Abstract
Acute hepatopancreatic necrosis disease (AHPND) caused by Vibrio parahaemolyticus resulted in great economic losses in global shrimp aquaculture. There is an urgent need for development of novel strategies to combat AHPND-causing V. parahaemolyticus (Vp AHPND), given that one of the greatest challenges currently is the widespread use of antibiotics and subsequent emergence of multidrug-resistant bacteria. Here, we proposed a broad-spectrum antivirulence approach targeting a conserved histidine kinase, QseC, which has been demonstrated to activate virulence expression in several Gram-negative pathogens. Our results showed that QseC mediated the catecholamine stimulated effects on growth and flagellar motility of Vp AHPND. Transcriptome analysis revealed that QseC was involved in the global regulation of the virulence of Vp AHPND as the ΔqseC mutant exhibited a decreased expression of genes related to type IV pilin, flagellar motility, and biofilm formation, while an overexpression of type VI secretion system and cell wall biosynthesis. Subsequently, the bacterial catecholamine receptor antagonist LED209 not only neutralized the stimulatory effects of host catecholamines on the growth and motility of Vp AHPND in vitro, but also attenuated the virulence of Vp AHPND towards brine shrimp larvae and white shrimp in vivo. Additionally, LED209 presented no interference with pathogen growth, nor the toxicity to the experimental animals. These results suggest that QseC can be an attractive antivirulence therapy target, and LED209 is a promising candidate for development of broad-spectrum antivirulence agents. This is the first study that demonstrated the role of QseC in the global regulation of Vp AHPND infection and demonstrated the antivirulence potential of LED209, which provides insight into the use of an antivirulence approach for targeting not only Vp AHPND, but also a much larger collection of pathogenic bacteria.
Keywords: acute hepatopancreatic necrosis disease, Vibrio parahaemolyticus, antivirulence therapy, QseC, LED209
Introduction
Vibrio parahaemolyticus is a gram-negative, halophilic bacterium that is disseminated worldwide in marine and estuarine environments (Jones, 2014). It is frequently isolated from environmental, seafood and clinical samples, and is considered to be the leading cause of human foodborne illness in the United States and Asian countries (Newton et al., 2012; Yu et al., 2013). Specific virulent strains of V. parahaemolyticus, which contain a ~70 kb plasmid with genes encoding homologues of the Photorhabdus insect-related (Pir) binary toxin PirABVP, have recently been identified as the causative agent of acute hepatopancreatic necrosis disease (AHPND) in global shrimp industry (Tran et al., 2013). AHPND can lead to mass mortalities (up to 100%) within the first 30–35 days after stocking shrimp post-larvae or juveniles in grow-out ponds (FAO, 2013). Till now, multiple shrimp species have been listed as being susceptible to AHPND-causing vibrios, including giant tiger prawn (Penaeus monodon), whiteleg shrimp (Penaeus vannamei), and fleshy prawn (Penaeus chinensis) (Joshi et al., 2014; Hong et al., 2016; Reantaso, 2016).
Research in terms of AHPND has been mostly focused on understanding its pathogenesis and identifying potential pathogens. Due to the current shortage of effective drugs in the shrimp farming practices, the widespread use of antibiotics and subsequently emergence of multidrug-resistant bacteria are increasing at an alarming rate and constitute one of our greatest challenges in combating AHPND-causing V. parahaemolyticus (Vp AHPND). Therefore, the development of alternative antimicrobial therapies to remedy this disease is urgently needed.
Among the novel therapeutic strategies, antivirulence therapy has emerged as a promising alternative. Instead of directly killing the bacteria, this strategy is aimed at depriving them from their essential virulence factors and preventing the attack on the host. Thus, it would exert a relatively mild selective pressure over pathogens than that exerted by the classical bactericidal or bacteriostatic antibiotic activities (Defoirdt, 2013; Vale et al., 2016; Dickey et al., 2017; Munguia and Nizet, 2017). In addition, the spread of resistance determinants through horizontal gene transfer would also be reduced since most virulence factors are usually restricted to a single few closely related species. Moreover, another encouraging potential of antivirulence agents is that they have less impact on the normal microbiota of the host, overcoming undesirable side effects of traditional antibiotic therapy (Buroni and Chiarelli, 2020).
A variety of virulence factors have been currently under investigation as potential antivirulence targets, such as adhesion and motility, biofilm formation, siderophores, and secretion systems (Donlan and Costerton, 2002; Gerlach and Hensel, 2007; Skaar, 2010; Maier and Wong, 2015). Since the production of virulence factors is energetically costly, the expression of virulence genes is usually controlled by complex regulatory mechanisms; therefore, disruption of these regulatory mechanisms could affect the production of several virulence factors (Defoirdt, 2018). Furthermore, the signal receptor proteins of bacteria are usually highly conserved, indicating that the same signal inhibitor can inhibit multiple bacteria and strains, reflecting the broad-spectrum nature of this method (Defoirdt, 2014). Given these properties, the possibility to interfere with the regulatory networks of multiple virulence determinants would be one of the most promising targets for antivirulence therapies. Among them, signaling pathways such as quorum sensing and two-component sensing have gained particular attention (Defoirdt, 2018).
A broad spectrum of bacterial pathogens rely on QseC, which is a conserved membrane-bound histidine sensor kinase belonging to the QseBC two-component system, to recognize the host environment by sensing and responding to the host catecholamine stress hormones in order to promote the expression of different virulence factors (Rasko et al., 2008; Weigel and Demuth, 2016). The catecholamine stress hormones epinephrine (EPI), norepinephrine (NE), and dopamine (DA) are an integral part of the stress response in humans (Reiche et al., 2004) and stimulate the growth of several pathogens in serum-based media (Lyte and Ernst, 1992; Coulanges et al., 1997; Yang et al., 2014; Boyanova, 2017). Suong et al. (2017) also demonstrated that the catecholamines NE and DA could significantly increase the motility and virulence of Vp AHPND.
Catecholamines exert their effects by binding to specific receptors. Several bacterial catecholamine receptors have also been reported, including the histidine sensor kinases QseC (Rasko et al., 2008). QseC homologs are widely present in at least 25 important human and plant pathogens, such as enterohemorrhagic Escherichia coli (EHEC; O157:H7), Salmonella enterica, uropathogenic E. coli, Haemophilus influenza, Aeromonas hydrophila, Edwardsiella tarda, and Francisella tularensis (Weiss et al., 2007; Bearson and Bearson, 2008; Kostakioti et al., 2009; Wang et al., 2011; Khajanchi et al., 2012; He et al., 2018). Thus, QseC represents a novel target for antivirulence strategy, and small molecule inhibitors have already been reported to show promise as broad-spectrum antimicrobials.
LED209 [N-phenyl-4-(3-phenylthioureido)benzenesulfonamide] is the first reported bacterial catecholamine receptor antagonist (Rasko et al., 2008). It blocks the autophosphorylation of QseC and thus blocks its binding to catecholamines, thereby inhibiting activation of the expression of several virulence genes mediated by QseC. LED209 has exhibited good potential for antivirulence activity of several bacterial pathogens, including S. enterica ser. Typhimurium, F. turalensis, adherent-invasive E. coli, and V. harveyi (Rasko et al., 2008; Curtis et al., 2014; Yang et al., 2014).
Catecholamine sensing has been reported to affect the virulence of a Vp AHPND strain isolated from outbreaks in Vietnam and LED209 has been proved to be effective for the inhibition of this moderate-virulence strain (LD50 = 6 × 106 CFU/ml-1) (Suong et al., 2017). In this article, we examined i) whether QseC mediates catecholamine-induced effects in Vp AHPND; and ii) whether QseC blockade with LED209 could be a promising antivirulence therapy for a high virulent Vp AHPND strain (LD50 = 5 × 104 CFU/ml-1). This work may shed lights on a better understanding of factors involved in AHPND epidemiology and provide insight into the development of potential antivirulence agents for the prevention and control of AHPND.
Materials and Methods
Bacterial Strains and Growth Conditions
A Vp AHPND strain 123 was used in this study, which previously isolated from an AHPND outbreak and caused 100% shrimp mortality in Hebei province of China. Bacterial strains and plasmids used in this study were listed in Table 1 . Unless otherwise stated, Vp AHPND strains were cultured at 28°C in Tryptic Soy Broth containing 30 g/L−1 NaCl (TSB30) under constant agitation (100 min−1), while Escherichia coli strains were cultured in Luria broth (LB; Difco) at 37°C. Cell densities were measured spectrophotometrically at 600nm.When required, ampicillin (Amp), chloramphenicol (Cm), IPTG, X-Gal and L-arabinose were supplemented at final concentrations of 100, 34, 100, 50 µg ml−1 and 2 mg ml−1, respectively.
Table 1.
Strains and plasmids used in this study.
| Strains | Description | Source |
|---|---|---|
| Vibrio parahaemolyticus | ||
| Vp AHPND 123 | Wild type, isolated from Penaeus vannamei, China, Cms, Ampr | This study |
| ΔqseC | Vp AHPND 123, in-flame deletion of qseC | This study |
| qseC+ | Vp AHPND 123, ΔqseC, containing pCqseC | This study |
| Escherichia coli | ||
| SY327 (λpir) | Host strain for π-dependent plasmids | Miller and Mekalanos, 1988 |
| Plasmids | ||
| pRE112 | Suicide vector, π-dependent, sacB, Cmr | Edwards et al., 1998 |
| pUCm-T | Cloning vector, Ampr | Sangon, Shanghai |
| pdCas9-bacteria | Expressing vector, Cmr | Addgene 44249 |
| pRE112ΔqseC | pRE112 derivative for qseC in-frame deletion | This study |
| pCqseC | QseC expressing plasmid, Cmr | This study |
Catecholamines and Bacterial Catecholamine Receptor Antagonist
Epinephrine hydrochloride (EPI) and Dopamine hydrochloride (DA) were dissolved in distilled water at 10 mM and were sterilized using a 0.22 μM filter, while norepinephrine (NE) and LED209 were dissolved in DMSO at 10 mM. EPI and DA were purchased from Sigma-Aldrich, Missouri, USA. NE and LED209 were obtained from Cayman Chemical, Michigan, USA. The reagents were stored at -20°C.
Construction of qseC In-Frame Deletion Mutant Vp AHPND 123ΔqseC
A qseC in-frame deletion (ΔqseC) mutant of Vp AHPND strain 123 was constructed by double crossover allelic exchange using suicide vector pRE112. In brief, two pairs of primers, qseC-LF/qseC-LR and qseC-RF/qseC-RR ( Table 2 ), were applied to amplify fragments upstream (797 bp) and downstream (668 bp) of qseC gene from chromosomal DNA, respectively. Both fragments were purified and fused in a subsequent overlap PCR using primers qseC-LF and qseC-RR. The fused segment (ΔqseC) was ligated into pRE112 at HindIII site and introduced into E. coli SY327 (λpir). The resulting plasmid, pRE112ΔqseC, was verified by PCR and sequencing and then introduced into Vp AHPND 123 by electroporation. The positive clone with pRE112ΔqseC integrated into the chromosome by a single crossover event was selected on TSA30 (Tryptic Soy Agar with 30g L-1 NaCl) containing ampicillin and chloramphenicol with primers Check-F/R. Then the excision of pRE112 from the chromosome was achieved by a second crossover with counter-selection on TSA30 containing 10% sucrose. The resulting ΔqseC mutant was verified by PCR and sequencing.
Table 2.
Primers used in this study.
| Primers | Sequences (5’-3’) | References |
|---|---|---|
| Construction of mutant and complementary strain | ||
| qseC-LF | TCGAAGCTTAGAAGCGAAAGTGTATTTGAC | This study |
| qseC-LR | CTATTGTTGCGCGTTTAATGGAGTAAGGTTT | |
| qseC-RF | CATTAAACGCGCAACAATAGAGCTGATTCC | This study |
| qseC-RR | CATAAGCTTTTTCGGGTAGATGAGGTATTC | |
| Check-F | GCAGAAGAAGTCATTCAAGTT | This study |
| Check-R | GGATGAACAACCAGCATAAC | |
| C-F | CATCTTAAGGAACTATTTTTATGGAGACTTCCTTG | This study |
| C-R | CTTCTCGAGCTAGTACTTTGGGAATGCGAC | |
| Quantitative real-time reverse transcription PCR | ||
| recA-F | GCTAGTAGAAAAAGCGGGTG | Ma et al., 2015 |
| recA-R | GCAGGTGCTTCTGGTTGAG | |
| qseC-F | TCAGCCAGAACAGCAACA | This study |
| qseC-R | AGACCGAGCATGAGAACG | |
| flaC-F | ACCGTATCGCAGAAACCACA | This study |
| flaC-R | GCCAGGAAGCGTGATTTTTA | |
| flaF-F | TTTGGATGTGGCGGTTCG | This study |
| flaF-R | GCTCGGCTTGGCTGTTTG | |
| flaK-F | GCGTATGGTCAGGCTGTATC | This study |
| flaK-R | TGTGGGTAATTTAGCGGTTG | |
| fliA-F | CCGACGAGCAAGATTATTACA | This study |
| fliA-R | ATCCAAGACGAGGGCTATTT | |
| motB-F | CTTTACGGTGCGGTGTTG | This study |
| motB-R | GCGGTTAAGCGTTTCTTG | |
| cpsA-F | GTATTACTCCGTTTGGTGG | This study |
| cpsA-R | CTGGTTTGACTTTGTGGC | |
| vasD-F | ATGCTCCGCCGCTAAT | This study |
| vasD-R | ACAACTGGTGATGGTCTTC | |
Construction of Complementary Strain Vp AHPND 123qseC+
To construct a complementary strain of Vp AHPND 123ΔqseC, the intact gene containing the putative promoter region was amplified by primers C-F and C-R and introduced into pdCas9-bacteria (addgene 44,249) at AflII/XhoI sites. The resulting QseC-expression plasmid, designated pCqseC, was then introduced into Vp AHPND 123ΔqseC by electroporation and verified by PCR and sequencing, resulting the complementary strain Vp AHPND 123qseC+.
Genome-Wide Transcriptomic Analysis of the Wild Type and ΔqseC Mutant
Wild type and ΔqseC mutant strains were grown in Dulbecco’s modified Eagle medium (DMEM; Invitrogen Life Technologies, Carlsbad, CA) with shaking at 100 rpm for 24 h. The total RNA was extracted using the RNeasy Protect Bacteria Mini Kit (Qiagen, Hilden, Germany) as per the manufacturer’s recommendation, and paired-end sequencing was performed on the Illumina Hi-Seq 2000 platform of the LC-Bio Technology Co., Ltd. (Hangzhou, Zhejiang, China). The raw transcriptomic sequencing data were submitted to GenBank (NCBI) under the BioProject No. PRJNA668022. Raw reads of transcriptome sequencing were mapped to genome 123 by Bowtie2-2.2.3. Differential expression analysis was performed for three biological replicates of two strains using the DESeq R package (1.18.0). Genes were considered differentially expressed with over 1.5-fold change and p-values <0.01 in samples.
Quantitative Reverse Transcription PCR (RT-qPCR)
To analyze the gene expression of seven genes, including qseC, cpsA (relating to biofilm formation), flaF and flaC (encoding polar flagellins), flaK (encoding σ54-interacting regulator), fliA (encoding polar flagellin specific chaperon), motB (encoding Na+ motor component), and vasD (associating with T6SS) between wild type and mutant strains, we performed RT-qPCR using methods described by Fu et al. (2020). The RT-qPCR assay was performed on three technical and three biological replicates for each sample. The expression levels of these genes were normalized with the house keeping gene recA. The fold changes relative to recA were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). RT-qPCR data of three independent experiments are showed as the Mean ± SD (standard deviation).
Bacterial Growth Assays
Standard American Petroleum Institute (SAPI) medium with serum was employed in this study to mimic the in vivo conditions within a host (Lyte and Ernst, 1992). Serum-SAPI medium contains limited nutrients, including 6.25 mM NH4NO3, 1.84 mM KH2PO4, 3.35 mM KCl, 1.01 mM MgSO4, and 2.77 mM glucose, supplemented with 30% (v/v) adult bovine serum (Biological industries, Israel). The medium pH was adjusted to 7.5 and filter-sterilized.
Different Vp AHPND strains were grown overnight in TSB30 at 28°C, after which the cultures were re-inoculated at a concentration of 102 CFU ml−1 into fresh serum-SAPI medium, with and without EPI (100 μM), NE (50 μM), or DA (50 μM). Additionally, different concentrations of bacterial catecholamine receptor antagonist were added in conjunction with the catecholamines to determine whether it could neutralize catecholamine-induced growth responses. The cultures were grown statically in 30 ml volumes at 28°C for 48 h in a 5% CO2 humidified incubator, after that the number of bacteria was enumerated by standard pour-plate analysis using TSA30 (Freestone and Lyte, 2008). Controls comprised equivalent volumes of the solvent used to dissolve the catecholamines or the antagonists. Growth response assays were determined for three independent cultures, and all experiments were conducted on at least three separate occasions.
Growth curves of different strains were also determined. Overnight cultures were prepared for all strains (i.e., Vp AHPND 123 wild type, ΔqseC mutant and complementary strain qseC+), then the cultures were re-inoculated into fresh TSB 30. Additionally, catecholamine receptor antagonist LED209 and equivalent volume of the solvent (DMSO) were added to evaluate whether they could affect the growth of Vp AHPND 123. The cultures were grown at 28°C for 36 h, and the turbidity at 600 nm was monitored every three hours. Growth curves were determined for three independent cultures.
Swimming Motility Assay
The swimming motility assay was performed on soft agar (TSA 30 plates containing 0.3% agar) as described previously (Yang and Defoirdt, 2015). The catecholamines and antagonist were directly added to the autoclaved agar. Vp AHPND strains were grown in TSB30 until OD600 = 1.0, and 2 μl aliquots were spotted in the center of the soft agar plates. The diameters of the motility halos were measured after incubation at 28°C for 18 h. All assays were conducted with freshly prepared plates in three replicates.
Biofilm Formation Assay
Biofilm formation assay was quantified using crystal violet staining as described by Yang et al. (2014). In brief, overnight cultures of Vp AHPND strains were diluted to an OD600 of 0.1 in TSB30, and were pipetted into a 96-well plate with 200 μl aliquots of each well. Then the plate was incubated without agitation for 24 h at 28°C to allow the bacteria adhere and grow. After then the cultures were removed and the wells were washed three times with 300 μl sterile physiological saline. The remaining attached bacteria were fixed with 200 μl of methanol per well for at least 20 min, after which the methanol was removed and plates were air-dried. Subsequently, biofilms were stained with 200 μl of a 0.1% crystal violet solution (Sigma-Aldrich, Missouri, USA) per well for 15 min. Excess stain was rinsed off by placing the plate under running tap water, and washing was continued until the washings were free of the stain. After the plates were air dried, the dye bound to the adherent cells was resolubilized with 200 μl of 95% ethanol per well, and absorbance was measured at 570 nm. Sterile medium served as negative control.
Axenic Hatching of Brine Shrimp Larvae and Brine Shrimp Challenge Test
Five hundred milligrams of high-quality hatching cysts of Artemia franciscana (EG® Type; INVE Aquaculture, Baasrode, Belgium) were hydrated in 45 ml of sterile distilled water for 1 h. Sterile cysts were obtained by decapsulation according to the method described by Marques et al. (2005). Briefly, 1.65 ml of NaOH (32%; w/v) and 25 ml of NaOCl (50% available chlorine) were added to the hydrated cyst suspension to facilitate decapsulation. The process was stopped after 2 min by adding 35 ml of Na2S2O3 (10 g L−1). Filtered (0.22 μm) aeration was provided during the reaction. The decapsulated cysts were washed with filtered (passed through 0.45 μm membrane filter) and autoclaved (moist heat at 121°C for 15 min) artificial seawater (containing 35 g/L-1 of instant ocean synthetic sea salt, Aquarium Systems, Sarrebourg, France). Afterwards, the cysts were re-suspended in 500 ml of filtered, autoclaved seawater and hatched for at least 28 h at 28°C with constant illumination (c. 2000 lux). Air was bubbled through the suspension by a sterile glass tube extending to the bottom of the hatching vessel to keep all the cysts in continuous motion. The axenity of cysts was verified by inoculating 1 ml of culture water into 9 ml of TSB30 and incubating at 28°C for 24 h. After 28 h of hatching, batches of 30 larvae were counted and transferred to fresh, sterile 50 ml glass tubes containing 30 ml of filtered and autoclaved seawater. All manipulations were performed in a laminar flow to maintain sterility of the cysts and larvae. Finally, the tubes were incubated on a rotator (4 min−1) under 28°C.
The effects of the bacterial catecholamine antagonist on the virulence of Vp AHPND strains were determined in a standardized challenge test with gnotobiotic brine shrimp larvae. The challenge tests were performed as described by Yang et al. (2014) with some modifications. Vp AHPND was incubated overnight and cultures were washed with phosphate-buffered saline (pH 7.4) prior to inoculation into the brine shrimp rearing water at 106 CFU ml−1, the bacterial catecholamine antagonist was added directly into the rearing water. Brine shrimp cultures without challenging with Vp AHPND strains were used as controls. The survival of the larvae was counted 48 h after the addition of the pathogens. Each treatment was carried out in triplicate and each experiment was repeated three times to verify the reproducibility. In each test, the sterility of the control treatments was checked at the end of the challenge by inoculating 1 ml of rearing water to 9 ml of TSB30 and incubating the mixture for 2 days at 28°C.
Conventional White Shrimp Challenge Test
Before the challenge test, healthy white shrimp (Penaeus vannamei; 4.6 ± 0.48 g) were transferred to 50 L seawater in plastic tanks, and were acclimated for one week under the following conditions: 30 shrimp/tank, natural photoperiod, seawater at salinity 30, pH 7.8, constant aeration and controlled temperature at 28 ± 1°C. Shrimp were fed with a commercial shrimp diet and water was exchanged at a rate of 30% per day to remove feces and food that was not ingested.
The immersion challenge tests were conducted according to Tran et al. (2013) with some modifications. Different Vp AHPND strains were cultured in TSB30 at 28°C under constant agitation (100 min−1) with or without different catecholamines and corresponding antagonists until the OD600 reached 0.8, afterwards, the bacterial suspension was adjusted to 1×108 CFU ml−1. The immersion procedure was carried out by immersing 20 shrimp in 6 L of this suspension for 15 min with aeration. Shrimp in the negative control group were immersed in sterile TSB30 broth. Following the immersion, the shrimp and bacterial suspension were transferred directly into an experimental tank containing 20 L of clean seawater to obtain an approximate bacterial density of 1×106 CFU ml−1. The cumulative mortality was recorded every 2 h within 48 h.
Activity Measurement of Immune-Related Enzymes
The activities of superoxide dismutase (SOD) and catalase (CAT) in the shrimp tissues from the bacteria challenge experiment were measured by the commercial kit (Nanjing, Jiancheng, A001-1 and A007) according to the operation instruction. Total SOD activity was determined by the hydroxylamine method. One SOD activity unit was defined as the enzyme amount causing 50% inhibition in 1 ml reaction solution. Total CAT activity was determined by using the spectrophotometry to measure yellowish complex compound yielded from the reaction between hydrogen peroxide and ammonium molybdate. One CAT enzyme activity unit was defined as the amount of protein to degradate 1 mmol hydrogen peroxide per second. The concentration of total protein in the supernate was quantified by BCA method. The SOD and CAT activities in supernate sample were the ratio of total enzyme activity unit to the total protein respectively, and the results were expressed as U mg−1 protein.
Statistical Analysis
All data were presented as the mean ± standard deviation (SD). Data analysis was carried out using the SPSS statistical software (version 26); t-test or one-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests was performed for between-group analyses. The survival of white shrimp was analyzed and expressed as a Kaplan–Meier survival curve. A probability (p) value <0.05 was considered as statistically significant, and a probability (p) value <0.01 was considered as extremely significant.
Results
QseC Mediates Catecholamine-Induced Effects on Growth and Motility in Vp AHPND
The bacterial receptor QseC, which is a membrane-bound histidine sensor kinase, has been reported to sense the bacterial signal autoinducer-3 (AI-3) and the host stress catecholamines epinephrine and/or norepinephrine (Clarke et al., 2006). Various bacterial pathogens have exploited the QseC signaling cascade to mediate virulence, such as enterohemorrhagic E. coli and S. enterica serovar Typhimurium (Rasko et al., 2008; Curtis et al., 2014; Moreira et al., 2016). Therefore, to verify the role of QseC in the pathogen response to catecholamines, we generated qseC in-frame deletion mutant (ΔqseC) and a complemented strain (qseC+).
The growth response of Vp AHPND strains to different catecholamines was investigated in a minimal salts medium supplemented with 30% (v/v) adult bovine serum (serum-SAPI), which exposed Vp AHPND to conditions similar to those inside a host, including limited nutrient availability, iron limitation, and immune defense antibodies. The results revealed that the addition of all three kinds of catecholamines tested (i.e., EPI, NE, and DA) could significantly increase the growth of Vp AHPND 123 wild type and complemented strain, but not of ΔqseC mutant ( Figure 1A ). However, the catecholamines had no effect on the growth of Vp AHPND strains in medium without serum (data not shown).
Figure 1.
QseC mediates catecholamine-induced growth and flagellar motility in Vp AHPND strains. (A) Viable counts of Vp AHPND wild type 123, ΔqseC mutant and complementary strain qseC+ after 48 h of incubation from an inoculum of 102 CFU ml-1 in serum-SAPI medium with catecholamines; (B) Sizes of motility halos after 18 h of incubation on TSA30 plates containing 0.3% agar with different catecholamines. Epinephrine (EPI), norepinephrine (NE) and dopamine (DA) were used at 100, 50, and 50 µM, respectively. Error bars represent the standard deviation of three independent cultures. Asterisks indicate significant differences when compared to untreated Vp AHPND (independent samples t- test; *p < 0.05; **p < 0.01; ***p < 0.001).
Flagellar motility is considered essential to enhance the initial interaction of a bacterium with a surface by enabling the cell to overcome negative electrostatic forces (Josenhans and Suerbaum, 2002). Several pathogenic species have been convincingly shown to require motility to infect a host, such as V. parahaemolyticus, V. harveyi, and V. cholerae (Krukonis and DiRita, 2003; Zhang and Orth, 2013; Yang and Defoirdt, 2015). Thus, we investigated the effect of QseC on catecholamines-induced swimming motility of Vp AHPND. All three catecholamines enhanced the flagellar motility of the wild type and complemented strain, while no stimulatory effect was observed in ΔqseC mutant ( Figure 1B ). These observations confirmed that QseC mediates the effects of catecholamines on Vp AHPND growth and motility.
Global Transcriptional Profile of ΔqseC Mutant
The difference between Vp AHPND 123 wild type and ΔqseC mutant strain was also analyzed at the whole transcriptome level. In total, 503 unigenes displayed different expression levels, of which 227 were up-regulated and 276 were down-regulated ( Figure 2A ). To analyze and characterize the specific differentially expressed genes (DEGs), a false discovery rate less than 5% and fold change >1.5 was selected as limitation for significant difference. Results revealed that there were two groups of genes with distinct expression profiles. Group 1 (136 genes) was up-regulated in the mutant strain, 51.4% of which distributed in nine gene clusters ( Table 3 ). Notably, 22 genes belonging to T6SS (Cluster 1) were up-regulated in the mutant strain, suggesting the deletion of qseC might result in a higher capacity to outcompete with other bacterial species. Another example of “unexpected” up-regulated response concerned the genes related to cell division (Cluster 4). The remaining seven clusters contained numerous genes controlling various cellular functions, three of which (clusters 1, 2, and 3) included a number of genes (see Table S1 ) primarily related to chromosome partition, while one cluster was involved with Copper-sensing two-component system (clusters 8), and four clusters associated with transport (clusters 5, 6, 7, and 9) were also over-expressed. In addition, a large number of genes in mutant strain involved in lipid metabolism were also up-regulated but without significant difference relative to wild type (fold change between 1.0 and 1.5) ( Table S1 ).
Figure 2.

The difference between wild type and mutant strain at the whole transcriptome level. (A) Volcano plot of differentially expressed genes (DEGs). In total, 503 unigenes present different expression levels. Red indicates upregulated expression, green indicates downregulated expression and blue indicates no significant differential expression when comparing. (B) Functional enrichment of differentially expressed genes on KEGG categorization in mutant strain.
Table 3.
Genes up-regulated (> 1.5-fold change) in the mutant strain.
| Cluster | Gene | logFC | P value | Function |
|---|---|---|---|---|
| 1 | VP1448 | 1.8407 | 8.78E-05 | Anaerobic dimethyl sulfoxide reductase chain B |
| VP1449 | 2.346482 | 1.05E-15 | Anaerobic dimethyl sulfoxide reductase chain A | |
| VP1450 | 3.442818 | 2.86E-32 | hypothetical protein | |
| VP1451 | 1.867141 | 1.08E-11 | hypothetical protein | |
| VP1452 | 2.885653 | 2.51E-23 | Outer membrane protein ImpK/VasF | |
| VP1453 | 2.951883 | 4.14E-25 | Uncharacterized protein ImpJ/VasE | |
| VP1454 | 2.990298 | 1.34E-23 | Type VI secretion lipoprotein/VasD | |
| VP1455 | 2.964036 | 9.35E-25 | Uncharacterized protein ImpI/VasC | |
| VP1456 | 2.786061 | 6.17E-22 | hypothetical protein | |
| VP1457 | 3.429694 | 2.33E-25 | Uncharacterized protein ImpA | |
| VP1458 | 4.096527 | 7.38E-41 | IcmF-related protein | |
| VP1459 | 2.94613 | 3.15E-19 | Transcriptional regulator | |
| VP1460 | 1.667593 | 9.05E-10 | Uncharacterized protein ImpH/VasB | |
| VP1461 | 2.279961 | 2.73E-16 | Protein ImpG/VasA | |
| VP1462 | 2.129158 | 6.08E-14 | Uncharacterized protein | |
| VP1463 | 2.518079 | 6.29E-20 | Uncharacterized protein ImpC | |
| VP1464 | 2.393249 | 4.34E-16 | Uncharacterized protein ImpB | |
| VP1465 | 2.476941 | 1.97E-19 | Uncharacterized protein ImpA | |
| VP1466 | 2.440396 | 6.30E-19 | Serine/threonine protein kinase | |
| VP1467 | 1.832535 | 6.93E-12 | Hcp | |
| VP1468 | 2.980778 | 3.95E-25 | ClpB-like protein | |
| VP1469 | 2.628548 | 1.12E-17 | sigma-54-dependent transcriptional regulator | |
| VP1470 | 1.536542 | 0.003818 | Uncharacterized peptidase YqjE | |
| 2 | VP1740 | 2.30968 | 1.75E-15 | FIG01199787: hypothetical protein |
| VP1741 | 2.394921 | 2.28E-17 | Chromosome segregation ATPase | |
| VP1742 | 2.438215 | 1.55E-05 | hypothetical protein | |
| 3 | VP1043 | 1.70837 | 3.81E-10 | hypothetical protein |
| VP1044 | 1.889812 | 7.64E-08 | hypothetical protein | |
| VP1045 | 1.791027 | 1.62E-11 | Chromosome partition protein MukB | |
| VP1046 | 1.945746 | 3.68E-12 | Chromosome partition protein MukE | |
| VP1047 | 2.002129 | 2.34E-13 | Chromosome partition protein MukF | |
| VP1048 | 2.21627 | 1.06E-15 | S-adenosylmethionine-dependent methyltransferase | |
| VP1049 | 1.637211 | 5.65E-10 | Positive regulator of competence TfoX | |
| 4 | VP0461 | 1.533797 | 5.29E-09 | UDP-N-acetylmuramate–L-alanine ligase |
| VP0462 | 1.756452 | 9.66E-11 | UDP-N-acetylglucosamine–N-acetylmuramyl | |
| VP0463 | 1.780125 | 2.75E-11 | Cell division protein FtsW | |
| VP0464 | 1.740744 | 8.28E-11 | UDP-N-acetylmuramoyl | |
| VP0465 | 1.743484 | 6.24E-11 | UDP-N-acetylmuramoylalanyl-D-glutamate–2 | |
| VP0466 | 1.889101 | 2.15E-12 | Cell division protein FtsI | |
| VP0467 | 2.073686 | 1.80E-14 | Penicillin-binding protein activator | |
| VP0468 | 1.984658 | 8.80E-13 | Predicted endonuclease | |
| 5 | VP0091 | 3.804372 | 1.85E-36 | UPF0118 membrane protein SMc00793 |
| VP0092 | 3.821178 | 2.33E-32 | FIG01201038: hypothetical protein | |
| VP0093 | 3.492542 | 8.14E-30 | putative membrane protein | |
| VP0094 | 3.575819 | 8.18E-22 | FIG01201571: hypothetical protein | |
| VP0095 | 4.757663 | 2.03E-50 | DUF2383 domain-containing protein | |
| VP0096 | 2.97958 | 2.04E-23 | FIG01200706: hypothetical protein | |
| VP0097 | 3.848639 | 1.97E-38 | Methylglyoxal synthase | |
| VP0098 | 3.289054 | 3.31E-26 | Hemerythrin domain protein | |
| VP0099 | 4.747694 | 5.81E-34 | hypothetical protein | |
| VP0100 | 4.948999 | 8.06E-56 | FIG01201373: hypothetical protein | |
| VP0101 | 4.774471 | 3.22E-54 | inducible periplasmic protein | |
| VP0102 | 2.187439 | 4.88E-15 | sigma-54 interacting response regulator | |
| 6 | VPA0220 | 3.603139 | 3.24E-05 | Protein ytfJ precursor |
| VPA0221 | 3.013183 | 2.07E-22 | Carbonic anhydrase alpha class | |
| VPA0222 | 3.386627 | 1.93E-13 | Long-chain fatty acid transport protein | |
| VPA0223 | 4.883209 | 1.43E-48 | Transcriptional regulator | |
| VPA0224 | 2.249278 | 6.81E-05 | hypothetical protein | |
| VPA0225 | 2.317688 | 1.16E-11 | Hemolysin | |
| 7 | VP2662 | 1.671946 | 3.19E-10 | Phospholipid ABC transporter |
| VP2663 | 1.973801 | 2.06E-13 | Phospholipid ABC transporter MlaC | |
| VP2664 | 1.781821 | 4.77E-11 | Phospholipid ABC transporter MlaB | |
| 8 | VPA0918 | 1.893487 | 1.55E-12 | FIG01199656: hypothetical protein |
| VPA0919 | 1.953746 | 2.73E-13 | Copper-sensing two-component system response | |
| VPA0920 | 2.052521 | 2.29E-14 | Signal transduction histidine kinase | |
| 9 | Unknown | 1.629797 | 1.10E-08 | hypothetical protein |
| Unknown | 1.715916 | 2.12E-10 | Outer membrane protein | |
| Unknown | 1.505204 | 1.19E-08 | hypothetical protein | |
| Unknown | 1.50863 | 7.42E-09 | hypothetical protein | |
| Unknown | 1.942236 | 4.04E-13 | hypothetical protein |
In contrast, 135 genes in group 2 were down-regulated, 47.4% of which belonged to eight gene clusters ( Table 4 ), including six genes encoding Sigma-fimbriae usher proteins, three genes encoding Arginine ABC transporter, three genes encoding Cytochrome c551 peroxidase, 15 genes related to Type II/IV secretion system, seven genes associated with flagellin biosynthesis, 10 genes associated with Glycosyltransferase, 15 genes encoding flagellin proteins, and five genes with unknown function. Genes related to biofilm formation such as sypN (encoding Glycosyltransferase), and cpsA (Capsular polysaccharide synthesis) were also down-regulated. Besides, the expression of several genes encoding Methyl-accepting chemotaxis protein, DNA protection during starvation protein and transcriptional regulator were also decreased.
Table 4.
Genes down-regulated (> 1.5-fold change) in the mutant strain.
| Cluster | Gene | logFC | pvalue | function |
|---|---|---|---|---|
| 1 | VPA1504 | -2.15582 | 1.68E-11 | Sigma-fimbriae tip adhesin |
| VPA1505 | -1.83097 | 3.17E-10 | Sigma-fimbriae usher protein | |
| VPA1506 | -1.85957 | 5.85E-10 | Sigma-fimbriae usher protein | |
| VPA1507 | -1.80196 | 6.91E-10 | Sigma-fimbriae usher protein | |
| VPA1508 | -1.66659 | 2.43E-09 | Sigma-fimbriae usher protein | |
| VPA1509 | -1.76698 | 1.29E-09 | Sigma-fimbriae chaperone protein | |
| 2 | VPA0636 | -1.55671 | 1.22E-07 | Arginine ABC transporter |
| VPA0637 | -1.69805 | 1.73E-09 | Arginine ABC transporter | |
| VPA0638 | -1.5677 | 3.61E-08 | Arginine ABC transporter | |
| 3 | VPA1099 | -1.95092 | 7.45E-08 | Predicted redox protein |
| VPA1100 | -2.42168 | 1.19E-14 | Sensor histidine kinase | |
| VPA1101 | -2.11042 | 2.77E-12 | Cytochrome c551 peroxidase | |
| 4 | VP2423 | -4.58784 | 2.59E-33 | hypothetical protein |
| VP2424 | -4.46142 | 4.30E-42 | Type IV prepilin peptidase TadV/CpaA | |
| VP2425 | -4.65177 | 3.56E-42 | Type IV prepilin peptidase TadV/CpaA | |
| VP2426 | -3.03105 | 1.31E-25 | Predicted ATPase with chaperone activity | |
| VP2427 | -3.02567 | 1.85E-24 | Flp pilus assembly protein RcpC/CpaB | |
| VP2428 | -2.55322 | 7.99E-19 | Type II/IV secretion system secretin RcpA | |
| VP2429 | -1.75485 | 0.000238 | hypothetical protein | |
| VP2430 | -1.97074 | 2.91E-12 | Von Willebrand factor type A domain protein | |
| VP2431 | -1.86147 | 6.08E-11 | Similar to TadZ/CpaE | |
| VP2432 | -1.78401 | 5.98E-10 | Similar to TadZ/CpaE | |
| VP2433 | -1.76889 | 2.37E-10 | Type II/IV secretion system ATPase TadZ/CpaE | |
| VP2434 | -1.73857 | 5.29E-10 | Type II/IV secretion system ATP hydrolase TadA/VirB11/CpaF | |
| VP2435 | -1.80475 | 2.63E-10 | Flp pilus assembly protein TadB | |
| VP2436 | -1.74251 | 1.77E-09 | Type II/IV secretion system protein TadC | |
| VP2437 | -1.7222 | 9.38E-10 | TPR repeat protein | |
| 5 | VP2261 | -3.03897 | 1.46E-21 | Flagellin protein FlaF |
| VP2262 | -3.36217 | 2.86E-28 | Flagellin protein FlaD | |
| VP2263 | -3.51179 | 1.21E-32 | Flagellin protein FlaA | |
| VP2264 | -3.24318 | 1.29E-28 | Flagellar protein FlaG | |
| VP2265 | -3.29545 | 2.24E-29 | Flagellar cap protein FliD | |
| VP2266 | -3.3816 | 2.61E-19 | Polar flagellar rod protein FlaI | |
| VP2267 | -2.97178 | 1.54E-22 | Flagellar biosynthesis protein FliS | |
| 6 | VP1657 | -1.99881 | 3.80E-12 | Type III secretion host injection protein (YopB) |
| VP1658 | -2.12616 | 3.16E-13 | Type III secretion negative regulator protein (YopD) | |
| 7 | VP1476 | -2.68838 | 9.64E-13 | Anti-sigma regulatory factor SypA |
| VP1477 | -1.58128 | 1.64E-07 | Outer membrane protein SypB | |
| VP1478 | -1.53419 | 1.10E-06 | Predicted protein SypD | |
| VP1479 | -1.6465 | 8.60E-05 | Signal transduction histidine kinase SypF | |
| VP1480 | -1.75662 | 7.30E-08 | Oligosaccharide translocase SypK | |
| VP1481 | -1.72636 | 5.83E-07 | Membrane protein SypL | |
| VP1482 | -2.82179 | 3.63E-14 | Glycosyltransferase SypN | |
| VP1483 | -3.18243 | 2.64E-15 | SypO | |
| VP1484 | -1.66319 | 4.82E-06 | Glycosyltransferase SypQ | |
| VP1485 | -3.20651 | 8.60E-05 | Sugar transferase SypR | |
| VPA1403 | -2.2481 | 0.000113 | Capsular polysaccharide synthesis enzyme CpsA | |
| VPA1406 | -1.54633 | 9.98E-06 | Capsular polysaccharide synthesis enzyme CpsD | |
| 8 | VP0791 | -2.52929 | 5.65E-11 | Flagellin protein FlaE |
| VP0792 | -3.17949 | 3.71E-24 | Flagellin protein FlaD | |
| VP0793 | -2.61879 | 1.70E-19 | Flagellin protein FlaC | |
| VP0794 | -2.43402 | 5.55E-16 | Flagellar hook-associated protein FlgL | |
| VP0795 | -2.2913 | 1.66E-14 | Flagellar hook-associated protein FlgK | |
| VP0796 | -2.39126 | 1.93E-13 | Flagellar protein FlgJ [peptidoglycan hydrolase] | |
| VP0797 | -2.70585 | 7.15E-18 | Flagellar P-ring protein FlgI | |
| VP0798 | -2.1387 | 7.01E-11 | Flagellar L-ring protein FlgH | |
| VP0799 | -2.49074 | 2.69E-16 | Flagellar basal-body rod protein FlgG | |
| VP0800 | -2.39727 | 4.95E-14 | Flagellar basal-body rod protein FlgF | |
| VP0801 | -1.86615 | 2.83E-11 | Flagellar hook protein FlgE | |
| VP0802 | -1.86572 | 3.50E-11 | Flagellar basal-body rod protein FlgD | |
| VP0803 | -1.88728 | 2.29E-11 | Flagellar basal-body rod protein FlgC | |
| VP0804 | -1.92001 | 8.89E-12 | Flagellar basal-body rod protein FlgB | |
| VP0805 | -3.55641 | 1.93E-07 | Flagellar protein FlgP | |
| VP0806 | -2.28843 | 9.85E-15 | Flagellar protein FlgO |
Overall, group 1 and group 2 comprised the majority of DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that a large number of DEGs primarily related to ABC transporter, flagellar assembly, secretion systems, and two-component system ( Figure 2B ).
Since strain 123 harbored one pirABVP-negative and one pirABVP-positive plasmid, we further examined the effect of qseC mutation on the expression of genes in these two plasmids. For pirABVP-negative plasmid, 12 genes were differentially expressed. However, none of them had significant difference (fold change < 1.5) except for ORF2 (encoding hypothetical protein), ORF3 (encoding outer membrane protein) and ORF9 (encoding hypothetical protein), indicating the deletion of qseC did not have significant impacts on the expression of genes in pirABVP-negative plasmid ( Table S2 ). Likewise, no genes including pirABVP was up- or down-regulated significantly on pirABVP-positive plasmid in ΔqseC mutant.
QseC Is Involved in Bacterial Motility, Biofilm Formation, and Pathogenesis of Vp AHPND
Based on the transcriptomic analysis, several genes associated with flagellar motility, biofilm formation, chemotaxis, and T6SS were found to be differentially expressed in ΔqseC mutant. To confirm whether QseC regulates these virulence factors of Vp AHPND 123, we further validate the results by RT-qPCR and also by a range of phenotypic screens. The expression of eight genes were verified in the wild type and mutant strains, including qseC, cpsA (relating to biofilm formation), flaC and flaF (encoding polar flagellins), flaK (encoding σ54-interacting regulator), fliA (encoding polar flagellin specific chaperon), motB (encoding Na+ motor component), and and vasD (associating with T6SS). Consistent with the transcriptomic results, all these genes were down-regulated in ΔqseC mutant (p < 0.01) except for vasD, which was up-regulated to 2.7-fold expression ( Figure 3A ). In addition, QseC expression was not detected in ΔqseC.
Figure 3.
QseC mediates virulence-related gene expression, flagellar motility, biofilm formation, and T6SS in Vp AHPND. (A) RT-qPCR detection of expression of the virulence-related genes in wild type (WT) and ΔqseC mutant cultured to exponential phase. Genes normalized to recA, and fold change relative to wild type strain. All values were expressed as mean ± standard deviation (SD) of three biological replicates. (B) Swimming motility assays performed on soft agar plates after 21 h of incubation. The data were replotted from Figure 1B . Error bars indicated the SD of three independent experiments. Different letters indicated significant differences (one way ANOVA with Tukey’s post hoc test; p < 0.01). (C) Biofilm formation in different Vp AHPND strains. Error bars indicated the SD of three independent experiments. Different letters indicated significant differences (one way ANOVA with Tukey’s post hoc test; p < 0.01). (D) Impact of qseC deletion and catecholamine receptor antagonist LED209 on the growth of Vp AHPND 123. Error bars represented SD of three independent cultures.
In the following experiments, we used soft agar plates and crystal violet staining to investigate the phenotypic differences in Vp AHPND strains and to corroborate our transcriptional expression data. According to the results, the ΔqseC mutant exhibited motility defects based on a significantly smaller swimming halo compared with the wile type strain, while the deficiencies were restored in the qseC+ complemented strain ( Figure 3B ). Similar results were observed in the biofilm formation assay, there was marked reduction of biofilm formation in ΔqseC mutant compared with the parental and complemented strains ( Figure 3C ), and substantiating the idea that QseC is involved in the regulation of Vp AHPND flagellar motility and biofilm formation. Additionally, the presence and absence of qseC gene showed no effect on the growth of Vp AHPND ( Figure 3D ).
Overall, QseC has been shown to regulate several virulence factors in Vp AHPND. However, the role it plays in pathogenesis in vivo has not been addressed. Given the fact that flagellar motility and biofilm formation are required for virulence of V. parahaemolyticus, we further assessed the virulence of ΔqseC mutant using a standard brine shrimp challenge test. According to the result, ΔqseC mutant is attenuated for virulence towards brine shrimp larvae ( Table 5 ), thereby establishing that QseC is involved in pathogenesis of Vp AHPND.
Table 5.
The impacts of qseC deletion and LED209 on the virulence of Vp AHPND towards gnotobiotic brine shrimp larvae (Artemia franciscana).
| Strains | Survival (%)1 | |
|---|---|---|
| Control | 100 ± 0 | |
| Vp AHPND WT | 32 ± 8 | a |
| Vp AHPND ΔqseC | 57 ± 3 | c |
| Vp AHPND [5 pM LED209] | 38 ± 3 | a |
| Vp AHPND [5 nM LED209] | 47 ± 8 | b |
| Vp AHPND [50 nM LED209] | 60 ± 5 | c |
1Survival rate of brine shrimp larvae after challenging with VpAHPND strain for 48 h.
Survival in the negative control was set at 100% and the other treatments were normalized accordingly. Batches of 30 brine shrimp larvae were used for each replicate, and each treatment was carried out in triplicate. Square brackets refer to pretreatment. LED209 was directly added into the Artemia rearing water. Data are average ± standard deviation of three independent experiments, and were analyzed by One-way ANOVA with Tukey’s post hoc test (p < 0.01). Values with a different letter are significantly different from each other.
LED 209 Inhibition of QseC as a Promising Antivirulence Strategy
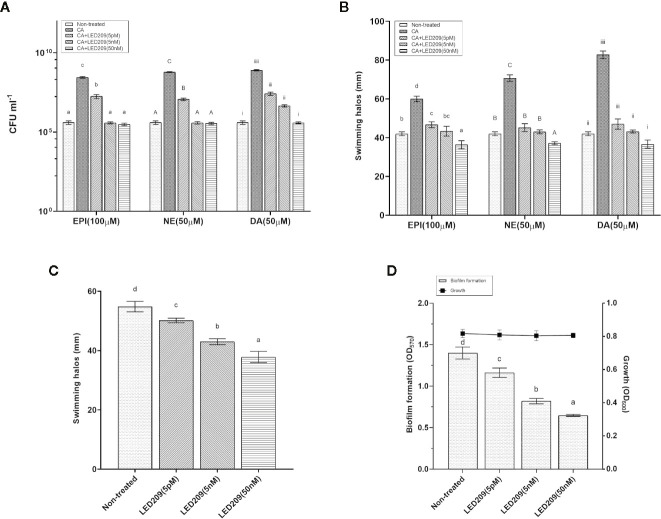
LED209 is a specific antagonist that can inhibit the binding of signaling molecules to QseC, thus prevents its autophosphorylation and consequently blocks QseC-mediated virulence gene expression (Rasko et al., 2008). According to our results, LED209 was able to neutralize the catecholamine-induced effects of EPI, NE, and DA on Vp AHPND 123 growth and motility ( Figures 4A, B ), which confirms its role on QseC inhibition. Besides, the addition of LED209 could also significantly decrease swimming motility and biofilm formation in Vp AHPND 123 without exogenous catecholamines ( Figures 4C, D ), this result echoes our previous observation showing that ΔqseC mutant exhibited motility and biofilm defects. Most importantly, our results revealed that LED209 reduced the virulence of Vp AHPND 123 towards brine shrimp larvae ( Table 5 ), while did not influence its growth in vitro ( Figure 3D ).
Figure 4.
LED209 neutralizes the catecholamine enhanced growth (A) and swimming motility (B) in Vp AHPND 123. Three kinds of catecholamine including Epinephrine (EPI), norepinephrine (NE), and dopamine (DA) were used at 100, 50, and 50 µM, respectively. (C) Treatment with LED209 diminished swimming motility of Vp AHPND 123. Diameters of motility halos after 18 h of incubation on TSA30 plates containing 0.3% agar were measured in the presence (5 pM, 5 nM, and 50 nM, respectively) or absence (DMSO only) of LED209. (D) Administration of LED209 reduced biofilm formation of Vp AHPND 123. Strain was grown statically in TSB30 in 96-well plate for 24 h in the presence (5 pM, 5 nM, and 50 nM, respectively) or absence (DMSO only) of LED209. Biofilm formation was determined by measuring crystal violet binding at OD570. The error bars represented standard deviation (SD) of three independent experiments. Different letters indicate significant differences (One-way ANOVA with Tukey’s post hoc test; p < 0.01).
Based on these promising results, we went further to investigate the effect of LED209 on the survival of challenged white shrimp (P. vannamei), a commercially important aquaculture species. According to our previous research, only 8% of shrimp challenged with untreated Vp AHPND 123 remained alive at 48 hpi (Yang et al., 2010), while in this study, 40 and 47% of shrimp were still alive after challenging by the Vp AHPND treated with 50 and 100 nM LED209, respectively ( Figure 5A ). Moreover, LED209 showed no evidence of toxicity towards the shrimp, revealed by the survival rate of the group treated with LED209 reached 92% after 48 hpi.
Figure 5.
Dynamics of mortalities and temporal activities of the immune-related enzymes after Vp AHPND challenge. (A) Treatment of LED209 protects white shrimp (Penaeus vannamei) from Vp AHPND infection. LED209 was used at 50 and 100 nM, respectively. The survival curves were recorded for 48 h after challenge, p < 0.0001 based on Kaplan-Meier survival analysis with a log rank test with Prism software (Graphpad Software). (B) Superoxide dismutase (SOD) activity. (C) Catalase (CAT) activity. Data were presented as mean ± standard deviation (SD) of three independent experiments. (*Note: the survival curve data of Vp AHPND 123 without treatment have been published in another article by the author (Yang et al., 2020, (J) Appl. Microbiol.), and the relevant data were obtained from the same batch of experiments.)
Furthermore, mortality was delayed in LED209 treatment groups after infection with Vp AHPND in comparison with the onset of the disease in the group without LED209. Mortality in the groups treated with LED209 began to increase at 10 hpi, and reached 56 and 52% in the group treated with 50 and 100 nM of LED209 during the following 24 h, respectively. After 30 hpi, no additional mortalities were observed. In contrast, for the group without LED209, mortality began to increase at 4 hpi and continued for an additional 40 h until reached a maximum of 92%. These observations demonstrated that LED209 can attenuate bacterial virulence without interfering with the growth, which makes it a potential antivirulence agent.
The Activity Change of Immune-Related Enzymes in the Shrimp After Bacterial Challenge
We also monitored the activity change of immune-related enzymes including SOD and CAT in the shrimp after challenge. The activities of CAT in the challenged group increased significantly during 24 h, and reached the peak (0.677 U mg-1, p < 0.05) at 24 h after challenge ( Figure 5B ). For the groups treated with LED209 (100 nM), it began to increase (0.401 U mg-1, p < 0.05) at 12 h and maintained the peak at 24 h after challenge. Similar observation was also identified for SOD activity ( Figure 5C ). The activities of SOD in the challenged group reached the peak (84 U mg-1, p < 0.05) at 12 h and were significantly higher than that in the treatment group with LED209 (100 nM) after challenge. There was no significant difference in the activities of SOD and CAT between the control group and the group treated with LED209 (100 nM) during the whole experiment.
Discussion
It is widely acknowledged that the extensive use of antibiotics in shrimp farming practices contributes to the development of multidrug-resistant bacteria, which has constituted one of the greatest challenges in combating bacterial infection, and posed high risks to the environment and human health (WHO, 2018). Therefore, antivirulence therapy - the use of antimicrobial agents that specifically target the regulation pathways required for the pathogenesis, but not for microbial viability—has received particular attention. QseC is a conserved histidine sensor kinase of the QseBC two-component system presented in at least 25 important pathogens, and it has been demonstrated to activate the expression of virulence genes by sensing and responding to the host catecholamine hormones (Rasko et al., 2008). Given these facts, further investigation of QseC in the pathogenesis of Vp AHPND may be important for developing novel strategies to combat AHPND.
In this study, we found that the wild type strain Vp AHPND 123 but not ΔqseC, exhibited catecholamine enhanced growth and flagellar motility, indicating that QseC functions as a bacterial catecholamine receptor in Vp AHPND. These results manifest that QseC might play a role in the regulation of host adaptation processes of Vp AHPND by utilizing the catecholamines or other hormones.
Primary metabolism is one of the essential phenotypes that significantly contribute to the virulence, depending on the availability of food sources and environmental dynamics (Peyraud et al., 2018). Transcriptome analysis revealed that QseC plays an essential role in the global gene regulation of Vp AHPND. The up-regulation of genes encoding lipid biosynthesis and transportation could imply that Vp AHPND might try to deal with an excess of lipids accumulated due to the trade-off between virulence and primary metabolism (Peyraud et al., 2016). Besides, transcriptome analysis showed that T6SS gene cluster was strongly up-regulated. In addition to the alterations in expression of T6SS, another affected gene cluster associated with cell wall biosynthesis was identified. The underlying mechanism for the up-regulation of these genes remains unclear and is worth further attention. Taken together, these results suggest that QseC is a bacterial adrenergic receptor for Vp AHPND that is crucial for environmental adaptation.
Transcriptome analysis also revealed the involvement of qseC in pathogenesis of Vp AHPND. Flagellum is an important surface organelle required for bacterial motility and host invasion. In the early stage of infection, flagellar motility promotes the translocation and colonization of pathogens within the host (Josenhans and Suerbaum, 2002). Elevated motility also helps the pathogen to more efficiently acquire nutrients and avoid toxic substances and facilitates dispersal in the environment during the course of transmission (Ottemann and Miller, 1997). In consistent with the findings in other pathogenic bacteria (Wang et al., 2011; Yang et al., 2014), our data indicated that expressions of various flagellar related genes were down-regulated in ΔqseC mutant. The investigation on soft agar plates confirmed that ΔqseC mutant displayed significantly impaired motilities and the deficiencies were rescued by complementation of the qseC gene.
Furthermore, similar results were observed in another assay indicating that ΔqseC showed weaker biofilm formation ability than the wild type and complementary strain. Transcriptome analysis also showed that genes related to capsular polysaccharide synthesis and glycosyltransferase were down-regulated in the mutant, suggesting the potential role of qseC in regulating biofilm formation. Bacterial biofilm formation is an important pathogenic mechanism that is involved in adherence, colonization, competence, metabolism, and stress response (Jefferson, 2004). Previous studies have demonstrated that QseC controls biofilm formation of different pathogens, such as Haemophilus parasuis and enteroaggregative E.coli (Curtis et al., 2014; He et al., 2018). At last, the observation that the virulence of ΔqseC mutant towards gnotobiotic brine shrimp larvae is attenuated, further underscores the role of QseC in Vp AHPND pathogenesis.
Thus, our results verified the idea that QseC is a modulator of the host-pathogen interaction, and also an important regulator of Vp AHPND virulence, all of which emphasize the need for further investigation into disturbing QseC as an approach to prevent Vp AHPND infection. LED209 has been identified as an inhibitor that is highly specific for QseC by Rasko et al. (2008). It acts as a prodrug that can allosterically modify QseC, impeding virulence in several pathogens without interfere with the growth. More excitingly, it possesses desirable pharmacokinetics and does not exhibit toxicity in vitro and in vivo (Curtis et al., 2014). Therefore, we investigated whether inhibition of QseC with LED209 could reduce virulence of Vp AHPND in two models, i.e., gnotobiotic brine shrimp larvae (Artemia franciscana) and conventional cultured white shrimp (P. vannamei). Intriguingly, we observed an attenuation of virulence by LED209 in both assays, even at a low concentration (nM). Unlike conventional antibiotics, LED209 does not kill or hinder Vp AHPND growth, which makes it a promising antivirulence agent (Rasko et al., 2008).
In general, as bacterial histidine sensor kinases, the class of QseC signal transduction molecules are absent in mammals (Roychoudhury et al., 1993; Lyon and Muir, 2003), which makes it an attractive antivirulence therapy targets. QseC allows pathogens to sense and respond to catecholamine host hormones, and subsequently modulate their virulence gene expression for optimal host colonization. Meanwhile, it also helps the pathogens to detect the health state of the host, thereby invading or leaving (Curtis et al., 2014). Since these processes are not involved in bacterial growth, possibly leading to a lower evolutionary pressure for the drug resistance development. Furthermore, given the broad distribution of QseC in various Gram-negative pathogens and its inherent role in bacterial pathogenesis, LED209 may have promising application prospect as a broad-spectrum antivirulence agent.
A practical strategy is to combine LED209 with other available antibacterial drugs to design targeted dual-function antibacterial drugs. This novel dual-function antibacterial molecule can block QseC and reduce bacterial pathogenicity while targeting bacteria to improve treatment efficacy and reduce adverse reactions. Moreover, the likelihood of developing resistance to this virulence-regulating drug is relatively low. In the future, we will further investigate how LED209 reduces the pathogenicity of Vp AHPND and clarify specific molecules and signal pathways in order to deepen our understanding of the mechanism of the QseC inhibition and provide a new strategy for treatment of AHPND.
Conclusion
In conclusion, our study showed that QseC mediated the catecholamine stimulated effects on growth and flagellar motility of Vp AHPND. Transcriptome analysis of the wild type and ΔqseC mutant of Vp AHPND confirmed the deletion of qseC suppressed flagellar motility, biofilm formation and virulence, but enhanced the expression of genes associated with T6SS and cell wall biosynthesis. The bacterial catecholamine receptor antagonist LED209 not only neutralized the effects of catecholamine but also attenuated the virulence of Vp AHPND towards brine shrimp larvae and white shrimp, highlighting that interfering QseC with LED209 is a promising strategy to develop broad-spectrum antivirulence agents. These results provide insight into the use of an antivirulence approach for targeting not only Vp AHPND, but also a much larger collection of pathogenic bacteria.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Author Contributions
JH and QY designed the study and coordinated the project. PZ, ZC, QW, and GX conducted the experiments. SF and QY conducted the transcriptome analysis. SF and QY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31702385), the Special Research Fund of Ghent University (BOF-UGent), the National Natural Science Foundation of China (Grant No. 81903372), and the China Agriculture Research System (CARS-48).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.594652/full#supplementary-material
References
- Bearson B. L., Bearson S. M. D. (2008). The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278. 10.1016/j.micpath.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Boyanova L. (2017). Stress hormone epinephrine (adrenaline) and norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe 44, 13–19. 10.1016/j.anaerobe.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Buroni S., Chiarelli L. R. (2020). Antivirulence compounds: a future direction to overcome antibiotic resistance? Future Microbiol. 15, 299–301. 10.2217/fmb-2019-0294 [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Hughes D. T., Zhu C., Boedeker E. C., Sperandio V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. States America 103, 10420–10425. 10.1073/pnas.0604343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulanges V., Andre P., Ziegler O., Buchheit L., Vidon D. J. (1997). Utilization of iron-catecholamine complexes involving ferric reductase activity in Listeria monocytogenes . Infect. Immun. 65, 2778–2785. 10.1128/IAI.65.7.2778-2785.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. M., Russell R., Moreira C. G., Adebesin A. M., Wang C., Williams N. S., et al. (2014). QseC inhibitors as an antivirulence approach for Gram-negative pathogens. mBio 5, e02165. 10.1128/mBio.02165-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt T. (2013). Antivirulence therapy for animal production: filling an arsenal with novel weapons for sustainable disease control. PloS Pathog. 9, e1003603–e1003603. 10.1371/journal.ppat.1003603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt T. (2014). Virulence mechanisms of bacterial aquaculture pathogens and antivirulence therapy for aquaculture. Rev. Aquacult. 6, 100–114. 10.1111/raq.12030 [DOI] [Google Scholar]
- Defoirdt T. (2018). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26, 313–328. 10.1016/j.tim.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Dickey S. W., Cheung G. Y. C., Otto M. (2017). Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discovery 16, 457–471. 10.1038/nrd.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A., Keller L. H., Schifferli D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- FAO (2013). Report of the FAO/MARD Technical Workshop on Early Mortality Syndrome (EMS) or Acute Hepatopancreatic Necrosis Syndrome (AHPNS) of Cultured Shrimp (under TCP/VIE/3304), 2013. Hanoi, Viet Nam, 25–27 June 2013. FAO Fisheries and Aquaculture Report No. 1053, Rome, Italy, 54 p.
- Freestone P. P. E., Lyte M. (2008). Microbial Endocrinology: Experimental Design Issues in the Study of Interkingdom Signalling in Infectious Disease. Adv. Appl. Microbiol. 64, 75–105. 10.1016/S0065-2164(08)00402-4 [DOI] [PubMed] [Google Scholar]
- Fu S., Wei D., Yang Q., Xie G., Pang B., Wang Y., et al. (2020). Horizontal plasmid transfer promotes the dissemination of asian acute hepatopancreatic necrosis disease and provides a novel mechanism for genetic exchange and environmental adaptation. mSystems 5, e00799-19. 10.1128/mSystems.00799-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach R. G., Hensel M. (2007). Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297, 401–415. 10.1016/j.ijmm.2007.03.017 [DOI] [PubMed] [Google Scholar]
- He L., Dai K., Wen X., Ding L., Cao S., Huang X., et al. (2018). QseC mediates osmotic stress resistance and biofilm formation in Haemophilus parasuis . Front. Microbiol. 9, 212. 10.3389/fmicb.2018.00212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X. P., Xu D., Zhuo Y., Liu H. Q., Lu L. Q. (2016). Identification and pathogenicity of Vibrio parahaemolyticus isolates and immune responses of Penaeus (Litopenaeus) vannamei (Boone) . J. Fish Dis. 39, 1085–1097. 10.1111/jfd.12441 [DOI] [PubMed] [Google Scholar]
- Jefferson K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. 10.1111/j.1574-6968.2004.tb09643.x [DOI] [PubMed] [Google Scholar]
- Jones J. L. (2014). “VIBRIO | Introduction, Including Vibrio parahaemolyticus, Vibrio vulnificus, and Other Vibrio Species,” in Encyclopedia of Food Microbiology, 2nd ed. Eds. Batt C. A., Tortorello M. L. (Oxford: Academic Press; ), 691–698. [Google Scholar]
- Josenhans C., Suerbaum S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. 10.1078/1438-4221-00173 [DOI] [PubMed] [Google Scholar]
- Joshi J., Srisala J., Truong V. H., Chen I. T., Nuangsaeng B., Suthienkul O., et al. (2014). Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture 428-429, 297–302. 10.1016/j.aquaculture.2014.03.030 [DOI] [Google Scholar]
- Khajanchi B. K., Kozlova E. V., Sha J., Popov V. L., Chopra A. K. (2012). The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila . Microbiol. 158, 259–271. 10.1099/mic.0.051805-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M., Hadjifrangiskou M., Pinkner J. S., Hultgren S. J. (2009). QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli . Mol. Microbiol. 73, 1020–1031. 10.1111/j.1365-2958.2009.06826.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukonis E. S., Dirita V. J. (2003). From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae . Curr. Opin. Microbiol. 6, 186–190. 10.1016/S1369-5274(03)00032-8 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lyon G. J., Muir T. W. (2003). Chemical signaling among bacteria and its inhibition. Chem. Biol. 10, 1007–1021. 10.1016/j.chembiol.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Lyte M., Ernst S. (1992). Catecholamine induced growth of gram negative bacteria. Life Sci. 50, 203–212. 10.1016/0024-3205(92)90273-R [DOI] [PubMed] [Google Scholar]
- Ma Y.-J., Sun X.-H., Xu X.-Y., Zhao Y., Pan Y.-J., Hwang C.-A., et al. (2015). Investigation of reference genes in Vibrio parahaemolyticus for gene expression analysis using Quantitative RT-PCR. PloS One 10, e0144362. 10.1371/journal.pone.0144362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B., Wong G. C. L. (2015). How Bacteria Use Type IV Pili Machinery on Surfaces. Trends Microbiol. 23, 775–788. 10.1016/j.tim.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Marques A., Dinh T., Ioakeimidis C., Huys G., Swings J., Verstraete W., et al. (2005). Effects of bacteria on Artemia franciscana cultured in different gnotobiotic environments. Appl. Environ. Microbiol. 71, 4307–4317. 10.1128/AEM.71.8.4307-4317.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR . J. Bacteriol. 170, 2575–2583. 10.1128/JB.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Russell R., Mishra A. A., Narayanan S., Ritchie J. M., Waldor M. K., et al. (2016). Bacterial Adrenergic Sensors Regulate Virulence of Enteric Pathogens in the Gut. mBio 7, e00826-16. 10.1128/mBio.00826-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munguia J., Nizet V. (2017). Pharmacological Targeting of the Host-Pathogen Interaction: Alternatives to Classical Antibiotics to Combat Drug-Resistant Superbugs. Trends Pharmacol. Sci. 38, 473–488. 10.1016/j.tips.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Kendall M., Vugia D. J., Henao O. L., Mahon B. E. (2012). Increasing Rates of Vibriosis in the United State–2010: Review of Surveillance Data From 2 Systems. Clin. Infect. Dis. 54, S391–S395. 10.1093/cid/cis243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann K. M., Miller J. F. (1997). Roles for motility in bacterial–host interactions. Mol. Microbiol. 24, 1109–1117. 10.1046/j.1365-2958.1997.4281787.x [DOI] [PubMed] [Google Scholar]
- Peyraud R., Cottret L., Marmiesse L., Gouzy J., Genin S. (2016). A resource allocation trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathogens 12 (10), e1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud R., Cottret L., Marmiesse L., Genin S. (2018). Control of primary metabolism by a virulence regulatory network promotes robustness in a plant pathogen. Nat. Commun. 9, 418. 10.1038/s41467-017-02660-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Moreira C. G., Li De R., Reading N. C., Ritchie J. M., Waldor M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Sci. 321, 1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantaso M. B. (2016). Acute hepatopancreatic necrosis disease (AHPND): A game changer in aquaculture. FAO Aquacult. Newslett. 55, 50–51. [Google Scholar]
- Reiche E. M., Nunes S. O., Morimoto H. K. (2004). Stress, depression, the immune system, and cancer. Lancet Oncol. 5, 617–625. 10.1016/S1470-2045(04)01597-9 [DOI] [PubMed] [Google Scholar]
- Roychoudhury S., Zielinski N. A., Ninfa A. J., Allen N. E., Jungheim L. N., Nicas T. I., et al. (1993). Inhibitors of two-component signal transduction systems: inhibition of alginate gene activation in Pseudomonas aeruginosa . Proc. Natl. Acad. Sci. U. States America 90, 965–969. 10.1073/pnas.90.3.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PloS Pathog. 6, e1000949. 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suong N. T., Van Hao N., Van Sang N., Hung N. D., Tinh N. T. N., Phuoc L. H., et al. (2017). The impact of catecholamine sensing on the virulence of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease (AHPND). Aquaculture 470, 190–195. 10.1016/j.aquaculture.2016.12.030 [DOI] [Google Scholar]
- Tran L., Nunan L., Redman R. M., Mohney L. L., Pantoja C. R., Fitzsimmons K., et al. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ 105, 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- Vale P. F., Mcnally L., Doeschl-Wilson A., King K. C., Popat R., Domingo-Sananes M. R., et al. (2016). Beyond killing: Can we find new ways to manage infection? Evol. Med. Public Health 2016, 148–157. 10.1093/emph/eow012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Q., Yang M., Xiao J., Liu Q., Wu H., et al. (2011). QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda . Fish Shellfish Immunol. 30, 944–953. 10.1016/j.fsi.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Weigel W. A., Demuth D. R. (2016). QseBC, a two-component bacterial adrenergic receptor and global regulator of virulence in Enterobacteriaceae and Pasteurellaceae . Mol. Oral. Microbiol. 31, 379–397. 10.1111/omi.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. States America. 104, 6037–6042. 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2018). Antibiotic Resistance. Antibiotic Resistance [Online]. Available at: http://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (Accessed 16 September 2018).
- Yang Q., Defoirdt T. (2015). Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi . Environ. Microbiol. 17, 960–968. 10.1111/1462-2920.12420 [DOI] [PubMed] [Google Scholar]
- Yang Q., Anh N. D., Bossier P., Defoirdt T. (2014). Norepinephrine and dopamine increase motility, biofilm formation, and virulence of Vibrio harveyi . Front. Microbiol. 5, 584. 10.3389/fmicb.2014.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Fu S., Zou P., Hao J., Wei D., Xie G., et al. (2020). Coordination of primary metabolism and virulence factors expression mediates the virulence of Vibrio parahaemolyticus towards cultured shrimp (Penaeus vannamei). J. Appl. Microbiol. 10.1111/jam.14922 [DOI] [PubMed] [Google Scholar]
- Yu W.-T., Jong K.-J., Lin Y.-R., Tsai S.-E., Tey Y. H., Wong H.-C. (2013). Prevalence of Vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int. J. Food Microbiol. 160, 185–192. 10.1016/j.ijfoodmicro.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Zhang L., Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16, 70–77. 10.1016/j.mib.2013.02.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.