Abstract
Molecular docking studies were done to show the inhibitory effect of two naturally occurring biflavone based anti-HIV agents, hinokiflavone and robustaflavone against the SARS-CoV-2 spike (S) protein mediated attack on the human ACE2 receptors via membrane fusion mechanism. Nefamostat, a FDA approved drug, well-known as a serine protease inhibitor for MERS-CoV infection, was used as the reference compound. Both the biflavones, showed potential as inhibitors for SARS-CoV-2 S protein-mediated viral entry. The binding affinities of these naturally occurring biflavones for RBD-S2 subunit protein of SARS-CoV-2 were explored for the first time. Such binding affinities play a critical role in the virus-cell membrane fusion process. These biflavones are able to interact more strongly with the residues of heptad repeat 1 and 2 (HR1 and HR2) regions of S2 protein of SARS-CoV-2 compared to nefamostat, and thus, these biflavones can effectively block the formation of six-helix bundle core fusion structure (6-HB) leading to the inhibition of virus-target cell-membrane fusion.
Keywords: SARS-CoV-2, Spike (S) glycoprotein, Flavonoids, Biflavones, Molecular docking
Graphical abstract
1. Introduction
By the end of 2019, scientists came to know about a novel Corona virus, SARS-CoV-2 [Severe Acute Respiratory Syndrome-Corona virus-2] causing COVID-19 (Corona Virus Disease-19). This initially affected people of Wuhan city of China. Later, this virus became the root cause of deaths and untold sufferings of millions of people around the globe due to the unavailability of specific medicine/vaccine or therapeutic strategies.
Corona viruses (CoVs) are a family of RNA viruses, responsible for mild as well as a range of severe respiratory disease outbreaks and epidemics in human in last two decades e.g. Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (World Health Organization, 2019; Masters, 2006; Corman et al., 2019; Lu et al., 2015; WHO, 2004; WHO, 2016). Like, SARS-CoV and MERS-CoV, the very deadly SARS-CoV-2 belongs to β genus of CoVs containing positive-strand RNA (Wu et al., 2020). The size of the genome of SARS-CoV-2 falls in the range of ∼30 kb involving 6 to 11 open ring frames (ORFs) (Song et al., 2019). Approximately, 67% of the entire genome is mainly located in the first ORF (ORF1a/ORF1b) which processes two polyproteins, pp1a and pp1ab and also encodes 16–17 non-structural proteins (NSPs) e.g. 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), helicase and RNA-dependent RNA polymerase (RdRp) (Dömling and Gao, 2020). The remaining ORFs encode accessory and structural proteins (Cui et al., 2019). Though SARS-CoV-2 genome has large size (characteristic of RNA virus), it genome encodes for fewer structural proteins; among which four major structural proteins are worth of mentioning: the structural spike (S) glycoprotein, small envelop (E) protein, nucleocapsid (N) protein and membrane (M) protein. These are essential for reproduction of a structurally complete virus particle (Dömling and Gao, 2020).
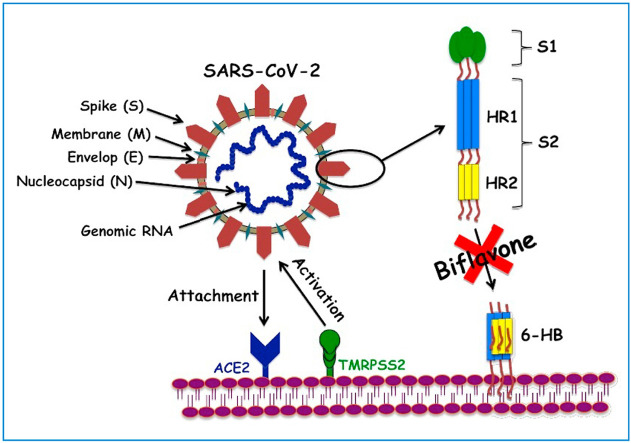
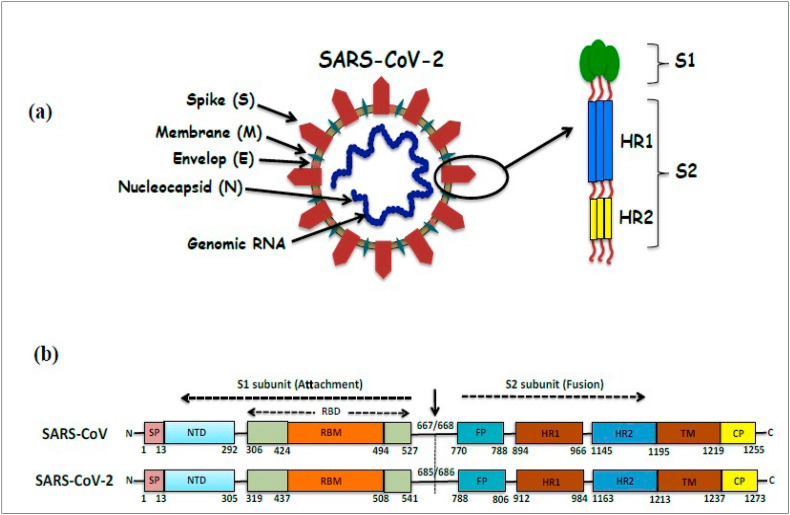
The spike (S) glycoprotein of CoVs, is responsible for the crown-like shape of the virus (Scheme 1 (a)) and belong to class-I viral fusion proteins, which facilitates the viral entry process into host cells through the binding with the receptors of the host cells, host tropism and pathogenesis (Lu et al., 2015; Millet and Whittaker, 2014). The binding of viral S protein through its receptor-binding domain (RBD) to the host cells instigates various vital steps necessary for viral infections e.g. fusion of viral and host membranes (Li, 2016; Zhu et al., 2018). The S proteins attacks the angiotensin-converting enzyme2 (ACE2) receptors of the host via its RBD and triggers a cascade of inflammation in the lower respiratory tract (Ksiazek et al., 2003; Kuba et al., 2005). Trimeric spike (S) glycoprotein is comprised of two functional subunits proteins, among them S1 subunit is responsible for binding to the host cell ACE2 receptors and induces concomitant changes in the conformation in the S2 subunit; which in turn, facilitates the infection of human cells via membrane fusion mechanism (Scheme 1) (Jaimes et al., 2020a).
Scheme 1.
(a) Representation of structure of SARS-CoV-2 showing its different structural proteins, viz., Spike (S), Membrane (M), Envelop (E) and Nucleocapsid (N) proteins. Enlarged view of SARS-CoV-2 spike proteins (at pre-fusion stage) shows its receptor-binding subunit S1 and the membrane-fusion subunit S2 [constituted of HR1 (heptad repeat 1) and HR2 (heptad repeat 2)]. (b) A comparison of SARS-CoV and SARS-CoV-2 S proteins. The residue numbers of each of the subunits and their position in S protein of SARS-CoV and SARS-CoV-2 are shown schematically. S1 subunit of SARS-CoV-2 S proteins contains NTD (14–305 aa), RBD (319–541 aa), and RBM (437–508 aa) residues; whereas its S2 subunit contains FP (788–806 aa), HR1 (912–984 aa), HR2 (1163–1213 aa), TM (1214–1237 aa) and CP (1238–1273 aa) residues.
Recent structural and biophysical data showed the evidence of the binding affinity of SARS-CoV-2 S protein with ACE2 receptors of host cells (Hoffmann et al., 2020; Wrapp et al., 2020). Furthermore, such effect is much more pronounced in case of SARS-CoV-2 S protein. Because the binding affinity of S1 subunit of SARS-CoV-2 is higher than that of the SARS-CoV. This is attributed to the higher infectivity of novel SARS-CoV-2 compared to SARS-CoV (Hoffmann et al., 2020; Wrapp et al., 2020). Comparative analysis of spike (S) glycoprotein by protein sequence alignment of SARS-CoV-2 with SARS-CoV shows 76% of sequence identity [Scheme 1(b)] (Zhou et al., 2020; Jaimes et al., 2020b). Therefore, to develop specific SARS-CoV-2 fusion inhibitors, it is very much necessary to study the fusion capacity of SARS-CoV-2 compared to that of SARS-CoV. As an alternate strategy, various research groups target the viral S protein for the inhibition of the membrane fusion and entry processes of SARS-CoV-2 in host cells with ACE2 receptors (Dömling and Gao, 2020; Jordan et al., 2018).
Heptad repeat 1 (HR1) and 2 (HR2) domains of S2 subunit play a crucial task in the SARS-CoV fusion with target cells (Scheme 1). Upon binding of S protein through RBD in S1 to the ACE2 receptor on the target cell, HR1 and HR2 domains combine to form a six-helix bundle core fusion structure (6-HB) and bring the viral envelop and the cellular membranes into close proximity; necessary for effective fusion and infection (Bosch et al., 2004). Therefore, FDA approved anti-viral drugs target the HR1 and HR2 regions in the S2 subunit domains and such drugs are now being extensively explored as the potential therapeutic option for COVID-19.
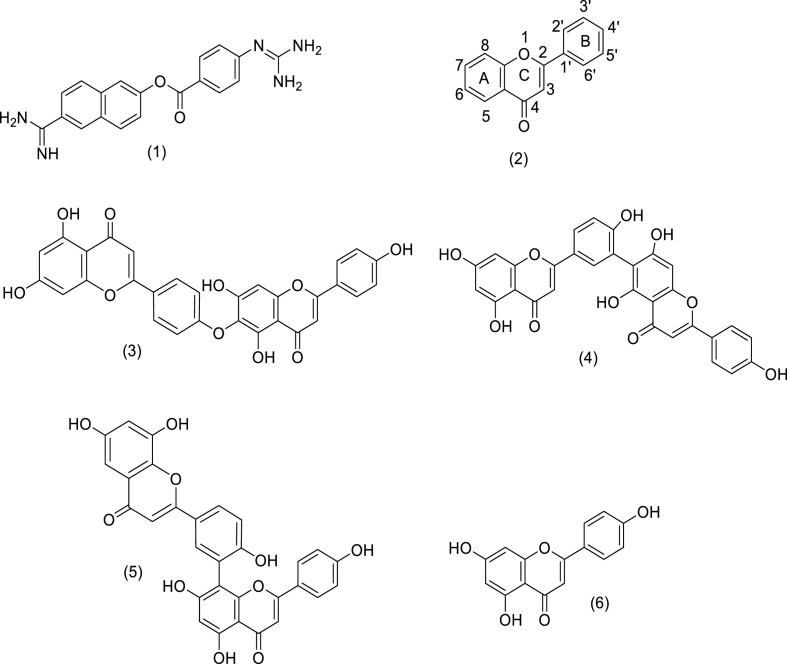
Identification of the genome sequence, 3D-structure and mechanism of action/pathogenesis of SARS-CoV-2 is necessary for developing effective treatment strategies to combat COVID-19 (Masters, 2006; Corman et al., 2019; Cui et al., 2019; Zhang et al., 2020; Guan et al., 2003; Al-Tawfiq and Memish, 2014). One of such therapeutic strategies targets the main protease (Mpro) of SARS-CoV-2 i.e. 3CLpro, having high genomic sequence similarity with SARS-CoV and plays a crucial role in COVID-19 pathogenesis. In this direction, a large number of U.S. Food and Drug Administration (FDA) approved protease inhibitors (showing efficacy in case of SERS, MERS and HIV) are put into trials (Dömling and Gao, 2020; Ryu et al., 2010a, 2010b; Shamsi et al., 2020; Cinatl et al., 2005; Jo et al., 2019, 2020). In this connection, it is worth to mention the efficacy of nefamostat (1), a serine protease inhibitor [Fig. 1 ] which is a FDA approved drug for the treatment of cystic fibrosis and acute pancreatitis (Yamamoto et al., 2016). During the in-vitro screening (with the help of Dual Split Protein (DSP) reporter fusion assay) of more than 1000 FDA-approved drugs to find out effective drug to combat MERS disease, it was observed that compound (1) has the potential to inhibit effectively the MERS-CoV S protein initiated membrane fusion and this study also proposed that (1) could be an effective candidate for inhibiting MERS-CoV infection (Yamamoto et al., 2016). However, despite having potentials, such protease inhibitors are not extensively explored for their inhibitory activity towards the SARS-CoV-2 S protein triggered membrane fusion and related infections. In this connection, plants sources and their active components used in traditional Chinese medicine and having antiviral activity are also being extensively explored in China (Dömling and Gao, 2020; Ryu et al., 2010a; Li et al., 2020; Xu and Zhang, 2020; Ang et al., 2020; Ling, 2020). Medicinal plants are the largest and the best combinational libraries of natural products. Although many drugs are made by synthetic chemistry but most of the core structures or scaffolds for synthetic drugs are based upon the natural products which are used in traditional medicines (Newman and Cragg, 2007, 2016; Harvey et al., 2015). The flavones represent a sub-class of one of the most diverse and wide-spread of plant-derived natural products i.e. flavonoids. Structure of parent flavone moiety (2) is shown in Fig. 1. These compounds have wide spectrum of biological and pharmacological applications as cardio protective, antimicrobial, antiviral, anti-allergic and hepato-protective agents (Kanakis et al., 2007; Pietta, 2000; Silva et al., 2002; Tsutomu et al., 1993; Vitorino and Sottomayor, 2010). It is believed that most of these bioactivities of flavonoids are originated from their behaviors as antioxidants. The presence of different number of varied substituents e.g. hydroxyl, alkoxy and/or glycosidic groups at various positions of basic skeleton of these compounds gives rise to their remarkable structural diversity which in turn control their biological activities. Wang et al. isolated a series of antioxidant biflavone compounds [having (C6–C3–C6)2 system] e.g. amentoflavone, hinokiflavone and robustaflavone from the plant, Selaginella doederleinii Hieron plant which are reported to show antiviral activity (Wang et al., 2015). Recently, an appreciable number of studies are being done to evaluate the viral (MERS-CoV and SERS-CoV) protease inhibiting activity of various plant derived flavonoinds (Dömling and Gao, 2020; Ryu et al., 2010a; Jo et al., 2019, 2020; But et al., 2001; Wilsky et al., 2012; Li et al., 2019; Zembower et al., 1998; Lin et al., 1997a, 1997b; Coulerie et al., 2012; Konoshima et al., 1988; Miki et al., 2007). Ryu et al. reported that the SARS-CoV 3CLpro inhibitory activity of some hydroxyflavones, like apigenin, luteolin and quercetin in addition to four biflavones e.g. amentoflavone, bilobetin, ginkgetin and sciadopitysin, isolated from the plant, Torreya nucifera. Among these biflavones, amentoflavone (5) [Fig. 1], was found to be the most potent inhibitor (Ryu et al., 2010a). Besides, SARS-CoV, the biflavones like amentoflavone showed antiviral activity towards a range of other viruses (Ryu et al., 2010a; But et al., 2001; Wilsky et al., 2012; Li et al., 2019). Apart from amentoflavones, the antiviral activity of biflavones like, hinokiflavone (3) (isolated from the plants, Rhus succedanea, Garcinia multiflora or Dacrydium balansae) and robustaflavone (4) (constituent of the plants, Rhus succedanea or Garcinia multiflora) are note-worthy. Hinokiflavone (3) showed efficacy as RNA polymerase (DV-NS5 RdRp) inhibitors of dengue 2 virus; in addition to this, the HIV-1 inhibitory activity of hinokiflavone is also reported (Lin et al., 1997b; Coulerie et al., 2012; Konoshima et al., 1988). On the other hand, robustaflavone (4) showed viral inhibitory activity against various viral species e.g. Hepatitis B virus (HBV) and HIV-1 (Zembower et al., 1998; Lin et al., 1997a, 1997b). However, in spite of the potent viral inhibitory activity of (3) and (4) towards other viral species, these biflavones were not explored for their inhibitory activity towards the SARS-CoV-2 S protein triggered membrane fusion and related COVID-19 infections. This prompted us to perform the present study involving the biflavones, hinokiflavone (3) and robustaflavone (4) [Fig. 1]; both of them showed similar inhibitory activity against HIV-1 reverse transcriptase (RT) and thus, these were identified as anti-HIV agents of natural product origin (Lin et al., 1997b). The inhibitory effect of these two plant derived anti-HIV or anti-viral biflavones, (3) and (4) against the SARS-CoV-2 S-mediated membrane fusion was explored with the help of molecular docking studies. Binding interactions of these compounds with RBD of S protein of SARS-CoV-2 have been studied. Such interactions are very much necessary to prohibit the exposure of RBD-S of SARS-CoV-2 towards ACE2 receptors of host cells, which is a key step for preventing viral infection initiation. In this study, nefamostat (1), showing potential towards inhibiting the MERS-CoV S protein initiated membrane fusion, was employed as the reference compound so that we can make a comparative assessment of (3) and (4) with a known RNA virus inhibitor, (1).
Fig. 1.
Chemical structures of nefamostat (1), hinokiflavone (3) and robustaflavone (4), used as the test drug/ligands against the spike protein of SARS-CoV-2 in the present molecular docking studies. Structures of parent flavone moiety (2) and two 3CLpro inhibitors for SARS-CoV i.e. amentoflavone (5) and apigenin (6) are also shown.
Our molecular docking study suggests that the biflavones, (3) and (4) can function as anti-viral inhibitors against SARS-CoV-2 S protein-mediated viral entry. Interestingly, these showed higher anti-SARS-CoV-2 efficacy over the reference, (1). Furthermore, this study, for the first time, sheds light on the role of two anti-HIV biflavones, (3) and (4) in controlling a critical step of the SARS-CoV-2 infection i.e. their affinity and the binding interactions with the S2 subunit spike protein of SARS-CoV-2 which indirectly blocks the propagation of virus at an early stage by inhibiting SARS-CoV-2 fusion with the host cells and thus, minimizing the chance for the entry of the virus into the host cells. Therefore, (3) and (4) may be explored as the fusion inhibitor-based potent therapeutics and prophylactics for the treatment and prevention of infection related to SARS-CoV-2. However, for this, higher levels of molecular simulations as well as in vivo/clinical studies are needed to be done which are beyond the scope of the present work.
2. Method for molecular docking studies
Molecular Graphics Laboratory (MGL) and AutoDock Vina tools were utilized to set up and perform blinded molecular docking studies on the intermolecular interactions of ligand molecules (Trott et al., 2010; Xu et al., 2018) [i.e. either nefamostat (1)/hinokiflavone (3) or robustaflavone (4)] with the receptor binding domain (RBD) of spike (S) glycoproteins i.e. termed as RBD-S.
The crystal structure of post fusion core of SARS-CoV-2 S2 subunit (PDB ID: 6LXT; https://www.rcsb.org/structure/6lxt) was obtained from Protein Data Bank (PDB) (Xia et al., 2020). At the beginning of the docking study, water molecules were separated from the PDB files and polar hydrogens were added simultaneously to the same PDB file. On the other hand, PDB structures of ligand molecules were created using ACD chemsketch software (https://www.acdlabs.com/resources/freeware/chemsketch/), which were subsequently changed into pdbqt format with the help of AutoDock Vina software. RBD-S of SARS-CoV-2 was enclosed in a grid box and the dimensions of the grid box were fixed at 114 x 108 x 126 with grid spacing value of 0.375 Å. The centre of the grid was positioned at −12.508, −0.032 and −7.904 Å respectively. The Lamarckian genetic algorithms (GA) modules available with AutoDock were employed to execute present ligand docking calculations. The default settings available with AutoDock programme were selected to denote other parameters (Sanner, 1999; Morris et al., 1998). The lowest energy docked conformations, obtained from the AutoDock Vina scoring function were considered as the preferred binding mode between the S protein receptor and the selected ligand molecule. The output of the AutoDock calculation was viewed by Python Molecular Viewer software (Stefano Forli, Olson Molecular Graphics Laboratory, The Scripps Research Institute, San Diego, CA).
3. Results and discussion
Molecular docking studies of biflavones, hinokiflavone (3) and robustaflavone (4) along with a known RNA virus inhibitor, nefamostat (1) with spike (S) glycoproten of SARS-CoV-2 were done to understand binding interactions of these ligands towards the RBD-S of this deadly virus and showing their potentials as anti-viral therapeutic agents for SARS-CoV-2. As shown in Scheme 1(b), its S1 subunit (14–685 aa) consists of N-terminal domain (NTD, 14–305 aa), receptor-binding domain (RBD, 319–541 aa), and receptor-binding motif (RBM, 437–508 aa) and S2 subunit (686–1273 aa) is made of fusion peptide (FP, 788–806 aa), heptad repeat 1 (HR1, 912–984 aa), heptad repeat 2 (HR2, 1163–1213 aa), transmembrane domain (TM, 1214–1237 aa) and cytoplasmic domain (CP, 1238–1273 aa). The residue numbers of each region are related to their corresponding positions in the S proteins of SARS-CoV-2 as it was also observed for SARS-CoV. The literature data indicates that the S1/S2 cleavage site for SARS-CoV-2 S protein located at R685/R686 (Xia et al., 2020).
Present molecular docking study focused on the repurposing of FDA approved drug, (1) and two naturally occurring anti-HIV biflavones, (3) and (4) against the main spike (S) glycoprotein mediated attack of SARS-CoV-2 to ACE2 receptors of host cells through membrane fusion mechanism. Post fusion core of SARS-CoV S2 subunit that has been repositioned and structured in PDB file recently (Xia et al., 2020) and we selected to a potential target to predict the mode of interaction between this target protein and the ligands/drug. Because such interactions may block this active site of the viral protein and thus, the spread of the infection by SARS-CoV-2 can be prevented. Results of the docking studies are summarized in Table 1, Table 2, Table 3, Table 4, Table 5 . Strength and the affinity of a specific ligand, which binds to the pocket of a target protein, can be expressed in terms of binding energy (ΔG) and the ligand having lower binding energy is preferred as a potential drug candidate. This parameter was compared for each of the test compounds in order to understand their effectiveness as the active antiviral agents for SARS-CoV-2 mediated attack on human ACE2 receptor cells.
Table 1.
Lengths of selected bonds between the interacting active site residues of SARS-CoV-2 S2 subunit (6LXT) and nefamostat (1) in the docked structure.
| The interacting active site residues of |
Bonds lengths (Å) | |
|---|---|---|
| 6LXT | Nefamostat (1) | |
| ASN1194 (C O) | -NH2 | 2.65 |
| ASN1194 (N–H) | -NH2 | 3.12 |
| GLN935 (N–H) | -NH- | 2.33 |
| GLU1195 (C O) | -NH | 1.98 |
| LYS1191 (C O) | -NH2 | 1.95 |
| GLN1201 (N–H) | -NH2 | 2.94 |
| ASN928 (N–H) | -O- | 3.00 |
Table 2.
Lengths of selected bonds between the interacting active site residues of SARS-CoV-2 S2 subunit (6LXT) and hinokiflavone (3) in the docked structure.
| The interacting active site residues of |
Bonds lengths (Å) | |
|---|---|---|
| 6LXT | Hinokiflavone (3) | |
| GLN1201 (N–H) | -O- | 1.94 |
| GLN926 (N–H) | -OH | 2.34 |
| GLN926 (N–H) | -OH | 2.57 |
| GLN926 (N–H) | -OH | 2.34 |
| GLN926 (N–H) | -OH | 2.94 |
| GLU1195 (C O) | -OH | 2.90 |
| LEU1197 (C O) | -OH | 2.96 |
| ASN928 (N–H) | (C O) | 3.06 |
| GLU918 (C O) | -OH | 2.45 |
| ASN919 (N–H) | -OH | 2.52 |
Table 3.
Lengths of selected bonds between the interacting active site residues of SARS-CoV-2 S2 subunit (6LXT) and robustaflavone (4) in the docked structure.
| The interacting active site residues of |
Bonds lengths (Å) | |
|---|---|---|
| 6LXT | Robustaflavone (4) | |
| ASP936 (COOH) | -OH | 2.76 |
| GLN926 (N–H) | (C O) | 3.14 |
| LYS1191 (N–H) | --OH | 2.68 |
| LYS1191 (N–H) | (C O) | 2.71 |
| GLU1195 (C O) | -OH | 3.82 |
| ASN928 (N–H) | -OH | 2.43 |
| ASN928 (N–H) | -OH | 3.37 |
| ASN928 (N–H) | -OH | 2.25 |
| ASN928 (C O) | -OH | 2.87 |
Table 4.
A comparison of binding free energies of test drug/ligands for potential binding domain of SARS-CoV-2 S2 (PDB: 6LXT) as obtained from molecular docking studies.
| Test drug/ligands | -ΔG (Gibb's free energy) (kcal mol−1) |
|---|---|
| Nefamostat (1) | 8.40 |
| Hinokiflavone (3) | 9.60 |
| Robustaflavone (4) | 9.40 |
Table 5.
ADME Properties of selected SARS-CoV-2 S2 fusion inhibitor [SWISSADME prediction (http://www.swissadme.ch/)]5.8.
| Test drug/ligands | ADME properties (Lipinki's Rule of Five) |
Drug likeliness | |
|---|---|---|---|
| Properties | Value | ||
| Nefamostat (1) | Molecular weight | 347 | YES |
| LogP | 1.35 | ||
| H-bond donor | 4 | ||
| H-bond acceptor | 4 | ||
| Molar Refractivity | 101.24 | ||
| Violations | NO | ||
| Hinokiflavone (3) | Molecular weight | 538 | YES |
| LogP | 3.06 | ||
| H-bond donor | 6 | ||
| H-bond acceptor | 10 | ||
| Molar Refractivity | 147 | ||
| Violations | NO | ||
| Robustaflavone (4) | Molecular weight | 538 | YES |
| LogP | 3.53 | ||
| H-bond donor | 5 | ||
| H-bond acceptor | 10 | ||
| Molar Refractivity | 146 | ||
| Violations | NO | ||
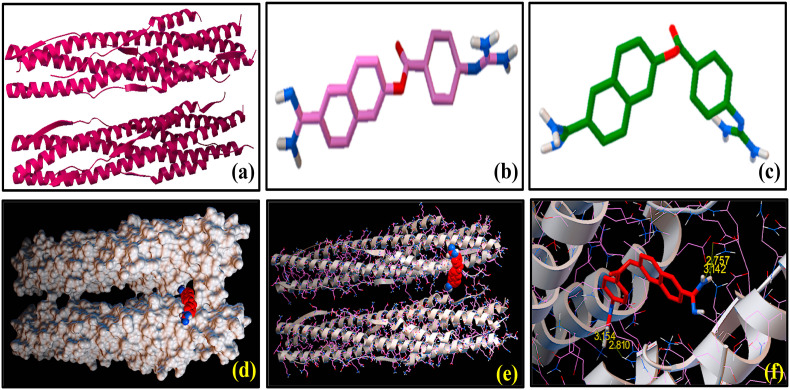
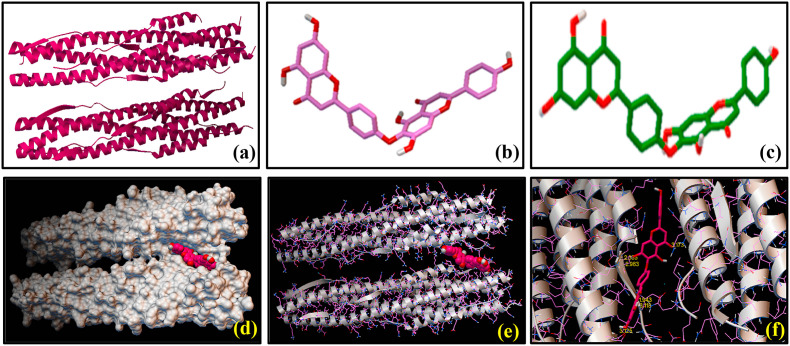
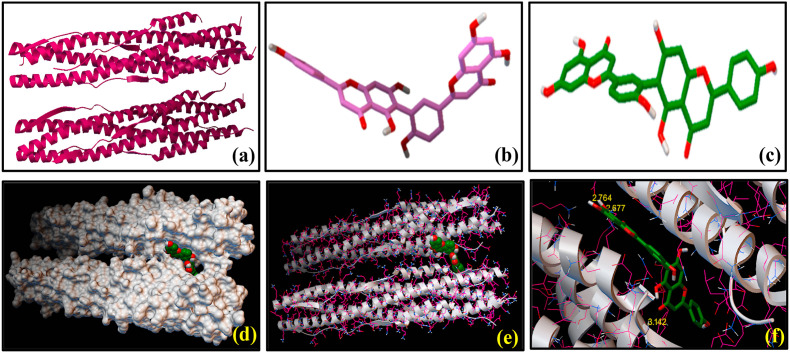
We have performed molecular docking studies with several FDA approved antiviral drugs of natural product origin using AutoDock Vina tools on the basis of their highest binding affinity with S2 domain of the spike protein of SARS-CoV-2 and these three compounds (1), (3) and (4) were selected for the present work. Nefamostat (1) was considered as the reference compound, which exhibited docked score of binding energy (ΔG) as −8.4 kcal mol−1 with SARS-CoV-2 S2 protein. Fig. 2 shows the SARS-CoV-2 S2 protein docked structures of (1) along with the residues of the protein with which the molecule (1) interact. Selected bond lengths of SARS-CoV-2 S2 protein-(1) docked structure are shown in Table 1. These results reveal that SARS-CoV-2 S2 protein interacts with (1) mostly via its aromatic amino acid residues e.g. ASN 1194, GLN 935, GLU 1195, LYS 1191, GLN 1201 and ASN 928. These amino acid residues are found to participate in the interactions with (1) through non-covalent bond formation, like hydrogen bonds (H-bonds) with the binding pocket of SARS-CoV-2 S2 protein [Fig. 2]. Results of the molecular docking of (3) and (4) with SARS-CoV-2 S2 protein are shown in Fig. 3, Fig. 4 respectively. As evidenced from the SARS-CoV-2 S2 protein-(3) docked structure (Fig. 3), (3) forms H-bonds with GLN 1201, GLN 926, GLU 1195, LEU 1197, ASN 928, GLU 918, ASN 919 amino acid residues of S2 domain of the spike protein of SARS-CoV-2. Selected bond lengths of SARS-CoV-2 S2 protein-(3) docked structures are shown in Table 2. Fig. 4 denotes the SARS-CoV-2 S2 protein docked structures of (4). It was observed that (4) interacts with the ASP 936, GLN 926, LYS 1191, GLU 1195, ASN 928 amino acid residues the S2 domain of SARS-CoV-2 through H-bonding interaction. Selected bond lengths of SARS-CoV-2 S2 protein-(4) docked structures are shown in Table 3. Both (3) and (4) showed relatively higher binding affinity compared to the reference compound (1) as evidenced from the comparison of their binding energy (ΔG) values (Table 4), obtained from molecular docking studies. It was interesting to note that (3) as well as (4) displayed lowest and almost identical binding energy values of −9.6 and −9.4 kcal mol−1 respectively. Result indicates that these reported anti-HIV agents showed similar binding preferences for S2 domain of the spike protein of SARS-CoV2. Despite of having similar preferences for the S2 domain, the presence of additional –OH group on the chromone ring of (3) and (4) affected the extent of their hydrogen bonding interactions compared to (1) (Fig. 1). This has been reflected in ΔG value of (1) indicating its lowest binding affinity for the binding site of SARS-CoV-2 S2. Moreover, these results strengthen our observation that the selected biflavones, (3) and (4), can interact strongly with the residues of the HR1 and HR2 regions of S2 protein of SARS-CoV-2, thereby they can be as active as to block the formation of six-helix bundle core fusion structure (6-HB). This ultimately prompts the inhibition of the fusion between the virus and target cell membranes. The plausible route of this inhibitory effect of plant-derived biflavones (3) and (4) is shown in Scheme 2 .
Fig. 2.
Molecular docking showing the interactions between SARS-CoV-2 S2 subunit [6LXT] and nefamostat (1): 3-dimensional structures of (a) SARS-CoV-2 S2 subunit [6LXT] and (b) nefamostat (1) before docking study. (c) 3-dimensional representation of [6LXT]-docked structure of (1). (d) Surface representation of the 6LXT-nefamostat binding, showing the active sites. (e) Ribbon docking model showing interactions between the active site residues of 6LXT and (1) (f) Close-up view.
Fig. 3.
Molecular docking showing the interactions between SARS-CoV-2 S2 subunit [6LXT] and hinokiflavone (3): 3-dimensional structures of (a) SARS-CoV-2 S2 subunit [6LXT] and (b) hinokiflavone (3) before docking study. (c) 3-dimensional representation of [6LXT]-docked structure of (3). (d) Surface representation of the 6LXT-hinokiflavone binding, showing the active sites. (e) Ribbon docking model showing interactions between the active site residues of 6LXT and (3) (f) Close-up view.
Fig. 4.
Molecular docking showing the interactions between SARS-CoV-2 S2 subunit [6LXT] and robustaflavone (4): 3-dimensional structures of (a) SARS-CoV-2 S2 subunit [6LXT] and (b) robustaflavone (4) before docking study. (c) 3-dimensional representation of [6LXT]-docked structure of (4). (d) Surface representation of the 6LXT-robustaflavone binding, showing the active sites. (e) Ribbon docking model showing interactions between the active site residues of 6LXT and (4) (f) Close-up view.
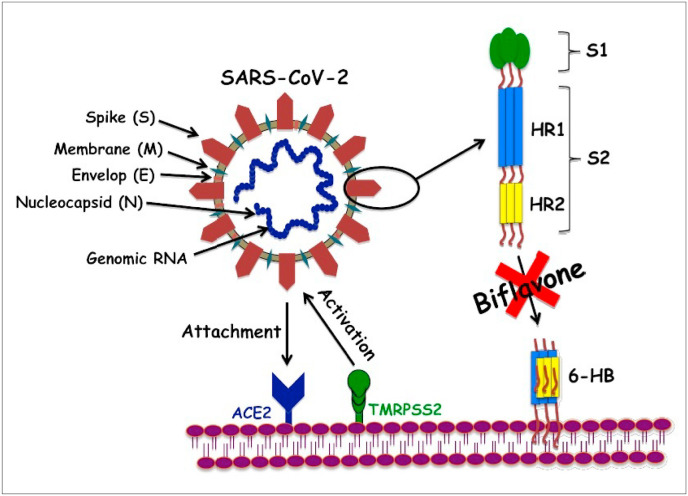
Scheme 2.
Schematic representation of the plausible route of naturally occurring biflavones, hinokiflavone (3) and robustaflavone (4) mediated inhibition of SARS-CoV-2 S protein attack on human ACE2 receptors. During the process of virions attachment to the receptors, type-2 transmembrane protease (TMPRSS2) cleaves the S proteins from ACE2 leading to the activation of the S proteins. HR1 and HR2 regions of S2 protein of SARS-CoV-2 interactions with ACE2 receptors instigating six-helix bundle core fusion structure (6-HB) formations is blocked by biflavones and hence, inhibition of membrane fusion can be achieved.
All the naturally occurring biflavones used in this study satisfied the Lipinski's rule of five that mainly determines the prime requisition of a compound to be a potential drug on the basis of its different molecular properties e.g. absorption, distribution, metabolism and excretion (ADME) (Lipinski, 2004). The ADME characteristics of our selected drug candidates i.e. (1, 3 and 4) shown in Table 5, indicates the fulfillment of Lipinski's rule of five by these compounds.
4. Conclusion
Molecular docking based in-silico studies were done to find out naturally occurring bio-friendly compounds based novel anti-COVID agents which can bind the SARS-CoV-2 and thus, inhibit its binding affinity towards the ACE2 receptors of human cells. Present study showed that two naturally occurring biflavone based anti-HIV agents, hinokiflavone (3) and robustaflavone (4) can be effective against the SARS-CoV-2 spike (S) protein mediated attack of the human ACE2 receptors via membrane fusion mechanism. Both (3) and (4) strongly bind with SARS-CoV-2 S protein which can prohibit its attack to ACE2 receptors of the host cells. The biflavones, (3) and (4) can interact more strongly with the residues of HR1 and HR2 regions of S2 protein of SARS-CoV-2 compared to (1) due to the presence of additional OH group on the chromone ring of (3) and (4) largely influence their hydrogen bonding interactions. Hence, these can effectively block the formation of six-helix bundle core fusion structure (6-HB) leading to the inhibition of virus-target cell-membrane fusion. Structures of any flavonoid class of compounds greatly control their pharmacological activity. Amentoflavone (5), showed most potent 3CLpro inhibitory effect in case of SARS-CoV, could be attributed to the presence of an apigenin moiety (6) [Fig. 1] at 3′ position of ring-B of the flavone skeleton (Ryu et al., 2010a). Interestingly, in our present study, both (3) and (4), have apigenin moiety on their parent flavone skeletons, which showed inhibitory activity against the SARS-CoV-2 spike protein invade of target cells by membrane fusion. It is note-worthy that huge number of structural analogs of various flavonoids with diverse substituent ranges/structural patterns can also be synthesized in the laboratory with the help of easily implementable synthetic protocols. On the other hand, flavones are well known antioxidants and reported that such antioxidant substances promote improvement in complications induced by human coronavirus in mice. This protective effect occurred through the reduction of the oxidative stress, cerebral lipid peroxidation, and regulation of inflammation (Do Carmo et al., 2008; Ganfornina et al., 2008). So, with the help of higher level molecular simulations, more task specific biflavones with desired structural patterns can be designed and their subsequent synthesis can be done as well as their in-vivo/clinical studies will help us to develop potential fusion inhibitor-based therapeutics and prophylactics to fight against SARS-CoV-2.
Credit author statement
S.M., N.A.B., A.K. and T.M. designed the study; S.M., A.K. and T.M. carried out the study; S.M., A.K., T.M. and N.A.B. wrote the manuscript; All authors have given approval to the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by DSTBT, GoWB, India [Grant No. ST/P/S&T/15G-20/2019 to N.A.B. and S.M.], DST-PURSE Program [SR/PURSE Phase2/42(G) & Phase2/42(C)] of Visva-Bharati as well as DST-FIST and UGC-SAP (Phase-II) programs of the Dept. of Chemistry, Visva-Bharati. A.K. and T.M. thank UGC for their fellowships [UGC-CSIR (NET) and MANF respectively]. S.M. acknowledges Rammohan College, Kolkata.
References
- Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus: transmission and phylogenetic evolution. Trends Microbiol. 2014;22:573–579. doi: 10.1016/j.tim.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L., Lee H.W., Choi J.Y., et al. Herbal medicine and pattern identification for treating COVID-19: a rapid review of guidelines. Integr. Med. Res. 2020:100407. doi: 10.1016/j.imr.2020.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E., Van Der Zee R., et al. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- But P.P.H., Ooi V.E.C., He Y.H., et al. Antiviral amentoflavone from Selaginella sinensis. Biol. Pharm. Bull. 2001;24:311–312. doi: 10.1248/bpb.24.311. [DOI] [PubMed] [Google Scholar]
- Cinatl J., Jr., Michaelis M., Hoever G., et al. Development of antiviral therapy for severe acute respiratory syndrome. Antivir. Res. 2005;66:81–97. doi: 10.1016/j.antiviral.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Lienau J., Witzenrath M. Coronaviruses as the cause of respiratory infections. Internist. 2019;60:1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulerie P., Eydoux C., Hnawia E., et al. Biflavonoids of Dacrydium balansae with potent inhibitory activity on dengue 2 NS5 polymerase. Planta Med. 2012;78:672–677. doi: 10.1055/s-0031-1298355. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Carmo S., Jacomy H., Talbot P.J., Rassart E. Neuroprotective effect of apolipoprotein D against human coronavirus OC43-induced encephalitis in mice. J. Neurosci. 2008;28(41):10330–10338. doi: 10.1523/JNEUROSCI.2644-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dömling A., Gao L. Chemistry and biology of SARS-CoV-2. Inside Chem. 2020;6:1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina M.D., Do Carmo S., Lora J.M., et al. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008;7(4):506–515. doi: 10.1111/j.1474-9726.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Harvey A.L., Edrada-Ebel R., Quinn R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., et al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically-sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim H., Kim S., et al. Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., et al. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.C., Stevens S.K., Deval J. Nucleosides for the treatment of respiratory RNA virus infections. Antivir. Chem. Chemother. 2018;26 doi: 10.1177/2040206618764483. 2040206618764483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakis C.D., Tarantilis P.A., Polissiou M., et al. An overview of DNA and RNA bindings to antioxidant flavonoids. Cell Biochem. Biophys. 2007;49:29–36. doi: 10.1007/s12013-007-0037-2. [DOI] [PubMed] [Google Scholar]
- Konoshima T., Takasaki M., Kozuka M., et al. Studies on inhibitors of skin tumor promotion-4-inhibitory effects of flavonoids on epstein-barr virus activation-1. Yakugaku Zasshi. 1988;42:343–346. [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Song X., Su G., et al. Amentoflavone inhibits HSV-1 and ACV-resistant strain infection by suppressing viral early infection. Viruses. 2019;11:466–481. doi: 10.3390/v11050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu X., Guo L., et al. Traditional Chinese herbal medicine for treating novel coronavirus (COVID-19) pneumonia: protocol for a systematic review and meta-analysis. Syst. Rev. 2020;9:1–6. doi: 10.1186/s13643-020-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.M., Zembower D.E., Flavin M.T., et al. Robustaflavone, a naturally occurring biflavanoid, is a potent non-nucleoside inhibitor of hepatitis B virus replication in vitro. Bioorg. Med. Chem. Lett. 1997;7:2325–2328. [Google Scholar]
- Lin Y.M., Anderson H., Flavin M.T., et al. In vitro anti-HIV activity of biflavonoids isolated from Rhus succedanea and Garcinia multiflora. J. Nat. Prod. 1997;60:884–888. doi: 10.1021/np9700275. [DOI] [PubMed] [Google Scholar]
- Ling C.Q. Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-CoV-2) J. Integr. Med. 2020;18:87–88. doi: 10.1016/j.joim.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]; Lu et al., 2015 Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K., Nagai T., Suzuki K., et al. Anti-influenza virus activity of biflavonoids. Bioorg. Med. Chem. Lett. 2007;17:772–775. doi: 10.1016/j.bmcl.2006.10.075. [DOI] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Pietta P.G. Flavonoids as antioxidants. J. Nat. Prod. 2000;63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- Ryu Y.B., Jeong H.J., Kim J.H., et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y.B., Park S.-J., Kim Y.M., et al. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorg. Med. Chem. Lett. 2010;20:1873–1876. doi: 10.1016/j.bmcl.2010.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- Shamsi A., Mohammad T., Anwar S., et al. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40(6) doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.M., Santos M.R., Caroço G., et al. Structure-antioxidant activity relationships of flavonoids: a re-examination. Free Radic. Res. 2002;36:1219–1227. doi: 10.1080/198-1071576021000016472. [DOI] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59–86. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutomu N., Munetaka Y., Toshihiko O., et al. Suppression of active oxygen-induced cytotoricity by flavonoids. Biochem. Pharmacol. 1993;45:265–267. doi: 10.1016/0006-2952(93)90402-i. [DOI] [PubMed] [Google Scholar]
- Vitorino J., Sottomayor M. DNA interaction with flavone and hydroxyflavones. J. Mol. Struct. 2010;975:292–297. [Google Scholar]
- Wang G., Yao S., Zhang X.X., Song H. Rapid screening and structural characterization of antioxidants from the extract of Selaginella doederleinii Hieron with DPPH-UPLC-Q-TOF/MS method. International Journal of Analatical Chemistry. 2015;849769 doi: 10.1155/2015/849769. [DOI] [PMC free article] [PubMed] [Google Scholar]; WHO, 2004 WHO . WHO; 2004. Summary of Probably SARS Cases with Onset of Illness from 1st November (2002) to 31st July 2003.http://www.who.int/csr/sars/country/table2004_04_21/en/ [Google Scholar]; WHO, 2016 WHO . WHO; 2016. Coronavirus Infections: Disease Outbreak News.http://www.who.int/csr/don/26-april-2016-mers-saudi-arabia/en/ [Google Scholar]
- Wilsky S., Sobotta K., Wiesener N., et al. Inhibition of fatty acid synthase by amentoflavone reduces coxsackievirus B3 replication. Arch. Virol. 2012;157:259–269. doi: 10.1007/s00705-011-1164-z. [DOI] [PubMed] [Google Scholar]
- World Health Organization Coronavirus disease (COVID-19) situation report‐207. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., et al. bioRxiv; China: 2020. Complete Genome Characterisation of a Novel Coronavirus Associated with Severe Human Respiratory Disease in Wuhan. [DOI] [Google Scholar]
- Xia S., Liu M., Wang C., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang Y. Traditional Chinese medicine treatment of COVID-19, Complement. Ther. Clin. Pract. 2020:101165. doi: 10.1016/j.ctcp.2020.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Hu Y.X., Li Y.C., et al. In vitro DNA binding studies of lenalidomide using spectroscopic in combination with molecular docking techniques. J. Mol. Struct. 2018;1154:9–18. [Google Scholar]
- Yamamoto M., Matsuyama S., Li X., et al. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zembower D.E., Lin Y.M., Flavin M.T., et al. Robustaflavone, a potential non-nucleoside anti-hepatitis B agent. Antivir. Res. 1998;39:81–88. doi: 10.1016/s0166-3542(98)00033-3. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang Z., Chen W., et al. Predicting the receptor-binding domain usage of the coronavirus based on kmer frequency on spike protein. Infect. Genet. Evol. 2018;61:183–184. doi: 10.1016/j.meegid.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]