Abstract
Ubr1 is a conserved ubiquitin ligase involved in the degradation of aberrant proteins in eukaryotic cells. The human enzyme is found mutated in patients with Johanson-Blizzard syndrome. We hypothesized that Ubr1 is necessary for optimal cellular fitness in conditions associated with elevated abundance of aberrant and misfolded proteins. Indeed, we found that loss of Ubr1 in the model eukaryotic microorganism Saccharomyces cerevisiae strongly sensitizes cells to hygromycin B, which reduces translational fidelity by causing ribosome A site distortion. Our results are consistent with a prominent role for Ubr1 in protein quality control. We speculate that disease manifestations in patients with Johanson-Blizzard syndrome are linked, at least in part, to defects in protein quality control caused by loss of Ubr1 function.
Introduction
The structure and function of the Ubr1 ubiquitin ligase are conserved across diverse eukaryotic organisms (8). In humans, mutation of UBR1 causes Johanson-Blizzard syndrome, a disorder characterized by multiorgan dysfunction, physical malformations, and cognitive impairment (19). Ubr1 has been extensively investigated in the model unicellular eukaryote Saccharomyces cerevisiae (budding yeast), where it contributes to multiple aspects of protein quality control. Experiments performed with yeast have been foundational in the discovery of molecular mechanisms of quality control conserved among eukaryotes (3). Among other roles, Ubr1 promotes turnover of substrates of the N-end rule (17), endoplasmic reticulum-associated degradation (ERAD) (12, 13), stress-induced homeostatically regulated protein degradation (SHRED) (14), and cytoplasmic quality control (CytoQC) (7, 11) pathways. Yeast lacking Ubr1 exhibit divergent responses to pharmacologic interventions expected to increase the abundance of misfolded proteins. For example, ubr1Δ yeast display enhanced sensitivity to the Hsp90 inhibitor geldanamycin, while they are resistant to the proline analog L-azetidine-2-carboxylic acid (15).
Materials
All yeast strains used in this study are presented in Table 1. These strains have been constructed in previous reports (6, 16). The entire coding sequences of UBR1, HRD1, and DOA10 have been replaced with the kanMX4 allele in gene knockout strains (16). Yeast were cultured in yeast extract-peptone-dextrose medium (1% yeast extract, 2% peptone, 2% glucose, 0.002% adenine, 2% agar) (5) with the indicated concentrations of hygromycin B (Corning).
Table 1. Yeast strains used in this study.
All strains used in this study are congenic with BY4741 (16).
| Name | Alias | Genotype | Source |
|---|---|---|---|
| VJY476 | BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | (16) |
| VJY22 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 hrd1Δ::kanMX4 | (16) | |
| VJY102 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 doa10Δ::kanMX4 | (16) | |
| VJY305 | SKY252 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 hrd1Δ::kanMX4 doa10Δ::kanMX4 | (6) |
| VJY469 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 ubr1Δ::kanMX4 | (16) |
Methodology and Results
We tested the hypothesis that Ubr1 is necessary for optimal growth in conditions associated with elevated abundance of aberrant proteins. The aminoglycoside hygromycin B reduces translational fidelity by causing ribosome A site distortion and is toxic to yeast at 200 μg/ml (1, 4, 9). We analyzed the growth of yeast lacking Ubr1 in the presence of sublethal doses of hygromycin B, which are expected to increase the cellular concentration of misfolded proteins (Figure 1). Wild type yeast, ubr1Δ yeast, and yeast lacking one or both genes encoding the primary ERAD ubiquitin ligases (HRD1 and DOA10) (10) were subjected to six-fold serial dilution, beginning with an optical density at 600 nm of 0.2. Each dilution (4 μl) was spotted onto agar plates containing rich yeast growth medium with no drug or increasing concentrations of hygromycin B. Plates were incubated at 30°C and imaged at the indicated times. A detailed explanation of the yeast growth assay procedure can be found in (18). All data were analyzed using Prism software (GraphPad Software Inc., San Diego, CA). Because of highly variable data, values of zero were normalized to 1 to make all data positive and positive data were log-transformed. All means between groups were compared by one-way ANOVA followed by Tukey post-hoc analysis. A P value less than 0.05 was designated as statistically significant.
Figure 1. Loss of UBR1 sensitizes yeast to hygromycin B.
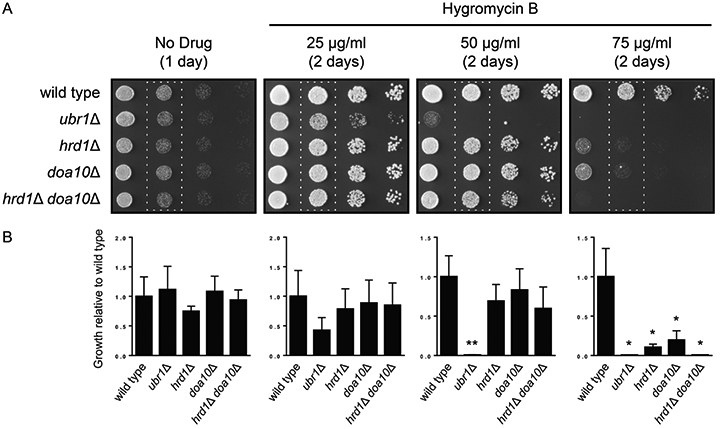
(A) Six-fold serial dilutions of yeast of the indicated genotypes were spotted onto agar plates containing rich medium (No Drug) or rich medium containing increasing concentrations of hygromycin B. Plates were imaged after 1-2 days (as indicated) of incubation at 30°C. (B) Growth in the second column of each plate (dashed rectangles) from three replicate experiments was quantified by densitometry. Data were analyzed by one-way ANOVA followed by Tukey post-hoc analysis (*, less than wild type; **, less than wild type, hrd1Δ, doa10Δ, and hrd1Δ doa10Δ; p < 0.05). Error bars represent standard error of the mean.
In the absence of hygromycin B, all yeast strains exhibited similar growth. Consistent with previous results (2), yeast lacking both HRD1 and DOA10 exhibited a pronounced growth defect in the presence of 75 μg/ml hygromycin B. Individual deletion of HRD1 or DOA10 also impaired growth in the presence of hygromycin B, but to a lesser extent than the double mutant. Finally, loss of UBR1 impaired growth in the presence of the compound more severely than any of the other mutations tested; this is most evident at 50 μg/ml hygromycin B.
This experiment was piloted by undergraduate students in the Methods in Cell Biology (BIO 315) Course at Ball State University and has been validated by three replicates in the research laboratories of EMR and PJS.
Discussion
As we hypothesized, our results indicate Ubr1 is crucial for optimal growth of yeast in conditions associated with elevated abundance of aberrant proteins, consistent with Ubr1 function in protein quality control. Mutations in UBR1 are found in patients with Johanson-Blizzard syndrome. A previous study demonstrated that homologous mutations also reduce Ubr1 function in yeast (8). We speculate that disease phenotypes present in patients with Johanson-Blizzard syndrome harboring mutations in UBR1 are linked, at least in part, to defects in protein quality control. One limitation of the present work is that hygromycin B is expected to trigger the accumulation of a large and heterogeneous population of aberrant proteins. Therefore, it is difficult to determine which types of protein aberrancies present the most substantial challenge to cellular health in the absence of Ubr1. Future biochemical experiments will be necessary to characterize the substrate range of Ubr1 in yeast and human cells. Such biochemical analyses may also provide insight into the divergent responses of ubr1Δ yeast to different forms of cellular stress.
Acknowledgements
This work was funded by NIH grant R15 GM111713 (EMR). This project was conceived while EMR was supported in part by a Ball State University Excellence in Teaching award. Research in the lab of PJS is supported by Grant 18-IIA-406 from the Amyotrophic Lateral Sclerosis Association. We thank the Ball State University Department of Biology (particularly Kemuel Badger, Clare Chatot, and Susan McDowell) for material and moral support for the design and implementation of an inquiry-based research course, where this experiment was piloted. We thank Jacob Price for serving as a Teaching Assistant in that course. We thank Bryce Buchanan and Courtney Broshar for technical assistance.
References
- 1.Brodersen DE, Clemons WM Jr., Carter AP, Morgan-Warren RJ, Wimberly BT, & Ramakrishnan V (2000). The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell, 103(7), 1143–1154. https://www.cell.com/fulltext/S0092-8674(00)00216-6 [DOI] [PubMed] [Google Scholar]
- 2.Crowder JJ, Geigges M, Gibson RT, Fults ES, Buchanan BW, Sachs N, … Rubenstein EM (2015). Rkr1/Ltn1 Ubiquitin Ligase-Mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J Biol Chem, 290(30), 18454–18466. http://www.jbc.org/content/290/30/18454.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finley D, Ulrich HD, Sommer T, & Kaiser P (2012). The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics, 192(2), 319–360. https://www.genetics.Org/content/192/2/319.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganoza MC, & Kiel MC (2001). A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob Agents Chemother, 45(10), 2813–2819. https://aac.asm.org/content/45/10/2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guthrie C, & Fink GR (2004). Guide to Yeast Genetics and Molecular and Cell Biology. San Diego: Elsevier. [Google Scholar]
- 6.Habeck G, Ebner FA, Shimada-Kreft H, & Kreft SG (2015). The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J Cell Biol, 209(2), 261–273. http://jcb.rupress.org/content/209/2/261.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heck JW, Cheung SK, & Hampton RY (2010). Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A, 107(3), 1106–1111. https://www.pnas.org/content/107/3/1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang CS, Sukalo M, Batygin O, Addor MC, Brunner H, Aytes, … Zenker M (2011). Ubiquitin ligases of the N-end rule pathway: assessment of mutations in UBR1 that cause the Johanson-Blizzard syndrome. PLoS One, 6(9), e24925 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0024925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaster KR, Burgett SG, & Ingolia TD (1984). Hygromycin B resistance as dominant selectable marker in yeast. Curr Genet, 8(5), 353–358. https://link.springer.com/article/10.1007/BF00419824 [DOI] [PubMed] [Google Scholar]
- 10.Mehrtash AB, & Hochstrasser M (2018). Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope. Semin Cell Dev Biol. https://www.sciencedirect.com/science/article/pii/S1084952118300673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, … Caplan AJ (2010). Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell, 21(13), 2102–2116. https://www.molbiolcell.org/doi/full/10.1091/mbc.e10-02-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggiano A, Mora G, Buxo L, & Carvalho P (2016). Spatial control of lipid droplet proteins by the ERAD ubiquitin ligase Doa10. EMBO J, 35(15), 1644–1655. https://www.embopress.org/doi/full/10.15252/embj.201593106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolz A, Besser S, Hottmann H, & Wolf DH (2013). Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc Natl Acad Sci U S A, 110(38), 15271–15276. https://www.pnas.org/content/110/38/15271.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szoradi T, Schaeff K, Garcia-Rivera EM, Itzhak DN, Schmidt RM, Bircham PW, … Schuck S (2018). SHRED Is a Regulatory Cascade that Reprograms Ubr1 Substrate Specificity for Enhanced Protein Quality Control during Stress. Mol Cell, 70(6), 1025–1037 e1025. https://www.cell.com/molecular-cell/fulltext/S1097-2765(18)30349-6 [DOI] [PubMed] [Google Scholar]
- 15.Theodoraki MA, Nillegoda NB, Saini J, & Caplan AJ (2012). A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J Biol Chem, 287(28), 23911–23922. http://www.jbc.org/content/287/28/23911.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, … Boone C (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science, 294(5550), 2364–2368. https://science.sciencemag.org/content/294/5550/2364.full [DOI] [PubMed] [Google Scholar]
- 17.Varshavsky A (2011). The N-end rule pathway and regulation by proteolysis. Protein Sci, 20(8), 1298–1345. https://onlinelibrary.wiley.com/doi/full/10.1002/pro.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts SG, Crowder JJ, Coffey SZ, & Rubenstein EM (2015). Growth-based determination and biochemical confirmation of genetic requirements for protein degradation in Saccharomyces cerevisiae. Journal of Visualized Experiments, (96), e52428 https://www.jove.com/video/52428/growth-based-determination-biochemical-confirmation-genetic [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, … Reis A (2005). Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat Genet, 37(12), 1345–1350. https://www.nature.com/articles/ng1681 [DOI] [PubMed] [Google Scholar]


