Abstract
Purpose
The goal of this study is to construct a mortality prediction model using the XGBoot (eXtreme Gradient Boosting) decision tree model for AKI (acute kidney injury) patients in the ICU (intensive care unit), and to compare its performance with that of three other machine learning models.
Methods
We used the eICU Collaborative Research Database (eICU-CRD) for model development and performance comparison. The prediction performance of the XGBoot model was compared with the other three machine learning models. These models included LR (logistic regression), SVM (support vector machines), and RF (random forest). In the model comparison, the AUROC (area under receiver operating curve), accuracy, precision, recall, and F1 score were used to evaluate the predictive performance of each model.
Results
A total of 7548 AKI patients were analyzed in this study. The overall in-hospital mortality of AKI patients was 16.35%. The best performing algorithm in this study was XGBoost with the highest AUROC (0.796, p < 0.01), F1(0.922, p < 0.01) and accuracy (0.860). The precision (0.860) and recall (0.994) of the XGBoost model rank second among the four models.
Conclusion
XGBoot model had obvious advantages of performance compared to the other machine learning models. This will be helpful for risk identification and early intervention for AKI patients at risk of death.
Introduction
Acute kidney injury (AKI) is a common condition with a high mortality rate, morbidity, high cost, and risk of developing chronic kidney disease [1]. It is also a global health issue [2]. Although the level of diagnosis and treatment has improved in recent years, the burden of disease caused by AKI is still very high, especially in the intensive care unit [1]. In clinical practice, the estimation of the mortality risk is helpful for triage and resource allocation, to determine the appropriate level of care, and even to discuss the expected outcomes with patients and their families [3].
In recent years, machine learning has been widely used to predict disease risk. Risk adjustment and mortality prediction are critically important for comparing outcomes across interventions and health systems. For example, Sevag Demirjian et al. (2011) constructed an LR (logistic regression) model to predict mortality in AKI patients and compared it with the prediction results of APACHE II (Acute Physiology and Chronic Health Evaluation II) score, SOFA (Sequential Organ Failure Assessment) score and CCF (Cleveland Clinic Foundation) score [4]. Ke Lin et al. (2019) used the RF (random forest) algorithm to build a mortality prediction model, and predicted the mortality risk of AKI patients in ICU. Their model was compared with SVM (support vector machine), ANN (artificial neural network), and Customized SAPS-II (Simplified Acute Physiology Score-II) scores [5]. These studies showed LR and RF exhibited good discrimination, and remarkable accuracy [4, 5]. Many advanced AI models, such as deep learning techniques, have shown remarkable accuracy in mortality prediction [6, 7]. However, in clinical real-world scenarios, the inability to provide sufficient data for model training has prevented AI models from performing well. The AI models perform poorly when dealing with relatively small datasets and cannot be widely used in clinical practice [8]. While machine learning models have a good predictive performance on smaller datasets. However, a single machine learning approach often leads to overfitting and difficulty in dealing with the large number of unbalanced datasets that occur in actual problems. To compensate for the shortcomings of a single machine learning method, the ensemble learning technique based on the GBDT (gradient boosting decision tree) algorithm was developed and has gradually become the mainstream approach in the field of machine learning research [9, 10]. XGBoost is a highly efficient boosting ensemble learning model that originated in the decision tree model, which uses the tree classifier for better results of prediction and higher operation efficiency [11, 12].
The purpose of this study is to use XGBoost to construct a predictive mortality model for AKI patients in the ICU, and to use the publicly available database eICU Collaborative Research Database V2.0 as a data source [13]. In addition, the performance of the XGBoost model was compared with LR, SVM, and RF model.
Method
Dataset
This study used the eICU-CRD V2.0 with 200,859 admissions between 2014 and 2015 at 208 hospitals of the United States (https://eicu-crd.mit.edu/). The database was a multicenter ICU database with a high granularity of data. It included data on patient vital sign measurements, care plan documentation, nurse charting, disease severity measures, laboratory variables, diagnostic information, and treatment information [13].
Patients
All patients in eICU-CRD version v 2.0 databases were eligible for inclusion in the present investigation. The following inclusion criteria were used: (1) All AKI patients (ICD-9, 584.x) admitted to the ICU with a length of stay> 24 hours; (2) age 18 years or more; and (3) patients with more than 30% missing values were excluded from the analysis [5]. As for those patients who were admitted to ICU for more than once, only data of the first ICU stay were used. The patients’ selection process was shown in Fig 1.
Fig 1. The patients’ selection process.
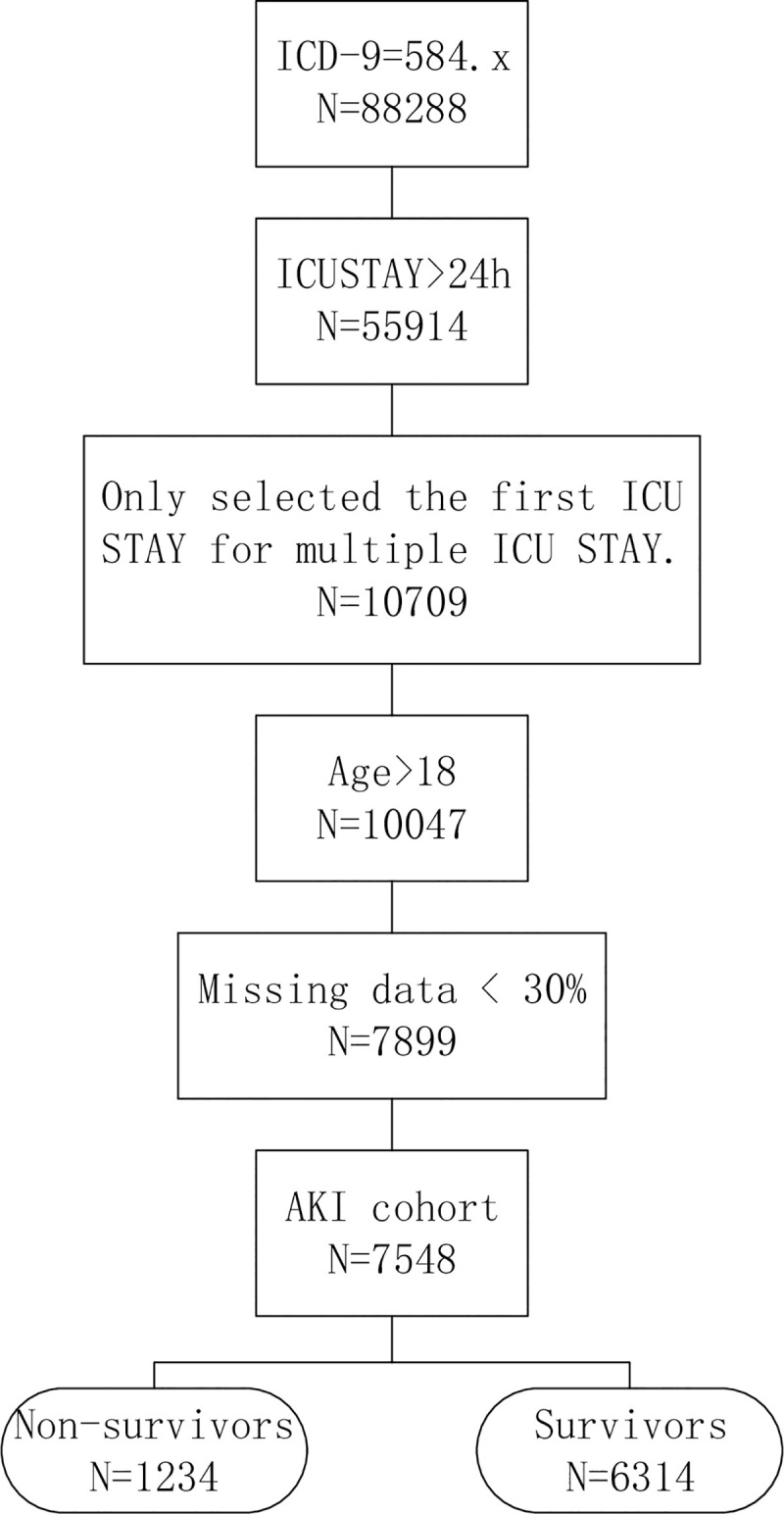
ICD-9: International Classification of Diseases, Ninth Revision.
Predictor variables
The variables used to predict the mortality of AKI include various demographic, clinical, and laboratory variables. These variables were based on experts’ opinion and roughly matched the variables used in the Acute Physiological and Chronic Health Assessment II (APACHE II) [14]. These variables were collected at admission within the first 24 hours of ICU admission. These variables in the specified period were collected, and in case of missing variables, the mean variable was assigned. After extracting all the characteristic variables, the Lasso (least absolute shrinkage and selection operator) regression method was used to select and filter the variables with the top 25 importance [15, 16].
Prediction models
To confirm the effectiveness of the XGBoost model in predicting AKI mortality, we used the following widely used machine learning models (LR, SVM, RF) for comparison and summarized the advantages and disadvantages of each of these models (Table 1).
Table 1. Advantages and disadvantages of each models.
| Models | Advantages | Disadvantages |
|---|---|---|
| LR [24] | LR is easier to implement, interpret, very fast calculations at classification and very efficient to train. | When the number of observations is less than the number of features, overfitting may result. |
| SVM [25, 26] | SVM can be used for linear and non-linear classification and regression problems. It provides a good out-of-sample generalization. | Kernel models can be quite sensitive to over-fitting the model selection criterion. |
| RF [27] | It can be used for both regression and classification tasks, and no need for feature normalization. | Not interpretable. |
| Performance is not good when there is class imbalance. | ||
| Avoids over-fitting. | ||
| XGBoost [28] | Do not require feature engineering (missing values imputation, scaling and normalization). | For numeric features only. |
| Leads to overfitting if hyperparameters are not adjusted correctly. | ||
| It can be used for classification, regression or ranking. | ||
| Extremely fast (parallel computation), and highly efficient. |
LR is a widely used statistical model. It is used to calculate the probability of occurrence of binary events and deal with classification problems. LR allows for multivariate analysis and modeling of a binary dependent variable. The multivariate analysis estimates the coefficients of each predictor included in the final model (e.g., log odds or hazard ratios) and adjusts them based on other predictors in the model. These coefficients quantify the contribution of each predictor to the risk estimate of the outcome [17].
SVM is a supervised machine learning algorithm. It is a binary classification method that separates two classes by a linear boundary and relies on extended linearity. In this algorithm, the main goal is to find the farthest distance between two classes, leading to more accurate classification and a reduction in generalization error [18].
RF is an ensemble algorithm, which combines multiple decorrelated decision tree prediction variables based on each subset of data samples [19]. RF is not only fast, easy to implement, and produces precise predictions, but it can also handle a large number of input variables without overfitting [20].
XGBoost is an improved algorithm based on the gradient boosting decision tree, which can efficiently construct boosted trees and run in parallel. The boosted trees in XGBoost is divided into regression trees and classification trees. The core of the algorithm is to optimize the value of the objective function [21]. XGBoost has the advantages of scalability in all scenarios, and fast [22]. The model works by combining a set of weaker machine learning algorithms to obtain an improved machine learning algorithm as a whole [23].
In model development and comparison, we employed 5-fold cross-validation, which provides a more stable and reliable way to measure the performances of models.
Model parameter setting
Based on the literature review and our experience, we chose the tuning parameter. For Lasso, we used alpha: ‘0.01’ to select the top 25 important variables. For the LR model, we set penalty: ‘l2’, solver: ‘liblinear’; In the SVM model, we used ‘rbf’ kernel and used ‘auto’ for gamma to train the classifier; For RF model, we set criterion: ‘gini’, and used the default parameter for other model parameters; For XGBoost model, we set learning rate: ‘0.1’, max_depth: ‘3’, objective: ‘binary:logistic’, booster:‘gbtree’, gamma: ‘0’.
Model evaluation
Each model was evaluated according to precision, recall, accuracy, F1 score, and AUROC (area under the receiver operating characteristic) curve. In this study, accuracy is the ratio of correctly predicted observations to the total number of observations. Precision refers to the ratio of correctly predicted positive observations to the total number of predicted positive observations. The recall is the ratio of correctly predicted positive observations to all actually positive observations. F1 Score is a harmonic mean of precision and recall. AUROC is a probability curve that graphically displays the trade-off between recall and specificity [29, 30].
Results
Participant characteristics
A total of 7,548 patients with AKI were included in the final cohort for this study, among which 1,234 (16.35%) died. In the 7,548 AKI patients, the proportion of male sex in the death group (57.7%) was higher than that in the survival group (55.7%). It was statistical significantly (P <0.01). The average age of the non-survival group and surviving group patients was 67.4 (SD ± 14.4) and 65.9 (SD ± 15.5) years, respectively. The non-survival group patients were statistically significantly older than survival group patients (P <0.01). The predominantly white population accounted for 76% of these patients. Patients in the non-survival group (7.5±8.2) had marked longer days of ICU stay than those in the survival group (6.6±9.4), and had statistical difference (P <0.01). Demographics of patients with AKI are shown in Table 2.
Table 2. Demographics of patients with AKI.
| Variable | Non-survivors (n = 1234) | Survivors (n = 6314) | P |
|---|---|---|---|
| Gender | − | − | − |
| Female | 522(42.3%) | 2794(44.3%) | <0,001 |
| Male | 712(57.7%) | 3520(55.7%) | |
| Age(year) | 67.4±14.4 | 65.9±15.5 | <0.01 |
| Height(cm) | 169.4±11.5 | 169.3±12.3 | 0.91 |
| Weight(kg) | 91.2±28.0 | 89.7±29.7 | 0.237 |
| BMI | 39.65±11.81 | 30.58±9.91 | 0.140 |
| Ethnicity | − | − | − |
| Caucasian | 951(77.1%) | 4816(76.%) | 0.5477 |
| African-American | 120(9.7%) | 736(11.7%) | 0.0502 |
| Hispanic | 72(5.8%) | 347(5.5%) | <0.001 |
| Other | 91(7.4%) | 415(6.6%) | 0.3024 |
| ICU days (mean,SD) | 7.5±8.2 | 6.6±9.4 | 0.003 |
Variable selection
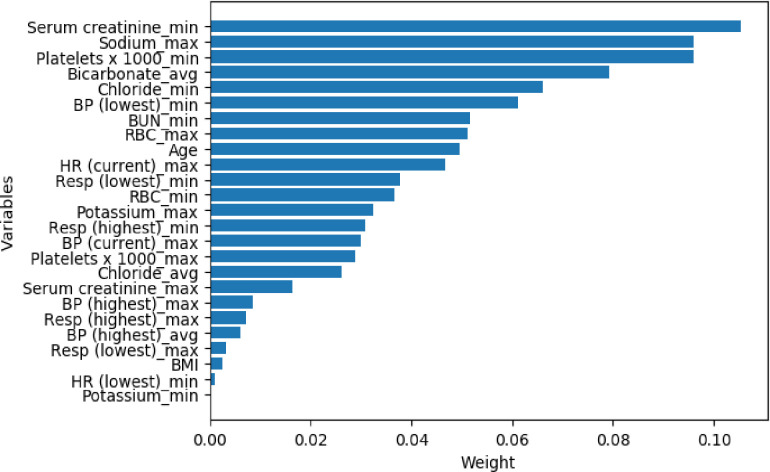
To detect the importance of variables in predicting mortality in AKI patients, Lasso (least absolute shrinkage and selection operator) was applied for feature selection. Lasso is a regression analysis method that uses L1 constraint to perform variable selection and regularization, providing a base to select a subset of the available covariates for use in the final model [12]. The Lasso selected the top 25 predictor variables (among 64 total variables) and weight (Fig 2). Creatinine (min) was the most important predictor variables for all prediction horizons, followed very closely by Sodium (max), markers of Platelets, Bicarbonate (average), and Chloride (min) (Tables 2 and 3).
Fig 2. The weights of variables importance.
The variables were collected from the eICU-CRD V2.0 database with the AKI patients’ admission date from 2014 to 2015. BP: blood pressure; BUN: blood urea nitrogen; RBC: red blood cell; HR: heart rate; Resp: respiratory rate; min: minimum; max: maximum; avg: average.
Table 3. All predictor variables for non-survivors and survivors.
| Variable(SD) | Non-survivors (n = 1234) | Survivors (n = 6314) | P |
|---|---|---|---|
| Serum creatinine min | 1.70(1.19) | 1.45(1.11) | <0.001 |
| Sodium max | 140.46(6.85) | 139.62(6.32) | <0.001 |
| Platelets x1000 min | 163.39(105.37) | 185.67(99.72) | <0.001 |
| Bicarbonate avg | 21.21(5.37) | 22.35(5.20) | <0.001 |
| Chloride min | 104.26(7.85) | 104.36(7.70) | <0.001 |
| BP Lowest min | 92.91(28.63) | 99.50(28.23) | 0.926 |
| BUN min | 42.47(26.99) | 43.13(27.27) | 0.066 |
| RBC max | 3.65(0.80) | 3.60(0.74) | 0.002 |
| Age | 67.41(14.37) | 65.96(15.50) | <0.001 |
| HR Current max | 97.42(22.32) | 92.55(21.12) | <0.001 |
| Resp Lowest min | 18.67(6.17) | 17.42(6.05) | <0.001 |
| RBC min | 3.38(0.82) | 3.44(0.76) | 0.411 |
| Potassium max | 4.61(0.85) | 4.50(0.84) | 0.001 |
| Resp Highest min | 25.89(7.56) | 24.20(7.92) | <0.001 |
| BP Current max | 115.37(25.57) | 120.12(26.09) | <0.001 |
| Platelets x1000 max | 186.75(111.91) | 199.44(104.19) | 0.002 |
| Chloride avg | 105.66(7.64) | 105.51(7.50) | 0.201 |
| Serum creatinine max | 3.42(1.85) | 3.17(2.39) | <0.001 |
| BP Highest max | 132.38(31.72) | 133.32(31.02) | 0.416 |
| Resp Highest max | 27.40(8.21) | 25.57(8.72) | <0.001 |
| BP Highest avg | 128.49(30.03) | 130.02(29.28) | 0.914 |
| Resp Lowest avg | 20.01(6.55) | 18.56(6.29) | 0.002 |
| BMI | 39.65(11.81) | 30.58(9.91) | 0.140 |
| HR Lowest min | 85.76(21.53) | 83.37(20.57) | 0.359 |
| Potassium min | 3.99(0.77) | 4.02(0.70) | 0.032 |
BP: blood pressure; BUN: blood urea nitrogen; RBC: red blood cell; HR: heart rate; Resp: respiratory rate.
Model performance
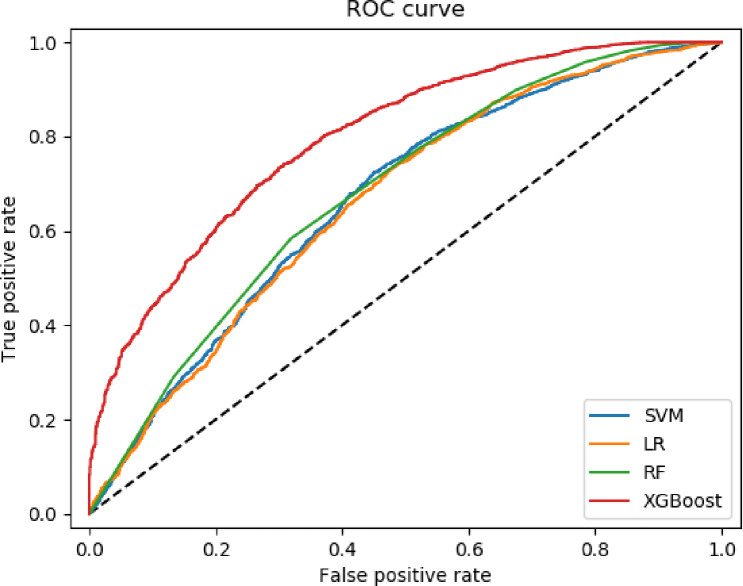
The results in the four machine learning methods found in the 5-fold cross-validation are shown in Table 4. The AUROC (0.796), accuracy (0.860), and F1 score (0.922) of XGBoost were higher than all other models. The precision and recall of the XGBoost model were the second-best among the four models. XGboost was superior to other models in terms of AUROC and F1, and had statistical significance (P<0.01). The lowest F1 score (0.910) and AUROC (0.662) were LR and RF, respectively (Table 4). The AUROC curves of these predictive models were shown in Fig 3.
Table 4. Comparison of mortality prediction performance among the four models of AKI patients.
| AUROC | Precision | Recall | Accuracy | F1 | P(AUROC) | P(F1 score) | |
|---|---|---|---|---|---|---|---|
| LR | 0.662 | 0.842 | 0.992 | 0.837 | 0.911 | - | - |
| SVM | 0.667 | 0.837 | 0.999 | 0.837 | 0.911 | <0.01vs.LR | <0.01vs.LR |
| RF | 0.692 | 0.862 | 0.956 | 0.836 | 0.910 | <0.01vs.LR | <0.01vs.LR |
| <0.01vs.SVM | <0.01vs.SVM | ||||||
| XGBoost | 0.796 | 0.860 | 0.994 | 0.860 | 0.922 | <0.01vs. LR | <0.01vs. LR |
| <0.01vs SVM | <0.01vs SVM | ||||||
| <0.01vs RF | <0.01vs RF |
Fig 3. The ROC curve among the four models of AKI patients.
Discussion
This study found a higher proportion of male sex in the AKI non-survival group patients than the survival group patients, and there was a statistical difference between the two groups (P<0.001). Elderly patients (average age 67.4 years old) were associated with an increased risk of death, and there was a statistical difference (P<0.01). Some researchers showed that there was a significant increase in old age and males in deceased AKI patients [31, 32]. At eGFR (estimated glomerular filtration rate) 80 ml/min/1.73 m2, the older age itself was linked with a higher risk of AKI [33].
Using Lasso, we could identify some important variables associated with AKI non-survival patients and survival patients. The most important variable for Lasso in this study was the minimum creatinine (non-survivors: 1.07±1.19; survivors: 1.45±1.11, P<0.01). Other research showed the slope of the minimum creatinine (30.32%) was the most important variable for predicting AKI [34]. This indicates that the minimum creatinine was more useful in predicting AKI mortality than any of the other laboratory measurements or vital signs [34]. Since the study used only data available in the eICU-CRD, the result had some implications and require further research.
In this study, four machine learning methods (RF, LR, SVM, and XGBoost) were used to predict the mortality of AKI. Performance comparison results showed the XGBoost achieved the highest scores in AUROC, accuracy, and F1 score, and the second-highest score in recall and precision. XGBoost performed better than other machine learning models, and the advantages were statistically significant in AUROC and F1 score (P<0.01). While the XGBoost model has outstanding advantages, the XGBoost model has not been externally validated against other databases. Inconsistencies between different databases may limit the applicability and generalizability of the XGBoost prediction model, as each algorithm is limited by the quality of the data used for training and testing purposes. Although the clinical applicability of the XGBoost mortality prediction model still needs to be tested in actual clinical practice. However, due to its performance and clinical interpretability, we believe that the model may help clinicians avoid treatment delays in high-risk AKI patients. The XGBoost model can play an auxiliary role for clinicians in clinical decision-making.
Meanwhile, there were some limitations to this study. Firstly, although the data quality of the eICU-CRD database is high, the results obtained had certain limitations due to geographical limitations. For example, in this study, 76.6% of the included patients were Caucasian. The applicability of the predictive model to other populations or regions still requires external verification. Second, though the eICU database is considered tele-ICU data, the data collection mode and data source are not well defined. Third, the terminology variations across institutions and health information systems constitute additional obstacles [35]. The next step would be to explore the intrinsic relationships between features and further validate the model results using additional clinical data sets.
Conclusions
The better prediction performance of XGBoost facilitates risk identification and early intervention of AKI patients at risk of death. It may be helpful to aid clinicians in making timely clinical intervention decisions for AKI patients, which is essential to help reduce the in-hospital mortality of AKI patients.
Data Availability
The eICU Collaborative Research Database is a third party data, which is a multi-center intensive care unit database with high granularity data for over 200,000 admissions to ICUs monitored by eICU Programs across the United States. We did not have any special access privileges. Other researchers could also access this database in the same manner. Details of the data access process are available online (https://eicu-crd.mit.edu). Use of the data requires proof of completion of the CITI “Data or Specimens Only Research” course (https://www.citiprogram.org/index.cfm?pageID=154&icat=0&ac=0) and signing of a data use agreement mandating responsible handling of the data and adhering to the principle of collaborative research. Once approved, data can be directly downloaded from the eICU Collaborative Research Database project on PhysioNet (https://physionet.org/login/).
Funding Statement
Jialin Liu, Sichuan Science and Technology Program under Grant No. 2020YFS0162. Ke Li,Special project of central government guiding local science and technology development under Grant No.2020ZYD001.Sichuan Science and technology support plan project NO.2019JDPT0008. The funders had a role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoste EAJ, Kellum JA, Selby NM, Zarbock A, Palevsky PM, Bagshaw SM, et al. Global epidemiology and outcomes of acute kidney injury. Nat Rev Nephrol. 2018;14(10):607–625. 10.1038/s41581-018-0052-0 [DOI] [PubMed] [Google Scholar]
- 2.Holmes J, Roberts G, Meran S, Williams JD, Phillips AO, Welsh AKI Steering Group. et al. Understanding Electronic AKI Alerts: Characterization by Definitional Rules. Kidney Int Rep. 2016;2(3):342–349. 10.1016/j.ekir.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J, Dubin JA, Maslove DM. Mortality Prediction in the ICU In: MIT Critical Data, ed. Secondary Analysis of Electronic Health Records. Cham (CH): Springer; 2016315–324. [PubMed] [Google Scholar]
- 4.Demirjian S, Chertow GM, Zhang JH, O’Connor TZ, Vitale J, Paganini EP, et al. Model to predict mortality in critically ill adults with acute kidney injury. Clin J Am Soc Nephrol. 2011;6(9):2114–2120. 10.2215/CJN.02900311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin K, Hu Y, Kong G. Predicting in-hospital mortality of patients with acute kidney injury in the ICU using random forest model. Int J Med Inform. 2019;125:55–61. 10.1016/j.ijmedinf.2019.02.002 [DOI] [PubMed] [Google Scholar]
- 6.Caicedo-Torres W, Gutierrez J. ISeeU: Visually interpretable deep learning for mortality prediction inside the ICU. J Biomed Inform. 2019;98:103269 10.1016/j.jbi.2019.103269 [DOI] [PubMed] [Google Scholar]
- 7.Mayampurath A, Sanchez-Pinto LN, Carey KA, Venable LR, Churpek M. Combining patient visual timelines with deep learning to predict mortality. PLoS One. 2019;14(7):e0220640 10.1371/journal.pone.0220640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon JM, Jeon KH, Kim HM, Kim MJ, Lim S, Kim KH, et al. Deep- learning-based risk stratification for mortality of patients with acute myocardial infarction. PLoS One. 2019;14(10):e0224502 10.1371/journal.pone.0224502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogunleye AA, Qing-Guo W. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans Comput Biol Bioinform. 2019;10:1109. [DOI] [PubMed] [Google Scholar]
- 10.Rufaida SI, Leu JS, Su KW, Haniz A, Takada JI. Construction of an indoor radio environment map using gradient boosting decision tree. Wireless Networks. 2020;26: 6215–6236. [Google Scholar]
- 11.Lee HC, Yoon SB, Yang SM, Kim WH, Ryu HG, Jung CW, et al. Prediction of Acute Kidney Injury after Liver Transplantation: Machine Learning Approaches vs. Logistic Regression Model. J Clin Med. 2018;7(11):428 10.3390/jcm7110428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu CN, Liu CL, Tain YL, Kuo CY, Lin YC. Machine Learning Model for Risk Prediction of Community-Acquired Acute Kidney Injury Hospitalization From Electronic Health Records: Development and Validation Study. J Med Internet Res. 2020;22(8):e16903 10.2196/16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard TJ, Johnson AEW, Raffa JD, et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018;5:180178 10.1038/sdata.2018.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kothiwale VA, Patil P, Gaur S. Correlation of Thyroid Hormone Profile with the Acute Physiology and Chronic Health Evaluation II Score as a Prognostic Marker in Patients with Sepsis in the Intensive Care Unit. J Assoc Physicians India. 2018;66(7):59–62. [PubMed] [Google Scholar]
- 15.Algamal ZY, Lee MH, Penalized logistic regression with the adaptive LASSO for gene selection in high-dimensional cancer classification. Expert Systems with Applications. 2015; 42 (23): 9326–9332. [Google Scholar]
- 16.Muthukrishnan R, Rohini R. LASSO: A feature selection technique in predictive modeling for machine learning. 2016 IEEE International Conference on Advances in Computer Applications (ICACA). 2016; 18–20.
- 17.Shipe ME, Deppen SA, Farjah F, et al. Developing prediction models for clinical use using logistic regression: an overview. J Thorac Dis. 2019;11(Suppl 4):S574–S584 10.21037/jtd.2019.01.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahrambeygi B, Moeinzadeh H. Comparison of support vector machine and neutral network classification method in hyperspectral mapping of ophiolite mélanges-A case study of east of Iran. Egypt J Remote Sens Space Sci. 2017; 20:1–10. [Google Scholar]
- 19.Mueller SQ. Pre-and within-season attendance forecasting in Major League Baseball: a random forest approach. Applied Economics. 2020; 52(41): 4512–4528. [Google Scholar]
- 20.Biau G. Analysis of a Random Forests Model. Journal of Machine Learning Research. 2010;13:1063–1095 [Google Scholar]
- 21.Zheng HT, Yuan JB, Chen L. Short-Term Load Forecasting Using EMD-LSTM Neural Networks with a Xgboost Algorithm for Feature Importance Evaluation. Energies. 2017;10(8): 1168. [Google Scholar]
- 22.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), San Francisco, CA. 2016: 785–794.
- 23.Chen X, Wang ZX, Pan XM. HIV-1 tropism prediction by the XGboost and HMM methods. Sci Rep. 2019;9(1):9997 10.1038/s41598-019-46420-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ottenbacher KJ, Smith PM, Illig SB, Linn RT, Fiedler RC, Granger CV. Comparison of logistic regression and neural networks to predict rehospitalization in patients with stroke. J Clin Epidemiol. 2001;54(11):1159–1165. 10.1016/s0895-4356(01)00395-x [DOI] [PubMed] [Google Scholar]
- 25.Hsu CW, Lin CJ. A comparison of methods for multiclass support vector machines. IEEE Trans Neural Netw. 2002;13(2):415–425. 10.1109/72.991427 [DOI] [PubMed] [Google Scholar]
- 26.Cawley GC, Talbot NLC. On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. The Journal of Machine Learning Research. 2010;11:2079–2107. [Google Scholar]
- 27.Hongyan L, Qian F. A review of random forests algorithm. Journal of the Hebei Academy of Sciences.2019; 36(3):37–41. [Google Scholar]
- 28.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. arXiv:1603.02754 (cs).2016.
- 29.Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Information Processing & Management 2009;45(4):427–437 [Google Scholar]
- 30.Ampomah EK, Qin ZG, Nyame G. Evaluation of Tree-Based Ensemble Machine Learning Models in Predicting Stock Price Direction of Movement. Information. 2020;11(6):332 [Google Scholar]
- 31.Kane-Gill SL, Sileanu FE, Murugan R, Trietley GS, Handler SM, Kellum JA. Risk factors for acute kidney injury in older adults with critical illness: a retrospective cohort study. Am J Kidney Dis. 2015;65(6):860–869. 10.1053/j.ajkd.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safari S, Hashemi B, Forouzanfar MM, Shahhoseini M, Heidari M. Epidemiology and Outcome of Patients with Acute Kidney Injury in Emergency Department; a Cross-Sectional Study. Emerg (Tehran). 2018;6(1):e30. [PMC free article] [PubMed] [Google Scholar]
- 33.Grams ME, Sang Y, Ballew SH, Gansevoort RT, Kimm H, Kovesdy CP, et al. A Meta-analysis of the Association of Estimated GFR, Albuminuria, Age, Race, and Sex With Acute Kidney Injury. Am J Kidney Dis. 2015;66(4):591–601. 10.1053/j.ajkd.2015.02.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parreco J, Soe-Lin H, Parks JJ, Byerly S, Chatoor M, Buicko JL, et al. Comparing Machine Learning Algorithms for Predicting Acute Kidney Injury. Am Surg. 2019;85(7):725–729. [PubMed] [Google Scholar]
- 35.Essay P, Shahin TB, Balkan B, Mosier J, Subbian V. The Connected Intensive Care Unit Patient: Exploratory Analyses and Cohort Discovery From a Critical Care Telemedicine Database. JMIR Med Inform. 2019;7(1):e13006 10.2196/13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The eICU Collaborative Research Database is a third party data, which is a multi-center intensive care unit database with high granularity data for over 200,000 admissions to ICUs monitored by eICU Programs across the United States. We did not have any special access privileges. Other researchers could also access this database in the same manner. Details of the data access process are available online (https://eicu-crd.mit.edu). Use of the data requires proof of completion of the CITI “Data or Specimens Only Research” course (https://www.citiprogram.org/index.cfm?pageID=154&icat=0&ac=0) and signing of a data use agreement mandating responsible handling of the data and adhering to the principle of collaborative research. Once approved, data can be directly downloaded from the eICU Collaborative Research Database project on PhysioNet (https://physionet.org/login/).