Abstract
Pancreatic cysts are common and often pose a management dilemma, because some cysts are precancerous, whereas others have little risk of developing into invasive cancers. We used supervised machine learning techniques to develop a comprehensive test, CompCyst, to guide the management of patients with pancreatic cysts. The test is based on selected clinical features, imaging characteristics, and cyst fluid genetic and biochemical markers. Using data from 436 patients with pancreatic cysts, we trained CompCyst to classify patients as those who required surgery, those who should be routinely monitored, and those who did not require further surveillance. We then tested CompCyst in an independent cohort of 426 patients, with histopathology used as the gold standard. We found that clinical management informed by the CompCyst test was more accurate than the management dictated by conventional clinical and imaging criteria alone. Application of the CompCyst test would have spared surgery in more than half of the patients who underwent unnecessary resection of their cysts. CompCyst therefore has the potential to reduce the patient morbidity and economic costs associated with current standard-of-care pancreatic cyst management practices.
INTRODUCTION
Pancreatic cysts are fluid-containing lesions located within the pancreas. These cysts are common, found in 4% of individuals in their 60s and 8% of people over the age of 70 (1). A conservative estimate is that about 800,000 people per year with a pancreatic cyst are identified in the United States alone (2). Mucin-producing pancreatic cysts called intraductal papillary mucinous neoplasms (IPMNs) or mucinous cystic neoplasms (MCNs) are precursors to pancreatic ductal adenocarcinoma (hereafter termed “pancreatic adenocarcinoma”), which is the third leading cause of cancer death. One of the key reasons for the abysmal prognosis of these cancers is the inability to identify them early, before they become widely metastatic or locally advanced (1, 3). Identification of precancerous mucin-producing cysts thereby offers the potential for the early detection and prevention of an important subset of pancreatic cancers. As a result, many expert groups recommend lifelong surveillance with imaging modalities (magnetic resonance imaging or computed tomography) to identify early-stage cancer or high-grade dysplasia in individuals with cysts (4–7).
Two dilemmas make pancreatic cyst clinical management challenging. First, it is difficult to differentiate IPMNs and MCNs, collectively termed “mucin-producing cysts,” from cysts that have no malignant potential and do not require any follow-up. Second, it can be difficult to differentiate patients with mucin-producing cysts that harbor early invasive cancer or high-grade dysplasia from patients with less advanced mucin-producing cysts. Surgery is recommended for patients with advanced cysts, whereas intermittent surveillance with imaging, rather than surgery, is considered appropriate for patients with less advanced cysts (4). Currently available clinical tools, however, are imperfect at assigning the most appropriate management strategies for patients with cysts. This is highlighted by the fact that 25% of cyst patients who undergo surgical resection have a pancreatic cyst with no malignant potential (8), and up to 78% of mucin-producing cysts referred for surgical resection are ultimately found not to be advanced, that is, they do not harbor high-grade dysplasia or cancer (9). Compounding this situation is the fact that pancreatic surgery is associated with a morbidity of more than 30% and a mortality of up to 5% in patients undergoing a pancreaticoduodenectomy (10, 11). Thus, identifying those individuals who truly require, and will likely benefit, from surgery is critical to avoid unnecessary iatrogenic morbidity.
Sequencing of the DNA isolated from pancreatic cyst fluid has identified somatically mutated genes and chromosomal copy number alterations that are strongly correlated with cyst type (12). The identification of DNA alterations in cyst fluid could therefore potentially be used to improve the evaluation of pancreatic cysts. However, the utility of this approach has yet to be determined in a large study in which cyst fluid analysis is compared with the gold standard—the final pathology of a surgically resected cyst. Here, we report the results of an international multicenter study of patients who had pancreatic cyst fluid analysis and surgery for a pancreatic cyst. There were three aims of this study: First, we evaluated the molecular profiles of a large number of pancreatic cysts and correlated these molecular profiles with the histopathology of the resected pancreatic cysts. Second, we developed a comprehensive test that incorporated clinical, imaging, and molecular features to classify patients into three clinically relevant management groups. Last, we compared the performance characteristics of the test with current methods of clinical evaluation.
RESULTS
Patient characteristics
A total of 875 patients were enrolled in the study between January 2012 and February 2016, including 130 patients who had been included in a previous study (13). All patients underwent surgical resection so that the histopathology of the cysts was known. Sixteen centers with expertise in pancreatic cancer from Asia, Europe, and the United States participated in this study. Thirteen (1.4%) patients were excluded from the final analysis because their cyst fluid DNA was too low to assess, resulting in 862 analyzable patients. The median patient age was 64 years, and 65% were female (Table 1). There were 148 nonmucin-producing cysts, 600 mucin-producing cysts (153 MCNs and 447 IPMNs), and 114 other types of malignant pancreatic cysts. The clinical and imaging features associated with each type of cyst are shown in Fig. 1 and listed in Table 1 and table S1.
Table 1.
General demographics and imaging features for all 862 patients with surgically resected pancreatic cysts. IQR, interquartile range; N/A, not applicable.
| All cysts (n = 862) | Nonmalignant cysts (n = 148) |
Cysts with malignant potential (n = 600) |
Malignant cysts (n = 114) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Benign other* (n = 11) | Pseudocyst (n = 22) | Serous cystic neoplasms (n = 115) | Mucin-producing cysts (n = 600) | Mucinous cystic neoplasm (n = 153) | IPMN† (n = 447) | Pancreatic adenocarcinoma (n = 62) | Pancreatic neuroendocrine tumor (n = 29) | Solid pseudopapillary tumor (n = 23) | ||
| Age at surgery, median (IQR) | 64 (52–72) | 56 (50–76) | 53.5 (45–61) | 57 (44–67) | 66 (54–73) | 47 (37–57) | 69 (63–74) | 72 (65–78) | 61 (50–71) | 28 (23–42) |
| Gender | ||||||||||
| Female, no. (%) | 560 (65) | 8 (73) | 13 (59) | 86 (75) | 386 (64) | 151 (99) | 235 (53) | 31 (50) | 14 (49) | 22 (96) |
| Missing data, no. (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Racial category | ||||||||||
| Caucasian, no. (%) | 681 (79) | 8 (73) | 16 (73) | 81 (70) | 476 (79) | 124 (81) | 352 (79) | 56 (90) | 27 (93) | 17 (74) |
| Asian, no. (%) | 107 (12) | 1 (9) | 0 | 18 (16) | 81 (13) | 10 (7) | 71 (16) | 3 (5) | 0 | 4 (17) |
| African American, no. (%) | 41 (5) | 1 (9) | 5 (23) | 7 (6) | 22 (4) | 8 (5) | 14 (3) | 2 (3) | 2 (7) | 2 (9) |
| Other, no. (%) | 17 (2) | 0 | 0 | 6 (5) | 10 (2) | 5 (3) | 5 (1) | 1 (2) | 0 | 0 |
| Missing data, no. (%) | 16 (2) | 1 (9) | 1 (4) | 3 (3) | 11 (2) | 6 (4) | 5 (1) | 0 | 0 | 0 |
| Symptoms | ||||||||||
| Present, no. (%) | 335 (39) | 2 (18) | 15 (68) | 34 (30) | 227 (38) | 67 (44) | 160 (36) | 42 (68) | 7 (24) | 8 (35) |
| Nonspecific abdominal pain, no. (%) | 251 (75) | 1 (50) | 15 (100) | 30 (88) | 175 (77) | 63 (94) | 112 (70) | 18 (43) | 4 (57) | 8 (35) |
| Pancreatic abdominal pain, no. (%) | 167 (50) | 1 (50) | 10 (67) | 13 (38) | 126 (56) | 35 (52) | 92 (58) | 11 (26) | 2 (29) | 3 (13) |
| Acute pancreatitis‡, no. (%) | 78 (23) | 1 (50) | 9 (60) | 2 (6) | 64 (28) | 12 (18) | 52 (33) | 1 (2) | 0 | 1 (4) |
| Chronic pancreatitis, no. (%) | 29 (8) | 1 (50) | 4 (27) | 1 (3) | 21 (9) | 3 (4) | 18 (11) | 2 (4) | 0 | 0 |
| Jaundice, no. (%) | 31 (9) | 0 | 0 | 2 (6) | 15 (6) | 0 | 15 (9) | 13 (31) | 0 | 1 (4) |
| Weight loss, no. (%) | 140 (42) | 2 (100) | 8 (53) | 8 (24) | 85 (4) | 15 (22) | 70 (45) | 30 (71) | 5 (71) | 2 (9) |
| Missing data, no. (%) | 12 (1) | 0 | 0 | 0 | 12 (2) | 3 (2) | 9 (2) | 0 | 0 | 0 |
| No. of cysts | ||||||||||
| Multiple, no. (%) | 254 (30) | 2 (18) | 5 (23) | 20 (17) | 197 (33) | 12 (8) | 185 (41) | 21 (34) | 8 (28) | 1 (4) |
| Missing data, no. (%) | 27 (3) | 0 | 0 | 2 (2) | 21 (4) | 5 (3) | 16 (4) | 2 (3) | 0 | 2 (9) |
| Cyst size cm, median (IQR)§ | 3.5 (2.5–5.2) | 3.7 (3.1–4.7) | 4.5 (3.3–6.3) | 4.6 (3.2–7.5) | 3.5 (2.6–5.2) | 4.3 (3.1–7.0) | 3.3 (2.3–4.0) | 3.9 (2.2–5.1) | 2.2 (2.0–3.2) | 5.2 (3.5–9.0) |
| Missing data, no. (%) | 34 (4) | 3.7 (3.1–4.7) | 1 (5) | 2 (2) | 27 (5) | 4 (3) | 23 (5) | 3 (5) | 0 | 1 (4) |
| Location‖ | ||||||||||
| Head/uncinate, no. (%) | 352 (41) | 4 (36) | 8 (36) | 37 (32) | 258 (43) | 1 (1) | 257 (57) | 32 (52) | 5 (17) | 8 (35) |
| Neck, no. (%) | 12 (1) | 0 | 0 | 5 (4) | 5 (1) | 1 (1) | 4 (1) | 0 | 1 (4) | 1 (4) |
| Body/tail, no. (%) | 485 (56) | 7 (64) | 14 (64) | 73 (63) | 324 (54) | 149 (97) | 175 (39) | 30 (48) | 23 (79) | 14 (61) |
| Whole gland, no. (%) | 8 (1) | 0 | 0 | 0 | 8 (1) | 0 | 8 (2) | 0 | 0 | 0 |
| Missing data, no. (%) | 5 (1) | 0 | 0 | 0 | 5 (1) | 2 (1) | 3 (1) | 0 | 0 | 0 |
| Mural nodule/solid component¶ | ||||||||||
| Present, no. (%) | 287 (33) | 4 (36) | 7 (32) | 25 (22) | 182 (3) | 42 (28) | 140 (31) | 37 (60) | 13 (45) | 19 (83) |
| Missing data, no. (%) | 45 (5) | 0 | 1 (4) | 6 (5) | 32 (5) | 8 (5) | 24 (6) | 2 (3) | 0 | 4 (17) |
| Communication with main pancreatic duct¶ | ||||||||||
| Present, no. (%) | 267 (31) | 2 (18) | 1 (4) | 21 (18) | 223 (37) | 8 (5) | 215 (48) | 15 (24) | 4 (14) | 1 (4) |
| Absent, no. (%) | 250 (29) | 6 (55) | 9 (41) | 45 (39) | 148 | 69 (45) | 79 (18) | 12 (19) | 16 (55) | 14 (61) |
| Unclear, no. (%) | 279 (32) | 3 (27) | 7 (32) | 42 (37) | 182 | 63 (41) | 119 (27) | 31 (50) | 9 (31) | 5 (22) |
| Missing data, no. (%) | 66 (8) | 0 | 5 (23) | 7 (6) | 47 (8) | 13 (9) | 34 (7) | 4 (7) | 0 | 3 (13) |
| Type of IPMN | ||||||||||
| Main or mixed-duct IPMN | N/A | N/A | N/A | N/A | N/A | N/A | 165 (37) | N/A | N/A | N/A |
| Branch-duct IPMN | N/A | N/A | N/A | N/A | N/A | N/A | 177 (40) | N/A | N/A | N/A |
| Missing data, no. (%) | N/A | N/A | N/A | N/A | N/A | N/A | 105 (23) | N/A | N/A | N/A |
| Dilated main pancreatic duct (mm) | ||||||||||
| 5 to 9 mm no. (%) | 144 (13) | 1 (9) | 3 (13) | 10 (9) | 115 (19) | 2 (1) | 113 (25) | 13 (21) | 2 (7) | 0 |
| ≥10, no. (%) | 100 (12) | 0 | 1 (5) | 3 (3) | 80 (13) | 2 (1) | 78 (17) | 14 (23) | 2 (7) | 0 |
| Missing data, no. (%) | 165 (19) | 1 (9) | 1 (5) | 16 (14) | 126 (21) | 21 (14) | 105 (24) | 9 (14) | 1 (3) | 11 (48) |
Includes seven retention cysts, two lymphoepithelial cysts, one simple mucinous cyst, and one duplication cyst.
Includes one intraductal tubulopapillary neoplasm.
Acute pancreatitis within the last year.
If more than one cyst largest cyst size noted.
Based on evaluation of preoperative imaging data.
Defined as communication between the main pancreatic duct and the cyst on any imaging.
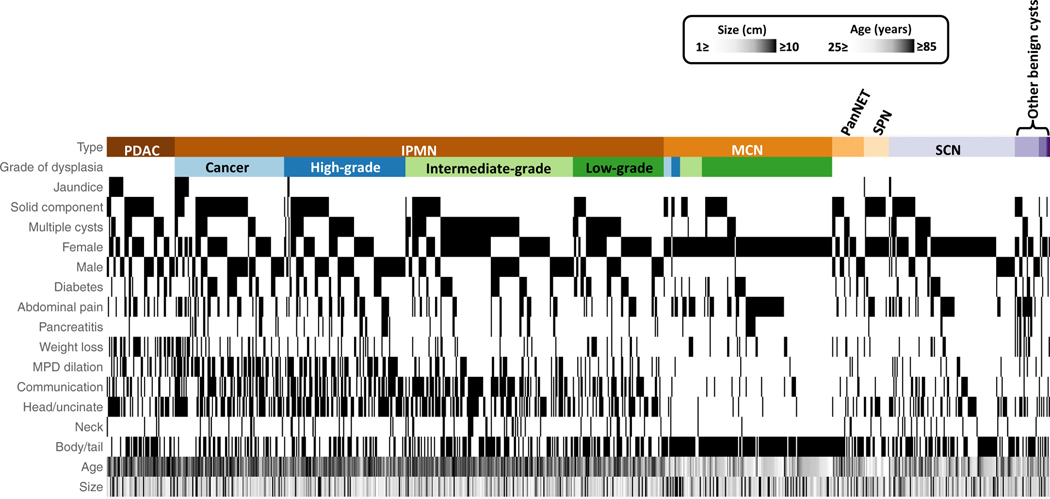
Fig. 1. Clinical features of all of the patients with pancreatic cysts.
This heatmap shows the clinical features of the 862 patients with pancreatic cysts. Areas highlighted in black show the features present in the different types of cysts. For example, one can easily see that almost all the patients with MCNs were female, with cysts located in the body or tail of the pancreas. MPD, main pancreatic duct; PDAC, pancreatic ductal adenocarcinoma; MCN, mucinous cystic neoplasm; PanNET, pancreatic neuroendocrine tumor; SCN, serous cystic neoplasm; SPN, solid pseudopapillary neoplasm.
Molecular features
We evaluated cyst fluid for four types of molecular abnormalities: (i) mutations in 11 genes associated with specific cyst types; (ii) losses of heterozygosity of chromosome regions containing tumor suppressor genes known to be involved in specific cyst types; (iii) aneuploidy, which is known to increase with grade of cyst dysplasia and with an associated invasive carcinoma; and (iv) two protein markers—the conventional mucin-producing cyst protein marker carcinoembryonic antigen (CEA, often found in mucin-producing cysts) and vascular endothelial growth factor A [VEGF-A, often elevated in serous cystic neoplasms (12, 14–17)]. The molecular features associated with each type of cyst are shown in Fig. 2 and listed in Table 2 and tables S1 and S2.
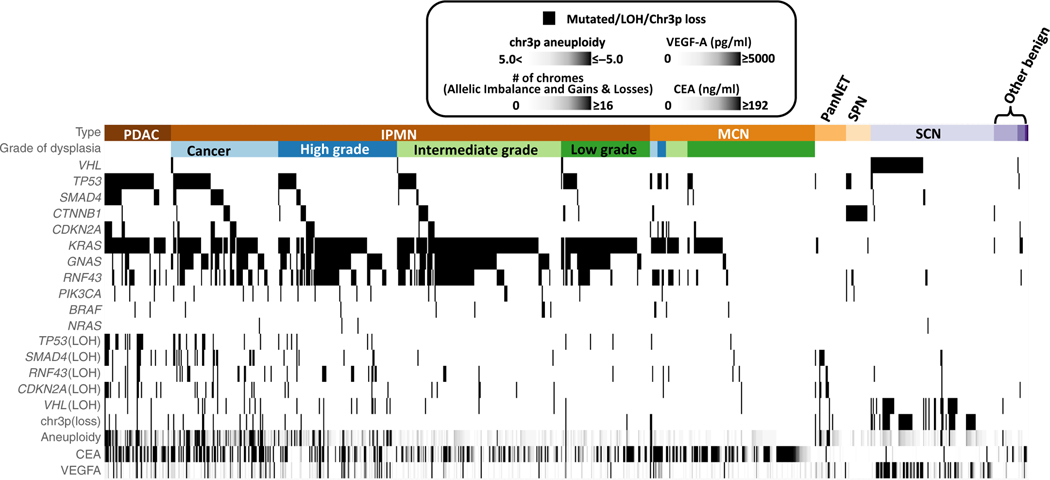
Fig. 2. Molecular features of all of the pancreatic cysts.
This heatmap shows the molecular features of the 862 patients with pancreatic cysts. Areas highlighted in black show the features present in the different types of cysts. For example, one can see that GNAS mutations occur almost exclusively in patients with IPMN and PDAC and that they occur in cysts with all grades of dysplasia. In contrast, SMAD4 mutations occur far more commonly in patients with PDAC or IPMNs with cancer or high-grade dysplasia than in patients with low- or intermediate-grade dysplasia.
Table 2.
Frequency of molecular features in different types of pancreatic cyst.
| All cysts (n = 862) | Nonmucinous cysts (n = 148) |
Mucinous cysts (n = 600) |
Malignant cysts (n = 114) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Retention cyst (n = 7) | Other nonmalignant cysts (n = 26)* | Serous cysts (n = 115) | Mucin-producing cysts (600) | Mucinous cystic neoplasm (n = 153) | IPMN† (n = 447) | Pancreatic adenocarcinoma (n = 62) | Pancreatic neuroendocrine tumor (n = 29) | Solid pseudopapillary tumor (n = 23) | ||
| Mutations | ||||||||||
| KRAS, no. (%) | 451 (52) | 3 (43) | 2 (8) | 0 | 390 (65) | 53 (35) | 337 (76) | 53 (85) | 2 (7) | 1 (4) |
| GNAS, no. (%) | 260 (30) | 0 | 0 | 0 | 252 (42) | 3 (2) | 249 (56) | 8 (13) | 0 | 0 |
| KRAS or GNAS, no. (%) | 502 (58) | 3 (43) | 2 (8) | 0 | 440 (73) | 56 (37) | 384 (86) | 54 (87) | 2 (7) | 1 (4) |
| RNF43, no. (%) | 213 (25) | 1 (14) | 0 | 2 (2) | 192 (32) | 20 (13) | 172 (39) | 15 (24) | 0 | 3 (13) |
| CDKN2A, no. (%) | 47 (5) | 1 (14) | 0 | 0 | 36 (6) | 8 (5) | 28 (6) | 10 (16) | 0 | 0 |
| CTNNB1, no. (%) | 50 (6) | 0 | 1 (5) | 1 (1) | 27 (5) | 2 (1) | 25 (6) | 1 (2) | 0 | 20 (87) |
| SMAD4, no. (%) | 50 (6) | 0 | 0 | 3 (3) | 26 (4) | 2 (1) | 24 (5) | 21 (34) | 0 | 0 |
| TP53, no. (%) | 151 (18) | 1 (14) | 0 | 2 (2) | 95 (16) | 13 (8) | 83 (19) | 46 (74) | 1 (3) | 5 (22) |
| VHL, no. (%) | 55 (6) | 1 (14) | 0 | 49 (43) | 5 (1) | 0 | 5 (1) | 0 | 0 | 0 |
| BRAF - no. (%) | 14 (2) | 0 | 0 | 0 | 12 (2) | 4 (3) | 8 (2) | 2 (3) | 0 | 0 |
| NRAS, no. (%) | 4 (0) | 0 | 0 | 1 (1) | 3 (1) | 0 | 3 (1) | 0 | 0 | 0 |
| PIK3CA, no. (%) | 24 (3) | 0 | 0 | 0 | 19 (3) | 1 (1) | 18 (4) | 3 (5) | 0 | 2 (9) |
| Missing data, no. (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Loss of heterozygosity | ||||||||||
| LOH chr3 (VHL), no. (%) | 61 (7) | 0 | 1 (5) | 30 (26) | 24 (4) | 1 (1) | 23 (5) | 4 (6) | 2 (7) | 0 |
| LOH chr9 (CDKN2A), no. (%) | 42 (5) | 1 (14) | 0 | 1 (1) | 21 (4) | 2 (1) | 19 (4) | 13 (21) | 6 (21) | 0 |
| LOH chr17 (RNF43), no. (%) | 49 (6) | 0 | 0 | 2 (2) | 38 (6) | 5 (3) | 33 (7) | 6 (10) | 3 (10) | 0 |
| LOH chr17 (TP53), no. (%) | 41 (5) | 0 | 0 | 0 | 24 (4) | 2 (1) | 22 (5) | 17 (27) | 0 | 0 |
| LOH chr18 (SMAD4), no. (%) | 48 (6) | 0 | 0 | 2 (2) | 27 (5) | 1 (1) | 26 (6) | 13 (21) | 6 (21) | 0 |
| VHL or LOH chr3 (VHL), no. (%) | 102 (12) | 1 (14) | 1 (5) | 65 (59) | 29 (5) | 1 (1) | 28 (6) | 4 (6) | 2 (7) | 0 |
| Missing, no. (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aneuploidy | ||||||||||
| COUNT_GL_Z score 3 (Range) | 104 (12) | 1.4 (0–3) | 1.8 (0–17) | 3.5 (−1–34) | 1.0 (0–16) | 5.2 (0–37) | 8.9 (0–33) | 0 | 1.5 (0–6) | |
| Missing data, no. (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Protein data | ||||||||||
| CEA | ||||||||||
| >192 ng/ml, no. (%) | 237 (27) | 1 (14) | 3 (12) | 3 (2) | 201 (30) | 70 (46) | 131 (29) | 28 (45) | 1 (3) | 0 |
| <192 ng/ml, no. (%) | 548 (64) | 3 (43) | 12 (46) | 94 (82) | 220 (37) | 58 (38) | 162 (36) | 21 (34) | 26 (90) | 17 (74) |
| Missing data, no. (%) | 77 (9) | 3 (43) | 11 (42) | 18 (16) | 178 (30) | 25 (16) | 154 (34) | 13 (21) | 2 (7) | 6 (26) |
| VEGF-A | ||||||||||
| >5000 pg/ml, no. (%) | 60 (7) | 0 | 0 | 33 (29) | 20 (3) | 3 (2) | 17 (4) | 5 (8) | 2 (7) | 0 |
| ≤5000 pg./ml, no. (%) | 725 (84) | 7 (100) | 26 (100) | 74 (64) | 515 (86) | 145 (95) | 370 (83) | 57 (92) | 27 (93) | 19 (83) |
| Missing data, no. (%) | 77 (9) | 0 | 0 | 8 (7) | 65 (11) | 5 (3) | 60 (13) | 0 | 0 | 4 (17) |
Includes simple mucinous cysts (n = 1), lymphoepithelial cyst, (n = 2), duplication cyst (n = 1), and pseudocyst (n = 22).
Includes one intraductal tubulopapillary neoplasm.
Molecular features associated with benign cysts
Serous cystic neoplasms are the commonest type of benign cyst and often have mutations in VHL or loss of heterozygosity in chromosome 3, where the VHL gene is located (12). We found that 59% (n = 65) of the serous cystic neoplasms in our study had a mutation of VHL or loss of heterozygosity of chromosome 3 (Fig. 2 and Table 2). Additional mutations were present in 5% (n = 8) of the serous cystic neoplasms and included mutations in RNF43, TP53, CTNNB1, and SMAD4, as well as loss of heterozygosity in chromosome 9, 17, or 18. One unexpected finding was the number of mutations found in the retention cysts. These are considered benign cysts that require no surveillance. However, five (71%) of seven retention cysts contained a mutation in CDKN2A, KRAS, RNF43, TP53, or VHL or loss of heterozygosity of chromosome 9, suggesting that these were not simple benign cysts and that these patients may require surveillance. Mutations were found in only two other benign cysts. One was a simple mucinous cyst, and the other was a pseudocyst, which had mutations in KRAS and CTNNB1, as well as loss of heterozygosity of chromosome 3. No follow-up was available in this patient; however, the presence of these mutations was unusual and suggests that some other process may be present in the nonresected pancreas.
VEGF-A has previously been reported as a promising marker for identifying serous cysts, the commonest type of benign pancreatic cyst (15, 16). In our study, elevated VEGF-A concentrations were found in serous cystic neoplasms, as well as some mucin-producing cysts and malignant cysts (Table 2 and fig. S2). A concentration of greater than 5000 pg/ml has previously been used to identify serous cystic neoplasm (16); in our study, this threshold was highly specific but had a sensitivity of only 32% (fig. S2). One possible reason for the differences between this and previous studies is that different platforms were used to evaluate the VEGF-A concentration.
Molecular features associated with mucin-producing pancreatic cysts
IPMNs and MCNs are together classified as mucin-producing cysts. Mutations were present in 390 (65%) of the mucin-producing cysts. The most frequent mutations were in KRAS or GNAS, with mutation in one or other of these genes present in 440 (73%) of the mucin-producing cysts. Mutations were more prevalent in IP MNs (76%), the commonest type of mucin-producing cyst, compared with MCNs (43%), and 384 (86%) of the cysts had a mutation in either KRAS or GNAS. Previous studies reported GNAS to be exclusively found in IPMNs. In our study, more than half (n = 249; 56%) of the IPMNs were found to have a mutation in GNAS, and this gene was also mutated in 2% (n = 3) of the MCNs. GNAS mutations were highly specific for mucin- producing cysts and were not identified in any other cyst type. In contrast, KRAS mutations were found in a number of other types of pancreatic cysts including 3% of benign cysts and 85% of pancreatic adenocarcinomas. A cyst fluid CEA value of greater than 192 ng/ml is currently believed to distinguish differentiating mucin-producing cysts from other types of cysts (37). In our study, cyst fluid CEA had 38% sensitivity and 96% specificity for identifying mucin-producing cysts from other cyst types (Table 2 and fig. S3) using this threshold.
Molecular features associated with high-grade dysplasia
The identification of patients with mucin-producing cysts harboring high-grade dysplasia or early invasive cancer is one of the key aims of clinical management, because these patients typically benefit from surgical resection. We evaluated 600 mucin-producing cysts in this study. The presence of mutations in CDKN2A, SMAD4, and TP53 in mucin-producing cysts associated with an odds ratio of between 2.8 and 7.2 of high-grade dysplasia or cancer (table S9). The number of chromosomal arms lost or gained had a strong correlation with the presence of high-grade dysplasia or cancer in mucin-producing cysts or in pancreatic adenocarcinomas (Table 2). For example, 36% of cysts with high-grade dysplasia or invasive cancers exhibited losses of at least eight chromosome arms, whereas this degree of aneuploidy was found in only 4% of cysts with low- or intermediate- grade dysplasia (fig. S4).
Molecular features associated with pancreatic adenocarcinomas
There were 62 pancreatic adenocarcinomas presenting as pancreatic cysts. Mutations occurred in nearly all (n = 58; 94%) of these and included mutations in KRAS, GNAS, RNF43, CDKN2A, CTNNB1, SMAD4, TP53, BRAF, or PIK3CA. GNAS mutations were previously thought to occur only in IPMNs. However, we identified mutations in GNAS in cyst fluid from 13% (n = 8) of the pancreatic adenocarcinomas that had no pathological evidence of an associated IPMN in the surgical resection specimen. These data suggest that the pancreatic adenocarcinomas arose from IPMNs. It is possible that the adenocarcinoma replaced the IPMN, explaining its absence on surgical histopathology. The majority of the pancreatic adenocarcinomas without GNAS mutations presumably arose through cystic degeneration of the cancers (38).
Molecular features associated with other malignant pancreatic cysts
Solid pseudopapillary neoplasms are rare malignant pancreatic cysts that occur in young women (39). The majority (n = 20; 87%) of the solid pseudopapillary neoplasms had a mutation in CTNNB1. In contrast to previous studies where other mutations were not identified, 35% (n = 8) of the solid pseudopapillary neoplasms had additional mutations in KRAS, RNF43, CTNNB1, TP53, or PIK3CA (Fig. 2).
Pancreatic neuroendocrine tumors can occasionally present with cystic degeneration. A mutation or loss of heterozygosity was present in 41% (n = 12) of the pancreatic neuroendocrine tumors and included mutations in KRAS and TP53, as loss of heterozygosity of chromosome 3, 9, 17, or 18.
Developing a combined clinical, imaging, and molecular test
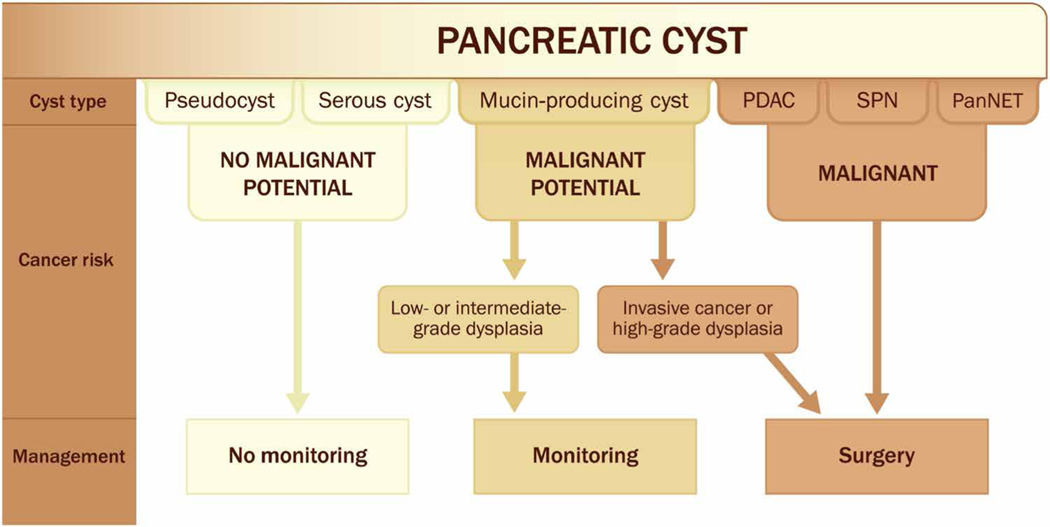
The clinical management of patients with pancreatic cysts is based on the potential of a pancreatic cyst to develop or harbor invasive cancer and requires classifying a cyst into one of three groups (Fig. 3). First are pancreatic cysts with essentially no malignant potential, such as pseudocysts and serous cystic neoplasms (18). Patients with these cysts can be reassured that little, if any, periodic monitoring is required (6, 7). The second group includes mucin-producing cysts without invasive cancer or high-grade dysplasia (19). These cysts have a small risk of progressing to cancer over the patient’s lifetime, with an incidence of 0.72% per year (20), and monitoring is recommended for these patients at regular intervals (6, 7). The third group includes cysts for which surgery is recommended, because invasive cancer is present or there is a high likelihood of progression to cancer. These include mucin-producing cysts with high-grade dysplasia or an associated invasive cancer and other malignant pancreatic neoplasms with a degenerative cystic component including pancreatic adenocarcinoma, neuroendocrine tumors, and solid pseudopapillary neoplasms (6, 7).
Fig. 3. Clinical management of patients with pancreatic cysts.
This figure shows how the type of pancreatic cyst determines the risk of the cyst developing cancer, which in turn dictates clinical management. Serous cystic neoplasms and pseudocysts have essentially no malignant potential and therefore require no monitoring. In contrast, cystic degeneration of a PDAC, PanNET, or solid pseudopapillary neoplasm are, or have a high risk for becoming, malignant, and therefore should undergo surgical resection. IPMNs and MCNs are mucin-producing cysts. A small number of these harbor high-grade dysplasia or cancer and should be surgically resected, while the remaining mucin-producing cysts simply need surveillance.
We used a stepwise, supervised machine learning-based approach to stratify patients into one of these three clinically relevant groups: those who required surgery, those who did not require surgery but who should periodically undergo surveillance, and those who could be safely discharged without the need for continuing surveillance (fig. S1). The combinatorial markers for this stratification were deliberately developed with either a high sensitivity or a high specificity. For example, we required the marker to have a very high specificity when identifying patients who could be discharged from follow-up, because we considered falsely classifying lesions with malignant potential to be unacceptable. Similarly, a high sensitivity was required when identifying patients who should be referred for surgery to minimize the risk of advising a patient with high-grade dysplasia or cancer not to have surgery. In total, there were 862 patients whom we divided into separate training and validation cohorts, such that the distribution of cyst type and clinical management category were the same in the training half of the data (436 patients) and validation half (426 patients). We assessed the performance of markers selected from the training cohort by classifying each patient in the validation cohort, provided that the patient being classified had the requisite data available to be assessed by a particular marker (table S3). As an example, if a composite marker was defined by the presence of aneuploidy, only patients with aneuploidy data could be assessed by that marker. Throughout, performance estimates are derived from the patient subset for which the marker could be tested on, and comparisons with physician’s diagnosis were always calculated from the exact same patient subset.
In the first step of this three-step process, we identified a combinatorial marker for serous cystic neoplasms (table S4). This marker achieved 46% sensitivity and 100% specificity for serous cystic neoplasms in the validation cohort, thus achieving the high specificity noted above. Twenty-six patients from the validation cohort tested positive for this marker, defined by the presence of VHL mutation but the absence of GNAS mutation. These 26 patients were thus removed before validating subsequent markers. In the second step, we derived combinatorial markers to identify cyst patients who should be referred for surgical resection. We exploited the fact that cysts requiring surgical resection have a large number of genetic alterations relative to nonmalignant cysts, but that the specific combination of alterations can vary greatly (Table 2). The marker panel used to identify cysts that required surgery included a solid component observed upon imaging, aneuploidy, and the presence of mutations in various genes (table S4). In the validation cohort, this combinatorial marker achieved 91% sensitivity and 54% specificity for patients who should have surgery. Eighty-one patients were missing the data required to test with this marker (owing mainly to missing aneuploidy and protein expression data; see table S3) and were removed at this stage. Patients who were negative for both the first and second combinatorial markers were tested with a third combinatorial marker. The purpose of the third marker was to distinguish patients who should undergo monitoring from those who could be safely discharged. This marker, defined by VEGF-A protein expression less than 1000 pg/ml, was optimized for high sensitivity to ensure that all patients who required monitoring were identified (table S4). In the validation cohort, this third marker achieved 99% sensitivity and 30% specificity in aggregate, meaning that only 1% of patients with cysts that should undergo continued monitoring would receive a recommendation that surveillance was not needed. We termed the successive application of these three composite markers the CompCyst test.
Comparing the performance of CompCyst with the current standard of care
Physicians currently use a variety of clinical features, imaging, and cyst fluid analysis to classify patients with a cyst into one of the three groups described above (Fig. 3). The current standard of care (see Materials and Methods) was compared with CompCyst-based recommendations for cyst management in the validation cohort (Fig. 4 and table S5). Because the histopathology of all cysts was known from surgical specimens, we could determine in retrospect what the management should have been. As noted above, the patients in this validation cohort were distinct from those used for training the CompCyst algorithm.
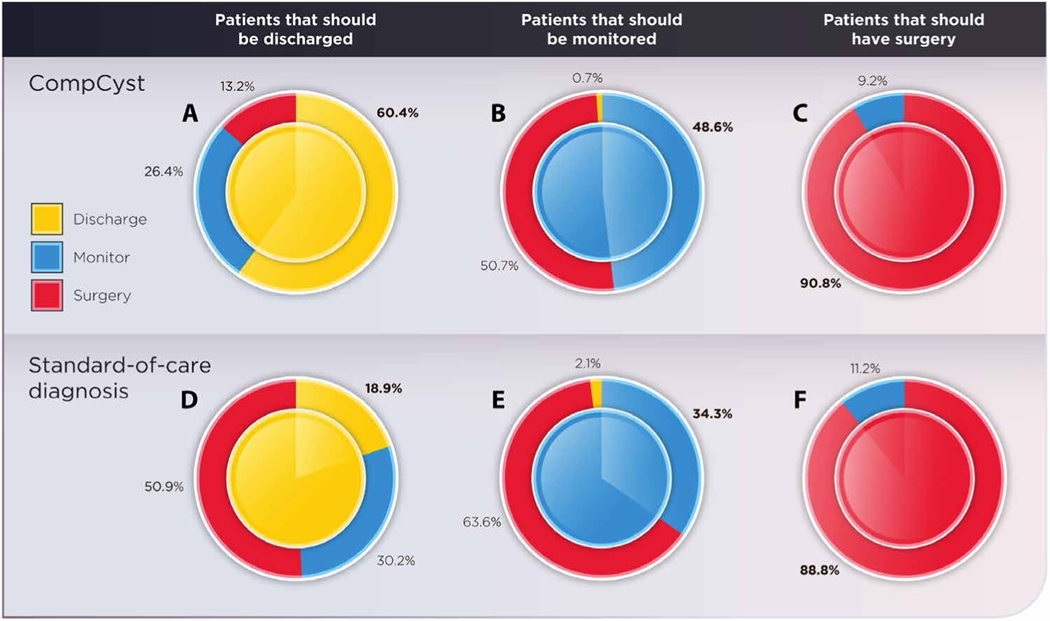
Fig. 4. Management of pancreatic cysts.
These donut charts show the management recommendations based on CompCyst and standard of care compared with the gold standard, pathology. The center of the circle indicates the management recommendation based on the final surgical pathology classification. A fully solid circle—one where the inner and outer circles are fully the same color—would indicate 100% accuracy. The performance of CompCyst compared with surgical pathology for cysts in whom the correct management was discharge, monitoring, or surgery is shown in (A), (B), or (C), respectively. The performance of standard of care compared with surgical pathology is shown in (D), (E), or (F), respectively.
On the basis of pathology of the resected specimens, 53 patients in the validation cohort had a benign, nonmucin-producing cyst and did not require resection or surveillance (in other words, they could have been discharged). Current clinical management correctly identified only 10 (19%) of these 53 patients as suitable for discharge. The CompCyst test performed significantly better, correctly identifying 32 (60%) of 53 patients (P = 1.3 × 10−4; McNemar’s test for comparing classifiers). On the basis of cyst histopathology, 140 patients had mucin-producing cysts without invasive cancer or high-grade dysplasia. Monitoring, rather than surgery or discharge, was appropriate for these patients. Current clinical management correctly recommended surveillance in 48 (34%) of these patients, whereas the CompCyst test correctly recommended surveillance in 68 (49%) patients (P = 0.02; McNemar’s test). In sum, more than 193 patients in the validation cohort who underwent surgical resection did not require surgery when it was performed. Relative to the current standard of care, CompCyst would have decreased the number of unnecessary operations (P = 3.5 × 10−5; McNemar’s test), with the difference most marked for benign, nonmucin-producing cysts, where it could have decreased the number of unnecessary operations by 74% (table S5). Overall, the use of CompCyst would have avoided surgery in 60% of the 193 patients who did not require surgery (Fig. 4).
On the basis of histopathology, surgery was indicated in the remaining 152 patients in the validation cohort. The current standard of care correctly identified 135 (89%) of these patients, similar to that identified by CompCyst (138 patients, 91%). Neither the current standard of care nor the CompCyst test discharged any patient for whom surgery was indicated. Overall, CompCyst had a significantly higher accuracy (69%) for classifying patients into one of the three groups (surgery, surveillance, or discharge) compared with the current standard of care (56%) (P = 7.3 × 10−5).
Prediction of cyst type
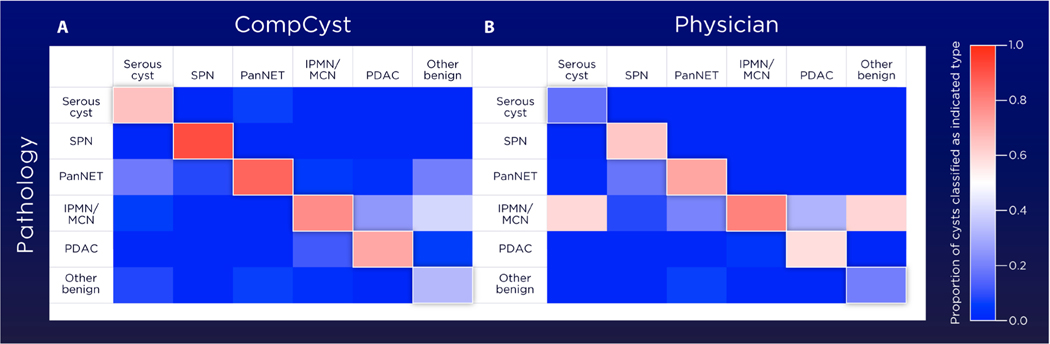
Although the main purpose of this study was to inform the management of patients with cysts, we also generated a composite marker panel for determining the most likely cyst type harbored by each patient. These categories included serous cystic neoplasm, “other nonmalignant cysts,” mucin-producing cysts, pancreatic adenocarcinomas, solid pseudopapillary neoplasms, and pancreatic neuroendocrine tumors. The approach to designing these markers was conceptually similar to that used for designing the management panels. Half of the patients were used to train the multivariate organization of combinatorial alterations (MOCA) algorithm to identify the most distinctive composite markers for each cyst type. The other half of the patients was then used to test the composite markers. The output of this test was the fraction of markers testing positive for each of the six cyst types in a given patient (Fig. 5 and tables S3 and S6). For example, the first patient (no. 15093) listed in table S3 tested positive for 98% of serous cystic neoplasm markers, whereas this patient tested positive for a far smaller fraction of other cyst type markers. This patient was believed to have a mucin-producing cyst based on conventional clinical and imaging criteria, explaining why she underwent surgery. In general, the CompCyst prediction of cyst type was more accurate than the preoperative diagnosis based on conventional clinical and imaging criteria (table S6). For example, the sensitivity of CompCyst for identifying serous cystic neoplasm was 65%, whereas only 18% of serous cystic neoplasms were correctly identified by clinical and imaging criteria (table S6). At the other end of the spectrum, CompCyst correctly identified 71% of pancreatic adenocarcinomas with cystic degeneration, whereas clinical and imaging criteria correctly identified 58% of pancreatic adenocarcinomas (table S6). Note that though the sensitivity of CompCyst for identifying pancreatic cancers was higher than conventional clinical and imaging criteria, the specificity of CompCyst was lower (90% versus 96%). The reason for this is that we designed the CompCyst algorithms to minimize the chance of missing a pancreatic cancer with cystic degeneration, that is, to achieve high sensitivity rather than specificity. In general, the CompCyst diagnosis of cyst types was significantly more accurate than that achieved by conventional clinical and imaging criteria (P = 0.01; McNemar’s test).
Fig. 5. Classification of the type of pancreatic cyst.
These two heatmaps compare the CompCyst classification (A) and physician’s preoperative diagnosis based on clinical and imaging features (B) with surgical pathology for classifying the type of pancreatic cyst. The fraction of cysts classified to be of the indicated type is shown in the color bar.
DISCUSSION
We compared the performance of a cyst classifier test based on clinical features, imaging characteristics, and genetic and biochemical markers with the standard of care for cyst management. We found that CompCyst was more accurate than conventional clinical tools for identifying patients with cysts that required surgery, cysts that should be monitored, and cysts that were benign, nonmucin producing, and did not require monitoring. Serous cystic neoplasms exemplify the challenge that clinicians face in making the correct diagnosis of cysts. “Typical” serous cystic neoplasms are single cysts that have a small main pancreatic duct, which does not communicate with the cyst. In our study, 9% of the serous cystic neoplasms were associated with an enlarged main pancreatic duct, 18% had communication between the cyst and the pancreatic duct, and 17% had more than one cyst. Thus, many serous cystic neoplasms were “atypical,” meaning that their clinical and imaging characteristics are not homogeneous, as has been observed in other large series (18, 21). Given the clinical and imaging features of the atypical cysts, it is not surprising that many cysts in our study were clinically mistaken for mucin-producing cysts, and that many patients underwent unnecessary surgery. The CompCyst test used the presence of a VHL mutation and other markers to identify serous cystic neoplasm more accurately, correctly identifying 65% of them with 99% specificity. By comparison, in our study, the preoperative diagnosis of a serous cystic neoplasm based on clinical and imaging criteria was correct only 18% of the time. It is likely that the performance of both the standard of care and of CompCyst would improve if applied to serous cystic neoplasms that presented with typical clinical features. Our study highlights the potential role of CompCyst as a complement to existing clinical and imaging criteria when evaluating atypical cysts. It could provide a greater degree of confidence for physicians, based on 99% specificity, when they advise patients whom they do not require follow-up and can be discharged from surveillance. Although CompCyst is not perfect, it represents an important advance over currently available tools for identifying cyst types and guiding their management.
A similar conundrum is posed by mucin-producing cysts. More than 60% of patients with mucin-producing cysts who underwent resection did not harbor high-grade dysplasia or an associated invasive cancer and, in hindsight, did not require surgery at the time they underwent surgery. These statistics are consistent with those found in other large surgical series (10, 22, 23). One of the unique features of our study is that we developed a set of clinical features, imaging characteristics, and molecular markers to identify not only high-grade mucin-producing cysts but all cysts that required surgery. These composite biomarkers were specifically developed to have a high sensitivity so as to minimize the risk of missing a patient with high-grade dysplasia or invasive cancer while maintaining reasonable specificity. The result was a considerably more accurate approach for identifying patients who actually required surgery while minimizing unnecessary surgeries. Our results suggest that if CompCyst were applied in general to the management of patients with cysts, 60% of unnecessary surgeries for these cysts types could be avoided. Given the high cost, morbidity, and even mortality associated with surgical procedures for pancreatic cyst removal (10, 11), this result has important implications for patients.
The molecular analyses performed in this study add to our understanding of the pathogenesis of certain cyst types. For example, cysts with nonserous flat epithelial lining are classified pathologically as retention cysts. These are considered to have no malignant potential and do not require monitoring or intervention. However, we found that more than 70% of the cysts classified pathologically as retention cysts had a mutation, including mutations in KRAS, RNF43, CTNNB1, and TP53. These mutations are similar to those that we observed in mucin-producing cysts. Although we cannot rule out that a lesion elsewhere in the pancreas drained its fluid into the cyst, the finding of clonal mutations raises the possibility that these lesions are in fact neoplastic and that patients with them should continue to be monitored.
Several studies have shown that adequate cyst fluid for cyst fluid CEA analysis is obtained in less than 50% of patients who undergo endoscopic ultrasonography (EUS)–guided fine-needle aspiration (24). One of the advantages provided by the sequencing technology (Safe-SeqS) used in this study is that it requires very little DNA. This allowed us to successfully analyze samples with as little as 250 μl of cyst fluid for this study. This volume of fluid is less than what is typically required for standard CEA analysis in most clinical laboratories (0.5 to 1 ml). Assuming that 250 μl was the entire volume contained in a pancreatic cyst and assuming that a pancreatic cyst were perfectly spherical (where V = 4/3πr3), CompCyst could be performed on DNA obtained from cysts of >0.8 cm in size.
Our study has several limitations, which should be acknowledged. The first is that for most cases, pancreatic cyst fluid was obtained at the time of surgical resection rather than during preoperative endoscopic ultrasound. We have previously shown that the genetic alterations in cyst fluid collected at the time of surgery is similar to that of cyst fluid collected endoscopically (13); however, this conclusion must be tested in a prospective study that rigorously compares both methods of collection. A second limitation is that the cysts that we studied are not representative of those seen in routine clinical practice. Rather, they are biased toward those that are atypical and thought to be most concerning for cancer, thereby warranting surgery. We expect that more typical cysts seen in routine clinical practice would be even more accurately diagnosed with CompCyst than those studied here, in part because CompCyst relies on clinical and imaging parameters in addition to biomarkers in cyst fluids. However, this expectation must be rigorously tested.
In conclusion, the use of a comprehensive test that evaluates clinical, imaging, and molecular features is imperfect but appears to offer substantial improvements over standard-of-care management of patients with pancreatic cysts. CompCyst does not replace conventional clinical tools. Instead, it contributes additional information, allowing clinicians to make more informed decisions. How and when tests like CompCyst can be implemented in routine clinical settings remains to be determined, but our results represent the next stage of research required for such implementation. An important next test of the markers presented here could be their validation in a follow-up, prospective study.
MATERIALS AND METHODS
Study design
The study was approved by the Institutional Review Boards for Human Research at each institution and complied with Health Insurance Portability and Accountability Act. In this retrospective study, patients were enrolled at 1 of 16 sites between January 2012 and February 2016. A sample size was not prespecified. Instead, we included the largest possible number of patients with resected pancreatic cysts to ensure that all unusual or rare cyst types were included. The inclusion criteria for the study were as follows: (i) age 18 years or older with the ability to given informed consent; (ii) availability of cyst fluid obtained either at the time of EUS or surgical resection; and (iii) surgical resection of a pancreatic cyst with final pathology available for review. Pathological diagnosis was used as the gold standard against which both the clinical standard of care and CompCyst recommendations were compared. General demographics, the presence of pancreas-related symptoms, computed tomography, magnetic resonance imaging, endoscopic ultrasound features, and cytology were documented. The preoperative cyst diagnosis (table S1) was based on evaluation of the clinical history, imaging, cyst fluid CEA, and cytology by the patient’s physician. In cases where the diagnosis was ambiguous (for example, the differential diagnosis included pancreatic adenocarcinoma or a serous cystic neoplasm), a diagnosis of cyst type “unclear” was recorded, and the cysts were assigned to “surgery” with respect to the classification for management based on standard of care (25).
Pathological evaluation
The pathology of surgically resected lesions was reviewed by one of three pancreatic pathologists (R.H.H., D.S.K., or E.T.). Some previous studies recommended combining IPMNs with low-grade and intermediate-grade dysplasia under the designation “low-grade IPMN” (26). In our study, we classified mucin-producing cysts as having low-grade, intermediate-grade, or high-grade dysplasia based on the 2010 World Health Organization classification of tumors of the digestive system (27), because maintaining this separation provided additional information.
Cyst fluid collection and DNA purification
Pancreatic cyst fluid was collected at the time of endoscopic ultrasound (n = 125) or from the resected specimen in the surgical pathology laboratory (n = 737) (13). DNA was purified from cyst fluid (0.25 to 1.0 ml) by adding 3 ml of RLTM buffer (Qiagen) and then binding to an AllPrep column (Qiagen) according to the manufacturer’s instructions. DNA was quantified using SYBR Green I, as specified by the manufacturer (Thermo Fisher Scientific). In 13 (1.4%) patients, very low amounts of DNA were recovered from the cysts, and these patients were excluded from analysis. “Very low” was defined as median uniquely identified reads (UIDs), that is, reads containing the same unique molecular tag, per amplicon of less than 600 in the assay for mutations or less than 50,000 total UIDs in the assay for loss of heterozygosity.
Assessment of mutations
Massively parallel sequencing allows rapid DNA mutation analysis of multiple samples. However, sample preparation and sequencing steps introduce artifactual mutations into analyses at a low but substantial frequency. To better discriminate genuine mutations from artifactual sequencing variants introduced during these processes, we used Safe-SeqS, a sequencing error reduction technology (28, 29). Safe-SeqS amplification primer pairs were designed to amplify 109- to 141–base pair (bp) segments each containing a region of interest. These regions of interest were derived from the following genes known to be drivers of neoplastic pancreatic cysts: KRAS, GNAS, RNF43, CDKN2A, CTNNB1, SMAD4, TP53, VHL, BRAF, NRAS, and PIK3CA, with primer sequences described in table S7. These primers were used to amplify DNA in 25-μl multiplex polymerase chain reactions (PCRs) as described previously (13). For each sample, three multiplex PCRs were performed, with each multiplex PCR containing 22 to 50 primer pairs. Reactions were purified with AMPure XP beads (Beckman Coulter) and eluted in 100 μl of Buffer EB (Qiagen). The purified PCR products (0.25%) were then amplified in a second round of PCR, as described in (13). The PCR products were purified with AMPure and used for sequencing on a MiSeq or HiSeq instrument. All experiments were performed in a blinded fashion, without previous knowledge of cyst diagnosis.
A mutant allele fraction (MAF)–based approach was used for the classification analysis. Mutations were defined as either insertions or deletions, pathologic single-base substitutions in tumor suppressor genes, or mutations in known hotspots of oncogenes. Pathogenic single-base substitutions in tumor suppressors were determined by comparing them to validated mutations in the COSMIC database. For each mutation identified, the MAF was determined by dividing the number of UIDs with mutations by the total number of UIDs (28). The MAF in the sample of interest was first normalized on the basis of how the distribution of MAFs for the same mutation in the control group, which consisted of DNA from 188 healthy donors sequenced concurrently with the rest of the samples, compared to the distribution of MAFs of every other mutation in the control group. Specifically, the empirical distribution of the MAF for each mutation found in the control group was obtained, and its median, mi, was estimated, resulting in a vector of medians. The 0.25 quantile of the values in that vector was calculated, termed q0.25, and the ratio q0.25/mi was used as a multiplier to normalize the specific mutation MAF, that is, normalized MAF = MAF × q0.25/mi. After this mutation-specific normalization, a P value was obtained by comparing the normalized MAF of each mutation in each well with a reference distribution of normalized MAFs built from normal controls where all mutations were included. The Stouffer Z score was then calculated from the P values of two independent wells, each weighted by their number of UIDs.
Analysis for loss of heterozygosity
This was performed in a fashion similar to that described above for mutations, but different primer sets were used (13). The primer sets amplified genomic regions of ~120 bp that contained common single-nucleotide polymorphisms (SNPs) that were within or closely surrounding (within 1 Mb) the tumor suppressor genes CDKN2A, RNF43, SMAD4, TP53, or VHL. Analogously to the mutation protocol, each DNA sample was used for two multiplex PCRs, each containing 44 primer pairs (table S8). The analysis was also carried out similarly, with the goal of identifying independent template molecules, defined by their UIDs that were informative for the analyzed SNPs. The 88 primer pairs used in this analysis were chosen from 111 primer pairs from the same genes on the basis of their ability to produce PCR products that could be uniquely mapped to the human genome and could be amplified robustly within multiplex reactions using the PCR-cycling conditions described above.
An analytical approach was developed to assess loss of heterozygosity by assigning a score to each gene in a test sample as follows. First, only SNPs with allele ratios of at least 10% and at most 90% with at least 300 UIDs were considered for detecting the loss of heterozygosity in the sample. A gene was required to have at least two qualified SNPs to be included in the analysis. Second, for each qualified SNP in the test sample, we estimated a P value using the distributions of the ratio observed for the same SNP among 188 normal training samples comprising DNA from peripheral white blood cells. Only normal training samples with the same qualified SNP were used to fit a Gaussian kernel to estimate the P value. P values were bounded from below, at 10−6, to avoid having a single qualified SNP dominate the analysis. Last, all SNP P values were aggregated using the Stouffer Z score method (30) to assign a single score to each gene in the test sample. The kernel fitting and Stouffer Z scores were weighted on the basis of UID counts of the normal training samples and the test samples, respectively.
Assessment of aneuploidy
Aneuploidy was assessed with FastSeqS, a technology that uses a single PCR to amplify about 38,000 loci of long interspersed nucleotide elements scattered throughout the genome (31). After massively parallel sequencing, single chromosomal arm gains or losses, as well as allelic imbalances on 39 chromosome arms were calculated and analyzed. For this analysis, we used WALDO (Within- Sample AneupLoidy DetectiOn) software (32). WALDO incorporates a support vector machine (SVM) to discriminate between aneuploid and euploid samples. The SVM was trained using 3150 synthetic aneuploid samples with low neoplastic content and 677 euploid peripheral white blood cell samples (32). Chromosome arm–specific aneuploid scores were defined using |Z score| ≥ 3.0 for gains (Z score ≥ 3.0) or losses (Z score ≤ −3.0) of each arm. For example, an aneuploid value of “6” indicates that a patient had six different chromosome arms meeting this Z score threshold.
Protein analysis
The Bio-Plex 200 platform (Bio-Rad) was used to determine the concentration of CEA and VEGF-A in cyst fluid (16). The Luminex bead-based immunoassay was performed following the manufacturer’s protocol with the samples diluted 1:20 in serum matrix buffer. Target concentrations were determined using five-parameter log curve fits (Bio-Plex Manager 6.0) with vendor provided standards and quality controls.
Deriving markers for the “CompCyst” test
Composite markers are those that combine multiple individual parameters into a single marker. For composite marker selection, we used the MOCA algorithm (33–35). MOCA selects random collections of parameters, derives every combination of the selected parameters using the Boolean Set union, intersection, and difference operations, and tests the ability of each composite marker to correctly classify the category under consideration. This process of randomly selecting parameters and comparing every parameter combination with the category of interest is repeated 10,000 times. During the optimization process, the top 1% of composite markers is defined by a user-provided diagnostic criterion (for example, sensitivity, specificity, balanced accuracy, and predictive value); different diagnostic criteria are useful in different clinical scenarios. After every 1000 iterations, the top 1% of composite markers is decomposed, and the individual parameters are appended back to the initial parameter pool (in other words, if a composite marker comprising KRAS mutation, TP53 loss, and CEA overexpression was a top-performing composite marker, then those three individual genetic/molecular features are duplicated in the parameter pool, thereby increasing the probability that these informative features will be sampled with high frequency during successive random samplings). Thus, as the algorithm progresses, the probability of selecting the most informative parameters increases, ultimately resulting in composite markers that are optimized for correctly classifying each target category. Only markers with a false discovery rate–corrected (Benjamini and Hochberg) P <0.05 (two-tailed Fisher’s exact test) were considered for validation and subsequent analysis.
We divided our dataset into independent training (436 patients) and validation cohorts (426 patients); the “true state” was known for all patients, and there was no overlap between the training and validation cohorts. The data were split before any marker selection or assessment, and the training and validation cohorts remained “locked down” for the duration of the study (patients were never removed, and cohorts were never reshuffled or mixed in any way). The classifiers/hypotheses were “prespecified” in the sense that markers were selected from the training cohort and assessed in the validation cohort without further optimization. Because this was a retrospective study that used machine learning to derive de novo markers, the exact composition of the composite markers was not known at the beginning of the study (data collection had to precede marker selection). The data were divided such that the relative distribution of cyst type and grade was the same in the marker- selection and validation datasets. This data split preceded all marker selection and assessment, and all model development resulted from assessment of training performance while the clinical and pathological status of the validation cohort remained blinded. It is essential for the derivation of composite markers with high balanced accuracy to use highly specific clinical parameters. The presence of a solid component within the cyst and jaundice are highly specific clinical parameters and were therefore used for composite marker selection process. Other clinical features, including patient age, main pancreatic duct dilation, cyst size, pancreatitis, and diabetes, were not as specific (Fig. 1) and were therefore not included in the composite marker selection process. All molecular features, including DNA mutations, aneuploidy, loss of heterozygosity, and protein biomarkers, were included in the composite marker selection process.
Selection of composite markers for clinical management
We selected composite markers to stratify patients into three categories relevant to clinical management: patients who could be discharged, patients who warrant periodic monitoring, and patients who require surgery (Fig. 3). For each of these three classifications, composite markers were selected from the training cohort, and the top-performing marker from that selection was tested in the remaining patients (the validation cohort).
Selection of composite markers for classifying cyst type
We also attempted to predict the type of cyst using composite markers derived in a fashion similar to that described for clinical management above. For each of six cyst types (serous cystic neoplasms, mucin-producing cysts, pancreatic adenocarcinomas, pancreatic neuroendocrine tumors, solid pseudopapillary neoplasms, or other nonmalignant cyst types), the top composite markers were selected from the training cohort. To assign a cyst to a specific cyst type, we calculated the fraction of those markers testing positive in each patient in the validation cohort. For example, if a patient tested positive for 75% of the top composite markers for the solid pseudopapillary neoplasm cyst type and 10% of markers for each of the other cyst types, the cyst was predicted to be a solid pseudopapillary neoplasm. Table S3 includes the detailed results for each patient with respect to its predicted cyst type. The actual cyst type was determined by histopathological examination, as described above.
Current standard of care
For each patient, we determined the appropriate management based on conventional clinical and imaging data (table S3). Indications for pancreatic resection were obstructive jaundice secondary to the cyst, a preoperative clinical diagnosis of a pancreatic adenocarcinoma, a solid pseudopapillary neoplasm, a pancreatic neuroendocrine tumor that was functional or measured greater than 20 mm, and a mucin- producing cyst that met the guideline criteria for surgical resection (the presence of any of the following criteria: jaundice, main pancreatic duct dilation of 6 mm or greater, a mural nodule, cyst size of greater than 40 mm, or the presence of high-grade dysplasia or adenocarcinoma on cytology). Cysts were considered to require monitoring if the preoperative clinical diagnosis was a mucin- producing cyst that did not meet the guidelines for surgical resection described previously. Patients whose cysts were classified as benign and nonmucin-producing on the basis of the preoperative cyst diagnosis were considered to be suitable for discharge.
Statistical analyses
Estimates of marker performance are provided in tables S4 to S6 and include the two-tailed Fisher’s exact test, sensitivity (true-positive rate), specificity (false-positive rate), and effect size. We computed the effect size as the difference of proportions from a 2×2 contingency table (36), which yielded a value between “0” (no effect) and “1” (difference between classes fully captured by marker). To compare the performance of our combinatorial markers with that of the physician’s diagnosis, we used McNemar’s test. McNemar’s test uses the false-negative and true-positive rates to estimate a test statistic that can be used to compare two classifiers (36); McNemar’s P values were calculated using the R statistical computing language.
Supplementary Material
Classification of patients into management groups.
Cyst fluid VEGF-A.
Cyst fluid CEA.
Association between aneuploidy and high-grade dysplasia or cancer.
Clinical, imaging, molecular, and pathological data for all 862 patients with surgically resected pancreatic cysts.
Genetic characteristics of the IPMNs based on histological subtype.
CompCyst and the preoperative clinical diagnosis and management recommendations.
Performance of the three-step approach to classify cysts into management groups.
CompCyst and preoperative clinical management recommendations compared with surgical pathology.
Identification of cyst type: comparison of CompCyst, the preoperative clinical diagnosis, and surgical pathology.
Primer sequences used in Safe-SeqS.
Primer sequences used in Safe-SeqS for loss of heterozygosity.
Frequency of molecular features associated with different grades of dysplasia in mucin-producing cysts.
Acknowledgments:
We are grateful to C. Blair and K. Judge from The Ludwig Center at Johns Hopkins University for expert technical and administrative assistance.
Funding: This work was supported by the Lustgarten Foundation for Pancreatic Cancer Research (to B.V.), S. Wojcicki and D. Troper (to M.G.G.), The Virginia and D.K. Ludwig Fund for Cancer Research (to K.W.K.), The Sol Goldman Pancreatic Cancer Research Center (to R.H.H.), The Michael Rolfe Pancreatic Cancer Research Foundation (to M.G.G.), Benjamin Baker Scholarship (to A.M.L.), and the National Institutes of Health grants P50-CA062924 (to A.P.K.), CA176828 (to M.G.G.), and CA210170 (to M.G.G.). All sequencing analyses were performed in the Sol Goldman Sequencing Center at Johns Hopkins.
Footnotes
Competing interests: N.P., K.W.K., and B.V. are founders of Personal Genome Diagnostics Inc. and PapGene Inc. These companies and others have licensed technologies from Johns Hopkins, and N.P., K.W.K., and B.V. receive equity or royalties from these licenses. The terms of these arrangements are being managed by the university in accordance with its conflict of interest policies. B.V. is a member of the Scientific Advisory Boards of Eisai-Morphotek, Sysmex-Inostics, Nexus (Camden Partners), NeoPhore, and CAGE. The first four of these companies have licensed technologies from Johns Hopkins University, on which B.V. is an inventor. These licenses and relationships are associated with equity or royalty payments to B.V. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. B.V. is an inventor on multiple patents filed by Johns Hopkins University covering the underlying technology and specific genes analyzed in this paper. K.W.K. is also a member of the Scientific Advisory Boards of Eisai-Morphotek, Syxmex-Inostics, CAGE, and NeoPhore. These companies, as well as other companies, have licensed technologies from Johns Hopkins University, on which K.W.K. is an inventor. These licenses and relationships are associated with equity or royalty payments to K.W.K. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. D.S.K. is a consultant and equity holder in PAIGE. AI. C. Douville is a paid consultant to PapGene and has previously developed and licensed intellectual property. The licenses are through Johns Hopkins University in accordance with its conflict of interest policies. C.T. is an inventor on multiple patents (CancerSEEK, PapSEEK, and UroSEEK) filed by Johns Hopkins University PapGene Inc. has licensed technology from Johns Hopkins University, on which C.T. is an inventor. This license is associated with royalty payments to C.T. The terms of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. A.M.L. is an inventor on the CancerSEEK patent. M.D.M. is an inventor on the Differential identification of pancreatic cysts patent (US9637796B2). W.P. is a consultant for Interpace Diagnostics. Additional patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. The following patents are related to this work: Safe Sequencing System US201161476150P, Rapid Aneuploidy Detection US201261615535P, Mutations in pancreatic neoplasms US9976184B2, and Differential identification of pancreatic cysts US9637796B2.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. The raw sequencing data is uploaded to Sequence Read Archive (SRP150837). The mutation code and the LOH code are on GitHub at https://github.com/cristomasetti; the MOCA algorithm is available at https://zenodo.org/record/3235801; and the WALDO code is available at https://zenodo.org/record/3234941. Questions relating to the following topics should be directed to: molecular data, B.V. (bertvog@gmail.com); clinical data, A.M.L. (amlennon@jhmi.edu) or C.L.W. (cwolfga2@jhmi.edu); and pathology, R.H.H. (rhruban@jhmi.edu).
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Lee KS, Sekhar A, Rofsky NM, Pedrosa I, Prevalence of incidental pancreatic cysts in the adult population on MR imaging. Am. J. Gastroenterol 105, 2079–2084 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Laffan TA, Horton KM, Klein AP, Berlanstein B, Siegelman SS, Kawamoto S, Johnson PT, Fishman EK, Hruban RH, Prevalence of unsuspected pancreatic cysts on MDCT. Am. J. Roentgenol 191, 802–807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lennon AM, Wolfgang CL, Canto MI, Klein AP, Herman JM, Goggins M,Fishman EK, Kamel I, Weiss MJ, Diaz LA, Papadopoulos N, Kinzler KW,Vogelstein B, Hruban RH, The early detection of pancreatic cancer: What will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res. 74,3381–3389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Fernández-del Castillo C, Kamisawa T, Jang JY, Levy P, Ohtsuka T, Salvia R,Shimizu Y, Tada M, Wolfgang CL, Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 17, 738–753 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Del Chiaro M, Verbeke C, Salvia R, Klöppel G, Werner J, McKay C, Friess H, Manfredi R,Van Cutsem E, Löhr M, Segersvärd R; European Study Group on Cystic Tumours of the Pancreas, European experts consensus statement on cystic tumours of the pancreas. Dig. Liver Dis 45, 703–711 (2013). [DOI] [PubMed] [Google Scholar]
- 6.European Study Group on Cystic Tumours of the Pancreas, European evidence-based guidelines on pancreatic cystic neoplasms. Gut 67, 789–804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elta GH, Enestvedt BK, Sauer BG, Lennon AM, ACG clinical guideline: Diagnosis and management of pancreatic cysts. Am. J. Gastroenterol 113, 464–479 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Valsangkar NP, Morales-Oyarvide V, Thayer SP, Ferrone CR, Wargo JA, Warshaw AL, Fernandez-del Castillo C, 851 resected cystic tumors of the pancreas: A 33-year experience at the Massachusetts General Hospital. Surgery 152, S4–S12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahora K, Mino-Kenudson M, Brugge W, Thayer SP, Ferrone CR, Sahani D, Pitman MB, Warshaw AL, Lillemoe KD, Fernandez-del Castillo CF, Branch duct intraductal papillary mucinous neoplasms: Does cyst size change the tip of the scale? A critical analysis of the revised international consensus guidelines in a large single-institutional series. Ann. Surg 258, 466–475 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Sahora K, Ferrone CR, Brugge WR, Morales-Oyarvide V, Warshaw AL, Lillemoe KD, Fernández-del Castillo C, Effects of comorbidities on outcomes of patients with intraductal papillary mucinous neoplasms. Clin. Gastroenterol. Hepatol 13, 1816–1823 (2015). [DOI] [PubMed] [Google Scholar]
- 11.de Wilde RF, Besselink MGH, van der Tweel I, de Hingh IHJT, van Eijck CHJ, Dejong CHC, Porte RJ, Gouma DJ, Busch ORC, Molenaar IQ; Dutch Pancreatic Cancer Group, Impact of nationwide centralization of pancreaticoduodenectomy on hospital mortality. Br. J. Surg 99, 404–410 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI, Schulick RD, Edil BH, Choti MA, Adsay V, Klimstra DS, Offerhaus GJ, Klein AP, Kopelovich L, Carter H, Karchin R, Allen PJ, Schmidt CM, Naito Y, Diaz LA Jr., Kinzler KW, Papadopoulos N, Hruban RH, Vogelstein B, Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl. Acad. Sci. U.S.A 108, 21188–21193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer S, Wang Y, Dal Molin M, Masica DL, Jiao Y, Kinde I, Blackford A, Raman SP, Wolfgang CL, Tomita T, Niknafs N, Douville C, Ptak J, Dobbyn L, Allen PJ, Klimstra DS, Schattner MA, Schmidt CM, Yip-Schneider M, Cummings OW, Brand RE, Zeh HJ, Singhi AD, Scarpa A, Salvia R, Malleo G, Zamboni G, Falconi M, Jang JY, Kim SW, Kwon W, Hong SM, Song KB, Kim SC, Swan N, Murphy J, Geoghegan J, Brugge W, Fernandez-Del Castillo C, Mino-Kenudson M, Schulick R, Edil BH, Adsay V, Paulino J, van Hooft J, Yachida S, Nara S, Hiraoka N, Yamao K, Hijioka S, van der Merwe S, Goggins M, Canto MI, Ahuja N, Hirose K, Makary M, Weiss MJ, Cameron J, Pittman M, Eshleman JR, Diaz LA Jr., Papadopoulos N, Kinzler KW, Karchin R, Hruban RH, Vogelstein B, Lennon AM, A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 149, 1501–1510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones M, Zheng Z, Wang J, Dudley J, Albanese E, Kadayifci A, Dias-Santagata D, Le L, Brugge WR, Fernandez-del Castillo C, Mino-Kenudson M, Iafrate AJ, Pitman MB, Impact of next-generation sequencing on the clinical diagnosis of pancreatic cysts. Gastrointest. Endosc. 83, 140–148 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Partyka K, McDonald M, Maupin KA, Brand R, Kwon R, Simeone DM, Allen P, Haab BB, Comparison of surgical and endoscopic sample collection for pancreatic cyst fluid biomarker identification. J. Proteome Res 11, 2904–2911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yip-Schneider MT, Wu H, Dumas RP, Hancock BA, Agaram N, Radovich M, Schmidt CM, Vascular endothelial growth factor, a novel and highly accurate pancreatic fluid biomarker for serous pancreatic cysts. J. Am. Coll. Surg 218, 608–617 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Carr RA, Yip-Schneider MT, Dolejs S, Hancock BA, Wu H, Radovich M, Schmidt CM, Pancreatic cyst fluid vascular endothelial growth factor a and carcinoembryonic antigen: A highly accurate test for the diagnosis of serous cystic neoplasm. J. Am. Coll. Surg, S1072–7515(17)30452–0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jais B, Rebours V, Malleo G, Salvia R, Fontana M, Maggino L, Bassi C, Manfredi R, Moran R, Lennon AM, Zaheer A, Wolfgang C, Hruban R, Marchegiani G, Fernandez Del Castillo C, Brugge W, Ha Y, Kim MH, Oh D, Hirai I, Kimura W, Jang JY, Kim SW, Jung W, Kang H, Song SY, Kang CM, Lee WJ, Crippa S, Falconi M, Gomatos I, Neoptolemos J, Milanetto AC, Sperti C, Ricci C, Casadei R, Bissolati M, Balzano G, Frigerio I, Girelli R, Delhaye M, Bernier B, Wang H, Jang KT, Song DH, Huggett MT, Oppong KW, Pererva L, Kopchak KV, Del Chiaro M, Segersvard R, Lee LS, Conwell D, Osvaldt A, Campos V, Aguero Garcete G, Napoleon B, Matsumoto I, Shinzeki M, Bolado F, Fernandez JM, Keane MG, Pereira SP, Acuna IA, Vaquero EC, Angiolini MR, Zerbi A, Tang J, Leong RW, Faccinetto A, Morana G, Petrone MC, Arcidiacono PG, Moon JH, Choi HJ, Gill RS, Pavey D, Ouaissi M, Sastre B, Spandre M, De Angelis CG, Rios-Vives MA, Concepcion-Martin M, Ikeura T, Okazaki K, Frulloni L, Messina O, Lévy P, Serous cystic neoplasm of the pancreas: A multinational study of 2622 patients under the auspices of the International Association of Pancreatology and European Pancreatic Club (European Study Group on Cystic Tumors of the Pancreas). Gut 65, 305–312 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Matthaei H, Schulick RD, Hruban RH, Maitra A, Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol 8, 141–150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheiman JM, Hwang JH, Moayyedi P, American gastroenterological association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 148, 824–848.e22 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Kimura W, Moriya T, Hanada K, Abe H, Yanagisawa A, Fukushima N, Ohike N, Shimizu M, Hatori T, Fujita N, Maguchi H, Shimizu Y, Yamao K, Sasaki T, Naito Y, Tanno S, Tobita K, Tanaka M, Multicenter study of serous cystic neoplasm of the Japan pancreas society. Pancreas 41, 380–387 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Griffin JF, Page AJ, Samaha GJ, Christopher A, Bhaijee F, Pezhouh MK, Peters NA, Hruban RH, He J, Makary MA, Lennon AM, Cameron JL, Wolfgang CL, Weiss MJ, Patients with a resected pancreatic mucinous cystic neoplasm have a better prognosis than patients with an intraductal papillary mucinous neoplasm: A large single institution series. Pancreatology 17, 490–496 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Crippa S, Bassi C, Salvia R, Malleo G, Marchegiani G, Rebours V, Levy P, Partelli S, Suleiman SL, Banks PA, Ahmed N, Chari ST, Fernández-Del Castillo C, Falconi M, Low progression of intraductal papillary mucinous neoplasms with worrisome features and high-risk stigmata undergoing non-operative management: A mid-term follow-up analysis. Gut 66, 495–506 (2017). [DOI] [PubMed] [Google Scholar]
- 24.de Jong K, Poley J-W, van Hooft JE, Visser M, Bruno MJ, Fockens P, Endoscopic ultrasound-guided fine-needle aspiration of pancreatic cystic lesions provides inadequate material for cytology and laboratory analysis: Initial results from a prospective study. Endoscopy 43, 585–590 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Lennon AM, Manos LL, Hruban RH, Ali SZ, Fishman EK, Kamel IR, Raman SP, Zaheer A, Hutfless S, Salamone A, Kiswani V, Ahuja N, Makary MA, Weiss MJ, Hirose K, Goggins M, Wolfgang CL, Role of a multidisciplinary clinic in the management of patients with pancreatic cysts: A single-center cohort study. Ann. Surg. Oncol 21, 3668–3674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basturk O, Hong SM, Wood LD, Adsay NV, Albores-Saavedra J, Biankin AV, Brosens LA, Fukushima N, Goggins M, Hruban RH, Kato Y, Klimstra DS, Kloppel G, Krasinskas A, Longnecker DS, Matthaei H, Offerhaus GJ, Shimizu M, Takaori K, Terris B, Yachida S, Esposito I, Furukawa T; Baltimore Consensus Meeting,A revised classification system and recommendations from the baltimore consensus meeting for neoplastic precursor lesions in the pancreas. Am. J. Surg. Pathol 39, 1730–1741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosman FT, Carneiro F, Hruban RH, Theise ND, WHO Classification of Tumours of the Digestive System (World Health Organization, ed. 4, 2010). [Google Scholar]
- 28.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B, Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A 108, 9530–9535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih Ie I-M, Kurman R, Dao F, Levine DA, Giuntoli R, Roden R, Eshleman JR, Carvalho JP, Marie SK, Papadopoulos N, Kinzler KW, Vogelstein B, Diaz LA Jr., Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med 5, 167ra4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM Jr., The American Soldier: Adjustment during Army Life (Princeton Univ. Press, 1949), vol. 1. [Google Scholar]
- 31.Kinde I, Papadopoulos N, Kinzler KW, Vogelstein B, FAST-SeqS: A simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLOS ONE 7, e41162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douville C, Springer S, Kinde I, Cohen JD, Hruban RH, Lennon AM, Papadopoulos N, Kinzler KW, Vogelstein B, Karchin R, Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl. Acad. Sci. U.S.A 115, 1871–1876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masica DL, Dal Molin M, Wolfgang CL, Tomita T, Ostovaneh MR, Blackford A, Moran RA, Law JK, Barkley T, Goggins M, Irene Canto M, Pittman M, Eshleman JR, Ali SZ, Fishman EK, Kamel IR, Raman SP, Zaheer A, Ahuja N, Makary MA, Weiss MJ, Hirose K, Cameron JL, Rezaee N, He J, Joon Ahn Y, Wu W, Wang Y, Springer S, Diaz LL Jr., Papadopoulos N, Hruban RH, Kinzler KW, Vogelstein B, Karchin R, Lennon AM, A novel approach for selecting combination clinical markers of pathology applied to a large retrospective cohort of surgically resected pancreatic cysts. J. Am. Med. Inform. Assoc 24, 145–152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masica DL, Karchin R, Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 71, 4550–4561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masica DL, Karchin R, Collections of simultaneously altered genes as biomarkers of cancer cell drug response. Cancer Res. 73, 1699–1708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agresti A, An Introduction to Categorical Data Analysis (Wiley, ed. 2, 2006). [Google Scholar]
- 37.Brugge WR, Lewandrowski K, Lee-Lewandrowski E, Centeno BA, Szydlo T, Regan S, del Castillo CF, Warshaw AL, Diagnosis of pancreatic cystic neoplasms: A report of the cooperative pancreatic cyst study. Gastroenterology 126, 1330–1336 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Kosmahl M, Pauser U, Anlauf M, Kloppel G, Pancreatic ductal adenocarcinomas with cystic features: Neither rare nor uniform. Mod. Pathol 18, 1157–1164 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Law JK, Ahmed A, Singh VK, Akshintala VS, Olson MT, Raman SP, Ali SZ, Fishman EK, Kamel I, Canto MI, Dal Molin M, Moran RA, Khashab MA, Ahuja N, Goggins M, Hruban RH, Wolfgang CL, Lennon AM, A systematic review of solid-pseudopapillary neoplasms: Are these rare lesions? Pancreas 43, 331–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Classification of patients into management groups.
Cyst fluid VEGF-A.
Cyst fluid CEA.
Association between aneuploidy and high-grade dysplasia or cancer.
Clinical, imaging, molecular, and pathological data for all 862 patients with surgically resected pancreatic cysts.
Genetic characteristics of the IPMNs based on histological subtype.
CompCyst and the preoperative clinical diagnosis and management recommendations.
Performance of the three-step approach to classify cysts into management groups.
CompCyst and preoperative clinical management recommendations compared with surgical pathology.
Identification of cyst type: comparison of CompCyst, the preoperative clinical diagnosis, and surgical pathology.
Primer sequences used in Safe-SeqS.
Primer sequences used in Safe-SeqS for loss of heterozygosity.
Frequency of molecular features associated with different grades of dysplasia in mucin-producing cysts.