Abstract
Fast photochemical oxidation of proteins (FPOP) is a hydroxyl radical protein footprinting method that covalently labels solvent-accessible amino acids by photolysis of hydrogen peroxide. Recently, we expanded the use of FPOP for in vivo (IV-FPOP) covalent labeling in C. elegans. In initial IV-FPOP studies, 545 proteins were oxidatively modified in all body systems within the worm. Here, with the use of chemical penetration enhancers (CPEs), we increased the number of modified proteins as well as the number of modifications per protein to gain more structural information. CPEs aid in the delivery of hydrogen peroxide inside C. elegans by disturbing the highly ordered lipid bilayer of the worm cuticle without affecting worm viability. IV-FPOP experiments performed using the CPE azone showed an increase in oxidatively modified proteins and peptides. This increase correlated with greater hydrogen peroxide uptake by C. elegans quantified using a chemical fluorophore demonstrating the efficacy of using CPEs with IV-FPOP. Mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD019290.
Keywords: in vivo, protein footprinting, FPOP, C. elegans, chemical penetration enhancers
Introduction
Mass spectrometry (MS) has emerged in recent years as a powerful method for analyzing protein structures.1 Specifically, MS-based protein footprinting methods have been used to study protein conformational changes as well as protein–protein and protein–ligand interactions.2−4 These methods utilize a chemical label to monitor changes in solvent accessibility in response to binding events or conformational changes. Depending on the chemical label used, these interactions can monitor changes in solvent accessibility at various time scales extending from nanoseconds to minutes. Hydroxyl radical protein footprinting (HRPF) is a labeling method that utilizes hydroxyl (OH) radicals to label solvent-accessible side chains in proteins on the nanosecond to second time scale.5 In HRPF, OH radicals are generated by several means including water radiolysis, fenton chemistry, or laser photolysis of hydrogen peroxide.6−8 These OH radicals irreversibly label solvent-accessible amino acid side chains. Subsequent downstream proteomic analysis by liquid chromatography tandem MS is then used to identify labeled amino acids and quantify the extent of labeling.
Fast photochemical oxidation of proteins (FPOP) is an HRPF method that utilizes a 248 nm KrF excimer laser for photolysis of hydrogen peroxide to label proteins on the microsecond time scale in the absence of secondary radicals.8−11In vitro applications of FPOP include the study of protein conformers, conformational changes, and protein–ligand interactions.12−14 Recently, we expanded the use of FPOP for the study of protein structure in vivo in Caenorhabditis elegans (C. elegans), in a new method entitled in vivo FPOP (IV-FPOP).15C. elegans, who share a common ancestor with humans, have been used as a model system for the study of many human diseases.16 While we were able to show the capabilities of IV-FPOP for in vivo protein structural analysis, protein oxidative coverage and the total number of peptides per single protein modified were less than desirable. In total, 545 proteins were oxidatively modified within the worm with an average of one peptide modified per protein. An increase in both the number of modified proteins and the number of modifications per protein would increase the structural information obtained by IV-FPOP.
HRPF methods are susceptible to radical scavenging by different buffer compositions and chemical additives. This scavenging can have a significant impact on the extent of oxidative modifications during in vitro FPOP experiments by suppressing the final radical concentration available in the solution after hydrogen peroxide photolysis.5,17 Significant advancements have been made on utilizing dosimeters in FPOP experiments to account for these scavenging effects and to compensate for them.18,19 In the case of in vitro FPOP, radical scavenging and quenching effects can be overcome by increasing the hydrogen peroxide concentration to increase the final concentration of hydroxyl radicals.17 However, for in vivo FPOP, increasing the final hydrogen peroxide concentration prior to photolysis is less desirable since it can have negative effects on sample viability as well as unwanted background oxidative stress in the worms. For IV-FPOP, hydrogen peroxide enters C. elegans both by ingestion and penetration through the skin. Skin penetration is limited by the worm cuticle, a highly structured extracellular matrix that protects C. elegans from the environment. The method also relies on diffusion of H2O2 throughout the worm body. Rather than increasing the concentration of hydrogen peroxide mixed with the worms, we aim to increase the uptake of hydrogen peroxide by the worms.
Here, we report the use of chemical penetration enhancers (CPEs) to increase hydrogen peroxide uptake by C. elegans. The use of CPEs has long demonstrated the ability to increase the permeability of drugs and other chemical compounds in humans and animals by disrupting the highly ordered lipid bilayer of the skin.20 The CPEs used should be nontoxic to the organism of interest and prove to be effective within the necessary experimental time scale. In the case of IV-FPOP, the CPE used should also not scavenge the OH radicals. We tested five different CPEs of varying chemistries to determine their efficacy in increasing peroxide uptake while not scavenging the OH radicals. These five CPEs, azone (AZ), dimethylacetamide (DMAC), dimethylformamide (DMF), oleic acid (OA), and propylene glycol (PG) (Figure 1), were selected based on literature reports describing their penetration-enhancing activity.20,21 These CPEs are from different chemical classifications including amides, fatty acids, glycols, and sulfoxide-like molecules.21 We chose to test CPEs with varying chemistries because studies have demonstrated that CPE potency is difficult to predict and appears to be drug-specific.20 We demonstrate that CPEs can increase hydrogen peroxide uptake by C. elegans, which leads to an increase in the number of oxidatively modified proteins by IV-FPOP.
Figure 1.
Chemical structures of the five CPEs tested. Each CPE has varying chemical properties.
Experimental Section
Materials
Unless otherwise noted, all materials were purchased from Thermo Fisher Scientific and used without any further purification.
In Vitro FPOP
The protocol was performed as described by Hambly and Gross with minor modifications.8 Ubiquitin from bovine erythrocytes (Sigma Aldrich) was prepared in 10 mM phosphate-buffered saline (PBS; Sigma Aldrich) at a final concentration of 0.18 mg/mL in a 50 μL final volume. l-glutamine was added to the protein sample as the hydroxyl radical scavenger at a final concentration of 40 mM. The effect of five different CPEs (azone (AZ), dimethylacetamide (DMAC), dimethylformamide (DMF), oleic acid (OA), or propylene glycol (PG); Sigma Aldrich) on the extent of FPOP modification was tested at various concentrations (0, 0.1, 0.5, 1, and 2%). Immediately prior to FPOP, hydrogen peroxide was added to a final concentration of 7.5 mM. The 50 μL sample was infused using a syringe pump through a 150 μm inner diameter (i.d.) fused silica capillary (Polymicro Technologies) using a 34.19 μL/min flow rate, 20% exclusion fraction, 2.58 mm spot width, and 10 Hz laser frequency. A 248 nm KrF excimer laser (GAM Laser, Inc.) was used to irradiate the sample and photolyze hydrogen peroxide at 161 mJ/pulse. Samples were collected in a vial containing a final concentration of 30 mM methionine and 500 nM catalase to quench excess OH radicals and hydrogen peroxide, respectively. A total of three laser-irradiated samples and three controls (no laser irradiation) were prepared for each condition.
Global MS Analysis
Intact mass spectra analysis was completed using a nanoAcquity UPLC in line with a Synapt G2S mass spectrometer (Waters) operating in resolution mode. Each sample was loaded onto a MassPREP Micro Desalting column (Waters), washed for 10 min with 0.1% formic acid (FA) in water (Solvent A), and eluted with 60% acetonitrile (ACN) in 0.1% FA for 10 min at a rate of 100 μL/min. The mass spectrometer electrospray ionization (ESI) source capillary voltage was set at +3.0 kV and 100 °C temperature. The acquisition mass range was 500–2000 m/z. Mass spectra were deconvoluted, and the area of the first +16 modification peak was calculated using UniDec.22
Nematode and Bacterial Culture
C. elegans strain BY250 (Pdat-1::GFP) was maintained following standard methods23 on either 8P plates with NA22 Escherichia coli or nematode growth media (NGM) plates with OP50 E. coli as a food source at 20 °C.
Viability Assay
Worm viability assays were performed on synchronized L4 larvae in a 96-well plate as previously described with minor modifications.15,24 Hydrogen peroxide at a final concentration of 200 mM with the indicated CPE was added to approximately 1000 worms. After 30 s, the reaction was quenched using the IV-FPOP quench solution at a final concentration of 20 mM N,N′-dimethylthiourea (DMTU; Acros Organics) and 20 mM N-tert-butyl-α-phenylnitroe (PBN; Sigma Aldrich). Worms were pelleted by centrifugation at 2000 rpm and washed with M9 buffer (0.02 M KH2PO4, 0.08 M Na2HPO4, 0.08 M NaCl, 1 mM MgSO4). Worms were suspended in M9 with 40 μM propidium iodine (PI; Sigma-Aldrich) and incubated at room temperature for 15 min before fluorescence imaging using a Nikon Eclipse TI microscope.
Hydrogen Peroxide Uptake
Hydrogen peroxide uptake was quantified as described by Fu et al. with some modifications.25 Peroxy Orange 1 (PO1; Tocris Bioscence) was dissolved in DMSO at a stock concentration of 5 mM and then freshly diluted in M9 buffer to prepare 50 μM PO1 working solution. L4 larvae were incubated with 200 mM hydrogen peroxide mixed with the indicated CPE in M9 buffer at room temperature for 30 s. Immediately after, the reaction was quenched using the IV-FPOP quench solution. Worms were collected by centrifugation at 2000 rpm and washed twice with M9 buffer before bathing in the PO1 working solution for 2 h at room temperature with mild shaking in the dark. Afterwards, worms were washed twice with M9 buffer, placed in glass slides with coverslips, and subjected to fluorescence imaging. The images were collected using a Nikon Eclipse TI microscope (10 × 0.3 air Nikon objective) with a Texas Red filter (560 nm excitation and emission in the range of 604–644 nm). Imaging data was processed and analyzed using ImageJ (NIH).
IV-FPOP, Protein Extraction, and Proteolysis
The procedure was performed with minor modifications as previously described.15 Prior to IV-FPOP, worms were kept separate from 200 mM hydrogen peroxide and the indicated CPE, mixed using a homemade flow system,26 and incubated for approximately 30 s just prior to IV-FPOP. Samples were flowed through a 250 μm i.d. capillary at a final flow rate of 379 μL/min, an irradiation window of 2.58 mm, and 50 Hz laser frequency.and the emission in the range of 604 Afterwards, each sample was pelleted, the quench solution was removed, and lysis buffer was added (8 M urea, 0.5% SDS, 50 mM HEPES, 50 mM NaCl, 1 mM EDTA, 1 mM PMSF). Worms were homogenized by sonication, and the protein lysate was separated from the worm debris by centrifugation at 400 g for 5 min at 4 °C. Total protein concentration was quantified using a BCA assay, and 100 μg of protein was obtained for proteolysis. The lysate was reduced with 10 mM dithiothreitol (DTT; AmericanBio) for 45 min at 50 °C, cooled down at room temperature for 10 min, and alkylated with 20 mM iodoacetamide (IAA; Sigma Aldrich) for 20 min in the dark at room temperature. The lysate was purified by acetone precipitation overnight, resuspended in 25 mM Tris-HCl, pH 8, proteolyzed overnight with trypsin at a 1:50 enzyme-to-protein ratio, and quenched with 5% formic acid (FA). The total peptide concentration was quantified using the Pierce Quantitative Colorimetric Peptide Assay (Thermo Scientific) following the manufacturer’s protocol. Peptides (10 μg) were dried by cold trap centrifugation and resuspended in 0.1% FA in water at a final concentration of 0.5 μg/μL.
LC-MS/MS Analysis
Peptides (0.5 μg) were loaded onto an M-class C18 trap column (Waters) and washed for 15 min with 0.1% FA in water at 15 μL/min using an M-class Acquity liquid chromatography (LC) UPLC system (Waters). Retained peptides were eluted and separated using a C18 reverse-phase in-house packed column (0.075 × 20 mm, 5 μm, 125 Å, Phenomenex). The gradient was pumped at 0.3 μL/min for 130 min as follows: 0–1 min, 3% solvent B (0.1% FA in ACN); 2–100 min, 10–45% B; 100–110 min, 45–100% B; held at 100% B for 15 min; and the column was re-equilibrated at 3% B for an additional 15 min. Eluted peptides were analyzed by nano electrospray (nESI) in positive ion mode using an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) using previously described parameters.15
Data Analysis
All mass spectra raw files were searched using Proteome Discoverer 2.2 against a C. elegans database (4039 proteins, downloaded 04/11/2019) using the Sequest 1.4 node allowing for one missed cleavage of tryptic peptides as described previously.27 The precursor mass tolerance was set at 10 ppm with a fragment match tolerance of 0.02 Da. All known side-chain hydroxyl radical modifications5,28 were searched as dynamic modifications using a multisearch node workflow. Carbamidomethylations of cysteine residues were set as fixed modifications. The extent of peptide oxidation was determined using extracted ion chromatograms (XIC) as previously done.15,27
Results and Discussion
Effect of CPEs on In Vitro FPOP Oxidation
A first step in demonstrating the potential of the five CPEs in increasing hydrogen peroxide uptake for IV-FPOP is determining whether they scavenge hydroxyl radicals. To test the effect of CPEs on the oxidation of the pattern of FPOP, in vitro FPOP of ubiquitin was performed in the presence and absence of five different CPEs. The modification distribution and the peak area for the +16 modification for each condition were compared (Figure 2).
Figure 2.
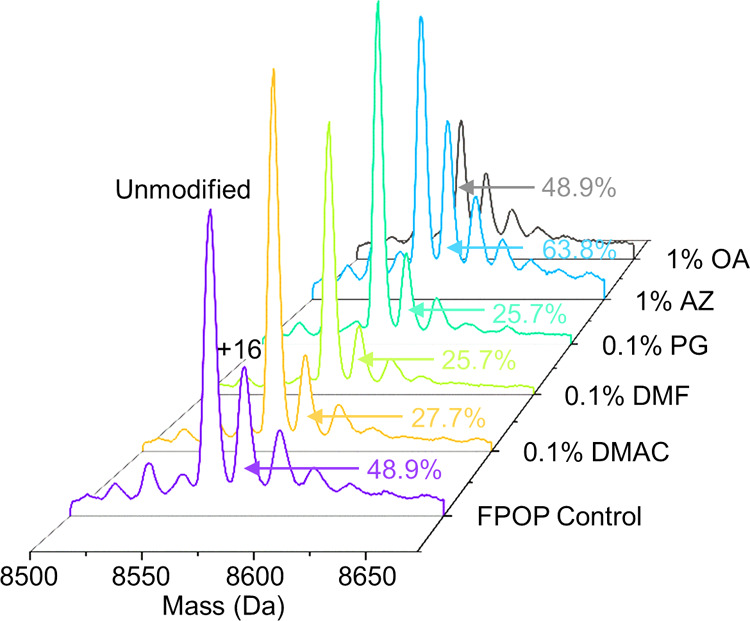
Intact MS analysis of ubiquitin. The peak area for the +16 FPOP modification peak for each CPE condition tested is shown.
Under standard FPOP conditions, the +16 modification peak was 48% of the unmodified peak of ubiquitin (Figure 2). This is typical of FPOP on ubiquitin and indicates a quantifiable level of modification without over labeling the protein. The +16 modification for DMAC, DMF, and PG showed a significant decrease when compared to the FPOP control (no CPE added). The use of 0.1% DMAC showed a 21.2% decrease for the +16 oxidation peak, while a decrease in 23.2% for the +16 oxidation peak was observed for both 0.1% DMF and 0.1% PG (Figure 2). This was not unexpected for DMAC and DMF, which are similar to sulfoxides, known scavengers of hydroxyl radicals, and are used as CPEs in place of dimethylsulfoxide (DMSO), a potent but problematic CPE. For PG, some glycols, specifically ethylene glycol, have also been shown to be efficient OH radical scavengers.29 In contrast, the +16 modification peaks of ubiquitin modified in the presence of AZ or OA did not show any statistical significance decrease at concentrations between 0.5 and 1% (Figure 2 and Supporting Information, Figure S1) when compared to the FPOP control. This suggests that for in vivo FPOP 0.5–1% of either AZ or OA can be used as possible CPEs without observed radical scavenging properties.
C. elegans Viability in the Combined Presence of Hydrogen Peroxide with CPEs and Hydrogen Peroxide Uptake
The use of certain CPEs, such as sulfoxides, at low concentrations (0.5–2%) has shown to increase the lifespan of C. elegans by up to 50%, but worm death has been observed at higher concentrations (>6%).30,31 Previous viability experiments from our group have shown < 2% loss in C. elegans viability when incubated in 200 mM hydrogen peroxide for up to 30 s.15 For IV-FPOP, an important parameter to consider is the additive toxic effects of the combined presence of hydrogen peroxide and a CPE could have on C. elegans viability. Since only two CPEs, AZ and OA, did not show significant radical scavenging properties from the initial five candidates, viability studies using 200 mM hydrogen peroxide were performed in the presence and absence of AZ and OA at varying concentrations (0.5–1%) (Figure 3).
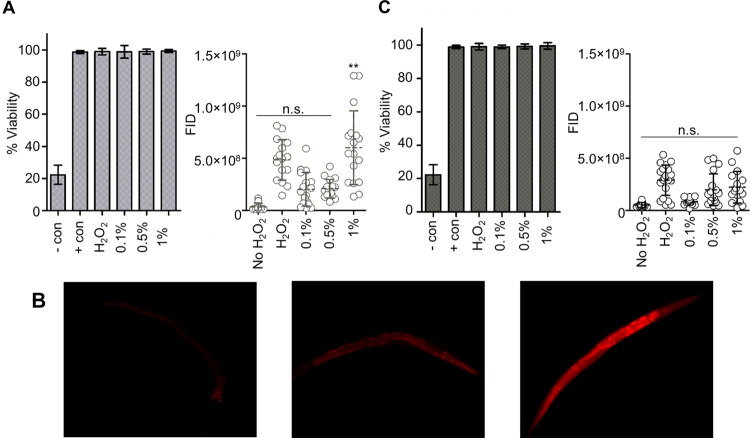
Figure 3.
C. elegans viability and hydrogen peroxide uptake. (A) Percent viability of C. elegans using 200 mM hydrogen peroxide in the presence of 0, 0.1, 0.5, and 1% AZ. Negative control is C. elegans in the presence of 50% methanol, and positive control is C. elegans in M9 buffer. Error bars are calculated across technical triplicates (left). Fluorescence integrated densities (FID) for hydrogen peroxide uptake in the presence of 0–1% AZ (right). (B) Representative C. elegans fluorescence images in response to intercellular hydrogen peroxide: no hydrogen peroxide control (left), 200 mM hydrogen peroxide (middle), and 200 mM hydrogen peroxide with 1% azone (right). (C) Percent viability of C. elegans using 200 mM hydrogen peroxide in the presence of 0, 0.1, 0.5, and 1% OA. Negative control is C. elegans in the presence of 50% methanol, and positive control is C. elegans with no hydrogen peroxide. Error bars are calculated across technical triplicates (left). FID for hydrogen peroxide uptake in the presence of 0–1% OA (right).
The presence of 0.1–1% AZ plus 200 mM hydrogen peroxide did not show any statistically significant loss in C. elegans viability when compared to the 200 mM hydrogen peroxide control after a 30 s incubation (Figure 3A, left). Additionally, neither the 200 mM control nor the 0.1–1% AZ plus 200 mM hydrogen peroxide showed a loss in viability in comparison to the positive control samples composed of C. elegans in the IV-FPOP quench solution. Similarly, 0.1–1% OA plus 200 mM hydrogen peroxide did not show significant changes in viability when compared to samples either in the presence of 200 mM or the absence of hydrogen peroxide controls (Figure 3C, left).
In addition to minor changes in viability, the presence of AZ or OA in combination with hydrogen peroxide should also demonstrate the ability to increase hydrogen peroxide uptake in C. elegans. To quantify hydrogen peroxide uptake in the presence and absence of either AZ or OA in vivo, the fluorophore PO1 was used (Figure 3). PO1 has been shown to linearly quantify in vivo hydrogen peroxide uptake and diffusion in cells and C. elegans.(25,32) For AZ, no statistically significant changes were observed in the uptake of 200 mM hydrogen peroxide in the presence of 0.1–0.5% AZ. Only 1% AZ showed a significant increase in hydrogen peroxide uptake quantified by PO1 fluorescence (Figure 3A, right). The uptake of hydrogen peroxide by C. elegans using 1% AZ was 1.24-fold higher compared to the samples containing 200 mM hydrogen peroxide in the absence of 1% AZ. Fluorescence imaging of hydrogen peroxide-treated worms in the presence and absence of 1% AZ shows the difference in uptake in the presence of AZ (Figure 3B). These imaging studies also demonstrate diffusion of hydrogen peroxide throughout the full body length of the worm enabling the modification of proteins across all body systems within the worm. Unforeseen, for OA, none of the concentrations tested, 0.1–1%, showed an increase in hydrogen peroxide uptake (Figure 3C, right). These results indicated that only 1% AZ can have advantageous effects in increasing hydrogen peroxide uptake, thus increasing the number of hydroxyl radicals generated inside the worm and ultimately increasing the number of oxidatively modified proteins and peptides by IV-FPOP. For subsequent IV-FPOP experiments, 200 mM hydrogen peroxide in the presence and absence of 1% AZ was used.
IV-FPOP in the Presence of the CPE Azone
For AZ to be considered a good CPE, the number of oxidatively modified proteins and peptides by IV-FPOP should be greater than the hydrogen peroxide only control. LC-MS/MS analysis of oxidatively modified proteins by IV-FPOP in the absence of AZ showed 1084 ± 81 total modified proteins, while 1140 ± 207 total modified proteins were observed in the presence of AZ (Figure 4A and Tables S1 and S2).
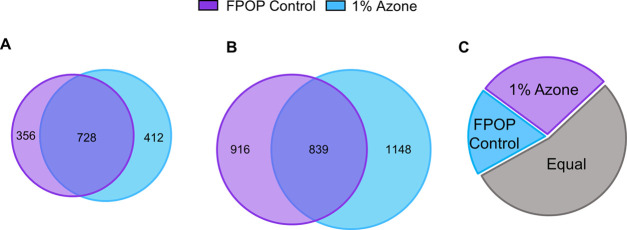
Figure 4.
IV-FPOP oxidatively modified proteins and peptides. (A) Oxidatively modified proteins using 200 mM hydrogen peroxide in the presence and absence of 1% AZ. (B) Oxidatively modified peptides using 200 mM hydrogen peroxide in the presence and absence of 1% AZ. (C) Pie chart of oxidatively modified peptides per single protein. Purple represents the number of proteins that show more oxidatively modified peptides per protein using 1% AZ (28%). Blue represents the number of proteins that show more oxidatively modified peptides per protein in the absence of 1% AZ (18%). Gray is the number of proteins with equal oxidatively modified peptides per protein in both conditions (54%).
Across two biological replicates, the use of hydrogen peroxide in combination with AZ increased the total number of oxidatively modified proteins by 1.05-fold. Likewise, an increase in the total number of oxidatively modified peptides was observed in the presence of AZ across two biological replicates (Figure 4B and Table S1). In the IV-FPOP control group, only 1755 ± 291 peptides were oxidatively modified, while 1987 ± 556 oxidatively modified peptides were quantified in the presence of AZ (Tables S1 and S2), equal to a 1.13-fold increase in modified peptides across two biological replicates. While the final number of oxidatively modified proteins and peptides did not increase dramatically, the increase fold differences are consistent with the hydrogen peroxide uptake quantified above (Figure 3A, right). The increase is also seen in the total number of oxidatively modified proteins in the background oxidation control samples in the presence and absence of 1% AZ (Supporting Information, Figure S2).
To further demonstrate the advantage of using 1% AZ as a CPE against the IV-FPOP control group, the extent of IV-FPOP oxidation at the peptide level for the protein myosin was calculated (Supporting Information, Figure S3). Myosin is a highly abundant protein in C. elegans and responsible for muscle contraction and intracellular transport. In the presence of 1% AZ, one additional peptide was detected in comparison to the IV-FPOP control group without AZ. This increase in the total number of oxidatively modified peptides per single protein was observed in 203 (28%) proteins when AZ was used (Figure 4C). Still, 399 (54%) proteins showed an equal number of oxidatively modified peptides per single protein in the presence and absence of AZ, while only 132 (18%) proteins showed more modified peptides in the absence of AZ (Figure 4C).
IV-FPOP in the Presence of Two CPEs
The use of single CPEs has been shown to offer limited permeability in drug delivery. As a result, a common practice is to employ a mixture of CPEs to enhance drug delivery.33 To test if these synergistic interactions can increase the uptake of hydrogen peroxide in IV-FPOP, the combined use of two CPEs was implemented. Since the initial screening demonstrated 1% AZ and 1% OA did not affect the labeling pattern of in vitro FPOP, two different combinations of AZ in addition to OA were tested (Figure 5).
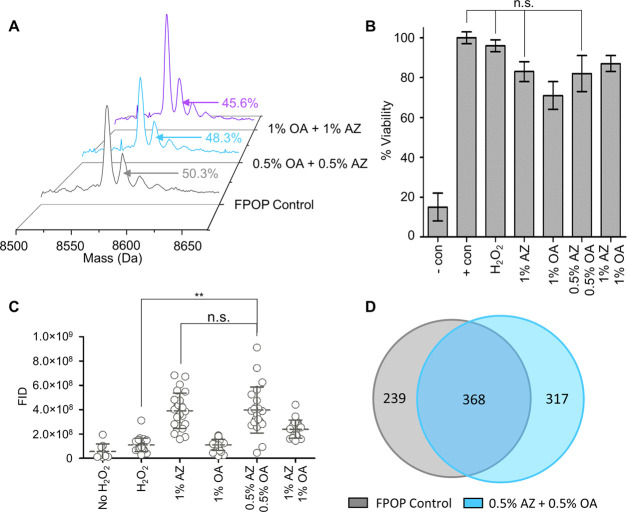
Figure 5.
In vitro and IV-FPOP using two CPEs. (A) Intact MS analysis of ubiquitin. The peak area for the +16 FPOP modification peak for each 2CPE condition tested is shown. (B) Percent viability of C. elegans using 200 mM hydrogen peroxide in the presence of AZ, OA, and 2CPEs. Negative control C. elegans are in the presence of 50% methanol, and positive control were not exposed to hydrogen peroxide. Error bars are calculated across technical triplicates. (C) FID for hydrogen peroxide uptake in the presence of AZ, OA, and 2CPEs. (D) Oxidatively modified proteins using 200 mM hydrogen peroxide in the presence and absence of 0.5% 2CPEs.
When comparing the peak area for the +16 modification of in vitro FPOP on ubiquitin, a 2% decrease in oxidation was observed when using 0.5% AZ plus 0.5% OA (0.5% 2CPEs), while a 4.7% decrease in oxidation was observed when using 1% AZ plus 1% OA (1% 2CPEs) (Figure 5A). However, the decrease in oxidation was not statistically significant, indicating that either combination of both CPEs is applicable for IV-FPOP.
Viability experiments in the presence of either 0.5% 2CPEs or 1% 2CPEs showed no statistically significant decrease in C. elegans viability when exposed to 200 mM hydrogen peroxide in comparison to the hydrogen peroxide control (Figure 5B). Interestingly, hydrogen peroxide uptake quantitation showed only 0.5% 2CPEs to significantly increase the absorption of hydrogen peroxide by C. elegans, while 1% 2CPEs did not show an increase (Figure 5C). Across two biological replicates, IV-FPOP experiments using 0.5% 2CPEs showed 685 ± 70 oxidatively modified proteins (Table S3), while only 607 ± 13 proteins were modified in the absence of 0.5% 2CPEs (Table S4), equivalent to a 1.13-fold increase in oxidatively modified proteins (Figure 5D and Tables S1 and S2). Unexpectedly, the fold increase in oxidatively modified proteins by 0.5% 2CPEs is equal to the increase in modified proteins when only 1% AZ is present. Although both conditions show an increase in hydrogen peroxide uptake when compared to hydrogen peroxide control (Figure 5C), 0.5% 2CPEs and 1% AZ are not statistically significant when compared to each other. These results suggest that AZ is majorly responsible for the increase in oxidatively modified proteins by IV-FPOP, and the combined use of AZ and OA is not additive.
Conclusions
Here, we report the use of CPEs to increase the total number of oxidatively modified proteins and peptides by IV-FPOP. Through in vitro FPOP, viability, and hydrogen peroxide uptake quantitation, only one CPE, AZ, showed to be advantageous to increase oxidatively modified proteins by IV-FPOP. This amide molecule did not scavenge OH radicals but did increase the uptake of hydrogen peroxide through the skin leading to a higher number of oxidatively modified proteins. This first attempt at using CPEs indicates the effectiveness of using this as an approach to increase the number of oxidatively modified proteins and modified peptides by IV-FPOP. This will increase the structural information obtained by IV-FPOP, thus increasing its utility as a method for structural biology. Further optimizations such as increased incubation time with the CPE may be useful to further increase the number of modified proteins. However, this must be carefully controlled as to not structurally damage the cuticle of the worms. There are strains of C. elegans that have disruptions in the low permeable cuticle, but these strains have other phenotypes that alter the biological function and may not be desirable when studying certain disease states.34,35 Perhaps the most useful approach would be to test more CPEs to determine which works best with hydrogen peroxide. Since it has been demonstrated that CPE efficacy is drug- or chemical compound-specific, a trial and error study of multiple CPEs would be useful in identifying the best CPE for hydrogen peroxide uptake. Our data demonstrates this with the OA CPE, which was not compatible with H2O2, even though its CPE activity for other compounds is well documented. Studies have shown that the mechanisms of enhancement between OA and AZ are different. The cis double bond in OA contributes to the mechanism of enhancement of this molecule and differentiates OA from AZ.36 It is highly probable that H2O2 is reducing this double bond lowering the efficacy of this molecule as a penetration enhancer.37,38 Literature reports show that the cis double bound in OA is oxidizable by reactive oxygen species (ROS) to synthesize azelaic and pelargonic acid.39,40 AZ, which worked well as an H2O2 enhancer, does not have this double bond. Here, we have tested CPEs classified as sulfoxide-like, fatty acids, amides, and glycols. Other chemistries available include alcohols, esters, glycol esters, pyrrolidones, surfactants, and terpenes. Undoubtedly, some of the molecules in these categories will be radical scavengers but others may be useful as CPEs for hydrogen peroxide uptake.
The IV-FPOP study testing the 2CPE combination had significantly fewer proteins modified than the original AZ study. Across two biological replicates in the AZ study, 1084 proteins were modified in the FPOP control sample, while only 607 were modified in the FPOP control for the 2CPE study. This difference in modification could be due to unforeseen scavenging and underscores the need for dosimetry in IV-FPOP to detect and compensate for scavenging. The increase in oxidatively modified proteins shown in the AZ study was concurrent with the quantified increase in hydrogen peroxide uptake using the PO1 fluorophore. This demonstrates the utility of PO1 in quantifying hydrogen peroxide uptake and the potential to use this molecule to compare hydrogen peroxide conditions across replicate worm samples. PO1 is highly specific for H2O2 over other reactive oxygen species,32 so it is sensitive to hydrogen peroxide quantification though is short of a true dosimeter for FPOP studies, where quantification of OH radicals is most desirable. It is unclear whether PO1 would be a good dosimeter for IV-FPOP because radical generation would lead to a decrease in fluorescence potentially leading to a loss in sensitivity of the measurement and because the molecule is not a direct measure of radical generation. However, the usefulness of PO1 quantifying hydrogen peroxide uptake does suggest a fluorescence-based dosimeter may be successful for IV-FPOP.
This initial CPE study does demonstrate the effectiveness that these molecules have in increasing the hydrogen peroxide uptake of C. elegans. IV-FPOP in the presence of 1% AZ showed an increase in the final number of oxidatively modified peptides per single protein, thus demonstrating the ability to gain higher structural information in vivo. Nonetheless, a higher increase in oxidatively modified proteins and peptides is still desired, requiring further screening of compatibility of other CPE categories with IV-FPOP, like alcohols, amides, or esters.
Acknowledgments
The authors thank Paige N. Zambrana and Dr. Audra L. Stinchcomb for the helpful discussion and Emily E. Chea and Dr. Alan McKenzie-Coe for help with editing.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00245.
Intact mass analysis of ubiquitin (Figure S1); background oxidation (Figure S2); C. elegans myosin extent of oxidation (Figure S3).
IV-FPOP oxidatively modified proteins using 200 mM hydrogen peroxide plus 1% AZ (Table S1); IV-FPOP oxidatively modified proteins using 200 mM hydrogen peroxide only (Table S2); IV-FPOP oxidatively modified proteins using 200 mM hydrogen peroxide plus 2CPEs (Table S3); IV-FPOP oxidatively modified proteins using 200 mM hydrogen peroxide (2CPE FPOP Control) (Table S4) (XLSX)
Author Contributions
L.M.J. devised the project, Z.Z. performed hydrogen peroxide absorption imaging, and J.A.E. performed viability, IV-FPOP experiments, and data analysis. L.M.J. and J.A.E. wrote the manuscript.
This work was supported by a grant from the NIH 1R01 GM 127595.
The authors declare no competing financial interest.
Notes
† Frontida BioPharm, Inc., 1100 Orthodox Street, Philadelphia, Pennsylvania 19124, United States.
Notes
This article was written in partial fulfillment of J.A.E. Ph. D. thesis.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE41 partner repository with the data set identifier PXD019290.
Supplementary Material
References
- Kaur U.; Johnson D. T.; Chea E. E.; Deredge D. J.; Espino J. A.; Jones L. M. Evolution of Structural Biology through the Lens of Mass Spectrometry. Anal. Chem. 2019, 91, 142–155. 10.1021/acs.analchem.8b05014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpikirati P.; Pan X.; Vachet R. W. Covalent Labeling with Diethylpyrocarbonate: Sensitive to the Residue Microenvironment, Providing Improved Analysis of Protein Higher Order Structure by Mass Spectrometry. Anal. Chem. 2019, 91, 8516–8523. 10.1021/acs.analchem.9b01732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Moniz H.; Wang S.; Ramiah A.; Zhang F.; Moremen K. W.; Linhardt R. J.; Sharp J. S. High structural resolution hydroxyl radical protein footprinting reveals an extended Robo1-heparin binding interface. J. Biol. Chem. 2015, 290, 10729–40. 10.1074/jbc.M115.648410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Rempel D. L.; Zhang J.; Sharma A. K.; Mirica L. M.; Gross M. L. Pulsed hydrogen–deuterium exchange mass spectrometry probes conformational changes in amyloid beta (Aβ) peptide aggregation. Proc. Natl. Acad. Sci. 2013, 110, 14604–14609. 10.1073/pnas.1309175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.; Chance M. R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007, 107, 3514–43. 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- Chance M. R.; Sclavi B.; Woodson S. A.; Brenowitz M. Examining the conformational dynamics of macromolecules with time-resolved synchrotron X-ray ‘footprinting’. Structure 1997, 5, 865–9. 10.1016/S0969-2126(97)00241-4. [DOI] [PubMed] [Google Scholar]
- Leser M.; Chapman J. R.; Khine M.; Pegan J.; Law M.; Makkaoui M. E.; Ueberheide B. M.; Brenowitz M. Chemical Generation of Hydroxyl Radical for Oxidative ‘Footprinting’. Protein Peptide Lett. 2019, 26, 61–69. 10.2174/0929866526666181212164812. [DOI] [PubMed] [Google Scholar]
- Hambly D. M.; Gross M. L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am. Soc. Mass Spectrom. 2005, 16, 2057–63. 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Chen J.; Rempel D. L.; Gau B. C.; Gross M. L. Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J. Am. Chem. Soc. 2012, 134, 18724–31. 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. S.; Shi L.; Gross M. L. Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization. Acc. Chem. Res. 2018, 51, 736–744. 10.1021/acs.accounts.7b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahidi S.; Konermann L. Probing the Time Scale of FPOP (Fast Photochemical Oxidation of Proteins): Radical Reactions Extend Over Tens of Milliseconds. J Am. Soc. Mass Spectrom. 2016, 27, 1156–64. 10.1007/s13361-016-1389-x. [DOI] [PubMed] [Google Scholar]
- Chea E. E.; Deredge D. J.; Jones L. M. Insights on the Conformational Ensemble of Cyt C Reveal a Compact State during Peroxidase Activity. Biophys. J. 2020, 118, 128–137. 10.1016/j.bpj.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B.; Gross M. L. MS-Based Hydroxyl Radical Footprinting: Methodology and Application of Fast Photochemical Oxidation of Proteins (FPOP). In Mass Spectrometry-Based Chemical. Proteomics 2019, 363–416.30779878 [Google Scholar]
- Johnson D. T.; Di Stefano L. H.; Jones L. M. Fast photochemical oxidation of proteins (FPOP): A powerful mass spectrometry-based structural proteomics tool. J. Biol. Chem. 2019, 294, 11969–11979. 10.1074/jbc.REV119.006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino J. A.; Jones L. M. Illuminating Biological Interactions with in Vivo Protein Footprinting. Anal. Chem. 2019, 91, 6577–6584. 10.1021/acs.analchem.9b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum-Krammer C. I.; Morimoto R. I. Caenorhabditis elegans as a model system for studying non-cell-autonomous mechanisms in protein-misfolding diseases. Dis. Models Mech. 2014, 7, 31–39. 10.1242/dmm.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S. K.; Orlando R.; Weinberger S. R.; Sharp J. S. Compensated Hydroxyl Radical Protein Footprinting Measures Buffer and Excipient Effects on Conformation and Aggregation in an Adalimumab Biosimilar. AAPS J. 2019, 21, 87. 10.1208/s12248-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. S.; Misra S. K.; Persoff J. J.; Egan R. W.; Weinberger S. R. Real Time Normalization of Fast Photochemical Oxidation of Proteins Experiments by Inline Adenine Radical Dosimetry. Anal. Chem. 2018, 90, 12625–12630. 10.1021/acs.analchem.8b02787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B.; Mackness B. C.; Rempel D. L.; Zhang H.; Cui W.; Matthews C. R.; Zitzewitz J. A.; Gross M. L. Incorporation of a Reporter Peptide in FPOP Compensates for Adventitious Scavengers and Permits Time-Dependent Measurements. J. Am. Soc. Mass Spectrom. 2017, 28, 389–392. 10.1007/s13361-016-1552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. C.; Barry B. W. Penetration enhancers. Adv. Drug Delivery Rev. 2004, 56, 603–18. 10.1016/j.addr.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Lane M. E. Skin penetration enhancers. Int. J. Pharm. 2013, 447, 12–21. 10.1016/j.ijpharm.2013.02.040. [DOI] [PubMed] [Google Scholar]
- Marty M. T.; Baldwin A. J.; Marklund E. G.; Hochberg G. K.; Benesch J. L.; Robinson C. V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015, 87, 4370–6. 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira S. R.; Mendes T. A.; Bueno L. L.; de Araujo J. V.; Bartholomeu D. C.; Fujiwara R. T. A new methodology for evaluation of nematode viability. Biomed. Res. Int. 2015, 2015, 879263 10.1155/2015/879263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.; Tang Y.; Dickinson B. C.; Chang C. J.; Chang Z. An oxidative fluctuation hypothesis of aging generated by imaging H2O2 levels in live Caenorhabditis elegans with altered lifespans. Biochem. Biophys. Res. Commun. 2015, 458, 896–900. 10.1016/j.bbrc.2015.02.055. [DOI] [PubMed] [Google Scholar]
- Espino J. A.; Jones L. M. In Vivo Hydroxyl Radical Protein Footprinting for the Study of Protein Interactions in Caenorhabditis elegans. JoVE 2020, e60910 10.3791/60910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinas A.; Espino J. A.; Jones L. M. An efficient quantitation strategy for hydroxyl radical-mediated protein footprinting using Proteome Discoverer. Anal. Bioanal. Chem. 2016, 408, 3021–31. 10.1007/s00216-016-9369-3. [DOI] [PubMed] [Google Scholar]
- Gau B. C.; Chen H.; Zhang Y.; Gross M. L. Sulfate radical anion as a new reagent for fast photochemical oxidation of proteins. Anal. Chem. 2010, 82, 7821–7. 10.1021/ac101760y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. G.; Raleigh J. A. Action of some hydroxyl radical scavengers on radiation-induced haemolysis. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1983, 43, 411–9. 10.1080/09553008314550471. [DOI] [PubMed] [Google Scholar]
- Frankowski H.; Alavez S.; Spilman P.; Mark K. A.; Nelson J. D.; Mollahan P.; Rao R. V.; Chen S. F.; Lithgow G. J.; Ellerby H. M. Dimethyl sulfoxide and dimethyl formamide increase lifespan of C. elegans in liquid. Mech. Ageing Dev. 2013, 134, 69–78. 10.1016/j.mad.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Wang X.; Wang X.; Li L.; Wang D. Lifespan extension in Caenorhabditis elegans by DMSO is dependent on sir-2.1 and daf-16. Biochem. Biophys. Res. Commun. 2010, 400, 613–8. 10.1016/j.bbrc.2010.08.113. [DOI] [PubMed] [Google Scholar]
- Dickinson B. C.; Huynh C.; Chang C. J. A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J. Am. Chem. Soc. 2010, 132, 5906–15. 10.1021/ja1014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karande P.; Jain A.; Mitragotri S. Insights into synergistic interactions in binary mixtures of chemical permeation enhancers for transdermal drug delivery. J. Controlled Release 2006, 115, 85–93. 10.1016/j.jconrel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Dodd W.; Tang L.; Lone J. C.; Wimberly K.; Wu C. W.; Consalvo C.; Wright J. E.; Pujol N.; Choe K. P. A Damage Sensor Associated with the Cuticle Coordinates Three Core Environmental Stress Responses in Caenorhabditis elegans. Genetics 2018, 208, 1467–1482. 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.; Pears C.; Woollard A. An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 2017, 7, 9839 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongpipattanakul B.; Burnette R. R.; Potts R. O.; Francoeur M. L. Evidence that oleic acid exists in a separate phase within stratum corneum lipids. Pharm. Res. 1991, 8, 350–4. 10.1023/A:1015845632280. [DOI] [PubMed] [Google Scholar]
- Loidl-Stahlhofen A.; Kern W.; Spiteller G. Gas chromatographic-electron impact mass spectrometric screening procedure for unknown hydroxyaldehydic lipid peroxidation products after pentafluorobenzyloxime derivatization. J. Chromatogr. B Biomed. Appl. 1995, 673, 1–14. 10.1016/0378-4347(95)00244-D. [DOI] [PubMed] [Google Scholar]
- Pradenas G. A.; Paillavil B. A.; Reyes-Cerpa S.; Perez-Donoso J. M.; Vasquez C. C. Reduction of the monounsaturated fatty acid content of Escherichia coli results in increased resistance to oxidative damage. Microbiology 2012, 158, 1279–1283. 10.1099/mic.0.056903-0. [DOI] [PubMed] [Google Scholar]
- Turnwald S. E.; Lorier M. A.; Wright L. J.; Mucalo M. R. Oleic Acid Oxidation Using Hydrogen Peroxide in Conjunction with Transition Metal Catalysis. J. Mater. Sci. Lett. 1998, 17, 1305–1307. 10.1023/A:1006532314593. [DOI] [Google Scholar]
- Brenna E.; Colombo D.; Di Lecce G.; Gatti F. G.; Ghezzi M. C.; Tentori F.; Tessaro D.; Viola M. Conversion of Oleic Acid into Azelaic and Pelargonic Acid by a Chemo-Enzymatic Route. Molecules 2020, 25, 1882 10.3390/molecules25081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Perez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaino J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.