Abstract
DEAD-box helicase 41 (DDX41) is an RNA helicase and accumulating evidence has suggested that DDX41 is involved in pre-mRNA splicing during tumor development. However, the role of DDX41 in tumorigenesis remains unclear. In order to determine the function of DDX41, the human DDX41 gene was cloned and overexpressed in HeLa cells. The present study demonstrated that DDX41 overexpression inhibited proliferation and promoted apoptosis in HeLa cells. RNA-sequencing analysis of the transcriptomes in overexpressed and normal control samples. DDX41 regulated 959 differentially expressed genes compared with control cells. Expression levels of certain oncogenes were also regulated by DDX41. DDX41 selectively regulated the alternative splicing of genes in cancer-associated pathways including the EGFR and FGFR signaling pathways. DDX41 selectively upregulated the expression levels of five antigen processing and presentation genes (HSPA1A, HSPA1B, HSPA6, HLA-DMB and HLA-G) and downregulated other immune-response genes in HeLa cells. Additionally, DDX41-regulated oncogenes and antigen processing and presentation genes were associated with patient survival rates. Moreover, DDX41 expression was associated with immune infiltration in cervical and endocervical squamous cancer. The present findings showed that DDX41 regulated the cancer cell transcriptome at both the transcriptional and alternative splicing levels. The DDX41 regulatory network predicted the biological function of DDX41 in suppressing tumor cell growth and regulating cancer immunity, which may be important for developing anticancer therapeutics.
Keywords: DEAD-box helicase 41, RNA-sequencing, gene expression, alternative splicing
Introduction
RNA helicases are enzymes that unwind nucleic acids in an energy-dependent manner via hydrolysis of nucleotide triphosphate (1); these enzymes are implicated in processes involving RNA, including transcription, splicing, RNA export, ribosome biogenesis, mRNA translation and RNA decay (2–5). Moreover, novel functions of RNA helicases include their activity as transcriptional coregulators and regulators of post-translational modifications, and their ability to modulate cellular signaling pathways such as Wnt-ß-catenin, NF-κB and MAPK (6–9). Deregulation of these RNA helicase functions may contribute to tumor development and progression (10–12). Dysregulation of RNA helicase expression levels, its mutation status in tumors and its role in the regulation of molecules have been implicated in cancer (13). High-throughput sequencing technology, with its rapidly decreasing costs and increasing applications, allows researchers to investigate the biological function of RNA helicases in cancer, which may be the key to ‘unwind’ cancer in the future.
DEAD-box helicase 41 (DDX41) is a highly conserved protein considered essential for cell growth and viability (14), which is mutated in primary tumor and relapse samples from patients with acute myeloid leukemia (AML) (15). Polprasert et al (16) identified somatic DDX41 mutations in myeloid neoplasms that resulted in loss of tumor suppressor function due to altered pre-mRNA splicing and RNA processing. RNA-sequencing (seq) analysis on peripheral bold mononuclear cells of DDX41 mutation carriers with hematological malignancy (HM) revealed altered expression levels of genes involved in hemoglobin complex and innate immunity (17). The majority of germline mutations are frameshift mutations, suggesting loss of function, with DDX41 serving as a tumor suppressor, which may impact initiation, maintenance or progression of tumorigenesis (18). The identification of DDX41 mutations has improved understanding of the potential mechanisms underlying HM; however, the expression and mechanism by which DDX41 contributes to tumorigenesis is still unknown.
As an upstream DNA sensor of stimulator of interferon gene (STING), DDX41 may serve dual roles in various types of cancer. The preventive role of DDX41 in HM is associated with inhibition of proliferation and promotion of apoptosis and DDX41 negative regulates p21 at the translational level (19); in addition, DDX41 silencing promotes apoptosis of HCT116 cells in a p21-dependent manner (19). These studies suggest that the role of DDX41 may be tumor type-dependent. However, the precise function of DDX41, and how it influences the formation and development of tumors remains poorly understood.
DDX41 is also associated with the immune response. Recent studies revealed that DDX41 primarily senses the viral DNA/RNA hybrid and is required for the anti-retroviral innate immune response to murine leukemia and HIV in primary mouse macrophages and dendritic cells (DCs) (20,21). Zhang et al (22) identified DDX41 as an intracellular DNA sensor in myeloid DCs; silencing of DDX41 was shown to block the activation of STING-TANK-binding kinase 1 signaling and the transcription factors NF-κB and interferon regulatory factor 3, resulting in decreased production of type I interferon. As a pattern-recognition receptor, DDX41 also recognizes bacterial di-GMP and di-AMP to activate the immune response via the STING-dependent signaling pathway (23). Intravenous administration of c-di-GMP into mice efficiently activates DDX41-STING signaling, production of interferon and activation of natural killer (NK) cells, and also facilitates antigen-specific cytotoxic T cell activity, resulting in a significant antitumor effect in a lung metastasis mouse model using malignant melanoma cells (24,25). Thus, DDX41 may trigger the production of molecules involved in the immune response.
Given the involvement of helicases in RNA splicing (26), Polprasert et al (16) expressed epitope-tagged versions of wild-type (WT) and R525H-mutated DDX41 in 293 cells. Epitope pull-down coupled with peptide sequencing analysis revealed that spliceosome proteins constitute a DDX41-associated group, which includes pre-mRNA processing factor 8 and splicing factor 3B subunit 1 (16). Furthermore, DDX41 mutation in the DEAD domain (such as R525H) alters this interaction, especially for major components in the U2 and U5 spliceosomes (16). In addition, DDX41 has been reported to be involved in pre-RNA splicing and DDX41 silencing may be associated with increased exon skipping (16). DDX41 is specifically recruited to the catalytically active C complex, which performs the second step of splicing, in which the 5′ and 3′ exons are ligated and an intronic lariat is released (27,28). These findings indicated that DDX41 may lead to the impairment of RNA processing and pre-mRNA splicing, which may partially contribute to tumorigenesis.
The present study aimed to investigate the function of DDX41 in HeLa cells and cervical cancer. DDX41 was cloned and overexpressed in HeLa cells. RNA-sequencing (RNA-seq) analysis of the effect of DDX41 on the transcriptome and alternative splicing of the overexpressed and normal control cell samples was performed. These results may improve understanding of the biological role of DDX41 in cancer.
Materials and methods
DDX41 cloning and plasmid construction
Primer pairs used for Hot Fusion were designed using CE Design v1.04 (Vazyme Biotech Co., Ltd.) with gene-specific sequences and oligonucleotides encoding 3× FLAG tag were added before the TAG termination codon, along with a portion of the vector pIRES-hrGFP-1a sequence, each with a 17–30 bp overlap. The following primers were used: Forward, 5′-agcccgggcggatccgaattcATGGAGGAGTCGGAACCCG-3′; and reverse, 5′-gtcatccttgtagtcctcgagGAAGTCCATGGAGCTGTGGGC-3′. Non-capitalized letters indicate sequence for homologous recombination during cloning, whereas capitalized letters indicate sequence paired with DDX41 gene. Vector pIRES-hrGFP-1a was digested by EcoRI and XhoI (New England BioLabs, Inc.) at 37°C for 2–3 h. The enzyme-digested vector was then run on a 1.0% agarose gel and purified using a QIAquick Gel Extraction kit (cat. no. 28704; Qiagen, Inc.). Total RNA from HeLa cells was isolated using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Then, 1 µg total RNA was transcribed into cDNA using a PrimeScript™ RT Reagent kit (Takara Bio, Inc.), according to the manufacturer's protocol. The thermocycling condition were as follows; 98°C for 30 sec; 28 cycles of 98°C for 10 sec and 60°C for 30 sec; final extension at 72°C for 1 min and holding at 4°C. Linearized vector digested by EcoRI and XhoI (New England BioLabs, Inc.) and the PCR insert (1,866 bp) were added to a PCR microtube and cloned using the ClonExpress® II One Step Cloning kit (Vazyme Biotech Co., Ltd.), according to the manufacturer's protocol. Plasmids were introduced into Escherichia coli by chemical transformation. Cells (5×106) were plated onto LB plates containing ampicillin and incubated overnight at 37°C. Colonies were screened using a TaKaRa Taq™ HS PCR kit (Takara Bio, Inc.) according to the manufacturer's protocol, with M13 primers (forward, 5′-TGTAAAACGACGGCCAGT-3′ and reverse, 5′-CAGGAAACAGCTATGACC-3′). The PCR insert was verified by Sanger sequencing.
Cell culture and transfection
The human HeLa cell line was purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. Cells were seeded into a petri dish (100 mm) at a density of 1×105 cells/ml and cultured at 37°C with 5% CO2 in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences), penicillin (100 U/ml) and streptomycin (100 g/ml). Transfection of 1×105 HeLa cells with 0.5 µg DDX41 overexpression or empty plasmid was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Transfected cells were harvested after 48 h for subsequent experimentation.
Assessment of DDX41 overexpression
GAPDH was used as an internal control. cDNA synthesis was performed according to standard procedure (29) and reverse transcription-quantitative (RT-q)PCR was performed on the Bio-Rad S1000 system with Bestar SYBR Green RT-PCR Master Mix kit according to the manufacturer's instructions. (DBI Bioscience; Shanghai Xinghan Biotechnology Co., Ltd.). The concentration of each transcript was then normalized to the GAPDH mRNA levels using the 2−ΔΔCq method (30). Comparisons were performed by one-way ANOVA using GraphPad Prism 8 software (GraphPad Software, Inc.).
Western blot analysis
In brief, for preparation of total cell lysates, normal and DDX41-overexpressing HeLa cells were lysed in RIPA buffer containing 50.0 mM Tris-HCl (pH, 7.4), 150.0 mM NaCl, 1.0% deoxycholate, 1.0% Triton X-100, 1.0 mM EDTA and 0.1% SDS. The samples were centrifuged (10,000 × g; 5 min; 4°C). The protein level was determined by Pierce™ BCA protein assay kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Then, 50 µg protein was incubated for 10 min in boiling water with 1X SDS sample buffer, separated by SDS-PAGE on a 10% gel, then transferred to a PVDF membrane (EMD Millipore, Inc.). The membranes were blocked with 5% skimmed milk [in buffer containing 10 mM Tris (pH, 8.0), 150 mM NaCl and 0.05% Tween-20] for 1 h. DDX41 was detected using monoclonal Flag antibody (1:2,000; cat. no. F7425; Sigma-Aldrich; Merck KGaA) with actin as a control (1:2,000; cat. no. AC038; ABclonal Biotech Co., Ltd.) diluted in TBST (0.05% Tween-20) at 4°C overnight. The blots were incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG; 1:10,000; cat. no. ab7090; Abcam) for 1 h at room temperature. Bands were detected using enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.).
MTT assay
MTT assay was used to measure cell proliferation. Briefly, cells (1×104) were seeded in 96-well culture plates containing 200 µl cell growth medium. Upon reaching 70% confluence, the HeLa cells were transfected with DDX41 overexpression vector or control vector using Lipofectamine 2000 according to the manufacturer's protocol. The cells were then incubated at 37°C for 48 h. Subsequently, 0.025 ml MTT solution (5 mg/ml) was added to each well and cells were incubated for 4 h. Following centrifugation (5,000 × g; 5 min; room temperature), the supernatant was removed from each well. The colored formazan crystals produced from MTT in each well were dissolved in 0.15 ml DMSO and the optical density values were measured at 490 nm.
Flow cytometric analysis of cell apoptosis
HeLa cells (5×104) were seeded in 24-well culture plates. At 70% confluence, the cells were transfected with the DDX41 overexpression or control vector using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cells were then incubated at 37°C for 48 h and viable cells were harvested and washed twice with PBS. The cell pellets were resuspended in 400 µl ice-cold 1X binding buffer at ~1×106 cells/ml, then incubated with 10 µl FITC-conjugated Annexin V/7-AAD (Beijing 4A Biotech Co., Ltd.) for 10 min in the dark at room temperature. Samples were analyzed within 1 h of staining using a A00-1-1102 CytoFLEX flow cytometer (Beckman Coulter, Inc.) and Flow-Jo software (v8.8.6; Tree Star, Inc.).
Library preparation and sequencing
Total RNA was extracted from HeLa cells using TRIzol. The RNA was further purified with two phenol-chloroform treatments and then treated with RQ1 DNase (Promega Corporation) at 37°C for 10 min to remove DNA. The quality and quantity of the purified RNA were determined by measuring the absorbance at 260/280 nm using Smartspec Plus (Bio-Rad Laboratories, Inc.). The integrity of RNA was further verified by 1.5% agarose gel electrophoresis. For each sample, 1 µg total RNA was used for RNA-seq library preparation using the VAHTS Stranded mRNA-seq Library Prep kit (cat. no. NR612-01; Vazyme Biotech Co., Ltd.). Polyadenylated mRNA was purified and fragmented, and then converted into double stranded cDNA. Following end repair and A tailing, DNA was ligated to VAHTS RNA Adapters (Vazyme Biotech Co., Ltd.). Purified ligation products corresponding to 200–500 bp were digested with heat-labile uracil-DNA glycosylase and the single stranded cDNA was amplified, purified, quantified and stored at −80°C before sequencing. Next, the 3M library was sequenced on an Illumina HiSeq X Ten platform in a 2 × 150 bp paired-end mode using HiSeq X Ten reagent kit v2.5 (cat. no. FC-501-2501; Illumina, Inc.). The fastq files were produced using the CASAVA pipeline v1.8.2 (Illumina, Inc.). Preliminary quality control analysis of the fastq files was performed using FASTQC software v 1.11.4 (fastqc.software.informer.com/).
Data processing and alignment
Raw reads containing >2-N bases were first discarded. Adaptors and low-quality bases were then trimmed from the raw sequencing reads using the FASTX-Toolkit (Version 0.0.13; hannonlab.cshl.edu/fastx_toolkit/). Short reads (<16 nucleotides in length) were also excluded. Clean reads were aligned to the GRCh38 genome by Tophat2 2.0.0 (ccb.jhu.edu/software/tophat/index.shtml) allowing four mismatches. Uniquely mapped reads were used to calculate the read number and fragments per kilobase of transcript per million fragments mapped for each gene.
Differentially expressed genes (DEGs) analysis
The R Bioconductor package edgeR 3.12 (31) (bioconductor.org/packages/release/bioc/html/edgeR.html) was utilized to screen out DEGs. A false discovery rate (FDR) <0.05 and fold change >1.5 or <0.7 were set as the cut-off criteria for identifying DEGs. In order to predict the gene function and calculate the functional category distribution frequency, Gene Ontology (GO) and enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified using the KOBAS 2.0 server (kobas.cbi.pku.edu.cn/) (32). Hypergeometric tests and the Benjamini-Hochberg FDR controlling procedure were used to define the enrichment of each pathway (corrected P-value <0.05).
Alternative splicing analysis
The alternative splicing events (ASEs) and regulated ASEs (RASEs) in the samples were defined and quantified using the ABLas pipeline, as described previously (33). In brief, detection of eight types of ASE was based on the splice junction reads. The eight types of ASE included cassette exon (CassetteExon), exon skipping (ES), mutually exclusive ES (MXE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), MXE combined with an alternative 5′ promoter (5pMXE) and MXE combined with an alternative polyadenylation site (3pMXE). After detecting the ASEs in each RNA-seq sample, the paired t-test was chosen to calculate the P-value, with the alternative reads and model reads of samples as input data. The changed ratio of alternatively spliced reads and constitutively spliced reads between compared samples was defined as the RASE ratio. P<0.05 and RASE ratio >0.2 were set as the threshold for RASE detection.
RT-qPCR validation of DEGs and ASEs
In order to confirm the validity of the RNA-seq data, RT-qPCR was performed for selected DEGs and normalized to the reference gene GAPDH. The primer sequences are presented in Table SI. The same RNA samples for RNA-seq were used for qPCR. The PCR conditions consisted of denaturation at 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 sec and annealing and extension at 60°C for 1 min. A total of three independent repeats was performed for each sample. RT-qPCR assay was also used to analyze ASEs in HeLa cells. PCR primer pairs (Table SI) were designed to amplify the long and short splicing isoforms in the same reaction. In order to detect one of the alternative isoforms, one primer was designed for the alternative exon, and an opposing primer was designed for the constitutive exon; to detect the other alternative isoforms, a boundary-spanning primer was designed for the sequence encompassing the exon-exon junction, with the opposing primer in a constitutive exon. The changed ratio of AS events in RNA-seq was calculated using the formula: AS1 junction reads/AS1 junction reads + AS2 junction reads. The altered ratio of AS events in RT-qPCR was calculated using the formula: AS1 transcripts level/AS2 transcripts level.
Downloading data
The RNA-seq data of cervical cancer samples were downloaded from The Cancer Genome Atlas (TCGA) database (xenabrowser.net/datapages/) to analyze the expression levels of DDX41-regulated genes. Tumor suppressor genes were downloaded from TSGene 2.0 database (bioinfo.mc.vanderbilt.edu/TSGene/). The oncogenes were downloaded from an oncogene database (ongene.bioinfominzhao.org/index.html).
Gene expression profiling interactive analysis (GEPIA)
GEPIA was used to analyze RNA-seq expression data of 9,736 tumors and 8,587 normal samples from TCGA and Genotype-Tissue Expression (GTEx) projects, using a standard processing pipeline (gepia.cancer-pku.cn/). GEPIA provides customizable functions, such as tumor or normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis.
Immunofluorescence assay
HeLa cells were placed on glass slides using a cytospin (Shandong Junteng Medical Technology Co., Ltd.). Cells were fixed in 4% paraformaldehyde and permeabilized in 1% Triton X-100/PBS at room temperature for 15 min. Cells were incubated with primary antibody for DDX41 (1:100; cat. no. A6576; ABclonal) for 1 h at room temperature in a humidified chamber, washed with 0.1% Tween-20/PBS and incubated with species-appropriate FITV-conjugated secondary antibody (1:500; cat. no. AS011; ABclonal) for 1 h at room temperature. Slides were mounted with 100 ng/ml 4,6 diamidino-2-phenylindole Vectashield (Vector Laboratories, Inc.; Maravai Life Sciences) to stain nuclei at room temperature for 15 min and cells were visualized using an Olympus AX70 epifluorescence microscope (Olympus Corporation) with triple color filters and a 20× air objective. Numeric aperture varied automatically according to the light emitted. Images were captured with a Zeiss Axiocam camera (Zeiss GmbH).
Statistical analysis
Data are presented as the mean ± SD of ≥3 independent repeats. The statistical difference between groups was analyzed using one-way ANOVA with SPSS software (version 17.0; SPSS, Inc.). Patient survival analysis was conducted using the Kaplan-Meier method and analyzed using the log-rank test. The heatmap analysis compared the data obtained from DEG of RNA-seq (34) and sashimi plot representation of RNA-seq data was used in the Integrative Genomics Viewer (35). P<0.05 was considered to indicate a statistically significant difference.
Results
DDX41 inhibits proliferation and promotes apoptosis of HeLa cells
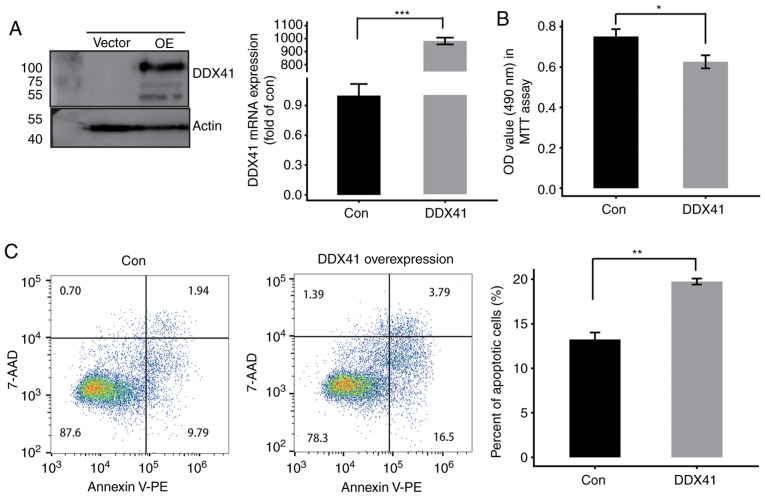
Previous studies have reported that in AML cell line xenografts in a mouse model, DDX41 knockdown accelerates tumor growth compared with control cells (16,48). In order to investigate the potential biological function of DDX41 in cervical cancer, DDX41-3×Flag plasmids was stably overexpressed in HeLa cells and compared with empty vector-transfected cells. The results showed significantly increased expression levels of DDX41, as assessed by western blotting and RT-qPCR (Fig. 1A). MTT assay was performed to determine cell proliferation; overexpression of DDX41 suppressed HeLa cell proliferation (Fig. 1B). In order to investigate the function of DDX41 in cell apoptosis, flow cytometry was performed. The results revealed that DDX41 overexpression significantly induced apoptosis of HeLa cells (Fig. 1C). By immunostaining with DDX41 antibody, DDX41 protein was detected both in nucleus and cytoplasm (Fig. S2B). Therefore, DDX41 may serve as a tumor suppressor in HeLa cells.
Figure 1.
DDX41 inhibits proliferation and promotes apoptosis. (A) HeLa cells were transiently transfected with DDX41 OE or Con vectors followed by determination of Flag-tagged DDX41 protein levels by western blot analysis. The mRNA expression levels were measured by reverse transcription-quantitative PCR. (B) Cell proliferation was measured by MTT assay. Results are presented as the mean ± SD. (C) Effects of DDX41 on HeLa cell apoptosis were determined by flow cytometric analysis (n=3). *P<0.05, **P<0.01 and ***P<0.001. DDX41, DEAD-box helicase 41; Con, control; OE, overexpression; 7-AAD, 7-amino actinomycin D; OD, optical density; PE, phycoerythrin.
RNA-seq analysis of the impact of DDX41 overexpression on the HeLa cell transcriptome
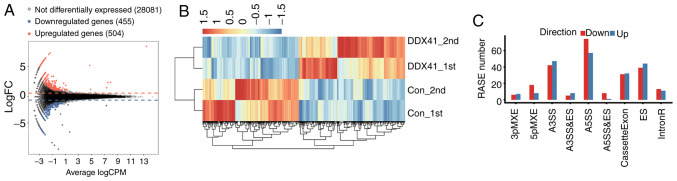
In order to analyze the gene expression profile of DDX41, RNA-seq was performed for 83±4 million total raw reads per sample (Table SII). Filtered reads were aligned to the human GRCH38 genome by Tophat2 (36) and expression patterns of 28,914 known annotated genes were detected and characterized (Table SIII). edgeR (31) and (cutoff, fold change ≥1.5 or ≤0.7; P<0.01) was used to identify the DEGs between the DDX41-overexpressing cells and controls. An M-versus-A plot was constructed to display the DEGs that were associated with DDX41 overexpression and identified 504 up- and 455 downregulated genes (Fig. 2A; Table SIV). Heatmap analysis of the expression patterns of all DEGs in each sample was performed; the results showed high consistency for DDX41-mediated transcription in all datasets (Fig. 2B).
Figure 2.
Differential gene expression and ASEs in response to DDX41 overexpression. (A) Detection of DDX41-regulated genes on the M-versus-A plot. Red, upregulated (FC≥1.5, P-value<0.01); blue, downregulated (FC≤0.7, P-value<0.01). (B) Hierarchical clustering of the expression levels of 959 DEGs regulated by DDX41 overexpression in HeLa cells expressing either the control or DDX41 plasmid. (C) Classification of different alternative spicing types regulated by DDX41 protein. FC, fold change; RASE, regulated alternative splicing events. 3pMXE, mutually exclusive 3′ untranslated regions; 5pMXE, mutually exclusive 5′ untranslated regions; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; ES, exon skipping; IntronR, intron retention.
The ABLas software tool (under permission) was used to analyze global changes in DDX41-mediated ASEs in HeLa cells. A total of 20,532 annotated ASEs were detected when comparing these uniquely mapped reads to the reference genome, as well as 60,654 novel ASEs (Table SV). A stringent cutoff of P≤0.05 was set to identify high-confidence RASEs and 707 RASEs were identified (Tables SVI and SVII). DDX41-regulated ASEs included 24 known intron retention events, 63 CassetteExons, 83 ES, 89 A3SS and 131 A5SS. The other event types included 11 3pMXE, 26 5pMXE, 13 A3SS + ES, 9 A5SS + ES and 18 MXE (Fig. 2C). These results suggested that DDX41 primarily regulated A5SS, A3SS and ES events in HeLa cells.
DDX41 regulates transcription and alternative splicing of certain tumorigenesis-associated genes
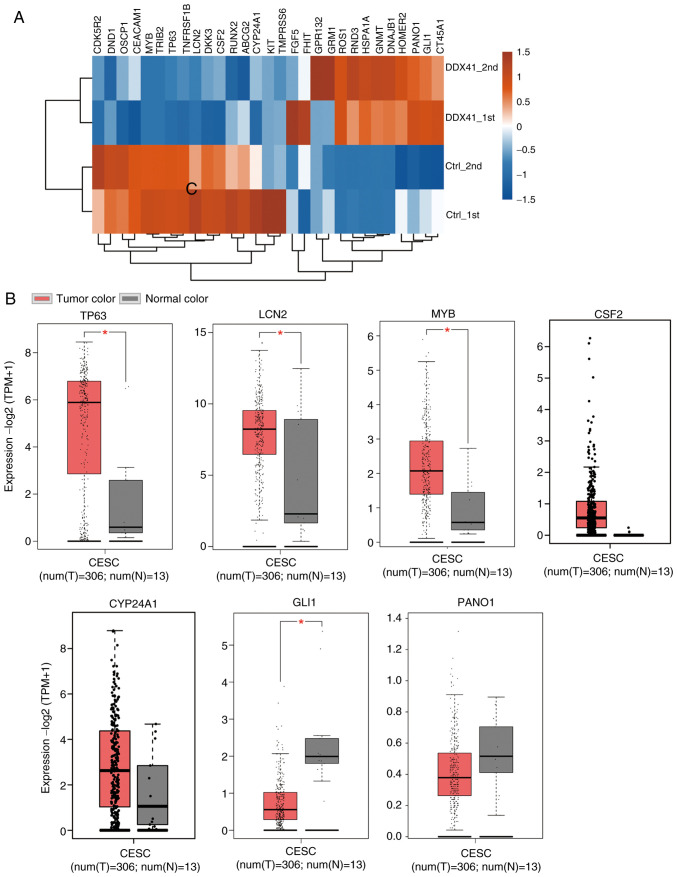
In order to test the hypothesis that DDX41 may regulate tumorigenesis-associated gene transcription, a gene dataset was constructed by downloading tumor suppressor genes from TSGene 2.0 (bioinfo.mc.vanderbilt.edu/TSGene/) (37) and oncogenes from an oncogene database (ongene.bioinfominzhao.org/index.html). The tumorigenesis-associated gene dataset was overlapped with DDX41-regulated genes (RNA-seq-identified DEGs). Heatmap analysis of the expression patterns of the overlapped genes was performed and showed a high consistency for DDX41-regulated gene expression in each sample (Fig. 3A). Certain oncogenes were downregulated by DDX41, such as LCN2, TP63, KIT, MYB, CDK5R2 and TNFRSF1B, whereas others were upregulated, including PANO1, GLI1, HOMER2 and RND3 (Fig. 3A). In order to assess the association between DDX41-regulated expression levels of oncogenes and tumor suppressors, expression data from CESC and control samples were downloaded (xenabrowser.net/datapages/) and analyzed. Expression levels of five oncogenes, TP63, LCN2, MYB, CSF2 and CYP24A1, were upregulated (Fig. 3B), whereas the expression levels of tumor suppressors GLI1 and PANO1 were downregulated in cancer samples (Fig. 3C).
Figure 3.
Analysis of tumorigenesis-associated gene expression levels regulated by DDX41. (A) Hierarchical clustering of the expression of tumorigenesis-associated differentially expressed genes regulated by DDX41 overexpression in HeLa cells compared with controls. (B) Gene expression profiling of tumorigenesis-associated genes in the CESC dataset. *P<0.05. DDX41, DEAD-box helicase 41; CESC, cervical and endocervical squamous cancer.
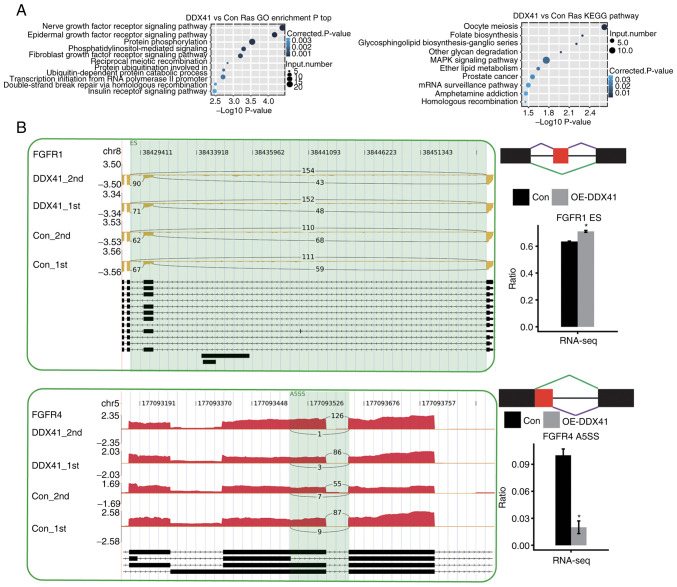
In order to identify the cellular function of DDX41-regulated alternatively spliced genes, high-confidence regulated alternatively spliced genes were analyzed by performing GO and KEGG enrichment analysis. DDX41-regulated genes with alternative splicing were enriched in a number of tumorigenesis-associated pathways, including ‘EGFR signaling’, ‘FGFR signaling’, ‘MAPK signaling’ and ‘prostate cancer’ (Fig. 4A). DDX41-regulated tumorigenesis-associated genes included FGFR1 and FGFR4 (Fig. 4B).
Figure 4.
Functional clustering of alternatively spliced genes regulated by DDX41. (A) Top ten GO biological processes and KEGG functional pathways of DDX41-regulated alternatively spliced genes. (B) Results for FGFR1 ES and FGFR4 A5SS. Integrative Genomics Viewer-sashimi plots showing alternative splicing changes in Con or DDX41-OE HeLa cells. Schematic diagram depicts the structure of model splicing variant AS1 (purple) and alternative splicing variant AS2 (green). Boxes indicate exon sequences; horizontal line indicates intron sequences. RNA-seq quantification of alternative splicing regulation. *P<0.05. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes ES, exon skipping; A5SS, alternative 5′ splice site; RNA-seq, RNA-sequencing; Con, Control; OE-DDX41, overexpression DEAD-box helicase 41.
DDX41 selectively regulates transcription of immunity-associated genes in HeLa cells
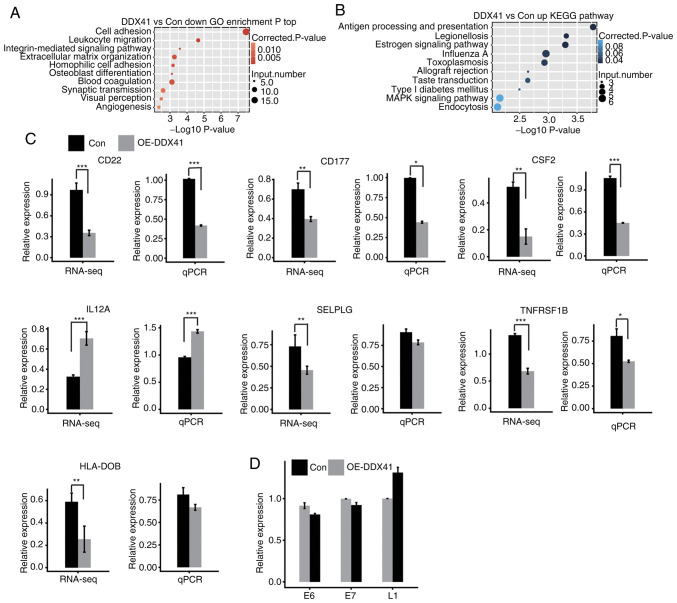
In order to assess the functional role of DDX41, all 959 DEGs were subjected to GO and KEGG annotation to identify their potential biological roles. Based on the cut-off criteria, the results showed that upregulated DEGs were enriched in six GO terms and downregulated DEGs were enriched in 30 GO terms (Tables SVIII and SIX). In the GO analysis, the upregulated genes were enriched in ‘regulation of transcription’ (Fig. S1A), while the downregulated genes were enriched in ‘cell adhesion’, ‘extracellular matrix organization’ and ‘angiogenesis’ (Fig. 5A). KEGG analysis showed that upregulated genes were associated with ‘antigen processing and presentation’, ‘endocytosis’ and ‘MAPK signaling pathway’ (Fig. 5B; Table SX). Moreover, the downregulated genes were enriched in ‘cell adhesion molecules’, ‘cytokine-cytokine receptor interaction’ and ‘B cell receptor signaling pathway’ (Fig. S1B; Table SXI. DDX41-downregulated immune-associated genes included CD22, CD177, CSF2, C4A, SELPLG, TNFRSF1B, HLA-DOB, HLA-DMA, GH1 and PDGFC.
Figure 5.
Analysis of immune-associated gene expression and functional clustering of differentially expressed genes regulated by DDX41. (A) Top ten GO biological processes of DDX41-downregulated genes. (B) Top ten KEGG functional pathways of DDX41-upregulated genes. (C) Transcription of genes was assessed by RNA-seq and qPCR. (D) HPV gene expression levels measured by qPCR. *P<0.05, **P<0.01 and ***P<0.001. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RNA-seq, RNA-sequencing; qPCR, quantitative PCR; Con, Control; OE-DDX41, overexpression DEAD-box helicase 41.
In order to investigate the DDX41-regulated transcription of immune-associated genes, heatmap analysis of these genes was performed, demonstrating a high consistency for DDX41-regulated transcription in both replicates (Fig. S1C). Expression levels of these genes were confirmed via qPCR analysis, showing a significant change in expression levels of CD22, CD177, SCF2, IL12A and TNFRSF1B, in agreement with results of RNA-seq analysis (Fig. 5C). HeLa is a well-characterized HPV+ cervical cancer cell line, so activated expression of immune response genes by DDX41 overexpression may have resulted from an effort to reactivate expression of the HPV genome (38,39). In order to assess this possibility, expression levels of HPV genes E6, E7 and L1 were evaluated via qPCR with the primer pairs shown in Table I, in ref. 40. None of these HPV genes showed significantly increased expression upon DDX41 overexpression (Fig. 5D). From these results, it was concluded that DDX41 regulated immune-associated gene expression.
Antigen processing genes upregulated by DDX41 in HeLa cells are associated with survival rate in patients with cervical cancer
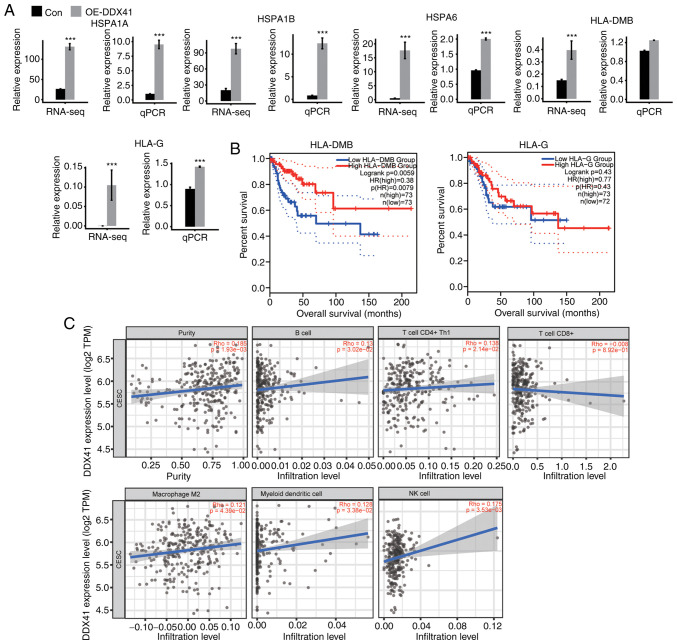
Previous studies have reported that antigen processing presents antigen-derived peptides into MHC-I and MHC-II molecules to be recognized by T cell receptors expressed by CD8+ and CD4+ T cells, respectively (41–43). The top KEGG pathway in analysis of DDX41 overexpression was ‘antigen processing and presentation’, with significant upregulation of HSPA1A, HSPA1B, HSPA6, HLA-DMB and HLA-G detected (Fig. 6A). Expression levels of these genes were confirmed via RT-qPCR, and the results were consistent with the RNA-seq analysis (Fig. 6A).
Figure 6.
Transcription of antigen processing and presentation genes regulated by DDX41. (A) Transcription of several genes were measured by RNA-seq and qPCR. (B) Survival analysis of two genes in the Gene Expression Profiling Interactive Analysis databases of CESC (cutoff point based on the quartile method). (C) Association between DDX41 expression and immunity in CESC, reproduced from TIMER2.0 database. ***P<0.001. Con, control; OE-DDX41, overexpression DEAD-box helicase 41; RNA-seq, RNA-sequencing; qPCR, quantitative PCR; CESC, cervical and endocervical squamous carcinoma.
DDX41 regulation of the expression of immune-associated genes in HeLa cells may reflect an important role of DDX41 in cervical cancer. To test the hypothesis, the association between the expression levels of DDX41-regulated genes and the survival rates of patients with CESC was analyzed in the GEPIA databases. Expression levels of HLA-DMB were positively associated with survival rate of patients with CESC in the whole disease course of >200 months (Fig. 6B). The expression levels of HLA-G, HLA-1A and HLA-1B were positively (though not significantly) associated with survival rate in either the early or late disease stage, while those of HSPA6 were negatively associated with survival rate (Figs. 6B and S2A).
Infiltration of tumor immune cell populations in solid tumors can be estimated from the transcriptomic changes in the bulk tumor samples (44). TIMER2.0 (timer.cistrome.org/) is a user-friendly web interface for dynamic exploring and visualizing the interaction between tumor-infiltrating immune cells and important tumoral transcriptome changes using deconvolution-based approaches (45). The TIMER2.0 database was next searched to estimate the correlation between DDX41 mRNA expression and immune cell infiltration in CESC. As illustrated in the scatter plot (Fig. 6C), expression of DDX41 was positively correlated with the population of B, CD4+ Th1 and NK cells, macrophage M2 and myeloid DCs. The correlation between DDX41 expression and CD8+ T cells was not significant. Collectively, the association between DDX41 expression levels and tumor immune cell populations supported the hypothesis that DDX41-regulated expression of immune-associated and antigen presentation genes evident in HeLa may regulate immune cell infiltration and cancer immunity in patients with CESC.
DDX41 regulates the alternative splicing of certain immune-response genes
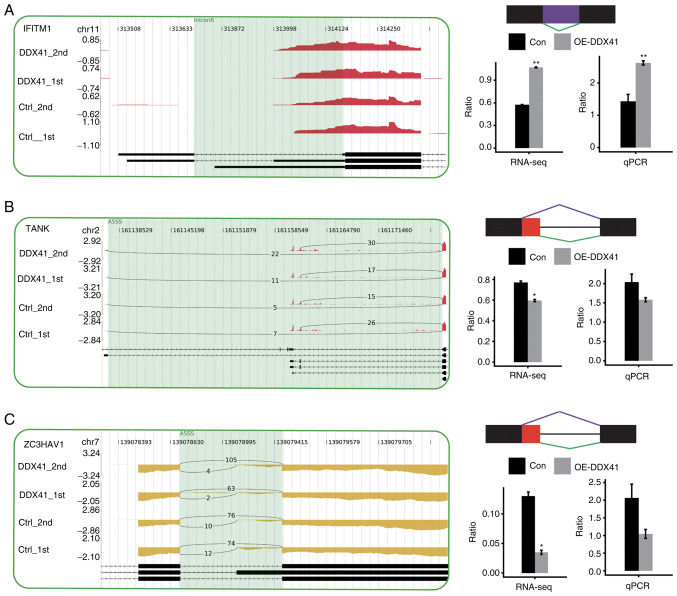
Genes regulated by DDX41-mediated alternative splicing were enriched in the ‘innate immune response’ GO pathway (MAP4K2, PGLYRP1, NFATC3, TNRC6B, RAF1, MAPKAP1, TANK, NLRX1, PLA2G6, NOD1, ZC3HAV1, PML, ATG9A, ERBB3, NR4A1, FGFR4, MLST8, FGFR1, LRRFIP1, CFD, UBA52, GRB2, PRKACB, CHID1, AGO3 and ATF2; Table SXII). Enriched KEGG pathways included those involved in ‘B cell receptor signaling pathway’ (GRB2, IFITM1, PPP3CC, RAF1 and NFATC3; Table SXIII). In order to validate the ASEs identified in the RNA-seq data, 10 potential ASEs were analyzed by RT-qPCR. Out of 11 tested events, four ASEs validated by RT-qPCR showed a significant change, which was in agreement with the RNA-seq results. The four validated splicing events were located in the genes IFITM1, CFD, NLRX1 and NFATC3 (Figs. 7 and S3).
Figure 7.
Validation of DDX41-regulated ASEs in the innate immune response. Integrative Genomics Viewer-sashimi plots showing alternative splicing changes that occurred in the control or DDX41 overexpression groups of HeLa cells. The results for IFITM1 (A), TANK (B) and ZC3HAV1 (C) are shown. Transcript isoforms and the density map of the distribution all RNA-seq reads around the gene regions involved in the ASEs from the four samples are shown. Alternative splicing isoforms of each ASE are depicted with the number of reads supporting each isoform shown. Schematic diagram depicts the structure of model splicing variant AS1 (purple) and alternative splicing variant AS2 (green). Boxes indicate exon sequences; horizontal line indicates intron sequences. RNA-seq quantification of alternative splicing regulation is shown. The changed ratio of AS events in RNA-seq was calculated using the formula: AS1 junction reads/AS1 junction reads + AS2 junction reads. The altered ratio of AS events in RT-PCR was calculated using the formula: AS1 transcripts level/AS2 transcripts level. Transcripts for genes are shown. *P<0.05 and **P<0.01. Con, control; OE-DDX41, overexpression DEAD-box helicase 41; RNA-seq, RNA-sequencing; qPCR, quantitative PCR; IntronR, intron retention; A5SS, alternative 5′ splice site.
Discussion
The present study demonstrated that DDX41 acted as a tumor suppressor by inhibiting cell proliferation and promoting apoptosis in HeLa cells. To the best of our knowledge, the present study is the first to profile changes in the HeLa cell transcriptome following DDX41 overexpression. A total of 959 DEGs were identified; GO/KEGG analysis of these DEGs revealed significant enrichment in ‘cell adhesion’, ‘angiogenesis’, ‘MAPK signaling pathway’ and ‘immune response’. Genes regulated by DDX41-mediated alternative splicing were highly enriched in ‘EGFR signaling pathway’ and ‘FGFR signaling pathway’. Identification and characterization of DDX41-regulated genes in cancer cells provided mechanistic insights into the link between tumorigenesis and cancer immunity, which may aid the development of novel therapeutic strategies against cervical and endocervical cancer (46,47).
A number of previously published works have indicated the diverse tumor cell type-specific roles of DDX41 in regulating tumorigenesis. For example, knockdown (KD) of DDX41 by short hairpin RNA promoted the proliferation of K562 cells, whereas DDX41 overexpression in U937 cells inhibited growth (16,48). Compared with that of WT DDX41 or DDX41-knockdown 293 cells, overexpression of DDX41R525H mutant increased cell soft agar colony formation (48). Similarly, DDX41 knockdown in CD34+ hematopoietic progenitor cells significantly enhanced colony formation compared with controls (16). An in vivo xenograft experiment of K562 cells in which DDX41 was knocked down showed accelerated tumor growth (16). Taken together, these studies support DDX41 as a tumor suppressor gene in multiple cell lines. The present results revealed that DDX41 inhibited proliferation and promoted apoptosis of HeLa cells and expression of DDX41 was associated with immune infiltration in CESC; this extends knowledge of the tumor suppressor function of DDX41 in cancer.
In the present study, DDX41 downregulated the expression levels of oncogenes LCN2, TP63, KIT, MYB and CDK5R2. Moreover, certain tumor suppressor genes were upregulated by DDX41, such as PANO1, GLI1 and HOMER2; expression of these genes was also deregulated and associated with survival rate in patients with CESC. p63 protein is a homologue of transcription factor p53, a tumor suppressor which serves key functions in tumorigenesis and oncogenesis (49,50). p63 protein is a useful immunohistochemical marker of differentiation of squamous neoplasms within the cervix and is a marker of premalignant lesions of the cervix used to predict malignant potential (51–54). The oncogenic roles of transcription factor MYB have been widely studied in different types of cancer including leukemia, colon and breast cancer and adenoid cystic carcinoma (55–57). Overexpression of MYB has been reported to be associated with poor prognosis in leukemogenesis (58), colorectal (59) and breast cancer (60) and Burkitt lymphoma (61). Therefore, DDX41 may function in tumorigenesis by regulating the expression levels of certain oncogenes and tumor suppressors.
In the present study, DDX41 regulated alternative splicing pre-mRNAs from genes enriched in cancer progression pathways, including ‘EGFR signaling’, ‘FGFR signaling’, ‘MAPK signaling’ and ‘prostate cancer’ pathways. Tumorigenesis-associated genes were FGFR1, FGFR4, NR4A1, CSK, MLST8, IGF1R and RRAF1. Alternative inclusion of one of two unique exons results in three versions of Ig-like domain III in FGFR 1–3 (62–64), but FGFR4 is not alternatively spliced in this region (65). Two FGFR1 IIIc splice variants (FGFR1a and FGFR1β) have been shown to be expressed at similar levels in normal urothelial cells, but FGFR1β is more highly expressed in tumor cells and results in increased proliferation (66). The FGFR IIIb isoform is primarily expressed in epithelial tissue, whereas the IIIc isoform is primarily expressed in mesenchymal tissue (67). Therefore, the FGF/FGFR axis may serve an important role in cancer progression (68,69). The present finding of DDX41-regulated alternative splicing of FGFR1 and FGFR4 suggested that DDX41 may regulate tumor progression via the FGF/FGFR axis.
GO/KEGG analysis revealed that DDX41 regulated the expression of genes enriched in immune-associated pathways including ‘leukocyte migration’, ‘cytokine-cytokine receptor interaction’, ‘antigen processing and presentation’ and ‘autoimmune disease’. This finding suggested a potential function of DDX41 in regulating cancer immunity. Among these pathways, the mRNA expression levels of CD22, CD177 and CSF2 were significantly downregulated and IL-12A was upregulated by DDX41. It has been reported that administration of IL-12 promotes the development of T helper type 1 CD4+ T cells, inducing rapid onset of insulin-dependent diabetes mellitus in non-obese diabetic mice (70). The interaction of CD22 with α2,6-linked sialic acid ligands has been proposed to regulate B lymphocyte function and migration (71). DDX41 selectively upregulated antigen processing and presentation genes including HSPA1A, HSPA1B and HSPA6. When these HSPs are released into the extracellular space, they act as a source of antigens due to their ability to chaperone peptides and induce DCs to cross-present antigens to T cells, and also stimulate the innate immune system independently of peptides (72). Castelli et al (73) reported that HSP70 purified from human melanoma activated T cells recognizing melanoma differentiation antigens by gaining access to the class I HLA presentation pathway. Noessner et al (74) reported that tumor-derived HSP70-peptide complexes have the immunogenic potential to instruct cross-presentation of antigenic peptide for T cell recognition by DCs. It has been suggested that combinatorial approaches encompassing DCs and tumor-associated antigens, such as HSP70, may be useful options for cancer therapies (75,76). The unique immune properties of HSP70 have enabled development of innovative prophylactic and therapeutic vaccines, particularly those for cervical and endocervical cancer (77).
DDX41 is an RNA helicase, which also acts as a dsDNA sensor in the cytoplasm, thus defending against pathogen invasion and activating the type 1 IFN response (22). Bruton's tyrosine kinase has been shown to phosphorylate Tyr414 of DDX41, which is critical for DDX41 recognition of DNA and binding to STING (78). In a zebrafish model, DDX41 contributed to STING-STAT6-mediated chemokine production via its DEADc domain (79). STING signaling is crucial for activation of transcription of type 1 IFN production genes, and therefore protects cells against pathogen invasion and the development of cancer by promoting antitumor immune response (80–82). The present results revealed that DDX41 regulated the expression of alternative splicing of hundreds of genes. Cytoplasmic and nuclear localization of DDX41 was also demonstrated. DDX41 may mediate gene regulation indirectly by affecting the function of its protein partners and/or directly by its RNA and DNA binding activity.
In summary, the present findings suggested that DDX41 selectively regulated the alternative splicing of numerous important genes involved in tumorigenesis, as well as expression of genes involved in the immune response in HeLa cells. The expression levels of DDX41-regulated cancer-associated genes were deregulated in cervical cancer samples and the expression of DDX41-regulated antigen processing and presentation genes was associated with survival rate of patients with CESC. These findings identified a DDX41-regulatory network connecting transcription and alternative splicing that predicted the biological functions of DDX41 in suppressing tumor cell growth, as well as regulating cross-presentation of antigens and other antitumor immune responses. Further study of DDX41 may lead to novel vaccine strategies in the treatment of cancer.
Supplementary Material
Acknowledgements
The authors would like to thank Professor. Yi Zhang (ABLife, Inc., Wuhan, China) for helpful discussions.
Funding Statement
The present study was supported by the Health Commission of Hubei Province Scientific Research Project (grant no. WJ2019M118) and ABLife (grant no. ABL-7702157).
Funding
The present study was supported by the Health Commission of Hubei Province Scientific Research Project (grant no. WJ2019M118) and ABLife (grant no. ABL-7702157).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information Gene Expression Omnibus repository (series accession no. GSE122986).
Authors' contributions
KQ, YZ and XLY designed and supervised the experiments. KQ, XLY, DNJ, YC, PZ, JZ, and HHX performed the experiments. YQX, YXW and YZ analyzed the data. KQ, YQX and YZ drafted the manuscript. All authors reviewed, read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jankowsky E, editor. RNA helicases. 1st edition. Vol. 511. Academic Press; 2012. [Google Scholar]
- 2.Tanner NK, Linder P. DExD/H Box RNA helicases: From generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol. 2016;17:426–438. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 4.Rocak S, Linder P. DEAD-Box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 5.Jankowsky E. RNA helicases at work: Binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in wnt-β-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 8.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Mosallanejad K, Sekine Y, Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A, Takeda K, Naguro I, Ichijo H. The DEAH-Box RNA helicase DHX15 activates NF-κB and MAPK signaling downstream of MAVS during antiviral responses. Sci Signal. 2014;7:ra40. doi: 10.1126/scisignal.2004841. [DOI] [PubMed] [Google Scholar]
- 10.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121–132. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis R, Jerry P. Perturbations of RNA helicases in cancer. Wiley Interdiscip Rev RNA. 2013;4:333–349. doi: 10.1002/wrna.1163. [DOI] [PubMed] [Google Scholar]
- 12.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Cai W, Chen ZX, Rane G, Singh SS, Choo Z, Wang C, Yuan Y, Tan TZ, Arfuso F, Yap CT, et al. Wanted DEAD/H or alive: Helicases winding up in cancers. J Natl Cancer Inst. 2017;25:109. doi: 10.1093/jnci/djw278. [DOI] [PubMed] [Google Scholar]
- 14.Chlon TM, Stepanchick E, Choi K, Zheng Y, Hueneman K, Davis A, Starczynowski DT. The inherited MDS gene DDX41 is required for ribosome biogenesis and cell viability. Blood. 2019;134(Suppl 1):S773. doi: 10.1182/blood-2019-127471. [DOI] [Google Scholar]
- 15.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venugopal P, Cheah JJC, Eshraghi L, Shahrin NH, Homan C, Feng J, Schreiber AW, Fine M, Phillips K, Poplawski N, et al. An integrative genomic approach to examine mutations and biological pathways associated with hematological malignancy development in DDX41 mutated families. Blood. 2019;134(Suppl 1):S2686. doi: 10.1182/blood-2019-131649. [DOI] [Google Scholar]
- 18.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol. 2017;106:163–174. doi: 10.1007/s12185-017-2260-y. [DOI] [PubMed] [Google Scholar]
- 19.Peters D, Radine C, Reese A, Budach W, Sohn D, Jänicke RU. The DEAD-box RNA helicase DDX41 is a novel repressor of p21 WAF1/CIP1 mRNA translation. J Biol Chem. 2017;292:8331–8341. doi: 10.1074/jbc.M116.772327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stavrou S, Aguilera AN, Blouch K, Ross SR. DDX41 recognizes RNA/DNA retroviral reverse transcripts and is critical for in vivo control of murine leukemia virus infection. mBio. 2018;9:e00923–18. doi: 10.1128/mBio.00923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan Y, Zeng J, Fan S, Liao Y, Feng M, Wang L, Zhang Y, Li Q. Herpes simplex virus type 1-encoded miR-H2-3p manipulates cytosolic DNA-stimulated antiviral innate immune response by targeting DDX41. Viruses. 2019;15:756. doi: 10.3390/v11080756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura T, Miyabe H, Hyodo M, Sato Y, Hayakawa Y, Harashima H. Liposomes loaded with a STING pathway ligand, cyclic di-GMP, enhance cancer immunotherapy against metastatic melanoma. J Control Release. 2015;216:149–157. doi: 10.1016/j.jconrel.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 25.Miyabe H, Hyodo M, Nakamura T, Sato Y, Hayakawa Y, Harashima H. A new adjuvant delivery system ‘cyclic di-GMP/YSK05 liposome’ for cancer immunotherapy. J Control Release. 2014;184:20–27. doi: 10.1016/j.jconrel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol. 2013;10:83–95. doi: 10.4161/rna.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/S1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 29.Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39((Web Server Issue)):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech. 2017;1860:911–921. doi: 10.1016/j.bbagrm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Pryke A, Mostaghim S, Nazemi A. Heatmap visualization of population based multi objective algorithms. In: Obayashi S, Deb K, Poloni C, Hiroyasu T, Murata T, editors. Evolutionary multi-criterion optimization. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. pp. 361–375. [DOI] [Google Scholar]
- 35.Katz Y, Wang ET, Silterra J, Schwartz S, Wong B, Thorvaldsdóttir H, Robinson JT, Mesirov JP, Airoldi EM, Burge CB. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015;31:2400–2402. doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44(D1):D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 2013;7:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Qi H, Li X, Hou X, Lu X, Xiao X. A novel dithiocarbamate derivative induces cell apoptosis through p53-dependent intrinsic pathway and suppresses the expression of the E6 oncogene of human papillomavirus 18 in HeLa cells. Apoptosis. 2015;20:787–795. doi: 10.1007/s10495-015-1114-4. [DOI] [PubMed] [Google Scholar]
- 41.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 42.Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat Immunol. 2018;19:923–931. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- 43.Rock KL, Reits E, Neefjes J. Present yourself! by MHC class I and MHC class II molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48((W1)):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flood BA, Higgs EF, Li S, Luke JJ, Gajewski TF. STING pathway agonism as a cancer therapeutic. Immunol Rev. 2019;290:24–38. doi: 10.1111/imr.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneyama-Hirozane M, Kondo M, Matsumoto SI, Morikawa-Oki A, Morishita D, Nakanishi A, Kawamoto T, Nakayama M. High-throughput screening to identify inhibitors of DEAD box helicase DDX41. SLAS Discov. 2017;22:1084–1092. doi: 10.1177/2472555217705952. [DOI] [PubMed] [Google Scholar]
- 48.Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen BP, Hosono N, Singh J, Padgett RA, Gu X, Jankowsky E, et al. DDX41 is a tumor suppressor gene associated with inherited and acquired mutations. Blood. 2014;124:125. doi: 10.1182/blood.V124.21.125.125. [DOI] [Google Scholar]
- 49.Nekulova M, Holcakova J, Coates P, Vojtesek BJC, Letters MB. The role of P63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett. 2011;16:296–327. doi: 10.2478/s11658-011-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graziano V, De Laurenzi V. Role of p63 in cancer development. Biochim Biophys Acta. 2011;1816:57–66. doi: 10.1016/j.bbcan.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Wang TY, Chen BF, Yang YC, Chen H, Wang Y, Cviko A, Quade BJ, Sun D, Yang A, McKeon FD, Crum CP. Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: A study of 250 cases. Hum Pathol. 2001;32:479–486. doi: 10.1053/hupa.2001.24324. [DOI] [PubMed] [Google Scholar]
- 52.McCluggage WG. Immunohistochemistry as a diagnostic aid in cervical pathology. Pathology. 2007;39:97–111. doi: 10.1080/00313020601123961. [DOI] [PubMed] [Google Scholar]
- 53.Houghton O, McCluggage WG. The expression and diagnostic utility of p63 in the female genital tract. Adv Anat Pathol. 2009;16:316–321. doi: 10.1097/PAP.0b013e3181b507c6. [DOI] [PubMed] [Google Scholar]
- 54.Saritha VN, Veena VS, Jagathnath KM, Somanathan T, Sujathan K. Significance of DNA replication licensing proteins (MCM2, MCM5 and CDC6), p16 and p63 as markers of premalignant lesions of the uterine cervix: Its usefulness to predict malignant potential. Asian Pac J Cancer Prev. 2018;27:141–148. doi: 10.22034/APJCP.2018.19.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pattabiraman D, Gonda T. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 56.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 57.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, Carey CD, Rodig SJ, Sholl LM, Afrogheh AH, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48:265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarvaiya PJ, Schwartz JR, Hernandez CP, Rodriguez PC, Vedeckis WV. Role of c-myb in the survival of pre B-cell acute lymphoblastic leukemia and leukemogenesis. Am J Hematol. 2012;87:969–976. doi: 10.1002/ajh.23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biroccio A, Benassi B, D'Agnano I, D'Angelo C, Buglioni S, Mottolese M, Ricciotti A, Citro G, Cosimelli M, Ramsay RG, et al. C-Myb and bcl-x overexpression predicts poor prognosis in colorectal cancer: Clinical and experimental findings. Am J Pathol. 2001;158:1289–1299. doi: 10.1016/S0002-9440(10)64080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knopfová L, Biglieri E, Volodko N, Masařík M, Hermanová M, Garzón JF, Dúcka M, Kučírková T, Souček K, Šmarda J, et al. Transcription factor c-myb inhibits breast cancer lung metastasis by suppression of tumor cell seeding. Oncogene. 2017;37:1020–1030. doi: 10.1038/onc.2017.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma M, Zhao R, Yang X, Zhao L, Liu L, Zhang C, Wang X, Shan B. Low expression of Mda-7/IL-24 and high expression of C-myb in tumour tissues are predictors of poor prognosis for burkitt lymphoma patients. Hematology. 2018;23:448–455. doi: 10.1080/10245332.2018.1435046. [DOI] [PubMed] [Google Scholar]
- 62.Werner S, Duan DS, de Vries C, Peters KG, Johnson DE, Williams LT. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol Cell Biol. 1992;12:82–88. doi: 10.1128/MCB.12.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson DE, Lu J, Chen H, Werner S, Williams LT. The human fibroblast growth factor receptor genes: A common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/MCB.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz M. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–11627. doi: 10.1016/S0021-9258(19)78170-8. [DOI] [PubMed] [Google Scholar]
- 65.Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C, Jaye M, Alitalo K. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;12:4273–4280. doi: 10.1002/j.1460-2075.1992.tb05526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomlinson DC, Knowles MA. Altered splicing of FGFR1 is associated with high tumor grade and stage and leads to increased sensitivity to FGF1 in bladder cancer. Am J Pathol. 2010;177:2379–2386. doi: 10.2353/ajpath.2010.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang S, Hao Y, Yuan Y, Liu R, Chen Q. Role of fibroblast growth factor receptor 4 in cancer. Cancer Sci. 2018;109:3024–3031. doi: 10.1111/cas.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Touat M, Ileana E, Postel-Vinay S, André F, Soria JC. Targeting FGFR signaling in cancer. Clin Cancer Res. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 69.Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17:318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 70.Trembleau S, Penna G, Bosi E, Mortara A, Gately MK, Adorini L. Interleukin 12 administration induces T helper type 1 cells and accelerates autoimmune diabetes in NOD mice. J Exp Med. 1995;181:817–821. doi: 10.1084/jem.181.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poe JC, Fujimoto Y, Hasegawa M, Haas KM, Miller AS, Sanford IG, Bock CB, Fujimoto M, Tedder TF. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nat Immunol. 2004;5:1078–1087. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 72.Milani V, Noessner E, Ghose S, Kuppner M, Ahrens B, Scharner A, Gastpar R, Issels RD. Heat shock protein 70: Role in antigen presentation and immune stimulation. Int J Hyperthermia. 2002;18:563–575. doi: 10.1080/02656730210166140. [DOI] [PubMed] [Google Scholar]
- 73.Castelli C, Ciupitu AM, Rini F, Rivoltini L, Mazzocchi A, Kiessling R, Parmiani G. Human heat shock protein 70 peptide complexes specifically activate antimelanoma T cells. Cancer Res. 2001;61:222–227. [PubMed] [Google Scholar]
- 74.Noessner E, Gastpar R, Milani V, Brandl A, Hutzler PJ, Kuppner MC, Roos M, Kremmer E, Asea A, Calderwood SK, Issels RD. Tumor-Derived heat shock protein 70 peptide complexes are cross-presented by human dendritic cells. J Immunol. 2002;169:5424–5432. doi: 10.4049/jimmunol.169.10.5424. [DOI] [PubMed] [Google Scholar]
- 75.Banchereau J, Palucka AK. Dendritic cells as therapeutic vaccines against cancer. Nat Rev Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 76.Noessner E. Thermal stress-related modulation of tumor cell physiology and immune responses. Cancer Immunol Immunother. 2006;55:289–291. doi: 10.1007/s00262-005-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, Sapozhnikov AM. HSP70 multi-functionality in cancer. Cells. 2020;9:587. doi: 10.3390/cells9030587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee KG, Susana SY, Kui L, Chih-Cheng Voon D, Mauduit M, Bist P, Bi X, Pereira NA, Liu C, Sukumaran B, et al. Bruton's tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep. 2015;10:1055–1065. doi: 10.1016/j.celrep.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 79.Ma JX, Li JY, Fan DD, Feng W, Lin AF, Xiang LX, Shao JZ. Identification of DEAD-box RNA helicase DDX41 as a trafficking protein that involves in multiple innate immune signaling pathways in a zebrafish model. Front Immunol. 2018;9:1327. doi: 10.3389/fimmu.2018.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woo SR, Corrales L, Gajewski TF. The STING pathway and the T cell-inflamed tumor microenvironment. Trends Immunol. 2015;36:250–256. doi: 10.1016/j.it.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barber GN. STING-Dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Barber GN. STING: Infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information Gene Expression Omnibus repository (series accession no. GSE122986).