Abstract
Upward (local growth and invasion of the base of skull), downward (distant metastasis) and mixed progressing types of nasopharyngeal carcinoma (NPC) have been identified and are distinctly different with respect to clinical symptoms, therapeutic strategies and prognosis. The present study aimed to identify the genetic difference and collagen expression levels in the upward and downward progressing types of NPC. Whole exon sequencing (WES) was used to detect genes differentially mutated between the upward and downward progressing types of NPC. Collagen deposition in the upward and downward progressing types of NPC was determined using Masson trichromatic staining, while the protein expression level of COL4A3 was detected using immunohistochemistry. Survival analysis was also performed using the Kaplan-Meier Plotter database to examine the role of COL4A3 expression level in the prognosis of head and neck squamous cell carcinoma. Knockdown of COL4A3 was performed using short interfering (si)RNA-COL4A3 in a 5-8F NPC cell line. Reverse transcription-quantitative PCR and western blot analyses were utilized to analyze the mRNA and protein expression levels of COL4A3, respectively. The roles of COL4A3 in the migration and invasion of the 5-8F cell line were examined using wound-healing Transwell and Matrigel assays, respectively. A total of 21 genes were differentially mutated between the upward and downward progressing types of NPC. The COL4A3 was investigated further, as it was found to be associated with extracellular matrix deposition and cancer metastasis. The COL4A3 gene was markedly downregulated in the downward progressing type compared with that in the upward progressing type (2.161±1.306 vs. 5.077±3.619; P<0.05). In addition, the deposition of collagen in the downward progressing type was also significantly decreased compared with that in the upward progressing type (5.63±6.83 vs. 10.94±9.60; P<0.05). Kaplan-Meier analysis indicated that high expression level of COL4A3 was positively associated with a favorable prognosis of head and neck squamous cell carcinoma (HR, 0.69; 95% CI, 0.49- 0.97; P=0.031). To confirm the role of COL4A3, the expression level of COL4A3 was knocked down using siRNA in the 5-8F cell line and the results showed that the invasion and migration was significantly increased when the expression of COL4A3 was inhibited (P<0.0001). In conclusion, the gene mutation patterns were significantly different between the upward and downward progressing types of NPC. In addition, the expression level of the COL4A3 gene was decreased in the downward progressing type, which might promote NPC metastasis through the downregulation of extracellular collagen expression.
Keywords: nasopharyngeal carcinoma, COL4A3, collagen deposition, metastasis, molecular classification
Introduction
Nasopharyngeal carcinoma (NPC) is a common malignant tumor of the head and neck in southern China (1,2). Histopathologically, unkeratinized squamous cell and undifferentiated carcinoma are more prevalent. According to the development of local invasion and distant metastasis, NPC can be categorized into three distinct types: Upward (local growth and invasion at the base of the skull), downward (distant metastasis) and mixed progressing types (3,4). For the upward progressing type of NPC, local invasion is dominant, and metastasis is rare, and the prognosis is relatively good. On the other hand, up to 90% of patients with the downward progressing type of NPC develop cervical lymph node metastasis or distant metastasis, even when the primary tumor was caught at an early stage. The extent of cervical lymph node metastases is one of the most important risk factors of distant metastasis (5) and the prognosis of downward progressing type of NPC is significantly worse compared with that in the upward progressing type (3,6). However, biomarkers associated with the upward and downward progressing types of NPC have not been fully elucidated. These two different biological behaviors of NPC have not been significantly associated with clinical characteristics, such as sex, age and pathological classification. Therefore, the investigation into genetic aberration and molecular mechanisms may lead to the identification of novel valuable biomarkers.
Type IV collagen, one of the main components of the extracellular matrix and basement membrane, has been found to interact with tumor cells and regulate tumor growth, proliferation, differentiation, adhesion and metastasis (7–9), which is important for the study of metastasis of NPC. Type IV collagen contains three highly similar collagen precursors namely, (α1)2α2(IV), α3α4α5(IV) and (α5)2α6(IV) (10). The genes, COL4A1, COL4A2, COL4A3, COL4A4, COL4A5 and COL4A6, which synthesize α1, α2, α3, α4, α5 and α6 peptide chains, respectively, are located in pairs on human chromosomes 13, 2 and X, respectively (11). In addition, hypermethylation of the COL4A5/COL4A6 genes in colon cancer led to a decrease in the expression level of the COL4A5/COL4A6 chain and the destruction of the structural integrity of the basement membrane (12). Similarly, it was found that the expression level of the COL4A5/COL4A6 genes was decreased or deleted in the invasive stage of basal cell carcinoma and prostate cancer (13,14). However, the role of the collagen-encoding genes in different types of NPC remains undetermined.
In the present study, whole exon sequencing (WES) was used to detected gene mutations associated with NPC biological behaviors and to determine the key genes associated with the invasion and metastasis potential of NPC. The role of COL4A3 gene expression and extracellular collagen, deposition in both the upward and downward progressing types of NPC, were also investigated. The inhibition of COL4A3 could promote invasion and migration of the 5-8F NPC cell line. The present study may provide new information in identifying novel biomarkers and establish a molecular classification of NPC.
Materials and methods
Cell culture and transfection
The human 5-8F NPC cell line was donated by Sun Yat-Sen University Cancer Center and was cultured in RPMI-1640 medium (supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 mg-ml) (all from Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5% CO2 (Sanyo Eletcric Co., Ltd). Cells in the logarithmic growth phase were placed in a 6-well plate and incubated until 70-90% confluence.
Cells were transfected with specific short interfering (si)RNA targeting COL4A3 (designed and synthesized by Guangzhou Ribobio Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to manufacturers instructions. The following siRNA was used: 5′-GGGTAATCCTGGATTTCTA-3′. A scramble siRNA was also purchased from (Guangzhou Ribobio Co., Ltd) and used as a control siRNA (siNC). The cells were assigned to three experimental groups: Non-transfected group (control), siRNA negative control group (si-NC) and COL4A3 siRNA transfected group (si-COL4A3).
Western blot analysis
Cells were lysed in RIPA lysis buffer, containing protease and phosphatase inhibitors (Sigma-Aldrich; Merck KGaA) on ice for 30 min. The total protein concentration was measured using a Bradford protein assay kit (Bio-Rad Laboratories, Inc.). Then, 60 µg per well of protein samples were separated using a 12% SDS-PAGE (Bio-Rad Laboratories, Inc.) and transferred to a PVDF membrane (EMD Millipore). After blocking with TBS-Tween-20 (TBST) containing 5% skimmed milk for 1 h at room temperature, the PVDF membrane was washed with TBST for 10 min, three times. The membrane was then incubated with primary antibodies diluted in TBST overnight at 4°C. Antibody information: COL4A3 (cat. no. PA5-39876, 1:500 dilution) and GAPDH (cat. no. MA5-15738-D800, 1:500 dilution) antibodies were from Thermo Fisher Scientific, Inc. Subsequently, the membrane was washed with TBST for 10 min thrice. Following incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (cat. no. A32731; 1:10,000 dilution; Thermo Fisher Scientific, Inc.) and HRP-conjugated goat anti-mouse secondary antibody (cat. no. 35518; 1:10,000 dilution; Thermo Fisher Scientific, Inc.) for 2 h at room temperature, respectively. After another three washes with TBST for 10 min, the proteins were visualized using an ECL substrate (Pierce™ ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.). ImageJ software version 1.48; National Institutes of Health) was used to quantify the optical density of each protein. The relative optical density was calculated by comparing with that in GAPDH.
Reverse transcription-quantitative (RT-q)PCR assay
Total RNA was extracted from treated cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) Then, 2.0 µg total RNA was used for RT using a TaqMan RT kit according to the manufacturers instructions (Applied Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, the cDNA was amplified using qPCR, in a 20 µl total reaction volume and the Go Tag Green Master Mix/Platinum SYBR Super mix (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR was performed using the following conditions: Initial denaturation at 95°C for 5 min, denaturation at 95°C for 15 sec, 60°C for 30 sec, followed by 40 cycles at 72°C for 30 sec and 72°C for 10 min. The following primers were used: COL4A3 forward, 5′-GGACTCACGGGTTCCAAAGGT-3′ and COL4A3 reverse, 5′-CCTGCTCACCCTTAGAACCACT-3′; GAPDH forward, 5′-CAATGACCCCTTCATTGACC-3′ and GAPDH reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′. GAPDH was used as the internal control.
Transwell and Matrigel assays
For the Transwell assay, 24 h following siRNA transfection, the 5-8F cell line was harvested with trypsin and seeded into Transwell chambers (8 µm pore size; Costar; Corning, Inc.), at a density of 2×104 cells per well. The upper chambers contained 200 µl serum-free RPMI-1640 medium, while the lower chambers were filled with 600 µl RPMI-1640 medium, containing 10% FBS. For the Matrigel assay, 100 µl Matrigel (Sigma-Aldrich; Merck KGaA) was added to the Transwell chamber, at a concentration of 200 µg/ml. The cells on the upper surface of the membrane were removed using a cotton swab. The invaded cells were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 30 min at room temperature. The numbers of migrated cells were counted in five random fields of view under a microscope at ×100 magnification using a Shunyu ICX41 inverted light microscope (Zhejiang Kangchen Biotech Co., Ltd.) in each membrane. Finally, cell counts were determined using the ImageJ software (National Institutes of Health). All experiments were repeated at least three times.
Wound-healing assay
A wound-healing assay was performed to determine the effect of COL4A3 on cell migration. The 5-8F cell line was seeded into 6-well plates, at a density of 2×105 cells/well, in RPMI-1640 medium, supplemented with 10% FBS, 24 h following transfection. When the cells had reached 90–100% confluence, a 200 µl pipette tip was used to scratch a wound on the cells. The medium was removed, and the cells were gently washed twice with PBS. Next, the cells were incubated with RPMI-1640 medium containing 1% FBS. Images were captured at 0 and 24 h under a light microscope at ×40 magnification and the location and migration of the cells was measured by determining the wound area. The wound healing rate was quantified by using ImageJ software and calculated with the help of following formula (Aw - At)/Aw × 100. Where Aw is an area of the wound at 0 h (control) and At is the area of the wound at 24 h after scratching. All the experiments were repeated at least three times.
Human specimens
The present study was approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (approval no. 2019084). Written informed consent was obtained from each living patient or from their relatives for deceased patients in the study. Biopsies were collected from 20 patients with upward and downward progressing types of NPC. In total, 17 patients were male and three were female. The median age was 57 years (range, 42–83 years). The following inclusion criteria were used: i) Newly diagnosed patients with NPC from January 2003 to December 2018; ii) between 18 and 85 years of age; iii) confirmed pathological diagnosis; and iv) stages of T3-4N0-1M0 (upward) and Tis-1N2-3M0-1 (downward) (based on the 8th edition of American Joint Committee on Cancer for nasopharyngeal carcinoma staging) (15). The following exclusion criteria were used: i) Recurrent NPC; ii) no pathological diagnosis or pathological specimens; and iii) a history of other malignant tumors. At the same time, the demographical and pathological information, and imaging data were collected.
Hematoxylin and eosin (H&E) staining
The histopathological examination was performed by a senior pathologist at Zhongnan Hospital. The biopsies of two groups of patients with NPC were stained with H&E, which was routinely used in the pathology laboratory using the following procedure: Deparaffinization with xylene, twice for 10 min each; re-hydration with absolute ethanol, twice, 5 min each; followed by 95% ethanol for 2 min and 70% ethanol for 2 min, followed by washing in distilled water for 1 min; staining in H solution for 8 min at room temperature, followed by washing in tap water for 5 min; differentiation in acid ethanol for 30 sec, followed by washing in tap water for 1 min; bluing in 0.2% ammonia water for 1 min, followed by washing in tap water for 5 min. Finally, slides were dehydrated with 85% ethanol and 95% ethanol for 5 min each and counterstaining with E Y solution for 5 min at room temperature. Dehydration with absolute ethanol, three times, 5 min each. Transparency with xylene, twice, 5 min each time. The slides were visualized under ×100 magnification using a light microscope. The compactness of cells between upward and downward progressing types of NPC was observed.
Masson trichromatic staining
The expression level and distribution of collagen in the upward and downward progressing types of NPC specimens was detected using Masson trichrome staining. The NPC specimens of the two groups were routinely dewaxed with water by successively putting into xylene for 20 min at room temperature, anhydrous ethanol for 10 min, an ethanol series (95, 90, 80 and 70%), washed with distilled water thrice, and then fixed with picric acid mixed fixation solution for 10 min at room temperature. The nucleus was subsequently stained with H for 5–8 min, rinsed with distilled water thrice, and then stained with Lichun red acid Fuhong solution for 5–7 min at room temperature. After washing with distilled water, specimens were stained with 1% phosphomolybdic acid solution for 5 min at room temperature and then stained directly with bright green dye for 5 to 10 min at room temperature. Subsequently, specimens were differentiated with 1% glacial acetic acid for 30–60 sec, dehydrated with 95% ethanol for 3 min and then dehydrated with anhydrous ethanol for 5 min. Finally, all the specimens were made transparent with xylene and sealed with neutral gum. The slides were scanned using a NanoZoomer S360 Digital slide scanner (C13220-01; Hamamatsu Co., Ltd.) and analyzed under ×10 magnification using NDP.view2 Viewing software (U12388-01; Hamamatsu Co., Ltd.). The expression level of collagen was quantified using Image-Pro Plus v6.0 software (Media Cybernetics, Inc.).
Immunohistochemistry (IHC)
For IHC staining, paraffin-embedded NPC tissue sections at 4–5 µm were deparaffinized with xylene I for 20 min, xylene II for 20 min, and rehydrated with 100% ethanol for 10 min, and 95, 85 and 80% ethanol for 5 min, each. Then the sections were incubated with anti-COL4A3 antibody (cat. no. sc-52317; 1:100 dilution; Santa Cruz Biotechnology, Inc.) overnight at 4°C. The next day, the slides were then incubated with a biotin-labeled secondary antibody (cat. no. sc-2018; 1:100 dilutions; Santa Cruz Biotechnology, Inc.) for 40 min at room temperature. Subsequently, the secondary antibody was washed off with TBST (0.1% Tween-20). The cells were then visualized using a REAL™ EnVision™Detection System, (Peroxidase/DAB+; Rabbit/Mouse; cat. no. K5007; Dako; Agilent Technologies, Inc.). The slides were scanned by NanoZoomer S360 Digital slide scanner and analysed under ×300 magnification using NDP.view2 Viewing software. The expression level of COL4A3 was quantified using Image-Pro Plus version 6.0 software (Media Cybernetics).
WES
Sequencing and bioinformatic analysis of whole exons from the upward and downward progressing types of NPC specimen were completed in cooperation with Shenzhen Chengqi Biotechnology Co., Ltd. DNA was fragmented and hybridized using the SureSelect Human All Exome kit V5 (Agilent Technologies, Inc.). Exome shotgun libraries were sequenced on the SureSelect Human All Exon v.6 enrichment, Illumina NovaSeq 6000 platform (Illumina, Inc.), generating paired end reads of 150 bp at each end. The cloud analysis platform with FANSe algorithm (Chi-Cloud NGS Analysis Platform) was applied for single-nucleotide variant calling (16). Image analysis and base calling were performed with CAVSAVR (Illumina, Inc.) using default parameters. Adapter sequences were removed to obtain high-quality reads. These were aligned to the NCBI human reference genome hg19 using the Burrows-Wheeler Aligner alignment algorithm. The high frequency mutant genes associated with metastasis of NPC were screened out. The associated amino acid replacement events and the changes in protein coding sequence and function were predicted.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of putative signaling pathways
The designated genes were uploaded into the function annotation portal of The Database for Annotation, Visualization, and Integrated Discovery (DAVID), online bioinformatics resources for investigating the biological signaling pathways enriched by input genes (17). GO (http://geneontology.org/) and KEGG (https://www.kegg.jp/) corpus were adopted to identify putative signaling pathways which were visualized via R clusterProfiler package version 3.18.0 (18).
Survival analysis
COL4A3 gene expression, as determined using RNA sequencing and survival data from patients with head and neck squamous cell carcinoma were obtained and analyzed using the Kaplan-Meier Plotter tool (http://kmplot.com/analysis) (19). Due to a lack of NPC data, the data of head and neck squamous cell carcinoma (N=499) were used instead. Patients were divided by selecting the auto select best cut-off. When the auto select best cut-off is selected, all possible cut-off values between the lower and upper quartiles are computed, and the best performing threshold is used as a cut-off. The auto select best cut-off value was 35. Patients of both sexes, different ethnicities, disease stages and grades were included. Hazard ratio (HR) with 95% CI were analyzed and the median survivals times were compared between the high and low expression level groups.
Statistical analysis
Quantitative results are expressed as the mean ± SD. A Wilcoxon rank sum test was performed to analyze data obtained from experiments on biopsy samples (IHC and Masson trichromatic staining data). One-way ANOVA test was used to compare quantitative data from in vitro experiments containing multiple groups. Post-hoc test between si-NC and si-COL4A3 groups were adjusted with Bonferroni correction. Kaplan-Meier survival plots were generated, and a log-rank test was used for comparisons between survival curves. GraphPad Prism version 6.0 (GraphPad Software, Inc.) was used to analyze all the data. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical and pathological characteristics of patients with the upward and downward progressing types of NPC
Between January 2003 and December 2018, 20 patients with NPC were eligible for analysis and were included in the present study. A total of 17 patients were male. The median age was 57 years (range, 42–83 years) (Table I). A total of 8 patients with the upward progressing type of NPC and 4 patients with the downward progressing type of NPC were included in the WES analysis. These patients and another eight patients were also included to investigate COL4A3 expression level and collagen deposition.
Table I.
Clinical characteristic of patients with nasopharyngeal carcinoma.
| A, Upward | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| ID | WES | Sex | Age, years | T stagea | N stagea | M stagea | WHO typeb |
| Patient 1 | Yes | Male | 66 | 4 | 0 | 0 | II |
| Patient 2 | Yes | Male | 42 | 3 | 1 | 0 | II |
| Patient 3 | Yes | Male | 59 | 3 | 1 | 0 | II |
| Patient 4 | Yes | Male | 55 | 3 | 1 | 0 | II |
| Patient 5 | Yes | Male | 68 | 4 | 1 | 0 | II |
| Patient 6 | Yes | Male | 53 | 4 | 1 | 0 | II |
| Patient 7 | Yes | Male | 54 | 4 | 0 | 0 | II |
| Patient 8 | Yes | Male | 67 | 4 | 1 | 0 | II |
| Patient 9 | No | Male | 49 | 4 | 1 | 0 | II |
| Patient 10 | No | Male | 78 | 4 | 1 | 0 | II |
| Patient 11 | No | Male | 62 | 4 | 1 | 0 | II |
| Patient 12 | No | Female | 54 | 3 | 1 | 0 | II |
| Patient 13 | No | Male | 62 | 3 | 1 | 0 | II |
| Patient 14 | No | Female | 44 | 4 | 1 | 0 | I |
| Patient 15 | No | Male | 73 | 4 | 0 | 0 | II |
|
| |||||||
| B, Downward | |||||||
|
| |||||||
| ID | WES | Sex | Age, years | T stagea | N stagea | M stagea | WHO typeb |
|
| |||||||
| Patient 16 | Yes | Male | 63 | 1 | 2 | 0 | II |
| Patient 17 | Yes | Male | 53 | 1 | 3 | 0 | II |
| Patient 18 | Yes | Male | 54 | 1 | 3 | 1 | II |
| Patient 19 | Yes | Male | 83 | 1 | 3 | 0 | II |
| Patient 20 | No | Female | 44 | 1 | 3 | 0 | I |
Based on the 8th edition of AJCC nasopharyngeal carcinoma staging
WHO types: I, keratinized squamous cell carcinoma; II, non-keratinized carcinoma. WES, whole exon sequencing; T, tumor; N, node; M, metastasis.
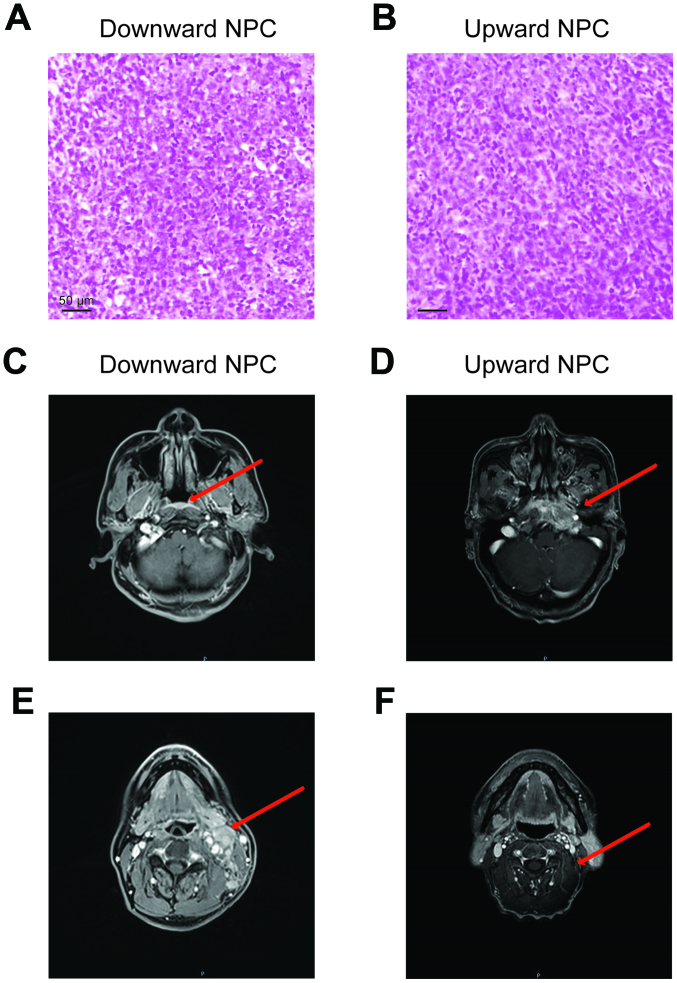
With respect the pathological features of the patients, the arrangement of the tumor cells in the downward progressing type of NPC was loose, as opposed to that in the upward progressing type of the NPC cells (Fig. 1A and B). In the magnetic resonance imaging (MRI) analysis, the downward progressing type of NPC showed small primary tumors (Tis-T1), confined in the nasopharyngeal cavity, and did not invade into the surrounding tissue (Fig. 1C and E). However, notable cervical lymph node metastasis or distant metastasis occurred even when the primary tumor was in early stages. On the other hand, the upward progressing type of NPC (T3-T4) showed significant infiltration and destruction of the surrounding tissue by the primary tumor, with documented clinical manifestations of space and nerve involvement associated with tumor invasions (Fig. 1D and F). Nonetheless, cervical lymph node metastasis of the upward progressing type was not significant (N0-N1) and no evidence of distant metastasis was detected.
Figure 1.
Pathological and imaging results of the downward and upward progressing types of NPC. H&E staining of (A) downward and (B) upward progressing types of NPC. (C) The primary tumor of downward progressing type of NPC was confined to the nasopharyngeal cavity (arrowhead). (D) The primary tumor of upward progressing type of NPC was prone to extensive invasion of the surrounding tissue (the arrow shows the invasion of tensor palatine sail muscle, levator palatine sail muscle, parapharyngeal space, clivus, foramen lacerum, internal pterygoid muscle and other structures). (E) There was notable cervical lymph node metastasis in the downward progressing type of NPC (the arrow shows multiple metastatic lymph nodes with marked enlargement, enhancement and local necrosis in the left cervical region). (F) The lymph node metastasis of upward progressing type of NPC was not notable (arrowhead). H&E, hematoxylin and eosin; NPC, nasopharyngeal carcinoma.
COL4A3 was identified as the target gene
A total of 8 upward NPC specimens and 4 downward NPC specimens were sequenced, from which 4,125 differential mutation sites were found, involving 2,511 genes. The molecular events such as single nucleotide variations, insertions/deletions, copy number variations and gene fusion, with significant differences in mutation frequency between the upward and downward progressing types of NPC were detected. After harmful predictive filtration, there were 21 residual differential mutation sites, involving 21 genes (Table II; Fig. 2A). Single nucleotide variations between upward and downward progressing types of NPC, detected by WES and Genome-Wide Association Studies, were used to analyze differential gene expression. P<0.05 was considered statistically significant and eight single nucleotide variations showed significant difference between the two types of NPC (Table II). The genes associated with tumor metastasis, in the mutant genes were further investigated by GO analysis and it was found that the differentially mutated genes between the upward and downward progressing types of NPC were enriched in the genes associated with collagen expression (Fig. 2B). KEGG signaling pathway analysis found that the differentially mutated genes were significantly enriched in extracellular matrix-associated signaling pathways (Fig. 2C). Within these identified genes, the target gene COL4A3, which was found to be associated with tumor metastasis, was investigated further with respect to its role in the metastasis of NPC.
Table II.
Whole exon sequencing results of patients with nasopharyngeal carcinoma.
| Chromosome | Position (1-based) | Gene name | Original | Mutation | Original (peptide) | Mutation (peptide) | Annotation | P-valuea |
|---|---|---|---|---|---|---|---|---|
| chr10 | 135440216 | FRG2B | G | A | H | Y | FSHD region gene 2 family member B | 0.87870 |
| chr10 | 17147521 | CUBN | G | T | P | T | Cubilin | 0.01335 |
| chr1 | 12939765 | PRAMEF4 | A | G | L | P | PRAME family member 4 | 0.00000 |
| chr12 | 31242081 | DDX11 | G | A | R | Q | DEAD/H-box helicase 11 | 0.27880 |
| chr12 | 8327016 | ZNF705A | T | C | A | T | zinc finger protein 705A | 0.01544 |
| chr15 | 102463117 | OR4F4 | T | G | H | P | Olfactory receptor family 4 subfamily F member 4 | 0.49690 |
| chr15 | 28197037 | OCA2 | T | C | H | R | OCA2 melanosomaltrans-membrane protein | 0.01397 |
| chr15 | 45392075 | DUOX2 | G | A | S | L | Dual oxidase 2 | 0.21210 |
| chr15 | 74374822 | GOLGA6A | T | G | H | P | golgin A6 family member A | 0.00665 |
| chr16 | 48258198 | ABCC11 | C | T | G | R | ATP binding cassette subfamily C member 11 | 0.07139 |
| chr17 | 76433898 | DNAH17 | T | C | H | R | Dyneinaxonemal heavy chain 17 | 0.20260 |
| chr17 | 78178893 | CARD14 | C | T | R | W | Caspase recruitment domain family member 14 | 0.07064 |
| chr2 | 21231524 | APOB | G | A | P | L | Apolipoprotein B | 0.30960 |
| chr2 | 228135631 | COL4A3 | C | T | P | L | Collagen type IV alpha 3 chain | 0.00149 |
| chr2 | 87214281 | RGPD1 | T | G | V | G | RANBP2-like and GRIP domain containing 1 | 0.05126 |
| chr3 | 172835082 | SPATA16 | C | T | G | E | Spermatogenesis associated 16 | 0.08735 |
| chr3 | 37574951 | ITGA9 | G | A | G | E | Integrin subunit alpha 9 | 0.28760 |
| chr4 | 120241902 | FABP2 | T | C | T | A | Fatty acid binding protein 2 | 0.03159 |
| chr4 | 190881957 | FRG1 | G | T | D | Y | FSHD region gene 1 | 0.05745 |
| chr9 | 33386465 | AQP7 | A | G | Y | H | Aquaporin 7 | 0.04698 |
| chr9 | 42376286 | ANKRD20A2 | C | T | A | V | Ankyrin repeat domain 20 family member A2 | 0.08193 |
Single nucleotide variations between upward and downward progressing types of nasopharyngeal carcinoma were detected by whole exome sequencing and Genome-Wide Association Studies were used to analyze differential gene expression. P<0.05 was considered as statistically significant.
Figure 2.
Sequencing results of the whole exons in nasopharyngeal carcinoma. (A) The differentially mutated genes between the upward and downward progressing types of NPC were identified using sequencing of the whole exons. The color block in the figure indicated the frequency of gene mutation. (B) GO analysis showed that some of the differentially mutated genes were enriched in the genes associated with collagen expression. (C) KEGG bubbles signal pathway analysis showed that the differentially mutated genes were involved in the extracellular matrix-associated signaling pathway. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Expression of COL4A3 and deposition of collagen in NPC
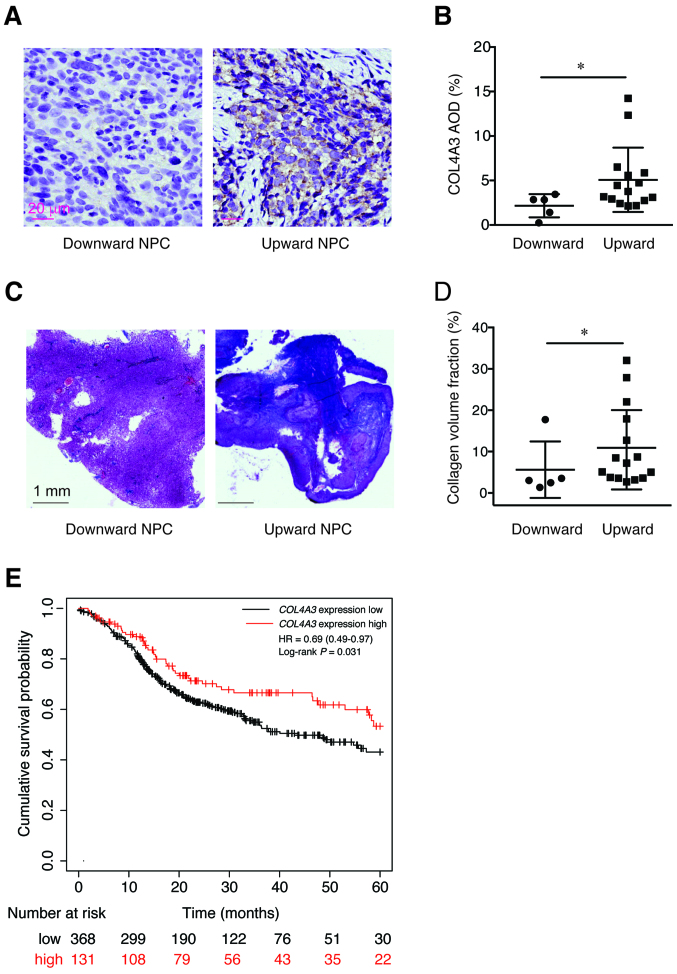
The protein expression level of COL4A3, in patients with the two types of NPC, was detected using IHC methods. It was found that the expression level of COL4A3 in the downward progressing type of NPC was significantly lower compared with that in the upward progressing type of NPC (2.161±1.306 vs. 5.077±3.619; P<0.05; Fig. 3A and B). Next, the extracellular collagen deposition was determined using Masson trichromatic staining. Notably, the deposition of blue precipitate (collagen) in the downward progressing type of NPC was significantly lower compared with that in the upward progressing type of NPC (5.63±6.83 vs. 10.94±9.60; P<0.05; Fig. 3C and D). In addition, the high expression level of COL4A3 was associated with favorable prognosis of head and neck squamous cell carcinoma (HR, 0.69; 95% CI, 0.49–0.97; P=0.031; Fig. 3E) by analyzing the gene expression level and patient survival data using the Kaplan-Meier plotter database.
Figure 3.
Expression of COL4A3 and the deposition of collagen fibers in NPC. (A and B) The expression of COL4A3 in the downward progressing type of NPC was significantly lower compared with that in upward progressing type of NPC. *P<0.05. (C and D) The deposition of collagen in the downward progressing type of NPC was significantly lower compared with that in the upward progressing type of NPC. *P<0.05. (E) The association between the expression level of COL4A3 and the prognosis of head and neck squamous cell carcinoma using the Kaplan-Meier plotter database found that the high expression level of COL4A3 was associated with good prognosis (hazard ratio, 0.69; 95% CI, 0.49–0.97; P=0.031). NPC, nasopharyngeal carcinoma.
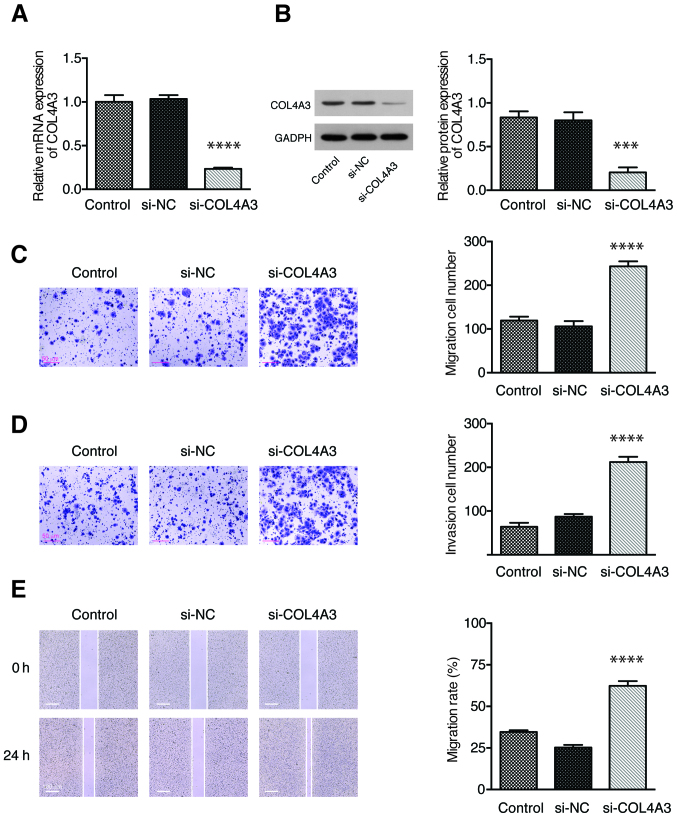
Knockdown of COL4A3 promotes the migration and invasion of NPC cells
To further confirm the role of COL4A3 in cell migration and invasion, the expression of COL4A3 was inhibited in 5-8F cells by siRNA. The expression of COL4A3 was significantly downregulated in the si-COL4A3 group compared with the si-NC group, both at mRNA (Fig. 4A) and protein (Fig. 4B) levels. The Transwell assay showed that the number of cell migration (Fig. 4C) and invasion (Fig. 4D) in the si-COL4A3 group was increased significantly compared with that in the si-NC group. A wound-healing assay was used to detect the migration ability of each group of 5-8F cell line. The migration rate of the cells in the si-COL4A3 group was significantly higher than that in the si-NC group (Fig. 4E). These results demonstrated that the inhibition of COL4A3 could promote the migration and invasion abilities of the 5-8F cell line.
Figure 4.
Knockdown of COL4A3 promoted the migration and invasion of the 5-8F cell line in vitro. The relative COL4A3 (A) mRNA and (B) protein expression levels in each group as determined by reverse transcription-quantitative PCR and western blot assay, respectively. (C) Cell migration and (D) invasion abilities were examined using Transwell assay in each group. (E) The migration rate of 5-8F cell line in each group, as detected by a wound-healing assay. Data are presented as the mean ± SD from 3 independent experiments. NC, negative control; si, short interfering. ***P<0.001 and ****P<0.0001 vs. si-NC.
Discussion
Metastasis is the major cause of death in patients with cancer. Cancer metastasis is considered to be primarily associated with changes in the tumor microenvironment, particularly in the remodeling of extracellular matrix characterized by the degradation, deposition and cross-linking of type IV collagen (20–22). The extracellular matrix is a barrier for tumor cell metastasis. Type IV collagen, from the extracellular matrix, is the primary component of the basement membrane. The expression of type IV collagen affects the deposition of the tumor extracellular matrix; thus, affecting the potential of tumor metastasis. The investigation into the effect of type IV collagen on extracellular matrix deposition and its function is beneficial for understanding the molecular mechanism of tumor metastasis and provide novel ideas for identifying molecular markers and effective treatment for tumor metastasis.
The COL4A3 domain binds and inhibits the proliferation of melanoma and other epithelial tumor cell lines in vitro (23). It is well known that the α3 (IV) chain encoded by COL4A3, is lysed by matrix metalloproteinase-9 (MMP9) to produce the primary bioactive fragment, tumstatin, which can inhibit the formation of blood vessels in vivo and inhibit the proliferation and metastasis of tumors (24). In the early stage of tumorigenesis, the MMP-mediated mechanism promotes the release of tumstatin (also an endogenous angiogenic inhibitor), by separating it from the basement membrane, to permit its anti-angiogenesis and antitumor activities (25). These two antitumor properties have been found to be regulated by an ITGB3-mediated RGD-independent mechanism. Once tumstatin has been dissociated from the basement membrane, it interacts with integrin αVβ3 in endothelial cells, which leads to the arrest of the cell cycle or apoptosis (26), and has been found to cause apoptosis (27,28) and block the proliferation of endothelial cells (29). Overexpression of the tumstatin domain, in a mouse melanoma model, inhibited tumor cell invasive properties (30). Tumstatin is also the most effective type IV collagen family and is expected to become a potentially beneficial therapeutic molecule to inhibit tumor growth. Therefore, the expression level and the association of COL4A3 and typeIV collagen play an important role in tumor metastasis; however, the role of COL4A3 in the metastasis of NPC is not clear, and associated studies have not been reported.
Previous studies have demonstrated that specific COL4A3 mutations could lead to loss of function in several diseases. For example, a novel missense mutation (3725G>A, G1242D) in exon 42 of COL4A3 played a causative role in Alport syndrome and thin basement membrane nephropathy (31). Other mutations were also recently identified in a consanguineous family with autosomal recessive Alport syndrome, including IVS 22-5 T>A in the COL4A3 gene, and R1677C and R1682Q in the COL4A4 gene (32), confirming that COL4A3 and COL4A4 mutations might cause different forms of nephropathy. However, the role of COL4A3 mutations in NPC pathogenesis remains undetermined. In the present study, whole exons were sequenced and analyzed in the upward and downward progressing types of NPC. It was found that the mutation rate of the COL4A3 gene was significantly increased and the expression level of the COL4A3 protein was significantly decreased in the downward progressing type of NPC. In the KEGG signaling pathway analysis, six associated signaling pathways were identified: PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, PPAR signaling pathway, cell adhesion molecules and NF-κB signaling pathway. Among these pathways, ECM-receptor interaction was considered to be the most effective signaling pathway for the regulation of expression of COL4A3. It has been demonstrated that MMP-9 can degrade COL4A3 in the basement membrane and extracellular matrix, resulting in decreased collagen in the cornea, thinning the central cornea and forming keratoconus (33). Indeed, there are significant association between genotypic and allelic distributions of COL4A3 (G/T) and MMP-9 (A/G) polymorphisms (34). Reciprocally, the major gene variant COL4A3 may also affect the expression of associated collagens or ECM proteins, thus decreasing the amount of ECM in corneas and resulting in keratoconus (35). Therefore, ECM-receptor interaction pathway is potentially involved in the regulation of COLA43. Nonetheless, it requires more mechanistic studies to confirm the hypothesis of the present study. Currently, there is still no data on how gene mutations in COL4A3 would impact on the protein expression level. It is hypothesized that the mutation in the COL4A3 gene, in metastatic NPC might lead to the decrease in COL4A3 protein expression level and the deposition of type IV collagen. Therefore, the biopsies of the upward and downward progressing types of NPC were analyzed using IHC methods. COL4A3 protein expression was significantly lower in the downward progressing type of NPC, suggesting that abnormal COL4A3 expression could be associated with the metastasis of NPC. The results from the present study were in accordance with previous studies showing that abnormal expression of COL4A3 was found in different types of tumor (36,37). At the same time, the deposition of the extracellular matrix in downward progressing type of NPC was significantly decreased. The results indicated that the knockdown of COL4A3 might promote the invasion and migration of NPC cells. The low expression level of COL4A3 in the downward progressing type of NPC was associated with increased risk of distant metastasis, suggesting that COL4A3 expression might be a good biomarker for NPC metastasis.
However, there are still some limitations in the present study. Firstly, a small number of matched samples were collected in this study. This was partly due to availability of eligible samples, since samples that were suitable for WES should be relatively recent to ensure DNA integrity. Therefore, the number of samples was relatively small in the current study. A larger number of samples should be performed to verify the results of the present study and further clarify the gene mutation spectrum affecting the metastatic risk of NPC by WES. Next, the function of certain genetic mutation should be examined in detail. The mutation status of the aforementioned genes should be verified by PCR and Sanger sequencing. Differentially expressed genes between upward and downward progressing types of NPC could potentially be detected by performing transcriptomic sequencing. Thirdly, in vivo experiments were lacking in the present study, which help validate the hypothesis, prove the essential role of COL4A3 in NPC metastasis and uncover potential molecular mechanisms. A plasmid harboring targeted gene mutation will be constructed in the future for the detection of protein expression. The in vivo effect of COL4A3 should also be studied by manipulating its expression and functions.
In conclusion, although several limitations exist, the present study showed that the COL4A3 gene was highly expressed in the upward progressing type of NPC, but was low in the downward progressing type. This was associated with the degree of collagen fibrosis, thus affecting the migration and invasion of NPC. COL4A3 may be a novel biomarker in the diagnosis and treatment of metastatic NPC, which could be beneficial to establish a potential molecular typing method and identify the target for individualized therapy for metastatic NPC.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Scientific Research Project of Hubei Provincial Health and Family Planning Commission (grant no. WJ2019H064) and from the National Natural Science Foundation of China (grant no. 81803061).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors contributions
XY, QW and YZ conceived the study and drafted the manuscript. XY collected the clinical tissues, was responsible for the study design and provided the data. QW performed the statistical analysis. FW performed the bioinformatic data collection, integration analysis and figure processing. All authors have made substantial contributions to this manuscript. All authors confirmed and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (Ethical number: 2019084). Written informed consent was obtained from each living patient or from their relatives for deceased patients in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Carioli G, Negri E, Kawakita D, Garavello W, La Vecchia C, Malvezzi M. Global trends in nasopharyngeal cancer mortality since 1970 and predictions for 2020: Focus on low-risk areas. Int J Cancer. 2017;140:2256–2264. doi: 10.1002/ijc.30660. [DOI] [PubMed] [Google Scholar]
- 3.Sun P, Chen C, Chen XL, Cheng YK, Zeng L, Zeng ZJ, Liu LZ, Su Y, Gu MF. Proposal of a clinical typing system and generation of a prognostic model in patients with nasopharyngeal carcinoma from Southern China. J BUON. 2014;19:474–483. [PubMed] [Google Scholar]
- 4.Liang WJ, Qiu F, Hong MH, Guo L, Qin HD, Liu QC, Zhang XS, Mai HQ, Xiang YQ, Min HQ, et al. Differentially expressed genes between upward and downward progressing types of nasopharyngeal carcinoma. Chin J Cancer. 2008;27:460–465. (In Chinese) [PubMed] [Google Scholar]
- 5.Mo L, Weng J, Zeng F, Li X, Liu B, Li Z, Kuang G. The relationship between extend types and distant metastasis of nasopharyngeal carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;24:554–555, 558. (In Chinese) [PubMed] [Google Scholar]
- 6.Chew MM, Gan SY, Khoo AS, Tan EL. Interleukins, laminin and Epstein - Barr virus latent membrane protein 1 (EBV LMP1) promote metastatic phenotype in nasopharyngeal carcinoma. BMC Cancer. 2010;10:574. doi: 10.1186/1471-2407-10-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissey MA, Jayadev R, Miley GR, Blebea CA, Chi Q, Ihara S, Sherwood DR. SPARC promotes cell invasion in vivo by decreasing type IV collagen levels in the basement membrane. PLoS Genet. 2016;12:e1005905. doi: 10.1371/journal.pgen.1005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanjore H, Kalluri R. The role of type IV collagen and basement membranes in cancer progression and metastasis. Am J Pathol. 2006;168:715–717. doi: 10.2353/ajpath.2006.051321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timpl R, Wiedemann H, van Delden V, Furthmayr H, Kühn K. A network model for the organization of type IV collagen molecules in basement membranes. Eur J Biochem. 1981;120:203–211. doi: 10.1111/j.1432-1033.1981.tb05690.x. [DOI] [PubMed] [Google Scholar]
- 10.Mundel TM, Kalluri R. Type IV collagen-derived angiogenesis inhibitors. Microvasc Res. 2007;74:85–89. doi: 10.1016/j.mvr.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech. 2008;71:357–370. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda K, Iyama K, Ishikawa N, Egami H, Nakao M, Sado Y, Ninomiya Y, Baba H. Loss of expression of type IV collagen alpha5 and alpha6 chains in colorectal cancer associated with the hypermethylation of their promoter region. Am J Pathol. 2006;168:856–865. doi: 10.2353/ajpath.2006.050384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, Iyama K, Kitaoka M, Ninomiya Y, Oohashi T, Sado Y, Ono T. Differential expression of alpha 1(IV), alpha 2(IV), alpha 5(IV) and alpha 6(IV) collagen chains in the basement membrane of basal cell carcinoma. Histochem J. 1997;29:563–570. doi: 10.1023/A:1026428010104. [DOI] [PubMed] [Google Scholar]
- 14.Dehan P, Waltregny D, Beschin A, Noel A, Castronovo V, Tryggvason K, De Leval J, Foidart JM. Loss of type IV collagen alpha 5 and alpha 6 chains in human invasive prostate carcinomas. Am J Pathol. 1997;151:1097–1104. [PMC free article] [PubMed] [Google Scholar]
- 15.Pan XX, Tong LH, Chen YF, Li FL, Tang WB, Liu YJ, Yang WA. simplified T classification based on the 8th edition of the UICC/AJCC staging system for nasopharyngeal carcinoma. Cancer Manag Res. 2019;11:3163–3169. doi: 10.2147/CMAR.S185860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Fedyunin I, Kirchner S, Xiao C, Valleriani A, Ignatova Z. FANSe: An accurate algorithm for quantitative mapping of large scale sequencing reads. Nucleic Acids Res. 2012;40:e83. doi: 10.1093/nar/gks196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei X, Li S, He J, Du H, Liu Y, Yu W, Hu H, Han L, Wang C, Li H, et al. Tumor-secreted PAI-1 promotes breast cancer metastasis via the induction of adipocyte-derived collagen remodeling. Cell Commun Signal. 2019;17:58. doi: 10.1186/s12964-019-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo S, Deng CX. Effect ofstromal cells in tumor microenvironment on metastasis initiation. Int J Biol Sci. 2018;14:2083–2093. doi: 10.7150/ijbs.25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur A, Ecker BL, Douglass SM, Kugel CH, III, Webster MR, Almeida FV, Somasundaram R, Hayden J, Ban E, Ahmadzadeh H, et al. Remodeling of the collagen matrix in aging skin promotes melanoma metastasis and affects immune cell motility. Cancer Discov. 2019;9:64–81. doi: 10.1158/2159-8290.CD-18-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chelberg MK, Tsilibary EC, Hauser AR, McCarthy JB. Type IV collagen-mediated melanoma cell adhesion and migration: Involvement of multiple, distinct domains of the collagen molecule. Cancer Res. 1989;49:4796–4802. [PubMed] [Google Scholar]
- 24.Hamano Y, Zeisberg M, Sugimoto H, Lively JC, Maeshima Y, Yang C, Hynes RO, Werb Z, Sudhakar A, Kalluri R. Physiological levels of tumstatin, a fragment of collagen IV alpha3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via alphaV beta3 integrin. Cancer Cell. 2003;3:589–601. doi: 10.1016/S1535-6108(03)00133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamano Y, Kalluri R. Tumstatin, the NC1 domain of alpha3 chain of type IV collagen, is an endogenous inhibitor of pathological angiogenesis and suppresses tumor growth. Biochem Biophys Res Commun. 2005;333:292–298. doi: 10.1016/j.bbrc.2005.05.130. [DOI] [PubMed] [Google Scholar]
- 26.Maeshima Y, Colorado PC, Kalluri R. Two RGD-independent alpha vbeta 3 integrin binding sites on tumstatin regulate distinct anti-tumor properties. J Biol Chem. 2000;275:23745–23750. doi: 10.1074/jbc.C000186200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang GM, Zhang YM, Fu SB, Liu XH, Fu X, Yu Y, Zhang ZY. Effects of cloned tumstatin-related and angiogenesis-inhibitory peptides on proliferation and apoptosis of endothelial cells. Chin Med J (Engl) 2008;121:2324–2330. doi: 10.1097/00029330-200811020-00020. [DOI] [PubMed] [Google Scholar]
- 28.Maeshima Y, Yerramalla UL, Dhanabal M, Holthaus KA, Barbashov S, Kharbanda S, Reimer C, Manfredi M, Dickerson WM, Kalluri R. Extracellular matrix-derived peptide binds to alphavbeta3 integrin and inhibits angiogenesis. J Biol Chem. 2001;276:31959–31968. doi: 10.1074/jbc.M103024200. [DOI] [PubMed] [Google Scholar]
- 29.Maeshima Y, Sudhakar A, Lively JC, Ueki K, Kharbanda S, Kahn CR, Sonenberg N, Hynes RO, Kalluri R. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science. 2002;295:140–143. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- 30.Pasco S, Ramont L, Venteo L, Pluot M, Maquart FX, Monboisse JC. In vivo overexpression of tumstatin domains by tumor cells inhibits their invasive properties in a mouse melanoma model. Exp Cell Res. 2004;301:251–265. doi: 10.1016/j.yexcr.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 31.Hou P, Chen Y, Ding J, Li G, Zhang H. A novel mutation of COL4A3 presents a different contribution to Alport syndrome and thin basement membrane nephropathy. Am J Nephrol. 2007;27:538–544. doi: 10.1159/000107666. [DOI] [PubMed] [Google Scholar]
- 32.Rana K, Tonna S, Wang YY, Sin L, Lin T, Shaw E, Mookerjee I, Savige J. Nine novel COL4A3 and COL4A4 mutations and polymorphisms identified in inherited membrane diseases. Pediatr Nephrol. 2007;22:652–657. doi: 10.1007/s00467-006-0393-y. [DOI] [PubMed] [Google Scholar]
- 33.Saravani R, Yari D, Saravani S, Hasanian-Langroudi F. Correlation between the COL4A3, MMP-9, and TIMP-1 polymorphisms and risk of keratoconus. Jpn J Ophthalmol. 2017;61:218–222. doi: 10.1007/s10384-017-0503-3. [DOI] [PubMed] [Google Scholar]
- 34.Saravani S, Yari D, Saravani R, Azadi Ahmadabadi C. Association of COL4A3 (rs55703767), MMP-9 (rs17576)and TIMP-1 (rs6609533) gene polymorphisms with susceptibility to type 2 diabetes. Biomed Rep. 2017;6:329–334. doi: 10.3892/br.2017.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao XD, Chen XN, Zhang YY, Chen P, Wei C, Shi WY, Gao H. Multi-level consistent changes of the ECM pathway identified in a typical keratoconus twins family by multi-omics analysis. Orphanet J Rare Dis. 2020;15:227. doi: 10.1186/s13023-020-01512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang CP, Wu BH, Chen SP, Fu MY, Yang M, Liu F, Wang BQ. High COL4A3 expression correlates with poor prognosis after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Tumour Biol. 2013;34:415–420. doi: 10.1007/s13277-012-0565-2. [DOI] [PubMed] [Google Scholar]
- 37.Nie XC, Wang JP, Zhu W, Xu XY, Xing YN, Yu M, Liu YP, Takano Y, Zheng HC. COL4A3 expression correlates with pathogenesis, pathologic behaviors, and prognosis of gastric carcinomas. Hum Pathol. 2013;44:77–86. doi: 10.1016/j.humpath.2011.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.