Abstract
Transposon sequencing (Tn-seq) is a powerful method that combines transposon mutagenesis and massive parallel sequencing to identify genes and pathways that contribute to bacterial fitness under a wide range of environmental conditions. Tn-seq applications are extensive and have not only enabled examination of genotype-phenotype relationships at an organism level but also at the population, community and systems levels. Gram-negative bacteria are highly associated with antimicrobial resistance phenotypes, which has increased incidents of antibiotic treatment failure. Antimicrobial resistance is defined as bacterial growth in the presence of otherwise lethal antibiotics. The “last-line” antimicrobial colistin is used to treat Gram-negative bacterial infections. However, several Gram-negative pathogens, including Acinetobacter baumannii can develop colistin resistance through a range of molecular mechanisms, some of which were characterized using Tn- seq. Furthermore, signal transduction pathways that regulate colistin resistance vary within Gram-negative bacteria. Here we propose an efficient method of transposon mutagenesis in A. baumannii that streamlines generation of a saturating transposon insertion library and amplicon library construction by eliminating the need for restriction enzymes, adapter ligation, and gel purification. The methods described herein will enable in-depth analysis of molecular determinants that contribute to A. baumannii fitness when challenged with colistin. The protocol is also applicable to other Gramnegative ESKAPE pathogens, which are primarily associated with drug resistant hospital-acquired infections.
Introduction
The discovery of antibiotics is undoubtedly one of the most impactful health-related events of the 20th century.Not only do antibiotics quickly resolve serious bacterial infections, they also play a pivotal role in modern medicine. Major surgeries, transplants and advances in neonatal medicine and chemotherapy leave patients susceptible to life threatening infections and these therapies would not be possible without antibiotics1, 2. However, rapid development and spread of antibiotic resistance among human pathogens has significantly decreased the efficacy of all clinically important classes of antibiotics3. Many bacterial infections that were once easily cleared with antibiotics treatment, no longer respond to classic treatment protocols, causing a serious threat to global public health1. Antimicrobial resistance (AMR) is where bacterial cells grow in otherwise lethal concentrations of antibiotics, regardless of the treatment duration4, 5. There is an urgent need to understand molecular and biochemical factors that regulate AMR, which will help guide alternative antimicrobial development. Specifically, ESKAPE pathogens are problematic in clinical settings and associated with extensive AMR. These include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp. While several mechanisms contribute to AMR in ESKAPE pathogens, the latter four organisms are Gram-negative.
Gram-negative bacteria assemble a defining outer membrane that protects them from adverse environmental conditions. The outer membrane serves as a permeability barrier to restrict entry of toxic molecules, such as antibiotics, into the cell. Unlike other biological membranes, the outer membrane is asymmetrical. The outer leaflet is enriched with surface- exposed lipopolysaccharide, while the inner leaflet is a mixture of phospholipids6. Lipopolysaccharide molecules are anchored to the outer membrane by a conserved lipid A moiety embedded within the lipid bilayer7. The canonical lipid A domain of Escherichia coli lipopolysaccharide is required for the growth of most Gram-negative bacteria and is synthesized by a nine-step enzymatic pathway that is one of the most fundamental and conserved pathways in Gram-negative organisms6, 7, 8.
Polymyxins are cationic antimicrobial peptides that target the lipid A domain of lipopolysaccharide to perturb the outer membrane and lyse the cell. The electrostatic interaction between positively charged residues of polymyxins and the negatively charged lipid A phosphate groups disrupt the bacterial cell membrane ultimately leading to cell death9, 10, 11, 12, 13. Colistin (polymyxin E) is a last-resort antimicrobial used to treat infections caused by multidrug resistant Gram-negative nosocomial pathogens, such as Acinetobacter baumanni14, 15, 16. First discovered in 1947, polymyxins are produced by the soil bacteria, Paenibacillus polymyxa17, 18, 19. Polymyxins were prescribed to treat Gram-negative infections for years before their clinical use was limited due to reports of significant nephro- and neurotoxicity20, 21.
A. baumannii is a nosocomial Gram-negative pathogen that has dramatically increased the morbidity and mortality of patient outcomes over recent decades22. What was once regarded as a low-threat pathogen, now poses a significant risk for hospital-acquired infection throughout the world due to its incredible ability to acquire AMR and high risk of epidemic23, 24. A. baumannii accounts for more than 10% of nosocomial infections in the United States. Disease manifests as pneumonia, bacteremia, urinary tract infections, skin and soft tissue infections, meningitis, and endocarditis25. Treatment options for A. baumannii infections have dwindled due to resistance against almost all antibiotic classes, including p-lactams, fluoroquinolones, tetracycline, and aminoglycosides23, 24. The prevalence of multidrug resistant, extensively drug-resistant and pan-drug resistant A. baumannii isolates has led to a resurgence in colistin treatment, which was thought to be one of the few remaining therapeutic options still effective against multidrug resistant A. baumannii. However, increased colistin resistance among A. baumannii isolates has further amplified its threat to the global public health10, 11, 12, 13, 27, 30, 31.
Recent advances in high-throughput sequencing technologies, such as transposon sequencing (Tn-seq), have provided important tools to advance our understanding of bacterial fitness in vitro and in vivo. Tn-seq is a powerful tool that can be leveraged to study genotype-phenotype interactions in bacteria. Tn-seq is broadly applicable across bacterial pathogens, where it combines traditional transposon mutagenesis with massive parallel sequencing to rapidly map insertion sites, which can be used to link DNA mutations to phenotypic variants on a genome-wide scale32, 33, 34, 35. While transposon mutagenesis methods have been previously described, the general steps are similar33. First, an insertion library is generated using transposon mutagenesis, where each bacterial cell within a population is restricted to a single transposon insertion within the genomic DNA (gDNA). Following mutagenesis, individual mutants are pooled. gDNA is extracted from the insertion mutant pool and the transposon junctions are amplified and subjected to high-throughput sequencing. The reads represent insertion sites, which can be mapped to the genome. Transposon insertions that reduce fitness quickly fall out of the population, while beneficial insertions are enriched. Tn-seq has been instrumental to advance our understanding of how genes impact bacterial fitness in stress33.
The Himar1 mariner transposon system encoded in pJNW684 was specifically constructed and optimized for the purpose of transposon mutagenesis. It includes a mariner-family transposon flanking the kanamycin resistance gene, which is used for the selection of transposon insertion mutants in A. baumannii. It also encodes an A. baumannii specific promoter that drives expression of the transposase encoding gene36. The mariner-based transposon also contains two translational terminators downstream of the kanamycin resistance gene, which prevents read-through downstream of the insertion37. pJNW684 also carries a RP4/oriT/oriR6K- conditional origin of replication which requires the λpir gene contributed by the donor strain to replicate38. In absence of the λpir gene, the pJNW684 vector carrying the transposition machinery will not be able to replicate in the A. baumannii recipient strain10, 36, 38. Therefore, during bacterial conjugation, only the transposon is inserted into the recipient genome without background insertion of the plasmid, which carries the transposase gene. This is significant because the loss of transposase activity along with the plasmid results in single, stable transposition event that prevents the transposon from moving to different locations once it inserts into the recipient genome.
pJNW648 has also been tested for activity in another Gram-negative organism, E. coli. Successful assembly of a saturating Tn-seq library in E. coli strain W3110 indicated the system is amenable to perform mutagenesis in a wide range of pathogens, including Enterobacteriaceae. Furthermore, the A. baumannii specific promoter that drives transposase expression can quickly be exchanged with a species-specific promoter. Lastly, the kanamycin resistance gene can be exchanged for other resistance cassettes, depending on the AMR phenotype of the organism being studied.
One factor that contributes to colistin resistance in A. baumannii is administration of insufficient doses, where bacteria are exposed to selective pressure at non-lethal levels39. Several reports showed that subinhibitory antimicrobial concentrations can induce regulated responses that alter cell physiology to reduce susceptibility of the entire bacterial population11, 12, 30, 31. Using Tn-seq, we discovered factors that regulate colistin resistance in A. baumannii strain ATCC 17978 after exposure to inhibitory10 and subinhibitory concentrations of colistin. This example details a Tn-seq method that streamlines the construction and enrichment of a saturated transposon mutant library using the mariner-based family of transposons40, 41. While several Tn-seq protocols generate 20,000 – 100,000 mutants35, 42, 43, 44, 45, 46, the protocol described herein can rapidly generate a transposon library of 400,000 + mutants, which roughly equates to a transposon insertion every 10-base pairs in A. baumannii10. Furthermore, the library size can be scaled up without significant additional effort. This method also eliminates the requirement for restriction endonucleases, adapter ligation and gel purification, which can reduce final library diversity.
Protocol
1. Bacterial strain preparation
Streak the “donor” strain (E. coli MFD DAP-/pJNW684, Table of Materials) for isolated colonies on Luria-Bertani agar supplemented with 600 μM diaminopimelic acid (DAP), 100 mg/L of ampicillin and 25 mg/L of kanamycin. Incubate overnight at 37 °C. Using a single isolated colony, inoculate 50 mL of Luria broth (LB) supplemented with 600 μM DAP, 100 mg/L of ampicillin and 25 mg/L of kanamycin in a 250 mL Erlenmeyer flask and label it as “donor”.
Streak the “recipient” strain (A. baumannii strain ATCC 17978, Table of Materials) for isolated colonies on Luria-Bertani agar. Incubate overnight at 37 °C. Using a single isolated colony, inoculate 50 mL of LB in a 250 mL Erlenmeyer flask and label it as “recipient”.
Incubate both cultures (“donor” and “recipient”) overnight at 37 °C with shaking.
2. Bacterial mating
Transfer overnight cultures to 50 mL conical tubes.
Pellet both recipient and donor cultures using centrifugation at 5,000 × g for 7 min.
Discard the supernatant and resuspend the “donor” strain pellet in 35 mL of LB supplemented with DAP to wash away residual antibiotics.
Pellet the “donor” strain cells using centrifugation at 5,000 × g for 7 min.
Discard the supernatant and resuspend the “donor” strain pellet in 4.5 mL of LB supplemented with DAP. Use a 10 mL serological pipette.
-
Transfer the resuspended “donor” strain into “recipient” strain tube, which contains the pelleted “recipient” cells. Use the same 10 mL serological pipette from step 2.5 to mix the cultures. Immediately move to the next step.
NOTE: The total volume of the final suspension should be 5 mL.
Distribute the mating suspension as individual 100 μL droplets on LB agar plates supplemented with DAP (5–7 droplets per plate) (Figure 1A).
Incubate plates at room temperature for 30 min.
-
Without disturbing the droplets, carefully transfer plates to a 37 °C incubator and allow cultures to mate for 1 h.
NOTE: Incubation periods exceeding 1 h risks generation of sister mutants.
Following incubation, add 1.5 mL of LB onto each plate and harvest by resuspending bacteria from the plates. Use a 1 mL micropipette for resuspension. Final volume should be approximately 12–15 mL.
Combine harvested cells in a 50 mL conical tube.
Pellet the mated cells using centrifugation at 5,000 × g for 7 min.
Discard the supernatant and resuspend cells in 50 mL of LB to remove residual DAP.
Pellet the mating using centrifugation at 5,000 × g for 7 min.
Repeat the wash step (steps 2.13 and 2.14).
Using a 10 mL serological pipette, resuspend the pellet in 10 mL of LB supplemented with 25% glycerol.
Using washed cells, make five serial dilutions in LB broth (1:10, 1:100, 1:1000, 1:10,000, 1:100,000).
Spread 100 μL of each dilution on 4 different plates using sterile glass beads: Luria-Bertani agar supplemented with kanamycin, agar supplemented with ampicillin, agar supplemented with DAP and agar only.
Incubate plates at 37 °C overnight.
Aliquot the remaining mating in 1 mL aliquots and store at-80 °C.
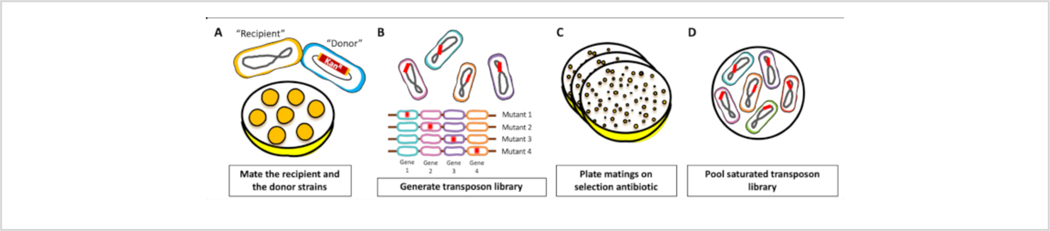
Figure 1: Schematic of transposon mutant library construction.
(A) Bacterial conjugation. The “donor” strain, which encodes the transposition machinery, was mixed with the “recipient” strain. The mixture was spotted on LB agar plates and allowed to mate for 1 h. (B) Generation of transposon library. The plasmid carrying the transposition machinery was transferred from the “donor” strain to the “recipient” strain and the transposon was randomly inserted throughout the genome of the “recipient” strain. (C) Selection. Resulting cells were plated on agar plates supplemented with kanamycin to select for transposon insertion mutants. (D) Pooled library. Colonies were scraped from plates, resuspended in LB and pooled. Please click here to view a larger version of this figure.
3. Determine the appropriate dilution of transposon library
Record colony-forming units (CFU) from overnight plates.
-
Image a plate with countable colonies for each different plate condition (Figure 2A).
NOTE: Both “donor” and “recipient” strains should grow on agar plates supplemented with DAP, so most plated dilutions will yield a lawn. Only the “recipient” strain can grow on agar plates. Neither the “donor” nor the “recipient” strain can grow on agar plates supplemented with ampicillin, so there should be none/minimal growth. Only target strain cells encoding the transposon insertion can grow on agar plates supplemented with kanamycin. Colonies should vary in size, indicating transposon insertions in genes that contribute to fitness on agar supplemented with kanamycin.
-
Calculate the number of transposon mutants in the frozen mating by counting the number of colonies on LB agar plates supplemented with kanamycin.
NOTE: For the A. baumannii genome (approximately 4 Mbps), the goal was to obtain about 400,000 colonies in order to generate a high-resolution mutant library (approximately one transposon insertion/10 base pairs). However, this number should be optimized based on genome size of the target species).
Figure 2: Representative bacterial conjugation results and a schematic of the colistin Tn-seq experiment.
(A) Representative kanamycin selection plate. The plate is divided into five equal sections. Blue dots represent colony counting for estimation of A. baumannii transposon insertion mutants. At least three separate plates were counted to calculate the final estimation. (B) Identification of fitness factors at subinhibitory colistin concentrations. The pooled transposon library was grown to logarithmic growth phase either in the absence (control) or in the presence (experimental) of colistin. Once the cultures reached appropriate optical density, the cells were pelleted and gDNA was extracted from each sample. Each condition was tested in duplicate for a total of four samples. Please click here to view a larger version of this figure.
4. Generation of final bacterial mutant library
Thaw an aliquot of the frozen mating on ice.
-
Plate mating on 150 mm Luria-Bertani agar plates supplemented with kanamycin based on CFUs calculated in step 3.1. Adjust the volume with LB to yield 13,333 colonies per 150 μL before plating.
NOTE: The CFU count here was determined to be about 105 CFU/mL, so the mating volume was adjusted to obtain 13,333 colonies per plate as this would provide an optimal number of colonies on 30 plates for a high-resolution mutant library without overcrowding the plates.
-
Use sterile glass beads to spread 150 μL of the dilution per plate on 30 × 150 mm Luria-Bertani agar plates supplemented with kanamycin to obtain 400,000 colonies (Figure 1C).
NOTE: Sterile rods or any kind of sterile spreader tool (i.e., glass beads) may be used to spread the bacteria on plates.
-
Dispose of the used tube containing excess mating.
NOTE: Freeze/thaw cycles adds selective pressures on the bacterial culture, which can skew the Tn-seq experiment results. Use a fresh aliquot each time.
-
Incubate plates at 37 °C for 14 h.
NOTE: The incubation time is optimized to prevent overgrowth (colonies touching). Minimizing growth by reducing the incubation time is suggested.
5. Estimating library density and pooling for storage
Count CFUs on each plate to estimate the total mutants in the transposon library. Count 20% of at least 3 plates to determine the colony count estimate for the entire group of plates (Figure 2A). Ensure that the colonies are not touching another colony.
-
After calculating the estimated colony yield, add 3–5 mL of LB (or more if needed) to each plate and scrape off the bacteria using a sterile scraping tool.
NOTE: Sterile inoculating loops were used to efficiently scrape plates.
Pool bacterial suspensions from all plates into 50 mL conical tubes (Figure 1D). This will require multiple 50 mL conical tubes, at least 3.
Pellet pooled bacterial suspension using centrifugation at 5,000 × g for 7 min.
Discard the supernatant and resuspend pellet in 5 mL of LB supplemented with 30% glycerol.
Aliquot 1 mL of the transposon library into cryovials and store at −80 °C.
6. Identification of factors that regulate colistin resistance in A. baumannii
- Prepare 4 × 250 mL Erlenmeyer flasks with each containing 50 mL LB broth and label as (Figure 2B)
- A. baumannii strain ATCC 17978 Tn-seq library; (−) colistin_1
- A. baumannii strain ATCC 17978 Tn-seq library; (−) colistin_2
- A. baumannii strain ATCC 17978 Tn-seq library; (+) colistin_1
-
A. baumannii strain ATCC 17978 Tn-seq library; (+) colistin_2NOTE: In the challenge growth described here, each condition, (−) colistin control and (+) colistin challenge, is being tested in duplicate. Therefore, the setup requires 4 × 250 mL Erlenmeyer flasks, two per condition.
Add 50 μL of 0.5 mg/L colistin to (+) colistin flasks (6.1.3 and 6.1.4) and 50 μL water to (−) colistin flasks (6.1.1 and 6.1.2).
Thaw a frozen Tn-seq library aliquot from step 5 on ice.
Pipette 1 μL of the thawed library into 1 mL of PBS.
-
Measure the optical density at 600 nm (OD600) and multiply by 1,000.
NOTE: This determines OD6OO of 1 μL of the Tn-library.
Based on the calculation in step 6.5, inoculate each flask containing 50 mL LB to a final OD600 0.001.
-
Grow cultures in a shaking incubator at 37 °C to OD600 0.5.
NOTE: It is important that cultures remain in logarithmic growth phase, so if your strain is at a different OD600 during exponential growth, the OD600 needs to be adjusted to ensure the culture replicates as many times as possible within logarithmic growth phase. If the culture only replicates 3 times, the power to detect fitness defects in mutants is capped at 3-fold differences, in theory. Since different bacteria have different doubling times, it is important to seed the cultures at a fixed OD600 to normalize the starting inoculum. This ensures consistent representation of the entire library in all cultures.
Harvest cultures using centrifugation at 5,000 × g at 4 °C for 7 min.
Remove the supernatant and wash with 50 mL of PBS.
Pellet using centrifugation at 5,000 × g at 4 °C for 7 min.
Remove the supernatant and resuspend the pellet in 1 mL of PBS. Aliquot ~ 200 μL into 5 microcentrifuge tubes.
Pellet using centrifugation at 5,000 × g at 4 °C for 5 min. Remove all of the supernatant using a pipette tip.
Store pellets at −20 °C or proceed with gDNA extraction.
7. gDNA extraction
Thaw one cell pellet on ice.
Add 0.6 mL lysis buffer (Table of Materials) and vortex to completely resuspend the pellet.
Incubate at 37 °C for 1 h.
Add 0.6 mL of phenol/chloroform/isoamyl alcohol to the sample and vortex vigorously.
Separate phases using centrifugation in a microcentrifuge at max speed at room temp for 5 min.
Transfer the upper aqueous phase to a new tube. Avoid disturbing the phase interface during transfer.
Add an equal volume of chloroform to the aqueous phase obtained above and vortex vigorously.
Separate phases using centrifugation in microcentrifuge at max speed at room temp for 5 min.
-
Transfer upper aqueous phase to a new tube.
NOTE: Be sure to avoid disturbing the interface during transfer.
Add 2.5x aqueous phase volume of cold 100 % ethanol and mix gently. Precipitated DNA will be visible.
Place tube at −80 °C for at least 1 h.
Pellet DNA using centrifugation at max speed at 4 °C for 30 min.
Carefully remove supernatant without disturbing the DNA pellet and wash with 150 μL of 70 % ethanol by pipetting.
Pellet DNA using centrifugation at max speed at 4 °C for 2 min.
Carefully remove the supernatant.
Repeat step 7.14 once. Carefully remove all remaining ethanol.
Dry DNA by incubating at room temp for 5–10 min.
Resuspend DNA pellet in 100 μL TE buffer by pipetting.
8. DNA shearing (Figure 3A)
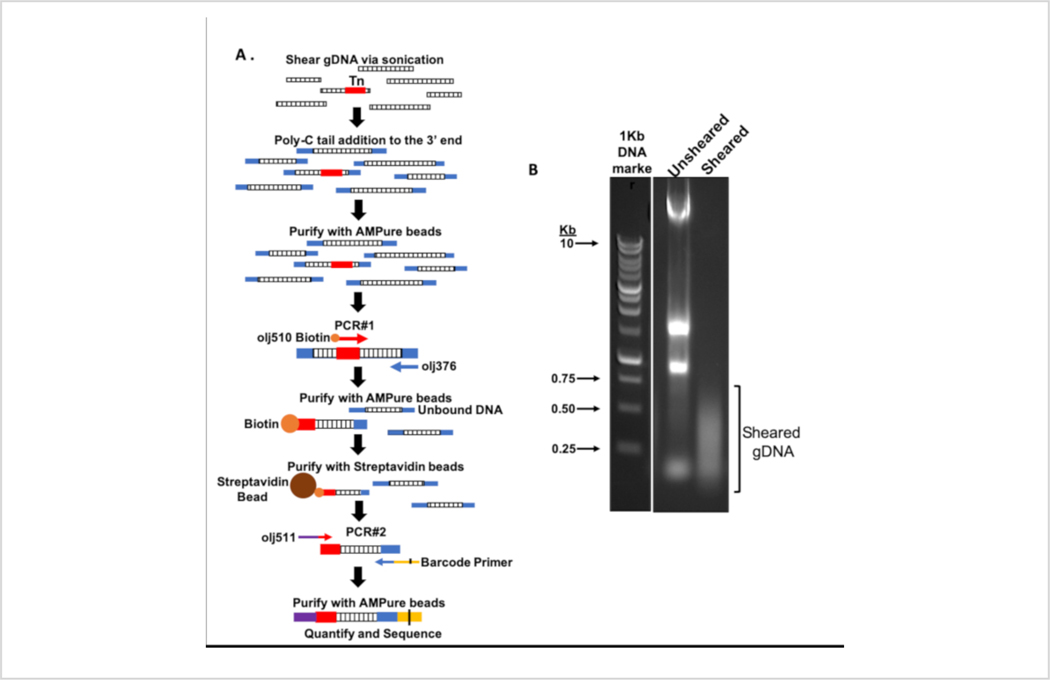
Figure 3: Flowchart of the DNA amplicon library build for massive parallel sequencing and representative sheared gDNA.
(A) Tn-seq DNA library build schematic. Following extraction, gDNA was fragmented via mechanical shearing. Terminal deoxynucleotidyl transferase was used to add a poly-C tail to the 3’ end of fragmented DNA before PCR amplification of the transposon-genome junctions and barcode addition. (B) 1% agarose gel of unsheared and sheared A. baumannii mutant libraries following gDNA shearing step. 1 Kb ladder was used as a DNA marker. Sheared gDNA smear primarily ranges between ~ 100 and 500 base pairs. Please click here to view a larger version of this figure.
Dilute gDNA with TE buffer to a concentration of 250 ng/ mL in a total volume of 200 μL.
Place tubes in a water bath sonicator.
Sonicate DNA to yield fragments of approximately 300 nucleotides. Power: 60 %, Total Time: 20 min, Cycles: 10 s ON and 10 s OFF at 4 °C (Figure 3A).
Confirm DNA is sheared appropriately by separating 10 μL of unsheared DNA and 10 μL of sheared DNA on a 1% agarose gel. Repeat sonication or optimize as needed (Figure 3B).
9. Poly-C tail addition to the 3’ end (Figure 3A)
Setup poly-C reaction according to the Table 1.
Incubate reaction at 37 °C for 1 h.
-
Purify poly-C reaction with 40 μL of size-selection paramagnetic beads (Figure 3A) by following the steps below.
Add 40 μL of size-selection paramagnetic beads to each sample. Vortex ~ 5 s or pipette up and down.
Incubate samples at room temperature for 5 min.
Briefly, centrifuge tubes to collect liquid in the bottom of the tube (~ 2 s).
Transfer tubes to a magnetic rack and incubate at room temperature for ~ 2 min until the solution is clear.
With tubes on magnetic rack, carefully remove the supernatant.
With tubes on magnetic rack, add 200 μL of freshly prepared 80% ethanol (do not disturb beads).
Incubate samples for at least 30 s until solution is clear.
With tubes on magnetic rack, carefully remove the supernatant.
Repeat the wash step (steps 9.3.6 – 9.3.8).
Collect liquid in the bottom of the tube using a brief centrifugation step (~ 2 s).
Transfer tubes to the magnetic rack and remove any remaining liquid.
Incubate at room temperature for 2–5 min to dry samples. Do not over dry.
Remove tubes from magnetic rack and add 25 μL of water to each. Vortex for ~ 5 s or pipette up and down.
Briefly, centrifuge tubes to collect liquid in the bottom of the tube (~ 2 s).
Transfer tubes to the magnetic rack and allow to sit for ~ 2 min until the solution is clear.
With tubes on the magnetic rack, remove liquid without disturbing the beads and transfer to a new tube (~23 μL of DNA).
Table 1: Reaction setup.
Setup and conditions for poly-C, PCR 1and PCR 2reactions.
| Reaction | Setup | Conditions |
|---|---|---|
| Poly-C reaction | 30 μL of sheared gDNA | Incubate at 37°C for 1 hour |
| 2.5 μL of 9.5 mM dCTP/0.5 mM ddCTP | ||
| 10 μL of 5X Terminal deoxynucleotidyl transferase (Tdt) reaction buffer | ||
| 1.25 μL of rTdt | ||
| 6.25 μL of water to 50 μL | ||
| PCR 1 | 23 μL 3’-poly-C purified DNA (entire sample from previous step) | 1 cycle: 2 min 94 °C |
| 15 cycles: 15 s 94 °C | ||
| 10 μL 10x high-fidelity DNA | 30 s 60 °C | |
| polymerase reaction mix | 2 min 68 °C | |
| 2 μL 10mM dNTPs | 1 cycle: 4 min 68 °C | |
| 2 μL 50 mM MgS04 | Hold: ∞ 4 °C | |
| 1 μL 30 μL 510 biotin | ||
| 3 μL 30 μL olj 376 | ||
| 0.5 μL high-fidelity DNA polymerase | ||
| 8.5 μL pure water to 50 μL total | ||
| PCR 2 | DNA bound beads from previous step | 1 cycle: 2 min 94 °C |
| 10 μL 10x high-fidelity DNA | 15 cycles: 15 s 94 °C | |
| polymerase reaction mix | 30 s 60 °C | |
| 2 μL 10mM dNTP | 2 min 68 °C | |
| 2 μL 50 mM MgSO4 | 1 cycle: 4 min 68 °C | |
| 1 μL 30 μM olj 511 | Hold: ∞ 4 °C | |
| 1 μL 30 μM barcode primer (Table 2) | ||
| 0.5 μL high-fidelity DNA polymerase | ||
| 33.5 μL pure water to 50 μL | ||
10. Transposon junction amplification (figure 3A)
Setup PCR 1 as described in Table 1 for the first nested PCR (Table 2).
Perform PCR 1 using conditions described in Table 1.
Purify PCR products with 40 μL of size-selection paramagnetic beads (steps 9.3.1 – 9.3–12). Elute in 50 μL of water (steps 9.3.13 – 9.3.16). At this point the samples can be stored at −20 °C.
-
Prepare Streptavidin-coupled paramagnetic beads:
Resuspend Streptavidin beads by shaking vigorously.
-
Add 32 μL of beads per jple to a fresh microcentrifuge tube.
NOTE: Beads for 6 + samples can be prepared in a single tube.
Transfer the tube to a magnetic rack. Incubate for~ 2 min until solution is clear.
With tube on magnetic rack, remove supernatant.
Remove the tube from magnetic rack. Wash beads by resuspending in 1 mL 1x B&W buffer by pipetting up and down.
Transfer the tube to magnetic rack. Incubate for ~ 2 min until solution is clear.
With the tube on magnetic rack, remove the supernatant.
Repeat wash step with 1 mL 1x B&W buffer (steps 10.4.5 – 10.4.7) twice more for a total of 3 washes.
Remove tube from magnetic rack and resuspend beads in 52 μL of 2x B&W buffer per sample.
Combine 50 μL of the prepared beads with 50 μL of purified PCR1 (from step 10.3). Pipette to mix.
Rotate at room temperature for 30 min.
-
Wash away unbound DNA:
Briefly, centrifuge to collect liquid at the bottom of the tube (~ 2 s).
2.Transfer the tube to magnetic rack. Incubate for ~ 2 min until the solution is clear.
With the tube on magnetic rack, remove the supernatant.
Remove the tube from magnetic rack, wash beads by resuspending in 100 μL 1x B&W buffer and pipetting up and down.
Transfer tube to magnetic rack. Incubate for ~ 2 min until the solution is clear.
With tube on magnetic rack, remove supernatant.
Repeat wash steps with 100 μL LoTE (steps 10.7.4 – 10.7.6) twice more for a total of 3 washes.
Setup PCR 2 as described in Table 1 to amplify transposon junctions and add a single barcode to each sample (Table 2 and Table 3).
Perform PCR 2 using conditions described in Table 1.
Purify with 40 μL of size-selection paramagnetic beads (steps 9.3.1 – 9.3.12). Elute in 17 μL of water (steps 9.3.13 – 9.3.16), collect ~ 15 μL.
Quantify DNA concentration using a fluorometer. The final concentrations should be ~ 50 to 250 ng/μL.
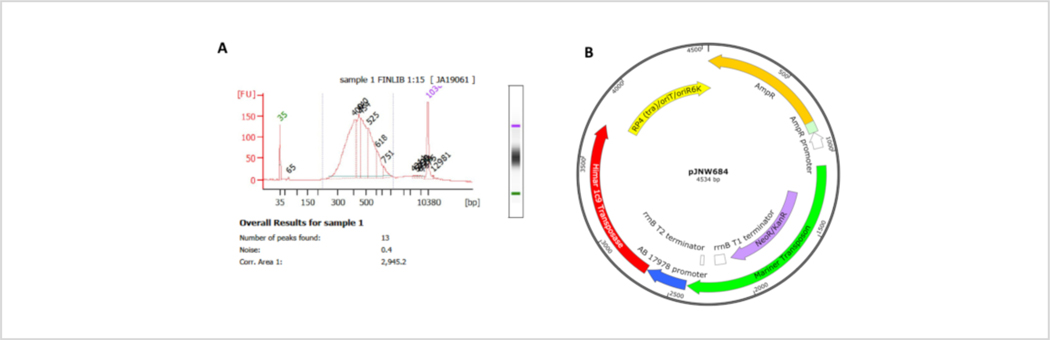
Assess DNA quality using chip-based capillary electrophoresis (Figure 4A).
Table 2: PCR amplification primers.
PCR amplification primers used in the protocol to amplify the transposon junctions. The purpose of each is listed in the first column.
| Purpose | Name | Sequence |
|---|---|---|
| Anneals to transposon | olj510-Biotin | Biotin-GATGGCCTTTTTGCGTTTCTACCTGCAGGGCGCG |
| Anneals to poly-C tail | olj376 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGG GGGGGGGGGGGGGG |
| Nested to transposon + P5 adapter: | olj511 | AATGATACGGCGACCACCGAGATCTACACTCTTT |
| P5 capture site − P5 sequencing site–N5–Tn | CCCTACACGACGCTCTTCCGATCTNNNNNGGGGACTTA TCATCCAACCTGTTAG |
|
| Nested to olj376: P7 sequencing site – barcode xref- P7 capture site) |
BC## | CAAGCAGAAGACGGCATACGAGATxxxxxxGTGA CTGGAGTTCAGACGTGTG |
| ***see Table 2 for specific barcodes*** |
Table 3: Barcode primers.
Barcode primers are used in the second PCR step to amplify the transposon junctions while adding a P7 sequencing site and barcodes to the amplicon. Not all barcode primers are needed to generate a library. Only barcode primers for the number of samples are required.
| Primer | Read | Barcode | Sequence |
|---|---|---|---|
| BC1 | ATCACG | CGTGAT | CAAGCAGAAGACGGCATACGAGATCGT GATGTGACTGGAGTTCAGACGTGTG |
| BC2 | CGATGT | ACATCG | CAAGCAGAAGACGGCATACGAGATAC ATCGGTGACTGGAGTTCAGACGTGTG |
| BC3 | TTAGGC | GCCTAA | CAAGCAGAAGACGGCATACGAGATGCC TAAGTGACTGGAGTTCAGACGTGTG |
| BC4 | TGACCA | TGGTCA | CAAG CAG AAGACGGCATACGAG ATT GGTCAGTGACTGGAGTTCAGACGTGTG |
| BC5 | ACAGTG | CACTGT | CAAGCAGAAGACGGCAT ACGAGAT CACTGTGTGACTGGAGTTCAGACGTGTG |
| BC6 | GCCAAT | ATTGGC | CAAGCAGAAGACGGCATACGAGATA TTGGCGTGACTGGAGTTCAGACGTGTG |
| BC7 | CAGATC | GATCTG | CAAGCAGAAGACGGCAT ACGAGAT GATCTGGTGACTGGAGTTCAGACGTGTG |
| BC8 | ACTTGA | TCAAGT | CAAGCAGAAGACGGCAT ACGAGAT TCAAGTGTGACTGGAGTTCAGACGTGTG |
| BC9 | GATCAG | CTGATC | CAAGCAGAAGACGGCATACGAGA TCTGATCGTGACTGGAGTTCAGACGTGTG |
| BC10 | TAGCTT | AAGCTA | CAAGCAGAAGACGGCAT ACGAGAT AAGCTAGTGACTGGAGTTCAGACGTGTG |
| BC11 | GGCTAC | GTAGCC | CAAGCAGAAGACGGCAT ACGAGAT GTAGCCGTGACTGGAGTTCAGACGTGTG |
| BC12 | CTTGTA | TACAAG | CAAG CAG AAGACGGCATACG AG ATT ACAAGGTGACTGGAGTTCAGACGTGTG |
| BC13 | AGTCAA | TTGACT | CAAGCAGAAGACGGCAT ACGAGAT TTGACTGTGACTGGAGTTCAGACGTGTG |
| BC14 | AGTTCC | GGAACT | CAAGCAGAAGACGGCATACGAGATG GAACTGTGACTGGAGTTCAGACGTGTG |
| BC15 | ATGTCA | TGACAT | CAAGCAGAAGACGGCATACGAGA TTGACATGTGACTGGAGTTCAGACGTGTG |
| BC16 | CCGTCC | GGACGG | CAAGCAGAAGACGGCAT ACGAGAT GGACGGGTGACTGGAGTTCAGACGTGTG |
| BC17 | GTAGAG | CTCTAC | CAAGCAGAAGACGGCAT ACGAGAT CTCTACGTGACTGGAGTTCAGACGTGTG |
| BC18 | GTCCGC | GCGGAC | CAAGCAGAAGACGGCAT ACGAGAT GCGGACGTGACTGGAGTTCAGACGTGTG |
| BC19 | GTGAAA | TTTCAC | CAAGCAGAAGACGGCAT ACGAGAT TTTCACGTGACTGGAGTTCAGACGTGTG |
| BC20 | GTGGCC | GGCCAC | CAAGCAGAAGACGGCATACGAGATG GCCACGTGACTGGAGTTCAGACGTGTG |
| BC21 | GTTTCG | CGAAAC | CAAGCAGAAGACGGCAT ACGAGAT CGAAACGTGACTGGAGTTCAGACGTGTG |
| BC22 | CGTACG | CGTACG | CAAGCAGAAGACGGCAT ACGAGAT CGTACGGTGACTGGAGTTCAGACGTGTG |
| BC23 | GAGTGG | CCACTC | CAAGCAGAAGACGGCATACGAGATC CACTCGTGACTGGAGTTCAGACGTGTG |
| BC24 | GGTAGC | GCTACC | CAAGCAGAAGACGGCAT ACGAGAT GCTACCGTGACTGGAGTTCAGACGTGTG |
| BC25 | ACTGAT | ATCAGT | CAAGCAGAAGACGGCAT ACGAGAT ATCAGTGTGACTGGAGTTCAGACGTGTG |
| BC26 | ATGAGC | GCTCAT | CAAGCAGAAGACGGCAT ACGAGAT GCTCATGTGACTGGAGTTCAGACGTGTG |
| BC27 | ATTCCT | AGGAAT | CAAGCAGAAGACGGCATACGAGATA GGAATGTGACTGGAGTTCAGACGTGTG |
| BC28 | CAAAAG | CTTTTG | CAAGCAGAAGACGGCATACGAGATC TTTTGGTGACTGGAGTTCAGACGTGTG |
| BC29 | CAACTA | TAGTTG | CAAGCAGAAGACGGCAT ACGAGAT TAGTTGGTGACTGGAGTTCAGACGTGTG |
| BC30 | CACCGG | CCGGTG | CAAGCAGAAGACGGCAT ACGAGAT CCGGTGGTGACTGGAGTTCAGACGTGTG |
| BC39 | CT AT AC | GTATAG | CAAGCAGAAGACGGCAT ACGAGAT GTATAGGTGACTGGAGTTCAGACGTGTG |
| BC40 | CTCAGA | TCTGAG | CAAGCAGAAGACGGCAT ACGAGAT TCTGAG GTGACTGGAGTTCAG ACGTGTG |
| BC42 | TAATCG | CGATTA | CAAGCAGAAGACGGCATACGAGATC GATTAGTGACTGGAGTTCAGACGTGTG |
| BC43 | TACAGC | GCTGTA | CAAGCAGAAGACGGCAT ACGAGAT GCTGTAGTGACTGGAGTTCAGACGTGTG |
| BC44 | TATAAT | ATTATA | CAAGCAGAAGACGGCAT ACGAGAT ATTATAGTGACTGGAGTTCAGACGTGTG |
| BC45 | TCATTC | GAATGA | CAAGCAGAAGACGGCAT ACGAGAT GAATGAGTGACTGGAGTTCAGACGTGTG |
| BC46 | TCCCGA | TCGGGA | CAAGCAGAAGACGGCAT ACGAGAT TCGGGAGTGACTGGAGTTCAGACGTGTG |
| BC47 | TCGAAG | CTTCGA | CAAGCAGAAGACGGCATACGAGATC TTCGAGTGACTGGAGTTCAGACGTGTG |
| BC48 | TCGGCA | TGCCGA | CAAGCAGAAGACGGCAT ACGAGAT TGCCGAGTGACTGGAGTTCAGACGTGTG |
Figure 4: Representative quality control (QC) results and map of the plasmid carrying the transposition genes.
(A) QC trace for a DNA library build. There is a peak at ~ 350 base pairs, indicating successful library build. If some larger DNA was detected in the QC results, the samples can be cleaned up further to remove large DNA fragments. (B) Plasmid pJNW684 consists of a Himar1 mariner transposon (green) with a kanamycin resistance cassette (purple) for mutant selection, a gene encoding the hyperactive mariner Himar1 C9 transposase (red) under control of an A. baumannii 17978 specific promoter (blue), an ampicillin resistance gene (bla, orange) and a RP4/oriT/oriR6K-conditional origin of replication (yellow). Please click here to view a larger version of this figure.
Representative Results
The outlined methods describe the generation of a high- density transposon library in A. baumannii strain ATCC 17978 through bacterial conjugation using E. coli MFD DAP-, which replicates the plasmid pJNW684 (Figure 4B). The detailed protocol uses bi-parental bacterial conjugation for transfer of pJNW684 from the E. coli λpir+ donor strain to the A. baumannii recipient strain. This is an efficient and inexpensive method for generating dense transposon mutant libraries. Bacteria were mixed at optimized ratios and matings were spotted on Luria-Bertani agar plates for 1 h (Figure 1A). During mating, the transposon was transferred from the donor to recipient strain, where it inserted into the gDNA (Figure 1B). Matings were collected, approximately 105 CFU/mL were calculated and plated on 150 mm × 15 mm agar plates supplemented with kanamycin. After 14 h of growth at 37 °C, plates contained thousands of colonies of varying size (Figure 1C) indicating successful generation of a transposon mutant library. The transposon insertion mutants were pooled (Figure 1D) and frozen in aliquots to prevent repeated freeze-thaw cycles, which could add selective pressure on the insertion library.
The pooled A. baumannii transposon mutant library was used to identify fitness factors important for colistin resistance under subinhibitory concentrations of the antimicrobial (Figure 2B). The mutant library was grown to logarithmic phase in the absence/presence of 0.5 mg/L colistin in duplicate to deplete mutant cells encoding insertions in genes that contribute to colistin resistance. The total gDNA of the remaining library pool was isolated from control and experimental cultures and processed to prepare DNA for sequencing (Figure 3A).
Isolated gDNA was fragmented using mechanical shearing and a poly-C tail was added on the DNA fragments (Figure 3A). Following the addition of poly-C tail, DNA was purified and the transposon-genome junctions were enriched using the poly-C specific primer followed by a second round of PCR using another poly-C specific primer that introduced the P7 sequencing site which is required for binding the lllumina flow cell and cluster generation, and a six-base barcode that is used to demultiplex libraries post sequencing37, 47. DNA concentrations were calculated and the samples were analyzed using chip-based capillary electrophoresis to confirm successful library builds (Figure 4A), which are ready for high-throughput sequencing.
DNA libraries were sequenced by next generation sequencing. A single-end, 50 cycle run was performed, which yielded 30 million high-quality reads/sample providing 62.5-fold coverage of the transposon library. Transposon junctions (reads) were mapped to the reference genome48, using commercially available bioinformatics analysis software. The number of reads at each insertion site in the input samples, (−) colistin control condition, were compared to the number of reads in the output samples, (+) colistin experimental condition, and fitness scores for each insertion site were calculated. The fitness scores were then grouped by gene. Genes demonstrating reduced scores when the library was grown in the presence of colistin relative to the input samples were considered fitness determinants for A. baumannii survival at subinhibitory concentrations of colistin. For example, transposon insertions within the PmrAB two- component system were present in the input sample, but were not found in the output sample. PmrAB directly regulates expression of pmrC, which transfers phosphoethanolamine onto lipid A to neutralize the charge on the cell surface12, 31. Neutralization of bacterial surface charge is thought to reduce the electrostatic potential required for colistin-mediated killing. Identification of depleted genes known to contribute to the resistance phenotype validated the method.
Discussion
A. baumannii is an emerging threat to global public health due to the rapid acquisition of AMR against “last- line” therapeutics, such as colistin10, 11, 12, 23, 24, 30, 31. In recent decades, Tn-seq has played a critical role in elucidating genotype-phenotype interactions across numerous bacterial species and expanding our understanding of bacterial genetics34, 35, 42, 43. Tn- seq protocols have been instrumental in identifying essential genes in diverse bacterial species such as Campylobacter jejuni, Staphylococcus aureus, the periodontal pathogen Porphyromonas gingivalis, and even Mycobacterium tuberculosis37, 49, 50, 51 Beyond identification of essential genes, Tn-seq has been used to identify antibiotic resistance genes in Pseudomonas aeruginosa, several conditionally essential genes in Salmonella typhimurium, and numerous genotype-phenotype relationships in Streptococcus pneumoniae52, 53, 54. More recently, transposon sequencing of Vibrio cholerae was employed in the infant rabbit model of Cholera to identify genes that are important for in vivo fitness during infection47. These studies demonstrate the versatility of Tn-seq as it can be utilized for both in vitro and in vivo studies.
The main advantage of Tn-seq over other methods, such as microarray technologies, 2D gel electrophoresis, and qPCR, is that it does not require prior knowledge of the genome or genomic information55. Therefore, transposon mutagenesis coupled with massive-parallel sequencing enables the study of known genes and genomes as well as discovery of novel genetic interactions. Here we have presented a comprehensive method for generating a highly dense transposon mutant library in A. baumannii to identify factors that are essential for bacterial fitness when exposed to subinhibitory concentrations of colistin. The described method has also been successfully used in E. coii (unpublished data), demonstrating the system is amenable to perform Tn-seq analysis in other Gram-negative pathogens, including Enterobacteriaceae.
Using mariner transposons for insertional mutagenesis has several advantages. The transposon family originated from eukaryotic hosts and have been widely used to generate saturating mutant libraries in diverse bacterial populations. Mariner transposons are host-independent, which means that stable random insertions can be achieved in the absence of specific host factors40, 41. Additionally, mariner transposons have a defined number of insertion events because they preferentially insert into thymine-adenine (“TA”) motifs, which reduces insertional bias and leads to more robust statistical analysis37, 56, 57, 58.
Several mariner-based Tn-seq methods use Mmei restriction digestion for gDNA fragmentation32, 42, 43. While enzymatic DNA fragmentation is a popular and successful method, it adds unnecessary steps to the procedure and increases potential bias37. Not only do these techniques require large quantities of starting materials, they can also potentially lead to unequal representation of insertion sequences in downstream analyses37, 59. Like some other methods that do not rely on Mmei nuclease activity52, 60, 61, the method outlined herein relies on mechanical shearing to fragment gDNA, and TdT to add a poly-C tail to the 3’ end of the DNA fragments. Compared to enzymatic DNA fragmentation and adapter ligation methods, this approach requires significantly smaller amounts of starting DNA while providing more consistent results, it also lowers the risk of DNA crosscontamination and reduces sample loss due to confinement in a sealed tube37, 59, 62. Furthermore, this method yields longer, high quality sequencing reads of 50 nucleotides which aid in more effective and precise mapping of sequences and a more robust downstream analysis37, 59. The addition of a synthetic poly-C tail disregards potential overhangs that may result from fragmentation as this method relies on the exogenously added poly-C tail as a recognition site for the reverse primer, which contains 16 dG nucleotides at its 3’ end and a sequence specific to next generation sequencing (e.g., Illumina sequencing) at the 5’ end, to prime synthesis47, 59. The use of a synthetic nucleotide tail expands the application of this method to many distinct genomes independent of their native content59. Subsequently, transposon-genome junctions are amplified using the poly-C specific primer37. This alternative simplifies the procedure by eliminating the need for expensive restriction enzymes, adapter ligation, formation of adapter dimers and gel purification steps. We have further optimized the protocol to efficiently generate highly saturated transposon insertion libraries in several Gram-negative ESKAPE pathogens, including Acinetobacter baumannii and can be used to study genetic interactions under diverse in vitro and in vivo conditions10.
One limitation of Tn mutagenesis is it relies on the antibiotic resistance markers for selection. However, many Gramnegative ESKAPE pathogens are multidrug or extensively drug resistant, meaning the user may need to exchange the resistance cassette according to the specific pathogen of interest. Furthermore, some clinical isolates are not amenable to transposon mutagenesis using the mariner- based transposon.
A critical step of the protocol is calculating the number of Tn mutants to plate. Plating too many colonies will result in a lawn that can complicate downstream analysis. If the colonies are too close or touching, they can add unwanted selective pressure on the library that can result in artifacts. Ideally, colonies would be not be touching and spaced evenly across the plate, as demonstrated (Figure 2A). Conversely, if too few colonies are plated, it will be difficult to isolate multiple Tn insertions in each gene.
It is also important to perform the controls listed in step 2.18. As stated in the Note of section of 3. 2, neither “donor” or “recipient” strain should grow on plates supplemented with Ampicillin. Since exogenous DAP is required for growth of the “donor” strain, any growth would indicate the “recipient” strain replicates pJNW684. This is a significant problem because if the transposon does not integrate into the gDNA, sequencing reads will only map to the plasmid, not an integration site. In this case the experiment will likely not yield useable data.
Acknowledgments
This work was supported by funding from the National Institute of Health (Grant AI146829 to J.M.B.) and is gratefully acknowledged.
Footnotes
Disclosures
The authors have nothing to disclose.
References
- 1.Gould IM, Bal AM New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence. 4 (2), 185–191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holzheimer RG Antibiotic induced endotoxin release and clinical sepsis: a review. Journal of Chemotherapy (Florence, Italy). 13 Spec No 1 (1), 159–172 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (U.S.) Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention (U.S.) (2019). [Google Scholar]
- 4.Scholar EM, Pratt WB The antimicrobial drugs. Oxford University Press; New York: (2000). [Google Scholar]
- 5.Brauner A, Fridman O, Gefen O, Balaban NQ Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature Reviews Microbiology. 14 (5), 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Henderson JC et al. The Power of Asymmetry: Architecture and Assembly of the Gram-Negative Outer Membrane Lipid Bilayer. Annual Review of Microbiology. 70, 255–278 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Whitfield C, Trent MS Biosynthesis and export of bacterial lipopolysaccharides. Annual Review of Biochemistry. 83, 99–128 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Raetz CRH, Whitfield C. Lipopolysaccharide Endotoxins. Annual Review of Biochemistry. 71 (1), 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang KN et al. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy- I -arabinose addition to lipid A. Molecular Microbiology. 111 (6), 1604–1616 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boll JM et al. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide- deficient Acinetobacter baumannii. Proceedings of the National Academy of Sciences. 113 (41), E6228–E6237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boll JM et al. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. mBio. 6 (3), e00478–00415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arroyo LA et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrobial Agents and Chemotherapy. 55 (8), 3743–3751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris TL et al. Small molecule downregulation of PmrAB reverses lipid A modification and breaks colistin resistance. /ACS Chemical Biology. 9 (1), 122–127 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Vaara M. Agents that increase the permeability of the outer membrane. Microbiological Reviews. 56 (3), 395–411 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaara M. Polymyxins and their novel derivatives. Current Opinion in Microbiology. 13 (5), 574–581 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Kassamali Z, Rotschafer JC, Jones RN, Prince RA, Danziger LH Polymyxins: wisdom does not always come with age. Clinical Infectious Diseases: An Official publication of the Infectious Diseases Society of America. 57 (6), 877–883 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Ainsworth GC, Brown AM, Brownlee G. Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature. 159 (4060), 263 (1947). [DOI] [PubMed] [Google Scholar]
- 18.Benedict RG, Langlykke AF Antibiotic activity of Bacillus polymyxa. Journal of Bacteriology. 54 (1), 24 (1947). [PubMed] [Google Scholar]
- 19.Stansly PG, Shepherd RG, White HJ Polymyxin: a new chemotherapeutic agent. Bulletin of the Johns Hopkins Hospital. 81 (1), 43–54 (1947). [PubMed] [Google Scholar]
- 20.Hermsen ED, Sullivan CJ, Rotschafer JC Polymyxins: pharmacology, pharmacokinetics, pharmacodynamics, and clinical applications. Infectious Disease Clinics of North America. 17 (3), 545–562 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Pedersen MF, Pedersen JF, Adsen PO A clinical and experimental comparative study of sodium colistimethate and polymyxin B sulfate. Investigative Urology. 9 (3), 234–237 (1971). [PubMed] [Google Scholar]
- 22.Falagas ME, Bliziotis IA Pandrug-resistant Gramnegative bacteria: the dawn of the post-antibiotic era? International Journal of Antimicrobial Agents. 29 (6), 630–636 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Peleg AY, Seifert H, Paterson DL Acinetobacter baumannii: Emergence of a Successful Pathogen. Clinical Microbiology Reviews. 21 (3), 538–582 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clinical Microbiology Reviews. 26 (2), 185–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliopoulos GM, Maragakis LL, Perl TM Acinetobacter baumannii: Epidemiology, Antimicrobial Resistance, and Treatment Options. Clinical Infectious Diseases. 46 (8), 1254–1263 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Lee A, Nolan A, Watson J, Tristem M. Identification of an ancient endogenous retrovirus, predating the divergence of the placental mammals. Philosophical Transactions of the Royal Society B: Biological Sciences. 368 (1626), 20120503–20120503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffatt JH et al. Colistin Resistance in Acinetobacter baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrobial Agents and Chemotherapy. 54 (12), 4971–4977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, Chai D, Wang R, Liang B, Bai N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. Journal of Antimicrobial Chemotherapy. 67 (7), 1607–1615 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Da Silva G, Domingues S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics. 6 (4), 28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin C-Y, Gregg KA, Napier BA, Ernst RK, Weiss DS A PmrB-Regulated Deacetylase Required for Lipid A Modification and Polymyxin Resistance in Acinetobacter baumannii. Antimicrobial Agents and Chemotherapy. 59 (12), 7911–7914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beceiro A. et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two- component regulatory system. Antimicrobial Agents and Chemotherapy. 55 (7), 3370–3379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Opijnen T, Lazinski DW, Camilli A. Genome-Wide Fitness and Genetic Interactions Determined byTn-seq, a High-Throughput Massively Parallel Sequencing Method for Microorganisms. Current Protocols in Microbiology.36, 1E.3.1–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon YM, Ricke SC, Mandal RK Transposon sequencing: methods and expanding applications. Applied Microbiology and Biotechnology. 100 (1), 31–43 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Yamaichi Y, Dorr T. Transposon Insertion Site Sequencing for Synthetic Lethal Screening. Methods in Molecular Biology (Clifton, N.J.). 1624, 39–49 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Goodman AL et al. Identifying Genetic Determinants Needed to Establish a Human Gut Symbiont in Its Habitat. Cell Host & Microbe. 6 (3), 279–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Ozer EA, Mandel MJ, Hauser AR Genome-Wide Identification of Acinetobacter baumannii Genes Necessary for Persistence in the Lung. mBio. 5 (3), e01163–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein BA et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 13 (1), 578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrieres L. et al. Silent Mischief: Bacteriophage Mu Insertions Contaminate Products of Escherichia coli Random Mutagenesis Performed Using Suicidal Transposon Delivery Plasmids Mobilized by Broad- Host-Range RP4 Conjugative Machinery. Journal of Bacteriology. 192 (24), 6418–6427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim LM et al. Resurgence of Colistin: A Review of Resistance, Toxicity, Pharmacodynamics, and Dosing. Pharmacotherapy. 30 (12), 1279–1291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampe DJ, Churchill ME, Robertson HM A purified mariner transposase is sufficient to mediate transposition in vitro. The EMBO Journal. 15 (19), 5470–5479 (1996). [PMC free article] [PubMed] [Google Scholar]
- 41.Mazurkiewicz P, Tang CM, Boone C, Holden DW Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nature Reviews. Genetics 7 (12), 929–939 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Goodman AL, Wu M, Gordon JI Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nature Protocols. 6 (12), 1969–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature Reviews. Microbiology. 11 (7), 3033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Bouillaut L, Sonenshein AL, Melville SB Use of a Mariner-Based Transposon Mutagenesis System To Isolate Clostridium perfringens Mutants Deficient in Gliding Motility. Journal of Bacteriology. 195 (3), 629–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singer JT, Finnerty WR Insertional specificity of transposon Tn5 in Acinetobacter sp. Journal of Bacteriology. 157 (2), 607–611 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subashchandrabose S. et al. Acinetobacter baumannii Genes Required for Bacterial Survival during Bloodstream Infection. mSphere. 1 (1), e00013–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shull LM, Camilli A. Transposon Sequencing of Vibrio cholerae in the Infant Rabbit Model of Cholera. Vibrio Cholerae. 1839, 103–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith MG et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes & Development. 21 (5), 601–614 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahl M, Stintzi A. Identification of essential genes in C. jejuni genome highlights hyper-variable plasticity regions. Functional & Integrative Genomics. 11 (2), 241–257 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Chaudhuri RR et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics. 10 (1), 291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffin JE et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathogens. 7 (9), e1002251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio. 2 (1), e00315–00310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khatiwara A. et al. Genome scanning for conditionally essential genes in Salmonella enterica Serotype Typhimurium. Applied and Environmental Microbiology. 78 (9), 3098–3107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Opijnen T, Camilli A. A fine scale phenotype- genotype virulence map of a bacterial pathogen. Genome Research. 22 (12), 2541–2551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurd PJ, Nelson CJ Advantages of next-generation sequencing versus the microarray in epigenetic research. Briefings in Functional Genomics and Proteomics. 8 (3), 174–183 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Barquist L, Boinett CJ, Cain AK Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biology. 10 (7), 1161–1169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry BJ, Yost CK Construction of a mariner- based transposon vector for use in insertion sequence mutagenesis in selected members of the Rhizobiaceae. BMC Microbiology. 14 (1), 298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plasterk RHA, Izsvak Z, Ivies Z. Resident aliens: the Tc1/ mariner superfamily of transposable elements. Trends in Genetics. 15 (8), 326–332 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Lazinski DW, Camilli A. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. BioTechniques. 54 (1), (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proceedings of the National Academy of Sciences of the United States of America. 106 (38), 16422–16427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langridge GC et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research. 19 (12), 2308–2316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quail MA, Swerdlow H, Turner DJ Improved Protocols for the lllumina Genome Analyzer Sequencing System. Current Protocols in Human Genetics. 62 (1), (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]