Abstract
Despite advances in transplant immunosuppression, long-term renal allograft outcomes remain suboptimal because of the occurrence of rejection, recurrent disease, and interstitial fibrosis with tubular atrophy. This is largely due to limitations in our understanding of allogeneic processes coupled with inadequate surveillance strategies. The concept of donor-derived cell-free DNA as a signal of allograft stress has therefore rapidly been adopted as a noninvasive monitoring tool. Refining it for effective clinical use, however, remains an ongoing effort. Furthermore, its potential to unravel new insights in alloimmunity through novel molecular techniques is yet to be realized. This review herein summarizes current knowledge and active endeavors to optimize cell-free DNA-based diagnostic techniques for clinical use in kidney transplantation. In addition, the integration of DNA methylation and microRNA may unveil new epigenetic signatures of allograft health and is also explored in this report. Directing research initiatives toward these aspirations will not only improve diagnostic precision but may foster new paradigms in transplant immunobiology.
INTRODUCTION
Kidney transplantation (KT) affords individuals with end-stage renal disease superior survival and quality of life over conventional dialytic therapies.1 Sustaining this survival advantage relies on effective allograft surveillance that consists of serial laboratory testing as well as percutaneous biopsy.2 Although the latter is regarded as the gold standard in defining the pathogenesis for allograft injury such as rejection, it has limitations such as sampling error, cost as well as carrying risks of bleeding and organ injury.3 This has led to the pursuit of noninvasive strategies including detection of donor-derived cell-free DNA (dd-cfDNA) as a marker of allogeneic injury. Since becoming Medicare reimbursable in the United States since October 2017, questions have arisen regarding the practical utility of dd-cfDNA in renal allograft management.4 This review discusses the role and limitations of this technology as well as offering insights into current challenges and future applications.
Causes of Allograft Injury and Current Surveillance Strategies
There has been progress in improving renal allograft survival in the first-year posttransplantation; however, efforts are still needed with respect to long-term graft outcomes.5,6 Nonsurgical causes of allograft failure are likely dependent on population characteristics, immunosuppression practice patterns along with allograft surveillance protocols.7 In a prospective cohort analysis of some 315 North American renal transplant recipients undergoing indication biopsies, allograft failure occurred in 54 patients because of rejection in (64%), glomerulonephritis (18%), polyomavirus nephropathy (7%), and intercurrent events (11%).8 Among these cases, antibody-mediated rejection (AMR) was identified universally in those with allograft loss from rejection, highlighting the significance of this allogeneic process in terms of clinical outcome. Interestingly, pure acute T cell–mediated rejection (ACR) was not seen in those with failed grafts in this study population. In a separate large UK cohort, aside from allograft failures due to death with functioning graft and allogeneic injury, recurrent primary disease and infection were also noted as potential drivers of allograft failure.9 Despite these advances in detecting and defining renal allograft injury, in a significant number of cases the causes of graft loss is not clearly identified. This has previously been referred to as a chronic allograft nephropathy and is now termed interstitial fibrosis and tubular atrophy (IFTA) following the eighth Banff conference in 2005.10,11 Hypothesized mechanisms for this entity may include ischemia-reperfusion injury (IRI), subclinical rejection, and calcineurin-inhibitor nephrotoxicity.12
Although these studies yield sobering results, it must be acknowledged that they reflect a surveillance strategy contingent on percutaneous allograft biopsy. Despite its risks, this approach using protocol biopsies was widely adopted upon recognition that early detection and treatment of subclinical rejection may be beneficial.13,14 Inflammation in areas of interstitial fibrosis was also subsequently demonstrated to predict allograft outcomes further supporting the role of surveillance biopsies in routine clinical practice.15 The cost and cumbersomeness of surveillance biopsies, however, remains a mitigating factor for many transplant programs, and has been cited at $3931 holistically per procedure according to the CMS Physician Fee Schedule from 2015.16-18
Principles of Cell-free DNA
Origins and Evolution in Clinical Care
Circulating cell-free DNA was originally observed by Mandel and Matais in 1948; however, its significance would not be realized till several decades later.19 Most plasma DNA is histone bound with a short half-life (10–15 min) and belongs to the Alu repeat family that may originate from apoptosis or via direct cellular secretion.20-22 Tissue necrosis is not believed to be a major source given that tumor derived cell-free DNA has been observed to decrease with cytotoxic therapy.23 Its presence is known to be associated with various disease states including autoimmunity, infections, malignancy, ischemia, and trauma.24-28 Cell-free DNA has thus found clinical value in oncology, prenatal care, and solid organ transplantation.29-31 The latter depends on delineating donor from host-circulating DNA to serve as a signal of allograft stress (refer to Figure 1).32 In 1998, Lo et al32 reported that donor-derived DNA is present in the plasma of kidney and liver transplant recipients. They envisioned that donor DNA might be used as a diagnostic tool for detecting transplant rejection, which became a reality with the advancement in molecular techniques and throughput sequencing.
FIGURE 1.
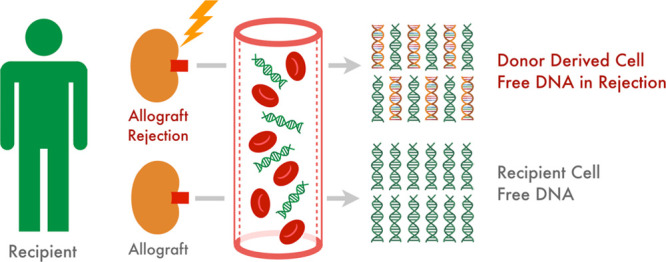
Concept of donor-derived cell-free DNA as a marker of allograft injury.
Kinetics of Cell-free DNA
In kidney transplant recipients, dd-cfDNA levels of <1% of total cfDNA appear to reflect the absence of active rejection.33 In the presence of rejection, donor cell injury results in its release into the blood stream. The majority of this cfDNA is likely a product of active apoptosis as fragmented DNA gets disseminated into the circulation.34 The half-life and the rate of clearance of cfDNA from blood remain unclear. A combination of nuclease degradation, clearance by the kidney, and uptake by the liver and spleen is likely to play a role.35 Thus, its half-life is likely to be prolonged in the context of impaired kidney function.
Cell-free DNA and the Immune Response
Aside from its prospects as a diagnostic tool, characterizing dd-cfDNA as a potential damage-associated molecular pattern (DAMP) raises some interesting possibilities. DAMPs encompass a range of endogenous biomolecules that are released upon cellular damage, which can activate the innate immune system via interaction with pattern recognition receptors such as Toll-like receptors.36 Indeed, the capacity of DNA to serve as a DAMP via interaction with TLR-9 to drive activation of the innate immune system has been reported.37 This may hold relevance in solid organ transplantation since the role of the innate immune response in allograft tolerance and rejection has been described.38 For instance, in a series of skin transplant experiments using a mouse model, Goldstein et al39 demonstrated the role of TLR signaling via MyD88 (a TLR signal adapter protein) in mediating allograft rejection. The significance of this pathway was similarly replicated in a murine renal transplant model.40 There is also evidence that TLR signaling via MyD88 is important in dendritic cell function and T-helper 1 cellular responses as part of the allogeneic process.41 Oetting et al42 have described the significance of donor polymorphisms in TLR4 in graft outcomes following liver transplantation, extending this concept to human subjects. Furthermore, DAMPs released during renal injury may also activate inflammasomes, which are cytoplasmic protein complexes triggered in infectious or sterile injuries.43
Technical Approaches and Currently Available Assays
There are various approaches for detecting dd-cfDNA ranging from copy number variation (CNV), molecular assays targeting the Y chromosome in female recipients of allografts from male donors, to more sophisticated genome transplant dynamic (GTD) strategies, which conventionally requires donor and recipient genomic data (2 genome-approach). The role of gene copy number in genetic disorders has been described by Inoue and Lupski and applying this concept solely to distinguish donor from host DNA is the basis of CNV.44 CNV is a DNA segment of 1 kb or larger that is present at a variable copy number in comparison with a reference genome. Organ transplantation is essentially genomic transplantation and the distinctive graft and recipient genotype single-nucleotide polymorphisms (SNPs) can be used to barcode donor DNA circulating in recipient serum. GTD utilizes SNPs distributed across the genome to discriminate donor from recipient DNA. It was first demonstrated as proof of concept in a retrospective analysis of heart transplant recipients in 2011.45 GTD methodology has subsequently been clinically validated in solid organ transplantation.46 A 1-genome strategy for quantifying dd-cfDNA in the absence of donor genotype through mathematical modeling was recently described by Sharon et al.47
At the time of publication, there are 3 dd-cfDNA assays widely available for clinical use. The Allosure Test (CareDx, Inc) exploits a panel of 266 SNPs known to be different among individuals based on work by Pakstis et al.48 Another method utilized by Sigdel et al which is based on work from cell-free DNA analytics involving massively multiplexed-PCR targeting 13 392 SNPs and is currently available as Prospera through Natera.49,50 A third assay—Viracor TRAC is also available through Eurofins Clinical Diagnostics and was recently combined with TruGraf (Eurofins—Transplant Genomics Inc.), a DNA microarray-based gene expression test. Table 1 compares widely used dd-cfDNA assays based on available information.33,46,51,52
Table 1.
| Allosure | Prospera | |
|---|---|---|
| Number of SNPs | 266 | 13 962 |
| Limit of detection | 0.16% (Grskovic et al) | 0.15% (related donors) 0.23% (unrelated donors) (Altug et al) |
| Limit of quantification | 0.2% (Grskovic et al) | 0.15% (related donors) 0.23% (unrelated donors) (Altug et al) |
| Sensitivity | 0.45 (Melancon) 0.59 (Bloom et al) | 0.55 (Melancon) 0.887 (Sigdel et al) |
| Specificity | 0.85 (Melancon) 0.85 (Bloom et al) | 0.69 (Melancon) 0.726 (Sigdel et al) |
| Coefficent of variation | 7.7% (dd-cfDNA < 2%) 4.5% (dd-cfDNA ≥ 2%) (Grskovic et al) | 4.29% (Altug et al) |
dd-cfDNA, donor-derived cell-free DNA.
Defining Cell-free DNA Using One-genome Strategy
Although an exhaustive description of 1-genome GTD methods are beyond the scope of this review, the basic steps in the detection of dd-cfDNA (as adopted in currently available assays) may be summarized.
Primer Design
The single-genome GTD approach exploits interindividual genomic variations as the basis of delineating donor from recipient DNA. Outside of the major histocompatibility complex region on the short arm of chromosome 6, the genome is highly conserved across the human population with approximately 99.9% similarity between individuals in their DNA sequence.53 Characterization of nonmajor histocompatibility complex genomic variations (for use in areas such as forensic genetics) has elucidated 3 types of polymorphisms: Microsatellites or short tandem repeats, variable number tandem repeats, and SNPs.54 Under this concept, primers may be designed according to a curated panel of SNPs that facilitates interindividual genomic distinction. Factors deemed important in SNP selection include minor allele frequency, polymerase error rate, heterozygosity, linkage disequilibrium, and fixation index, and the reader is directed to the review by Dengu for in-depth account of these considerations.55 For instance, 85 SNPs identified by Pakstis et al and an additional 181 SNPs incorporated by Grskovic et al form the basis of the Allosure assay made available through CareDx, Inc.46,48 In contrast, Prospera utilizes allelic frequency at 132 926 SNPs through prior work using SNP multiplexed polymerase chain reaction in the prenatal and oncology contexts.56,57
Cell-free DNA Amplification
Targeted amplification to sequence predefined SNPs of interest (as well described in the prenatal diagnostics literature), contrasts with quantitative whole genome sequencing.57 This entails a process of preamplification and amplification using multiplex PCR reactions.46 The generated PCR amplicons are then barcoded for next generation sequencing (NGS).51 The details of these steps obviously vary according to individual proprietary protocols along with their SNP selections as stated above.
Next Generation Sequencing and Data Analysis
PCR amplicons are sequenced using a next generation sequencer such as the Illumina MiSeq instrument.46,51 Customized bioanalytic pipelines are used to quality check and process raw data to yield allele counts, which may in turn be inputted in a computational model and was described in a proof of concept report by Sharon et al.47 This involves aligning amplicon data to the reference hg19 genome using an aligning tool such as Bowtie 2 followed by filtering unmapped or nonuniquely mapped reads using a program such as SamTools.47,58,59 This pipeline also incorporates barcode trimming, eliminating genotype-biased mapping, PCR duplicate removal, computing chromosomal coverage, and allele counting.46,47,60 The model henceforth gives an estimate of dd-cfDNA (as a percentage) based on the allele counts generated among other considerations.
Clinical Utility in Renal Transplantation: Current Evidence and Ongoing Efforts
In an endeavor to elucidate a role for dd-cfDNA in renal allograft management, numerous clinical studies have been undertaken to date (refer to Table 2).33,61-86 Observational data suggest that KT recipients with stable allograft function exhibit a wide range of dd-cfDNA levels immediately postsurgery with a median percentage of 10.02% at postoperative day 1 that decreases to a mean of 0.46% after 10 days.87 Bromberg et al elucidated the biological variation and clinical reference intervals of dd-cfDNA in kidney transplant recipients with stable allograft function and determined that a cutoff of 1.0% dd-cfDNA delineated the 96th percentile of test results.88 Most centers including ours, therefore postpone testing until 10–14 days posttransplantation given that IRI likely predominates till that point.
Table 2.
| Name | Study type | Organ(s) | Patients (N) | Sample type | dd-cfDNA method |
|---|---|---|---|---|---|
| Shen et al | Prospective multicenter | Kidney | 28 | Plasma | Informative SNP panel |
| Stites et al | Prospective multicenter | Kidney | 79 | Plasma | Informative SNP panel |
| Zhang et al | Prospective multicenter | Kidney | 37 | Plasma | Informative SNP panel |
| Gieles et al | Prospective multicenter | Kidney | 107 | Plasma | Informative SNP panel |
| Oellerich et al | Prospective single center | Kidney | 189 | Plasma | Informative SNP panel |
| Bloom et al | Prospective multicenter | Kidney | 384 | Plasma | Informative SNP panel |
| Brubaker et al | Prospective single center | Kidney | 27 | Plasma | Informative SNP panel |
| Gielis et al | Prospective multicenter | Kidney | 11 | Plasma | Informative SNP panel |
| Goh et al | Prospective multicenter | Kidney | 10 | Plasma | INDEL allele polyrnorphism |
| Huang et al | Prospective single center | Kidney | 25 | Plasma | Informative SNP panel |
| Jackson et al | Prospective single center | Kidney | 8 | Plasma | Informative SNP panel |
| Kollmann et al | Retrospective single center | Kidney | 20 | Plasma | INDEL allele polyrnorphism |
| Lada et al | Case report single center | Kidney | 1 | Plasma | NR |
| Lee et al | Prospective single center | Kidney | 34 | Plasma and urine | Informative SNP panel |
| Moreira et al | Prospective single center | Kidney | 17 | Plasma and urine | Sex misma tch (TSPY gene) |
| Sigdel et al | Retrospective single center | Kidney | 21 | Urine | Sex mismatch (SRY gene) |
| Sigdel et al | Retrospective single center | Kidney | NR | Plasma and urine | Custom primer sets |
| Stolz et al | Prospective single center | Kidney | 13 | Plasma | Informative SNP panel |
| Whitlam et al | Prospective single center | Kidney | 31 | Plasma | Deletion copy variation |
| Zhang et al | Prospective single center | Kidney | 61 | Urine | Sex mismatch (SRY gene) |
| Zhang et al | Retrospective single center | Kidney | 37 | Plasma | Cell-free -Alu DNA |
| Zhong et al | Prospective single center | Kidney | 25 | Urine | Sex mismatch (SRY gene) |
| Gadi et al | Retrospective single center | SPK | 42 | Plasma | HLA Mismatches |
| Beck et al | Retrospective single center | Kidney/liver/heart | 34 | Plasma | Informative SNP pane, l |
| Beck et al | Retrospective single center | Kidney/heart | NR | Plasma | Informative SNP pane, l |
| Woodward et al | Retrospective multicenter | Kidney/heart | 114 | Plasma | Sex Mismatch-TSPY1 gene |
| Mieczkowski et al | Retrospective single center | Kidney/liver | 16 | Plasma | Informative SNP pane, l |
dd-cfDNA, donor-derived cell-free DNA; SNP, single-nucleotide polymorphism.
Acute Cellular Rejection
The circulating donor-derived cell-free DNA in blood for diagnosing acute rejection in kidney transplant recipients (DART) study, which used the aforementioned Allosure assay, was a prospective observational undertaking of plasma dd-cfDNA levels among 102 KT recipients with 107 surveillance and for-cause biopsy samples.33 Of note were higher dd-cfDNA levels detected in cases with biopsy-proven ACR 1B (median 1.2%), whereas biopsies with ACR 1A (median 0.3%) acute tubular necrosis (ATN) and calcineurin-inhibitor toxicity had comparatively lower levels of dd-cfDNA. However, in another study dd-cfDNA levels measured via the Allosure test did not discriminate adult patients with ACR from those without rejection, as the AUC for ACR was 0.42 (95% CI, 0.17-0.66) in this particular analysis.89 However, in a group of 13 pediatric patients, 2 cases of ACR exhibited dd-cfDNA levels of >2% and were statistically higher than those without rejection.77 A recent prospective multicenter analysis found dd-cfDNA levels >0.5% to be predictive of adverse clinical outcomes among individuals with borderline rejection and ACR 1A on allograft biopsy.90 If corroborated in larger studies, this may be clinically relevant since borderline rejection is a heterogeneous diagnostic category with variable prognostic outcomes.91 In contrast to the DART study, in which relatively lower dd-cfDNA fractions were found in ACR compared with AMR using the Allosure test, the use of multiplexed-PCR with 13 392 SNP targets among 300 samples from 217 biopsy matched cases demonstrated higher levels among all types of rejection with 2.2%, 2.7%, 0.6%, 0.7%, and 0.4% in AMR, ACR, borderline changes, cases with “other injury,” and stable renal allograft biopsies, respectively.50 This assay was deemed applicable for clinical use in KT patients based on detection limits, linearity, accuracy, and precision within a validation cohort in a study funded and conducted trial by Natera, Inc.51
Antibody-mediated Rejection
Among the DART study population, higher levels of dd-cfDNA were also seen in those with biopsy-proven AMR (median 2.9%), which based on cutoff of 1% translated to an area under the receiver operating characteristic curve (AUC) of 0.74 (95% CI, 0.61-0.86) and positive and negative predictive values for active rejection of 61% and 84%, respectively.33 In another reanalysis of DART study samples by Weir et al, cell-free DNA was deemed to be more accurate than changes in serum creatinine for detecting rejection.92 As far as AMR is concerned, Jordan et al’s findings were consistent with the observations from the DART study in that dd-cfDNA of >1% derived through the same means was associated with the presence of donor-specific antibodies (DSAs), and that threshold conferred a positive and negative predictive value for AMR of 81% and 83%, respectively.93 Furthermore, incorporating levels of dd-cfDNA with levels of donor-specific antibody may also delineate ACR from AMR.33,93 Finally, dd-cfDNA has been associated with de novo donor-specific antibodies raising the possibility that donor-derived nucleic acids may independently elicit a humoral immune response.94 Such a causal relationship if present may construe dd-cfDNA a heralding signal of impending antibody-mediated injury.
Other Forms of Allograft Injury
It is clear that (mechanistically) dd-cfDNA release is not limited exclusively to allograft rejection. Nonalloimmune processes including IFTA, ATN, recurrent glomerulonephritis, and other disease states may also be associated with elevations in dd-cfDNA levels and are reviewed here.
Given its elusive causes, targeting therapies for IFTA is challenging, which can render the risk of allograft biopsy clinically unjustifiable. Nevertheless, it is understood mechanistically as a form of downstream cellular injury, which could conceivably lead to release of cfDNA. Although no dedicated study has been undertaken, a recent series comparing dd-cfDNA levels between 12 stable repeat- and 202 single-KT recipients from the DART cohort showed higher levels among those with a prior transplant.95 In a separate study, there was also a trend toward higher dd-cfDNA levels among subjects with IFTA on biopsies; however, this was not statistically significant and it was also in the setting of AMR.86 Taken together, it may be inferred that IFTA may be associated with somewhat higher dd-cfDNA levels but to what extent has yet to be defined.
As is the case for IFTA, there are no published data on dd-cfDNA levels specifically in the setting of ATN. This is not surprising since biopsy is rarely undertaken when suspicion for ischemic tubular injury is favored over rejection. In a study of total rather than donor-derived cfDNA, Moreira et al reported significantly higher levels in acute rejection and drug toxicity than ATN.63 Similarly, results of the DART study also suggested more significant dd-cfDNA release in the context of allogeneic injury than in ATN.33 Considering that elevated levels are seen perioperatively when IRI is most profound, dd-cfDNA is likely reflective of the extent and not solely the cause of allograft stress.
Recurrent disease remains an important source of death-censored graft loss with incidence varying depending on pathogenesis, population characteristics, and follow-up time periods.96,97 There is currently a paucity of data on dd-cfDNA levels in this setting; however, host-derived circulating DNA has been reported in systemic lupus erythematous, progressive diabetic kidney disease, and urinary mitochondrial DNA has also been described in IgA nephropathy.98-100 It should be noted that cell-free DNA may be extruded during the inflammatory process of neutrophil extracellular traps formation.101 NETosis involves the deployment of extracellular chromatin, which is speculated to entangle microbes as host-defense mechanism, and is seen in a number of autoimmune conditions.102,103 It is therefore plausible, although unproven that inflammatory glomerular diseases may be associated with higher cell-free DNA levels compared with noninflammatory conditions. Whether donor cells exhibit similar DNA release in recurrent disease states also needs to be investigated.
Although appraisals of these 2 available tests clearly show higher fractions of dd-cfDNA in cases of rejection compared with other allograft states, there has been no dedicated study to date looking at levels in the context of allograft infections, urologic, or hemodynamic-mediated insults. Nevertheless in a subsequent analysis of 11 individuals among the DART cohort with BK viremia, 7 had viral titers that correlated with elevated dd-cfDNA levels and histopathology findings; however, the sample size was inadequate to draw further conclusions regarding the detection of BK nephritis.104 In other forms of organ stress, it may also be speculated that greater dd-cfDNA is released as a result of cellular injury. Anecdotally, a markedly elevated level has been seen in a case of Page kidney following traumatic allograft biopsy at our center in a 28-year-old male, which normalized postsurgical intervention and with recovery in the acute kidney injury. To what extent dd-cfDNA levels correlate with specific disease states has yet to be determined.
Utility in Multiorgan Transplantation
Other questions regarding the usability of dd-cfDNA include interpretation in multiorgan transplant recipients. The aforementioned comparison of stable single- and repeat-KT recipients found that dd-cfDNA levels remained <1% despite the higher levels seen among the latter, and that levels were further elevated during rejection.95 Based on this work and the established threshold for dd-cfDNA levels, it is reasonable to utilize this assay in multiorgan transplant recipients. Nonetheless, it is important to recognize, however, that the currently available assays cannot discern which allograft is injured in the setting of an elevated dd-cfDNA level.
Current Challenges
Although cell-free DNA-based diagnostic tests have rapidly been accepted and implemented in the care of transplant patients, there are several issues and unanswered questions that need to be addressed.
Quantification of Donor-derived Cell-free DNA
The use of NGS-based platforms and indeed the method of dd-cfDNA quantification in itself has been questioned. Whitlam et al have pointed out potential bias arising from DNA preamplification along with advantages of CNV in absolute quantification of dd-cfDNA (as opposed to a percentage), which also overcomes changes in circulating recipient–derived cfDNA.105 The latter may be particularly important during physiologic stress in which the recipient’s leukocyte-derived cfDNA becomes the predominant source of total cfDNA, undermining the accuracy of a percentage.85,106 Using CNV and droplet digital PCR, Whitlam et al found absolute graft derived cf to be more specific for AMR, whereas fractionated estimates had more diagnostic utility for chronic active antibody-mediated rejection (CAAMR) and ACR.105 Furthermore in a recent blinded prospective study, absolute dd-cfDNA demonstrated superiority in discriminating the presence versus absence of rejection or other forms of graft injury compared with a fractionated metric.85 Although the findings of both reports suggest value in absolute quantification of dd-cfDNA, they still retain a role for fractionated dd-cfDNA in clinical practice. Further work is therefore necessary to refine this assay in the context of the highly dynamic host-derived cfDNA mileu.
Improving Predictability
The superiority of dd-cfDNA over serum creatinine in detecting rejection has been demonstrated.92 However, defining a threshold that can prompt therapy in advance of histologic changes continues to be a challenge given the risk of false positives. Based on work from Bromberg et al, normal biological variation of up to 61% may occur in serial laboratory values but the role of trending levels to predict rejection is not known.88 Adopting measures of DNA methylation and microRNA may help overcome these issues and are discussed below.
Role in Monitoring Therapy
In oncology cfDNA has been used for real-time molecular monitoring of treatment, detection of recurrence, and tracking resistance. If such “liquid biopsy” could be used to complement traditional markers to guide therapy, it could be transformative in transplant medicine. Although there is currently limited experience with cell-free DNA as a tool to monitor response to therapy, at time of publication, we note a registered trial examining the use of cfDNA to characterize therapeutic response with tocilizumab in CAAMR.107 This study, which is due for completion in December 2020, will define changes in dd-cfDNA in conjunction with histopathologic changes in response to monthly therapy with tocilizumab for CAAMR.
Future Directions: Enhancing Diagnostic Precision and Wider Applications
There is an ongoing quest to improve the diagnostic accuracy of existing cfDNA platforms through incorporation of new and novel molecular markers such as DNA methylation pattern and microRNAs. This may carry important implications with respect to renal allograft assessment, monitoring and transplant therapeutics. Furthermore, there is opportunity to expand the use of cfDNA (beyond that which is solely donor-derived) to detect infectious states that transplant recipients are typically predisposed to.
Integration of DNA Methylation
Although the mere presence of circulating dd-cfDNA is evidently significant, its characteristics and the role of host-derived cfDNA are also points of interest. In particular, the concept of epigenetic phenomenon as the interface between the genome and the environment potentially represents another biomarker of renal disease states.108 This essentially reflects the interplay between epigenetic modification including DNA methylation and gene expression in both physiological and pathophysiological conditions.109 DNA methylation is generally associated with gene silencing, whereas hypomethylation is associated with permissive gene expression. Indeed, there is evidence that genome-wide epigenetic modification are associated with decline in native renal function in those with established chronic kidney disease.110 Whether these observations hold true with respect to allograft health and function has yet to be explored. In addition, epigenetic factors have been described in the maturation and function of memory T cells, T-reg cells, B cells, and NK cells.111-113 Defining these epigenetic “signatures” may offer new insights into allogeneic processes and may foster novel diagnostic models of rejection.
Circulating microRNA
While this review primarily targets cfDNA, the prospect of enhancing diagnostic specificity through current insights in epigenetics warrants a brief review of such factors. Essentially gene expression may be influenced by noncoding RNA such as small interfering RNAs and microRNAs (miRNAs), which are linked with epigenetic processes such as DNA methylation and histone modifications.114 Extracellular miRNA is known to be present in plasma and serum, and its role in intercellular signaling is also recognized.115,116 In fact, altered expression of a specific miRNA is also believed to be associated with renal allograft rejection possibly through modifying the expression of certain genes in regulatory T cells.117 Thus, whether integration of miRNA signature to dd-cfDNA will improve the diagnostic accuracy needs to be studied.
Damp Signaling and Therapeutic Potential of Cell-free DNA
As alluded to above, characterizing dd-cfDNA as a candidate DAMP molecule may afford new insights in transplant immunology. The role of such mediators in sterile inflammation in autoimmunity, malignancy, and cardiovascular disease is well recognized.118-120 Moreover, the concept of DAMPs as a therapeutic target in a cancer model has also been proposed.121 Applying this theory using dd-cfDNA may therefore cultivate new knowledge in mechanisms and treatment of allograft rejection.
Cell-free DNA for Microbial Diagnostics Posttransplantation
Fragments of genomic DNA from pathogens causing infection at various locations in the body are found in purified plasma cfDNA. Microbial cfDNA can be detected by NGS and could identify the pathogens accurately.25 The Karius Test relies on sequencing of microbial cfDNA circulating in plasma to identify over various pathogens, including bacteria, viruses, and fungi.122 Generally, high concentrations of microbial cfDNA have been associated with true infections, whereas low concentrations could still be due to true infections versus commensal, colonizing, or contaminant states of unknown clinical significance. This is particularly relevant in the renal transplant recipient who is prone to opportunistic infections and colonization. The Karius assay has shown sensitivity in detecting CMV and BK virus in stem-cell transplant recipients, but it has yet to be confirmed if this is generalizable in renal transplantation.123 Given that these viruses can reflect overimmunosuppression but are also associated with allograft rejection makes clinical management particularly difficult.124,125 Integrating cfDNA of donor and microbial origin may offer an opportunity to navigate this challenging dilemma and warrants further investigation.
CONCLUSION
The adoption of noninvasive methods in renal allograft monitoring is evolving rapidly and the transplant community must recognize the strengths and caveats of these novel methods. In doing so, the field must also integrate other insights such as epigenetic analyses to further advance the diagnostic potential of cfDNA. This is particularly important given the cost, inconvenience, and limitations of percutaneous allograft biopsies. A practical question raised in this review includes the optimal surveillance interval of this assay, which has yet to be defined, particularly given the short half-life of cell-free DNA. Coupled with this issue is the assay’s usability following therapy, which will hopefully become better understood through the ongoing clinical trial described above. Another challenging scenario concerns the multiorgan transplant recipient or allograft monitoring in the setting of pregnancy where making finer-genomic delineations is necessary.
Although overcoming practical issues is important, ultimately ensuring that dd-cfDNA assay translates into overall benefit in patient and allograft outcomes is paramount. To date, there is no published report on how such testing impacts allograft survival or whether there is a cost-benefit compared with percutaneous tissue sampling. It must also be acknowledged that false positive results may lead to unnecessary allograft biopsies and paradoxically expose patients to discomfort or harm. Another important consideration will be the influence on immunosuppressive management decisions. Long-term risks related to overimmunosuppression in the form of infections, metabolic syndrome, and malignancy are real and balancing these issues in any allograft surveillance strategy is essential. In summary, we regard dd-cfDNA as a complex yet promising biomarker of allograft health and more work will be needed to optimize it for routine use in the renal transplant population.
Footnotes
Published online 4 February, 2021.
The authors declare no funding or conflicts of interest.
R.S.P., I.A., A.C., D.R., and M.J. participated in the writing of the article.
REFERENCES
- 1.Wolfe RA, Ashby VB, Milford EL, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999; 341:1725–1730 [DOI] [PubMed] [Google Scholar]
- 2.El Ters M, Grande JP, Keddis MT, et al. Kidney allograft survival after acute rejection, the value of follow-up biopsies. Am J Transplant. 2013; 13:2334–2341 [DOI] [PubMed] [Google Scholar]
- 3.Furness PN, Philpott CM, Chorbadjian MT, et al. Protocol biopsy of the stable renal transplant: a multicenter study of methods and complication rates. Transplantation. 2003; 76:969–973 [DOI] [PubMed] [Google Scholar]
- 4.Gielis EM, Ledeganck KJ, De Winter BY, et al. Cell-free DNA: an upcoming biomarker in transplantation. Am J Transplant. 2015; 15:2541–2551 [DOI] [PubMed] [Google Scholar]
- 5.Meier-Kriesche HU, Schold JD, Kaplan B. Long-term renal allograft survival: have we made significant progress or is it time to rethink our analytic and therapeutic strategies? Am J Transplant. 2004; 4:1289–1295 [DOI] [PubMed] [Google Scholar]
- 6.Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. Am J Transplant. 2011; 11:450–462 [DOI] [PubMed] [Google Scholar]
- 7.Naylor KL, Knoll GA, Shariff SZ, et al. Socioeconomic status and kidney transplant outcomes in a universal healthcare system: a population-based cohort study. Transplantation. 2019; 103:1024–1035 [DOI] [PubMed] [Google Scholar]
- 8.Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant. 2012; 12:388–399 [DOI] [PubMed] [Google Scholar]
- 9.Burton H, Iyamu Perisanidou L, Steenkamp R, et al. Causes of renal allograft failure in the UK: trends in UK Renal Registry and National Health Service blood and transplant data from 2000 to 2013. Nephrol Dial Transplant. 2019; 34:355–364 [DOI] [PubMed] [Google Scholar]
- 10.Paul LC. Chronic allograft nephropathy: an update. Kidney Int. 1999; 56:783–793 [DOI] [PubMed] [Google Scholar]
- 11.Solez K, Colvin RB, Racusen LC, et al. Banff ‘05 meeting report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’). Am J Transplant. 2007; 7:518–526 [DOI] [PubMed] [Google Scholar]
- 12.Chapman JR, O’Connell PJ, Nankivell BJ. Chronic renal allograft dysfunction. J Am Soc Nephrol. 2005; 16:3015–3026 [DOI] [PubMed] [Google Scholar]
- 13.Rush DN, Henry SF, Jeffery JR, et al. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994; 57:208–211 [DOI] [PubMed] [Google Scholar]
- 14.Rush D, Nickerson P, Gough J, et al. Beneficial effects of treatment of early subclinical rejection: a randomized study. J Am Soc Nephrol. 1998; 9:2129–2134 [DOI] [PubMed] [Google Scholar]
- 15.Mannon RB, Matas AJ, Grande J, et al. ; DeKAF Investigators. Inflammation in areas of tubular atrophy in kidney allograft biopsies: a potent predictor of allograft failure. Am J Transplant. 2010; 10:2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rush D. Protocol transplant biopsies: an underutilized tool in kidney transplantation. Clin J Am Soc Nephrol. 2006; 1:138–143 [DOI] [PubMed] [Google Scholar]
- 17.Centers for Medicare & Medicaid Services. 2015. Available at https://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/PhysicianFeeSched/PFS-Federal-Regulation-Notices-Items/CMS-1612-FC. Accessed May 30, 2020 [PubMed]
- 18.First MR, Lee D, Lewis P, et al. An economic analysis of the cost effectiveness of blood gene expression profiling in kidney transplant recipients. J Health Med Econ. 2017; 3:3 [Google Scholar]
- 19.MANDEL P, METAIS P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948; 142:241–243 [PubMed] [Google Scholar]
- 20.Rumore PM, Steinman CR. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J Clin Invest. 1990; 86:69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stroun M, Lyautey J, Lederrey C, et al. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells? Ann N Y Acad Sci. 2001; 945:258–264 [DOI] [PubMed] [Google Scholar]
- 22.Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001; 313:139–142 [DOI] [PubMed] [Google Scholar]
- 23.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37:646–650 [PubMed] [Google Scholar]
- 24.Lo YM, Rainer TH, Chan LY, et al. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000; 46:319–323 [PubMed] [Google Scholar]
- 25.Hong DK, Blauwkamp TA, Kertesz M, et al. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn Microbiol Infect Dis. 2018; 92:210–213 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka S, Sugimoto S, Kurosaki T, et al. Donor-derived cell-free DNA is associated with acute rejection and decreased oxygenation in primary graft dysfunction after living donor-lobar lung transplantation. Sci Rep. 2018; 8:15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duvvuri B, Lood C. Cell-free DNA as a biomarker in autoimmune rheumatic diseases. Front Immunol. 2019; 10:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiala C, Diamandis EP. New approaches for detecting cancer with circulating cell-free DNA. BMC Med. 2019; 17:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grace MR, Hardisty E, Dotters-Katz SK, et al. Cell-free DNA screening: complexities and challenges of clinical implementation. Obstet Gynecol Surv. 2016; 71:477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif. 2019; 17:100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight SR, Thorne A, Lo Faro ML. Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation. 2019; 103:273–283 [DOI] [PubMed] [Google Scholar]
- 32.Lo YM, Tein MS, Pang CC, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998; 351:1329–1330 [DOI] [PubMed] [Google Scholar]
- 33.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017; 28:2221–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kustanovich A, Schwartz R, Peretz T, et al. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019; 20:1057–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung F, Kulasingam V, Diamandis EP, et al. Circulating tumor DNA as a cancer biomarker: fact or fiction? Clin Chem. 2016; 62:1054–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. 2018; 18:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008; 8:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaRosa DF, Rahman AH, Turka LA. The innate immune system in allograft rejection and tolerance. J Immunol. 2007; 178:7503–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldstein DR, Tesar BM, Akira S, et al. Critical role of the Toll-like receptor signal adaptor protein MyD88 in acute allograft rejection. J Clin Invest. 2003; 111:1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Noordmans GA, O’Brien MR, et al. Absence of MyD88 signaling induces donor-specific kidney allograft tolerance. J Am Soc Nephrol. 2012; 23:1701–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tesar BM, Zhang J, Li Q, et al. TH1 immune responses to fully MHC mismatched allografts are diminished in the absence of MyD88, a toll-like receptor signal adaptor protein. Am J Transplant. 2004; 4:1429–1439 [DOI] [PubMed] [Google Scholar]
- 42.Oetting WS, Guan W, Schladt DP, et al. Donor polymorphisms of toll-like receptor 4 associated with graft failure in liver transplant recipients. Liver Transpl. 2012; 18:1399–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002; 10:417–426 [DOI] [PubMed] [Google Scholar]
- 44.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002; 3:199–242 [DOI] [PubMed] [Google Scholar]
- 45.Snyder TM, Khush KK, Valantine HA, et al. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011; 108:6229–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free dna in solid organ transplant recipients. J Mol Diagn. 2016; 18:890–902 [DOI] [PubMed] [Google Scholar]
- 47.Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. Plos Comput Biol. 2017; 13:e1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pakstis AJ, Speed WC, Fang R, et al. SNPs for a universal individual identification panel. Hum Genet. 2010; 127:315–324 [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann B, Hill M, Gemelos G, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn. 2012; 32:1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018; 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altuğ Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019; 103:2657–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melancon JK, Khalil A, Lerman MJ. Donor derived cell-free DNA: is it all the same? Kidney360. 2020; 1:1038–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006; 7:85–97 [DOI] [PubMed] [Google Scholar]
- 54.Kowalczyk M, Zawadzka E, Szewczuk D, et al. Molecular markers used in forensic genetics. Med Sci Law. 2018; 58:201–209 [DOI] [PubMed] [Google Scholar]
- 55.Dengu F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant Rev (Orlando). 2020; 34:100542. [DOI] [PubMed] [Google Scholar]
- 56.Jamal-Hanjani M, Wilson GA, Horswell S, et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann Oncol. 2016; 27:862–867 [DOI] [PubMed] [Google Scholar]
- 57.Ryan A, Hunkapiller N, Banjevic M, et al. Validation of an enhanced version of a single-nucleotide polymorphism-based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagn Ther. 2016; 40:219–223 [DOI] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alborelli I, Generali D, Jermann P, et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: a proof of concept and technical validation study. Cell Death Dis. 2019; 10:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Tong KL, Li PK, et al. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: urinary DNA chimerism. Clin Chem. 1999; 45:1741–1746 [PubMed] [Google Scholar]
- 62.Zhong XY, Hahn D, Troeger C, et al. Cell-free DNA in urine: a marker for kidney graft rejection, but not for prenatal diagnosis? Ann N Y Acad Sci. 2001; 945:250–257 [PubMed] [Google Scholar]
- 63.Gadi VK, Nelson JL, Boespflug ND, et al. Soluble donor DNA concentrations in recipient serum correlate with pancreas-kidney rejection. Clin Chem. 2006; 52:379–382 [DOI] [PubMed] [Google Scholar]
- 64.García Moreira V, Prieto García B, Baltar Martín JM, et al. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem. 2009; 55:1958–1966 [DOI] [PubMed] [Google Scholar]
- 65.Lada Z, Jerko B, Jasenka W, et al. Disappearance of circulating male donor-derived cell-free DNA in female recipient blood after rabbit anti-human thymocyte globulin treatment for acute cellular kidney transplant rejection—case report. Acta Med Croatica. 2011; 65:150 [Google Scholar]
- 66.Zhang L, Hu X, Yin L, et al. Early upsurge of cell-free Alu DNA predicts declining long-term graft survival. [abstract]. Am J Transplant. 2011; 11s2389 [Google Scholar]
- 67.Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013; 59:1732–1741 [DOI] [PubMed] [Google Scholar]
- 68.Sigdel TK, Vitalone MJ, Tran TQ, et al. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation. 2013; 96:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woodward R, Grskovic M, Dedrick R. Increased plasma levels of graft-derived cell-free DNA correlate with rejection in solid organ transplant recipients: Abstract# 594 Transplantation. 2014; 98:22224926826 [Google Scholar]
- 70.Beck J, Oellerich M, Schulz U, et al. Donor-derived cell-free DNA is a novel universal biomarker for allograft rejection in solid organ transplantation. Transplant Proc. 2015; 47:2400–2403 [DOI] [PubMed] [Google Scholar]
- 71.Sigdel T, Tran T, Hsieh S-C, et al. Urinary cell-free DNA is a sensitive marker of early renal transplant injury [abstract]. Am J Transplant. 2015; 15(Suppl 3). Available at https://atcmeetingabstracts.com/abstract/urinary-cell-free-dna-is-a-sensitive-marker-of-early-renal-transplant-injury/ [Google Scholar]
- 72.Gielis E, Ledeganck K, Wils H, et al. Quantification of plasma donor-derived cell-free DNA to monitor kidney transplant health: preliminary results of a single tube multiplex PCR assay. Nephrol Dial Transplant. 2016; 31:1–2. Presented at the 53rd ERA-EDTA Congress26627633 [Google Scholar]
- 73.Mieczkowski P, Malc E, Steele P, et al. Human blood cell-free circulating DNA (cfDNA) and miRNA as biomarkers of liver and kidney antibody mediated rejection (AMR) or cellular allograft rejection (ACR). Pilot study. Transplantation. 2016; 100:S483 [Google Scholar]
- 74.Goh Y, Ho S, Raman L, et al. Noninvasive renal transplant graft monitoring in single institution using cell-free DNA in recipient plasma via insertion-deletion allele polymorphism. [abstract]. Am J Transplant. 2017; 17(Suppl 3). Available at https://atcmeetingabstracts.com/abstract/noninvasive-renal-transplant-graft-monitoring-in-single-institution-using-cell-free-dna-in-recipient-plasma-via-insertion-deletion-allele-polymorphism/ [Google Scholar]
- 75.Kollmann D, Dauber EM, Fischer G, et al. Cell-free donor DNA circulating in recipient plasma: Indel-polymorphisms as non-invasive marker for acute rejection in kidney transplantation [Oral Abstract]. Transplant Int. 2017; 30S2111 [Google Scholar]
- 76.Lee H, Park YM, We YM, et al. Evaluation of digital PCR as a technique for monitoring acute rejection in kidney transplantation. Genomics Inform. 2017; 15:2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stoltz D, Brubaker A, Grskovic M, et al. Donor-derived cell-free DNA predicts biopsy-proven acute cellular rejection in pediatric kidney transplant recipients. [abstract]. Am J Transplant. 2017; 17(Suppl 3). Available at https://atcmeetingabstracts.com/abstract/donor-derived-cell-free-dna-predicts-biopsy-proven-acute-cellular-rejection-in-pediatric-kidney-transplant-recipients/ [Google Scholar]
- 78.Brubaker A, Stoltz D, Grskovic M, et al. Donor-derived cell-free DNA predicts development of clinically significant changes in donor specific antibody following kidney transplant [abstract]. 2018. Available at https://atcmeetingabstracts.com/abstract/title-donor-derived-cell-free-dna-predicts-development-of-clinically-significant-changes-in-donor-specific-antibody-following-kidney-transplant/. Accessed October 7, 2020
- 79.Huang E, Vo A, Choi J, et al. Early experience using donor-derived cell-free DNA for detection of rejection in kidney transplantation [abstract]. 2018. Available at https://atcmeetingabstracts.com/abstract/early-experience-using-donor-derived-cell-free-dna-for-detection-of-rejection-in-kidney-transplantation/. Accessed October 6, 2020
- 80.Jackson A, Bagnasco S, Desai N. Molecular monitoring for kidney allograft injury following hla incompatible transplantation [abstract].. 2018. Available at https://atcmeetingabstracts.com/abstract/molecular-monitoring-for-kidney-allograft-injury-following-hla-incompatible-transplantation/. Accessed October 7, 2020
- 81.Whitlam J, L.L., Ierino F, Bruno D, et al. 2018. Absolute quantification of plasma graft-derived cell-free DNA for diagnosis of kidney transplant rejection [abstract].. Available at https://atcmeetingabstracts.com/abstract/absolute-quantification-of-plasma-graft-derived-cell-free-dna-for-diagnosis-of-kidney-transplant-rejection/. Accessed October 7, 2020
- 82.Zhang H, Liu L, Zheng C, et al. The role of donor-derived cell-free DNA in the Identification of injury in kidney allografts with antibody-mediated rejection or de novo DSA Transplantation. 2018; 102:S5–S6 [Google Scholar]
- 83.Gielis EM, Ledeganck KJ, Dendooven A, et al. The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant. 2020; 35:714–721 [DOI] [PubMed] [Google Scholar]
- 84.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: results from a prospective observational study. Am J Transplant. 2019; 19:3087–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen J, Guo L, Yan P, et al. Prognostic value of the donor-derived cell-free DNA assay in acute renal rejection therapy: a prospective cohort study. Clin Transplant. 2020; 34:e14053. [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Zheng C, Li X, et al. Diagnostic performance of donor-derived plasma cell-free DNA fraction for antibody-mediated rejection in post renal transplant recipients: a prospective observational study. Front Immunol. 2020; 11:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gielis EM, Beirnaert C, Dendooven A, et al. Plasma donor-derived cell-free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay. PLoS One. 2018; 13:e0208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bromberg JS, Brennan DC, Poggio E, et al. Biological variation of donor-derived cell-free DNA in renal transplant recipients: clinical implications. J Appl Lab Med. 2017; 2:309–321 [DOI] [PubMed] [Google Scholar]
- 89.Huang E, Sethi S, Peng A, et al. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant. 2019; 19:1663–1670 [DOI] [PubMed] [Google Scholar]
- 90.Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020; 20:2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nankivell BJ, Agrawal N, Sharma A, et al. The clinical and pathological significance of borderline T cell-mediated rejection. Am J Transplant. 2019; 19:1452–1463 [DOI] [PubMed] [Google Scholar]
- 92.Weir M, Hiller D, Yee J, et al. Donor-derived cell-free DNA outperforms serum creatinine changes for identifying kidney transplant rejection [abstract]. 2018. Available at https://atcmeetingabstracts.com/abstract/donor-derived-cell-free-dna-outperforms-serum-creatinine-changes-for-identifying-kidney-transplant-rejection/ Accessed October 7, 2020
- 93.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018; 4:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jordan S, Sawinski D, Dholakia S. Donor derived cell-free DNA initiates de-novo donor specific antibody (DSA) responses [abstract]. Am J Transplant. 2019; 19(Suppl 3). Available at https://atcmeetingabstracts.com/abstract/donor-derived-cell-free-dna-initiates-de-novo-donor-specific-antibody-dsa-responses/ [Google Scholar]
- 95.Mehta SG, Chang JH, Alhamad T, et al. Repeat kidney transplant recipients with active rejection have elevated donor-derived cell-free DNA. Am J Transplant. 2019; 19:1597–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Briganti EM, Russ GR, McNeil JJ, et al. Risk of renal allograft loss from recurrent glomerulonephritis. N Engl J Med. 2002; 347:103–109 [DOI] [PubMed] [Google Scholar]
- 97.Gourishankar S, Leduc R, Connett J, et al. Pathological and clinical characterization of the ‘troubled transplant’: data from the DeKAF study. Am J Transplant. 2010; 10:324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan EM, Kunkel HG. Characteristics of a soluble nuclear antigen precipitating with sera of patients with systemic lupus erythematosus. J Immunol. 1966; 96:464–471 [PubMed] [Google Scholar]
- 99.Yu BC, Cho NJ, Park S, et al. IgA nephropathy is associated with elevated urinary mitochondrial DNA copy numbers. Sci Rep. 2019; 9:16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li X, Hu R, Luo T, et al. Serum cell-free DNA and progression of diabetic kidney disease: a prospective study. BMJ Open Diabetes Res Care. 2020; 8:e001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yipp BG, Kubes P. NETosis: how vital is it? Blood. 2013; 122:2784–2794 [DOI] [PubMed] [Google Scholar]
- 102.Clark SR, Ma AC, Tavener SA, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007; 13:463–469 [DOI] [PubMed] [Google Scholar]
- 103.He Y, Yang FY, Sun EW. Neutrophil extracellular traps in autoimmune diseases. Chin Med J (Engl). 2018; 131:1513–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brennan D, Bromberg J, Yee J, et al. Donor derived cell-free DNA (dd-cfDNA) may aid in the diagnosis of bk virus nephropathy [abstract]. Am J Transplant. 2019; 19(Suppl 3). Available at https://atcmeetingabstracts.com/abstract/donor-derived-cell-free-dna-dd-cfdna-may-aid-in-the-diagnosis-of-bk-virus-nephropathy/ [Google Scholar]
- 105.Whitlam JB, Ling L, Skene A, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant. 2019; 19:1037–1049 [DOI] [PubMed] [Google Scholar]
- 106.Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015; 112:E5503–E5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.ClinicalTrials.gov Identifier: NCT03859388. Longitudinal changes in donor-derived cell-free DNA with tocilizumab treatment for chronic antibody-mediated rejection. 2020. Available at https://clinicaltrials.gov/ct2/show/NCT03859388
- 108.Dressler GR, Patel SR. Epigenetics in kidney development and renal disease. Transl Res. 2015; 165:166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Murrell A, Hurd PJ, Wood IC. Epigenetic mechanisms in development and disease. Biochem Soc Trans. 2013; 41:697–699 [DOI] [PubMed] [Google Scholar]
- 110.Wing MR, Devaney JM, Joffe MM, et al. ; Chronic Renal Insufficiency Cohort (CRIC) Study. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant. 2014; 29:864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parra M. Epigenetic events during B lymphocyte development. Epigenetics. 2009; 4:462–468 [DOI] [PubMed] [Google Scholar]
- 112.Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013; 139:277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morikawa H, Ohkura N, Vandenbon A, et al. ; FANTOM Consortium. Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc Natl Acad Sci U S A. 2014; 111:5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oetting WS, Schladt DP, Dorr CR, et al. ; DeKAF Genomics and GEN03 Investigators. Analysis of 75 candidate SNPs associated with acute rejection in kidney transplant recipients: validation of rs2910164 in microRNA MIR146A. Transplantation. 2019; 103:1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011; 39:7223–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Turchinovich A, Samatov TR, Tonevitsky AG, et al. Circulating miRNAs: cell-cell communication function? Front Genet. 2013; 4:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Abdi R, Tran TB, Sahagun-Ruiz A, et al. Chemokine receptor polymorphism and risk of acute rejection in human renal transplantation. J Am Soc Nephrol. 2002; 13:754–758 [DOI] [PubMed] [Google Scholar]
- 118.Foell D, Wittkowski H, Roth J. Mechanisms of disease: a ‘DAMP’ view of inflammatory arthritis. Nat Clin Pract Rheumatol. 2007; 3:382–390 [DOI] [PubMed] [Google Scholar]
- 119.Frantz S, Monaco C, Arslan F. Danger signals in cardiovascular disease. Mediators Inflamm. 2014; 2014:395278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hernandez C, Huebener P, Schwabe RF. Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene. 2016; 35:5931–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hendriks LEL, Dingemans AC. Heat shock protein antagonists in early stage clinical trials for NSCLC. Expert Opin Investig Drugs. 2017; 26:541–550 [DOI] [PubMed] [Google Scholar]
- 122.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019; 4:663–674 [DOI] [PubMed] [Google Scholar]
- 123.Fung M, Seng H, Hollemon D, et al. The DISCOVER Trial: application of the karius plasma next-generation sequencing test for pathogen detection in stem-cell transplant patients. Open Forum Infect Dis. 2017; 4(Suppl 1):S616 [Google Scholar]
- 124.Dickenmann MJ, Cathomas G, Steiger J, et al. Cytomegalovirus infection and graft rejection in renal transplantation. Transplantation. 2001; 71:764–767 [DOI] [PubMed] [Google Scholar]
- 125.Sawinski D, Forde KA, Trofe-Clark J, et al. Persistent BK viremia does not increase intermediate-term graft loss but is associated with de novo donor-specific antibodies. J Am Soc Nephrol. 2015; 26:966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]


