ABSTRACT
The regulated expansion of chondrocytes within growth plates and joints ensures proper skeletal development through adulthood. Mutations in the transcription factor NKX3.2 underlie spondylo-megaepiphyseal-metaphyseal dysplasia (SMMD), which is characterized by skeletal defects including scoliosis, large epiphyses, wide growth plates and supernumerary distal limb joints. Whereas nkx3.2 knockdown zebrafish and mouse Nkx3.2 mutants display embryonic lethal jaw joint fusions and skeletal reductions, respectively, they lack the skeletal overgrowth seen in SMMD patients. Here, we report adult viable nkx3.2 mutant zebrafish displaying cartilage overgrowth in place of a missing jaw joint, as well as severe dysmorphologies of the facial skeleton, skullcap and spine. In contrast, cartilage overgrowth and scoliosis are absent in rare viable nkx3.2 knockdown animals that lack jaw joints, supporting post-embryonic roles for Nkx3.2. Single-cell RNA-sequencing and in vivo validation reveal increased proliferation and upregulation of stress-induced pathways, including prostaglandin synthases, in mutant chondrocytes. By generating a zebrafish model for the skeletal overgrowth defects of SMMD, we reveal post-embryonic roles for Nkx3.2 in dampening proliferation and buffering the stress response in joint-associated chondrocytes.
KEY WORDS: Nkx3.2, Chondrocyte, Proliferation, Joint, Spine, Zebrafish
Summary: Characterization of a zebrafish model of SMMD reveals that the transcription factor Nkx3.2 fine-tunes chondrocyte proliferation at skeletal joints versus endochondral growth plates by modulating stress-response and mTORC1 pathways.
INTRODUCTION
In much of the developing vertebrate body, cartilage is a transient tissue that progressively remodels to bone through a process known as endochondral ossification. In other contexts, such as cartilage of the nose and ear and the articular cartilage lining the bony surfaces of healthy joints, cartilage is permanent. The growth and function of both transient and permanent cartilage relies on the stratification of chondrocytes into distinct zones. In the growth plates of endochondral bones, chondrocytes are arranged into a resting zone containing stem cells (Mizuhashi et al., 2018; Newton et al., 2019), a zone of round proliferative chondrocytes, a zone of proliferative flattened chondrocytes that merges into a pre-hypertrophic zone, a hypertrophic zone in which chondrocytes enlarge and calcify, and a transitional zone in which chondrocytes undergo apoptosis or transdifferentiate into osteoblasts (Giovannone et al., 2019; Jing et al., 2015; Kronenberg, 2003; Yang et al., 2014; Zhou et al., 2014). Joint cartilage is also zonally arranged: superficial chondrocytes of a flattened morphology line the synovial cavity and produce specialized molecules such as Prg4 (lubricin) (Askary et al., 2016; Kozhemyakina et al., 2015), with deeper chondrocytes arranged in zones reminiscent of growth plates and transitioning into the underlying subchondral bone (Lui et al., 2015). Defects in the specification and maintenance of these cartilage zones can lead to dwarfism and other skeletal dysplasias in the context of the growth plate, and arthritis in the context of the joint. The mechanisms for maintaining the correct proportions and identities of cartilage layers at either growth plates or joints remain, however, incompletely understood. In particular, we still know little about how the proliferative expansion of cartilage is zonally regulated to meet the differing demands of endochondral bone and joint growth.
Humans with spondylo-megaepiphyseal-metaphyseal dysplasia (SMMD; OMIM #613330), a rare skeletal dysplasia linked to homozygous frameshift mutations in the homeobox transcription factor NKX3.2 (also called BAPX1 or NKX3-2), display skeletal overgrowth in the wrists and digits that is accompanied by supernumerary bones (pseudoepiphyses) as well as scoliosis (Hellemans et al., 2009). In these patients, the cartilaginous growth plates of long bones are abnormally wide. Whereas work in animal models has revealed requirements for Nkx3.2 in regulating chondrocyte development within both the growth plate and joints, the skeletal overgrowth seen in SMMD has not been observed. In chick and mouse, Nkx3.2 is expressed in the proliferating and pre-hypertrophic chondrocytes of the growth plate but largely excluded from the less proliferative hypertrophic zone and articular surfaces of joints (Church et al., 2005; Provot et al., 2006; Tribioli et al., 1997). Nkx3.2−/− mice have defects in their vertebrae and cranial skeleton, which have been attributed to decreased cartilage formation (Akazawa et al., 2000; Tribioli and Lufkin, 1999) and is exacerbated by further mutation of the paralog Nkx3.1 (Herbrand et al., 2002). However, despite expression of Nkx3.2 in the long bones of the developing limbs, Nkx3.2−/− mice do not display the elongated wrists and digits of SMMD patients at birth. As these mice die shortly after birth, possibly because of asplenia, the lack of limb overgrowth phenotypes could reflect postnatal roles for Nkx3.2 in restricting cartilage growth. Nkx3.2 is thought to form a positive regulatory loop with Sox9 (Yamashita et al., 2009; Zeng et al., 2002), a master regulator of chondrogenesis (Bi et al., 1999), consistent with their co-expression in growth plate chondrocytes (Provot et al., 2006). Studies in the chick limb and mammalian cell culture have suggested that Nkx3.2 may delay hypertrophic maturation of growth plate chondrocytes through repression of Col10a1 (Provot et al., 2006) and Runx2 (Kawato et al., 2011; Yamashita et al., 2009), a transcription factor required for the differentiation of cells into mineral-producing osteoblasts and hypertrophic chondrocytes (Takeda et al., 2001).
In addition to roles in growth plate regulation, work in zebrafish has revealed essential roles for Nkx3.2 in formation of the jaw joint (Miller et al., 2003). Inhibition of Nkx3.2 function with an antisense morpholino results in a complete loss of the zebrafish jaw joint, including the bony retroarticular process that is a site of insertion for the jaw adductor (adductor mandibulae). The role of Nkx3.2 in specifying the jaw joint appears to be shared between zebrafish and amphibians but not mammals, potentially due to positive regulation of joint-promoting Gdf5/6 family members only in non-mammalian species (Lukas and Olsson, 2018). Although the malleus-incus joint (the evolutionary homolog of the zebrafish jaw joint) or the temporomandibular joint (the mammalian jaw joint) are normal in Nkx3.2−/− mice, the malleus is narrower and the associated gonial bone is lost (Tucker et al., 2004).
Here, we characterize a zebrafish nkx3.2 mutant that lacks the jaw joint (see also Miyashita et al., 2020 for a detailed analysis of adult cranial defects). Similar to SMMD patients, mutants are adult viable and develop cartilage outgrowths, albeit in the jaw joint region rather than wrists and digits, as well as scoliosis. By comparing mutants to animals with transient, early knockdown of nkx3.2 function, we find that Nkx3.2 functions at post-embryonic stages to prevent cartilage overgrowth and pattern the spine. Single-cell RNA-seq of facial chondrocytes reveals a mutant-enriched population of chondrocytes with high levels of cell cycle and stress-induced pathway genes, including the prostaglandin D2 synthase gene ptgdsb.1 and the mTORC1 pathway genes sestrin1 (sesn1) (Budanov and Karin, 2008) and eukaryotic translation initiation factor 4E binding protein 3 (eif4ebp3) (Tsukumo et al., 2016; Yogev et al., 2013). We validate that, in place of the missing jaw joint, adult nkx3.2 mutants develop abnormally proliferative chondrocytes with elevated levels of ptgdsb.1 and sesn1, yet do not upregulate genes associated with hypertrophic maturation. Rather than suppressing hypertrophic maturation, our results indicate post-embryonic roles for Nkx3.2 in restricting chondrocyte proliferation, possibly linked to prostaglandin and mTORC1 regulation, to ensure proper skeletal proportions, including in the limbs of SMMD patients.
RESULTS
Mutant nkx3.2 zebrafish lack a jaw joint and display skull and spine defects as adults
Given concerns over efficacy and specificity of morpholino knockdown (Eve et al., 2017; Gentsch et al., 2018; Joris et al., 2017; Kok et al., 2015; Law and Sargent, 2014), we first aimed to confirm requirements for nkx3.2 in zebrafish jaw joint development (Miller et al., 2003) using genetic mutants. To do so, we generated two mutant alleles, nkx3.2el802 and nkx3.2ua5011 using TALEN (Huang et al., 2011; Sander et al., 2011) and CRISPR (Hwang et al., 2013) mutagenesis, respectively (Fig. 1A). nkx3.2el802 contains a 14 bp deletion that removes the ‘A’ of the start codon and is predicted to prevent translation of nkx3.2 mRNA, and nkx3.2ua5011 introduces a 20 bp deletion and frameshift mutation predicted to disrupt the DNA-binding homeobox domain. These are both predicted to be severe loss-of-function alleles; lack of a zebrafish-reactive Nkx3.2 antibody did not allow us to test this directly.
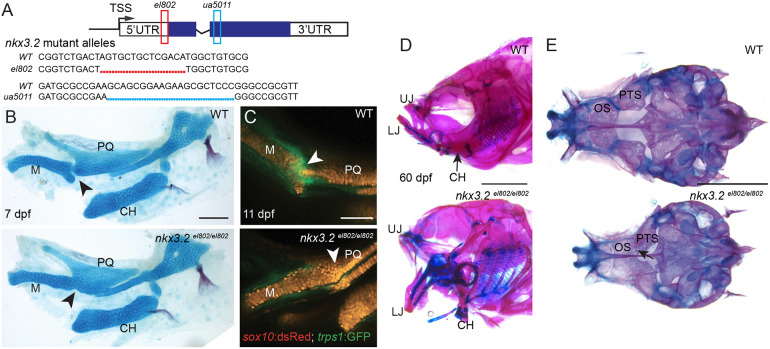
Fig. 1.
nkx3.2 mutant zebrafish are adult viable and develop craniofacial skeletal abnormalities. (A) Schematic of nkx3.2 mutant alleles: el802 – 14 bp deleted sequence including the ‘A’ of the ATG at the nkx3.2 translation start codon; ua5011 – 20 bp deleted sequence at the start of the homeobox DNA-binding domain. Coding regions of exons shown in blue and non-coding regions in white. (B) Lateral views of pharyngeal skeletal preparations stained with Alcian Blue (cartilage) and Alizarin Red (bone) show fusions of the jaw joint (arrowheads) in nkx3.2 mutants at 7 dpf. (C) Live imaging of transgenic juvenile zebrafish demonstrates loss of sox10:DsRed+/trps1:GFP+ jaw joint chondrocytes (arrowheads) in nkx3.2 mutants at 11 dpf. (D,E) Lateral views of the facial skeleton (D) and ventral views of the neurocranium (E) in adult nkx3.2 mutants at 60 dpf show craniofacial skeletal abnormalities including gaping jaw, fused jaw joint and displaced ceratohyal, and fusion of neurocranial orbitosphenoid and pterosphenoid bones (arrow in E). CH, ceratohyal; LJ, lower jaw; M, Meckel's cartilage; OS, orbitosphenoid; PQ, palatoquadrate; PTS, pterosphenoid; UJ, upper jaw; WT, wild type. Scale bars: 100 µm (B,C); 2 mm (D,E).
Consistent with the nkx3.2 morpholino knockdown phenotype (Miller et al., 2003), animals homozygous for either nkx3.2 mutant allele fail to develop a jaw joint, as assessed by Alcian Blue labeling of cartilage at 7 days post-fertilization (dpf) (n=3/3; Fig. 1B; Miyashita et al., 2020). In wild-type animals at 11 dpf, trps1:GFP marks articular chondrocytes that are flanked by sox10:dsRed+ chondrocytes of the upper and lower jaw cartilages. Live imaging of nkx3.2 mutant embryos revealed loss of trps1:GFP expression and a continuous field of sox10:dsRed+ chondrocytes across the presumptive jaw joint region (n=5/5; Fig. 1C). In contrast to the vast majority of nkx3.2 morpholino-treated embryos that die by 7 dpf, zebrafish homozygous for either mutant allele are viable and fertile into adulthood, especially when raised separately from their wild-type siblings, and display a fixed open jaw (n=6/6; Fig. 1D; Miyashita et al., 2020).
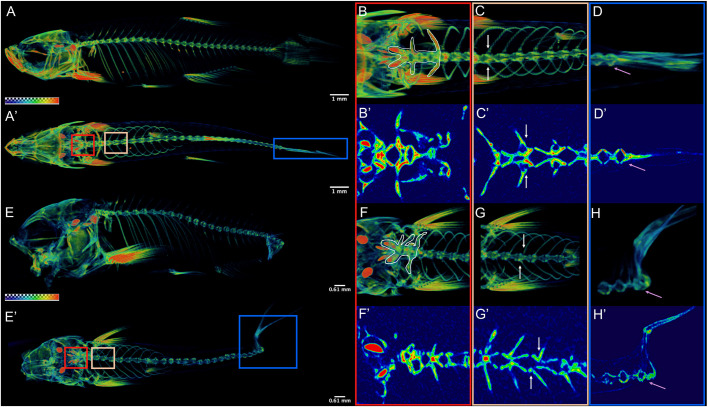
Adult nkx3.2 mutant zebrafish display a number of other cranial skeletal defects beyond jaw joint loss, as well as vertebral abnormalities. Mutants display altered orientation of facial bones (e.g. ceratohyal), possibly due to mechanical restriction from jaw joint fusion (n=6/6; Fig. 1D). The paired pterosphenoid and orbitosphenoid bones are articulating endochondral bones of the neurocranium (Cubbage and Mabee, 1996) and these are abnormally fused in nkx3.2 mutants (n=6/6; Fig. 1E). Micro-computed tomography (µCT) imaging further reveals that 11/11 nkx3.2 mutants have abnormal ossification and a shortening of the rostral spine (Fig. 2B,F), misalignment of ribs (Fig. 2C,G) and rotation and fusion of the caudal vertebrae (Fig. 2D,H) at 60 dpf, with spinal defects apparent as early as 30 dpf (n=11/11; Fig. S1). Overall, the combined rostral defects and abnormal curvature are associated with shortening of the spine (Fig. 2A,E). Spinal deformities are consistent with the reported expression of nkx3.2 in the neural and hemal arches that contribute to vertebrae (Crotwell et al., 2007), as well as the scoliosis and shortening of the neck in SMMD patients.
Fig. 2.
nkx3.2 mutant zebrafish develop axial skeletal abnormalities. (A,E) µCT imaging of 60 dpf wild types (A,A′) and nkx3.2ua5011/ua5011 mutants (E,E′) shown with gradient 3D rendering in sagittal (A,E) and dorsal (A′,E′) views. (B-D,F-H) Magnifications of three spinal regions: anterior vertebrae (red box, inclusive of the occiput and Weberian apparatus: B,B′,F,F′), precaudal vertebrae with rib articulations (tan box, white arrows indicate corresponding left and right ribs: C,C′,G,G′), and caudal fin vertebrae (blue box, pink arrow indicates caudal-most vertebra: D,D′,H,H′) shown in 3D (B,C,D,F,G,H) and 2D (B′,C′,D′,F′,G′,H′) slice. Scale bars: 1 mm (A,A′); 0.61 mm (E,E′).
Ectopic jaw cartilage overgrowth in nkx3.2 mutants
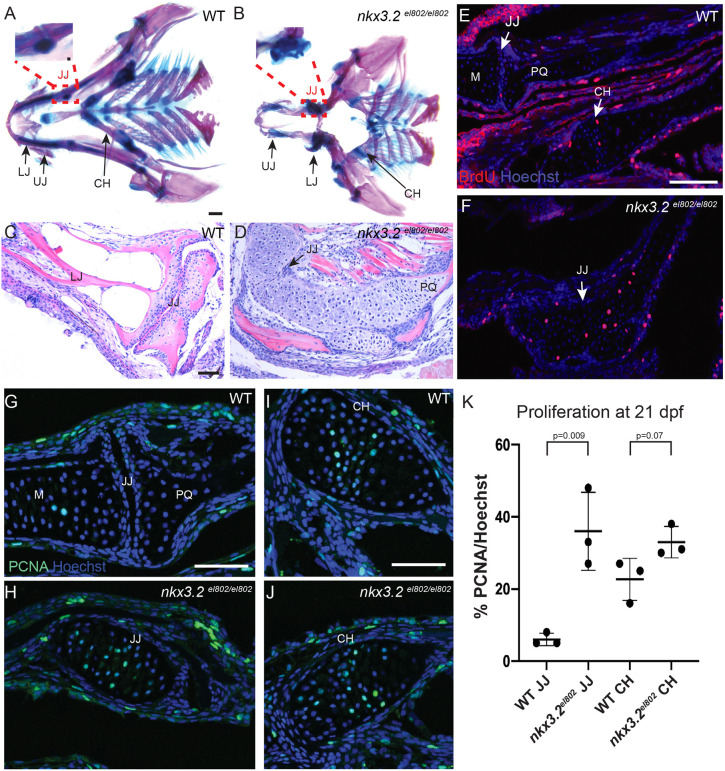
We next examined the long-term fate of nkx3.2 mutant cells that would have normally formed the jaw joint. In whole-mount preparations of the 60 dpf skull stained with Alcian Blue (cartilage) and Alizarin Red (bone), we observed greatly increased Alcian-positive tissue in the region where the jaw joint would have been (n=6/6; Fig. 1D). This cartilage overgrowth was particularly evident upon dissection of the presumptive jaw joint tissue (insets in Fig. 3A,B). Hematoxylin and Eosin (H&E) staining of frontal sections through the presumptive jaw joint region further demonstrated a dramatic expansion of cartilage tissue in mutants (n=3/3; Fig. 3C,D). We next assessed proliferation in the presumptive jaw joint region at 21 dpf by subjecting animals to a 1 h pulse of bromodeoxyuridine (BrdU), which is incorporated into newly replicating DNA (Fig. 3E,F). In 3/3 wild types, we observed very few BrdU+ cells near the jaw joint, and sparse BrdU+ chondrocytes in the ceratohyal growth plate. In 3/3 mutants, we observed increased numbers of BrdU+ chondrocytes in place of the missing jaw joint, consistent with cartilage overgrowth. To validate the BrdU incorporation analysis, we also assessed expression of Proliferating Cell Nuclear Antigen (PCNA), a marker for DNA replication, in chondrocytes (Fig. 3G-J). Around wild-type joints, we observed a few PCNA+ cells within the lower jaw Meckel's cartilage, but almost no PCNA+ cells at the joint articular surface. PCNA+ chondrocytes were more abundant in the proliferative zone of the ceratohyal growth plate. In nkx3.2 mutants, numerous PCNA+ chondrocytes are evident in the fused jaw joint region. Quantification of the percentage of chondrocytes expressing PCNA (Fig. 3K) revealed a dramatic increase in proliferative chondrocytes at the nkx3.2 mutant jaw joint region (identified by nkx3.2 expression in adjacent sections) compared with the wild-type jaw joint (n=3 per genotype, P=0.009), similar to the levels of proliferative chondrocytes within the wild-type ceratohyal growth plate. We also observed a modest increase in proliferation rates within the mutant ceratohyal growth plate (n=3 per genotype, P=0.07). These findings indicate that chondrocytes in the presumptive jaw joint region, and likely to a lesser extent in the ceratohyal growth plate, become increasingly proliferative in nkx3.2 mutants, likely contributing to the cartilage overgrowth seen in juvenile and adult mutants.
Fig. 3.
Cartilage overgrowth at the jaw joint in nkx3.2 mutant zebrafish. (A,B) Ventral views of dissected pharyngeal skeletons stained with Alcian Blue (cartilage) and Alizarin Red (bone) at 60 dpf. Insets with further dissections of the boxed regions in A and B demonstrate the wild-type retroarticular cartilage in A and the cartilage overgrowth at the site of the fused jaw joint in nkx3.2 mutants in B. (C,D) Frontal H&E-stained sections through 60 dpf jaw joints show fusion of the jaw joint and cartilage overgrowth in nkx3.2 mutants. (E-J) In wild types at 21 dpf (E,G,I), PCNA+ cells are present largely on the lower jaw side of the jaw joint, and PCNA+ and BrdU+ chondrocytes are present in the proliferative zone of the ceratohyal. In nkx3.2el802 mutants (F,H,J), proliferating chondrocytes marked by BrdU (E,F) and PCNA (G-J) are present across the fused jaw joint region. (K) Quantification of PCNA+ cells per total nuclei stained with Hoechst. Individual animals are plotted (dots) with mean±s.d. CH, ceratohyal; JJ, jaw joint; LJ, lower jaw; M, Meckel's cartilage; PQ, palatoquadrate; UJ, upper jaw; WT, wild type. Scale bars: 500 µm (A,B); 50 µm (C-J).
Nkx3.2 function is required in post-embryonic stages to suppress cartilage overgrowth
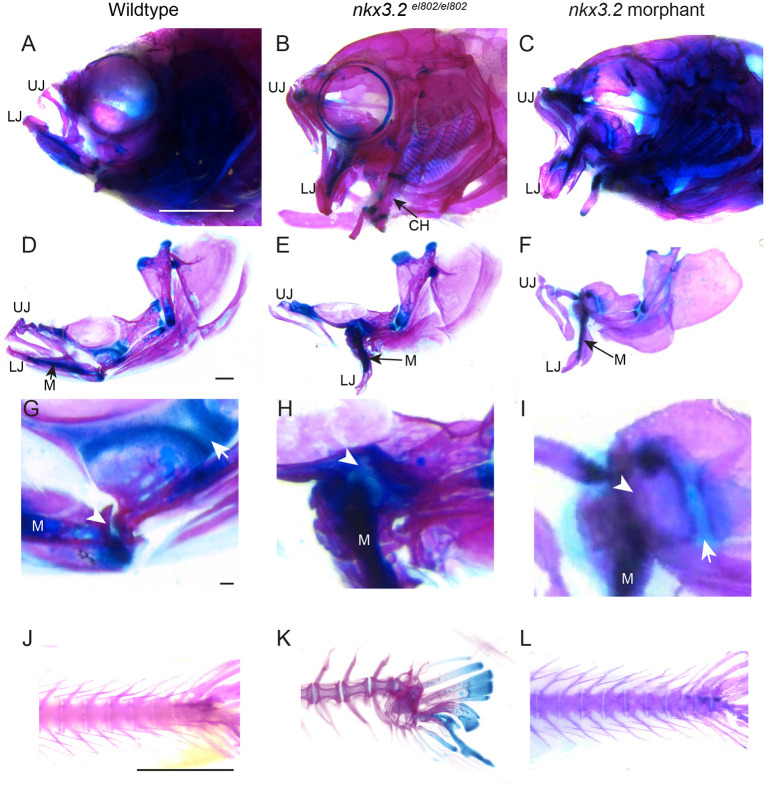
We next sought to determine whether the cartilage overgrowth seen in adult nkx3.2 mutants was solely the consequence of not forming the jaw joint, or alternatively reflected a continued requirement for nkx3.2 in chondrocyte regulation. To uncouple early versus continual requirements for nkx3.2, we took advantage of the transient nature of antisense morpholino knockdown – dilution during embryonic cell divisions is thought to limit the window of efficacy of most morpholinos to the first few days of embryogenesis (Eisen and Smith, 2008). As described earlier, the jaw joint loss seen in both nkx3.2 mutant alleles is nearly identical to that reported for nkx3.2 morpholino knockdown (Miller et al., 2003). We therefore injected one-cell-stage embryos with the same dose of nkx3.2 morpholino reported by Miller et al. (2003). Using sox10:dsRed to visualize chondrocytes in live animals, we confirmed loss of the jaw joint in the majority of morpholino-injected animals at 7 dpf and only raised animals lacking jaw joints to adulthood. From 225 animals lacking the embryonic jaw joint, only four inflated their swim bladders and survived until adulthood (n=2 at 60 dpf, n=2 at 90 dpf), in marked contrast to the viability of jaw-joint-less nkx3.2 mutants. The four viable morpholino-injected adults exhibited a similar open gape of the jaw as seen in nkx3.2 mutants (Fig. 4A-F). In wild types, permanent Meckel's cartilage within the lower jaw is separated from the cartilaginous growth plate of the upper jaw palatoquadrate bone by the jaw joint and associated subchondral bone (Fig. 4D,G). In mutants, we observed fusion of lower jaw cartilage to the palatoquadrate cartilage growth plate (Fig. 4H; see also Fig. 3D). In contrast, in morpholino-injected animals, bone continues to separate lower jaw cartilage and the palatoquadrate cartilage growth plate despite the lack of a jaw joint, and no cartilage overgrowth was observed in any of the four adult-viable animals (Fig. 4I). Whereas 5/6 nkx3.2el802 mutant animals displayed severe caudal tail curvature defects (Fig. 4K), all four morpholino-injected adults had normal spines and tails (Fig. 4J-L). These findings indicate post-embryonic roles for Nkx3.2 in restricting joint-associated cartilage growth and ensuring normal spine development.
Fig. 4.
Adult nkx3.2 morphants lack the cartilage overgrowth and spine defects of nkx3.2 mutants. (A-I) Lateral views of whole-mount (A-C) and dissected (D-F) facial skeletons at 60 dpf. Alcian Blue (cartilage) and Alizarin Red (bone) staining show similar gaping jaws in mutant and morphant animals. High magnification views of the jaw joint (G-I) show ectopic cartilage across the fused jaw joint domain (arrowheads) in mutants (H) but not morphants (I). Arrows indicate distinct palatoquadrate growth plates present in wild types and morphants but not in mutants. (J-L) Lateral views of the caudal spine stained with Alcian Blue and Alizarin Red show spinal curvature in mutants but not in morphants. CH, ceratohyal; LJ, lower jaw; M, Meckel's cartilage; UJ, upper jaw. Scale bars: 2 mm (A-C; J-L); 500 µm (D-F); 50 µm (G-I).
Single-cell RNA-seq reveals upregulated mitotic and stress response genes in nkx3.2 mutant chondrocytes
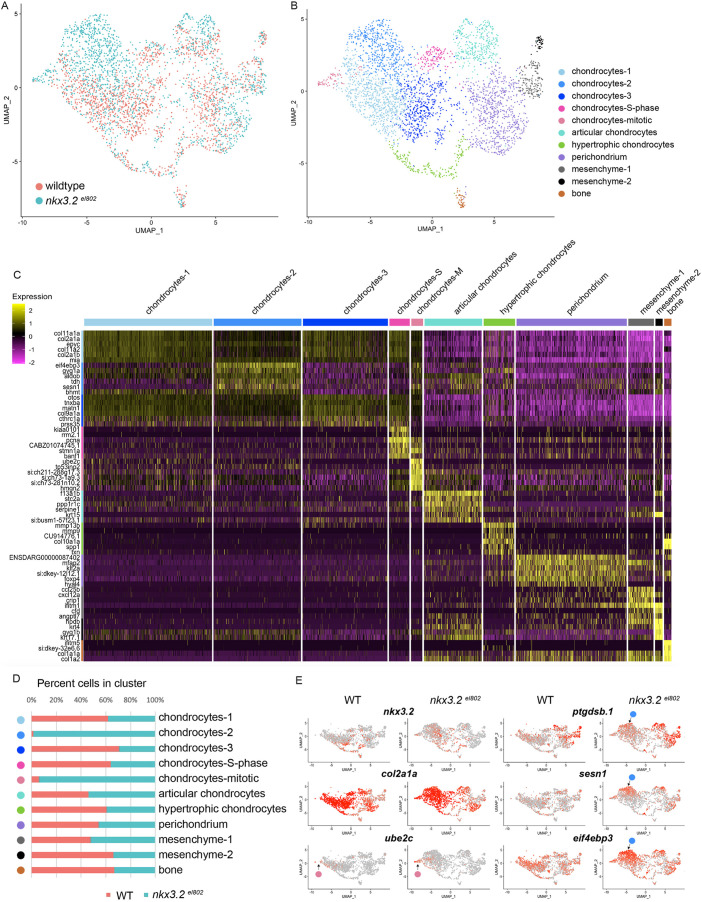
In order to understand how Nkx3.2 might regulate subtypes of joint-associated chondrocytes, we used single-cell RNA-seq to profile chondrocytes in wild-type and nkx3.2 mutant juvenile heads. In zebrafish, fli1a:GFP and sox10:dsRed transgenes co-localize predominantly in chondrocytes (Askary et al., 2017; Giovannone et al., 2019). We therefore performed fluorescence activated cell sorting (FACS) of fli1a:GFP+/sox10:DsRed+ cells (Fig. S2) from pooled 21 dpf zebrafish heads (n=5 per genotype), followed by single-cell barcoded cDNA synthesis using the 10x Genomics platform and Illumina sequencing. After quality control, we obtained 1641 cells from wild-types and 1699 cells from nkx3.2el802 mutants (Fig. 5A). We used Seurat (Butler et al., 2018) on aggregated data and Uniform Manifold Approximation and Projection (UMAP) for dimension reduction and visualization (McInnes et al., 2018) to identify 11 distinct cell clusters (Fig. 5B). Five clusters are defined by high levels of cartilage collagen-encoding genes, col2a1a and col9a1a (Col2hi/Col9hi), with two of these representing chondrocytes undergoing S-phase DNA replication (pcna+) or mitosis (ube2c+/pcna+) (Fig. 5C,E). We also observed clusters with hypertrophic chondrocyte features (col10a1a+/spp1+/col2a1alo), articular chondrocyte features (f13a1b+/prg4b+/col2a1alo), perichondrial features (foxp4+/col2a1alo), two with mesenchymal/fibroblast connective tissue features (ifitm1+/col5a1+/col2a1alo) and one with an osteoblast signature (col1a1a+/ifitm5+) (Askary et al., 2016; Moffatt et al., 2008; Zhao et al., 2015).
Fig. 5.
Single-cell analysis reveals altered facial chondrocyte subtypes in nkx3.2 mutants. (A,B) UMAP projections of the aggregate datasets colored according to dataset (A) or cluster identity (B). (C) Heatmap of the top 4-6 enriched genes per cluster. Yellow indicates high expression and magenta minimal. (D) Percent contribution within each cluster by control (WT) and mutant cells. (E) UMAP projections of gene expression split by genotype. Red indicates high expression and grey minimal. Pink circles indicate position of the mitotic chondrocytes cluster and blue circles indicate the chondrocytes-2 cluster.
All of the cell clusters present in wild types are also present in nkx3.2 mutants. However, we observed two clusters comprised almost entirely of mutant cells (Fig. 5D). The first represents mitotic chondrocytes (94% mutant cells), consistent with the abnormal chondrocyte proliferation seen in place of the missing jaw joint in mutants (Fig. 3E-K). A second chondrocyte cluster (98% mutant cells) was characterized by higher levels of sesn1 and eif4ebp3 (Fig. 5C, E), which encode inhibitors and repressed targets, respectively, of mTORC1-driven cellular growth and translation (Budanov and Karin, 2008; Tsukumo et al., 2016; Yogev et al., 2013). Both genes are induced by cellular stress in other systems (Budanov and Karin, 2008; Sukarieh et al., 2009; Taba et al., 2000). Compared with other chondrocytes, this mutant chondrocyte cluster also displayed higher levels of the prostaglandin D2 synthase gene ptgdsb.1, another stress-response pathway (Fig. 5E). However, many other regulators of chondrocyte biology, such as sox9a and hypertrophic genes runx2b and col10a1a, were unchanged in mutants.
Upregulation of ptgdsb.1 and sesn1 in nkx3.2 mutant chondrocytes
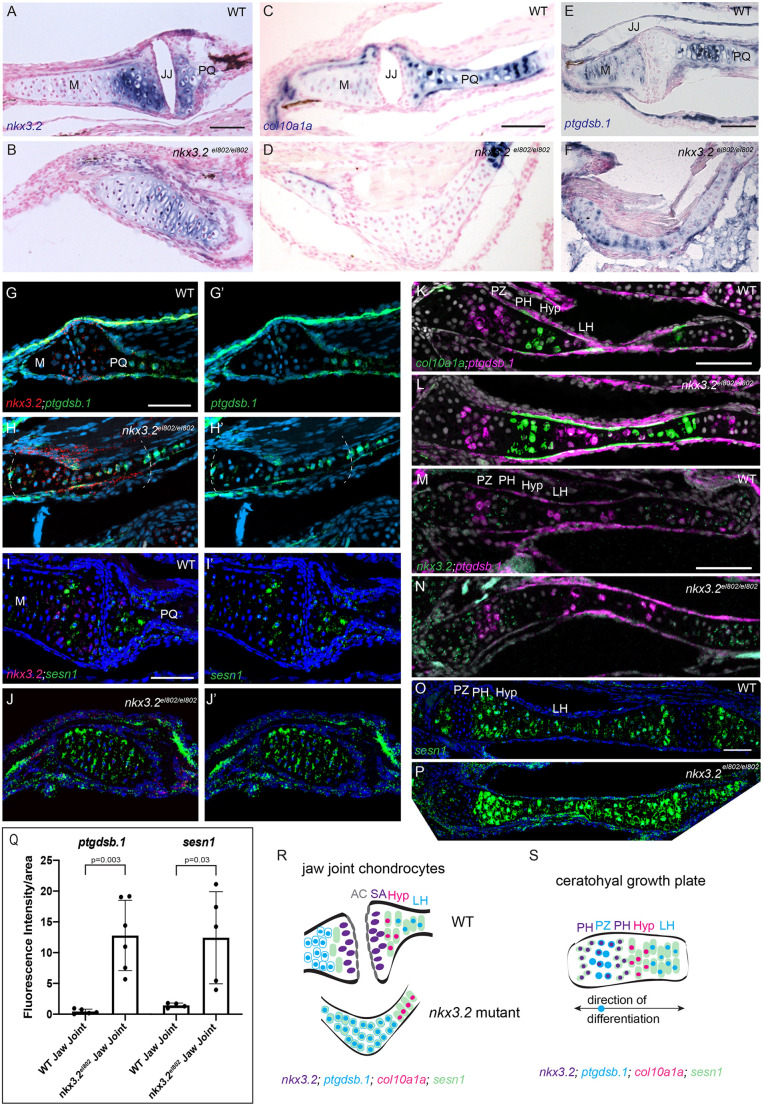
We next sought to understand the in vivo location of the mutant chondrocyte cluster identified in the single-cell analysis. In wild types at 21 dpf, we observed nkx3.2 expression in chondrocytes on either side of the jaw joint but not in the superficial zone (Fig. 6A,G,I). Expression of nkx3.2 was also seen in pre-hypertrophic chondrocytes of the bidirectional ceratohyal growth plates, in a largely non-overlapping pattern to expression of the hypertrophic marker col10a1a (Fig. 6C,K,M), similar to what has been observed in chick and mouse (Church et al., 2005; Provot et al., 2006; Tribioli et al., 1997). In nkx3.2el802 mutants, nkx3.2 mRNA transcripts were present in chondrocytes spanning the fused jaw joint (Fig. 6B,H,J), consistent with single-cell RNA-seq analysis showing nkx3.2 expression in the mutant-enriched chondrocyte cluster (Fig. 5E), yet col10a1a was not expressed (Fig. 6D). In wild types, ptgdsb.1 was expressed in the late hypertrophic zone in the upper jaw palatoquadrate and ceratohyal cartilages, in the proliferative zone of the ceratohyal growth plate, at modest levels in the lower jaw Meckel's cartilage and at lower levels in jaw joint articular cartilage, particularly on the upper jaw side of the joint (Fig. 6E,G,K,M). Although sesn1 was expressed on either side of the wild-type jaw joint, it was expressed at much lower levels at the articular surface, with only rare sesn1+/nkx3.2+ double-positive cells detected; in the ceratohyal growth plate sesn1 was expressed at high levels in all regions except the proliferative zone (Fig. 6I,O). In nkx3.2 mutants, ptgdsb.1 and sesn1 were upregulated across most chondrocytes within the fused jaw joint region, which we identified by co-expression of nkx3.2 (Fig. 6E-J). In the mutant ceratohyal growth plates, we observed largely normal zones of ptgdsb.1, col10a1a, sesn1 and nkx3.2 expression, with a potential modest upregulation of sesn1 in proliferative zones (Fig. 6K-P). Quantification of fluorescence intensity in the nkx3.2-positive jaw joint region confirmed upregulation of ptgdsb.1 and sesn1 in mutants (Fig. 6Q; ptgdsb.1: n=6, P=0.003; sesn1: n=4, P=0.03). These results show upregulation of stress response and mTORC1 pathway genes in the nkx3.2 mutant chondrocytes that fail to form a jaw joint and contribute to cartilage overgrowth (Fig. 6R).
Fig. 6.
Increased expression of ptgdsb.1 and sesn1 across the fused jaw joint of nkx3.2 mutant zebrafish. (A,B) Colorimetric in situ hybridization demonstrates nkx3.2 expression in the sub-articular zone of the jaw joint and across the fused jaw joint in nkx3.2el802 mutants at 21 dpf. (C-F) Colorimetric in situ hybridization shows expansion of ptgdsb.1 but not col10a1a across the fused jaw joint region of nkx3.2 mutants. (G-J′) RNAScope in situ hybridization shows ectopic expression of ptgdsb.1 (G-H′) and sesn1 (I-J′) across the fused jaw joint region of mutants (identified by nkx3.2 expression in red). DAPI labels nuclei in blue. Dashed lines in H,H′ indicate fused jaw joint region. (K-P) RNAscope in situ hybridizations show expression of col10a1a, ptgdsb.1, nkx3.2 and sesn1 in the growth plates of the ceratohyal. (Q) Quantification of fluorescence intensity at the jaw joint demonstrates upregulation of ptgdsb.1 and sesn1 in nkx3.2 mutants. Individual animals are plotted (dots) with mean±s.d. (R,S) Model of chondrocyte zones at the wild-type and fused mutant jaw joint (R) and within the bidirectional ceratohyal growth plate (S). AC, articular surface; Hyp, hypertrophic; JJ, jaw joint; LH, late hypertrophic; M, Meckel's cartilage; PH, pre-hypertrophic; PQ, palatoquadrate; PZ, proliferative zone; SA, sub-articular zone; WT, wild type. Scale bars: 50 µm.
DISCUSSION
Our results support temporally distinct roles for Nkx3.2 in embryonic specification of the zebrafish jaw joint and post-embryonic regulation of joint-associated cartilage growth. By comparing embryonic-only loss of nkx3.2 (morpholino knockdown) to constitutive loss (mutants), we uncovered post-embryonic requirements for Nkx3.2 in restricting subarticular cartilage growth and patterning the spine. In contrast to Nkx3.2−/− mice that die shortly after birth and do not display skeletal overgrowth (Akazawa et al., 2000; Tribioli and Lufkin, 1999), the cartilage overgrowth seen in the jaw region of adult viable nkx3.2 mutant zebrafish is a closer model to the supernumerary bones and lengthening of the wrist and digits in SMMD patients. As the number of cartilage segments in the digits appears to be proportional to cartilage length, with joint-derived signals such as Wnt9a inhibiting the formation of another joint within a certain distance (Hartmann and Tabin, 2001), the increased numbers of digit bones could be explained by cartilage overgrowth analogous to that seen in the zebrafish nkx3.2 mutant jaw. Spinal curvature defects are also more pronounced in zebrafish nkx3.2 than mouse Nkx3.2 mutants, again more closely aligning to the scoliosis seen in SMMD individuals (Hellemans et al., 2009; Tribioli and Lufkin, 1999).
Previous studies have focused on a potential role for Nkx3.2 in inhibiting hypertrophic maturation of chondrocytes. As in chick (Provot et al., 2006), we observed that nkx3.2 expression largely anti-correlates with col10a1a expression in hypertrophic chondrocytes. However, we observed no changes in hypertrophic cartilage maturation, or expansion of hypertrophic gene expression, in zebrafish nkx3.2 mutants, similar to what has been reported for mouse Nkx3.2 mutants (Akazawa et al., 2000; Tribioli and Lufkin, 1999). Instead, we observed a marked increase in chondrocyte proliferation within the mutant jaw joint region, and a more modest trend toward increased proliferation within the mutant growth plate of the ceratohyal endochondral bone. We did not, however, observe apparent cartilage overgrowth in the cranium and spine that are also affected in zebrafish nkx3.2 mutants, suggesting that Nkx3.2 has roles beyond restricting chondrocyte proliferation.
Our expression analysis also points to important differences in zones of chondrocytes between growth plates and joints. In the bidirectional growth plates of the ceratohyal (Fig. 6S), we observed a central proliferative zone of nkx3.2lo, sesn1lo, ptgdsb.1+ chondrocytes (Fig. 3E,I; Giovannone et al., 2019). Flanking these proliferative chondrocytes, we observed nkx3.2+, ptgdsb.1− zones, which we consider pre-hypertrophic based on the absence of col10a1a expression. Toward the middle of the ceratohyal we observed successive zones of col10a1a+ hypertrophic chondrocytes and then ptgdsb.1+; col10a1alo late hypertrophic chondrocytes. In contrast to the growth plate, we did not observe a prominent nkx3.2lo, ptgdsb.1+ proliferative zone close to the jaw joint. Relative to its expression in the pre-hypertrophic zones of the growth plates, nkx3.2 expression is also higher in subarticular chondrocytes of the juvenile jaw joint, consistent with a greater role in restricting chondrocyte proliferation. In nkx3.2 mutants, upregulation of ptgdsb.1 but not the hypertrophic marker col10a1a in the fused jaw joint region suggests that chondrocytes are adopting a partial ptgdsb.1+ proliferative identity rather than a late hypertrophic identity. This is supported by single-cell transcriptomic analysis showing that mitotic chondrocytes are greatly increased in abundance in nkx3.2 mutants, and our observations of increased proliferation and cartilage overgrowth at the fused jaw joint region. However, upregulation of sesn1 in the fused jaw joint region suggests that mutant chondrocytes are not equivalent to growth plate proliferative chondrocytes that normally lack sesn1. This is borne out by mutant chondrocytes forming a distinct cluster in our single-cell analysis. Our data therefore support a role for Nkx3.2 in limiting chondrocyte proliferation, particularly at the jaw joint where it is most abundantly expressed.
The higher levels of nkx3.2 expression at the wild-type jaw joint compared with the pre-hypertrophic zone of the growth plates may reflect slower growth of the subarticular zone relative to the expansive growth of the endochondral cartilage template; this is reflected by a fivefold lower proliferation rate at the jaw joint (Fig. 3K). These findings align with recent single-cell transcriptome analysis in mouse showing that chondrocyte zones in joints and growth plates are related but not identical (Lui et al., 2015). It will be interesting to test how Bmp, Fgf, and Shh signaling pathways regulate nkx3.2 expression at joints versus growth plates, given previous studies showing important roles for each of these pathways in regulating Nkx3.2 expression (Wilson and Tucker, 2004; Zeng et al., 2002). It will also be important to delete nkx3.2 only at post-embryonic stages to further confirm its requirements independent from initial specification of the fish jaw joint. Whereas we detected a modest increase in chondrocyte proliferation at the ceratohyal growth plate of nkx3.2 mutant zebrafish, we did not detect prominent cartilage overgrowth. This cannot be attributed to compensation by nkx3.1 (which we failed to detect in our single-cell RNA-seq analysis) or a broader genetic compensation (as we saw no evidence of the nonsense-mediated decay of nkx3.2 transcripts required for transcriptional adaptation). One possibility is that the preferential requirement for nkx3.2 at the zebrafish jaw joint reflects the lower levels of baseline proliferation there, and hence the greater magnitude of cartilage overgrowth when nkx3.2 is missing.
The increased proliferation of joint-associated chondrocytes in nkx3.2 mutants was linked to increased expression of stress-induced genes such as prostaglandin synthase and mTORC1 inhibitors. The presence of stress-induced pathways in late hypertrophic chondrocytes could reflect the hypoxic stress linked with their eventual apoptosis. However, strong expression of prostaglandin D2 synthase ptgdsb.1 in the proliferative zone of the wild-type growth plate was surprising, as it is not clear what types of stress these cells would experience. Expression of Ptgdsb proteins in the adult zebrafish skeleton has also been observed by proteomic analysis (Kessels et al., 2014). In mouse, single-cell transcriptomic analysis of the growth plate revealed expression of the prostaglandin E2 synthase Ptges3 in both the early proliferative zone and the late hypertrophic zone (Li et al., 2016), similar to ptgdsb.1 in zebrafish. The functions of prostaglandins in regulating chondrocyte biology, whether they are linked to proliferation, and why zebrafish would have D2 and mouse E2 subtypes remain outstanding questions. In the zebrafish testes, treatment with the D2 analogue BW-245C from 15 to 40 days of life resulted in upregulated expression of sox9a (Pradhan and Olsson, 2014), a master regulator of chondrogenesis in the skeletal system (Bi et al., 1999; Yan et al., 2005), though whether similar regulation occurs in chondrocytes has not been examined. In nkx3.2 mutants, it will be interesting to determine whether the upregulation of stress-induced pathways such as prostaglandin production, as well as modulators of mTORC1 signaling such as sesn1, are linked to the observed ectopic cartilage outgrowth, and possibly scoliosis and other skeletal defects. In the postnatal mouse, the proliferative zone of the growth plate undergoes an mTORC1-regulated transition to become a source of clonal stem cells that fuel cartilage growth (Mizuhashi et al., 2018; Newton et al., 2019). One possibility is that Nkx3.2 may function to normally regulate such an mTORC1-regulated transition below the joint surface, and to a certain extent in the pre-hypertrophic zone, to prevent cartilage overgrowth. The supernumerary bones and lengthening of the wrists and digits in SMMD patients may reflect an analogous role for Nkx3.2 in fine-tuning the rate of stem cell-mediated expansion of cartilage. In the future, it will be informative to determine how prostaglandins and other stress-induced pathways interact with mTORC1 signaling to regulate chondrocyte proliferation, and how Nkx3.2-mediated regulation of these pathways differentially fine-tunes stem cell-mediated chondrocyte expansion in joints versus growth plates.
MATERIALS AND METHODS
Zebrafish lines
The Institutional Animal Care and Use Committees of the University of Southern California, Columbia University, and the University of Alberta approved all use of zebrafish in this study. Previously reported zebrafish lines used in this study include: Tg(sox10:dsRED)el10 (Das and Crump, 2012), trps1j127aGt (RRID:ZFIN_ZBD-GENO-100809-11) (Talbot et al., 2010), Tg(fli1a:EGFP)y1 (ZIRC Cat# ZL1085 ZDB-ALT-011017-8) (Lawson and Weinstein, 2002). nkx3.2 mutant lines were generated by TALEN (el802) and CRISPR (ua5011) mutagenesis. TALEN constructs were generated using TALEN targets L: TAATGCTCCGCGACCCGGTC and R: GACATGGCTGTGCGCAGTAA. For genotyping, fin clips were performed at 14 or 90 dpf and genotyped by PCR using GoTaq DNA polymerase (Promega). Genotyping for allele el802 was performed using nkx32-F: TAACCCTAATGCTCCGCGAC and nkx32-R: TGGCGAGACTCCTCTTTTCG primers to generate 118 bp WT and 104 bp MUT bands. nkx3.2 mutants inflate their swim bladders and are recovered at expected rates for analyses at 7-21 dpf (39/161: 24.2% mutant). When raised with wild types, mutant adults are recovered at lower rates (6/90: 6.6% mutant) perhaps owing to food competition with phenotypically normal clutch-mates. To raise sufficient numbers of mutants to adulthood for experimental purposes, genotyping was performed at 14 dpf. Mutants and wild types were then raised at similar densities in different tanks and size matched for downstream analyses at the indicated ages with no survival defects noted.
µCT
Wild-type (AB; sox10:GFP) and nkx3.2ua5011/ua5011 zebrafish were scanned using MILabs μCT at the School of Dentistry, University of Alberta, Canada. For µCT scanning, 30 and 60 dpf individuals from two distinct F2 populations for each genotype (wild type: n=10; nkx3.2ua5011: n=11 at each age) were fixed in 4% paraformaldehyde (PFA) for 24 h then dehydrated in a graded ethanol series. Scanning parameters were as follows: voxel size=10 µm; voltage=50 kV; current=0.24 mA; exposure time=75 ms. Skeletal reconstruction was performed using AMIRA (Milabs) by selecting and delineating regions of high tissue densities at a voxel size of 10 μm.
Histology and proliferation analysis
Anaesthetized fish were transferred into system water containing 4.5 mg/ml BrdU (B5002, Sigma-Aldrich) and immersed for 1 h before euthanasia. Tissue was fixed overnight in 4% PFA, transferred to 20% EDTA at room temperature for decalcification for 3 days and then processed for paraffin embedding. Immunohistochemistry was performed on 5 µm sections with antigen retrieval by steaming for 20 min in citrate buffer (pH 6.0). Primary antibodies used were rat anti-BrdU (1:100, MCA2060GA, Bio-Rad), mouse anti-PCNA (1:1000, P8825, Sigma-Aldrich) diluted in serum-free antibody diluent (Dako, Agilent) overnight at 4°C. Primary antibodies were detected by incubating slides in secondary AlexaFluor antibodies (A21094, A11001, Invitrogen) diluted at 1:500 in antibody diluent for 1 h at room temperature in the dark with Hoechst 33342 nuclear stain. H&E staining was performed on 5 µm sections. Briefly, sections were de-paraffinized, stained for 2 min in Hematoxylin solution (Harris Modified, HHS16, Sigma-Aldrich), followed by a brief acid rinse and water wash and incubation for 2 min in Blueing Reagent (Scott's Tap Water Substitute, 11160, EK Industries). Slides were then stained in Eosin Y (E6003, Sigma-Aldrich) for 30 s followed by mounting using Cytoseal 60 (8310, Richard-Allan Scientific) for imaging.
In situ hybridization and RNAscope
In situ hybridization labeled probes for col10a1a (Askary et al., 2016) and ptgdsb.1 were generated by PCR. ptgdsb.1 primers: Fwd, CTGCAAACATGACGAGTGTC; Rev, AATGCTTTGCCATTCTTTCCC. Digoxigenin (DIG)-labeled antisense probes were synthesized using SP6 or T7 polymerase (Roche). In situ hybridization was performed as previously described (Askary et al., 2016). Briefly, after deparaffinization, 5 µm sections were digested in 7.5 µg/ml proteinase K for 5 min and post-fixed in 4% PFA/0.2% glutaraldehyde for 20 min. Then, 1 µg of DIG-labeled probe per slide was incubated overnight at 62°C. Following hybridization, slides were washed 3× in 50% Formamide, 1× saline sodium citrate (SSC), 0.1% Tween-20 Wash buffer and 3× in MABT [0.1M maleic acid, 0.15M NaCl, 0.1% Tween-20 (pH 7.5)]. Slides were blocked in 2% Roche Blocking buffer followed by 1 h incubation in anti-DIG-AP (Roche). Color reaction was developed with NBT/BCIP (Roche). Nuclei were counterstained with Fast Red and slides were mounted using Cytoseal for imaging. RNAscope staining was performed using the RNAscope multiplex fluorescent assay v2 using the manufacturer's protocol for formalin-fixed paraffin-embedded 5 µm sections (323100, Advanced Cell Diagnostics). RNAscope probes for col10a1a (C1), nkx3.2 (C2), ptgdsb.1 (C3) and sesn1 (C3) were synthesized by Advanced Cell Diagnostics. For RNAscope analysis, nuclei were stained with DAPI. For all in situ hybridizations and RNAscope stains we saw similar patterns of expression in at least three individuals for each experiment.
Morpholino injections
As per Miller et al. (2003), knockdown of nkx3.2 (bapx1) expression during early embryonic development was performed by injecting antisense morpholino oligos. bapx1-MO1 (5′-GCGCACAGCCATGTCGAGCAGCACT-3′; ATG start complementary sequence underlined) was purchased from GeneTools, diluted to 3 mg/ml and injected into the yolk of one-cell-stage Tg(sox10:DsRed)el10 embryos. Morpholino-injected animals to be raised to adulthood were screened for jaw joint fusion by imaging sox10:DsRed+ craniofacial cartilages at 7 dpf.
Skeletal staining
Adult specimens were fixed overnight at 4°C in 4% PFA. Following a 1 h wash in Tris/MgCl2, cartilage was stained overnight in 0.01% Alcian Blue in 10 mM MgCl2. Samples were re-hydrated through an ethanol/100 mM Tris (pH 7.5) series into 0.5% KOH and incubated overnight at 4°C. Samples were bleached in 3% H2O2/0.5% KOH for 6-8 h and then neutralized overnight in 35% NaBO4. Tissue was cleared using 1% Trypsin in 35% NaBO4 for 3-4 h followed by staining in 0.02% Alizarin Red (pH 7.5) overnight. Samples were washed in 50% glycerol/0.5% KOH to remove residual stain and stored in 100% glycerol before imaging using a Leica S8 APO stereomicroscope.
Single-cell preparation and sequencing
For single-cell profiling of juvenile chondrocytes, zebrafish double-positive for fli1a:GFP; sox10:DsRed were selected and screened for mutant joint fusion phenotype. PCR analysis to confirm genotypes was performed at 14 dpf. At 21 dpf, wild-type and nkx3.2el802 mutant craniofacial skeletons were micro-dissected and processed into a single-cell suspension using mechanical and chemical dissociation for 30 min at 28°C in protease solution [0.25% Trypsin, 1 mM EDTA (pH 8.0), PBS] containing collagenase-D (Roche Life Sciences) (n=5 animals/genotype). GFP+/DsRed+ live cells were sorted into suspension medium (1% calf serum, 0.8 mM CaCl2, 50 U/ml penicillin, 0.05 mg/ml streptomycin) and immediately loaded onto a 10x Genomics microfluidic chip. Barcoded cDNA libraries were generated using 10x Genomics Chromium Single Cell 3′ Library and Gel Bead Kit v.2 according to the manufacturer's instructions. Library sequencing on Illumina NextSeq was performed to a depth of at least 1,000,000 reads/cell for each library. Cell Ranger software (v. 3.1.0, 10x Genomics) was used for barcode recovery, genome alignment (Ensembl GRCz11) and to generate gene-by-cell count matrices with default parameters for each library. RNA-seq files have been deposited in the NCBI Gene Expression Omnibus and are available under accession number GSE151354.
Data analysis
R software with Seurat Version 3 was used for downstream analysis. Cells expressing <200 and >2500 unique RNA Features and >5% mitochondrial RNA were excluded. Datasets were aggregated using the merge function. Aggregate data was log normalized, centered and scaled using default settings of NormalizeData and ScaleData functions. Linear dimensional reduction was performed with principal component analysis using the RunPCA function. Cell clusters were determined using Find Neighbors and FindClusters functions. Cluster marker genes were identified using FindMarkers function. In order to focus the analysis on the craniofacial chondrocytes affected in nkx3.2 mutants, cells of the gill were identified based on the expression of ucmaa and ppp1r1c (P.F. and J.G.C., unpublished) and fin cells based on expression of posterior hox genes (hox9-13) (Fig. S3) (Ahn and Ho, 2008). We then excluded gill and fin cells from downstream analysis using the subset function and re-clustered the craniofacial-specific cells as above. UMAP non-linear dimensional reduction was used to visualize clusters and gene expression.
Imaging and statistical analysis
Fluorescence imaging of live zebrafish and sections was performed using a Zeiss LSM800 confocal microscope and Zen software or Leica SP8 confocal and Leica LAS software. Skeletal preparations were imaged using Leica S8APO and DM2500 microscopes. Image processing was performed with the same settings used among all images from each experiment using Fiji (Schindelin et al., 2012) and Adobe Photoshop. Proliferating PCNA+ cells and total Hoechst+ nuclei at the jaw joint and ceratohyal growth plates were counted using the cell counter function of Fiji and expressed as a %PCNA+/total cells. The regions selected for proliferation analysis were identified by wild-type joint morphology and/or by performing RNAscope staining for nkx3.2 in adjacent 5 µm sections (e.g. Fig. 3G,H PCNA staining versus Fig. 6I,J nkx3.2 expression). Fluorescence intensity of sesn1 and ptgdsb.1 transcripts was measured in at least two sections per animal in Fiji using the ‘Measure’ feature of a region of interest defined by co-expression with nkx3.2 and shown as mean intensity/area. Statistical analysis was performed in GraphPad Prism software using unpaired two-tailed t-test function (proliferation analysis) or unpaired two-tailed t-test with Welch's correction (fluorescence intensity analysis) and represented in the scatter dot plot as individual data points with mean±s.d.
Supplementary Material
Acknowledgements
We thank Megan Matsutani and Jennifer DeKoeyer Crump for fish care; Jeffrey Boyd at the University of Southern California Stem Cell Flow Cytometry Core for FACS; Nellie Nelson for single-cell RNA-seq library preparation; the University of Southern California's Center for High-Performance Computing (HPC) for single-cell RNA-seq data alignments; Amjad Askary for generating the nkx3.2el802 allele; and Ted Allison for his helpful feedback and facilitating our collaboration.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author Contributions
Conceptualization: J.S., J.G.C.; Methodology: J.S., N.N., T.M., P.B.; Software: J.S., P.F.; Validation: J.S., N.N., A.N.K., P.F.; Formal analysis: J.S., N.N., A.N.K., T.M., P.B., D.G., J.G.C.; Investigation: J.S., N.N., A.N.K., T.M., P.B., P.F.; Resources: P.F., D.G., J.G.C.; Data curation: J.S., P.F.; Writing - original draft: J.S., J.G.C.; Writing - review & editing: J.S., P.F., J.G.C.; Visualization: J.S., P.B.; Supervision: D.G., J.G.C.; Project administration: J.G.C.; Funding acquisition: J.S., D.G., J.G.C.
Funding
This work was supported by National Institutes of Health grants R00DE027218 (to J.S.) and R35DE027550 (to J.G.C), and Natural Sciences and Engineering Research Council of Canada grant RGPIN-2014-06311 (to D.G.). Deposited in PMC for release after 12 months.
Data availability
RNA-seq files have been deposited in GEO under accession number GSE151354.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193409.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193409.reviewer-comments.pdf
References
- Ahn, D. and Ho, R. K. (2008). Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220-233. 10.1016/j.ydbio.2008.06.032 [DOI] [PubMed] [Google Scholar]
- Akazawa, H., Komuro, I., Sugitani, Y., Yazaki, Y., Nagai, R. and Noda, T. (2000). Targeted disruption of the homeobox transcription factor Bapx1 results in lethal skeletal dysplasia with asplenia and gastroduodenal malformation. Genes Cells 5, 499-513. 10.1046/j.1365-2443.2000.00339.x [DOI] [PubMed] [Google Scholar]
- Askary, A., Smeeton, J., Paul, S., Schindler, S., Braasch, I., Ellis, N. A., Postlethwait, J., Miller, C. T. and Crump, J. G. (2016). Ancient origin of lubricated joints in bony vertebrates. eLife 5, e16415 10.7554/eLife.16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary, A., Xu, P., Barske, L., Bay, M., Bump, P., Balczerski, B., Bonaguidi, M. A. and Crump, J. G. (2017). Genome-wide analysis of facial skeletal regionalization in zebrafish. Development 144, 2994-3005. 10.1242/dev.151712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R. and de Crombrugghe, B. (1999). Sox9 is required for cartilage formation. Nat. Genet. 22, 85-89. 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- Budanov, A. V. and Karin, M. (2008). p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451-460. 10.1016/j.cell.2008.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E. and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, V., Yamaguchi, K., Tsang, P., Akita, K., Logan, C. and Francis-West, P. (2005). Expression and function of Bapx1 during chick limb development. Anat. Embryol. 209, 461-469. 10.1007/s00429-005-0464-z [DOI] [PubMed] [Google Scholar]
- Crotwell, P. L. and Mabee, P. M. (2007). Gene expression patterns underlying proximal-distal skeletal segmentation in late-stage zebrafish, Danio rerio. Dev. Dyn.236, 3111-3128. 10.1002/dvdy.21352 [DOI]
- Cubbage, C. C. and Mabee, P. M. (1996). Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J. Morphol.229, 121-160 [DOI] [PubMed]
- Das, A. and Crump, J. G. (2012). Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet.8, e1002710. 10.1371/journal.pgen.1002710 [DOI]
- Eisen, J. S. and Smith, J. C. (2008). Controlling morpholino experiments: don't stop making antisense. Development 135, 1735-1743. 10.1242/dev.001115 [DOI] [PubMed] [Google Scholar]
- Eve, A. M. J., Place, E. S. and Smith, J. C. (2017). Comparison of Zebrafish tmem88a mutant and morpholino knockdown phenotypes. PLoS ONE 12, e0172227 10.1371/journal.pone.0172227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch, G. E., Spruce, T., Monteiro, R. S., Owens, N. D. L., Martin, S. R. and Smith, J. C. (2018). Innate immune response and off-target mis-splicing are common morpholino-induced side effects in Xenopus. Dev. Cell 44, 597-610.e10. 10.1016/j.devcel.2018.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannone, D., Paul, S., Schindler, S., Arata, C., Farmer, D. T., Patel, P., Smeeton, J. and Crump, J. G. (2019). Programmed conversion of hypertrophic chondrocytes into osteoblasts and marrow adipocytes within zebrafish bones. eLife 8, e42736 10.7554/eLife.42736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, C. and Tabin, C. J. (2001). Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 104, 341-351. 10.1016/S0092-8674(01)00222-7 [DOI] [PubMed] [Google Scholar]
- Hellemans, J., Simon, M., Dheedene, A., Alanay, Y., Mihci, E., Rifai, L., Sefiani, A., van Bever, Y., Meradji, M., Superti-Furga, A.et al. (2009). Homozygous inactivating mutations in the NKX3-2 gene result in spondylo-megaepiphyseal-metaphyseal dysplasia. Am. J. Hum. Genet. 85, 916-922. 10.1016/j.ajhg.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrand, H., Pabst, O., Hill, R. and Arnold, H.-H. (2002). Transcription factors Nkx3.1 and Nkx3.2 (Bapx1) play an overlapping role in sclerotomal development of the mouse. Mech. Dev. 117, 217-224. 10.1016/S0925-4773(02)00207-1 [DOI] [PubMed] [Google Scholar]
- Huang, P., Xiao, A., Zhou, M., Zhu, Z., Lin, S. and Zhang, B. (2011). Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699-700. 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., Peterson, R. T., Yeh, J.-R. J. and Joung, J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, Y., Zhou, X., Han, X., Jing, J., von der Mark, K., Wang, J., de Crombrugghe, B., Hinton, R. J. and Feng, J. Q. (2015). Chondrocytes directly transform into bone cells in mandibular condyle growth. J. Dent. Res. 94, 1668-1675. 10.1177/0022034515598135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris, M., Schloesser, M., Baurain, D., Hanikenne, M., Muller, M. and Motte, P. (2017). Number of inadvertent RNA targets for morpholino knockdown in Danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors. Nucleic Acids Res. 45, 9547-9557. 10.1093/nar/gkx638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawato, Y., Hirao, M., Ebina, K., Tamai, N., Shi, K., Hashimoto, J., Yoshikawa, H. and Myoui, A. (2011). Nkx3.2-induced suppression of Runx2 is a crucial mediator of hypoxia-dependent maintenance of chondrocyte phenotypes. Biochem. Biophys. Res. Commun. 416, 205-210. 10.1016/j.bbrc.2011.11.026 [DOI] [PubMed] [Google Scholar]
- Kessels, M. Y., Huitema, L. F. A., Boeren, S., Kranenbarg, S., Schulte-Merker, S., van Leeuwen, J. L. and de Vries, S. C. (2014). Proteomics analysis of the zebrafish skeletal extracellular matrix. PLoS ONE 9, e90568 10.1371/journal.pone.0090568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok, F. O., Shin, M., Ni, C.-W., Gupta, A., Grosse, A. S., van Impel, A., Kirchmaier, B. C., Peterson-Maduro, J., Kourkoulis, G., Male, I.et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97-108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozhemyakina, E., Zhang, M., Ionescu, A., Ayturk, U. M., Ono, N., Kobayashi, A., Kronenberg, H., Warman, M. L. and Lassar, A. B. (2015). Identification of a prg4- expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 67, 1261-1273. 10.1002/art.39030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg, H. M. (2003). Developmental regulation of the growth plate. Nature 423, 332-336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Law, S. H. W. and Sargent, T. D. (2014). The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS ONE 9, e100268 10.1371/journal.pone.0100268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, N. D. and Weinstein, B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol.248, 307-318. 10.1006/dbio.2002.0711 [DOI]
- Li, J., Luo, H., Wang, R., Lang, J., Zhu, S., Zhang, Z., Fang, J., Qu, K., Lin, Y., Long, H.et al. (2016). Systematic reconstruction of molecular cascades regulating GP development using single-cell RNA-seq. Cell Rep. 15, 1467-1480. 10.1016/j.celrep.2016.04.043 [DOI] [PubMed] [Google Scholar]
- Lui, J. C., Chau, M., Chen, W., Cheung, C. S. F., Hanson, J., Rodriguez-Canales, J., Nilsson, O. and Baron, J. (2015). Spatial regulation of gene expression during growth of articular cartilage in juvenile mice. Pediatr. Res. 77, 406-415. 10.1038/pr.2014.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas, P. and Olsson, L. (2018). Bapx1 is required for jaw joint development in amphibians. Evol. Dev. 20, 192-206. 10.1111/ede.12267 [DOI] [PubMed] [Google Scholar]
- McInnes, L., Healy, J., Saul, N. and Großberger, L. (2018). UMAP: uniform manifold approximation and projection. J. Open Source Software 3, 861 10.21105/joss.00861 [DOI] [Google Scholar]
- Miller, C. T., Yelon, D., Stainier, D. Y. R. and Kimmel, C. B. (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130, 1353-1365. 10.1242/dev.00339 [DOI] [PubMed] [Google Scholar]
- Miyashita, T., Baddam, P., Smeeton, J., Oel, A. P., Natarajan, N., Gordon, B., Palmer, A. R., Crump, J. G., Graf, D. and Allison, W. T. (2020). nkx3.2 mutant zebrafish accommodate jaw joint loss through a phenocopy of the head shapes of Paleozoic jawless fish. J. Exp. Biol. 223, jeb216945 10.1242/jeb.216945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuhashi, K., Ono, W., Matsushita, Y., Sakagami, N., Takahashi, A., Saunders, T. L., Nagasawa, T., Kronenberg, H. M. and Ono, N. (2018). Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature 563, 254-258. 10.1038/s41586-018-0662-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, P., Gaumond, M.-H., Salois, P., Sellin, K., Bessette, M.-C., Godin, E., de Oliveira, P. T., Atkins, G. J., Nanci, A. and Thomas, G. (2008). Bril: a novel bone-specific modulator of mineralization. J. Bone Miner. Res. 23, 1497-1508. 10.1359/jbmr.080412 [DOI] [PubMed] [Google Scholar]
- Newton, P. T., Li, L., Zhou, B., Schweingruber, C., Hovorakova, M., Xie, M., Sun, X., Sandhow, L., Artemov, A. V., Ivashkin, E.et al. (2019). A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature 567, 234-238. 10.1038/s41586-019-0989-6 [DOI] [PubMed] [Google Scholar]
- Pradhan, A. and Olsson, P.-E. (2014). Juvenile ovary to testis transition in Zebrafish involves inhibition of Ptges. Biol. Reprod. 91, 33 10.1095/biolreprod.114.119016 [DOI] [PubMed] [Google Scholar]
- Provot, S., Kempf, H., Murtaugh, L. C., Chung, U.-I., Kim, D.-W., Chyung, J., Kronenberg, H. M. and Lassar, A. B. (2006). Nkx3.2/Bapx1 acts as a negative regulator of chondrocyte maturation. Development 133, 651-662. 10.1242/dev.02258 [DOI] [PubMed] [Google Scholar]
- Sander, J. D., Cade, L., Khayter, C., Reyon, D., Peterson, R. T., Joung, J. K. and Yeh, J.-R. J. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697-698. 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarieh, R., Sonenberg, N. and Pelletier, J. (2009). The eIF4E-binding proteins are modifiers of cytoplasmic eIF4E relocalization during the heat shock response. Am. J. Physiol. Cell Physiol. 296, C1207-C1217. 10.1152/ajpcell.00511.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taba, Y., Sasaguri, T., Miyagi, M., Abumiya, T., Miwa, Y., Ikeda, T. and Mitsumata, M. (2000). Fluid shear stress induces lipocalin-type prostaglandin D2 synthase expression in vascular endothelial cells. Circ. Res. 86, 967-973. 10.1161/01.RES.86.9.967 [DOI] [PubMed] [Google Scholar]
- Takeda, S., Bonnamy, J.-P., Owen, M. J., Ducy, P. and Karsenty, G. (2001). Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 15, 467-481. 10.1101/gad.845101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, J. C., Johnson, S. L. and Kimmel, C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development137, 2507-2517. 10.1242/dev.049700 [DOI]
- Tribioli, C., Frasch, M. and Lufkin, T. (1997). Bapxl: an evolutionary conserved homologue of the Drosophila bagpipe homeobox gene is expressed in splanchnic mesoderm and the embryonic skeleton. Mech. Dev. 65, 145-162. 10.1016/S0925-4773(97)00067-1 [DOI] [PubMed] [Google Scholar]
- Tribioli, C. and Lufkin, T. (1999). The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development 126, 5699-5711. [DOI] [PubMed] [Google Scholar]
- Tsukumo, Y., Alain, T., Fonseca, B. D., Nadon, R. and Sonenberg, N. (2016). Translation control during prolonged mTORC1 inhibition mediated by 4E-BP3. Nat. Commun. 7, 11776 10.1038/ncomms11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, A. S., Watson, R. P., Lettice, L. A., Yamada, G. and Hill, R. E. (2004). Bapx1 regulates patterning in the middle ear: altered regulatory role in the transition from the proximal jaw during vertebrate evolution. Development 131, 1235-1245. 10.1242/dev.01017 [DOI] [PubMed] [Google Scholar]
- Wilson, J. and Tucker, A. S. (2004). Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev. Biol. 266, 138-150. 10.1016/j.ydbio.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Yamashita, S., Andoh, M., Ueno-Kudoh, H., Sato, T., Miyaki, S. and Asahara, H. (2009). Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp. Cell Res. 315, 2231-2240. 10.1016/j.yexcr.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y.-L., Willoughby, J., Liu, D., Crump, J. G., Wilson, C., Miller, C. T., Singer, A., Kimmel, C., Westerfield, M. and Postlethwait, J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069-1083. 10.1242/dev.01674 [DOI] [PubMed] [Google Scholar]
- Yang, L., Tsang, K. Y., Tang, H. C., Chan, D. and Cheah, K. S. E. (2014). Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA 111, 12097-12102. 10.1073/pnas.1302703111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev, O., Williams, V. C., Hinits, Y. and Hughes, S. M. (2013). eIF4EBP3L acts as a gatekeeper of TORC1 in activity-dependent muscle growth by specifically regulating Mef2ca translational initiation. PLoS Biol. 11, e1001679 10.1371/journal.pbio.1001679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L., Kempf, H., Murtaugh, L. C., Sato, M. E. and Lassar, A. B. (2002). Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev. 16, 1990-2005. 10.1101/gad.1008002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Zhou, W., Yao, Z., Wan, Y., Cao, J., Zhang, L., Zhao, J., Li, H., Zhou, R., Li, B.et al. (2015). Foxp1/2/4 regulate endochondral ossification as a suppresser complex. Dev. Biol. 398, 242-254. 10.1016/j.ydbio.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., von der Mark, K., Henry, S., Norton, W., Adams, H. and de Crombrugghe, B. (2014). Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 10, e1004820 10.1371/journal.pgen.1004820 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.