Abstract
Microspore embryogenesis is a biotechnological process that allows us to rapidly obtain doubled-haploid plants for breeding programs. The process is initiated by the application of stress treatment, which reprograms microspores to embark on embryonic development. Typically, a part of the microspores undergoes cell death that reduces the efficiency of the process. Metacaspases (MCAs), a phylogenetically broad group of cysteine proteases, and autophagy, the major catabolic process in eukaryotes, are critical regulators of the balance between cell death and survival in various organisms. In this study, we analyzed the role of MCAs and autophagy in cell death during stress-induced microspore embryogenesis in Brassica napus. We demonstrate that this cell death is accompanied by the transcriptional upregulation of three BnMCA genes (BnMCA-Ia, BnMCA-IIa and BnMCA-IIi), an increase in MCA proteolytic activity and the activation of autophagy. Accordingly, inhibition of autophagy and MCA activity, either individually or in combination, suppressed cell death and increased the number of proembryos, indicating that both components play a pro-cell death role and account for decreased efficiency of early embryonic development. Therefore, MCAs and/or autophagy can be used as new biotechnological targets to improve in vitro embryogenesis in Brassica species and doubled-haploid plant production in crop breeding and propagation programs.
Keywords: Autophagy, Metacaspase, Microspore embryogenesis, Programmed cell death, Rapeseed, Stress
Introduction
Stress-induced microspore embryogenesis is an in vitro process, in which application of stress conditions reprograms the precursor cell of the pollen grain, the haploid microspore, into a totipotent cell. The reprogrammed cell develops into a haploid or doubled-haploid embryo, and eventually into a plant. Among various stress conditions, temperature stress (mild heat or cold conditions) is one of the most common external treatments used to induce embryogenic cell reprogramming (B�r�ny et�al. 2005, German� and Lambardi 2016, Testillano 2019).
In Brassica napus, as in other plant species, the vacuolated microspore is the most responsive developmental stage for cell reprogramming. After exposure to mild heat stress treatment at 32�C, the normal development of the vacuolated microspore switches to the embryogenic program, resulting in the production of a haploid embryo, which after spontaneous or chemically induced-diploidization will produce doubled-haploid plants (Custers et�al. 1994, Prem et�al. 2012). Being widely used by seed and horticulture companies, doubled-haploid plants are important biotechnological tools in plant breeding programs providing a unique source of new genetic variability, fully homozygous for each locus and fixed in only one generation (Maluszynski et�al. 2003, Forster et�al. 2007, Murovec and Bohanec 2012, Iba�ez et�al. 2020). Since the mechanisms that control microspore reprogramming upon exposure to stress are not yet fully understood, efficient protocols for microspore embryogenesis are not available for many species of agronomic interest that show recalcitrant responses to stress treatment. In particular, increased cell death is a common stress response shared by all microspore embryogenesis in vitro systems that impairs the developmental reprogramming process leading to decreased embryo yield (Satpute et�al. 2005, Rodriguez-Serrano et�al. 2012, Huang et�al. 2016, B�r�ny et�al. 2018, Berenguer et�al. 2019). Understanding the mechanisms controlling cell death in microspore cultures will help to identify new targets for improving biotechnological methods.
Programmed cell death (PCD) is an active, genetically controlled process that leads to the elimination of specific cells and tissues. In plants, PCD occurs as an integral part of development as well as in response to biotic and abiotic stresses (van Doorn et�al. 2011). Being widespread throughout both pro- and eukaryotic (except animals) lineages, a group of proteases called metacaspases (MCAs) (Uren et�al. 2000, Minina et�al. 2017, Minina et�al. 2020) was functionally linked to the regulation of both stress-induced (Coll et�al. 2010, Coll et�al. 2014, Lema Asqui et�al. 2018, Salguero-Linares and Coll 2019) and developmental (Su�rez et�al. 2004, Bozhkov et�al. 2005, Bollh�ner et�al. 2013, Escamez et�al. 2016) cell deaths in plants. Genomes of higher plants encode two types of MCAs, type I and type II, distinguished by domain composition (Tsiatsiani et�al. 2011). Although MCAs have a secondary structure similar to animal-specific caspases, their substrate specificity and other biochemical properties are different. In contrast to caspases, MCAs do not cleave their substrates after aspartate (Asp) but prefer arginine (Arg) or lysine (Lys) in the P1 position, and most of them require millimolar concentrations of calcium (Ca2+) for catalysis (Minina et�al. 2020).
One of the pathways that has been shown to contribute to plant PCD is autophagy, the ‘self-eating’ process conserved throughout eukaryotes for the degradation and recycling of cytoplasmic components (Minina et�al. 2014a; Tang and Bassham 2018). During autophagy, the cytoplasmic cargo is engulfed by autophagosomes, dedicated double-membrane vesicles, that deliver the cargo to the lytic compartment, in the case of plants the vacuoles. The outer membrane of autophagosomes fuses with a membrane of the lytic compartment releasing the cargo to the lumen (Li and Vierstra 2012, Avin-Wittenberg et�al. 2018). More than 40 autophagy-related genes (ATG) have been so far identified to be required for the sequential steps of the autophagy pathway. Autophagosome formation relies on two ubiquitin-like conjugation systems that involve the E1 ligase-like ATG7, E2 ligase-like ATG3 and ATG10, and E3 ligase-like ATG12–ATG5–ATG16 complex, which jointly confer ATG8 lipidation and its association with autophagosomal membranes (Li and Vierstra 2012, �st�n et�al. 2017). Finally, ATG8 facilitates cargo recruitment and incorporation into the autophagosome (Michaeli et�al. 2016). The best-characterized plant autophagy cargo receptor is NBR1 (neighbor of BRCA1 gene 1). Plant NBR1 is a hybrid protein that harbors functions of two mammalian proteins, such as p62 and NBR1, that can bind ATG8 and poly-ubiquitinated proteins (Zhou et�al. 2013, Hafr�n et�al. 2017, Yoshimoto and Ohsumi 2018). Both ATG8 and NBR1 are present in ripe autophagosomes and are subject to degradation, providing reliable reporters of autophagic activity.
Recent studies have revealed the participation of papain-like cysteine proteases and uncharacterized proteases with caspase 3-like proteolytic activity in cell death during microspore embryogenesis of B. napus and H. vulgare (Rodriguez-Serrano et�al. 2012, B�r�ny et�al. 2018, Berenguer et�al. 2019). However, the involvement of MCAs and autophagy remains elusive. Here, we have analyzed the possible role of MCAs and autophagy in stress-induced cell death of B. napus with two main purposes: (i) to identify new targets for improving microspore embryogenesis and (ii) to establish a new model system for studying the role of MCAs and autophagy in plant cell death. We show that both MCAs and autophagy are activated by heat stress in B. napus microspores and that inhibition of either component or both components leads to the suppression of cell death and concomitant increase in the frequency of embryonic development. These findings provide new evidence of the participation of MCAs and autophagy in plant cell death induced by mild heat stress and open the door for new strategies to enhance the efficiency of microspore embryogenesis.
Results
Cell death at early stages of stress-induced microspore embryogenesis
Vacuolated microspores of B. napus (Fig.�1A) were isolated from anthers, cultured in a liquid medium and subjected to the heat stress treatment at 32�C, as previously reported (Prem et�al. 2012). Four days after the treatment, the responsive microspores switched their developmental program and showed the first sign of embryogenesis initiation by producing multicellular microspores or proembryos still surrounded by the microspore wall, the exine (Fig.�1B). During the following days, proembryos developed further, passing through a stereotyped sequence of stages including globular (Fig.�1C), heart-shaped, torpedo (Fig.�1D) and, finally, reaching fully differentiated, cotyledonary embryo stage after 30 d in culture (Fig.�1E, F).
Fig. 1.
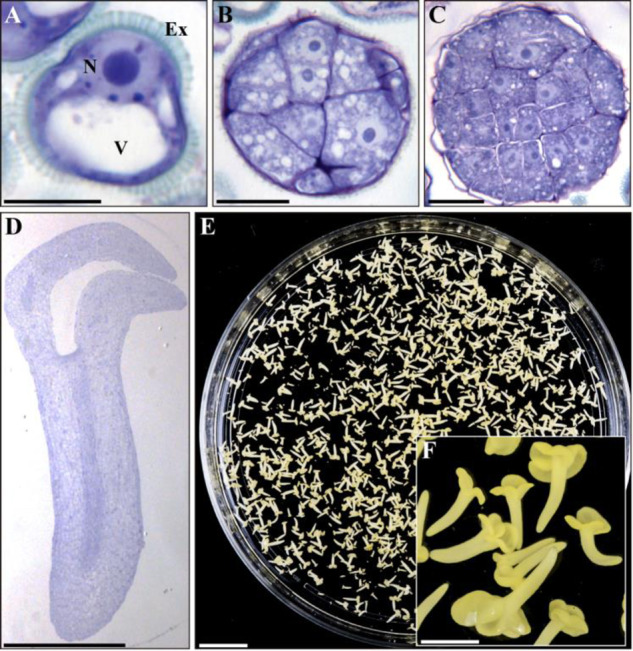
Stress-induced microspore embryogenesis in B. napus. Micrographs of toluidine blue-stained sections of (A) isolated vacuolated microspore, before stress (B) proembryo of stress-treated microspore culture, (C) globular embryo and (D) cotyledonary embryo. (E) Petry dish containing microspore-derived embryos and (F) higher magnification of cotyledonary embryos. Ex: exine; N: nucleus; V: vacuole. Scale bars: (A–C): 10 �m; (D): 500 �m; (E): 10 mm; and (F): 1 mm.
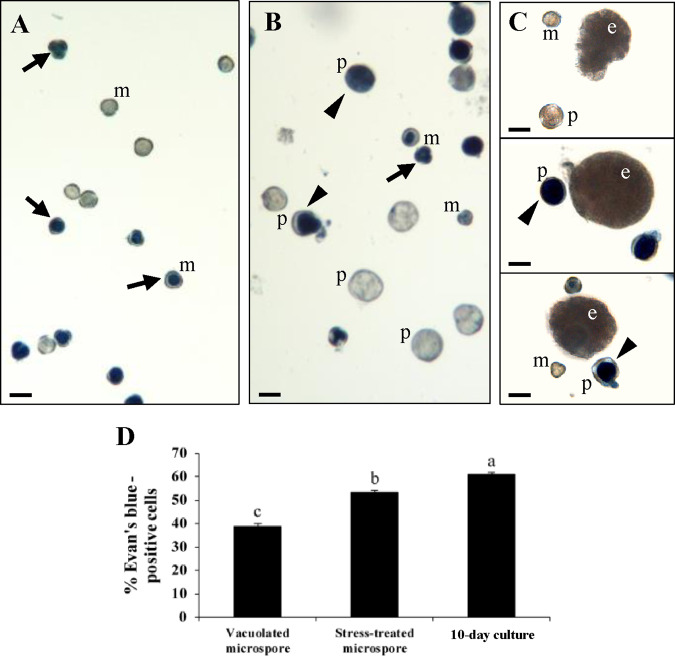
Not all microspores responded to stress treatment in the same way; some of them were reprogrammed and initiated an embryogenesis pathway, while a certain proportion underwent cell death. To analyze the cell death rate during microspore embryogenesis induction, we performed Evan’s blue staining of dead cells at three culture stages: (i) ‘vacuolated microspores’, i.e. freshly isolated microspores at the beginning of culture and before stress; (ii) ‘stress-treated microspores’, i.e. a microspore culture post-stress, when early proembryos were already formed and coexisted with microspores that did not respond to induction and degenerated; and (iii) ‘10-day culture’, i.e. stage when globular, heart-shaped and early torpedo embryos were visible, together with microspores and proembryos that had not progressed in development (Fig.�2). Some Evan’s blue-positive microspores were detected already before stress (Fig.�2A), apparently reflecting cell damage imposed by microspore isolation procedure, a common feature of isolated microspore cultures (Rodriguez-Serrano et�al. 2012, Berenguer et�al. 2019). Following stress treatment, Evan’s blue staining revealed the death of individual microspores and a significant part of proembryos (Fig.�2B), which were easily distinguished from microspores by their larger size and round shape. As embryogenesis proceeded, in 10-day cultures (Fig.�2C), individual microspores, proembryos (still present at this time point) and some embryos revealed Evan’s blue staining. The magnitude of cell death was estimated by calculating the percentage of Evan’s blue-positive structures (microspores, proembryos or embryos). As shown in Fig.�2D, at culture initiation, before stress, when the culture was only composed of vacuolated microspores, 39% of them were dead. In stress-treated microspore cultures, 53% of cells were Evan’s blue positive and these dead cells could be further characterized into microspores (51%) and proembryos (49%). Later, in 10-day cultures, the cell death level increased to 60% affecting microspores, proembryos and embryos (41%, 38% and 21% of dead cells, respectively).
Fig. 2.
Cell death in stress-induced microspore embryogenesis. Representative micrographs of Evan’s blue staining showing cell death in: (A) microspores (m) before stress; (B) stress-treated microspore culture, containing microspores (m) and proembryos (p); (C) 10-day culture composed by globular embryos (e), as well as microspores (m) and proembryos (p). Arrows: dead microspores; arrowheads: dead proembryos; Scale bars: 10 �m. (D) Quantification of cell death. The data represent mean � SEM. Different letters indicate significant differences (chi-square test at P < 0.05).
MCA activity during stress-induced microspore embryogenesis
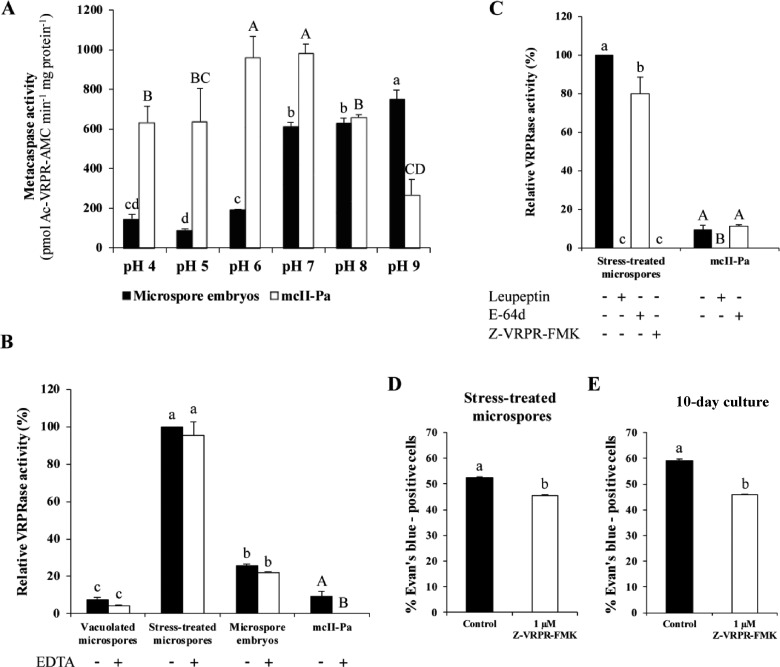
To evaluate whether MCAs play a role in stress-induced microspore embryogenesis, we analyzed the enzymatic activity of MCAs in protein extracts from microspore cultures using a dedicated substrate, a fluorogenic tetrapeptide acetyl-Val-Arg-Pro-Arg-amido-4-methylcoumarin (Ac-VRPR-AMC), under a wide pH range. Recombinant type II Norway spruce MCA mcII-Pa (or PaMCA-IIb according to recently accepted, unified nomenclature; Minina et�al. 2020) was used as a control (Minina et�al. 2014c). As shown in Fig.�3A, while recombinant mcII-Pa showed maximal proteolytic activity at neutral pH (6.0–7.0) (Bozhkov et�al. 2005), the highest levels of Ac-VRPR-AMC cleavage in B. napus microspore cultures were detected at more alkaline pH, between pH 7.0 and 9.0. Measurement of MCA activity at three successive stages of microspore embryogenesis revealed a sharp increase in the activity shortly after stress treatment, with a subsequent drop in microspore embryos (Fig.�3B).
Fig. 3.
MCA activity in microspore embryogenesis. Effect of pH (A), EDTA (B) and protease inhibitors at 10 �M (C) on VRPRase activity of protein extracts from microspore-derived embryos and recombinant mcII-Pa. The data represent mean � SEM. Different letters indicate significant differences within protein extracts or recombinant mcII-Pa series (Tukey’s t-test at P < 0.05). Quantification of cell death in control and Z-VRPR-FMK-treated microspore cultures, at two stages: stress-treated microspore culture (D) and 10-day culture (E). Different letters indicate significant differences (chi-square test at P < 0.05).
As the vast majority of MCAs studied so far require millimolar Ca2+ concentration for activation, we performed the MCA activity assay in the presence of Ca2+-chelating agent EDTA. In contrast to recombinant mcII-Pa, MCA activity in B. napus microspore cultures was not dependent on Ca2+ (Fig.�3B). To further characterize the MCA activity in cell extracts from stressed microspores, we assessed Ac-VRPR-AMC cleavage in the presence of protease inhibitors l-trans-epoxysuccinyl-leucylamide-(4-guanido)-butane (E-64d), leupeptin and Z-Val-Arg-Pro-Arg-fluoromethylketone (Z-VRPR-FMK) (MCA-specific inhibitor). While E-64d did not inhibit MCA activity of both cell extracts and recombinant mcII-Pa, the activity was completely abolished by leupeptin and Z-VRPR-FMK (Fig.�3C).
To evaluate whether the MCA activity was required for cell death during microspore embryogenesis, we compared the level of cell death in control cultures and cultures treated with the MCA-specific inhibitor Z-VRPR-FMK at two time points when MCA activity was detected: (i) after stress (stress-treated microspore culture) and (ii) in 10-day culture, where embryos at early developmental stages were already present. Inhibition of MCAs resulted in a significant decrease in the frequency of Evan’s blue-positive cells (chi-square test at P < 0.05) in stress-treated microspores (Fig.�3D). In the 10-day culture, inhibition of MCA activity reduced cell death level to a similar extent as in the case of stress-treated microspores (Fig.�3E). Taken together, these results link MCA activity to cell death occurring in both stress-treated microspore cultures and microspore-derived embryos.
Characterization of MCA family in Brassica napus and expression analysis of selected genes in stress-induced microspore embryogenesis
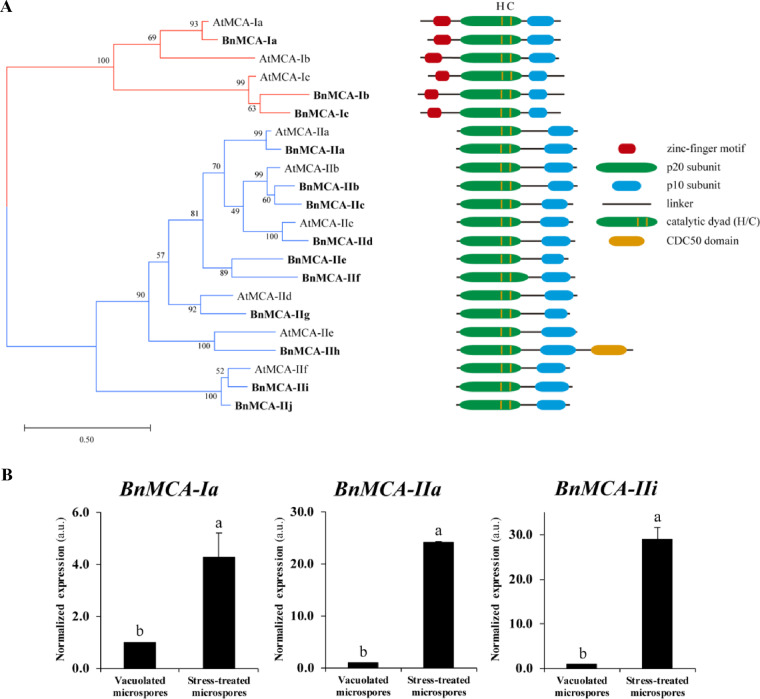
To investigate MCA gene expression during microspore embryogenesis, we first performed the phylogenetic analysis of the MCA family in B. napus. Nine MCA protein sequences of Arabidopsis thaliana were used as a query to search the Brassica rapa genome annotation database (v.3.0) because B. napus is an interspecific hybrid between B. rapa and Brassica oleracea. Altogether, 13 BnMCAs were identified, all containing a full-length caspase-like domain (PF00656) (Supplementary Fig. S1) composed of conserved subunits (p20 and p10) and harboring a catalytic dyad His-Cys in the p20 subunit (Supplementary Fig. S2). Three BnMCAs were closely related to type I AtMCAs possessing an N-terminal prodomain with a zinc finger LSD1-like region (Coll et�al. 2010). Ten other BnMCAs showed close resemblance to type II AtMCAs. A phylogenetic analysis of B. napus and A. thaliana MCAs revealed single B. napus homolog for AtMC1/AtMCA-Ia (according to recently accepted, unified nomenclature; Minina et�al. 2020), AtMC4/AtMCA-IIa, AtMC6/AtMCA-IIc, AtMC7/AtMCA-IId and AtMC8/AtMCA-IIe, while duplications had occurred resulting in two different B. napus homologs to AtMC3/AtMCA-Ic, AtMC5/AtMCA-IIb and AtMC9/AtMCA-IIf. We found no close homolog for AtMC2/AtMCA-Ib but identified a separate subgroup of type II BnMCAs composed of BnMCA-IIe and BnMCA-IIf (Fig.�4A). Brassica napus homolog of AtMC8/AtMCA-IIe, the BnMCA-IIh, contained a CDC50 domain, which is not a typical domain for MCAs (Fig.�4A). In A. thaliana, CDC50 proteins, also named ALIS [ALA (AminophosphoLipid ATPase)-Interacting Subunit], comprise the β-subunit of ALA/ALIS P4-ATPase that forms a heterodimeric αβ-complex with the catalytic α-subunit; this complex has been reported to contribute to vesicle formation as a phospholipid flippase (Poulsen et�al. 2008, L�pez-Marqu�s et�al. 2010, Zhang et�al. 2020). Since this is the first work describing the CDC50 domain in MCA proteins, further work is required to identify the functional relevance of this domain in BnMCA-IIh.
Fig. 4.
MCA protein family of B. napus and expression of selected genes during microspore embryogenesis. (A) Phylogenetic tree of Arabidopsis and B. napus MCAs inferred using the maximum-likelihood method with 1,000 bootstrap replicates using MEGA software. Percentages of taxa clustered together are shown next to the branches. The schematic domain structure of individual MCAs is shown to the right. (B) Relative changes in mRNA levels of BnMCA-Ia, BnMCA-IIa and BnMCA-IIi normalized to vacuolated microspore levels determined by RT-qPCR. The data represent mean � SEM. Different letters indicate significant differences (Student’s t-test at P < 0.05).
For the expression analysis, we selected three B. napus MCA genes based on spatio-temporal gene expression data available through the Arabidopsis eFP browser (http://bar.utoronto.ca/∼dev/eplant/, Winter et�al. 2007). Thus, we selected BnMCA-Ia (homolog of AtMC1/AtMCA-Ia) and BnMCA-IIa (homolog of AtMC4/AtMCA-IIa), which show the highest gene expression levels among Arabidopsis MCAs during male gametophytic development including the vacuolated microspore stage (Supplementary Fig. S3). Considering that the VRPRase activity of protein extracts isolated from microspore cultures was insensitive to EDTA (Fig.�3B), we have also included BnMCA-IIi as the closest homolog of AtMC9/AtMCA-IIf whose activity is independent of calcium (Vercammen et�al. 2004). The choice of BnMCA-II was also supported by a relatively high expression level of its homolog AtMC9/AtMCA-IIf during male gametophytic development (Supplementary Fig. S3). Quantitative real-time PCR (RT-qPCR) analysis revealed that expression of all three genes was strongly upregulated in stressed microspores, the effect being especially pronounced for the type II MCAs, BnMCA-IIa and BnMCA-IIi (Fig.�4B). These results correlate with both cell death (Fig.�2D) and VRPRase activity (Fig.�3B) measurements, indicating potential involvement of these MCAs in microspore stress response and/or embryogenesis.
Autophagy activation during stress-induced microspore embryogenesis
To examine whether autophagy has a role at the early stages of stress-induced microspore embryogenesis, as a first step, we performed RT-qPCR analysis of BnATG5, a core gene-regulating autophagosome formation. The analysis has revealed a significant increase in BnATG5 transcript levels in stress-treated microspores in comparison with non-stressed, vacuolated microspores (Fig.�5A). This result agreed with the immunolocalization analysis of ATG5, which revealed a significant increase in the protein abundance and punctate protein localization pattern in the microspore culture following stress treatment (Fig.�5B). The localization pattern of ATG5 was similar to that of ATG8 (Fig.�5C), whichwas also increased in abundance upon stress.
Fig. 5.
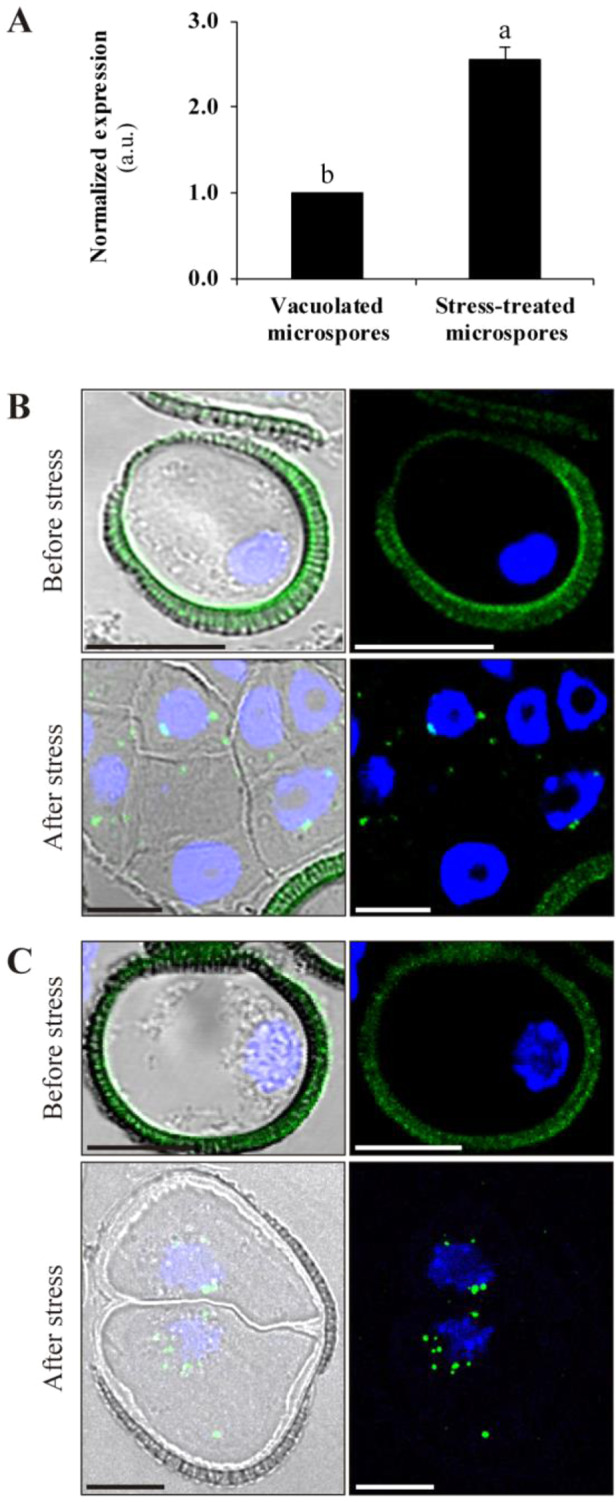
Expression and localization of autophagy-related proteins in microspore embryogenesis. (A) BnATG5 mRNA levels in stress-treated microspores compared to vacuolated microspores before stress, as determined by RT-qPCR. The data represent mean � SEM. Expression levels were normalized to vacuolated microspores. Different letters indicate significant differences (Student’s t-test at P < 0.05). Immunofluorescence and confocal microscopy analysis of ATG5 (B) and ATG8 (C) in vacuolated microspores (before stress) and stress-treated microspores (after stress). Left panels are merged images of Normarsky’s differential interference contrast, ATG5 or ATG8 immunofluorescence signal (green) and DAPI staining of nuclei (blue). Right panels are merged images of ATG5 or ATG8 immunofluorescence signal (green) and DAPI staining of nuclei (blue). Scale bars: 10 �m.
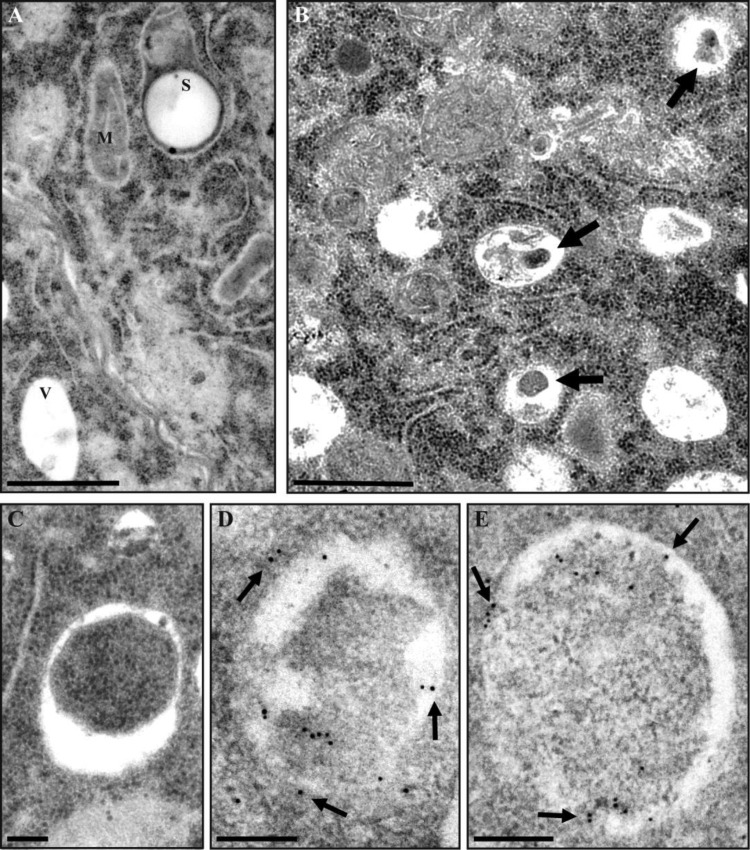
Next, ultrastructural analysis using transmission electron microscopy (TEM) revealed an increased number of autophagic bodies in the lumen of the vacuoles in stress-treated compared to untreated microspores (Fig.�6A, B, arrows). Similarly, stressed microspores were accumulating higher number of autophagosomes, engulfing material with electron density similar to the surrounding cytoplasm (Fig.�6C). We further used immunogold labeling to confirm that organelles ascribed to as autophagosomes indeed contained ATG8 protein (Fig.�6D, E, arrows).
Fig. 6.
Electron microscopy analysis of autophagosomes and ATG8 localization in microspores. (A) Cytoplasmic area of vacuolated microspore, before stress. (B) Cytoplasmic area of stress-treated microspore treated with E-64d to preserve autophagic bodies in the vacuoles (arrows). (C) Autophagosome in stress-treated microspore. (D and E) Immunogold labeling of ATG8 (arrows) is restricted to autophagosomes in stress-treated microspores. V, vacuole; M, mitochondria; S, starch granule. Scale bars: (A, B) 1 �m; (C–E) 0.2 �m.
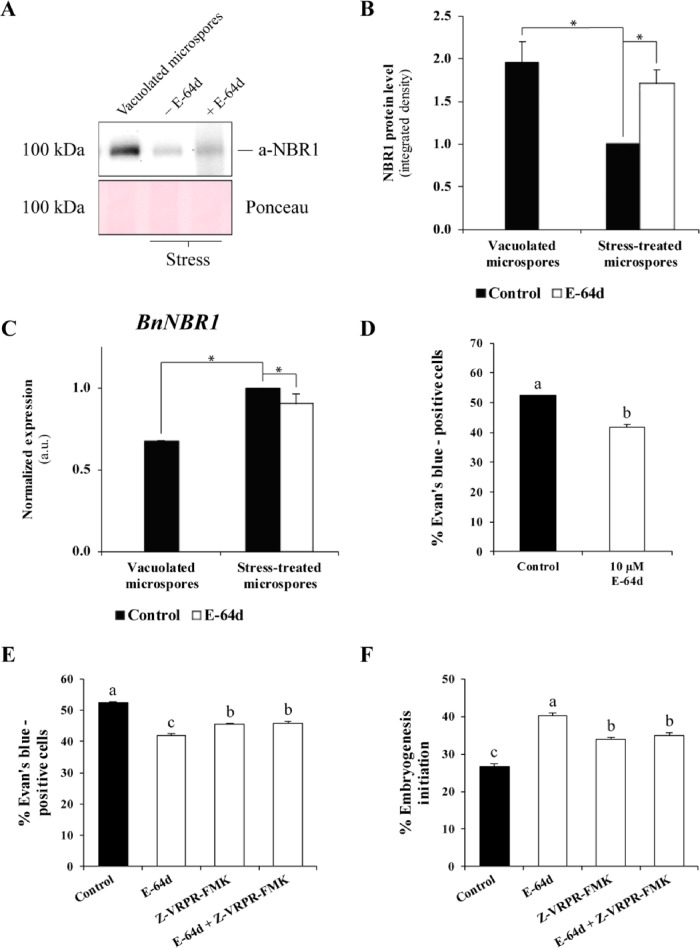
To evaluate the level of the autophagic flux in stressed and unstressed microspores, we evaluated the abundance of cargo adaptor protein NBR1 using the Western blot analysis. The abundance of NBR1 was significantly reduced following heat stress indicating elevated autophagic flux (Fig.�7A, B). Accordingly, inhibition of autophagic degradation in stressed microspores by the cysteine protease inhibitor E-64d reversed the abundance of NBR1 up to a level close to unstressed microspores (Fig.�7A, B), indicating that NBR1 amount was indeed regulated by autophagic activity. To confirm that differences in the abundance of NBR1 protein were not caused by transcriptional regulation, we measured the BnNBR1 gene expression in the corresponding samples. The RT-qPCR analysis detected an upregulation of BnNBR1 transcription under stress conditions and no differences in the transcript levels between control and E-64d-treated samples (Fig.�7C).
Fig. 7.
Autophagic flux analysis and effect of autophagy and MCA inhibitors on cell death and microspore embryogenesis. (A) Immunoblot analysis of NBR1 degradation in stress-induced microspores, with and without E-64d treatment (7.5 �M). Ponceau staining of total proteins was used as a loading control. (B) Quantification of bands in A using ImageJ and two-step normalization: first, to control stress-treated microspore and then to corresponding intensities of Ponceau staining (loading). (C) RT-qPCR analysis of BnNBR1 mRNA levels. The values were normalized to control, stress-treated microspore samples. The data represent mean � SEM. Asterisks in (B) and (C) indicate significant differences (Student’s t-test at P < 0.05). (D) Effect of E-64d on cell death in stress-induced microspore cultures. (E) Effect of individual and combined treatments with E64d and Z-VRPR-FMK on cell death in stress-induced microspores. (F) Quantification of proembryos (a marker of embryogenesis initiation) after stress treatment in control cultures and cultures treated with E-64d and/or Z-VRPR-FMK. The data represent mean � SEM. Different letters in (D)–(F) indicate significant differences (chi-square test at P < 0.05).
To further investigate whether autophagic activity was merely coinciding with or contributing to the stress-induced cell death during microspore embryogenesis, we compared the frequency of Evan’s blue-positive cells in heat-stressed microspore cultures incubated with and without E-64d. As shown in Fig.�7D, suppression of autophagic degradation by E-64d significantly reduced the frequency of Evan’s blue-stained cells and hence cell death level. Collectively, these data suggest that autophagy contributes to stress-induced cell death during the induction of microspore embryogenesis.
Effect of the combination of autophagy and MCA inhibitors on cell death and microspore embryogenesis
Since inhibition of either autophagy or MCA activity suppressed cell death in embryogenic cultures, we wondered whether the combination of the two types of inhibitors could have an additive effect. Microspore cultures were subjected to individual treatment with only one type of inhibitor, E64d or Z-VRPR-FMK, and to combined treatment with both E64d and Z-VRPR-FMK. Quantification of cell death in stress-treated microspore cultures showed that the combination of both autophagy and MCA inhibitors resulted in a significant decrease in cell death frequency in comparison with control, mock-treated, cultures (Fig.�7E). However, a combined application of the two inhibitors did not yield an additive inhibitory effect on cell death, when compared with individual treatments (Fig.�7E).
We were interested in evaluating whether the cell death suppression effect provoked by autophagy and/or MCA inhibition had an impact on early embryonic development. To this end, we evaluated the frequency of proembryo formation in stressed, E-64d- and/or Z-VRPR-FMK-treated cultures, compared to mock. Proembryos were clearly distinguished from microspores since they exhibited larger size, rounded morphology and higher density than microspores (Fig.�1A, B). We found that cultures treated with E-64d- and Z-VRPR-FMK, used either individually or in combination, outperformed mock cultures by the frequency of proembryo formation (Fig.�7F). No significant additive effect on embryogenesis was found in combined treatment with autophagy and MCA inhibitors, when compared with the individual ones (Fig.�7F). These findings provide strong evidence that autophagy- and MCA-dependent cell death induced by mild heat stress impedes microspore embryogenesis.
To evaluate the quality of microspore embryos produced in the presence of inhibitors, mature embryos regenerated from mock- and inhibitor-treated cultures were tested for germination ability. High germination frequencies were found in all the cases, ranging from 90% to 100%, as reported for B. napus microspore embryos (Prem et�al. 2012, data not shown). Furthermore, neither E-64d nor Z-VRPR-FMK or their combination had any effect on the phenotype of the germinated embryos (Supplementary Fig. S4).
Discussion
MCAs are activated during stress-induced microspore embryogenesis and involved in cell death
Our study is the first survey of MCAs in B. napus. This species has a complex allopolyploid genome, as a result of interspecific hybridization between B. rapa and B. oleracea (Liu et�al. 2018). We have identified 13 BnMCA candidate genes, although we cannot rule out the existence of additional MCA-encoding genes. Phylogenetic analysis revealed that BnMCAs share similar structures with their corresponding Arabidopsis homologs. We have found several duplications in both type I and type II BnMCAs, which agrees with high-frequency gene duplication events typical for Brassica spp. (Parkin et�al. 2003, Schranz et�al. 2006, Mand�kov� and Lysak 2008).
Gene expression analysis has revealed enhanced accumulation of BnMCA-Ia, BnMCA-IIa and BnMCA-IIi transcripts in stress-treated microspores, the effect being especially pronounced for type II MCA genes. Arabidopsis homologs of these MCAs have been shown to have cell death-related functions under different settings. Thus, AtMCA-Ia/AtMCA1 was established as a positive regulator of pathogen-triggered hypersensitive response (HR)-associated cell death (Coll et�al. 2010), whereas AtMCA-IIa/AtMCA4 was implicated in the HR-like response to fungal mycotoxin FB1 (Watanabe and Lam 2011). AtMCA-IIf/AtMCA9 and its poplar homologs PttMCA13 and PttMCA14 seem to be unrelated to stress responses but are essential for the autolysis of tracheary elements during xylem differentiation (Escamez et�al. 2016, Bollh�ner et�al. 2018).
Of note, analysis of VRPRase proteolytic activity in cell extracts from microspore-derived embryos revealed that BnMCAs might have a neutral-alkaline pH optimum and are calcium independent. Since BnMCAs are more active at pH 7.0–9.0, they most likely function in the cytoplasm, nucleus or organelles with neutral pH, but not in the lytic vacuole. Previous studies using recombinant proteins have established a neutral to slightly basic pH optimum (pH 7.0–8.0) for most type II MCAs, except for AtMCA-IIf/AtMCA9, which is active in an acidic environment (pH 5.0–6.0; Vercammen et�al. 2004, Bozhkov et�al. 2005, Tsiatsiani et�al. 2011, Tsiatsiani et�al. 2013). Unlike caspases, MCAs are calcium-dependent proteases, with the exception of AtMCA-IIf/MCA9, which does not require calcium for activation (Vercammen et�al. 2004). Zhu et�al. (2020) have established that the negatively charged segment of four residues 96EDDD99 located in the L5 loop of the p20 domain is a key determinant of calcium dependence of AtMCA-IIa/MC4 and possibly other type II MCAs. Correspondingly, AtMCA-IIf/AtMCA9 is the only Arabidopsis MCA lacking this negatively charged segment (Zhu et�al. 2020). The molecular determinant of calcium dependence of Arabidopsis MCAs established by Zhu et�al. (2020) appears to be strictly conserved in B. napus homologs. Indeed, all but BnMCA-IIi and BnMCA-IIj (two homologs of AtMCA-IIf/AtMCA9) display negatively charged segment situated in the position aligning with 96EDDD99 of AtMCA-IIa/AtMC4 (Supplementary Fig. S2).
Although the present study did not address the contribution of individual BnMCAs into VRPRase activity measured in total protein extracts, it is tempting to speculate that there might be some structural differences between B. napus and Arabidopsis homologs that account for calcium independence but neutral–alkaline pH activity profile. One possibility is low-affinity calcium-binding sites, as found in type II MCAs in other plant species (Klemenčič and Funk 2019). CRISPR-Cas9 technology has recently been applied in B. napus (Braatz et�al. 2017, Okuzaki et�al. 2018), although it has never been used in embryogenesis responsive genotypes. Future work to obtain MCA and autophagy mutants by this technique would be of great interest to understand their contribution to proteolytic activities and their mechanistic role in cell death and microspore embryogenesis.
In our study, enhanced expression of BnMCA genes in microspore cultures induced to form embryos by mild heat treatment correlated well with the increase in MCA-like proteolytic activity. Furthermore, inhibition of MCA-like activity by Z-VRPR-FMK partly suppressed cell death following heat stress treatment, indicating that MCAs may participate as pro-cell death regulators under the described conditions. Our results thus extend the role of MCAs in abiotic stress responses to include mild heat stress-induced cell death. Since this cell death antagonizes embryogenesis in microspore cultures of B. napus, its suppression by the MCA inhibitor Z-VRPR-FMK led to an increased frequency of early embryonic development.
Enhanced autophagy contributes to cell death during microspore embryogenesis induction by stress
Our results indicate that autophagy takes part in cell death induced by mild heat stress during microspore embryogenesis of B. napus. The abundance of both BnATG5 transcripts and immunolocalized ATG5 and ATG8 proteins increase following stress treatment. Electron microscopy has revealed a large number of ATG8-positive autophagosomes in stress-treated microspores. Precise and reliable quantification of plant autophagy activity has been demonstrated through the analysis of cargo receptors and the use of autophagy modulators (Bozhkov et�al. 2005, Luo et�al. 2017, Minina et�al. 2018). We show that the rate of NBR1 degradation, a measure of autophagic flux, is enhanced following heat stress, the effect abrogated by protease inhibitor E-64d. Taken together, these results provide strong evidence that mild heat stress activates autophagy in B. napus microspore cultures.
Autophagy contributes to cell death during abiotic stress conditions (Yang and Bassham 2015, Avin-Wittenberg 2019); however, little is known about its role in microspore embryogenesis, except for our previous report in barley where we showed that autophagy was activated after the cold treatment applied to trigger embryogenesis in this species (B�r�ny et�al. 2018). In B. napus, suppression of heat-induced cell death by E64d, an inhibitor of autophagic flux, indicates that autophagy has a pro-death role in the stress response of microspore. On the other hand, we cannot rule out that autophagy might play a dual role in microspore cultures, contributing to both cell survival and death (�st�n et�al. 2017, Avin-Wittenberg et�al. 2018). However, given the results showing that autophagy inhibition reduced cell death provoked by stress, we can hypothesize that although autophagy activation could have both pro-survival and pro-death functions, the pro-survival role would have a lower impact in the viability of the culture than the pro-death function. A possible explanation could be that induction of microspore embryogenesis would give rise to rapid stress-related responses, including activation of autophagy, which could initially play a homeostatic function by removing and recycling cellular damage caused by stress. However, in some microspores, accumulation of cellular damage could not be efficiently compensated by autophagy and other mechanisms, and high amounts of toxic metabolites, protein aggregates or defective organelles could lead to a runaway autophagy that would contribute to cell death. Inhibition of this autophagy-dependent cell death by E64d resulted in cell death suppression. This effect is relevant for biotech applications, as it leads to the increased frequency of embryogenesis initiation.
We have previously reported the activation of autophagy and its involvement in cell death using barley microspore embryogenesis as a model system (B�r�ny et�al. 2018). The present study extends these findings to B. napus, revealing a common feature of the participation of autophagy in stress-induced cell death in two crop species where embryogenesis is induced by two contrasting stress treatments, i.e. heat in rapeseed and cold in barley. Several proteolytic activities have been recently linked to cell death execution during microspore embryogenesis induction in barley and rapeseed (Rodriguez-Serrano et�al. 2012, B�r�ny et�al. 2018, Berenguer et�al. 2019). In barley, papain-like cysteine proteases (cathepsins) are activated and play a role in the stress-induced death of microspores (B�r�ny et�al. 2018), whereas proteases with caspase 3-like aspartate cleavage specificity have been found to participate in stress-induced cell death in both barley and rapeseed microspore embryogenesis (Rodriguez-Serrano et�al. 2012, Berenguer et�al. 2019). The present study has shown activation and involvement in stress-induced microspore death of MCAs, proteases known to crosstalk with autophagy in various cell death pathways (Coll et�al. 2014, Minina et�al. 2014b). Notably, when combined, inhibitors of autophagy and MCA activity did not have an additive effect on cell death suppression in rapeseed microspore cultures suggesting that the two components act epistatically rather than mediating independent pathways. Further work is required to investigate functional relationships between autophagy and MCAs in stress-induced microspore embryogenesis.
Conclusion
Since stress-induced cell death is one of the major factors reducing embryogenesis efficiency, its suppression should lead to the improvement of microspore embryogenesis. The use of small molecules as pharmacological modulators represents a potent future strategy to improve in vitro plant cell viability, reprogramming and development (Hicks and Raikhel 2012, Chuprov-Netochin et�al. 2016, Berenguer et�al. 2017, B�r�ny et�al. 2018, Testillano 2019). Our data demonstrate the involvement of both MCA activity and autophagy in cell death during microspore embryogenesis induction by stress. Accordingly, the application of E-64d and VRPR-FMK, either individually or in combination, yields similar phenotype characterized by reduced cell death and increased embryogenesis initiation rate, leading to mature embryos with high germination frequency, similar to embryos produced under standard in vitro conditions. We thus establish MCAs and autophagy as new biotechnological targets to improve doubled-haploid plant production in breeding and propagation programs of crop plant species. Further work is necessary for mechanistic comprehension of stress-induced cell death pathway in microspore embryogenesis, with an important role in plant biotechnology.
Materials and Methods
Plant growth and microspore culture
Plants of B. napus L. cv. Topas (line DH4079) were used for the study. This cultivar is an embryogenesis-responsive genotype widely used as a model system for fundamental research in microspore embryogenesis (Custers 2003). Plants were grown in a growth chamber (Sanyo MLR-351-H) in pots containing a mixture of organic substrate and vermiculite (2/1, v/v) under long-day photoperiod at 15�C/10�C (16 h light/8 h dark) and 60–70% relative humidity for 4–5 weeks. Continuous flowering was achieved by the regular harvest of open flowers. Young flower buds (3.25–3.35 mm length) containing vacuolated microspores were selected for microspore isolation and culture, which were performed as previously described (Prem et�al. 2012). Stress treatment at 32�C for 6 d was applied to microspore cultures for embryogenesis induction (Prem et�al. 2012). Germination capability of microspore embryos produced in culture was tested in fully developed cotyledonary embryos, which were subjected to in vitro germination conditions, as described previously (Prem et�al. 2012).
Chemical treatments
Autophagy and MCA activity inhibitors were applied, either individually or in combination, at the beginning of microspore cultures, before stress treatment. Stock solutions of 1 mM E-64d (Sigma-Aldrich-Merck KGaA, Darmstadt, Germany) in H2O and 10 mM Z-VRPR-FMK (Bachem, Bubendorf, Switzerland) in dimethyl sulfoxide (DMSO) were used. Corresponding volumes of each stock solution were added to the microspore culture medium to reach the final working concentration of 10 �M E64d and 1 �M Z-VRPR-FMK, keeping DMSO concentration below 0.2% (v/v).
Cell death detection
Microspore cultures were incubated with 0.25% (w/v) Evan’s blue aqueous solution for 45 min at 4�C, as previously described (Berenguer et�al. 2019). Cell death was quantified as the frequency (percentage) of blue-stained cells observed under bright-field in a light microscope Leitz Laborlux 12. Three independent experiments were performed, with at least 1,500 microspores or multicellular proembryos counted in control cultures and cultures treated with E-64d or Z-VRPR-FMK.
Embryogenesis efficiency quantification
Following stress treatment, embryogenesis efficiency was quantified as previously reported (Berenguer et�al. 2017) by counting the number of proembryos formed vs. the total number of microspores and proembryos present in the culture. Random micrographs of microspore cultures were imaged under the stereomicroscope Leica MZ16F and inverted microscope Leica DMI6000B. Three independent experiments were performed, each containing a count of at least 3,000 microspores and multicellular proembryos.
Sample processing for light microscopy
Samples from microspore cultures were collected and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 6.8) overnight at 4�C. Fixed samples were washed with PBS, embedded in gelatin, dehydrated in an acetone series and embedded in Technovit 8100 (KulzerGmbH, Wehrheim, Germany), which was polymerized at 4�C. Semithin sections were obtained with an ultramicrotome (LKB Ultratome III) and placed on either conventional slides for staining with 1% (w/v) toluidine blue or multiwell slides covered with (3-aminopropyl)triethoxysilane (APTES) for immunofluorescence assays.
Immunofluorescence and confocal analysis
Semithin sections on APTES-coated slides were blocked by 5% (w/v) bovine serum albumin (BSA) (Sigma) in PBS for 10 min, washed with PBS and incubated for 1 h with primary rabbit polyclonal anti-ATG5 (B�r�ny et�al. 2018) or anti-ATG8 (Abcam, Cambridge, UK, Cat. ab77003) diluted 1:50 in 1% (w/v) BSA in PBS. After washing with PBS, the signal was revealed with Alexa Fluor 488-labeled anti-rabbit IgG (Molecular Probes, Eugene, OR, USA) diluted 1:25 in 1% (w/v) BSA in PBS for 45 min in darkness. Finally, sections were counterstained with 1 mg/ml 4', 6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) for 10 min, mounted in Mowiol� 4-88 (Sigma-Aldrich-Merck, Darmstad, Germany) and imaged under the confocal microscope (Leica TCS-SP5-AOBS). Laser excitation (488-nm argon laser) and sample emission capture settings (�63 oil immersion objective) were the same in all immunofluorescence preparations of each antibody. Controls were performed by omitting the primary antibody.
Sample processing for electron microscopy
For ultrastructural analyses, samples were processed as previously described (Testillano et�al. 2004). Briefly, the cultures were fixed in Karnovsky fixative (4% formaldehyde + 5% glutaraldehyde in 0.025 M cacodylate buffer, pH 6.7), post-fixed in 2% osmium tetraoxide (OsO4), dehydrated in an ethanol series and embedded in Epon resin. Ultrathin sections were collected on copper grids, counterstained with uranyl acetate and lead citrate and observed under a JEOL 1400 TEM operating at 80 kV.
Sample cryoprocessing for immunogold labeling
Samples were fixed in 4% formaldehyde in PBS, pH 7.3, at 4 �C, overnight. After washing in PBS, samples were dehydrated through a methanol series by a progressive lowering of temperature (B�r�ny et�al. 2010), infiltrated and embedded in Lowicryl K4M acrylic resin (Polysciences, Warrington, PA, USA) at −30�C and polymerized under UV irradiation. Lowicryl ultrathin sections were mounted on nickel grids and used for immunogold labeling.
Immunogold labeling
Immunogold labeling was performed as previously described (B�r�ny et�al. 2010). Nickel grids carrying ultrathin Lowicryl sections were sequentially floated in PBS, 5% BSA in PBS (w/v), and incubated with rabbit polyclonal anti-ATG8 (Abcam, Cambridge, UK, Cat. ab77003), diluted 1:100 (v/v) in 1% BSA in PBS for 1 h. After several washes in PBS, grids were incubated with anti-rabbit IgG conjugated to 10-nm gold particles (Biocell, Cardiff, UK) diluted 1:25 in PBS, for 1 h at room temperature, washed in PBS and water and air-dried. Ultrathin sections were counterstained with uranyl acetate and lead citrate and observed under a JEOL 1400 TEM at 80 kV. Controls were performed excluding the primary antibody.
RT-qPCR analysis
Total RNA was extracted from in vitro samples using the RNeasy� Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction and subsequently treated with Turbo DNase (ThermoFisher, Waltham, MA, USA). The quality and concentration of RNA were analyzed using NanoDrop ND-1000 (ThermoFisher, Waltham, MA, USA). cDNAs were obtained from 1 μg of RNA using the SuperScript™ II Reverse Transcriptase enzyme (Invitrogen Life Technologies, Waltham, MA, USA) and RT-qPCR analyses were performed using the FastStart DNA Green Master (Roche Diagnostics, Basel, Switzerland) on the iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the gene-specific primers listed in Supplementary Table S1. The Arabidopsis HELICASE gene was used as an internal control. qPCR conditions and normalized expression were performed as previously described (P�rez-P�rez et�al. 2019).
MCA activity assay
Cell samples were ground in liquid nitrogen, total proteins were obtained using extraction buffer [50 mM HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.0, 0.1% (v/w) CHAPS, 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate, 5 mM DTT, 1,4-Dithiothreitol, 50 �M PMSF, Phenylmethylsulfonyl fluoride, and 5 �M pepstatin] and centrifuged for 5 min at 20,000 � g at 4�C. The supernatant was filtered through four layers of miracloth (Calbiochem, San Diego, CA, USA) and protein extracts were kept at 4�C. Protein concentration was determined by the Bradford (1976) method using the Bio-Rad Protein Assay (Quick-Start Bradford Dye Reagent; Bio-Rad).
The MCA activity assay was performed as previously described (Minina et�al. 2014c). For the determination of the MCA activity pH dependence profile, the pH values of 50 mM 2-(N-morpholino)ethanesulfonic acid, MES, and 50 mM HEPES buffers were adjusted in the ranges from pH 4.0 to 6.0 and from pH 7.0 to 9.0, respectively. The assay was performed in the presence or absence of 100 mM ethylenediaminetetraacetic acid (EDTA) or one of the inhibitory compounds, including E-64d, leupeptin and Z-VRPR-FMK (each at 10 �M final concentration). Fluorogenic substrate Ac-VRPR-AMC (50 �M final concentration; Bachem) was added to 25–50 �g of total protein. The time-dependent release of free 7-amino-4-methylcoumarin (AMC) was detected at the excitation/emission of 355/460 nm using the plate reader (Omega FLUOstar; BMG Labtech, Ortenberg, Germany) for 1 h at 28�C. A linear part of the reaction curve was determined using Omega MARS software (3.02 R2; BMG Labtech) and converted into picomoles of AMC released per minute per milligram of total protein based on the standard curve of AMC (Bachem). Three independent experiments, each including three technical replicates, were performed.
Immunoblotting
Cell samples were ground in liquid nitrogen, and total proteins were extracted in urea extraction buffer [4 M urea, 1% (v/v) Triton X-100 and 10 mM DTT] and kept on ice for 15 min. After adding 5� Laemmli buffer, protein extracts were mixed and boiled at 100�C for 10 min and centrifuged for 10 min at 20,000 � g and the supernatants were stored at −20�C. Prior to loading on a precast 4–15% gradient Sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS–PAGE, gel (Bio-Rad), the samples were preheated at 100�C for 2 min and quickly spun down in a microfuge. Equal sample volumes were loaded on a polyacrylamide gel, and proteins were separated and transferred onto a polyvinylidene difluoride membrane (Bio-Rad, 1620177). The NBR1 protein was detected using anti-NBR1 1:2,000 (Svenning et�al. 2011) and a-rabbit HRP conjugate 1:25,000 (Agrisera, AS09 602, V�nn�s, Sweden). The signal was developed using the ECL Prime kit (RPN2232, Amersham; GE Healthcare, Little Chalfont, UK). Chemiluminescence was detected using Chemidoc XRS+ (Bio-Rad). After the detection of chemiluminescence, membranes were stained with Ponceau S to visualize the total protein concentration in each sample.
The integrated density values for bands corresponding to NBR1 protein were measured using ImageJ software and normalized to the respective values of total protein for the same sample. The experiment was repeated three times, and statistical analysis to compare the amount of NBR1 in different samples was performed using Origin 2017 software.
Multiple sequence alignment and phylogenetic analysis
The genome resource of B. rapa (version 3.0) was downloaded from the BRAD database (http://brassicadb.org/index.php) (Zhang et�al. 2018). The published A. thaliana p20 and p10 MCA domain sequences were used to query putative MCA homologous proteins using the BlastP tool. Multiple sequence alignment of identified putative MCAs of B. rapa along with the A. thaliana MCAs was carried out using ClustalW with default parameters. Subsequently, the MEGA software (10.1.5 version) was used to construct a maximum-likelihood phylogenetic tree with 1,000 bootstrap replications.
Supplementary Data
Supplementary data are available at PCP online.
Funding
The Spanish National Agency of Research (Agencia Estatal de Investigaci�n, AEI) and European Regional Development Fund (ERDF/FEDER) (grant AGL2017-82447-R to P.S.T.) and the Swedish Foundation for Strategic Research and the research program ‘Crops for the future’ (to P.V.B.).
Supplementary Material
Acknowledgments
The authors thank Transautophagy COST Action CA15138 for the Short Term Scientific Mission grant to E.B. for carrying out a part of the project in E.A.M. and P.V.B. laboratory. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Disclosures
The authors have no conflicts of interest to declare.
References
- Avin-Wittenberg T. (2019) Autophagy and its role in plant abiotic stress management. Plant. Cell Environ. 42: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T., Baluška F., Bozhkov P.V., Elander P.H., Fernie A.R., Galili G., et al. (2018) Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69: 1335–1353. [DOI] [PubMed] [Google Scholar]
- B�r�ny I., Berenguer E., Sol�s M.T., P�rez-P�rez Y., Santamar�a M.E., Crespo J.L., et al. (2018) Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J. Exp. Bot. 69: 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B�r�ny I., Fad�n B., Risue�o M.C., Testillano P.S. (2010) Cell wall components and pectin esterification levels as markers of proliferation and differentiation events during pollen development and pollen embryogenesis in Capsicum annuum L. J. Exp. Bot. 61: 1159–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B�r�ny I., Gonz�lez-Melendi P., Fad�n B., Mityk� J., Risue�o M.C., Testillano P.S. (2005) Microspore-derived embryogenesis in pepper (Capsicum annuum L.): Subcellular rearrangements through development. Biol. Cell 97: 709–722. [DOI] [PubMed] [Google Scholar]
- Berenguer E., B�r�ny I., Sol�s M.T., P�rez-P�rez Y., Risue�o M.C., Testillano P.S. (2017) Inhibition of histone H3K9 methylation by BIX-01294 promotes stress-induced microspore totipotency and enhances embryogenesis initiation. Front. Plant Sci. 8: 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer E., Sol�s M.T., P�rez-P�rez Y., Testillano P.S. (2019) Proteases with caspase 3-like activity participate in cell death during stress-induced microspore embryogenesis of Brassica napus. EuroBiotech J. 3: 152–159. [Google Scholar]
- Bollh�ner B., Jokipii‐Lukkari S., Bygdell J., Stael S., Adriasola M., Mu�iz L., et al. (2018) The function of two type II metacaspases in woody tissues of Populus trees. New Phytol. 217: 1551–1565. [DOI] [PubMed] [Google Scholar]
- Bollh�ner B., Zhang B., Stael S., Denanc� N., Overmyer K., Goffner D., et al. (2013) Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 200: 498–510. [DOI] [PubMed] [Google Scholar]
- Bozhkov P.V., Su�rez M.F., Filonova L.H., Daniel G., Zamyatnin A.A., Rodr�guez-Nieto S., et al. (2005) Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA 102: 14463–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braatz J., Harloff H.J., Mascher M., Stein N., Himmelbach A., Jung C. (2017) CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Chuprov-Netochin R., Neskorodov Y., Marusich E., Mishutkina Y., Volynchuk P., Leonov S., et al. (2016) Novel small molecule modulators of plant growth and development identified by high-content screening with plant pollen. BMC Plant Biol. 16: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Smidler A., Puigvert M., Popa C., Valls M., Dangl J.L. (2014) The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 21: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., et al. (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Custers J.B.M. (2003). Microspore culture in rapeseed (Brassica napus L.) In Doubled Haploid Production in Crop Plants. Edited by Maluszynski M., Kasha J.K., Forster B.P., Szarejko I. pp. 185–193. Kluwer Academic Publishers, Dordrecht. [Google Scholar]
- Custers J.B.M., Cordewener J.H.G., N�llen Y., Dons H.J.M., Campagne M.M.V. (1994) Temperature controls both gametophytic and sporophytic development in microspore cultures of Brassica napus. Plant Cell Reports 13: 267–271. [DOI] [PubMed] [Google Scholar]
- Escamez S., Andr� D., Zhang B., Bollh�ner B., Pesquet E., Tuominen H. (2016) METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biology Open 5: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster B.P., Heberle-Bors E., Kasha K.J., Touraev A. (2007) The resurgence of haploids in higher plants. Trends Plant Sci. 12: 368–375. [DOI] [PubMed] [Google Scholar]
- Fortin J., Lam E. (2018) Domain swap between two type-II metacaspases defines key elements for their biochemical properties. Plant J. 96: 921–936. [DOI] [PubMed] [Google Scholar]
- German� M.A., Lambardi M. (2016). In Vitro Embryogenesis in Higher Plants. Humana Press, New York/Heidelberg. [Google Scholar]
- Hafr�n A., Macia J.L., Love A.J., Milner J.J., Drucker M., Hofius D. (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 114: E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks G.R., Raikhel N.V. (2012) Small molecules present large opportunities in plant biology. Annu. Rev. Plant Biol. 63: 261–282. [DOI] [PubMed] [Google Scholar]
- Huang S., Mira M.M., Stasolla C. (2016) Dying with Style: Death Decision in Plant Embryogenesis. Methods Mol. Biol. 1359: 101–115. [DOI] [PubMed] [Google Scholar]
- Iba�ez S., Carneros E., Testillano P.S., P�rez-P�rez J.M. (2020) Advances in plant regeneration: shake, rattle and roll. Plants 9: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenčič M., Funk C. (2019) Evolution and structural diversity of metacaspases. J. Exp. Bot. 70: 2039–2047. [DOI] [PubMed] [Google Scholar]
- Lema Asqui S., Vercammen D., Serrano I., Valls M., Rivas S., Van Breusegem F., et al. (2018) AtSERPIN1 is an inhibitor of the metacaspase AtMC1-mediated cell death and autocatalytic processing in planta. New Phytol. 218: 1156–1166. [DOI] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Liu S., Snowdon R., Chalhoub B. (2018). The Brassica Napus Genome. Springer International Publishing, New York. [Google Scholar]
- L�pez-Marqu�s R.L., Poulsen L.R., Hanisch S., Meffert K., Buch-Pedersen M.J., Jakobsen M.K., et al. (2010) Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA alpha-subunit. Mol. Biol. Cell 21: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Zhang P., Zhu R., Fu J., Su J., Zheng J., et al. (2017) Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 8: 1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynski M., Kasha K., Forster B., Szarejko I. (eds.) (2003). Doubled Haploid Production in Crop Plants: A Manual. Kluwer, Dordrecht. [Google Scholar]
- Mand�kov� T., Lysak M.A. (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S., Galili G., Genschik P., Fernie A.R., Avin-Wittenberg T. (2016) Autophagy in plants-what's new on the menu? Trends Plant Sci. 21: 134–144. [DOI] [PubMed] [Google Scholar]
- Minina E.A., Bozhkov P.V., Hofius D. (2014. a) Autophagy as initiator or executioner of cell death. Trends Plant Sci. 19: 692–697. [DOI] [PubMed] [Google Scholar]
- Minina E.A., Coll N.S., Tuominen H., Bozhkov P.V. (2017) Metacaspases versus caspases in development and cell fate regulation. Cell Death Differ. 24: 1314–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Moschou P.N., Vetukuri R.R., Sanchez-Vera V., Cardoso C., Liu Q., et al. (2018) Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 69: 1415–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Smertenko A.P., Bozhkov P.V. (2014. b) Vacuolar cell death in plants: metacaspase releases the brakes on autophagy. Autophagy 10: 928–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Staal J., Alvarez V.E., Berges J.A., Berman-Frank I., Beyaert R., et al. (2020) Classification and nomenclature of metacaspases and paracaspasas: no more confusion with caspases. Mol. Cell 77: 927–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E.A., Stael S., Van Breusegem F., Bozhkov P.V. (2014. c) Plant metacaspase activation and activity. Methods Mol. Biol. 1133: 237–253. [DOI] [PubMed] [Google Scholar]
- Murovec J., Bohanec B. (2012) Haploids and doubled haploids in plant breeding. In Plant Breeding. Edited by Abdurakhmonov I.Y.87–106. InTech; Europe, Rijeka, Croatia.�Available from: http://www.intechopen.com/books/plant-breeding/haploids-and-doubled-haploids-in-plant-breeding [Google Scholar]
- Okuzaki A., Ogawa T., Koizuka C., Kaneko K., Inaba M., Imamura J., et al. (2018) CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131: 63–69. [DOI] [PubMed] [Google Scholar]
- Parkin I.A.P., Sharpe A.G., Lydiate D.J. (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303. [DOI] [PubMed] [Google Scholar]
- P�rez-P�rez Y., Carneros E., Berenguer E., Sol�s M.T., B�r�ny I., Pintos B., et al. (2019) Pectin de-methylesterification and AGP increase promote cell wall remodelling and are required during somatic embryogenesis of Quercus suber. Front. Plant Sci. 9: 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L.R., Lopez-Marques R.L., McDowell S.C., Okkeri J., Licht D., Schulz A., Pomorski T., et al. (2008) The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20: 658–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prem D., Sol�s M.T., B�r�ny I., Rodr�guez-Sanz H., Risue�o M.C., Testillano P.S. (2012) A new microspore embryogenesis system under low temperature which mimics zygotic embryogenesis initials, expresses auxin and efficiently regenerates doubled-haploid plants in Brassica napus. BMC Plant Biol. 12: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Serrano M., Barany I., Prem D., Coronado M.-J., Risueno M.C., Testillano P.S. (2012) NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 63: 2007–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero-Linares J., Coll N.S. (2019) Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 70: 2087–2095. [DOI] [PubMed] [Google Scholar]
- Satpute G.K., Long H., Segu�-Simarro J.M., Risue�o M.C., Testillano P.S. (2005) Cell architecture during gametophytic and embryogenic microspore development in Brassica napus L. Acta Physiol. Plant. 27: 665–674. [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. [DOI] [PubMed] [Google Scholar]
- Su�rez M.F., Filonova L.H., Smertenko A., Savenkov E.I., Clapham D.H., von Arnold S., et al. (2004) Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 14: R339–R340. [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Bassham D.C. (2018) Autophagy in crop plants: what's new beyond Arabidopsis? Open Biol. 8: 180162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testillano P., Georgiev S., Mogensen H.L., Coronado M.J., Dumas C., Risueno M.C., et al. (2004) Spontaneous chromosome doubling results from nuclear fusion during in vitro maize induced microspore embryogenesis. Chromosoma 112: 342–349. [DOI] [PubMed] [Google Scholar]
- Testillano P.S. (2019) Microspore embryogenesis: targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 70: 2965–2978. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Timmerman E., De Bock P.-J., Vercammen D., Stael S., van de Cotte B., et al. (2013) The Arabidopsis metacaspase9 degradome. Plant Cell 25: 2831–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P.V. (2011) Metacaspases. Cell Death Differ. 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A.G., O'Rourke K., Aravind L., Pisabarro M.T., Seshagiri S., Koonin E.V., et al. (2000) Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6: 961–967. [DOI] [PubMed] [Google Scholar]
- �st�n S., Hafr�n A., Hofius D. (2017) Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40: 122–130. [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., Beers E.P., Dangl J.L., Franklin-Tong V.E., Gallois P., Hara-Nishimura I., et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ. 18: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D., van de Cotte B., De Jaeger G., Eeckhout D., Casteels P., Vandepoele K., et al. (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279: 45329–45336. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2011) Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J. Biol. Chem. 286: 10027–10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Bassham D.C. (2015) New insight into the mechanism and function of autophagy in plant cells. Int. Rev. Cell Mol. Biol. 320: 1–40. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Ohsumi Y. (2018) Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59: 1337–1344. [DOI] [PubMed] [Google Scholar]
- Zhang L., Cai X., Wu J., Liu M., Grob S., Cheng F., et al. (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Adamowski M., Marhava P., Tan S., Zhang Y., Rodriguez L., et al. (2020) Arabidopsis flippases cooperate with ARF GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. Plant Cell 32: 1644–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., et al. (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Yu X.H., Wang C., Zhang Q., Liu W., McSweeney S., et al. (2020) Structural basis for Ca2+-dependent activation of a plant metacaspase. Nat. Commun. 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.