Abstract
Background
Immunoglobulin A nephropathy (IgAN) is the most common form of primary glomerulonephritis. The intestinal microbial ecosystem and metabolic network of IgAN have not been systematically analyzed. The present study aims to improve understanding of the gut microbiota and its metabolic capabilities to facilitate the development of diagnostic, therapeutic, and prognostic methods for IgAN.
Methods
We characterized the gut microbiota and metabolic patterns of fecal and serum samples of IgAN patients and healthy controls from the south of China using 16s ribosomal RNA gene sequencing and liquid chromatography-tandem mass spectrometry, respectively, and bioinformatics approaches.
Results
We found that the relative abundances of Streptococcus and Enterococcus were higher in IgAN patients, whereas Bacteroidetes and Bacteroides were lower. Changes in the gut microbiota of IgAN affected the metabolism and absorbance of microbiota-associated metabolites, in particular polyunsaturated fatty acids, free amino acid, and oligopeptides, and activated the phenylalanine metabolism pathway, thereby constructing a unique metabolic system of IgAN. We identified six pivotal metabolites, including bilirubin, trimethoprim, stearamide, phenylalanine, cis-9,10-epoxystearic acid, and phosphatidylethanolamine 17:0, that connected the metabolic networks of the gut and blood. Additionally, 5-hydroxyeicosatetraenoic acid and 5-hydroxy-6E,8Z,11Z-eicosatrienoic acid were shown to be associated with the classification of glomerular sclerosis.
Conclusions
We establish a relational network between microbiota, fecal metabolites, and serum metabolites in IgAN. The core microbiota and metabolites identified have promising value in therapeutic applications.
Keywords: Immunoglobulin A nephropathy (IgAN), microbiome, metabolome, metabolic network
Introduction
Immunoglobulin A nephropathy (IgAN), the most common form of primary glomerulonephritis (PGN) worldwide, is particularly prevalent in Asia and accounts for almost 45% of all PGN cases in China (1,2). The etiology and pathogenesis of primary IgAN are complicated and not entirely clear, but are thought to involve abnormally glycosylated IgA1 deposition, which contributes to local inflammation, mesangial proliferation, and glomerular fibrosis (3-6). Changes in components of the mucosal environment, such as exogenous oral and intestinal antigens, have a potential impact on the occurrence and progression of IgAN by regulating the synthesis of IgA, as IgA is known to be produced mainly by the mucosal immune system (7,8).
With the development of 16S ribosomal RNA (rRNA) sequencing analysis, emerging evidence has linked the microbiome to IgAN (7,9). It was speculated that a shift in gut microbiota stimulated epithelial cells to secrete excessive B-cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL), which induced the overproduction of IgA and maintained tolerogenic immune responses (10). Additionally, microbial metabolites could be used to represent the function of microbial activities and act as intermediate phenotypes between the host and microbiota (11). For instance, indoxyl sulfate, p-cresyl sulfate, and trimethylamine-N-oxide are intestinal metabolites closely related to the progression of chronic kidney disease (CKD) (12). Enterotoxin-activated immune cells in the intestinal mucosa continue to produce inflammatory cytokines, leading to chronic systemic inflammatory responses (13).
The relational network among microbiota, fecal metabolites, and serum metabolites has not been systematically analyzed. Knowledge of the gut microbiota and metabolites of IgAN patients should contribute to the identification of potential biomarkers that correlate with clinical symptoms. Therefore, the present study aimed to identify a particular network between gut bacteria and metabolites in IgAN and to explore the influence of metabolites on clinical symptoms for application in clinical practice.
Methods
Study design
Forty-five participants, including 15 IgAN patients aged 22–56 years and 30 age-matched healthy controls (HCs) were enrolled in the present study. All participants were recruited from Shenzhen People’s Hospital. IgAN patients were diagnosed by postoperative pathological examinations, and patients with diabetes mellitus, recent infection, severe liver disease, malignancies, cardiac insufficiency, autoimmune diseases, secondary IgAN, or alcohol abuse were excluded. Participants recruited for the present study were treatment-naïve IgAN patients who had not taken any drugs (e.g., antibiotics or aspirin) or nutritional supplements for at least two months. All subjects were Han Chinese from southern China with comparable eating habits and lifestyles. The present study was conducted following the principles of the Declaration of Helsinki (as revised in 2013) and was authorized by the ethics board of Shenzhen People’s Hospital (LL-KY-2019514). All participating individuals provided signed informed consent. Clinical data (age, sex, body mass index, and hypertension), biochemical indices, and the Oxford classification of IgAN were recorded. The estimated glomerular filtration rate (eGFR) was assessed using CKD epidemiology (14,15).
Metabolite extraction from fecal and serum samples
Fecal and serum samples were collected after overnight fasting. After adding 1,000 µL extract solvent (2:2:1 acetonitrile-methanol-water containing 1 µg/mL internal standard), fecal samples were vortexed for 30 seconds, homogenized at 45 Hz for 4 minutes, and sonicated for 5 minutes in an ice-water bath. 300 µL of methanol containing 1 µg/mL internal standard was added to the serum samples, vortexed for 30 seconds, and sonicated for 10 minutes in an ice-water bath. The samples were then incubated at –20 °C for 1 hour and centrifuged at 12,000 rpm at 4 °C for 15 minutes. A quality control sample was acquired by mixing an equal amount of supernatant from each of the samples.
DNA extraction and 16S rRNA gene sequencing
A PowerSoil DNA Isolation Kit (Qiagen, Germany) was used to extract genomic DNA from feces. DNA size and integrity were verified using 1% agarose gel electrophoresis. NanoDrop spectrophotometry (NanoDrop) was used to determine DNA concentrations. Primers (319F: 5'-ACTCCTACGGGAGGCAGCAG-3'; 806R: 5'-GGACTACHVGGGTWTCTAAT-3') targeting the V3 and V4 hypervariable regions of the 16S rRNA gene were used for 16s rRNA-based amplification. After amplification, polymerase chain reaction products were mixed with AMPure XP beads (Beckman Coulter, UK) at a ratio of 1:1.5, and the fragments were screened and cleaned. After purification and library construction, the amplicons were quantified using a Qubit fluorometer (Invitrogen, USA) and sequenced on a HiSeq PE250 sequencing instrument (Illumina, USA).
16s rRNA data analysis
Paired-end reads were produced and allocated to each sample according to the marked barcodes. Reads were further merged using FLASH software version 1.2.11 (16). After high-quality filtering of the raw tags using Trimmomatic software version 0.33 (17), we used UCHIME software version 8.1 (18) to remove chimeric sequences and acquire high-quality clean tags. Sequences with >97% similarity were allocated to the same operational taxonomic unit (OTU) using USEARCH software version 10.0 (19). RDP Classifier version 2.2 was applied to annotate species based on the 16S SILVA online database release 128 (20,21). We used PyNAST (http://biocore.github.io/pynast/) to analyze phylogenetic differences in dominant OTUs (22). Alpha diversity analysis was performed using Mothur version v.1.30 (http://www.mothur.org/) (23). We used unweighted principal coordinate analysis (PCoA) to calculate beta diversity. Linear discriminant analysis (LDA) of effect size (http://huttenhower.sph.harvard.edu/lefse/) was used to compare species between groups, and LDA was used to estimate the influence of abundance of each species. Metastats analysis (http://metastats.cbcb.umd.edu/), an improved statistical method for the analysis of metagenomic data, was used to screen significant species.
Liquid chromatography-tandem mass spectrometry data acquisition
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was carried out using a UHPLC system (Agilent Technologies, USA). The liquid included 2 phases: mobile phase A (5 mmol/L ammonium acetate in water as the negative, and 0.1% formic acid in water as the positive) and mobile phase B (acetonitrile). The gradient program was as follows: 0 minutes, 1% B; 1 minute, 1% B; 8 minutes, 99% B; 10 minutes, 99% B; 10.1 minutes, 1% B; and 12 minutes, 1% B. A total of 2 µL of each sample was injected at a flow rate of 0.5 ml/minutes. A Q Exactive (QE) mass spectrometer was used to acquire MS/MS spectra. Operation of the electrospray ionization (ESI) source used the following parameters: sheath gas flow rate =45 arb, aux gas flow rate =15 arb, full MS resolution =70,000, capillary temperature =400 °C, collision energy =20/40/60 eV in a normalized collision energy model, MS/MS resolution =17,500, and spray voltage =4.0 kV (positive)/–3.6 kV (negative). ProteoWizard was used to convert the LC-MS/MS data to mzXML format, and the raw data were processed, cleaned, and aligned using MAPS software version 1.0 (24). Finally, we used the MS2 database for further analysis of metabolites.
LC-MS/MS-based metabolomics data analysis
The ionization source of the QE platform was ESI in the positive ion mode and negative ion modes. After relative standard deviation de-noising and standard internal normalization, the final dataset was imported to the SIMCA software package version 15.0.2 for further analysis (25). To visualize group separation and to highlight phenotypic differences between the groups, supervised orthogonal projections to latent structures-discriminant analysis (OPLS-DA) was performed based on principal component analysis (PCA) (26). The quality of the model was verified using 7-fold cross-validation. The validity of the model was evaluated using R2 (model’s interpretability to variables) and Q2 (predictability of the model) values obtained from 7-fold cross-validation. The variable importance in the projection (VIP) of the first principal component of the OPLS-DA analysis was determined to identify differential metabolites. Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and MetaboAnalyst (http://www.metaboanalyst.ca/) were used for pathway-enrichment analyses.
Statistical analyses
Differential analysis was performed using 2-tailed Student’s t-tests for measurement data or χ2-tests for enumeration data. Pearson’s correlation analysis was used to assess correlations between gut bacteria and metabolites. Metabolites with P<0.05 and VIP >1 were considered significantly changed metabolites. All data were analyzed with GraphPad Prism 7.0 software. Heat maps were created using R version 3.5.3 (https://www.r-project.org/), and Cytoscape version 3.7.1 (https://cytoscape.org/) was used to construct the relational network.
Results
Summary of clinical characteristics
All patients were diagnosed with IgAN by pathological examination. General condition surveys (e.g., age, body mass index, and blood pressure) of the two groups showed no statistically significant differences (P>0.05). However, there were significant differences in 24 hours proteinuria, serum albumin, creatinine, and uric acid between the HC and IgAN groups (P<0.05) (Table S1). Information on the histological grades of IgAN patients is shown in Table S2.
Bacterial OTUs and diversity analyses
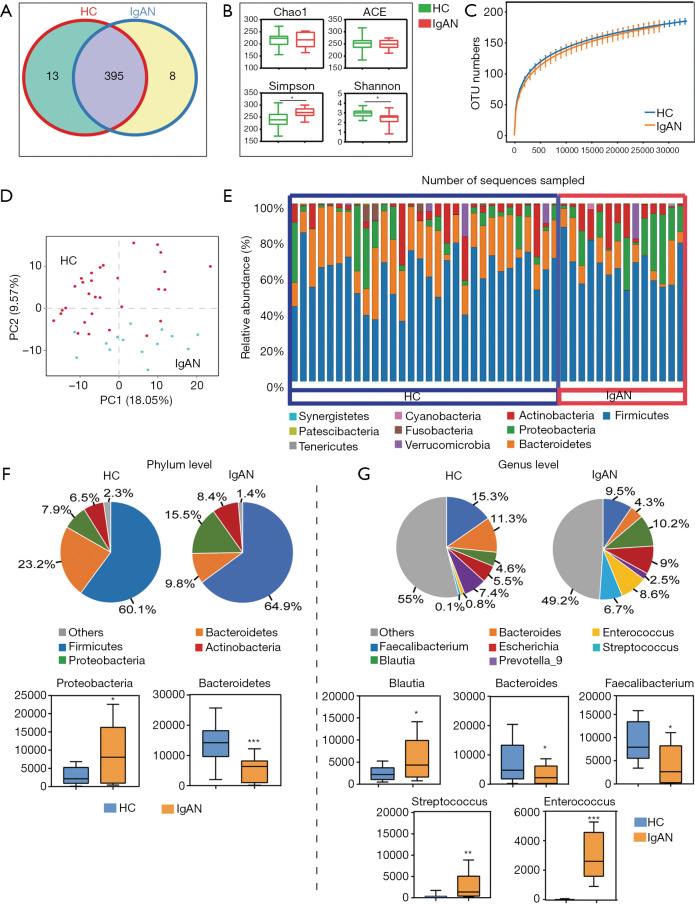
Total intestinal bacteria from 15 IgAN patients and 30 HCs were analyzed by 16S rRNA gene sequencing. After quality control, pyrosequencing analysis yielded an average of 58,188 effective tags (average length: 414 bp) per sample. Sequences with >97% similarity were classified into the same OTU. A total of 408 and 403 OTUs were identified in the HC and IgAN groups, respectively, including 395 shared OTUs (Figure 1A). Based on the OTUs, paired t-tests showed significant differences in bacterial diversity (Simpson and Shannon diversity indices), but not species abundance (Chao1 and abundance-based coverage estimator) between the HC and IgAN groups (Figure 1B). Rarefaction curves suggested that sequencing depth and coverage were sufficient (Figure 1C). PCoA plots were applied to evaluate similarities between microbial communities from each HC and IgAN patient and the result revealed an obvious variation between the two groups (Figure 1D).
Figure 1.
Microbiota diversity analysis. Operational taxonomic units (OTUs) of immunoglobulin A nephropathy (IgAN) and healthy control (HC) groups. (B) Richness estimator (Chao1), abundance-based coverage estimator (ACE), and diversity indices (Simpson and Shannon) of microbiota from the fecal samples of IgAN and HC groups. (C) Rarefaction curves of microbiota from fecal samples. Means ±95% confidence intervals are shown. (D) Community clustering using principal coordinate analysis of the unweighted UniFrac distance matrix. Change in the relative abundance of bacteria in fecal samples from IgAN and HC groups at the phylum level (E,F) and genus level (G). *, P<0.05; **, P<0.01; ***, P<0.001
Abundance distribution and differential analysis of different biologic classifications
Ten main bacterial phyla were identified (Figure 1E). Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria represented more than 97% of all 16S rRNA sequences in the two groups. The relative abundance of Proteobacteria was higher in IgAN patients (median value: 15%) than HCs (median value: 7.97%). The opposite trend was found for Bacteroidetes, which was lower in IgAN patients (9.81%) than in HCs (23.2%) (Figure 1F). The relative abundance of OTUs was assessed at the class, order, family, genus, and species levels, and the top 10 microbiota are shown in Figure S1. At the genus level, Blautia (HCs: 4.56%, IgAN patients: 10.2%) showed a remarkable upward trend in the IgAN group, whereas Bacteroides (HCs: 15.3%, IgAN patients: 9.5%) and Faecalibacterium (HCs: 15.3%, IgAN patients: 9.5%) showed a downward trend in the IgAN group. Furthermore, Streptococcus (HCs: 0.77%, IgAN patients: 5.75%) and Enterococcus (HCs: 0.1%, IgAN patients: 6.7%) were nearly absent among HCs, but were highly abundant among IgAN patients (Figure 1G).
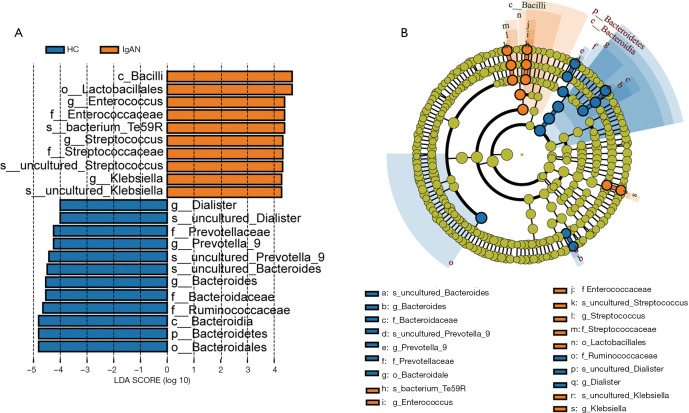
We compared taxa using Metastats analysis with a P-value cutoff <0.05 and an LDA cutoff >4 (Figure 2A) to predict the core microbiota for each bacterial classification (Table 1). Cladograms based on LDA >4 show the evolutionary relationship between each microbe (Figure 2B). The results showed that the phylum Bacteroidetes, the family Bacteroidaceae, and the genus Bacteroides were more abundant in the HC group and might be the principal taxa among healthy individuals. At the genus level, we found that Streptococcus and Enterococcus were more abundant in IgAN patients than HCs. Interestingly, Streptococcus and Enterococcus accounted for a low proportion of the microbes in HCs, but were highly abundant in the fecal samples of IgAN patients, indicating that the two microbes could be significant features of IgAN.
Figure 2.
Linear discriminant analysis (LDA) is integrated with effect size. (A) Differences in the relative abundance of bacteria in healthy control (HC) and immunoglobulin A nephropathy (IgAN) groups. (B) Cladogram of the phylogenetic distribution of microbes. Each circle represents a classification level from phylum to species from the inner to outer circles. The size of each circle is proportional to relative abundance. Microbes with no significant difference in abundance are shown in green. Microbes with an LDA value >4 in IgAN and HC groups are marked with orange and blue, respectively.
Table 1. Metastats and LEfSe analyses of differential microbes at different levels of biological classification.
| Bacteria | Metastats analysis | LEfSe analysis | ||||
|---|---|---|---|---|---|---|
| HC (mean) | IgAN (mean) | t-test | HC (LDA value) | IgAN (LDA value) | ||
| Phylum | ||||||
| Firmicutes | 60.1% | 64.9% | 0.210 | <2 | ||
| Bacteroidetes | 23.20% | 9.81% | <0.01 | 4.98 | – | |
| Proteobacteria | 7.97% | 15.00% | 0.045 | – | <2 | |
| Class | ||||||
| Clostridia | 51.8% | 47.4% | 0.425 | <2 | ||
| Bacteroidia | 23.20% | 9.81% | 0.001 | 4.98 | – | |
| Bacilli | 1.4% | 11.6% | 0.054 | 4.65 | ||
| Order | ||||||
| Clostridiales | 51.8% | 47.4% | 0.425 | <2 | ||
| Bacteroidiales | 23.20% | 9.81% | 0.001 | 4.98 | – | |
| Lactobacillales | 1.39% | 11.6% | 0.052 | 4.64 | ||
| Family | ||||||
| Bacteroidaceae | 11.30% | 4.48% | 0.018 | 4.532 | – | |
| Enterococcaceae | 0.03% | 5.08% | 0.009 | – | 4.48 | |
| Streptococcaceae | 0.78% | 5.76% | 0.021 | – | 4.415 | |
| Genus | ||||||
| Faecalibacterium | 15.3% | 9.5% | 0.048 | <2 | ||
| Streptococcus | 0.77% | 8.6% | <0.01 | – | 4.415 | |
| Blautia | 4.56% | 10.20% | 0.030 | – | <2 | |
| Bacteroides | 11.30% | 4.30% | 0.020 | 4.61 | – | |
| Enterococcus | 0.10% | 6.70% | <0.001 | – | 4.48 | |
HC, healthy control; IgAN, immunoglobulin A nephropathy; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis of effect size; Note: Microbes with LDA values >2 and t-test P values <0.05 were considered statistically significantly different.
Identification of metabolites in fecal and serum samples
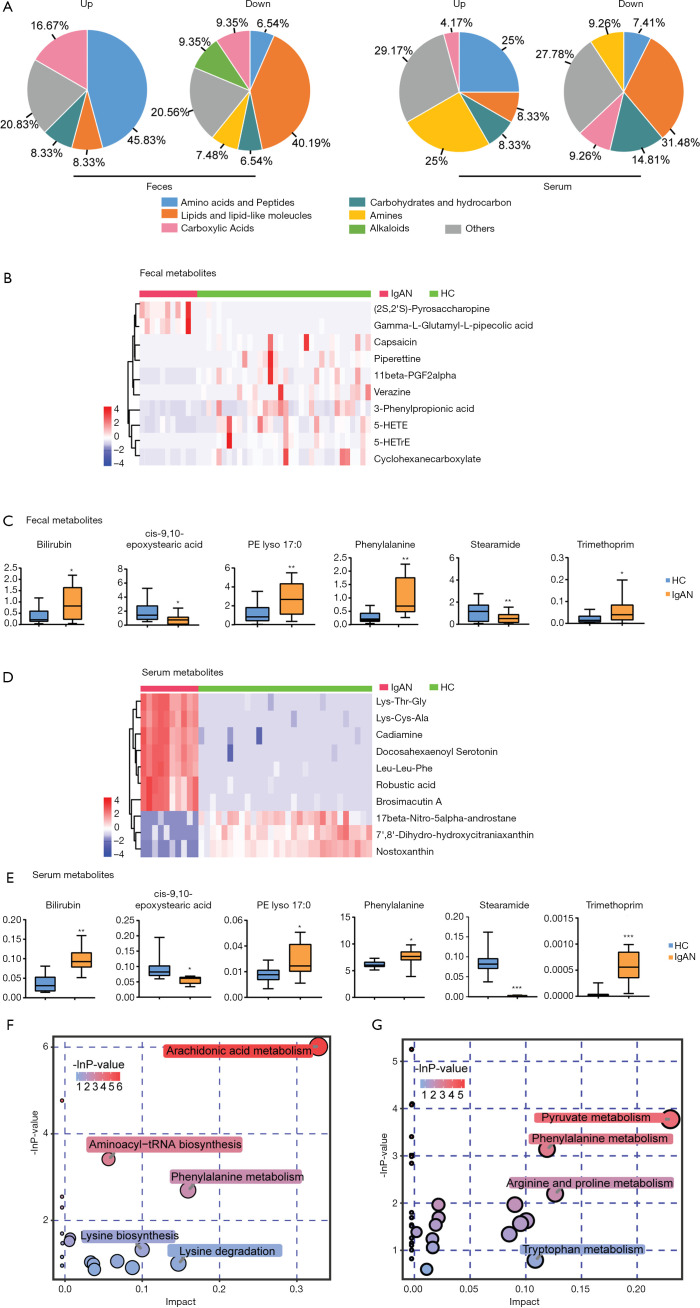
OPLS-DA models demonstrated that the metabolic profile of the IgAN group was significantly different from that of the HC group (Figure S2). Based on the screening criteria, VIP >1 and P<0.05, 131 and 80 differential metabolites were found in fecal and serum samples, respectively (Tables S3,S4). Lipids and lipid-like molecules (e.g., long-chain fatty acids and phospholipids), alkaloids, and fatty acid amides showed a downward trend in the fecal metabolites of IgAN patients, whereas free amino acids, oligopeptides, and carboxylic acids showed an upward trend. A decrease in lipids (medium-/long-chain fatty acids) and an increase in free amino acids and polypeptides were also observed in serum samples (Figure 3A). To identify core fecal metabolites, we determined the top 10 differential metabolites based on the criteria VIP >2 and log fold change >4 (Figure 3B). Notably, 6 metabolites–bilirubin, trimethoprim, stearamide, phenylalanine, cis-9,10-epoxystearic acid, and phosphatidylethanolamine (PE lyso 17:0)—were found in both fecal and serum samples, and could potentially serve as core metabolites for IgAN (Figure 3C). The top 10 serum metabolites and the expression levels of the 6 core metabolites in the serum samples are shown in Figure 3D and E. Furthermore, 2 pathways of metabolites from the fecal samples of IgAN patients, including the arachidonic acid metabolism pathway (impact value: 0.32) and the phenylalanine metabolism pathway (impact value: 0.16), were found to be statistically significant (P<0.05, impact value: >0.1) (Figure 3F). The phenylalanine metabolism pathway (impact value =0.12) was also enriched in the serum samples of IgAN patients (Figure 3G).
Figure 3.
Differential metabolite identification and pathway-enrichment analyses. (A) Altered metabolite class composition for feces and serum. (B) Differential metabolites with variable importance in the projection (VIP) >2 and log fold change >4. Expression of the 6 core metabolites in fecal samples (C) and serum samples (D,E). Metabolic pathway enrichment of fecal samples (F) and serum samples (G). *, P<0.05; **, P<0.01; ***, P<0.001.
Cross-talk among fecal metabolites, serum metabolites, and intestinal microbes
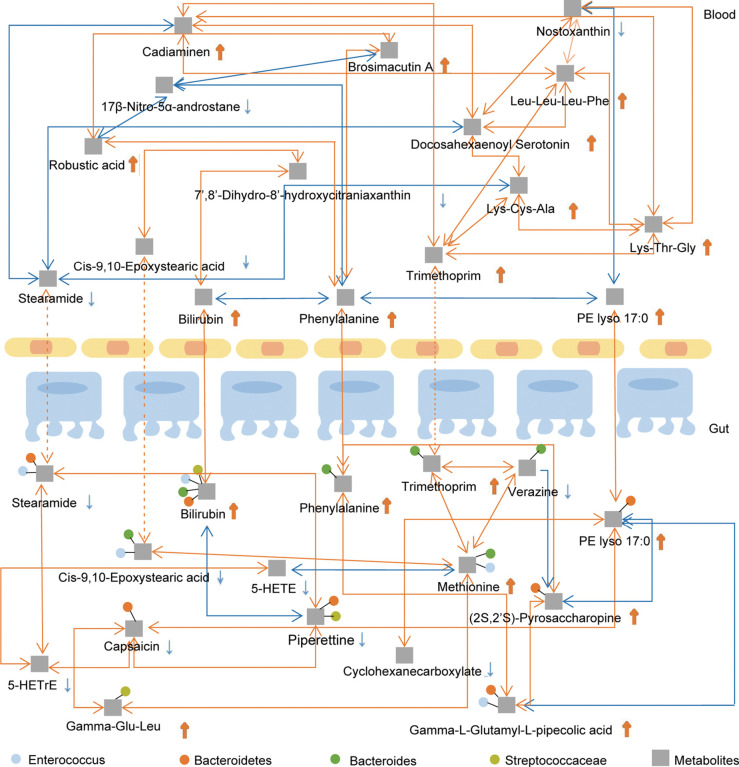
We performed Pearson correlation analysis to reveal the relationship between fecal bacteria and the essential metabolites identified. We found that Streptococcaceae was positively correlated with fecal and serum bilirubin, whereas Bacteroidetes was correlated with fecal and serum PE lyso 17:0 (Figure 4, Table S5). Significantly, the fecal metabolites bilirubin, phenylalanine, and PE lyso 17:0 were positively correlated with the serum metabolites bilirubin, phenylalanine, and PE lyso 17:0 (r>0.5, P<0.05), respectively, suggesting that these core metabolites might be significant mediators between the intestinal tract and blood circulation (Figure 4 and Figures S3,S4). Details of the microbe-metabolite network are shown in Figure 4.
Figure 4.
Metabolite network between gut and blood systems in immunoglobulin A nephropathy patients. Blue lines represent negative correlations (r<–0.5), and red lines represent positive correlations (r>0.5). Dotted lines represent |r|<0.5 with no significant correlation. r, Pearson correlation coefficient.
Correlation analysis between metabolites and clinical symptoms
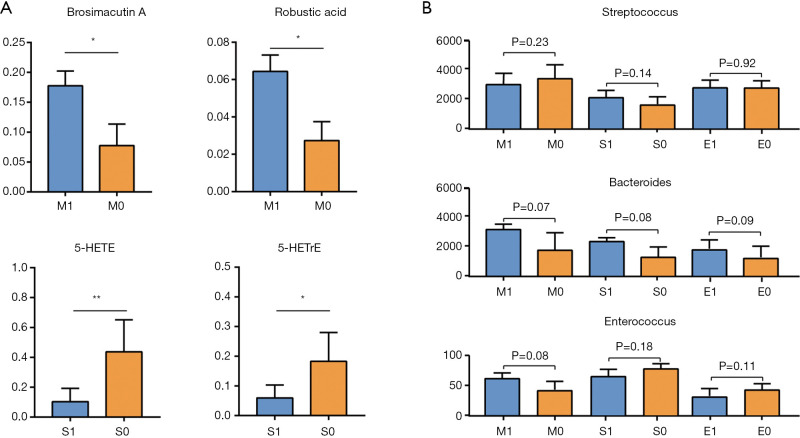
We performed t-tests to analyze the relationship between the core metabolites identified above and 24-hour proteinuria, eGFR, and pathological classification of IgAN. The results showed that the metabolites were not correlated with 24-hours proteinuria or eGFR. Using the Oxford classification of IgAN, we found that patients with mesangial hypercellularity had higher serum brosimacutin A and robustic acid levels (P<0.05). Also, levels of fecal metabolites 5-hydroxy-6E,8Z,11Z-eicosatrienoic acid (5-HETrE), and 5-hydroxyeicosatetraenoic acid (5-HETE) were lower in patients with severe segmental glomerulosclerosis (Figure 5A, Tables S6,S7). No significant correlation was found between the Oxford classification of IgAN and core gut microbes Streptococcus, Bacteroides, and Enterococcus (Figure 5B).
Figure 5.
Correlation between the Oxford classification of immunoglobulin A nephropathy (IgAN) and core metabolites and gut microbiota. (A) Significant metabolites associated with Oxford classification. (B) The relative abundance of core intestinal flora in patients with different Oxford classification. M, mesangial hypercellularity; S, segmental sclerosis. *, P<0.05; **, P<0.01.
Discussion
It is estimated that approximately 100 trillion microorganisms inhabit the human gut, and the total number of microbial genes is approximately 100 times that of the human genome (27). In addition to participating in the metabolism of nutrients and the synthesis of some vitamins, the intestinal microbial ecosystem helps maintain intestinal homeostasis and induces immune system maturation, as well as immune tolerance (28). Additionally, changes in the composition, abundance, and functional genetics of human micro-ecology are closely related to human health and disease (29). Therefore, the gut microbiota and its metabolites may provide feasible early detection tools for IgAN.
Unlike the results of De Angelis, which showed lower microbial diversity in IgAN patients compared with HCs (9), our study indicated that microbial diversity did not differ significantly between the two groups based on alpha and beta diversity analyses. However, the relative abundance of some specific bacteria was found to be significantly different. We suspect that the number of certain bacteria, rather than bacterial categories, changes during the early stages of IgAN. It is now known that approximately 1,000 different species inhabit the adult human gut, and the most abundant microbial phyla are Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (30,31). This is consistent with the results of our study, in which these microbial phyla accounted for >97% of all species in both the HC and IgAN groups. More importantly, at the genus level, a 2.6-fold decrease in Bacteroides, a 7.5-fold increase in Streptococcus, and a 67-fold increase in Enterococcus were observed in IgAN patients compared with HCs, indicating that a dramatic shift occurred in the fecal microbiome of IgAN patients. A previous study using both 16S DNA and 16S rRNA analyses revealed that Bacteroides was less abundant in IgAN patients, although these results were not supported by fecal bacterial culture (9). Our results also showed that the relative abundance of fecal Streptococcus was low in HCs, but was significantly increased in IgAN patients. Although a previous study showed no statistical difference in the number of cultivable Streptococcus between IgAN and HC groups (9), tissue deposits of IgA-binding streptococcal M proteins are considered an indicator of IgAN pathogenesis (32). Overall, we found that the abundance of Gram-positive bacteria showed an upward trend, whereas Gram-negative bacteria declined. Therefore, we conclude that the effects of the microbiome on IgAN are likely to be associated with holistic dysbiosis rather than specific bacterial species.
As mediators of the functional activity of bacteria, gut microbiota-generated metabolites play an essential role in the development and progression of kidney disease (33,34). In the present study, we identified a greater number of decreasing metabolites (medium-/long-chain fatty acids) and increasing metabolites (amino acids and oligopeptides) in both the fecal and serum samples of IgAN patients. A previous study also reported that some free amino acids (Met, Phe, and Gly) showed an upward trend in the serum or fecal samples of IgAN patients (9), which corresponds to the findings of our study and demonstrates the underlying role of the transformation of amino acids early in the pathogenesis of related diseases. Additionally, an increase in oligopeptides rather than polypeptides was found in our study, suggesting a possible effect of bacterial proteolytic fermentation. Furthermore, impaired protein assimilation has been reported in CKD patients (35), leading to easier absorption of amino acids and oligopeptides due to low energy consumption and non-competitive transport. We also detected a decrease in some fatty acids in IgAN patients, in particular, polyunsaturated fatty acids (e.g., 5-HETE and 5-HETrE). Studies have reported that polyunsaturated fatty acid supplementation can attenuate inflammation, oxidative stress, and tubulointerstitial fibrosis in the remnant kidney (36). Decreasing polyunsaturated fatty acids have not been reported to influence IgAN. It has also been speculated that a reduction in short-chain fatty acids contributes to the progression of CKD (37). Our results suggest that IgAN is characterized by a decrease in medium-chain fatty acids in fecal samples and long-chain fatty acids in serum samples, but not short-chain fatty acids. In general, we found differential metabolic features in the fecal and serum samples from IgAN patients and HCs.
The species richness of the microbial flora is closely related to metabolic diversity in feces (38). We found that changes in fecal metabolites were caused by a variety of bacteria rather than by a specific gut bacteria (Figure 4). Fecal metabolites further affect serum metabolites and lead to changes in body metabolism. for example, the increasing fecal metabolites phenylalanine and bilirubin are proportional to serum phenylalanine and bilirubin levels, respectively. KEGG analysis revealed that the phenylalanine metabolism pathway was enriched in both the fecal and serum samples of IgAN patients. Notably, gut microbiota can produce indoxyl sulfate, a toxin that causes damage to the kidney, by metabolizing phenylalanine (39). In the present study, no correlation was found between metabolites and the clinical measures 24-hours proteinuria or eGFR (7). We found that 5-HETE and 5-HETrE were associated with the classification of segmental sclerosis, indicating that polyunsaturated fatty acids might play a positive role in the kidney (36). To the best of our knowledge, the present study is the first to report an association between metabolites and renal function, and this relationship requires further investigation with larger sample sizes.
At present, systemic corticosteroids and immunosuppressive drugs are the main treatments for IgAN. However, the side-effects of these drugs and high recurrence rates should be taken into consideration. The complexity of the etiology of IgAN requires comprehensive treatment and management. Our findings provide a new therapeutic approach involving the reconstruction of the intestinal microenvironment and maintenance of metabolic balance. Fecal microbiota transplantation (FMT) is rapidly emerging due to its safety and stability (40). As mentioned earlier, Bacteroidetes, one of the earliest colonizing and most abundant constituents of the intestinal flora that may induce an anti-inflammatory milieu (41), was found to be significantly lower among IgAN patients. Food supplements, pharmaceutical products, and FMT may be promising therapeutic approaches to re-establish stable Bacteroidetes levels. Also, fat and protein metabolism disorders in IgAN patients, especially polyunsaturated fatty acid deficiency, should be considered. Maintaining metabolic balance and homeostasis is an essential therapeutic priority. Previous studies have reported that n-3 and n-6 long-chain polyunsaturated fatty acid supplements had a positive therapeutic effect in type 2 diabetes mellitus and coronary heart disease patients (42,43). A reasonable diet and wise supplementation of polyunsaturated fatty acids are essential for the prevention and treatment of IgAN.
Our study had the following limitations. Accurately assessing fecal metabolites, most of which are affected by diet and lifestyle factors, and distinguishing host-derived metabolites from microbially-generated metabolites is exceptionally challenging. Our findings revealed a significant alteration in the gut microbiota and metabolic phenotypes of IgAN patients. However, to improve the accuracy and specificity of molecular diagnoses and to identify molecular biomarkers, in-depth research on the characteristics of the gut microbiota and relative metabolites associated with different types of glomerulonephritis will be necessary. Additionally, 16S rRNA techniques can be used to assess the relative abundance, but not the absolute abundance of the microbial community. Therefore, the observed dynamics may not accurately reflect actual taxon densities (44).
Conclusions
A shift in the relative abundance of some bacteria was observed in IgAN patients compared with HCs. The changes in gut microbiota also affected metabolism and the absorbance of specific fatty acids, particularly polyunsaturated fatty acids (5-HETE and 5-HETrE), free amino acids, and oligopeptides, resulting in a unique metabolic system in IgAN patients. Six metabolites were identified–bilirubin, trimethoprim, stearamide, phenylalanine, cis-9,10-epoxystearic acid, and PE lyso 17:0—that connected the metabolic networks of the gut and blood and might be pivotal metabolites for IgAN. The activation of the phenylalanine metabolism pathway should be further examined. Furthermore, we found that 5-HETE and 5-HETrE were associated with the classification of segmental sclerosis but not 24-hour proteinuria or eGFR. This comprehensive analysis of microbial metabolites provides more functional insights than any single dataset alone.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank the many volunteers who participated in the study and AME Editing (http://editing.amegroups.cn/) for editing this manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (No. 81671596), The National Science Foundation for Young Scientists of China (No. 31700795), The Education Innovation Project for Graduates of Guangzhou, China (No. 2020XLLT10), The Twenty-one Teaching Reform Project of Jinan University (No. JG2019044).
Ethical Statement: The authors are accountable for all aspects of the work and ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was conducted following the principles of the Declaration of Helsinki (as revised in 2013) and was authorized by the ethics board of Shenzhen People’s Hospital (LL-KY-2019514). All participating individuals provided signed informed consent.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2506
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2506
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2506). The authors have no conflicts of interest to declare.
References
- 1.Li LS, Liu ZH. Epidemiologic data of renal diseases from a single unit in China: analysis based on 13,519 renal biopsies. Kidney Int 2004;66:920-3. 10.1111/j.1523-1755.2004.00837.x [DOI] [PubMed] [Google Scholar]
- 2.D’Amico G. Natural history of idiopathic IgA nephropathy and factors predictive of disease outcome. Semin Nephrol 2004;24:179-96. 10.1016/j.semnephrol.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 3.Robert T, Berthelot L, Cambier A, et al. Molecular insights into the pathogenesis of IgA nephropathy. Trends Mol Med 2015;21:762-75. 10.1016/j.molmed.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 4.Chen A, Yang SS, Lin TJ, et al. IgA nephropathy: clearance kinetics of IgA-containing immune complexes. Semin Immunopathol 2018;40:539-43. 10.1007/s00281-018-0708-7 [DOI] [PubMed] [Google Scholar]
- 5.Kiryluk K, Novak J. The genetics and immunobiology of IgA nephropathy. J Clin Invest 2014;124:2325-32. 10.1172/JCI74475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med 2013;368:2402-14. 10.1056/NEJMra1206793 [DOI] [PubMed] [Google Scholar]
- 7.Piccolo M, De Angelis M, Lauriero G, et al. Salivary microbiota associated with immunoglobulin A nephropathy. Microb Ecol 2015;70:557-65. 10.1007/s00248-015-0592-9 [DOI] [PubMed] [Google Scholar]
- 8.Floege J, Feehally J. The mucosa–kidney axis in IgA nephropathy. Nat Rev Nephrol 2016;12:147. 10.1038/nrneph.2015.208 [DOI] [PubMed] [Google Scholar]
- 9.De Angelis M, Montemurno E, Piccolo M, et al. Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One 2014;9:e99006. 10.1371/journal.pone.0099006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy DD, Kujawa J, Wilson C, et al. Mice overexpressing BAFF develop a commensal flora–dependent, IgA-associated nephropathy. J Clin Invest 2011;121:3991-4002. 10.1172/JCI45563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcobal A, Kashyap PC, Nelson T, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J 2013;7:1933-43. 10.1038/ismej.2013.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishima E, Fukuda S, Mukawa C, et al. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS–based metabolomics approach. Kidney Int 2017;92:634-45. 10.1016/j.kint.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 13.Sabatino A, Regolisti G, Brusasco I, et al. Alterations of intestinal barrier and microbiota in chronic kidney disease. Nephrol Dial Transplant 2015;30:924-33. 10.1093/ndt/gfu287 [DOI] [PubMed] [Google Scholar]
- 14.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604-12. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens LA, Schmid CH, Zhang YL, et al. Development and validation of GFR-estimating equations using diabetes, transplant and weight. Nephrol Dial Transplant 2010;25:449-57. 10.1093/ndt/gfp510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957-63. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261-7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Bittinger K, Bushman FD, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010;26:266-7. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537-41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adusumilli R, Mallick P. Data conversion with ProteoWizard msConvert. Methods Mol Biol 2017;1550:339-68. 10.1007/978-1-4939-6747-6_23 [DOI] [PubMed] [Google Scholar]
- 25.Wiklund S, Johansson E, Sjöström L, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem 2008;80:115-22. 10.1021/ac0713510 [DOI] [PubMed] [Google Scholar]
- 26.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemom 2002;16:119-28. 10.1002/cem.695 [DOI] [Google Scholar]
- 27.Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res 2017;179:24-37. 10.1016/j.trsl.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasubuchi M, Hasegawa S, Hiramatsu T, et al. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015;7:2839-49. 10.3390/nu7042839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez B, Delgado S, Blanco-Míguez A, et al. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res 2017;61:1600240. 10.1002/mnfr.201600240 [DOI] [PubMed] [Google Scholar]
- 30.D’Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta 2015;451:97-102. 10.1016/j.cca.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 31.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789-99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt R, Carlsson F, Mörgelin M, et al. Tissue deposits of IgA-binding streptococcal M proteins in IgA nephropathy and Henoch-Schönlein purpura. Am J Pathol 2010;176:608-18. 10.2353/ajpath.2010.090428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han L, Fang X, He Y. ISN forefronts symposium 2015: IgA nephropathy, the gut microbiota, and gut-kidney crosstalk. Kidney Int Rep 2016;1:189-96. 10.1016/j.ekir.2016.08.002 [DOI] [Google Scholar]
- 34.Meijers BK, Evenepoel P. The gut–kidney axis: indoxyl sulfate, p-cresyl sulfate and CKD progression. Nephrol Dial Transplant 2011;26:759-61. 10.1093/ndt/gfq818 [DOI] [PubMed] [Google Scholar]
- 35.Bammens B, Verbeke K, Vanrenterghem Y, et al. Evidence for impaired assimilation of protein in chronic renal failure. Kidney Int 2003;64:2196-203. 10.1046/j.1523-1755.2003.00314.x [DOI] [PubMed] [Google Scholar]
- 36.An WS, Kim HJ, Cho KH, et al. Omega-3 fatty acid supplementation attenuates oxidative stress, inflammation, and tubulointerstitial fibrosis in the remnant kidney. Am J Physiol Renal Physiol 2009;297:F895-903. 10.1152/ajprenal.00217.2009 [DOI] [PubMed] [Google Scholar]
- 37.Wang S, Lv D, Jiang S, et al. Quantitative reduction in short-chain fatty acids, especially butyrate, contributes to the progression of chronic kidney disease. Clin Sci (Lond) 2019;133:1857-70. 10.1042/CS20190171 [DOI] [PubMed] [Google Scholar]
- 38.Chen YY, Chen DQ, Chen L, et al. Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease. J Transl Med 2019;17:5. 10.1186/s12967-018-1756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mafra D, Barros AF, Fouque D. Dietary protein metabolism by gut microbiota and its consequences for chronic kidney disease patients. Future Microbiol 2013;8:1317-23. 10.2217/fmb.13.103 [DOI] [PubMed] [Google Scholar]
- 40.Vindigni SM, Surawicz CM. Fecal Microbiota Transplantation. Gastroenterol Clin North Am 2017;46:171-85. 10.1016/j.gtc.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 41.Xu M, Xu X, Li J, et al. Association between gut microbiota and autism spectrum disorder: A systematic review and meta-analysis. Front Psychiatry 2019;10:473-86. 10.3389/fpsyt.2019.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forouhi NG, Imamura F, Sharp SJ, et al. Association of plasma phospholipid n-3 and n-6 polyunsaturated fatty acids with type 2 diabetes: The EPIC-InterAct Case-Cohort Study. PLoS Med 2016;13:e1002094. 10.1371/journal.pmed.1002094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bird JK, Calder PC, Eggersdorfer M. The role of n-3 long chain polyunsaturated fatty acids in cardiovascular disease prevention, and interactions with statins. Nutrients 2018;10:775. 10.3390/nu10060775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Props R, Kerckhof FM, Rubbens P, et al. Absolute quantification of microbial taxon abundances. ISME J 2017;11:584-7. 10.1038/ismej.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as