regulators. These elements include: the poly-A signal itself, typically AAUAAA or its variants (termed PAS), which is recognized by cleavage and polyadenylation specificity factors (CPSFs),6,7 auxiliary UGUA sequence elements recognized by members of the mammalian cleavage factor I (CFIm),8 and GU-/U-rich downstream sequences bound by cleavage stimulation factors (CSTFs).9,10 While the full repertoire of proteins within the cleavage and polyadenylation (CPA) complex may exceed 80 members, the typical core APA regulation complex consists of CPSFs, CSTFs, CFIm and the mammalian cleavage factor complexes II (CFIIm).11 In addition to these complexes, several single factors are also required for the cleavage and polyadenylation process including poly(A) polymerase (PAP) and the scaffolding proteins poly(A)-binding protein (PAB) and Symplekin.11, 12, 13
Global PAS analyses using a variety of next-generation sequencing (NGS) approaches estimate that over one-third of mouse genes and two-thirds of human genes utilize more than one PAS, indicating that the generation of different APA transcripts from a single gene in different cellular contexts is not just a potential event but is a widespread phenomenon.14, 15, 16, 17 APA has been found to occur naturally during a variety of cellular processes including during development,18, 19, 20 in response to stress response,21,22 changes in cell growth status,23,24 in different tissues25 and pathologically in normal versus cancer cells/tissues.26,27 As compelling examples, well-differentiated cells like neurons have a general tendency to use PASs distally located relative to stop codons, resulting in the expression of transcripts with longer 3′UTR.28,29 On the other hand, faster growing cells tend to use the proximal PAS creating shorter 3′UTR resulting in potential higher expression levels.26,30, 31, 32 From a physiological perspective, Sandberg et al demonstrated that activated T cells with genes bearing shorter 3′UTR exhibit growth advantages relative to their counterparts with longer 3′UTR, and 3′UTR shortening was similarly observed after human monocytes or B cells were stimulated.23 In addition, proximal 3′UTR PAS usage is increased in multiple cancer cell lines and tissues and promotes cancer proliferation.26,33 These findings paint a picture that highlights the importance of APA in areas of cellular physiology ranging from stress response to cellular differentiation, and most importantly cancer progression. This review focuses on the general concept of APA and its roles in gene regulation as it pertains to tumorigenesis.
Mechanistic regulation of APA
Different types of APA
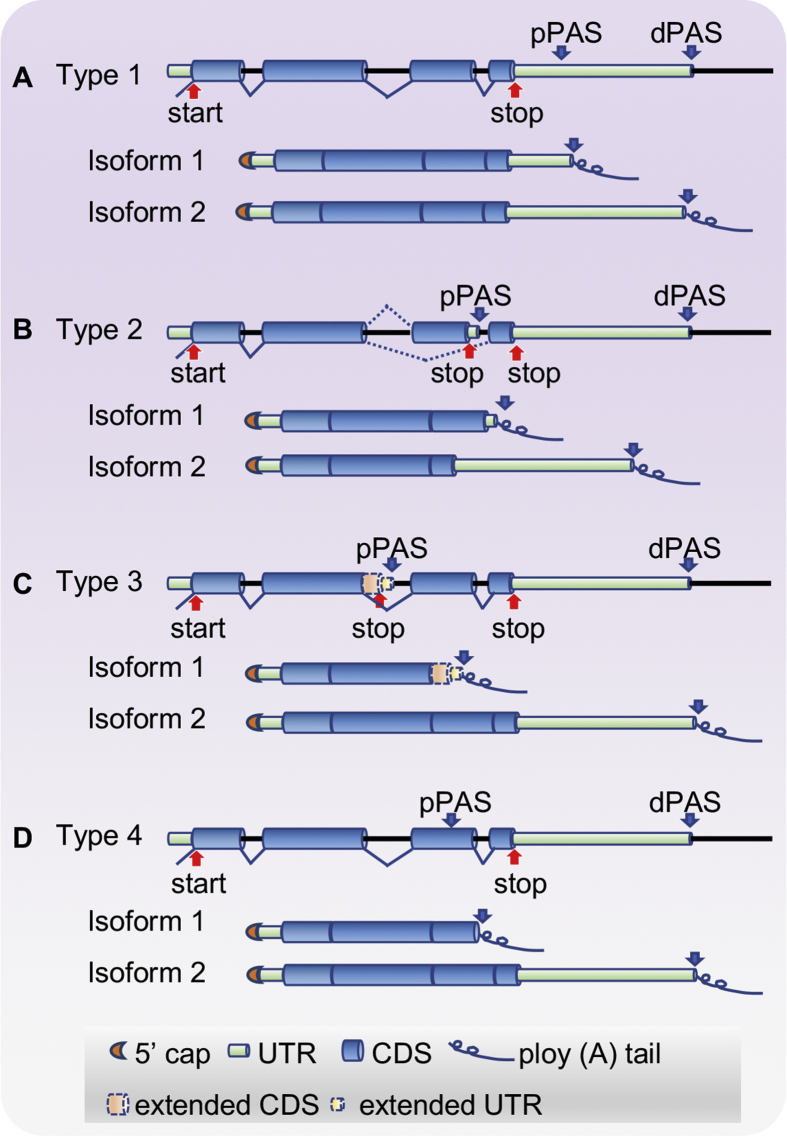
The distinct 3'terminal region of each APA-generated isoform is determined by the location of the PAS and, as such, APA can be broken down into four subclasses.2,34 ‘Tandem 3′UTR APA’ is the simplest and most common type, involving cleavage within the 3′UTR resulting in altered 3′UTR length but does not result in any change to coding potential. ‘Splicing-APA’ or alternative terminal exon polyadenylation is the second most widely-occurring type that leads to transcripts with entirely distinct 3′UTR sequences as well as different C-terminal amino acids of the encoded protein.35,36 The last two types, which are less common, are ‘intronic APA’ and ‘internal exon APA’ which occur upstream of the annotated stop codon and can lead to production of truncated proteins (Fig. 1). Importantly, as both intronic APA and internal exon APA can be subject to mRNA decay pathways such as nonsense-mediated decay (NMD) or non-stop decay, estimates of their frequency may be low. The alternative splicing machinery may also contribute APA further increasing transcript diversity generated from a single gene.37 Regardless of which APA type occurs, there is the potential to alter the presence of specific cis-regulatory elements located in the distal 3′UTR, which are known to determine mRNA stability, efficiency of translation, nuclear localization, and even the localization of the encoded protein.26,38, 39, 40, 41 One remarkable feature of 3′UTR APA is that it has been shown to change globally during cancer progression and in different cellular and tissue models of cancer.26,27,33,42 This seems surprising given the complexity of outcomes that altering APA on global scale would generate but clearly provides growth advantages to tumors. Regardless of the root cause, it is clear that understanding the mechanistic regulation of 3′UTR APA and identifying the key factors responsible for 3′UTR APA changes during tumorigenesis and cancer progression are of great importance for the discovery of potential alternative tumor therapies.
Figure 1.
Four types of alternative polyadenylation events. (A) Tandem 3′UTR APA occurs within transcripts that possess two or more cleavage/poly(A) sites within their 3′ UTR, resulting in 3′UTR length differences between APA isoforms that code for identical gene products. (B) Alternative terminal exon APA is caused by alternative splicing, which results in internal exon skipping and use of a distinct poly(A) site contained within the new terminal exon. (C) Intronic APA involves the use of cryptic alternative poly(A) sites found within introns. (D) Internal exon APA features Poly(A) site usage within upstream exons, resulting in a truncated mRNA isoform lacking both a stop codon and a 3′UTR.
Cis elements required for APA
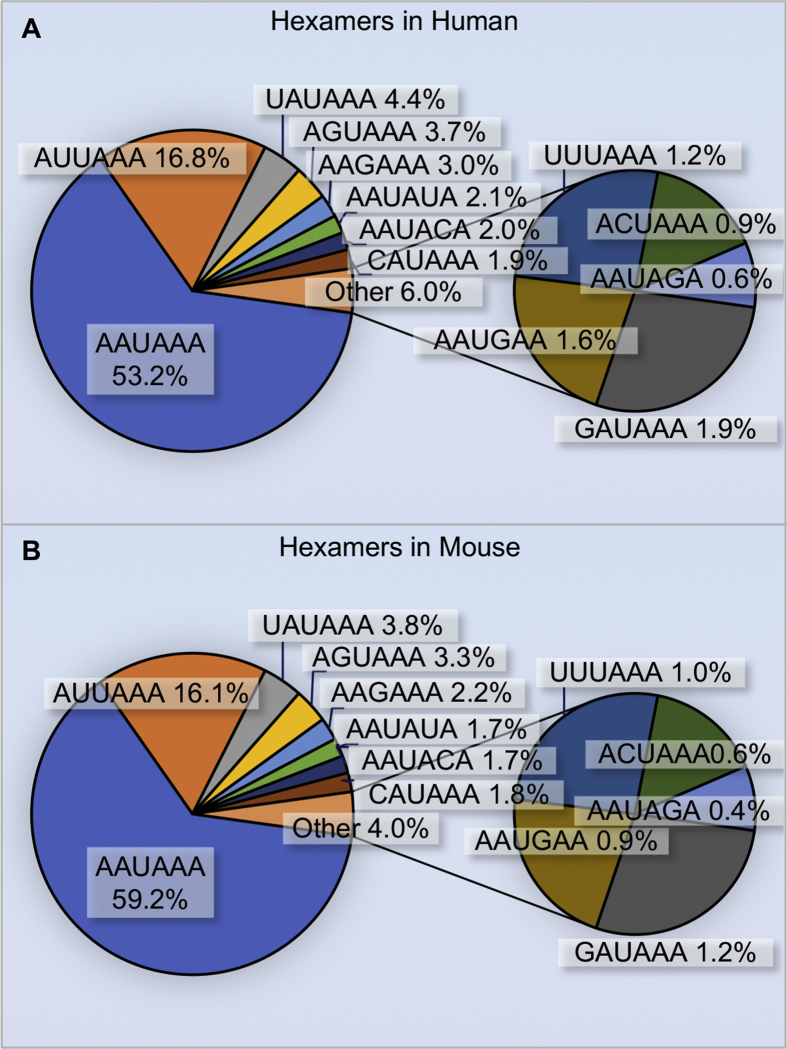
Alignment of human and mouse cDNA/EST sequences near their 3′ end allows for readily identifiable specific motifs potentially required for correct PAS recognition and processing (Fig. 2). This approach identifies that the most prominent elements in both human and mouse transcripts are PAS hexamers, which appear as AAUAAA in approximately 50% of human and 60% of mouse transcripts.6,7 Nine additional PAS variants have been identified and collectively appear in ~30% of PAS sites. These hexamer variants are functionally weaker at directing polyadenylation compared to the consensus AAUAAA hexamers and may even be inactive in vitro.43 Recently, several structural studies have uncovered the molecular basis for the AAUAAA preference and these structures underscore how just single variants in this motif can dramatically reduce affinity.44, 45, 46 Despite the relative intolerance of AAUAAA base alterations, variants such as AAGAAA are very common, especially for proximal/upstream APA sites. This suggests that other elements and protein factors may impact recognition of a suboptimal PAS. Indeed, several other cis elements located upstream and downstream of the PAS are also necessary for correct and efficient PAS recognition, cleavage and polyadenylation. Most commonly, UGUA elements are usually found within 40 nt upstream of AAUAAA and function to recruit and bind subunits of the CFIm complex.47 Moreover, the cleavage site itself is most commonly a CA dinucleotide that is often found about 15–30 bp downstream of PAS and is recognized and cut by CPSF73.48, 49, 50, 51, 52 Finally, the downstream sequence element (DSE) is a U-rich or GU-rich motif that appears frequently and is recognized by subunits of the CSTF complex.53 All of these sequence motifs are usually contained within a region of 100 nt, as downstream U/GU-rich elements and upstream UGUA elements are located near the cleavage site and usually within 40 nt of the PAS.47,54 Collectively, the overall adherence to consensus and number of UGUA motifs is thought to establish how efficiently a given PAS is utilized.
Figure 2.
Top detected PAS hexamers in human and mouse. (A and B) Human (A) and mouse (B) genomic sequences located between −40 and −1 nucleotides upstream of poly(A) sites were used to detect hexamers that might function as polyadenylation signals.
Trans-regulators involved in APA
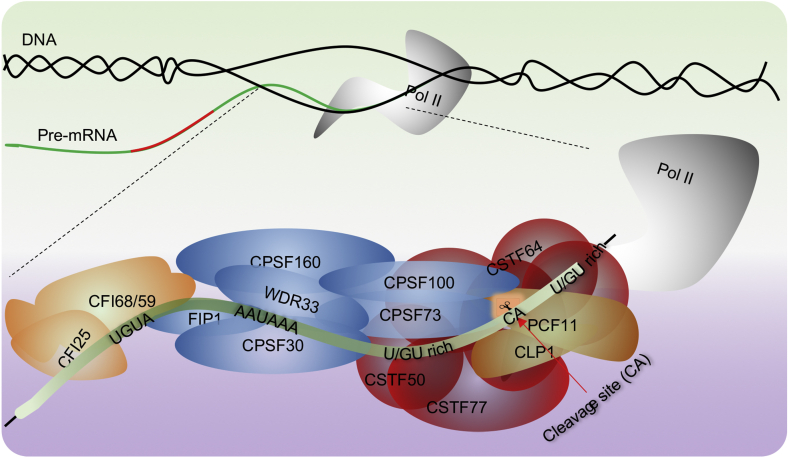
More than 80 APA-related factors have been identified in the human pre-mRNA 3'processing complex.11 Polyadenylation machinery in metazoans canonically consists of ~20 core proteins5 that make up the CPSF, CSTF, CFI and CFII complexes (Fig. 3).
Figure 3.
Cis elements and core factors involved in cleavage and polyadenylation. The PAS is recognized by the CPSF complex and is directly bound by CPSF30 and WDR33. The cleavage site, usually located ~15–30 nt downstream of the PAS, can be sheared by the endonuclease CSPF73 with the help of other regulators. U/GU-rich elements are often found within 100 nt downstream of PAS hexamers, and can be recognized by the CSTF complex. The CFI complex can specifically recognize the UGUA-rich element located within 40 nt upstream of the PAS. Several additional proteins not shown here are also required for APA including PAP, PABP1, RBBP6, symplekin and the Pol II CTD.
CPSF
The CPSF complex contains six subunits and includes: the CPSF 160 kDa subunit (CPSF160), CPSF100, CPSF73, CPSF30, WDR33 and FIP1.11,13,55,56 Overall, this complex is functionally separated into two submodules: 1) components that recognize the AAUAAA hexamer within the mammalian PAS and mainly act as an anchor for other 3′ processing factors; 2) components that function in that actual catalysis of RNA cleavage. More specifically, WDR33 and CPSF30 recognize and directly bind to the AAUAAA hexamer57, 58, 59 while CPSF160 coordinates the assembly of other CPSF components to facilitate AAUAAA binding.44,45,56 FIP1 binds to U-rich sequence upstream of the AAUAAA hexamer in vitro and in vivo via its Arg-rich C-terminal domain to further modulate PAS recognition. Consistent with a potential regulatory function, FIP1 was reported to regulate APA in embryonic stem cells (ESCs) and is important for ESC self-renewal.60 FIP1 and CPSF160 also function to directly facilitate the accumulation of poly(A) polymerase (PAP) at the mRNA 3' processing site.12,13 The cleavage reaction itself is carried out by CPSF100 and CPSF73, which form an obligate heterodimer with the endonuclease activity being housed within β-lactamase/β-CASP domain of CPSF73.
CSTF
The CSTF complex consists of three subunits: CSTF1 (also named CSTF50), either CSTF2 (also named CSTF64) or its paralog CSTF64τ (or another isoform βCSTF64 that is usually found specifically in the central nervous system61), and CSTF3 (also named CSTF77). CSTF64 or CSTF64τ directly binds to the downstream U/GU rich element and acts as a scaffold to recruit CSTF77 and CSTF50, each of which is thought to self-associate to form a homodimer.62, 63, 64 The function of the CSTF complex is to enhance recognition of the upstream PAS by the CPSF but the molecular basis of this potential stimulation is currently unknown. Consistent with this role, modulation of the strength of the downstream element or CSTF subunit expression can have significant impact on PAS selection. Consistently, while knockdown of either CSTF64 or CSTF64τ alone has little effect on global PAS selection, double knockdown of both CSTF64 and CSTF64τ leads to significant APA changes.65
CFIm and CFIIm complexes
The process of cleavage and polyadenylation typically requires two auxiliary complexes, termed CFIm and CFIIm. The better-characterized CFIm complex consists of the CFIm 25 kDa subunit (CFIm25) that forms homodimers that further heterodimerize with two copies of either CFIm68 or CFIm59 and is thought to stimulate cleavage reactions both in vitro and in vivo.8,66, 67, 68 The CFIm complex can directly bind to the UGUA motifs68 located nearby the PAS, through specific binding of its CFIm25. Both CFIm68 and CFIm59 also contain the ability to bind pre-mRNA but are thought to be non-specific.69 The CFIm complex plays an important role in APA selection, especially for the selection preference of distal polyadenylation sites.33,47,66,70,71 CFIm25 contains a critical Nudix domain that has been shown in other Nudix proteins to contain pyrophospho–hydrolase activity, but evolution has mutated/inactivated key catalytic residues rendering CFIm25 inert. Rather, this Nudix domain is responsible for interactions with the UGUA motif68 and may ultimately be critical for regulating PAS selection.68,72 Both CFIm59 and CFIm68 exhibit domain structures similar to those of SR proteins, a family of well-known splicing regulators.73 Recent work has shown that the CFIm complex functions early in the APA process and helps to stabilize CPSF binding to the PAS.69,74 Given the timing of this function, cleavage factor complexes are believed to play critical roles in PAS selection. Indeed, reduction in the expression of CFIm25 or CFIm68 (but not CFIm59) leads to global 3′UTR shortening, supporting the role of the CFIm complex in determining the terminal exon definition.70,75 As an example underscoring this property, glioblastoma cells exhibit decreased expression of CFIm25 accompanied by a global increase in proximal PAS usage and enhanced tumorigenicity.33,42 The CFIIm complex is the least characterized among 3′ end processing subcomplexes, consisting of the polyadenylation factor CLP1 (also known as hClp1) and PCF11.76 PCF11 preferentially binds to G-rich sequence elements of RNA with high affinity, and is necessary for the cleavage stimulation activity of CFIIm.76 The CFIIm complex differs from the CFIm complex in that it tends to promote proximal PAS usage, with knockdown of PCF11 leading to upregulation of distal PAS isoforms.35,77
Auxiliary APA factors
In addition to the core complexes described above, several other RNA binding proteins and other factors can regulate APA. These include the poly A polymerase (PAP) complex (which consists of PAPα and PAPγ), retinoblastoma-binding protein 6 (RBBP6), nuclear polyadenylate-binding protein 1 (PABPN1), Symplekin, heterogeneous nuclear RNP H (HNRNPH), and the phosphorylation status of the regulatory carboxy-terminal domain (CTD) of RPB1, the large subunit of RNA polymerase II (Pol II).3,11 Within this group, PAP is required for efficient cleavage and is recruited by FIP1 and CPSF160 to auxiliary upstream elements.13 The binding of PAP with U1 snRNP, a core component of the spliceosome,78 results in inhibition of polyadenylation as U1 snRNP can negatively regulate PAP polyadenylation activity.79, 80, 81 PABPN1 is a major poly(A) tail binding protein in the cytoplasm that shuttles back and forth to the nucleus and is important for synthesis of the appropriately sized poly(A) tail.82,83 PABPN1 also promotes distal PAS usage, as it inhibits mRNA proximal PAS cleavage in vitro, and its ablation induces global 3′UTR shortening.84 RBBP6 was first identified as a binding protein for p53 and Rb,85,86 and more recently has been implicated in APA regulation.87 This role appears to be mediated by the N-terminal region of RBBP6, which interacts with the CSTF complex and contains a DWNN/ubiquitin-like domain, a zinc knuckle and a RING finger. The zinc knuckle and RING finger domains function in RNA binding, and the N-terminal region is sufficient for stimulating 3′ processing activity in vitro. Knockdown of RBBP6 in mammalian cells leads to broad 3′UTR lengthening and preferential inhibition of usage of PAS containing AU-rich elements within their 3′UTRs.87 PAS usage is also determined in part by Pol II, with distinct PAS usage patterns resulting from alterations in Pol II elongation rate within the PAS region.51,88, 89, 90, 91, 92 Mutations in the transcription elongation factors TFIIS and Spt5 or in the Pol II subunit Rpb2 also promote proximal PAS usage in yeast.93 Finally, proteins such as Symplekin and the Pol II CTD do not bind RNA but function as scaffolds for 3′ processing.11,94
Molecular effects of APA
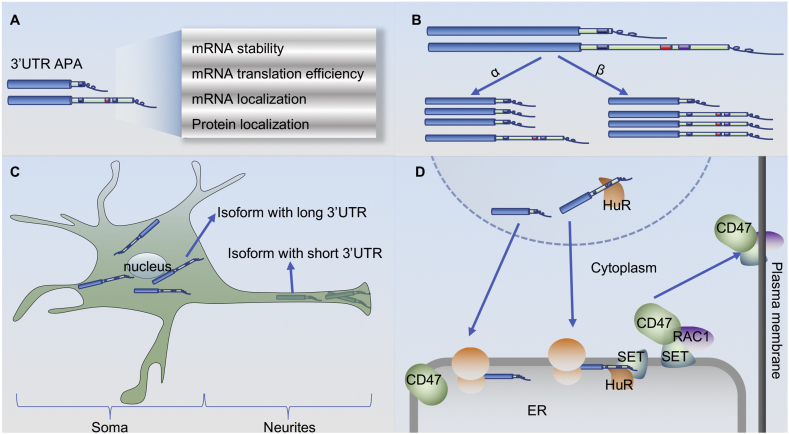
The most straightforward result of APA occurs when the alternative PAS is located within the protein coding region (Types 2–4 in Fig. 1) so as to alter the amino acid sequence generating a different protein isoform.35,36 For instance, Lee et al found that primary chronic lymphocytic leukemia (CLL) cells exhibit widespread upregulation of truncated mRNAs and proteins resulting from intronic polyadenylation.30 Aside from these less common instances, predicting the impact of 3′UTR APA (Type 1 in Fig. 1) is less intuitive as coding sequences are not altered, yet lengthening or shortening the 3′UTR affects the abundant cis elements that can be targeted by miRNA or RNA binding proteins (RBPs).23 Therefore, the ultimate output of 3′UTR APA depends on the cellular milieu in terms of whether miRNA or RBPs are expressed in a given cell type (Fig. 4). Here we will discuss the effects of 3′UTR APA in detail.
Figure 4.
Impact of 3′UTR APA on its target mRNA and protein. (A) Summary of potential consequences of 3′UTR alternative cleavage and polyadenylation on the affected mRNA and its encoded protein. (B) Long 3′UTRs contain more cis elements than short 3′UTRs, influencing both the stability and translation rate of the isoforms. The α pathway depicts the effect of cis elements such as miRNA binding sites in the long 3′UTR, promoting degradation of the lengthened isoform. The β pathway depicts the effect of 3′UTR shortening in increasing the miRNA binding efficiency in the middle region of the 3′UTR, thus decreasing the stability of the shortened isoform. In addition, some cis elements in the 3′UTR contribute to translation efficiency differences between transcripts bearing a longer or shorter 3′UTR. (C) Transcripts that undergo APA at the distal PAS remain in the soma of neurons, whereas those that use proximal PAS preferentially localize to neurites. (D) Cis-regulatory elements in 3′UTRs can direct the cellular localization of the encoded protein by altering interactions with RNA-binding proteins. The 3′UTR of the long isoform of CD47 can be bound by HuR and recruits SET to the translation site, facilitating the nascent CD47 protein to join active RAC1 and translocate to the plasma membrane.
APA impacts mRNA stability
The 3′UTR contains cis elements specifically recognized by RBPs, miRNAs and lncRNAs. miRNAs are small RNAs (~22 nt) that modulate the stability and/or translation of complementary target mRNAs, and were once hypothesized to be the main factors governing the differential mRNA stability of distinct mRNA isoforms produced by APA. This assumption is supported by the fact that most miRNA target sites are located within 3′UTRs and over 50% of conserved miRNA binding sites in mRNA transcripts are collectively located within 3′UTR regions impacted by APA.23,29 However, despite numerous studies that have observed that 3′UTR shortening results in a global increase in mRNA half-life, especially in cancer-derived cells and tissues,26,27 Nam et al found that miRNAs may influence only 10% of APA isoform expression differences between Hela, HEK293 and Huh7 cells.95 A more recent study has further demonstrated that although 3′UTR APA results in the loss of cis elements from the 3′UTR, only small fraction of shortened mRNA exhibit expression differences.96 One possible explanation is related to the observation that miRNAs bind more efficiently to target sites located in proximity to the location of the poly(A) tail. As a result, proximal APA usage resulting in 3′UTR shortening not only causes loss of miRNA binding elements but also increases the binding efficiency of some miRNAs that bind upstream of the proximal PAS. For example, 3′UTR shortening on antiproliferative and pro-differentiation genes during cell proliferation has been shown to enhance the targeting efficiency of miRNAs that bind upstream of the proximal PAS, decreasing mRNA stability and promoting cell proliferation.39 Taken together, these findings indicate that the impact of miRNA on the stability of different transcripts generated from 3′UTR APA can vary depending on the transcript and cellular context. 3′UTR shortening on the one hand may result in loss of miRNA binding sites and increased stability of the transcript, or it may enhance the binding efficiency of miRNAs targeting sites upstream of the proximal PAS and promote 3′UTR shortened mRNA degradation. Thus, the ultimate of effects of APA must be considered on a case-by-case format.
In addition to miRNA binding elements, 3′UTRs harbor mRNA destabilization elements that can be recognized by RBPs. These include GU-rich elements, AU-rich elements and PUF protein-binding elements.97,98 The inclusion or exclusion of these elements represents yet an additional mechanism by which 3′UTR APA can affect mRNA stability. In mammalian cells, transcripts with longer 3′UTRs containing predominant destabilizing elements are generally degraded more quickly than those with shorter 3′UTRs.99, 100, 101 However, the impact on mRNA stability of 3′UTR elements recognized by RBPs is far more complicated than previously thought. Other studies have found stabilizing elements, such as HuR binding element, specifically in longer 3′UTRs, which can substantially affect mRNA decay.100,102, 103, 104
In summary, the impact of 3′UTR APA on mRNA stability is highly heterogeneous and dependent on 3′UTR sequence contents and cellular conditions. This is consistent with transcriptome-wide studies that have found that although 3′UTR APA influences the decay rates of about one-third of mRNA isoforms, the isoforms with shorter 3′UTRs are only slightly more stable on average than those with longer 3′UTRs.100,102 Despite this unpredictability, it is clear that 3′UTR APA produces multiple mRNA isoforms that decay at different rates, highlighting the importance of 3ʹ UTR APA in regulating mRNA stability.
APA on mRNA translation
In addition to containing sequences that determine mRNA stability, the 3′UTR also includes elements that interact with factors regulating mRNA translation. Indeed, studies analyzing the effects of APA on mRNA translation have reported that longer isoforms are often associated in polysome fractions and with higher translation efficiency.41,100 In one example, unstimulated hippocampal neurons predominantly contain a brain-derived neurotrophic factor (BDNF) isoform with a shorter 3′UTR, while a longer, more efficiently translated 3′UTR isoform is exclusively present following neuronal activation or differentiation.41,105,106 The difference in translation efficiency enables neuronal activation/differentiation to trigger a rapid accumulation of BDNF protein levels. Similarly, the Drosophila transcript polo, which encodes Polo-like kinase,90,107 was demonstrated to exhibit higher translation efficiency from its longer 3′UTR isoform than for the shorter 3′UTR isoform. Loss of the distal PAS of polo is lethal for flies during development, while proximal PAS deletion has no phenotypic effect. These studies indicate that the higher translation rates of mRNA isoforms with longer 3′UTR are required in some cases for large-scale cell proliferation and differentiation.90 Transcripts with long 3′UTR, however, do not universally exhibit a higher translation efficiency than their short 3′UTR counterparts. In fact, a recent study has reported that in some cell lines, distal PAS usage resulting in lengthened 3′UTR decreases polysome occupancy and represses mRNA translation in comparison with shortened 3′UTR isoform.108 Therefore, the impact of 3′UTR APA on mRNA translation may vary in a gene- and 3′UTR sequence-dependent manner. Further work is required to delineate how various cis elements affect the translation efficiency of APA isoforms in different conditions and different cell types.
mRNA nuclear export and localization
Interactions between 3′UTR-associated RBPs and motor proteins can facilitate transcript mobilization to different subcellular compartments.109, 110, 111, 112 Recent studies have reported that ~10% of 3′UTR APA isoforms exhibit distribution differences between the nucleus and cytoplasm, with long 3′UTR isoforms generally enriched in the nucleus relative to the cytoplasm.113,114 However, it is difficult to assess whether 3′UTR length difference actively contributes to nuclear accumulation or simply mediates cytoplasmic degradation of the long 3′UTR isoform. 3′UTR APA also leads to localization differences within the cytoplasm, as long 3′UTRs facilitate mRNA localization to the endoplasmic reticulum (ER) and accelerate the expression of membrane proteins.115,116 In neuronal cells, symmetrically localized mRNAs can function to sequester proteins, establish cell polarity and direct asymmetric cell division.117,118 As an example, in neurons, the short 3′UTR isoform encoding BDNF is restricted to and translated within the soma, while the long 3′UTR isoform is required for appropriate localization of BDNF to the dendrites. The importance of this mechanism is seen in mice lacking the long the 3′UTR isoform, which present with altered dendritic spine morphology and decreased dendritic synapse plasticity.109 Similar isoform-specific distribution patterns between the axon and the cell body are observed for distinct isoforms encoding RAN119 and inositol monophosphates 1.120 Although the importance of 3′UTR APA in mRNA localization and downstream biological processes has been shown in these examples, further investigations are needed to delineate the underlying mechanisms. These future studies must identify the cis elements unique to long 3′UTR isoforms and the trans factors that bind to them to mediate mRNA localization differences. Since 3′UTR length simultaneously impacts mRNA stability, future studies are also necessary to clarify whether cis elements involved in mRNA stability also contribute to the apparent enrichment of different APA isoforms in various subcellular compartments.
Protein localization
Surprisingly, recent studies have reported that 3′UTR APA can impact the localization of proteins generated from mRNA transcripts of different 3′UTR length, independently of mRNA localization.38,121 A well-established example is the impact of 3′UTR APA on CD47 protein localization. CD47 isoforms with short and long 3′UTR encode the same protein. The protein encoded by the shorter isoform protein localizes to the ER, while the protein encoded by the longer isoform localizes to the plasma membrane. Mechanistic studies have revealed that the long 3′UTR can bind to the RBP human antigen R (HuR), which then recruits the effector protein SET to site of translation where it is transferred from the mRNA to the nascent CD47 protein.38 SET then binds and activates RAC1 to trigger translocation of the SET/CD47 complex to the plasma membrane. TIS11B, an AU-rich binding protein, is required for 3′UTR-mediated CD47 cell surface localization and for the interaction of SET with the long 3′UTR CD47 isoform.122 While plasma membrane-associated CD47 generated from the long 3′UTR isoform mainly functions to protect cells from phagocytosis by macrophages, CD47 translated from the short 3′UTR isoform functions in the regulation of apoptosis.38 Other proteins generated from alternative 3′UTR isoforms, such as CD44, TNF receptor superfamily member 13C (TNFRSF13C) and α1 integrin (ITGA1), exhibit similar protein distribution differences.38 Future studies may be needed to investigate the extent to which 3′UTR APA regulates protein localization in a global manner, leading to distinct functions of the same proteins localized at difference places in the cell.
APA modulation in cancer
Widespread APA in tumorigenesis
Next-generation sequencing technologies have facilitated global mRNA profiling of a variety of human disease states and their corresponding normal tissues.123, 124, 125 From the subset of those studies that permit evaluation of PAS usage, an increasing body of evidence has suggested that APA is an important regulatory mechanism for the activation of oncogenes. First, in proliferating cells, the upregulation of genes related to cell growth is accompanied by 3′UTR shortening.23,26,27,30, 31, 32, 33,126, 127, 128 Second, studies have observed preferential global truncation of 3′ UTR isoforms in different cancer cell lines, indicating a strong association of APA with cancers.26 Third, systems analysis of RNA-seq data from 358 sets of paired tumor and adjacent non-tumor tissues across seven tumor types from the Cancer Genome Atlas (TCGA) database has identified ~1350 genes with recurrent APA changes, with around ~90% occurring in the 3′UTR region and resulting in 3′UTR shortening. Importantly, selected APA events add strong prognostic power beyond regular clinical and molecular parameters, indicating that these APA events may serve as novel prognostic biomarkers.27,127 Two recent studies of triple-negative breast cancer found that the majority of 3′UTR APA in breast cancer tissues results in 3′UTR shortening relative to normal breast tissues. Most of these genes with shortened 3′UTR in patient tissues are proliferation-related transcripts correlated with recurrence-free survival times, supporting the notion of APA-based proto-oncogene activation.129,130 These data are complemented by evidence from mouse model studies supporting a critical role for APA in cancer. A study from a mouse model of B-cell leukemia/lymphoma has revealed that global 3′UTR length changes in cancer can serve as biomarkers to distinguish between similar tumor subtypes with different survival characteristics.31 In addition to the direct impact of individual genes with 3′UTR APA on cancer, the diversity of competing-endogenous RNA (ceRNA) networks generated from 3′UTR APA may also contribute to tumorigenesis by competitively altering the availability of shared miRNAs targeting them. A recent study has demonstrated that 3′UTR shortening can alter the distribution of ceRNA networks and repress tumor-suppressor genes such as PHF6 and LARP1.131
APA factors involved in cancer
Although global 3′UTR APA occurs in different cancer cells and tissues,26,30, 31, 32 the biological consequences remain largely unknown. A number of APA regulators implicated in cancer progression are discussed below, while further studies are required to uncover how APA generates cancer type-specific isoform profiles.
-
(1)
CFIm25: CFIm25 is encoded by the gene ‘NUDT21’, and is a subunit of the CFIm complex. CFIm25 binds directly to the UGUA element found in proximity of a subset of PASs. CFIm25 knockdown enhances proximal PAS usage, resulting in the expression of transcripts with shortened 3′UTRs. Interestingly, CFIm25 expression is down-regulated in many different cancer types, which is then accompanied by global 3′UTR shortening.33,42,127,132,133 In glioblastoma, down-regulation of CFIm25 results in 3′UTR shortening and upregulation of oncogenes such as CCND1 and Pak1,33,42 enhancing tumorigenic properties and increasing tumor size.33 Reduced expression of CFIm25 has been observed in hepatocellular carcinoma (HCC) tissue, and forced expression of CFIm25 inhibits HCC cell proliferation, metastasis, and tumorigenesis.132,134 Mechanistically, CFIm25 knockdown promotes proximal PAS usage in the CXXC5 and PSMB2 3′UTRs, leading to markedly increased expression of both genes and contributing to hepatocellular carcinoma suppression.134
-
(2)
PCF11: PCF11, a subunit of the CFIIm complex, is also involved in cancer progression. PCF11 controls APA of several hundred transcripts in neuroblastoma, including an operon consisting of multiple neurodifferentiation-regulating genes. PCF11 functions as a key APA regulator of WNT signaling involved in the regulation of proliferation, the cell cycle, apoptosis and neurodifferentiation. Low expression of PCF11 in neuroblastoma has been linked to extensive transcriptome APA and is associated with good prognosis and spontaneous tumor regression, while PCF11 knockdown induces aberrant neurodifferentiation.135 A recent mechanistic study has shown that PCF11 promotes intronic PAS usage and that down-regulation of PCF11 drives enhanced expression of long genes functioning in cell morphology, adhesion, and migration.136
-
(3)
hnRNPC: Fischl et al used subcellular fractionation of colon cancer cells to find that cancer progression is associated with distinct nuclear and cytoplasmic APA profiles. Further studies have shown that hnRNPC overexpression plays a critical role establishing APA profiles characteristic for metastatic colon cancer cells, while hnRNPC knockdown can reverse the cancer progression-linked APA profile.137
Conclusions and perspective
Our understanding of mRNA APA has benefitted greatly from the recent development of experimental approaches to purify mature RNAs and to study 3′ terminal regions through technologies such as 3′ end sequencing (3'seq),138 3'region extraction and deep sequencing (3′READS),139,140 Poly(A)-ClickSeq141 and sequencing APA sites (SAPAS),142 as well as bioinformatic methods to analyze APA.27,143, 144, 145, 146, 147 Characterization of APA profiles in different cell and tissue types and conditions has highlighted their functional importance in multiple physiological and disease states. Although proteomics methods have identified over 80 proteins that may be involved in mRNA 3'end processing, the roles of most of these factors in APA remain unclear.11 Furthermore, the impacts of 3′UTR APA on transcript stability and translation of regulated proteins are still controversial. Importantly, while changes in APA are associated with cancer onset and are observed in different cancer cell and tissue models, the mechanisms by which 3′UTR APA is dysregulated in cancer are poorly understood. Understanding the molecular mechanisms of abnormal APA regulation in cancers is critical for identifying new molecular targets for therapeutic intervention. Recent research targeting the APA of the androgen receptor (AR) to coordinately block the expression of resistance-associated pathogenic AR splice variants in prostate cancer is a good example of the cancer therapy applications of these findings.148 Finally, while manipulating the expression of polyadenylation machinery factors (e.g. CFIm25) can globally regulate 3′UTR length,33,96 current technologies are not able to directly manipulate the length of a desired 3′UTR. Developing such a novel method may help reveal the causal role of specific 3′UTR APA in regulating gene function and the larger process of tumorigenesis.
Conflict of Interests
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227(5258):561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 3.Gruber A.J., Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20(10):599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 4.Mayr C. What are 3’ UTRs doing? Cold Spring Harb Perspect Biol. 2019;11(10) doi: 10.1101/cshperspect.a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaudoing E., Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11(9):1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkataraman K., Brown K.M., Gilmartin G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagaki Y., Manley J.L. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17(7):3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F., Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26(12):2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y., Di Giammartino D.C., Taylor D. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murthy K.G., Manley J.L. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3'-end formation. Genes Dev. 1995;9(21):2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann I., Martin G., Friedlein A., Langen H., Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23(3):616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derti A., Garrett-Engele P., Macisaac K.D. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18(12):2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R., Zheng D., Yehia G., Tian B. A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res. 2018;28(10):1427–1441. doi: 10.1101/gr.237826.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyes A., Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018;46(2):582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang J.Y., Yu W.H., Juan H.F., Huang H.C. Dynamics of alternative polyadenylation in human preimplantation embryos. Biochem Biophys Res Commun. 2018;504(4):727–733. doi: 10.1016/j.bbrc.2018.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Hu W., Li S., Park J.Y. Dynamic landscape of alternative polyadenylation during retinal development. Cell Mol Life Sci. 2017;74(9):1721–1739. doi: 10.1007/s00018-016-2429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X., Zhang Y., Michal J.J. Alternative polyadenylation coordinates embryonic development, sexual dimorphism and longitudinal growth in Xenopus tropicalis. Cell Mol Life Sci. 2019;76(11):2185–2198. doi: 10.1007/s00018-019-03036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadek J., Omer A., Hall D., Ashour K., Gallouzi I.E. Alternative polyadenylation and the stress response. Wiley Interdiscipl Rev RNA. 2019;10(5) doi: 10.1002/wrna.1540. [DOI] [PubMed] [Google Scholar]
- 22.Hoque M., Ji Z., Zheng D. Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat Methods. 2013;10(2):133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Z., Tian B. Reprogramming of 3' untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4(12) doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr C. Evolution and biological roles of alternative 3'UTRs. Trends Cell Biol. 2016;26(3):227–237. doi: 10.1016/j.tcb.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayr C., Bartel D.P. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Z., Donehower L.A., Cooper T.A. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nat Commun. 2014;5 doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilgers V., Perry M.W., Hendrix D., Stark A., Levine M., Haley B. Neural-specific elongation of 3' UTRs during Drosophila development. Proc Natl Acad Sci U S A. 2011;108(38):15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S.H., Singh I., Tisdale S., Abdel-Wahab O., Leslie C.S., Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;561(7721):127–131. doi: 10.1038/s41586-018-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh P., Alley T.L., Wright S.M. Global changes in processing of mRNA 3' untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69(24):9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y., Sun Y., Li Y. Differential genome-wide profiling of tandem 3' UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21(5):741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masamha C.P., Xia Z., Yang J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner R.E., Pattison A.D., Beilharz T.H. Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin Cell Dev Biol. 2018;75:61–69. doi: 10.1016/j.semcdb.2017.08.056. [DOI] [PubMed] [Google Scholar]
- 35.Li W., You B., Hoque M. Systematic profiling of poly(A)+ transcripts modulated by core 3' end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11(4) doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian B., Pan Z., Lee J.Y. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17(2):156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q.Q., Liu Z., Lu W., Liu M. Interplay between alternative splicing and alternative polyadenylation defines the expression outcome of the plant unique OXIDATIVE TOLERANT-6 gene. Sci Rep. 2017;7(1):2052. doi: 10.1038/s41598-017-02215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berkovits B.D., Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522(7556):363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman Y., Bublik D.R., Ugalde A.P. 3'UTR shortening potentiates MicroRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet. 2016;12(2) doi: 10.1371/journal.pgen.1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taliaferro J.M., Vidaki M., Oliveira R. Distal alternative last exons localize mRNAs to neural projections. Mol Cell. 2016;61(6):821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau A.G., Irier H.A., Gu J. Distinct 3'UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF) Proc Natl Acad Sci U S A. 2010;107(36):15945–15950. doi: 10.1073/pnas.1002929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu Y., Elrod N., Wang C. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene. 2019;38(21):4154–4168. doi: 10.1038/s41388-019-0714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y., Zhang Y., Hamilton K. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci U S A. 2018;115(7):E1419–E1428. doi: 10.1073/pnas.1718723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clerici M., Faini M., Muckenfuss L.M., Aebersold R., Jinek M. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat Struct Mol Biol. 2018;25(2):135–138. doi: 10.1038/s41594-017-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casanal A., Kumar A., Hill C.H. Architecture of eukaryotic mRNA 3'-end processing machinery. Science. 2017;358(6366):1056–1059. doi: 10.1126/science.aao6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J., Lutz C.S., Wilusz J., Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11(10):1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacDonald C.C., Redondo J.L. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol Cell Endocrinol. 2002;190(1-2):1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 49.Tian B., Graber J.H. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscipl Rev RNA. 2012;3(3):385–396. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruber A.J., Schmidt R., Gruber A.R. A comprehensive analysis of 3' end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26(8):1145–1159. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dantonel J.C., Murthy K.G., Manley J.L., Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3' end of mRNA. Nature. 1997;389(6649):399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 52.Mandel C.R., Kaneko S., Zhang H. Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gil A., Proudfoot N.J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 54.Cheng Y., Miura R.M., Tian B. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22(19):2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 55.Murthy K.G., Manley J.L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267(21):14804–14811. [PubMed] [Google Scholar]
- 56.Clerici M., Faini M., Aebersold R., Jinek M. Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. Elife. 2017;6 doi: 10.7554/eLife.33111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan S.L., Huppertz I., Yao C. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3' processing. Genes Dev. 2014;28(21):2370–2380. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schonemann L., Kuhn U., Martin G. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28(21):2381–2393. doi: 10.1101/gad.250985.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grozdanov P.N., Masoumzadeh E., Latham M.P., MacDonald C.C. The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity. Nucleic Acids Res. 2018;46(22):12022–12039. doi: 10.1093/nar/gky862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lackford B., Yao C., Charles G.M. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33(8):878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shankarling G.S., Coates P.W., Dass B., Macdonald C.C. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol. 2009;10 doi: 10.1186/1471-2199-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takagaki Y., Manley J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20(5):1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreno-Morcillo M., Minvielle-Sebastia L., Fribourg S., Mackereth C.D. Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CF IA complex. Structure. 2011;19(4):534–545. doi: 10.1016/j.str.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Bai Y., Auperin T.C., Chou C.Y., Chang G.G., Manley J.L., Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell. 2007;25(6):863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 65.Yao C., Biesinger J., Wan J. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci U S A. 2012;109(46):18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown K.M., Gilmartin G.M. A mechanism for the regulation of pre-mRNA 3' processing by human cleavage factor Im. Mol Cell. 2003;12(6):1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 67.Coseno M., Martin G., Berger C., Gilmartin G., Keller W., Doublie S. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36(10):3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Q., Gilmartin G.M., Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3' processing. Proc Natl Acad Sci U S A. 2010;107(22):10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruegsegger U., Beyer K., Keller W. Purification and characterization of human cleavage factor Im involved in the 3' end processing of messenger RNA precursors. J Biol Chem. 1996;271(11):6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 70.Martin G., Gruber A.R., Keller W., Zavolan M. Genome-wide analysis of pre-mRNA 3' end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Rep. 2012;1(6):753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y., Wang X., Forouzmand E. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol Cell. 2018;69(1):62–74. doi: 10.1016/j.molcel.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Q., Allard P., Huang M., Zhang W., Clarke H.J. Proteasomal activity is required to initiate and to sustain translational activation of messenger RNA encoding the stem-loop-binding protein during meiotic maturation in mice. Biol Reprod. 2010;82(1):123–131. doi: 10.1095/biolreprod.109.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manley J.L., Tacke R. SR proteins and splicing control. Genes Dev. 1996;10(13):1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 74.Ruegsegger U., Blank D., Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1(2):243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 75.Gruber A.R., Martin G., Keller W., Zavolan M. Cleavage factor Im is a key regulator of 3' UTR length. RNA Biol. 2012;9(12):1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- 76.Schafer P., Tuting C., Schonemann L. Reconstitution of mammalian cleavage factor II involved in 3' processing of mRNA precursors. RNA. 2018;24(12):1721–1737. doi: 10.1261/rna.068056.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamieniarz-Gdula K., Gdula M.R., Panser K. Selective roles of vertebrate PCF11 in premature and full-length transcript termination. Mol Cell. 2019;74(1):158–172. doi: 10.1016/j.molcel.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papasaikas P., Valcarcel J. The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem Sci. 2016;41(1):33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Gunderson S.I., Polycarpou-Schwarz M., Mattaj I.W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1(2):255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 80.Berg M.G., Singh L.N., Younis I. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150(1):53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaida D., Berg M.G., Younis I. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468(7324):664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhn U., Gundel M., Knoth A., Kerwitz Y., Rudel S., Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem. 2009;284(34):22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tudek A., Schmid M., Jensen T.H. Escaping nuclear decay: the significance of mRNA export for gene expression. Curr Genet. 2019;65(2):473–476. doi: 10.1007/s00294-018-0913-x. [DOI] [PubMed] [Google Scholar]
- 84.Jenal M., Elkon R., Loayza-Puch F. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149(3):538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 85.Sakai Y., Saijo M., Coelho K., Kishino T., Niikawa N., Taya Y. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics. 1995;30(1):98–101. doi: 10.1006/geno.1995.0017. [DOI] [PubMed] [Google Scholar]
- 86.Simons A., Melamed-Bessudo C., Wolkowicz R. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene. 1997;14(2):145–155. doi: 10.1038/sj.onc.1200825. [DOI] [PubMed] [Google Scholar]
- 87.Di Giammartino D.C., Li W., Ogami K. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3' UTRs. Genes Dev. 2014;28(20):2248–2260. doi: 10.1101/gad.245787.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cevher M.A., Kleiman F.E. Connections between 3'-end processing and DNA damage response. Wiley Interdiscipl Rev RNA. 2010;1(1):193–199. doi: 10.1002/wrna.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280(5361):298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 90.Pinto P.A., Henriques T., Freitas M.O. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30(12):2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de la Mata M., Alonso C.R., Kadener S. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 92.Yu L., Rege M., Peterson C.L., Volkert M.R. RNA polymerase II depletion promotes transcription of alternative mRNA species. BMC Mol Biol. 2016;17(1):20. doi: 10.1186/s12867-016-0074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oktaba K., Zhang W., Lotz T.S. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell. 2015;57(2):341–348. doi: 10.1016/j.molcel.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan S., Choi E.A., Shi Y. Pre-mRNA 3'-end processing complex assembly and function. Wiley Interdiscipl Rev RNA. 2011;2(3):321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nam J.W., Rissland O.S., Koppstein D. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53(6):1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brumbaugh J., Di Stefano B., Wang X. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell. 2018;172(1-2):106–120. doi: 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Plass M., Rasmussen S.H., Krogh A. Highly accessible AU-rich regions in 3' untranslated regions are hotspots for binding of regulatory factors. PLoS Comput Biol. 2017;13(4) doi: 10.1371/journal.pcbi.1005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 99.Gupta I., Clauder-Munster S., Klaus B. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol Syst Biol. 2014;10(2) doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spies N., Burge C.B., Bartel D.P. 3' UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23(12):2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moqtaderi Z., Geisberg J.V., Struhl K. Extensive structural differences of closely related 3' mRNA isoforms: links to Pab1 binding and mRNA stability. Mol Cell. 2018;72(5):849–861. doi: 10.1016/j.molcel.2018.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Geisberg J.V., Moqtaderi Z., Fan X., Ozsolak F., Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156(4):812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tycowski K.T., Shu M.D., Steitz J.A. Myriad triple-helix-forming structures in the transposable element RNAs of plants and fungi. Cell Rep. 2016;15(6):1266–1276. doi: 10.1016/j.celrep.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee J.E., Lee J.Y., Wilusz J., Tian B., Wilusz C.J. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O'Neill K.M., Donohue K.E., Omelchenko A., Firestein B.L. The 3' UTRs of brain-derived neurotrophic factor transcripts differentially regulate the dendritic arbor. Front Cell Neurosci. 2018;12 doi: 10.3389/fncel.2018.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oe S., Miki H., Nishimura W., Noda Y. Mechanism of the dendritic translation and localization of brain-derived neurotrophic factor. Cell Struct Funct. 2016;41(1):23–31. doi: 10.1247/csf.15015. [DOI] [PubMed] [Google Scholar]
- 107.Oliveira M.S., Freitas J., Pinto P.A.B. Cell cycle kinase polo is controlled by a widespread 3' untranslated region regulatory sequence in Drosophila melanogaster. Mol Cell Biol. 2019;39(15) doi: 10.1128/MCB.00581-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu Y., Chen L., Chen C. Crosstalk between alternative polyadenylation and miRNAs in the regulation of protein translational efficiency. Genome Res. 2018;28(11):1656–1663. doi: 10.1101/gr.231506.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.An J.J., Gharami K., Liao G.Y. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 111.Niedner A., Edelmann F.T., Niessing D. Of social molecules: the interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA Biol. 2014;11(8):998–1009. doi: 10.4161/rna.29946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ephrussi A., Dickinson L.K., Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66(1):37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 113.Neve J., Burger K., Li W. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 2016;26(1):24–35. doi: 10.1101/gr.193995.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Loya A., Pnueli L., Yosefzon Y., Wexler Y., Ziv-Ukelson M., Arava Y. The 3'-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008;14(7):1352–1365. doi: 10.1261/rna.867208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reid D.W., Nicchitta C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16(4):221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taliaferro J.M., Lambert N.J., Sudmant P.H. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol Cell. 2016;64(2):294–306. doi: 10.1016/j.molcel.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jambhekar A., Derisi J.L. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13(5):625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yudin D., Hanz S., Yoo S. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59(2):241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Andreassi C., Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19(9):465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 121.Ciolli Mattioli C., Rom A., Franke V. Alternative 3' UTRs direct localization of functionally diverse protein isoforms in neuronal compartments. Nucleic Acids Res. 2019;47(5):2560–2573. doi: 10.1093/nar/gky1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma W., Mayr C. A membraneless organelle associated with the endoplasmic reticulum enables 3'UTR-mediated protein-protein interactions. Cell. 2018;175(6):1492–1506. doi: 10.1016/j.cell.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Curinha A., Oliveira Braz S., Pereira-Castro I., Cruz A., Moreira A. Implications of polyadenylation in health and disease. Nucleus. 2014;5(6):508–519. doi: 10.4161/nucl.36360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang J.W., Yeh H.S., Yong J. Alternative polyadenylation in human diseases. Endocrinol Metabol. 2017;32(4):413–421. doi: 10.3803/EnM.2017.32.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lin Y., Li Z., Ozsolak F. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40(17):8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin W.H., Gunay C., Marley R., Prinz A.A., Baines R.A. Activity-dependent alternative splicing increases persistent sodium current and promotes seizure. J Neurosci. 2012;32(21):7267–7277. doi: 10.1523/JNEUROSCI.6042-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Masamha C.P., Wagner E.J. The contribution of alternative polyadenylation to the cancer phenotype. Carcinogenesis. 2018;39(1):2–10. doi: 10.1093/carcin/bgx096. [DOI] [PubMed] [Google Scholar]
- 128.Xue Z., Warren R.L., Gibb E.A. Recurrent tumor-specific regulation of alternative polyadenylation of cancer-related genes. BMC Genomics. 2018;19(1) doi: 10.1186/s12864-018-4903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang L., Hu X., Wang P., Shao Z.M. The 3'UTR signature defines a highly metastatic subgroup of triple-negative breast cancer. Oncotarget. 2016;7(37):59834–59844. doi: 10.18632/oncotarget.10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang L., Hu X., Wang P., Shao Z.M. Integrative 3' untranslated region-based model to identify patients with low risk of axillary lymph node metastasis in operable triple-negative breast cancer. The Oncologist. 2019;24(1):22–30. doi: 10.1634/theoncologist.2017-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park H.J., Ji P., Kim S. 3' UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet. 2018;50(6):783–789. doi: 10.1038/s41588-018-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun M., Ding J., Li D., Yang G., Cheng Z., Zhu Q. NUDT21 regulates 3'-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410:158–168. doi: 10.1016/j.canlet.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 133.Zhu Z.J., Huang P., Chong Y.X. MicroRNA-181a promotes proliferation and inhibits apoptosis by suppressing CFIm25 in osteosarcoma. Mol Med Rep. 2016;14(5):4271–4278. doi: 10.3892/mmr.2016.5741. [DOI] [PubMed] [Google Scholar]
- 134.Tan S., Li H., Zhang W. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37(35):4887–4900. doi: 10.1038/s41388-018-0280-6. [DOI] [PubMed] [Google Scholar]
- 135.Ogorodnikov A., Levin M., Tattikota S. Transcriptome 3'end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-07580-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang R., Zheng D., Wei L., Ding Q., Tian B. Regulation of intronic polyadenylation by PCF11 impacts mRNA expression of long genes. Cell Rep. 2019;26(10):2766–2778. doi: 10.1016/j.celrep.2019.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fischl H., Neve J., Wang Z. hnRNPC regulates cancer-specific alternative cleavage and polyadenylation profiles. Nucleic Acids Res. 2019;47(14):7580–7591. doi: 10.1093/nar/gkz461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lianoglou S., Garg V., Yang J.L., Leslie C.S., Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27(21):2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng D., Liu X., Tian B. 3'READS+, a sensitive and accurate method for 3' end sequencing of polyadenylated RNA. RNA. 2016;22(10):1631–1639. doi: 10.1261/rna.057075.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hoque M., Li W., Tian B. Accurate mapping of cleavage and polyadenylation sites by 3' region extraction and deep sequencing. Methods Mol Biol. 2014;1125:119–129. doi: 10.1007/978-1-62703-971-0_10. [DOI] [PubMed] [Google Scholar]
- 141.Routh A., Ji P., Jaworski E., Xia Z., Li W., Wagner E.J. Poly(A)-ClickSeq: click-chemistry for next-generation 3-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 2017;45(12) doi: 10.1093/nar/gkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X., Li M., Yin Y. Profiling of alternative polyadenylation sites in luminal B breast cancer using the SAPAS method. Int J Mol Med. 2015;35(1):39–50. doi: 10.3892/ijmm.2014.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ha K.C.H., Blencowe B.J., Morris Q. QAPA: a new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 2018;19(1) doi: 10.1186/s13059-018-1414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 145.Ye C., Long Y., Ji G., Li Q.Q., Wu X. APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics. 2018;34(11):1841–1849. doi: 10.1093/bioinformatics/bty029. [DOI] [PubMed] [Google Scholar]
- 146.Chang J.W., Zhang W., Yeh H.S. An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic Acids Res. 2018;46(12):5996–6008. doi: 10.1093/nar/gky340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Arefeen A., Liu J., Xiao X., Jiang T. TAPAS: tool for alternative polyadenylation site analysis. Bioinformatics. 2018;34(15):2521–2529. doi: 10.1093/bioinformatics/bty110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Van Etten J.L., Nyquist M., Li Y. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res. 2017;77(19):5228–5235. doi: 10.1158/0008-5472.CAN-17-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]