Abstract
INTRODUCTION:
Epidemiological studies estimate that having a first-degree relative (FDR) with colorectal cancer (CRC) increases 2-fold to 3-fold the risk of developing the disease. Because FDRs of CRC patients are more likely to co-inherit CRC risk variants, we aimed to evaluate potential differences in genotype distribution of single nucleotide polymorphisms (SNPs) related to CRC risk between FDRs of patients with nonsyndromic CRC (cases) and individuals with no family history of CRC (controls).
METHODS:
We designed a case-control study comprising 750 cases and 750 Spanish Caucasian controls matched by sex, age, and histological findings after colonoscopy. Genomic DNA from all participants was genotyped for 88 SNPs associated with CRC risk using the MassArray (Sequenom) platform.
RESULTS:
Ten of the 88 SNPs analyzed revealed significant associations (P < 0.05) with a family history of CRC in our population. The most robust associations were found for the rs17094983G>A SNP in the long noncoding RNA LINC01500 (odds ratio = 0.72; 95% confidence interval: 0.58–0.88, log-additive model), and the rs11255841T>A SNP in the long noncoding RNA LINC00709 (odds ratio = 2.04; 95% confidence interval: 1.19–3.51, dominant model). Of interest, the observed associations were in the same direction than those reported for CRC risk.
DISCUSSION:
FDRs of CRC patients show significant differences in genotype distribution of SNPs related to CRC risk as compared to individuals with no family history of CRC. Genotyping of CRC risk variants in FDRs of CRC patients may help to identify subjects at risk that would benefit from stricter surveillance and CRC screening programs.
INTRODUCTION
Colorectal cancer (CRC) is a multifactorial disease resulting from complex interactions between environmental and genetic factors. Although most cases of CRC correspond to sporadic forms, approximately 25%–30% of all cases occur in patients with a family history of CRC. Only a small fraction (2%–8%) of those CRC arises in the setting of the highly penetrant inherited syndromes due to germline mutations in well-known genes. The main subtypes of hereditary CRC are hereditary nonpolyposis CRC and familial adenomatous polyposis. Aside from these, there is a significant number of CRCs in which a strong family aggregation is observed but do not show a defined Mendelian inheritance pattern. It is termed familial CRC to distinguish it from the above well-established hereditary syndromes (1).
Over the past 2 decades, genome-wide association studies (GWAS) (2–17) and candidate gene analysis (18,19) have identified multiple gene variants, mainly single nucleotide polymorphisms (SNPs), associated with susceptibility to CRC. The risk conferred by each of these variants located in low penetrance genes is usually modest. However, the combination of several risk variants in a polygenic model has been reported to increase the risk of CRC in an additive or exponential way. Despite the large number of association studies, very few have point out the relevance of gene polymorphisms in the so-called nonsyndromic familial CRC.
From a clinical point of view, the risk of developing familial CRC depends mainly on 3 factors: (i) the number of affected relatives, (ii) the degree of relationship, and (iii) the age at diagnosis of CRC in the affected family member (1). Epidemiological studies estimate that having a first-degree relative (FDR) with CRC increases 2-fold to 3-fold the lifetime risk of developing CRC (20–22). Taking into account that FDRs share the genome with a CRC patient in the same family (parents, offspring, and siblings), it is rational to think that FDRs of patients with CRC would have a higher probability of presenting a co-inheritance of multiple common risk variants in low penetrance genes that would provide them a greater risk of developing CRC and precancerous lesions as compared to individuals with no family history of CRC.
Trying to address this issue, we design a case-control study to evaluate potential differences in the distribution of genotypes and allele frequencies of a panel of selected SNPs related to CRC and adenoma risk between FDRs of patients with CRC (cases) and individuals with no family history of CRC (controls).
PATIENTS AND METHODS
Patients
The study design and data collection methods have been described in detail previously (23). Briefly, this is a case–control study simultaneously conducted in 2 general hospitals integrated into the Spanish National Health System. A total of 1,500 subjects (750 cases and 750 controls) were recruited at the University Hospital Lozano Blesa of Zaragoza and the University Hospital of the Canary Islands in Tenerife from May 2010 to May 2014.
Cases were selected from the CRC screening programs in Zaragoza and Tenerife and comprised 750 Spanish White FDRs of patients with nonsyndromic CRC. The control group consisted of 750 individuals with no family history of CRC recruited from those patients who were scheduled for colonoscopy at hospitals either by symptoms or by CRC screening in the average-risk population. Cases and controls were matched by sex, age (±5 years), and histological lesions found during colonoscopy. Patients were stratified in 3 groups based on the endoscopic findings and pathology review: (i) patients with no lesions or with no neoplastic lesions; (ii) patients with low-risk adenomas (LRAs), defined as <3 nonadvanced adenomas; and (iii) patients with high-risk adenomas (HRAs), defined as advanced adenomas or ≥3 non-advanced adenomas. Adenomas were classified as advanced if they were ≥10 mm in size or/and had ≥20% villous components or high-grade dysplasia. Designation as LRA or HRA was based on the likelihood of developing advanced neoplasia during surveillance after polypectomy as recommended by the American and European Societies of Gastrointestinal Endoscopy (24,25).
Information concerning demographic characteristics and potential risk factors including family history of CRC (any reported CRC in FDRs or 2 or more CRC cases in second-degree relatives [SDRs]), smoking habit, alcohol consumption, and chronic use of nonsteroidal anti-inflammatory drugs (NSAIDs) or acetylsalicylic acid (ASA) was obtained using a questionnaire administered by trained personnel as previously described (23).
After completion of the interview, 10 mL of peripheral blood from each patient was collected into Ethylenediaminetetraacetic acid tubes for subsequent DNA extraction. Once processed, whole blood samples were aliquoted and stored at −80 °C until use. All subjects gave written informed consent to the study, which was conducted in accordance with the Ethical Committee of the participating Hospitals.
SNP selection and genotyping methods
Genomic DNA was extracted from Ethylenediaminetetraacetic acid-preserved whole blood using the automated DNA isolation system AutoGenFlex 3000. DNA samples were aliquoted and stored at 4 °C until analysis.
The panel of 99 SNPs included in our study was selected from the NCBI data base (http://www.ncbi.nlm.nih.gov/snp) and the NHGRI-EBI GWAS Catalog (http://www.ebi.ac.uk/gwas) based on 3 main criteria: (i) published evidence of an association with CRC or adenoma risk by GWAS of candidate gene studies, (ii) having reported a minor allele frequency ≥ 1% in Whites, or (iii) having potential functional consequences leading to altered protein concentrations or protein functions. Genotyping was performed at the Spanish National Genotyping Centre (CEGEN-Santiago de Compostela) using the Sequenom MassARRAY iPLEX platform. For quality control, intraplate and interplate duplicates were included. Genotype concordance was 100% for all samples. The post-genotype quality control comprised the exclusion of 11 SNPs due to failure of genotyping (rs11632715, rs17730929, PTGS1 rs3842787, and PNMAL1 rs7248888), a call rate < 95% (TPH2 rs10879357, MYRF rs174537, PTGS2 rs20417, ERCC2 rs1799793, and HADC9 rs1919314), or lack of Hardy-Weinberg equilibrium in controls (Fisher test P < 10−4, rs11671104, rs2965667). Finally, 88 SNPs (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A497) were successfully genotyped in our population and remained available for statistical analysis.
Statistical analysis
Continuous variables were expressed as means with SD, whereas qualitative variables were expressed as frequencies and percentages. The relationship between qualitative variables was evaluated by contingency tables with χ2 tests.
Concerning gene polymorphisms, genotype frequencies for each SNP in controls were tested for Hardy-Weinberg equilibrium by a χ2 test with one degree of freedom (df). Genotype and allele frequencies between cases and controls were compared using the χ2 test with Yates' correction or Fisher exact tests. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for each genetic variant using the SNPassoc package implemented in R 3.2.2. Univariate and logistic regression analyses were evaluated under codominant, dominant, recessive, overdominant, and log-additive genetic models. The best-fitting genetic model was selected using the Akaike information criteria. For all tests, a 2-sided P value < 0.05 was considered statistically significant. To address the issue of conducting multiple comparisons, the False Discovery Rate method was applied. The statistical analysis was performed using the SPSS software v 22.0 for Windows (SPSS Ibérica, Madrid, Spain).
Based on the frequencies of the analyzed SNPs in our population, the size of the study was sufficient to detect ORs > 1.41 or <0.73 with a power of 80% and a α value of 0.05. For the less prevalent polymorphisms (minor allele frequency: 0.01–0.10), the study had a power of 80% to detect ORs > 4.85 in the whole data set. Power calculations were estimated using the program Epidat 4.1.
RESULTS
Clinical and demographic characteristics of patients
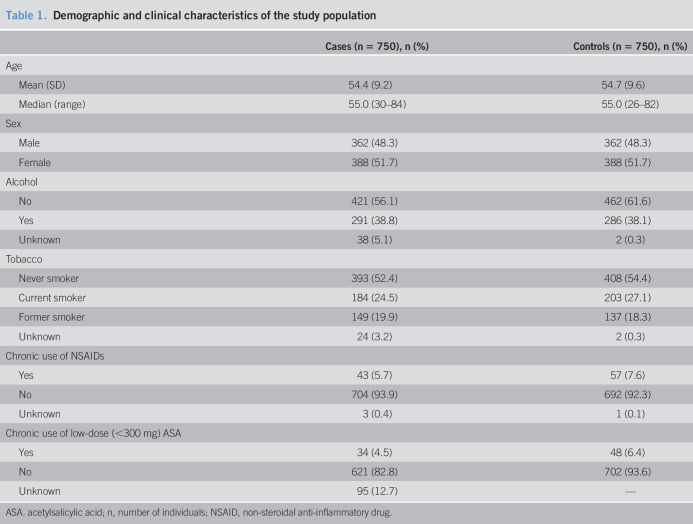
Table 1 summarizes the demographic and clinical characteristics of cases (FDRs of patients with CRC) and controls (individuals with no family history of CRC). No significant differences between cases and controls were observed regarding consumption of tobacco, alcohol, and chronic use of NSAIDs or low-dose ASA.
Table 1.
Demographic and clinical characteristics of the study population
| Cases (n = 750), n (%) | Controls (n = 750), n (%) | |
| Age | ||
| Mean (SD) | 54.4 (9.2) | 54.7 (9.6) |
| Median (range) | 55.0 (30–84) | 55.0 (26–82) |
| Sex | ||
| Male | 362 (48.3) | 362 (48.3) |
| Female | 388 (51.7) | 388 (51.7) |
| Alcohol | ||
| No | 421 (56.1) | 462 (61.6) |
| Yes | 291 (38.8) | 286 (38.1) |
| Unknown | 38 (5.1) | 2 (0.3) |
| Tobacco | ||
| Never smoker | 393 (52.4) | 408 (54.4) |
| Current smoker | 184 (24.5) | 203 (27.1) |
| Former smoker | 149 (19.9) | 137 (18.3) |
| Unknown | 24 (3.2) | 2 (0.3) |
| Chronic use of NSAIDs | ||
| Yes | 43 (5.7) | 57 (7.6) |
| No | 704 (93.9) | 692 (92.3) |
| Unknown | 3 (0.4) | 1 (0.1) |
| Chronic use of low-dose (<300 mg) ASA | ||
| Yes | 34 (4.5) | 48 (6.4) |
| No | 621 (82.8) | 702 (93.6) |
| Unknown | 95 (12.7) | — |
ASA. acetylsalicylic acid; n, number of individuals; NSAID, non-steroidal anti-inflammatory drug.
In relation to the endoscopic findings, 57% of the patients (429 cases and 429 controls) showed no neoplastic lesions, 288 patients (144 cases and 144 controls) had LRA, and 354 patients (177 cases and 177 controls) had HRA.
Family history of CRC
In our study, subjects referred as cases (n = 750) had at least one FDR affected with CRC. The distribution of cases according to the number of affected FDRs and the age at diagnosis of CRC in the affected relative is shown in Figure 1. Most cases (89.3%) had one FDR with CRC, 10.5% had 2 FDR with CRC, and 20.1% of cases had both, FDRs and SDRs with CRC. Parents were the most often affected FDRs (68.8%), followed by siblings (30.5%) and children (0.6%). Concerning SDRs, uncles were the most often affected (54.9%), followed by grandparents (21.5%) and cousins (11.5%).
Figure 1.
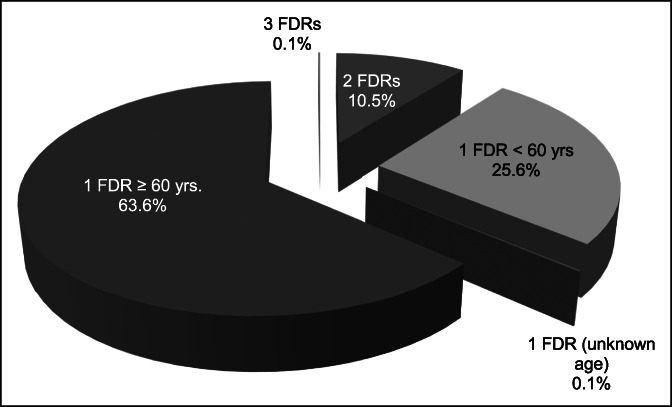
Distribution of cases according to the number of FDRs affected and the age at diagnosis of colorectal cancer (CRC). Most cases had one FDR with CRC diagnosed younger than or equal to 60 years (63.6%, 477/750) or younger than 60 years (25.6%, 192/750). Seventy-nine cases (10.5%) had 2 FDRs with CRC, and only one case had 3 FDRs with CRC (0.13%). In 20.1% of cases, patients had both, FDRs and second-degree relatives with CRC. Mean age at diagnosis of CRC in FDRs was 66 ± 12.6 years. Age at diagnosis was <60 years in nearly 30% of index cases (patients with CRC). FDR, first-degree relative.
When patients were stratified according to histological findings, we found that cases with 2 FDRs with CRC were significantly more frequent in the group of patients with adenomas (HRA or LRA) than that in the group of patients with non-neoplastic lesions (14.3% vs 7.9%, OR = 1.9; 95% CI: 1.2–3.1, P = 0.005).
Gene polymorphisms and family history of CRC
A total of 1,500 subjects (750 cases and 750 controls) were successfully genotyped for a panel of 88 SNPs previously related to CRC risk. Genotype frequencies did not deviate significantly from Hardy-Weinberg equilibrium in the control group (Fisher test, P > 10−4) (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A497).
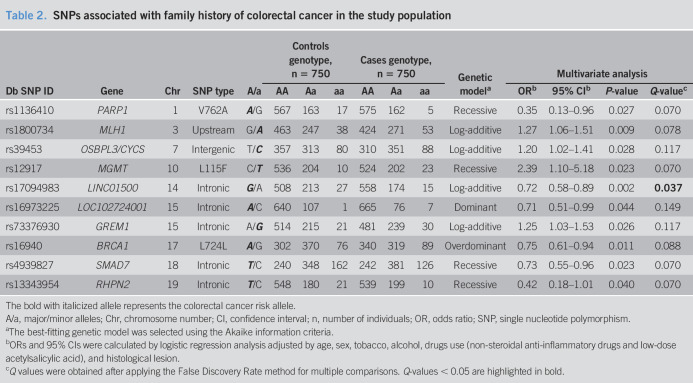
In our population, 10 SNPs (rs1136410, rs12917, rs13343954, rs16940, rs16973225, rs17094983, rs1800734, rs39453, rs4939827, and rs73376930) revealed significant associations (P < 0.05) with a family history of CRC in at least one of the 5 genetic models evaluated in the analysis (Table 2). After False Discovery Rate multiple test correction, only the rs17094983 SNP in the long noncoding RNA (lncRNA) LINCO1500 gene retained statistical significance. Thus, the frequency of the rs17094983 minor allele A was significantly lower in FDRs of CRC patients than that in controls (log-additive model, OR = 0.72; 95% CI: 0.58–0.89, Q value = 0.037) (Table 2).
Table 2.
SNPs associated with family history of colorectal cancer in the study population
| Db SNP ID | Gene | Chr | SNP type | A/a | Controls genotype, n = 750 | Cases genotype, n = 750 | Genetic modela | Multivariate analysis | |||||||
| AA | Aa | aa | AA | Aa | aa | ORb | 95% CIb | P-value | Q-valuec | ||||||
| rs1136410 | PARP1 | 1 | V762A | A/G | 567 | 163 | 17 | 575 | 162 | 5 | Recessive | 0.35 | 0.13–0.96 | 0.027 | 0.070 |
| rs1800734 | MLH1 | 3 | Upstream | G/A | 463 | 247 | 38 | 424 | 271 | 53 | Log-additive | 1.27 | 1.06–1.51 | 0.009 | 0.078 |
| rs39453 | OSBPL3/CYCS | 7 | Intergenic | T/C | 357 | 313 | 80 | 310 | 351 | 88 | Log-additive | 1.20 | 1.02–1.41 | 0.028 | 0.117 |
| rs12917 | MGMT | 10 | L115F | C/T | 536 | 204 | 10 | 524 | 202 | 23 | Recessive | 2.39 | 1.10–5.18 | 0.023 | 0.070 |
| rs17094983 | LINC01500 | 14 | Intronic | G/A | 508 | 213 | 27 | 558 | 174 | 15 | Log-additive | 0.72 | 0.58–0.89 | 0.002 | 0.037 |
| rs16973225 | LOC102724001 | 15 | Intronic | A/C | 640 | 107 | 1 | 665 | 76 | 7 | Dominant | 0.71 | 0.51–0.99 | 0.044 | 0.149 |
| rs73376930 | GREM1 | 15 | Intronic | A/G | 514 | 215 | 21 | 481 | 239 | 30 | Log-additive | 1.25 | 1.03–1.53 | 0.026 | 0.117 |
| rs16940 | BRCA1 | 17 | L724L | A/G | 302 | 370 | 76 | 340 | 319 | 89 | Overdominant | 0.75 | 0.61–0.94 | 0.011 | 0.088 |
| rs4939827 | SMAD7 | 18 | Intronic | T/C | 240 | 348 | 162 | 242 | 381 | 126 | Recessive | 0.73 | 0.55–0.96 | 0.023 | 0.070 |
| rs13343954 | RHPN2 | 19 | Intronic | T/C | 548 | 180 | 21 | 539 | 199 | 10 | Recessive | 0.42 | 0.18–1.01 | 0.040 | 0.070 |
The bold with italicized allele represents the colorectal cancer risk allele.
A/a, major/minor alleles; Chr, chromosome number; CI, confidence interval; n, number of individuals; OR, odds ratio; SNP, single nucleotide polymorphism.
The best-fitting genetic model was selected using the Akaike information criteria.
ORs and 95% CIs were calculated by logistic regression analysis adjusted by age, sex, tobacco, alcohol, drugs use (non-steroidal anti-inflammatory drugs and low-dose acetylsalicylic acid), and histological lesion.
Q values were obtained after applying the False Discovery Rate method for multiple comparisons. Q-values < 0.05 are highlighted in bold.
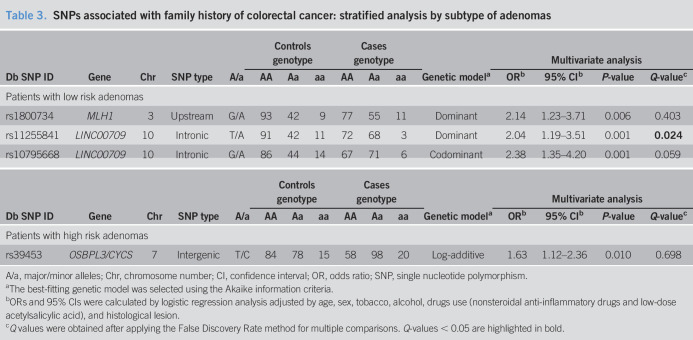
Stratified analysis by histological type of adenoma (LRA/HRA) showed additional differences in genotype distribution and allele frequencies between cases and controls. Table 3 summarizes those SNPs significantly associated with family history of CRC in the multivariate analysis adjusted by age, sex, tobacco, alcohol, and drug use (NSAIDs and low-dose ASA). Three SNPs located in the lncRNA LINC00709 (rs11255841T>A, rs10795668G>A) and MLH1 (rs1800734G>A) genes were significantly associated with susceptibility to family history of CRC in the group of patients with LRAs. Similarly, the rs39453T variant was found to be more frequent in FDRs of CRC patients than that in controls in the subgroup of patients with HRA (log-additive model, OR = 1.63; 95% CI: 1.12–2.36). However, after applying the False Discovery Rate method, only the rs11255841T>A in the lncRNA LINC00709 retained significance (Table 3). Carriers of the rs11255841A variant were significantly more frequent in cases than that in controls in the group of patients with LRA (49.7% vs 36.8%; dominant model, OR = 2.04; 95% CI: 1.19–3.51).
Table 3.
SNPs associated with family history of colorectal cancer: stratified analysis by subtype of adenomas
| Db SNP ID | Gene | Chr | SNP type | A/a | Controls genotype | Cases genotype | Genetic modela | Multivariate analysis | |||||||
| AA | Aa | aa | AA | Aa | aa | ORb | 95% CIb | P-value | Q-valuec | ||||||
| Patients with low risk adenomas | |||||||||||||||
| rs1800734 | MLH1 | 3 | Upstream | G/A | 93 | 42 | 9 | 77 | 55 | 11 | Dominant | 2.14 | 1.23–3.71 | 0.006 | 0.403 |
| rs11255841 | LINC00709 | 10 | Intronic | T/A | 91 | 42 | 11 | 72 | 68 | 3 | Dominant | 2.04 | 1.19–3.51 | 0.001 | 0.024 |
| rs10795668 | LINC00709 | 10 | Intronic | G/A | 86 | 44 | 14 | 67 | 71 | 6 | Codominant | 2.38 | 1.35–4.20 | 0.001 | 0.059 |
| Db SNP ID | Gene | Chr | SNP type | A/a | Controls genotype | Cases genotype | Genetic modela | Multivariate analysis | |||||||
| AA | Aa | aa | AA | Aa | aa | ORb | 95% CIb | P-value | Q-valuec | ||||||
| Patients with high risk adenomas | |||||||||||||||
| rs39453 | OSBPL3/CYCS | 7 | Intergenic | T/C | 84 | 78 | 15 | 58 | 98 | 20 | Log-additive | 1.63 | 1.12–2.36 | 0.010 | 0.698 |
A/a, major/minor alleles; Chr, chromosome number; CI, confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
The best-fitting genetic model was selected using the Akaike information criteria.
ORs and 95% CIs were calculated by logistic regression analysis adjusted by age, sex, tobacco, alcohol, drugs use (nonsteroidal anti-inflammatory drugs and low-dose acetylsalicylic acid), and histological lesion.
Q values were obtained after applying the False Discovery Rate method for multiple comparisons. Q-values < 0.05 are highlighted in bold.
DISCUSSION
Epidemiological studies estimate that having a FDR with CRC increases 2-fold to 3-fold the risk of developing CRC (20–22). Because FDRs share the genome and the genetic risk background of their affected relative (parents, offspring, and siblings), it is rational to think that FDRs of patients with CRC have a higher probability of presenting a co-inheritance of multiple risk variants that would provide them a greater risk of developing CRC as compared to subjects with no family history of CRC. Based on this hypothesis, we designed a case–control study to assess potential differences in genotype and allele distribution of 88 SNPs related to CRC risk between FDRs of patients with CRC (cases) and individuals with no family history of CRC (controls). The most robust association in our study was observed for the intronic variant rs17094983G>A (LINC01500) located in the 14q23.1 chromosomal region. This SNP was firstly reported as associated with CRC risk in a GWAS performed in 2013 by Peters et al. (11). According to the authors, the minor allele (A) of rs17094983 was associated with a lower risk of developing CRC (P < 3 × 10E−6), although this association did not reach genome-wide significance levels. A subsequent GWAS by Lemire et al. (26) corroborated the protective effect of the rs17094983A variant in the development of CRC in both, African and White populations (P = 2.5 × 10−10), reaching in the later, significance levels required in GWAS studies. Unlike the study of Peters, Lemire et al. (26) did not include as controls individuals having a family history of CRC because they considered FDRs of patients with CRC as an intermediate step between CRC and healthy subjects with no family history of CRC. In line with their findings, the frequency of the rs17094983 allele A in our study was significantly lower in FDRs of patients with CRC than that in controls (log-additive model, OR = 0.72; 95% CI: 0.58–0.89, Q value = 0.037). Based on these results, we could speculate that the rs17094983A variant, identified in GWAS as a protective factor of CRC, would be in turn inversely associated with the risk of having a positive family history of CRC. This hypothesis is also biologically plausible in the light of the putative functional effect of the rs17094983 SNP. Some studies have reported the link between rs17094983 genotypes and variations in the expression of the RTN1 (Reticulon 1) gene (26). The RTN1 gene encodes the synthesis of 3 proteins (RTN1-A, RTN1-B, and RTN1-C), localized in the endoplasmic reticulum membrane, that play a dominant regulatory role in the control of apoptosis and cell proliferation (27). It has been shown that individuals heterozygous for the rs17094971A>T variant in strong linkage disequilibrium with rs17094983 (r2 = 0.81) show higher RTN1 expression levels in colon tumors than common risk homozygous rs17094971AA subjects. This association is consistent with the minor rs17094983 A allele being inversely associated with CRC risk because reduction in the expression of RTN1 gene in patients carrying risk alleles would favor cell proliferation and survival of tumor colorectal cells. Further studies are required to conclusively assess the functional consequences of the rs17094983 polymorphism and its relevance in the susceptibility to the risk of family history of CRC.
Stratified analysis by histological lesions showed some additional differences in genotype distribution and allele frequencies between cases and controls. The most remarkable association was observed for the rs11255841T>A variant. Thus, individuals carrying the reported CRC risk rs11255841 allele A were significantly more frequent in FDRs of patients with CRC than that in controls among subjects with low-risk adenomas (dominant model, OR = 2.04; 95% CI: 1.19–3.51). The rs11255841T>A intergenic variant is located in the LINC00709 gene at the 10p14 chromosomal region. The LINC00709 gene belongs, such as LINC01500, to the new category of lncRNAs. Rather than to be transcriptional noise, recent studies suggest that lncRNAs are important players in cancer. It has been shown that lncRNAs regulate multiple functions in carcinogenesis including cell cycle, cell proliferation, and apoptosis through controlling gene transcription and posttranscriptional processing (28). Moreover, it has been observed that SNPs located in lncRNAs may influence gene expression through long-range cis-regulatory elements (29). In our population, the rs11255841T>A variant was in strong linkage disequilibrium (D′ = 0.96, r2 = 0.84) with the rs10795668G>A SNP, previously identified as CRC risk factor by Tomlinson et al. (5). Functionally, both SNPs are located near to the DD431424 and HV455515 genes, which are important regulators of the hTERT region that harbor several susceptibility loci for various types of cancers, including CRC (30).
In our study, only 2 of the 88 SNPs analyzed were significantly associated with family history of CRC after False Discovery Rate test correction (rs17094983G>A in LINC01500, and rs11255841T>A in LINC00709). However, we consider that some SNPs (Table 2) merit additional follow-up evaluation for risk of family history of CRC because Q values were close to required significance levels. Taking into account the prevalence of the SNPs evaluated in our population, the study has a power of 85% to detect ORs > 1.41 or <0.73. As a result, it is possible that we could have missed minor statistical differences, mainly in low-frequency variant polymorphisms and stratified analysis. Studies with larger populations and different ethnic groups are warranted to elucidate the contribution of genetic susceptibility on the risk of family history of CRC.
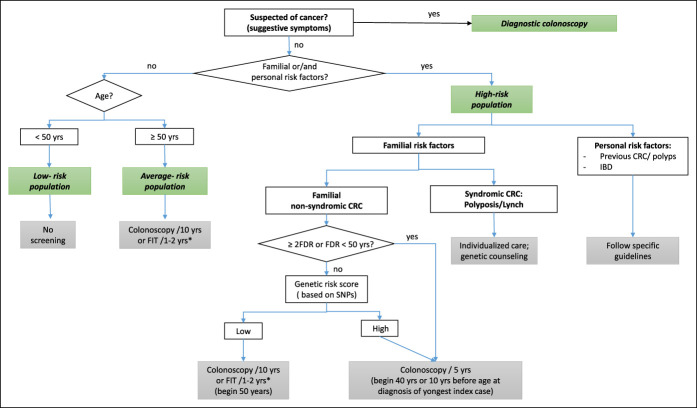
In summary, we found that some polymorphisms previously related to CRC risk (rs17094983 and rs11255841) show significant differences in genotype distribution and allele frequencies between FDRs of patients with CRC and individuals with no family history of CRC. Of interest, the observed associations were in the same direction than those reported for CRC risk. Our results suggest that FDRs of CRC patients carry risk variants that would provide them a greater susceptible to developing the disease as compared to individuals with no family history of CRC. A deeper knowledge of genetic factors related to familiar CRC may have significant implications for the identification of those FDRs at risk of CRC that would benefit from stricter surveillance and cancer screening programs (Figure 2).
Figure 2.
Proposal of colorectal cancer screening algorithm. *Positive results on FIT should be followed up with timely colonoscopy. CRC, colorectal cancer; FDR, first-degree relative; FIT, fecal immunochemical test; IBD, inflammatory bowel disease; SNP, single nucleotide polymorphism.
CONFLICTS OF INTEREST
Guarantor of the article: Carla J. Gargallo-Puyuelo, MD, PhD.
Specific author contributions: C.J.G.-P.: study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content. A.L.: study concept and design, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. P.C.-L.: statistical analysis. A.F.: study concept and design. E.Q., M.C., and I.A.-A.: acquisition of data. M.A.G.-G.: study concept and design, analysis and interpretation of data, drafting of the manuscript, and critical revision of the manuscript for important intellectual content. All authors approved the final draft submitted.
Financial support: (i) Research Grant of the Aragonese Society of Digestive Pathology, (ii) Research Grant of the Spanish Gastroenterology Association (AEG), (iii) IIS Aragón, (iv) Gobierno de Aragón. Grupo investigación DGA, (v) CIBERehd, and (vi) Institute of Health Carlos III (ISCIII). The genotyping service was carried out at CEGEN-PRB3-ISCIII; it is supported by grant PT17/0019, of the PE I+D+i 2013-2016, funded by ISCIII and ERDF. The work was independent of all these funding sources.
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ FDRs of patients with CRC have an increased risk of developing CRC and adenomas.
✓ GWAS studies have identified over 80 gene polymorphisms associated with CRC risk.
✓ The prevalence of CRC gene variants in FDRs of CRC patients as compared to individuals with no family history of CRC has been scarcely analyzed.
WHAT IS NEW HERE
✓ The protective rs17094983 variant (LINC01500) on the CRC risk was significantly less frequent in FDRs of CRC patients than that in individuals with no family history of CRC.
✓ The rs11255841 variant in the lncRNA LINC00709, associated with a higher CRC risk, was significantly more frequent in FDRs of patients with CRC among subjects with low-risk adenomas.
✓ The observed associations were in the same direction than those reported for CRC risk.
TRANSLATIONAL IMPACT
✓ FDRs of CRC patients have a higher probability of presenting a co-inheritance of multiple risk variants that would provide them a greater risk of developing CRC as compared to individuals with no family history of CRC.
✓ Genotyping of CRC risk variants in FDRs of CRC patients may have significant implications for the identification of FDRs at risk that would benefit from stricter surveillance and CRC screening programs.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A497
Contributor Information
Carla J. Gargallo-Puyuelo, Email: carlajerusalen@hotmail.com.
Ángel Lanas, Email: angel.lanas@gmail.com.
Patricia Carrera-Lasfuentes, Email: pcarreralasfuentes@gmail.com.
Ángel Ferrández, Email: angel.ferrandez@telefonica.net.
Enrique Quintero, Email: equinter@gmail.com.
Marta Carrillo, Email: martacarry@yahoo.es.
Inmaculada Alonso-Abreu, Email: macuaa@hotmail.com.
REFERENCES
- 1.Cubiella J, Marzo-Castillejo M, Mascort-Roca JJ, et al. Clinical practice guideline. Diagnosis and prevention of colorectal cancer. 2018 update. Gastroenterol Hepatol 2018;41:585–96. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson I, Wevv E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007;39:984–8. [DOI] [PubMed] [Google Scholar]
- 3.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007;239:989–94. [DOI] [PubMed] [Google Scholar]
- 4.Broderick P, Carvajal-Carmona L, Pitmann AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet 2007;39:1315–7. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 2008;40:623–30. [DOI] [PubMed] [Google Scholar]
- 6.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 2008;40:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COGENT Study, Houlston RS, Webb E, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 2008;40:1426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 2010;42:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui R, Okada Y, Jang SG, et al. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut 2011;60:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters U, Hutter CM, HSU L, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet 2012;131:217–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters U, Jiao S, Schumacher FR, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology 2013;144:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao S, Hsu L, Berndt S, et al. Genome-wide search for gene-gene interactions in colorectal cancer. PLoS One 2012;7:e52535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Rozadilla C, Cazier JB, Tomlinson IP, et al. A colorectal cancer genome-wide association study in a Spanish cohort identifies two variants associated with colorectal cancer risk at 1p33 and 8p12. BMC Genomics 2013;14:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Jia WH, Matsuo K, et al. Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in East Asians. Int J Cancer 2014;135:948–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiffin N, Hosking FJ, Farrington S, et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet 2014;23:4729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Tassan NA, Whiffin N, Hosking FJ, et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep 2015;5:10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher FR, Schmit SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun 2015;6:7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theodoratou E, Montazeri Z, Hawken S, et al. Systematic meta-analyses and field synopsis of genetic association studies in colorectal cáncer. J Natl Cancer Inst 2012;104:1433–57. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Zhang B, Zheng W. Genetic variants associated with colorectal cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut 2014;63:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baglietto L, Jenkins MA, Severi G, et al. Measures of familial aggregation depend on the definition of family history: Meta-analysis for colorectal cancer. J Clin Epidemiol 2006;59:114–24. [DOI] [PubMed] [Google Scholar]
- 21.Butterworth AS, Higgins JPT, Pharoah P. Relative and absolute risk of colorectal cancer for individuals with a family history: A meta-analysis. Eur J Cancer 2006;42:216–27. [DOI] [PubMed] [Google Scholar]
- 22.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol 2001;96:2992–3003. [DOI] [PubMed] [Google Scholar]
- 23.Gargallo CJ, Lanas A, Carrera-Lasfuentes P, et al. Genetic susceptibility in the development of colorectal adenomas according to family history of colorectal cancer. Int J Cancer 2019;144:489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassan C, Quintero E, Dumonceau JM, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy 2013;45:842–51. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman DA, Rex DK, Winawer SJ, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: A consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012;143:844–57. [DOI] [PubMed] [Google Scholar]
- 26.Lemire M, Qu C, Loo LWM, et al. A genome-wide association study for colorectal cancer identifies a risk locus in 14q23.1. Hum Genet 2015;134:1249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Sano F, Fazi B, Tufi R, et al. Reticulon-1C acts as a molecular switch between endoplasmic reticulum stress and genotoxic cell death pathway in human neuroblastoma cells. J Neurochem 2007;102:345–53. [DOI] [PubMed] [Google Scholar]
- 28.Silva A, Bullock M, Calin G. The clinical relevance of long non-coding RNAs. Cancers (Basel) 2015;7:2169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 2020;21:102–7. [DOI] [PubMed] [Google Scholar]
- 30.Pellatt AJ, Wolff Roger K, Herrick J, et al. TERT's Role in colorectal carcinogenesis. Mol Carcinog 2013;52:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.