Abstract
Apolipoproteins C-I, C-II, and C-III interact with ApoE to regulate lipoprotein metabolism and contribute to Alzheimer's disease pathophysiology. In plasma, apoC-I and C-II exist as truncated isoforms, while apoC-III exhibits multiple glycoforms. This study aimed to 1) delineate apoC-I, C-II, and C-III isoform profiles in cerebrospinal fluid (CSF) and plasma in a cohort of nondemented older individuals (n = 61), and 2) examine the effect of APOE4 on these isoforms and their correlation with CSF Aβ42, a surrogate of brain amyloid accumulation. The isoforms of the apoCs were immunoaffinity enriched and measured with MALDI-TOF mass spectrometry, revealing a significantly higher percentage of truncated apoC-I and apoC-II in CSF compared with matched plasma, with positive correlation between CSF and plasma. A greater percentage of monosialylated and disialylated apoC-III isoforms was detected in CSF, accompanied by a lower percentage of the two nonsialylated apoC-III isoforms, with significant linear correlations between CSF and plasma. Furthermore, a greater percentage of truncated apoC-I in CSF and apoC-II in plasma and CSF was observed in individuals carrying at least one APOE Ɛ4 allele. Increased apoC-I and apoC-II truncations were associated with lower CSF Aβ42. Finally, monosialylated apoC-III was lower, and disialylated apoC-III greater in the CSF of Ɛ4 carriers. Together, these results reveal distinct patterns of the apoCs isoforms in CSF, implying CSF-specific apoCs processing. These patterns were accentuated in APOE Ɛ4 allele carriers, suggesting an association between APOE4 genotype and Alzheimer's disease pathology with apoCs processing and function in the brain.
Supplementary key words: apolipoproteins, apoC-I, apoC-II, apoC-III, apoE, amyloid-β42, CSF, mass spectrometry, proteomics, plasma
Abbreviations: AD, Alzheimer's disease; BBB, blood-brain barrier; BCB, blood-cerebrospinal fluid barrier; CVD, cardiovascular disease; DPP-IV, dipeptidyl peptidase-IV; H/D, hydrogen/deuterium; PBS, phosphate buffered saline buffer; TG, triglyceride
Apolipoproteins C-I, C-II, and C-III (collectively termed apoCs) reside on lipoprotein particles where they take part in regulating lipid metabolism and transport. The three apoCs are small proteins: apoC-I is comprised of 57 amino acids, and both apoC-II and C-III contain 79 amino acids. All three apoCs exhibit isoforms in vivo. In addition to the full-length translated and processed protein, apoC-I also exists as a truncated form (apoC-Iʹ), lacking the two N-terminal amino acids (1), which is created by the action of dipeptidyl peptidase-IV (DPP-IV) (2). ApoC-II also undergoes proteolytic cleavage, resulting in the removal of its N-terminal six amino acids and yielding an isoform termed mature apoC-II (apoC-IIʹ) (3). On the other hand, apoC-III is glycosylated at Thr74, with the most common glycoforms having an O-linked N-acetylgalactosamine-Galactose disaccharide (-GalNAc-Gal), which can be further modified with up to two sialic acid (Sia) residues (1). All three apoCs are expressed predominantly in the liver, and upon entry into plasma, they are rapidly exchanged among the major classes of lipoproteins where they play an important role in the regulation of triglyceride (TG) metabolism (4, 5, 6).
While much is known about apoCs in plasma, their roles in the central nervous system (CNS) remain obscured. The CNS is separated from the peripheral tissues by the blood-brain barrier (BBB) and blood-cerebrospinal fluid barrier (BCB). CNS contains only HDL-like lipoprotein particles that are typically enriched with apolipoproteins A-I, E, and J (7, 8). The early evidence for the presence of apoC-I in the brain came in the form of apoC-I mRNA from marmoset brain tissues, which was present in the brain at only a fraction (<5%) of the mRNA expressed by the liver (9). Gene microarray data from postmortem prefrontal cortex tissues revealed that apoC-I mRNA is highest within the first 5 years of life, but still at much lower levels than the other highly expressed apolipoproteins in the CNS (10). The gene microarray data also revealed minimum amounts of apoC-II mRNA in the brain that increased in school-age children and then fell back below detection levels toward adulthood; the gene expression values for apoC-III were below the limit of reliable detection at all ages (10). Nevertheless, apoC-II and apoC-III were detected at the protein level in cerebrospinal fluid (CSF) using electroimmunoassays at concentrations that were <5% of their levels in plasma (11). These low CSF levels were confirmed in a recent study of 22 matched plasma and CSF samples that showed apoC-III in CSF to be 0.01% of its concentration in plasma (12). Interestingly, plasma apoA-I-containing apoC-III, but not total plasma apoC-III, correlated most strongly with CSF apoC-III. This suggests possible crossing of apoC-III from plasma into CSF along with apoA-I, which has been postulated to cross the BCB via cellular mediated transport (13) and the BBB through clathrin-independent and cholesterol-mediated endocytosis (14). Crossing of the apoCs from plasma into CNS has also been postulated, but has not been conclusively demonstrated.
APOE4 is the strongest genetic risk factor for late-onset Alzheimer's disease (AD). APOE4 influences the expression of the apoCs and how they are biologically processed. The genes for apoC-I and apoC-II are located on the long arm of chromosome 19 (19q13.32), along with the gene for apoE (15). The APOE ε4 allele is typically expressed with the APOC1 allele, H2. Indeed, APOC1 polymorphisms are associated with APOE, and the H2 allele is associated with an increased risk of developing AD (16). Apolipoprotein C-III is separately encoded by a region of the long arm of chromosome 11q23, known as the apoA-I/C-III/A-IV gene cluster (17). As an abundant apolipoprotein in TRL, apoC-III induces hypertriglyceridemia and promotes atherogenesis, and it is an important risk factor for cardiovascular disease (CVD) (18, 19, 20). In one study, APOE ε4 carriers had lower TG, ApoE, and ApoC-III levels, and the ApoC-III/ApoE ratio on HDL was reported to be the highest (21). Therefore, understanding how the 4 allele affects the apoCs' expression and their posttranslational modifications may further elucidate the biological mechanisms behind protein-related pathologies, such as AD and CVD. We have recently developed a mass spectrometry (MS)-based assay for detection of apoCs and their isoforms (22), and applied it to study apoC-III plasma glycoforms and how they are associated with clinical lipid measures and outcomes (23, 24, 25, 26). In this work, we have examined for the first time the distribution of the apoCs isoforms in CSF and compared it with paired plasma samples from a cohort of older, nondemented individuals grouped by APOE genotypes.
Materials and methods
Reagents
Polyclonal goat anti-human antibodies to apoC-I (Cat. No. 31A-G1b), apoC-II (Cat. No. 32A-G2b), and apoC-III (Cat. No. 33A-G2b) were obtained from Academy Biomedical (Houston, TX). Acetone (UN1090) was from JT Baker (Radnor, PA). Hydrochloric acid (HCl; AB06037), trifluoroacetic acid (TFA, AB02010), and acetonitrile (ACN; AB00120) were from AmericanBio (Natick, MA). N-methylpyrrolidinone (NMP; BP1172-4), 1,1ʹ carbonyldiimidazole (97%) (CDI, 115533), phosphate buffered saline buffer (PBS, 28372), 2-(N-morpholino) ethanesulfonic acid (MES) saline buffer (28390), and Mass Spectrometric Immunoassay (MSIA) Tips (991CUS01) were acquired from Thermo Fisher Scientific (Waltham, MA). Tween20 (Cat. No. P7949), sinapic acid (85429), and ethanolamine (ETA; 398136) were obtained from Sigma Aldrich (St. Louis, MO). ApoC-I (Cat. No. EA8011-1), C-II (EA8012-1), and C-III (EA8133-1) ELISA kits were acquired from Assay Pro (St. Charles, MO).
Human samples
A set of 61 paired human EDTA plasma and CSF samples were analyzed for the apoCs. Recruitment methods were directed at persons enrolled in the University of Southern California (27) Alzheimer Disease Research Center (ADRC) aged 60 years and older. Inclusion criteria included a Clinical Dementia Rating Scale (CDR) score of 0 (n = 48), 0.5 (n = 9), and 1 (n = 1). The study and procedures were approved by the Institutional Review Board of USC. All participants provided informed consent prior to enrollment in the study (USC IRB: HS-16-00888).
ApoC-III and apoC-I/C-II MSIA tips preparation
Activation and derivatization of the microcolumns inside the MSIA Tips were performed on a Multimek 96 automated 96-channel pipettor (Beckman Coulter, Brea, CA). The MSIA Tips were first rinsed with 200 mM HCl (10 aspiration/dispense cycles, 100 μL each), followed by water (10 cycles) and acetone (10 cycles). Then, the microcolumns inside the tips were activated with CDI (100 mg/mL in NMP (1,000 cycles, 50 μL each) followed by two rinses with NMP (10 cycles each, 100 μL). For the apoC-III tips, the activated tips were immediately immersed into the wells of a 96-microwell microplate containing 2.5 μg apoC-III antibody/well (in 100 μL of 10 mM MES buffer), and 1,000 cycles (50 μL each) were performed, allowing for antibody attachment to the activated microcolumns. For the apoC-I/C-II multiplex tips, the activated tips were immersed into wells containing 0.32 μg apoC-I antibody and 2.25 μg apoC-II antibody/well (in 100 μL of 10 mM MES buffer). Following the antibodies' attachment, the tips were rinsed with ETA and two rinses with PBS (50 cycles each, 100 μL). The total time taken for activation and derivatization of 96 MSIA Tips was 1.5 h. The apoC-III and apoC-I/C-II antibody-derivatized tips were stored at 4°C until use.
Analytical samples preparation
Plasma and CSF samples were thawed and diluted immediately prior to running the assays. For plasma analyses, the first dilution (S1) was prepared by mixing 3 μL plasma with 117 μL of PBS, 0.1% Tween (PBST). Then, 40 μL of the S1 dilution was mixed with 120 μL of PBST, yielding 160 μL analytical plasma samples (S2 dilution). Two S2 dilutions were prepared from one S1 dilution: the first S2 was for analysis of apoC-III, and the second S2 was for analysis of apoC-I/C-II. For the CSF analyses, 100 μL of CSF was mixed with 100 μL of PBST. The 200 μL analytical CSF samples were used first for apoC-III assay and then (sequentially) for the apoC-I/C-II assay. Higher volumes of CSF were utilized because the apoCs' concentrations in CSF are much lower compared with plasma.
Assays execution
The antibody-derivatized tips were mounted onto the head of the Multimek 96 pipettor and first rinsed with PBST (10 cycles, 100 μL). The tips were then immersed into the wells of a microplate containing the analytical samples, and 250 cycles (100 μL each) for plasma, and 500 cycles for CSF, were performed, allowing for affinity capture of the targeted proteins. Then, one rinse with PBST (100 cycles, 100 μL) and two rinses with water (10 cycles each, 100 μL) followed to wash off the nonspecifically bound proteins from the microcolumns. To elute the captured proteins, 5 μL of MALDI matrix (20 g/L sinapic acid in 33% (v/v) ACN and 0.4% (v/v) TFA) was aspirated into each tip, pushed up and down three times, and then dispensed directly onto a 96-well formatted MALDI target. Sample spots were dried on a hot plate at 50°C.
MALDI-TOF MS detection
Bruker Autoflex III MALDI-TOF instrument (Bruker, Billerica, MA) was utilized to acquire linear mass spectra. The instrument was operated in positive ion mode with 20.00 kV and 18.45 kV ion source voltages. The mass spectra were acquired in the mass range from 5 to 20 kDa, with 50 ns delay, and signal suppression of up to 4,500 Da. Total of 1,000 laser-shots were acquired and summed for each mass spectrum.
Quantification of human plasma and CSF apoCs
The concentrations of plasma and CSF apoCs were determined by Sandwich ELISAs, performed per manufacturer's instructions and as described (28). CSF apoCs concentrations were low but still above the detection limits of the ELISAs; CSF samples were assayed without any dilutions. Plasma samples were diluted 1:1,000 for the apoC-III ELISA and 1:200 for apoC-I and apoC-II ELISAs.
Data analysis
The mass spectra were first externally calibrated with protein calibration standards and then internally calibrated using the highest intensity apoCs signals. The spectra were baseline subtracted (Convex Hull algorithm, 0.8 flatness) and smoothed (Savizky Golay algorithm, 5 m/z width and 1 cycle) using Flex Analysis software (Bruker Daltonics). Areas under the peaks for all isoforms of apoC-I, apoC-II, and apoC-III signals were integrated using Zebra 1.0 software (Intrinsic Bioprobes Inc) and tabulated in a spreadsheet. To obtain the percent abundance of truncated apoC-I, the peak area of truncated apoC-I was divided with the summed peak areas of both truncated and full-length apoC-I. The percent abundance of truncated apoC-II was similarly calculated. The percent abundance of the individual apoC-III isoforms was calculated by dividing the peak area of each isoform with the summed peak areas of all apoC-III isoforms.
GraphPad Prism 7 was utilized for statistical analysis. Normality of the data sets was assessed with the Shapiro-Wilk test. To identify apoCs isoforms differences between the paired plasma and CSF samples, parametric t-test was performed for normally distributed data sets, and Wilcoxon matched-pairs signed rank test was applied to data sets that were not normally distributed. The correlations between the apoCs isoforms in the paired CSF and plasma samples, between CSF Aβ42 and the apoCs isoforms, and between the apoCs isoforms and the total concentrations of apoCs in both CSF and plasma, were assessed via Pearson's correlation (for parametric data sets) or Spearman's rank correlation (for nonparametric data sets). Individuals were further separated into two groups based on the presence of the APOE Ɛ4 allele: non-Ɛ4 carriers and Ɛ4 carriers (heterozygous or homozygous for Ɛ4). The percent abundance of the apoC-I, apoC-II, and apoC-III isoforms in plasma and CSF was compared between these two unpaired groups using an unpaired t-test with Welch's correction for the normally distributed data sets and the Mann-Whitney test at 5% false discovery rate for data sets that were not normally distributed. Differences among non-Ɛ4, homozygous Ɛ4, and heterozygous Ɛ4 allele carriers were examined with Kruskal-Wallis test with Dunn's multiple comparisons test for nonnormally distributed data and one-way ANOVA with post-hoc Tukey HSD test at a 5% false discovery rate for normally distributed data.
Results and discussion
The paired human CSF and plasma samples from 61 individuals were analyzed for total apoCs concentrations using sandwich ELISAs and for the isoform ratios using MS-based immunoassays. The MS-based immunoassays were comprised of two steps: 1) immuno capture of apoCs from the samples using antibodies immobilized within a porous microcolumn inside a pipettor tip, and 2) elution and detection of the captured intact apoCs with MALDI-TOF MS. The assays are fast (∼1 h) and high-throughput (96 samples at a time). The assays determine the relative abundance of the isoforms as a percentage of the total protein. These types of assays have been developed and employed for analysis of several plasma proteins (29), including other apolipoproteins (30). The apoCs assays were applied to CSF samples for the first time in this study. A summary of the 61 individuals cohort by APOE genotype subgroups and concentrations of the apoCs is shown in Table 1.
Table 1.
Differences in demographic characteristics, plasma apoCs levels, and CSF apoCs levels among APOE subgroups
| E2/E3 (N=5) | E2/E4 (N=2) | E3/E3 (N=27) | E3/E4 (N=16) | E4/E4 (N=11) | Total (N=61) | P-value | |
|---|---|---|---|---|---|---|---|
| Age | 0.042a | ||||||
| Mean (SD) | 66.8 (6.4) | 73.5 (2.1) | 67.7 (6.9) | 63.6 (5.6) | 61.9 (7.7) | 65.7 (7.0) | |
| Sex | 0.241 | ||||||
| female | 3 (60%) | 2 (100%) | 17 (63%) | 6 (38%) | 8 (73%) | 36 (59%) | |
| male | 2 (40%) | 0 (0%) | 10 (37%) | 10 (63%) | 3 (27%) | 25 (41%) | |
| Ethnicity | 0.510 | ||||||
| Hispanic/Latinos | 0 (0%) | 0 (0%) | 3 (11%) | 4 (25%) | 1 (9%) | 8 (13%) | |
| Not Hispanic/Latinos | 5 (100%) | 2 (100%) | 24 (89%) | 12 (75%) | 10 (91%) | 53 (87%) | |
| BMI | 0.700 | ||||||
| Median (IQR) | 27.4 (4.8) | 24.0 (NA) | 26.8 (5.7) | 25.5 (4.7) | 24.4 (3.7) | 25.4 (5.5) | |
| Years of Education | 0.658 | ||||||
| Median (IQR) | 16.0 (1.0) | 15.0 (3.0) | 17.5 (2.8) | 16.5 (4.5) | 17.0 (2.0) | 17.0 (3.0) | |
| CDR | 0.690 | ||||||
| 0 | 4 (100%) | 2 (100%) | 22 (88%) | 12 (75%) | 8 (73%) | 48 (83%) | |
| 0.5 | 0 (0%) | 0 (0%) | 2 (8%) | 4 (25%) | 3 (27%) | 9 (16%) | |
| 1 | 0 (0%) | 0 (0%) | 1 (4%) | 0 (0%) | 0 (0%) | 1 (2%) | |
| CSF ApoC-I (ng/ml) | 0.731 | ||||||
| Median (IQR) | 106 (57) | 96 (64) | 153 (115) | 169 (120) | 200 (50) | 158 (115) | |
| CSF ApoC-II (ng/ml) | 0.650 | ||||||
| Median (IQR) | 15 (11) | 25 (4) | 21 (14) | 19 (7) | 15 (6) | 20 (13) | |
| CSF ApoC-III (ng/ml) | 0.804 | ||||||
| Median (IQR) | 109 (31) | 81 (40) | 69 (69) | 76 (37) | 67 (16) | 79 (53) | |
| Plasma ApoC-I (μg/ml) | 0.641 | ||||||
| Mean (SD) | 116 (12) | 133 (15) | 113 (22) | 109 (20) | 111 (21) | 113 (20) | |
| Plasma ApoC-II (μg/ml) | 0.320 | ||||||
| Median (IQR) | 6.73 (2.32) | 10.37 (4.43) | 7.02 (8.45) | 7.71 (3.29) | 12.40 (9.92) | 7.36 (7.99) | |
| Plasma ApoC-III (μg/ml) | 0.043b | ||||||
| Median (IQR) | 198 (13) | 161 (4) | 107 (63) | 136 (42) | 136 (44) | 130 (73) |
no significant TukeyHSD pairwise comparisons
significant TukeyHSD pairwise comparison for Ɛ3/Ɛ3- Ɛ2/Ɛ3 (P adj = 0.022)
Apolipoproteins C-I and C-II truncations
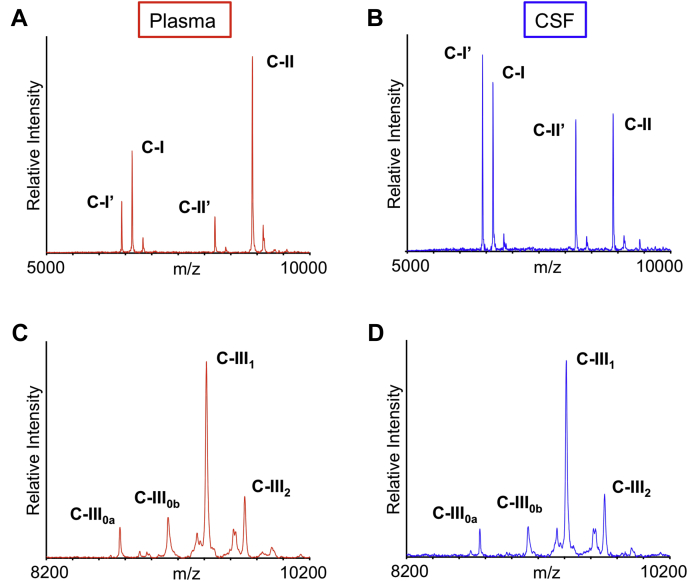
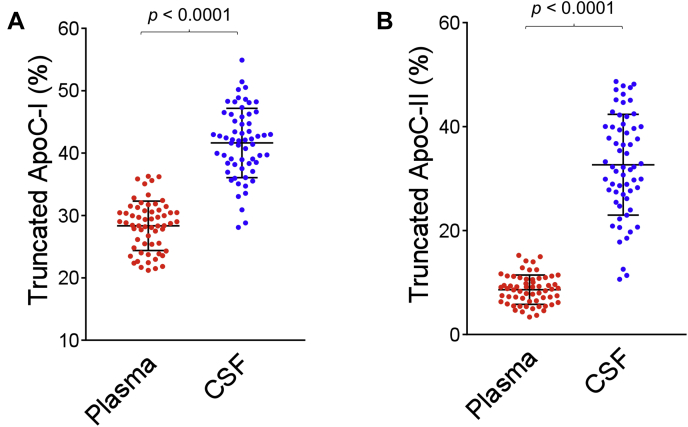
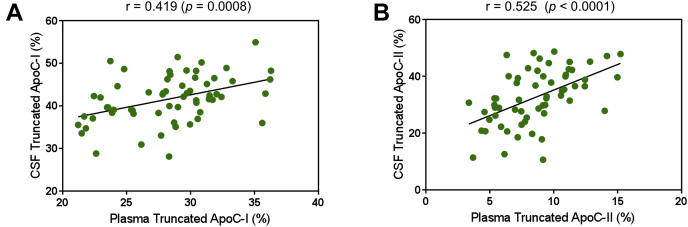
ApoC-I and apoC-II MALDI-TOF mass spectra resulting from the analysis of paired plasma and CSF samples from a single individual are shown in Fig. 1A, B. Strong signals for apoC-I (MW = 6630.58) and apoC-II (MW = 8914.92) are present in the mass spectra from both plasma and CSF, along with signals of their truncated isoforms (apoC-Iʹ MW = 6432.35 and apoC-IIʹ MW = 8204.17). Compared with the signals of the full-length proteins, the signals for the truncated isoforms are stronger in CSF than in plasma. The percent abundance for truncated apoC-I and apoC-II was obtained by dividing the peak area of the truncated isoform with the summed peak area of the truncated and full-length protein and is shown in Fig. 2 for all 61 plasma and CSF samples. Both proteins were truncated significantly more in CSF than in plasma. For truncated apoC-I, a mean of 41.6 ± 5.6% (SD) was observed in CSF compared with 28.3 ± 4.0% in plasma (P < 0.0001). For truncated apoC-II, a mean of 32.7 ± 9.7% was observed in CSF versus 8.62 ± 2.8% in plasma (P < 0.0001). The correlations of truncated apoC-I and apoC-II between plasma and CSF are shown in Fig. 3. A positive trend of increased truncations in CSF with increased truncations in plasma was observed, with Pearson correlation coefficients of r = 0.419 (P = 0.0008) for truncated apoC-I, and r = 0.525 (P < 0.0001) for truncated apoC-II. There was no significant correlation between the total concentrations of apoC-I and apoC-II and their respective truncated isoforms in both plasma and CSF, except for plasma apoC-II, with r = 0.352 (P = 0.013) (supplemental Fig. S1).
Fig. 1.
MALDI-TOF mass spectra for apoC-I and apoC-II resulting from the analysis of (A) plasma and (B) CSF paired samples obtained from a single individual. C-Iʹ, truncated apoC-I; C-IIʹ, truncated apoC-II. MALDI-TOF mass spectra for apoC-III resulting from the analysis of (C) plasma and (D) CSF paired samples obtained from the same individual. ApoC-III0a, no glycosylation; apoC-III0b, GalNac-Gal; apoC-III1, GalNac-Gal-Sia; apoC-III2, GalNac-Gal-Sia-Sia.
Fig. 2.
Percent abundance of (A) truncated apoC-I and (B) truncated apoC-II in plasma versus CSF. Percent truncation was computed by dividing the peak area of each truncated isoform with the summed peak areas of the truncated and full-length protein. Parametric paired t-tests were performed to identify differences between plasma and CSF samples between the normally distributed data sets.
Fig. 3.
Correlation between (A) truncated apoC-I and (B) truncated apoC-II in the paired plasma and CSF samples. Shown are the parametric Pearson's correlation coefficients.
These results suggest increased proteolytic processing of apoC-I and C-II in CSF, creating more truncated isoforms. The enzyme responsible for removing the two N-terminal amino acids of apoC-I is dipeptidyl peptidase-IV (DPP-IV) (2). DPP-IV is expressed throughout the body, including the CNS (31, 32) where it participates in the regulation of biologically active peptides. ApoC-I is a suitable substrate peptide for DPP-IV (33), because it contains the preferred sequence NH2-Thr-Pro, and the N terminus is in a flexible conformation (34, 35). The higher abundance of truncated apoC-I in CSF may therefore indicate a higher level of DPP-IV expression/activity in the brain.
The enzyme responsible for the cleavage of apoC-II is unknown, but it has been speculated that it is the same endopeptidase that cleaves the six N-terminal amino acids of apoA-I (36), because the cleavage sites of the two proteins share sequence homology (37). Interestingly, while almost all of apoA-I in circulation is in the form of the truncated isoform (termed mature apoA-I), the truncated apoC-II constitutes only a minor fraction (< 10%) of total apoC-II in plasma (1, 3). The significantly increased percentage of truncated apoC-II in CSF may also indicate increased expression/activity of this enzyme, similar to DPP-IV.
The linear trend of increased truncations in CSF with increased truncations in plasma could also indicate a connection between the two pools of apoC-I and C-II in plasma and CSF. Expression of apoC-I in astrocytes and endothelial cells within the hippocampus and frontal cortex has been demonstrated (38, 39), but there is very little evidence for apoC-II expression in the brain. Thus, it is possible that some apoC-I and C-II may cross the BCB barrier and enter the CSF from plasma, most likely carried by the HDL, which crosses the barrier by transcytosis (40, 41). It is also possible that that transport of the truncated apoC-I and apoC-II isoforms across the BCB is increased compared with the full-length proteins.
The observed differences in apoC-I and apoC-II proteolytic cleavages between plasma and CSF may also be caused by the different types of lipoprotein particles in the periphery and CNS. In plasma, apoC-I and C-II are rapidly interchanged among HDL, CM, and VLDL and may be somewhat shielded from extensive proteolysis. In CNS, apoC-I and C-II are most likely only associated with HDL-like particles and may be more prone to proteolysis as they disassociate from the HDL. Therefore, domain interactions between phospholipids and bound apolipoproteins may protect them against proteolysis. It was, however, shown that lipid-free apoC-II's N and C terminals had a considerable percentage of shielding from hydrogen/deuterium (H/D) exchange and proteolysis (42), suggesting some structured order that helps protect against proteolysis. Increased levels of protease inhibitors in plasma should also be considered (43) and together may provide a more comprehensive explanation for the present trends. Further studies are warranted to explore these differences and delineate the functionality of the truncated isoforms of apoC-I and C-II.
Apolipoprotein C-III isoforms
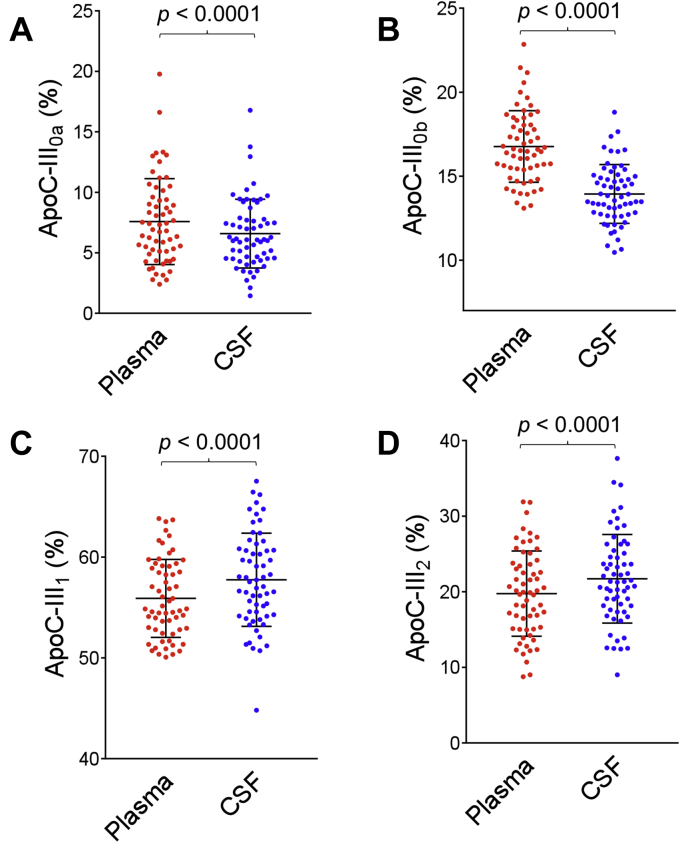
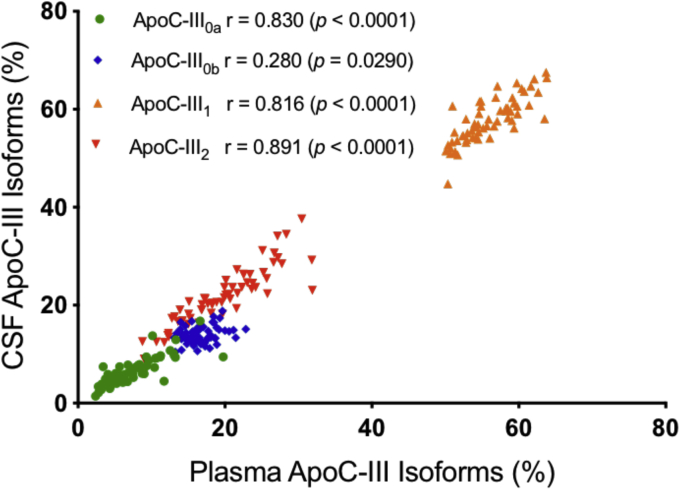
ApoC-III MALDI-TOF mass spectra resulting from the analysis of paired plasma and CSF samples from a single individual are shown in Fig. 1C, D. Signals were observed for all four major apoC-III isoforms in both plasma and CSF: apoC-III0a (no glycosylation at Thr74, MW = 8764.7); apoC-III0b (Thr74-GalNAc-Gal, MW = 9129.9), apoC-III1 (Thr74-GalNAc-Gal-Sia, MW = 9421.1), and apoC-III2 (Thr74-GalNAc-Gal-Sia-Sia, MW = 9712.4). The apoC-III1 signal was strongest in both plasma and CSF. The percent abundance of the individual apoC-III isoforms was calculated by dividing the peak area of each isoform with the summed peak areas of all the apoC-III isoforms and is shown in Fig. 4 for all 61 plasma and CSF samples. The percent abundance for apoC-III0a and apoC-III0b was decreased in CSF: a mean of 6.58 ± 2.8% in CSF versus 7.58 ± 3.5% in plasma was observed for apoC-III0a (P < 0.0001), and a mean of 13.9 ± 1.7 in CSF versus 16.8 ± 2.1% in plasma was observed for apoC-III0b (P < 0.0001). The opposite trend was detected for apoC-III1 and C-III2, with both isoforms showing increased percent abundance in CSF. A mean of 57.8 ± 4.6% in CSF versus 55.9 ± 3.9% in plasma was observed for apoC-III1 (P < 0.0001), and a mean of 21.7 ± 5.9% in CSF versus 19.7 ± 5.6% in plasma was observed for apoC-III2 (P < 0.0001). The relationships between the apoC-III isoforms in plasma and CSF are shown in Fig. 5, revealing positive linear relationships for apoC-III0a (r = 0.830, P < 0.0001), apoC-III0b (r = 0.280, P = 0.0290), apoC-III1 (r = 0.816, P < 0.0001), and C-III2 (r = 0.891, P < 0.0001). There was a correlation between the total plasma apoC-III concentrations and three of the apoC-III isoforms: apoC-III0b and apoC-III1 showed strong positive correlations, whereas apoC-III2 exhibited significant negative association with total plasma apoC-III concentrations (supplemental Fig. S2). In CSF, the correlations were not statistically significant, although similar trends were noticeable (supplemental Fig. S3).
Fig. 4.
Percent abundance of: (A) apoC-III0a (B) apoC-III0b (C) apoC-III1, and (D) apoC-III2 in plasma versus CSF. Percent abundance was computed by dividing the peak area of each isoform with the summed peak areas of all apoC-III isoforms. Parametric paired t-test was performed to identify differences between plasma and CSF samples for normally distributed data sets (apoC-III0b and apoC-III2). Nonparametric Wilcoxon matched-pairs signed rank test was applied to data sets that were not normally distributed (apoC-III0a and apoC-III1).
Fig. 5.
Correlation between apoC-III isoforms in the paired plasma and CSF samples. Shown are the parametric Pearson's correlation coefficients for apoC-III0b, apoC-III2, and the nonparametric Spearman correlation coefficients for apoC-III0a and apoC-III1.
This is the first study of the apoC-III isoforms in CSF. Previously, it was shown that total apoC-III concentration in CSF is only a small fraction of that measured in plasma (11, 12). We confirmed these findings by determining in this study that the concentration of apoC-III in CSF was <0.1% of that in plasma (Table 1). There is currently no evidence that apoC-III is expressed in the CNS; thus, the apoC-III detected in CSF most likely originates from plasma. The excellent linear correlation observed between plasma and CSF apoC-III isoforms in this study suggests a connection between the two pools of apoC-III.
One mechanism for the transport of apoC-III from plasma to CSF could be on small HDL. The discoidal shape of apoA-I containing small HDL may promote its brain delivery, in contrast to the large spherical HDL particles that are lipid-rich and have lower BBB transport (44). ApoA-I protein is abundant in CSF, but there is no apoA-I mRNA expression in the brain (7, 10, 13, 45). ApoA-I is believed to enter the CSF via the choroid plexus (13). In contrast to individual apolipoproteins, small HDL particles carry a large number of apolipoproteins (46) that include apoCs and can confer pleiotropic effects, allowing brain access for these apolipoproteins. This is further supported by recent findings that plasma apoA-I-associated apoC-III correlated most strongly with CSF apoC-III (12), suggesting that plasma apoC-III crosses into CSF along with apoA-I on HDL.
The percent abundance of the two sialylated apoC-III isoforms (apoC-III1 and C-III2) was increased in CSF compared with plasma. In plasma, apoC-III is distributed among all classes of lipoproteins, while CSF apoC-III is most likely associated only with HDL-like particles. It is possible that apoC-III1 and apoC-III2 are preferentially bound to the HDL, which can shield them from degradation and thus increase their CSF abundance. Furthermore, HDL-bound apoC-III1 and apoC-III2 crossing over from plasma across the BCB barrier would result in their enrichment in CSF. Supporting this hypothesis are results from studies with apoE, which has the same O-linked glycan structure as apoC-III. It was shown that the removal of the apoE sialic acids decreased the binding of apoE to HDL in plasma, leading to impaired reverse cholesterol transport (47). But unlike apoC-III, apoE is also expressed in the brain, and its terminal sialic acids are critically involved in the formation of the CSF lipoprotein particles (48). It is possible that the apoC-III sialic acids play similarly important role in the CSF lipoproteins.
ApoC-III is known to inhibit the very low density lipoprotein receptor/low density lipoprotein receptor (VLDLR/LDLR), but was not shown to affect VLDLs' binding to LDL receptor related protein 1 (LRP1) (49, 50). The presence of one sialic acid on apoC-III enabled TRLs' clearance through LDLR and LRP1, while apoC-III2 was shown to be preferentially cleared by heparan sulfate proteoglycan (HSPG)-type receptors (26). These receptors are also present in the brain (8) and mediate pathways toward endocytosis and lipid catabolism. Since nonsialylated apoC-III functions to inhibit HSPG and LRP1, the presence of negatively charged mono/disialic acids may serve to nullify apoC-III's interaction with HSPG's or LDLR's/LRP's by orientating these lipoproteins in a way that promotes endocytosis. This could allow the lipoproteins with an affinity for these receptors, such as apoA-I and apoE, to readily bind to them (51, 52). Furthermore, an increased rate of HDL endocytosis by neuronal cells may optimize neuronal cell maintenance, given that lipid metabolism is virtually isolated in the CNS (53).
Correlation of apoCs isoforms with APOE Ɛ4 allele and CSF Aβ42
The apoE phenotypes for the individuals that provided the 61 plasma and CSF samples were known from genotyping. Five allelic APOE combinations were present in the cohort: 5 Ɛ2/Ɛ3, 2 Ɛ2/Ɛ4, 27 Ɛ3/Ɛ3, 16 Ɛ3/Ɛ4, and 11 Ɛ4/Ɛ4. We investigated the correlations of the apoCs isoforms with the Ɛ4 allele, which is the strongest genetic risk factor for developing AD (54, 55, 56, 57). The samples were grouped into two groups: non-Ɛ4 (32 samples) and Ɛ4 (29 samples, heterozygous or homozygous for Ɛ4). ApoC-I and C-II truncationsand apoC-III isoforms percent abundance were compared between the two groups, in both plasma and CSF.
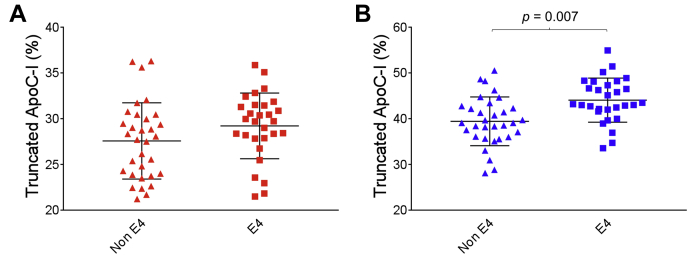
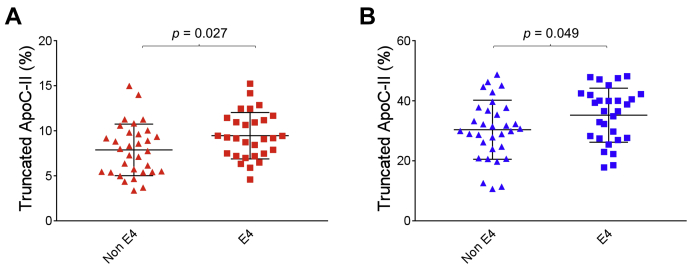
In plasma, the percent truncated apoC-I in the Ɛ4 group was 29.2 ± 3.6% versus 27.6 ± 4.2% in the non-Ɛ4 group (Fig. 6A). In CSF, the Ɛ4 group showed a significantly greater percentage of truncated apoC-I (44.1 ± 4.8%) when compared with the non-Ɛ4 group (39.4 ± 5.3%) (P = 0.007) (Fig. 6B). A statistically significant difference was also observed for the truncated apoC-II in both plasma and CSF, with the Ɛ4 group exhibiting higher percent truncation. In plasma, 9.45 ± 2.6% truncated apoC-II was observed in the Ɛ4 group versus 7.88 ± 2.9% in the non-Ɛ4 group (P = 0.027), while in CSF 35.2 ± 9.0% truncated apoC-II was observed in the Ɛ4 group versus 30.4 ± 9.9% the non-Ɛ4 group (P = 0.049) (Fig. 7). We also investigated the difference in apoC-I and apoC-II percent truncations between homozygous and heterozygous Ɛ4 carriers (supplemental Fig. S4). In plasma, these differences were not statistically significant, while in CSF only the truncated apoC-I exhibited similar differences between the homozygous Ɛ4 and the non-Ɛ4 group (P = 0.012) and the heterozygous Ɛ4 and the non-Ɛ4 group (P = 0.018). Total apoC-I and apoC-II concentrations were not significantly different among the various APOE genotypes, in both plasma and CSF (Table 1).
Fig. 6.
Percent abundance of truncated apoC-I in non-Ɛ4 versus Ɛ4 allele carriers, in (A) plasma and (B) CSF. Unpaired t-test with Welch's correction was utilized for the normally distributed data sets.
Fig. 7.
Percent abundance of truncated apoC-II in non-Ɛ4 versus Ɛ4 allele carriers, in (A) plasma and (B) CSF. Unpaired t-test with Welch's correction was utilized for the normally distributed data sets.
We also examined the association of the truncated apoC-I and C-II isoforms with CSF Aβ42, a surrogate of brain amyloid accumulation. There was a trend association between the % of truncated CSF apoC-I and CSF Aβ42 (r = −0.273, P = 0.055) and a significant association between truncated plasma apoC-II and CSF Aβ42 (r = −0.383, P = 0.006) (supplemental Fig. S5). This association persisted after adjusting for APOE genotype and did not differ between the two CDR groups (CDR = 0 and CDR > 0.5). Albeit modest, these associations may support a role of apoC truncations as biomarkers of brain amyloid plaques, reflecting a proteolytic milieu within the Aβ plaques that promotes greater enzymatic truncation of local (brain) or systemic apoC proteins. The nonsignificant associations of the other apoC-I and C-II truncations with CSF Aβ42 may be a function of the smaller sample size of this cohort.
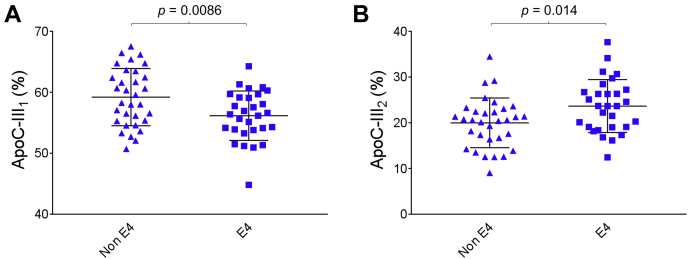
Of the four apoC-III isoforms, only apoC-III1 and C-III2 in CSF exhibited statistically significant differences between the Ɛ4 and non-Ɛ4 groups. The directions were opposite, with apoC-III1 decreased and apoC-III2 increased in the Ɛ4 group: 56.1 ± 4.1% apoC-III1 was observed in the Ɛ4 group versus 59.2 ± 4.7% in the non-Ɛ4 group (P = 0.0086); and 23.6 ± 5.8% apoC-III2 was observed in the Ɛ4 group versus 20.0 ± 5.4% in the non-Ɛ4 group (P = 0.014) (Fig. 8). The increased percent apoC-III2 suggests increased sialylation in the Ɛ4 group. ApoC-III1 and apoC-III2 did not show a statistically significant difference in plasma between the Ɛ4 and non-Ɛ4 groups, and neither did apoC-IIIoa and C-IIIob, in both plasma and CSF (supplemental Figs. S6 and S7). The difference in apoC-III isoforms between the homozygous and heterozygous Ɛ4 carriers in plasma was also not statistically significant (supplemental Fig. S8). In CSF, only the homozygous Ɛ4 carriers showed statistically significant difference from the non-Ɛ4 group for apoC-III1 (P = 0.033) and apoC-III2 (P = 0.020) (supplemental Fig. S9), suggesting that the observed differences between the Ɛ4 group and the non-Ɛ4 group shown in Fig. 8 were driven by the homozygous Ɛ4 carriers. The total apoC-III concentrations were not significantly different among the various APOE genotypes in both plasma and CSF (Table 1). Furthermore, there were no significant associations between the apoC-III isoforms and CSF Aβ42 (data not shown).
Fig. 8.
Percent abundance in CSF of (A) apoC-III1 and (B) apoC-III2 isoforms in non-Ɛ4 versus Ɛ4 allele carriers. Unpaired t-test with Welch's correction was utilized for the normally distributed data sets.
The interplay between apoE and apoCs is critical to the regulation of lipoprotein metabolism. The apoC-I and C-II genes are colocated with the apoE gene in a cluster on chromosome 19, and their expression is coregulated (5). In addition to the well-documented role of the APOE Ɛ4 allele as a risk factor for AD, apoC-I allelic variations were also suggested to be significant risk factor for AD (58, 59). ApoC-I was colocalized with amyloid-β (Aβ) in senile plaques from the brains of AD patients and shown to exacerbate soluble Aβ oligomer-induced neuronal death (39). Significantly reduced levels of ApoC-I mRNA were found in the frontal cortex of AD patients that carried the Ɛ4 allele (38), and apoC-I levels in CSF were found to vary with APOE genotype, with Ɛ4 carriers showing significantly less apoC-I (60). Increased truncation with the Ɛ4 allele was also observed with apoC-II, suggesting a common denominator to this phenomenon that could be modulated by the Ɛ4 allele and/or the expressed apoE4 isoform.
The truncations of apoC-I and apoC-II in the brain may reflect the plaque accumulation environment that is increased with APOE4. The emergence of protein deposits is linked to many pathologies like AD and CVD and therefore relevant to study. In general, the reactive oxygen species (ROS) (61, 62, 63, 64) produced from atherosclerotic and Aβ plaques may lead to a feedback loop that promotes apolipoprotein truncation (65). Indeed, apoE4 is more sensitive to proteolytic cleavage (66), and its truncated fragments have been found in the brains of AD patients (67). Specifically, apoE4[Δ(166–299)] was shown to promote the intracellular uptake of Aβ42, leading to an increased production of ROS (62). While it does play a critical role in AD pathogenesis, apoE4 was also shown to be associated with atherosclerosis (68). Therefore, there may be a correlation among some apolipoproteins and amyloid plaque formation. However, the biological route remains obscure, and whether increased proteolysis of related apolipoproteins is necessarily involved remains unclear. In a previous study, which demonstrated that some of the lipid-free apoC-II's amino acid sequences are protected from proteolytic cleavage, it was proposed that certain morphologies can promote amyloid formation: monomeric lipid-free apoC-II's regions that were less protected from H/D exchange and proteolysis corresponded to the core within its amyloid fibrils (42). Interestingly, apoC-II, apoA-I, and apoB aggregates were also found in atherosclerotic plaques (69), and apoA-I, apoA-II, apoC-I, apoB100, and apoE were found to colocalize with amyloid in vivo (39, 70, 71), although they were not necessarily truncated. The increased truncation of apoC-I and apoC-II in ε4 carriers and its relation to developing pathologies, such as AD, warrant further investigation.
While apoC-III's polymorphisms and relative abundance are affected by APOE4 expression, the present ε4 trends in apoC-III sialylation are perhaps a reaction to ε4 pathology that affects lipid metabolism. The observed divergent tendencies of apoC-III1 and C-III2 with the Ɛ4 allele in CSF are intriguing and may be specific only to CSF as those differences were not statistically significant in plasma. It was observed previously that carriers of the Ɛ4 allele had increased apoC-III/apoE plasma ratios compared with the other two apoE alleles (72). Our previous plasma apoC-III isoforms studies revealed a negative relationship between apoC-III2 and TG, which was the opposite of the positive correlation observed for the other three isoforms (23, 24). We also showed that plasma apoC-III2 was preferentially cleared by HSPG type of receptors, whereas apoC-III1 was cleared more rapidly through LDLR and LRP1 (26). In another study, apoC-III2 demonstrated diminished ability to inhibit VLDL binding to the lipolysis-stimulated receptor in rat liver plasma membranes, unlike apoC-III1 which showed greater inhibitory effect (73), which might explain apoC-III2's negative relationship with TG. In the current study, we demonstrate an increased percent abundance of apoC-III2 with the Ɛ4 allele in CSF. Based on the peripheral sink hypothesis, apoC-III on TG-rich lipoproteins can serve as a circulating Aβ binding protein that could facilitate the efflux of Aβ from the brain (74), and apoC-III sialylation may affect this process.
It has been shown that lipoprotein bound apoE4 does not preferentially bind to LRP1 like the other two apoE isoforms (E2 and E3), which was suggested to affect Aβ clearance (75, 76). Instead, Aβ and Aβ-apoE complexes are redirected to VLDLR in ε4 carriers and thus cleared at a much slower rate (77). This receptor also has a much slower endocytosis rate (78), which may affect clearance of HDL particles in the CNS. One possibility is that the increased presence of apoC-III2 within the CNS of Ɛ4 carriers facilitates endocytosis by offering an alternative route to HDL catabolism via HSPG type of receptors. Although apoE4 has been shown to bind with HSPG receptors with similar affinity as apoE3, it has also been demonstrated that apoE inhibits Aβ clearance by HSPG (79). Consequently, this would exacerbate Aβ accumulation as apoE and Aβ compete for HSPG type of receptors' binding. Therefore, the increased ratio of apoC-III/apoE in the HDL of ε4 carriers may serve to nullify this effect, allowing for increased Aβ clearance.
One of the limitations of our study is that the observed apoCs differences may result from a differential antibody-antigen capture from plasma and CSF. This is possible but less likely. The comparison within plasma and CSF by AD risk factors argues against this limitation. For example, the % apoCs truncation in APOE4 carriers was increased in both CSF and plasma (Figs. 6 and 7). Spike and recovery experiments were not feasible because apoCs standards commercially available do not contain the same isoforms (in the case of apoC-I and II) and are grossly oxidized (for all 3 apoCs), which creates additional signals in the mass spectra and makes the quantitative comparison using those standards inaccurate. For this assay, well-characterized plasma samples as QC points for all of our analyses, and not purified protein standards, were used.
A second limitation was that the study sample size was relatively small. This limited our ability to detect whether the % of apoC isoforms differed by early disease markers such as CDR 0 and CDR 0.5. Future studies will expand subgroup analysis to help understand the relationship of apoC processing with clinical disease onset.
Conclusion
Distinct patterns of apoCs isoforms were detected in CSF in a set of paired plasma and CSF samples obtained from a cohort of healthy individuals. Truncated apoC-I and C-II isoforms were elevated in CSF, which could be the result of increased enzymatic activity. Sialylated isoforms of apoC-III were also elevated in CSF, possibly indicating preferred binding to HDL. Some of the apoCs isoforms' changes were accentuated for individuals that were carriers of the APOE Ɛ4 allele. ApoC-I and C-II truncations were greater in Ɛ4 carriers. The doubly sialylated apoC-III isoform was also elevated in Ɛ4 carriers, in agreement with previous observations in plasma about the distinctive feature of this isoform. Future studies should evaluate how the APOE Ɛ4 allele affects the apoCs metabolism and regulation, which may lead to implications on protein-related pathologies, such as AD and CVD.
Data avilability
All data are contained within the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
D. N. and H. N. Y. study design; Y. H., C. M., A. M., and D. N. experiments; Y. H., C. M., and A. M. data analysis; Y. H., H. N. Y. and D. N. writing - original draft; Y. H., C. M., A. M., H. N. Y., and D. N. editing.
Funding and additional information
D. N. was supported by 1R43AG069552 from the National Institutes of Health, United States. H. N. Y. was supported by R21AG056518, R01AG055770, R01AG054434, and R01AG067063 from the National Institutes of Health, United States. This work was also supported by P50AG05142 (USC ADRC) from the National Institutes of Health, United States. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
This article contains supplemental data.
Contributor Information
Hussein N. Yassine, Email: hyassine@usc.edu.
Dobrin Nedelkov, Email: dobrin.nedelkov@isoformix.com.
Supplemental data
References
- 1.Bondarenko P.V., Cockrill S.L., Watkins L.K., Cruzado I.D., Macfarlane R.D. Mass spectral study of polymorphism of the apolipoproteins of very low density lipoprotein. J. Lipid Res. 1999;40:543–555. [PubMed] [Google Scholar]
- 2.Lambeir A.M., Durinx C., Scharpé S., De Meester I. Dipeptidyl-peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 2003;40:209–294. doi: 10.1080/713609354. [DOI] [PubMed] [Google Scholar]
- 3.Fojo S.S., Taam L., Fairwell T., Ronan R., Bishop C., Meng M.S., Hoeg J.M., Sprecher D.L., Brewer H.B. Human preproapolipoprotein C-II. Analysis of major plasma isoforms. J. Biol. Chem. 1986;261:9591–9594. [PubMed] [Google Scholar]
- 4.Sacks F.M., Zheng C., Cohn J.S. Complexities of plasma apolipoprotein C-III metabolism. J. Lipid Res. 2011;52:1067–1070. doi: 10.1194/jlr.E015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuior E.V., Gafencu A.V. Apolipoprotein C1: Its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20235939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolska A., Dunbar R.L., Freeman L.A., Ueda M., Amar M.J., Sviridov D.O., Remaley A.T. Apolipoprotein C-II: New findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis. 2017;267:49–60. doi: 10.1016/j.atherosclerosis.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch S., Donarski N., Goetze K., Kreckel M., Stuerenburg H.J., Buhmann C., Beisiegel U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J. Lipid Res. 2001;42:1143–1151. [PubMed] [Google Scholar]
- 8.Wang H., Eckel R.H. What are lipoproteins doing in the brain? Trends Endocrinol. Metab. 2014;25:8–14. doi: 10.1016/j.tem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauer S.J., Walker D., Elshourbagy N.A., Reardon C.A., Levy-Wilson B., Taylor J.M. Two copies of the human apolipoprotein C-I gene are linked closely to the apolipoprotein E gene. J. Biol. Chem. 1988;263:7277–7286. [PubMed] [Google Scholar]
- 10.Elliott D.A., Weickert C.S., Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin. Lipidol. 2010;51:555–573. doi: 10.2217/CLP.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roheim P.S., Carey M., Forte T., Vega G.L. Apolipoproteins in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 1979;76:4646–4649. doi: 10.1073/pnas.76.9.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch M., Furtado J.D., Falk K., Leypoldt F., Mukamal K.J., Jensen M.K. Apolipoproteins and their subspecies in human cerebrospinal fluid and plasma. Alzheimers Dement. (Amst) 2017;6:182–187. doi: 10.1016/j.dadm.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stukas S., Robert J., Lee M., Kulic I., Carr M., Tourigny K., Fan J., Namjoshi D., Lemke K., DeValle N., Chan J., Wilson T., Wilkinson A., Chapanian R., Kizhakkedathu J.N., Cirrito J.R., Oda M.N., Wellington C.L. Intravenously injected human apolipoprotein A-I rapidly enters the central nervous system via the choroid plexus. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou A.L., Swaminathan S.K., Curran G.L., Poduslo J.F., Lowe V.J., Li L., Kandimalla K.K. Apolipoprotein A-I crosses the blood-brain barrier through clathrin-independent and cholesterol-mediated endocytosis. J. Pharmacol. Exp. Ther. 2019;369:481–488. doi: 10.1124/jpet.118.254201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott J., Knott T.J., Shaw D.J., Brook J.D. Localization of genes encoding apolipoproteins CI, CII, and E to the p13----cen region of human chromosome 19. Hum. Genet. 1985;71:144–146. doi: 10.1007/BF00283370. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q., Zhao F., Lv Z.-p., Zheng C.-g., Zheng W.-d., Sun L., Wang N.-n., Pang S., de Andrade F.M., Fu M., He X.-h., Hui J., Jiang W., Yang C.-y., Shi X.-h., Zhu X.-q., Pang G.-f., Yang Y.-g., Xie H.-q., Zhang W.-d., Hu C.-y., Yang Z. Association between APOC1 polymorphism and Alzheimer's disease: a case-control study and meta-analysis. PLoS One. 2014;9:e87017. doi: 10.1371/journal.pone.0087017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallinga-Thie G.M., Bu X.D., van Linde-Sibenius Trip M., Rotter J.I., Lusis A.J., de Bruin T.W. Apolipoprotein A-I/C-III/A-IV gene cluster in familial combined hyperlipidemia: effects on LDL-cholesterol and apolipoproteins B and C-III. J. Lipid Res. 1996;37:136–147. [PubMed] [Google Scholar]
- 18.Wyler von Ballmoos M.C., Haring B., Sacks F.M. The risk of cardiovascular events with increased apolipoprotein CIII: A systematic review and meta-analysis. J. Clin. Lipidol. 2015;9:498–510. doi: 10.1016/j.jacl.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Ramms B., Gordts P.L.S.M. Apolipoprotein C-III in triglyceride-rich lipoprotein metabolism. Curr. Opin. Lipidol. 2018;29:171–179. doi: 10.1097/MOL.0000000000000502. [DOI] [PubMed] [Google Scholar]
- 20.Taskinen M.R., Packard C.J., Borén J. Emerging Evidence that ApoC-III Inhibitors Provide Novel Options to Reduce the Residual CVD. Curr. Atheroscler. Rep. 2019;21:27. doi: 10.1007/s11883-019-0791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivieri O., Martinelli N., Bassi A., Trabetti E., Girelli D., Pizzolo F., Friso S., Pignatti P.F., Corrocher R. ApoE ε2/ε3/ε4 polymorphism, ApoC-III/ApoE ratio and metabolic syndrome. Clin. Exp. Med. 2007;7:164–172. doi: 10.1007/s10238-007-0142-y. [DOI] [PubMed] [Google Scholar]
- 22.Trenchevska O., Schaab M.R., Nelson R.W., Nedelkov D. Development of multiplex mass spectrometric immunoassay for detection and quantification of apolipoproteins C-I, C-II, C-III and their proteoforms. Methods. 2015;81:86–92. doi: 10.1016/j.ymeth.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yassine H.N., Trenchevska O., Ramrakhiani A., Parekh A., Koska J., Walker R.W., Billheimer D., Reaven P.D., Yen F.T., Nelson R.W., Goran M.I., Nedelkov D. The Association of Human Apolipoprotein C-III Sialylation Proteoforms with Plasma Triglycerides. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koska J., Yassine H., Trenchevska O., Sinari S., Schwenke D.C., Yen F.T., Billheimer D., Nelson R.W., Nedelkov D., Reaven P.D. Disialylated apolipoprotein C-III proteoform is associated with improved lipids in prediabetes and type 2 diabetes. J. Lipid Res. 2016;57:894–905. doi: 10.1194/jlr.P064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendoza S., Trenchevska O., King S.M., Nelson R.W., Nedelkov D., Krauss R.M., Yassine H.N. Changes in low-density lipoprotein size phenotypes associate with changes in apolipoprotein C-III glycoforms after dietary interventions. J. Clin. Lipidol. 2017;11:224–233. doi: 10.1016/j.jacl.2016.12.009. e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kegulian N.C., Ramms B., Horton S., Trenchevska O., Nedelkov D., Graham M.J., Lee R.G., Esko J.D., Yassine H.N., Gordts P.L.S.M. ApoC-III glycoforms are differentially cleared by hepatic TRL (triglyceride-rich lipoprotein) receptors. Arterioscler. Thromb. Vasc. Biol. 2019;39:2145–2156. doi: 10.1161/ATVBAHA.119.312723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tudorache I.F., Trusca V.G., Gafencu A.V. Apolipoprotein E - a multifunctional protein with implications in various pathologies as a result of its structural features. Comput. Struct. Biotechnol. J. 2017;15:359–365. doi: 10.1016/j.csbj.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen J.-B., Fernández J.A., Notø A.-T.W., Deguchi H., Björkegren J., Mathiesen E.B. The apolipoprotein C-I content of very-low-density lipoproteins is associated with fasting triglycerides, postprandial lipemia, and carotid atherosclerosis. J. Lipids. 2011;2011:271062. doi: 10.1155/2011/271062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trenchevska O., Nelson R., Nedelkov D. Mass spectrometric immunoassays in characterization of clinically significant proteoforms. Proteomes. 2016;4 doi: 10.3390/proteomes4010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nedelkov D. Mass spectrometric studies of apolipoprotein proteoforms and their Role in lipid metabolism and type 2 diabetes. Proteomes. 2017;5 doi: 10.3390/proteomes5040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein H.G., Schön E., Ansorge S., Röse I., Dorn A. Immunolocalization of dipeptidyl aminopeptidase (DAP IV) in the developing human brain. Int. J. Dev. Neurosci. 1987;5:237–242. doi: 10.1016/0736-5748(87)90034-7. [DOI] [PubMed] [Google Scholar]
- 32.Busek P., Stremenova J., Sedo A. Dipeptidyl peptidase-IV enzymatic activity bearing molecules in human brain tumors--good or evil? Front. Biosci. 2008;13:2319–2326. doi: 10.2741/2846. [DOI] [PubMed] [Google Scholar]
- 33.Skinner N.E., Wroblewski M.S., Kirihara J.A., Nelsestuen G.L., Seaquist E.R. Sitagliptin Results in a Decrease of Truncated Apolipoprotein C1. Diabetes Ther. 2015;6:395–401. doi: 10.1007/s13300-015-0123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozek A., Sparrow J.T., Weisgraber K.H., Cushley R.J. Sequence-specific 1H NMR resonance assignments and secondary structure of human apolipoprotein C-I in the presence of sodium dodecyl sulfate. Biochem. Cell Biology. 1998;76:267–275. doi: 10.1139/bcb-76-2-3-267. [DOI] [PubMed] [Google Scholar]
- 35.Rozek A., Sparrow J.T., Weisgraber K.H., Cushley R.J. Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy. Biochemistry. 1999;38:14475–14484. doi: 10.1021/bi982966h. [DOI] [PubMed] [Google Scholar]
- 36.Edelstein C., Gordon J.I., Toscas K., Sims H.F., Strauss A.W., Scanu A.M. In vitro conversion of proapoprotein A-I to apoprotein A-I. Partial characterization of an extracellular enzyme activity. J. Biol. Chem. 1983;258:11430–11433. [PubMed] [Google Scholar]
- 37.Scanu A.M., Byrne R.E., Edelstein C. Proteolytic events affecting plasma apolipoproteins at the co- and post-translational levels and after maturation. J. Lipid Res. 1984;25:1593–1602. [PubMed] [Google Scholar]
- 38.Petit-Turcotte C., Stohl S.M., Beffert U., Cohn J.S., Aumont N., Tremblay M., Dea D., Yang L., Poirier J., Shachter N.S. Apolipoprotein C-I expression in the brain in Alzheimer's disease. Neurobiol. Dis. 2001;8:953–963. doi: 10.1006/nbdi.2001.0441. [DOI] [PubMed] [Google Scholar]
- 39.Abildayeva K., Berbee J.F., Blokland A., Jansen P.J., Hoek F.J., Meijer O., Lutjohann D., Gautier T., Pillot T., De Vente J., Havekes L.M., Ramaekers F.C., Kuipers F., Rensen P.C., Mulder M. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J. Lipid Res. 2008;49:856–869. doi: 10.1194/jlr.M700518-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Balazs Z., Panzenboeck U., Hammer A., Sovic A., Quehenberger O., Malle E., Sattler W. Uptake and transport of high-density lipoprotein (HDL) and HDL-associated alpha-tocopherol by an in vitro blood-brain barrier model. J. Neurochem. 2004;89:939–950. doi: 10.1111/j.1471-4159.2004.02373.x. [DOI] [PubMed] [Google Scholar]
- 41.Fung K.Y., Wang C., Nyegaard S., Heit B., Fairn G.D., Lee W.L. SR-BI mediated transcytosis of HDL in brain microvascular endothelial cells is independent of caveolin, clathrin, and PDZK1. Front. Physiol. 2017;8:841. doi: 10.3389/fphys.2017.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson L.M., Mok Y.F., Binger K.J., Griffin M.D., Mertens H.D., Lin F., Wade J.D., Gooley P.R., Howlett G.J. A structural core within apolipoprotein C-II amyloid fibrils identified using hydrogen exchange and proteolysis. J. Mol. Biol. 2007;366:1639–1651. doi: 10.1016/j.jmb.2006.12.040. [DOI] [PubMed] [Google Scholar]
- 43.Safi W., Maiorano J., Davidson W. A proteolytic method for distinguishing between lipid-free and lipid-bound apolipoprotein A-I. J. Lipid Res. 2001;42:864–872. [PubMed] [Google Scholar]
- 44.Dal Magro R., Simonelli S., Cox A., Formicola B., Corti R., Cassina V., Nardo L., Mantegazza F., Salerno D., Grasso G., Deriu M.A., Danani A., Calabresi L., Re F. The extent of human apolipoprotein A-I lipidation strongly affects the β-amyloid efflux across the blood-brain barrier in vitro. Front. Neurosci. 2019;13 doi: 10.3389/fnins.2019.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guyton J.R., Miller S.E., Martin M.E., Khan W.A., Roses A.D., Strittmatter W.J. Novel large apolipoprotein E-containing lipoproteins of density 1.006-1.060 g/ml in human cerebrospinal fluid. J. Neurochem. 1998;70:1235–1240. doi: 10.1046/j.1471-4159.1998.70031235.x. [DOI] [PubMed] [Google Scholar]
- 46.Holzer M., Kern S., Birner-Grunberger R., Curcic S., Heinemann A., Marsche G. Refined purification strategy for reliable proteomic profiling of HDL2/3: Impact on proteomic complexity. Sci. Rep. 2016;6:38533. doi: 10.1038/srep38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marmillot P., Rao M.N., Liu Q.H., Lakshman M.R. Desialylation of human apolipoprotein E decreases its binding to human high-density lipoprotein and its ability to deliver esterified cholesterol to the liver. Metabolism. 1999;48:1184–1192. doi: 10.1016/s0026-0495(99)90136-1. [DOI] [PubMed] [Google Scholar]
- 48.Kawasaki K., Ogiwara N., Sugano M., Okumura N., Yamauchi K. Sialic acid moiety of apolipoprotein E and its impact on the formation of lipoprotein particles in human cerebrospinal fluid. Clin. Chim. Acta. 2009;402:61–66. doi: 10.1016/j.cca.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 49.Agnani G., Bard J.M., Candelier L., Delattre S., Fruchart J.C., Clavey V. Interaction of LpB, LpB:E, LpB:C-III, and LpB:C-III:E lipoproteins with the low density lipoprotein receptor of HeLa cells. Arterioscler. Thromb. 1991;11:1021–1029. doi: 10.1161/01.atv.11.4.1021. [DOI] [PubMed] [Google Scholar]
- 50.Kowal R.C., Herz J., Weisgraber K.H., Mahley R.W., Brown M.S., Goldstein J.L. Opposing effects of apolipoproteins E and C on lipoprotein binding to low density lipoprotein receptor-related protein. J. Biol. Chem. 1990;265:10771–10779. [PubMed] [Google Scholar]
- 51.Gordts P., Esko J.D. The heparan sulfate proteoglycan grip on hyperlipidemia and atherosclerosis. Matrix Biol. 2018;71-72:262–282. doi: 10.1016/j.matbio.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordts P.L., Nock R., Son N.H., Ramms B., Lew I., Gonzales J.C., Thacker B.E., Basu D., Lee R.G., Mullick A.E., Graham M.J., Goldberg I.J., Crooke R.M., Witztum J.L., Esko J.D. ApoC-III inhibits clearance of triglyceride-rich lipoproteins through LDL family receptors. J. Clin. Invest. 2016;126:2855–2866. doi: 10.1172/JCI86610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meaney S., Hassan M., Sakinis A., Lutjohann D., von Bergmann K., Wennmalm A., Diczfalusy U., Bjorkhem I. Evidence that the major oxysterols in human circulation originate from distinct pools of cholesterol: a stable isotope study. J. Lipid Res. 2001;42:70–78. [PubMed] [Google Scholar]
- 54.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 55.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., van Duijn C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 57.Verghese P.B., Castellano J.M., Holtzman D.M. Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol. 2011;10:241–252. doi: 10.1016/S1474-4422(10)70325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poduslo S.E., Neal M., Herring K., Shelly J. The apolipoprotein CI A allele as a risk factor for Alzheimer's disease. Neurochem. Res. 1998;23:361–367. doi: 10.1023/a:1022409617539. [DOI] [PubMed] [Google Scholar]
- 59.Ki C.S., Na D.L., Kim D.K., Kim H.J., Kim J.W. Genetic association of an apolipoprotein C-I (APOC1) gene polymorphism with late-onset Alzheimer's disease. Neurosci. Lett. 2002;319:75–78. doi: 10.1016/s0304-3940(01)02559-9. [DOI] [PubMed] [Google Scholar]
- 60.Cudaback E., Li X., Yang Y., Yoo T., Montine K.S., Craft S., Montine T.J., Keene C.D. Apolipoprotein C-I is an APOE genotype-dependent suppressor of glial activation. J. Neuroinflammation. 2012;9:192. doi: 10.1186/1742-2094-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skjot-Arkil H., Barascuk N., Register T., Karsdal M.A. Macrophage-mediated proteolytic remodeling of the extracellular matrix in atherosclerosis results in neoepitopes: a potential new class of biochemical markers. Assay Drug Dev. Technol. 2010;8:542–552. doi: 10.1089/adt.2009.0258. [DOI] [PubMed] [Google Scholar]
- 62.Dafnis I., Stratikos E., Tzinia A., Tsilibary E.C., Zannis V.I., Chroni A. An apolipoprotein E4 fragment can promote intracellular accumulation of amyloid peptide beta 42. J. Neurochem. 2010;115:873–884. doi: 10.1111/j.1471-4159.2010.06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burtenshaw D., Kitching M., Redmond E.M., Megson I.L., Cahill P.A. Reactive oxygen species (ROS), intimal Thickening, and Subclinical Atherosclerotic Disease. Front. Cardiovasc. Med. 2019;6 doi: 10.3389/fcvm.2019.00089. 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohn T.T. Proteolytic cleavage of apolipoprotein E4 as the keystone for the heightened risk associated with Alzheimer's disease. Int. J. Mol. Sci. 2013;14:14908–14922. doi: 10.3390/ijms140714908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medeiros L.A., Khan T., El Khoury J.B., Pham C.L., Hatters D.M., Howlett G.J., Lopez R., O'Brien K.D., Moore K.J. Fibrillar amyloid protein present in atheroma activates CD36 signal transduction. J. Biol. Chem. 2004;279:10643–10648. doi: 10.1074/jbc.M311735200. [DOI] [PubMed] [Google Scholar]
- 66.Brecht W.J., Harris F.M., Chang S., Tesseur I., Yu G.-Q., Xu Q., Dee Fish J., Wyss-Coray T., Buttini M., Mucke L., Mahley R.W., Huang Y. Neuron-specific apolipoprotein e4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y., Liu X.Q., Wyss-Coray T., Brecht W.J., Sanan D.A., Mahley R.W. Apolipoprotein E fragments present in Alzheimer's disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zannis V.I., Kypreos K.E., Chroni A., Kardassis D., Zanni E.E. Lipoproteins and atherogenesis. In: Loscalzo J., editor. Molecular Mechanisms of Atherosclerosis. Taylor & Francis; New York, NY: 2004. pp. 111–174. [Google Scholar]
- 69.O'Brien K.D., Olin K.L., Alpers C.E., Chiu W., Ferguson M., Hudkins K., Wight T.N., Chait A. Comparison of apolipoprotein and proteoglycan deposits in human coronary atherosclerotic plaques: colocalization of biglycan with apolipoproteins. Circulation. 1998;98:519–527. doi: 10.1161/01.cir.98.6.519. [DOI] [PubMed] [Google Scholar]
- 70.Namba Y., Tomonaga M., Kawasaki H., Otomo E., Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer's disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541:163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- 71.Wisniewski T., Ghiso J., Frangione B. Biology of A beta amyloid in Alzheimer's disease. Neurobiol. Dis. 1997;4:313–328. doi: 10.1006/nbdi.1997.0147. [DOI] [PubMed] [Google Scholar]
- 72.Olivieri O., Martinelli N., Bassi A., Trabetti E., Girelli D., Pizzolo F., Friso S., Pignatti P.F., Corrocher R. ApoE epsilon2/epsilon3/epsilon4 polymorphism, ApoC-III/ApoE ratio and metabolic syndrome. Clin. Exp. Med. 2007;7:164–172. doi: 10.1007/s10238-007-0142-y. [DOI] [PubMed] [Google Scholar]
- 73.Mann C.J., Troussard A.A., Yen F.T., Hannouche N., Najib J., Fruchart J.C., Lotteau V., André P., Bihain B.E. Inhibitory effects of specific apolipoprotein C-III isoforms on the binding of triglyceride-rich lipoproteins to the lipolysis-stimulated receptor. J. Biol. Chem. 1997;272:31348–31354. doi: 10.1074/jbc.272.50.31348. [DOI] [PubMed] [Google Scholar]
- 74.Shih Y.H., Tsai K.J., Lee C.W., Shiesh S.C., Chen W.T., Pai M.C., Kuo Y.M. Apolipoprotein C-III is an amyloid-Œ≤-binding protein and an early marker for Alzheimer's disease. J. Alzheimers Dis. 2014;41:855–865. doi: 10.3233/JAD-140111. [DOI] [PubMed] [Google Scholar]
- 75.Bell R.D., Winkler E.A., Singh I., Sagare A.P., Deane R., Wu Z., Holtzman D.M., Betsholtz C., Armulik A., Sallstrom J., Berk B.C., Zlokovic B.V. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang D.S., Small D.H., Seydel U., Smith J.D., Hallmayer J., Gandy S.E., Martins R.N. Apolipoprotein E promotes the binding and uptake of beta-amyloid into Chinese hamster ovary cells in an isoform-specific manner. Neuroscience. 1999;90:1217–1226. doi: 10.1016/s0306-4522(98)00561-2. [DOI] [PubMed] [Google Scholar]
- 77.Deane R., Sagare A., Hamm K., Parisi M., Lane S., Finn M.B., Holtzman D.M., Zlokovic B.V. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J. Clin. Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Lu W., Marzolo M.P., Bu G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J. Biol. Chem. 2001;276:18000–18006. doi: 10.1074/jbc.M101589200. [DOI] [PubMed] [Google Scholar]
- 79.Fu Y., Zhao J., Atagi Y., Nielsen H.M., Liu C.C., Zheng H., Shinohara M., Kanekiyo T., Bu G. Apolipoprotein E lipoprotein particles inhibit amyloid-beta uptake through cell surface heparan sulphate proteoglycan. Mol. Neurodegener. 2016;11:37. doi: 10.1186/s13024-016-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.