Abstract
Purpose
Lamins are the major component of nuclear lamina, maintaining structural integrity of the nucleus. Lamin A/C variants are well established to cause a spectrum of disorders ranging from myopathies to progeria, termed laminopathies. Phenotypes resulting from variants in LMNB1 and LMNB2 have been much less clearly defined.
Methods
We investigated exome and genome sequencing from the Deciphering Developmental Disorders Study and the 100,000 Genomes Project to identify novel microcephaly genes.
Results
Starting from a cohort of patients with extreme microcephaly, 13 individuals with heterozygous variants in the two human B-type lamins were identified. Recurrent variants were established to be de novo in nine cases and shown to affect highly conserved residues within the lamin ɑ-helical rod domain, likely disrupting interactions required for higher-order assembly of lamin filaments.
Conclusion
We identify dominant pathogenic variants in LMNB1 and LMNB2 as a genetic cause of primary microcephaly, implicating a major structural component of the nuclear envelope in its etiology and defining a new form of laminopathy. The distinct nature of this lamin B–associated phenotype highlights the strikingly different developmental requirements for lamin paralogs and suggests a novel mechanism for primary microcephaly warranting future investigation.
Key words: LMNB1, LMNB2, laminopathy, primary microcephaly, neurodevelopmental disorder
INTRODUCTION
The nuclear lamina is a protein structure that lines the inner nuclear membrane and provides structural support to the nucleus.1 Lamins are the major component of the nuclear lamina, forming a meshwork of filaments; they interact with numerous proteins and also act as a signaling hub, linking the nuclear lamina to the cytoskeleton and chromatin. Consequently, as well as maintaining structural integrity of the nucleus, they influence chromatin organization, DNA transcription, repair, and replication.1
Vertebrate cells express two classes of lamins, A and B, grouped based on sequence homology. Lamins A and C (A-types) are splice isoforms encoded by the same gene, while the B-type lamins are the products of different genes.1
Over the past two decades many disorders have been linked to LMNA variants, collectively termed laminopathies. Four major disease categories have been described with overlapping features: striated muscle diseases, lipodystrophy syndromes, peripheral neuropathies, and accelerated aging (segmental progeroid) disorders.2
In contrast to LMNA, few reports have associated human disease with variants in B-type lamin genes. No pathogenic single-nucleotide variants in LMNB1 have been reported, although genomic duplications incorporating LMNB1 cause adult-onset leukodystrophy (MIM 169500).3 For LMNB2, a homozygous missense variant in a family with progressive myoclonic epilepsy and ataxia has been described.4 Enrichment of heterozygous LMNB2 variants in acquired partial lipodystrophy patients has also been reported.5
Microcephaly is a reduction in head size, defined by a head circumference at least three standard deviations below the mean for age and gender. The underlying reduction in brain volume has many etiologies: chromosomal, environmental, as well as single-gene syndromic and nonsyndromic causes. Primary microcephaly represents a recognizable monogenic form of microcephaly in which brain size is markedly reduced in the absence of other malformations and/or significant neurological deficits aside from intellectual disability.6 Many genes associated with centrosome and/or mitotic spindle function have been described, suggesting primary microcephaly to be a disorder of neural stem cell mitosis.7
Here we report the identification of recurrent heterozygous variants in LMNB1 and LMNB2, implicating the nuclear lamina in the etiology of microcephaly.
MATERIALS AND METHODS
Ethics statement
Ethical approval was obtained as follows: Scottish Multicentre Research Ethics Committee (05/MRE00/74); Deciphering Developmental Disorders (DDD) Study (10/H0305/83, Cambridge South REC; GEN/284/12 Republic of Ireland REC); 100,000 Genomes Project (100kGP), East of England–Cambridge South REC (14/EE/1112). Informed written consent obtained for all participating families. Parents provided written authorization for publication of clinical photographs.
Bioinformatics
DDD Study exome sequencing and variant calling were performed as previously described.8 De novo variants were identified using VASE (v0.2.4, https://github.com/david-a-parry/vase). For 100kGP, variants and Human Phenotype Ontology (HPO) terms were extracted from the Genomics England research environment.
For de novo variant calling criteria, structural modeling, and immunofluorescence experiment methodology, see Supplementary methods.
RESULTS
Identification of LMNB1 pathogenic variants
To identify novel microcephaly genes, DDD exome sequencing data were analyzed in a subset of 1056 trios and singletons where the proband had microcephaly (occipital frontal circumference [OFC] <−4 SD below age and sex-matched mean; ~43% cases primary, 57% secondary microcephaly). This identified two individuals (P1 and P2) with de novo variants in LMNB1 (Table S1, S2) resulting in substitution or deletion of the same amino acid residue (NM_005573.4:c.97A>G, p.Lys33Glu and c.97_99del, p.Lys33del). P3 was also heterozygous for the same c.97_99del variant but not confirmed de novo as the family was lost to follow up. In the 100kGP cohort further variants in LMNB1 were identified in microcephalic patients, with P9 and P11 also having the same de novo c.97A>G variant as P1 (Table S3). An additional recurrent substitution (c.269G>C, p.Arg90Pro) was identified in P10 and P13. This was confirmed to be de novo in P10; however, no parental data were available for P13.
In total, three recurrent LMNB1 variants were identified in seven individuals (Fig. 1a), none of which were reported in gnomAD (v2.1). All were at residues conserved in all lamin metazoan homologs (Fig. 1b) and predicted damaging by multiple in silico tools (Table S4).
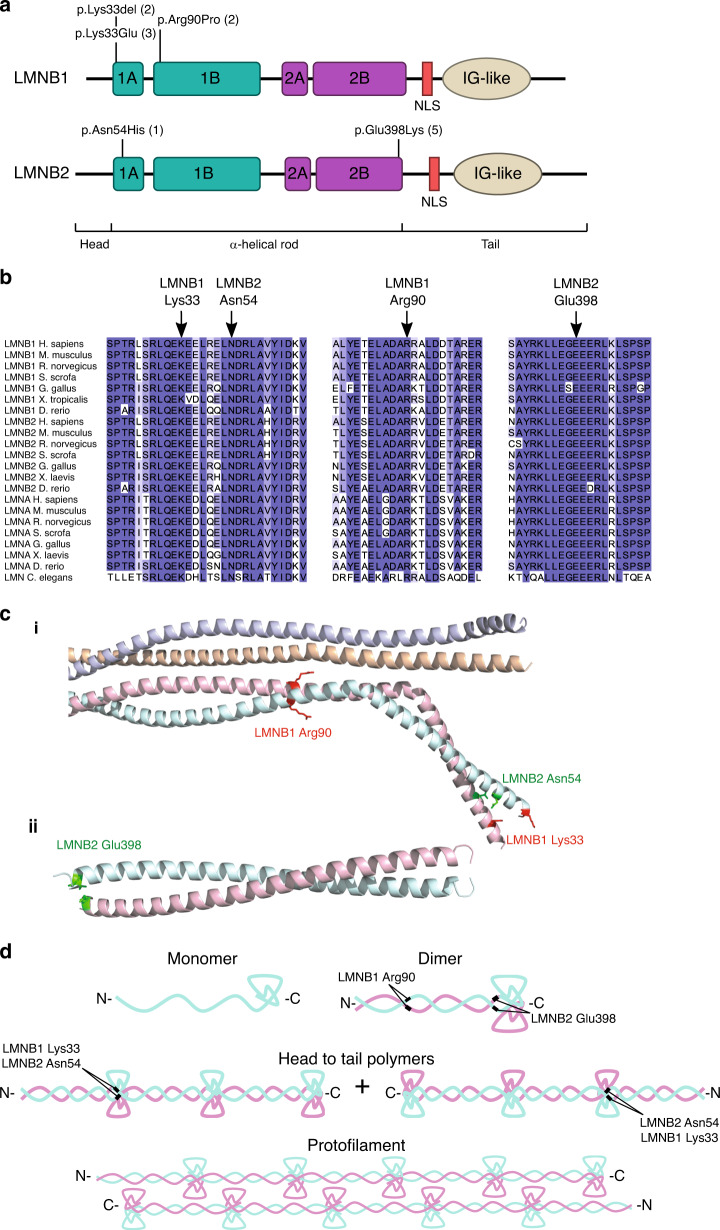
Fig. 1. Pathogenic LMNB1 and LMNB2 variants occur recurrently at highly conserved residues at intra- and interdimer filament interfaces.
(a) Schematic of B-type lamin protein structure. Locations of variants identified in microcephaly patients indicated, number of occurrences in parentheses. The ɑ-helical rod comprises coiled-coil domains 1A, 1B and 2A, 2C, indicated by green and purple boxes respectively. IG-like, immunoglobulin-like domain; NLS, nuclear localization signal. (b) Variants alter residues conserved in all vertebrate and invertebrate lamins. Lamins are specific to metazoa and the number of lamin genes has increased during evolution, with a single lamin gene in C. elegans compared with three mammalian lamin genes. Orthologs of both A- and B-type lamins aligned using Clustal X and colored by percent identity using Jalview. (c) Location of altered residues within lamin structures. While LMNB1/2 structures covering the mutated residues have not been published, as all residues are conserved with LMNA, we could use crystal structures of LMNA with Protein Data Bank (PDB) accessions 6JLB (i) and 1X8Y (ii) for molecular modeling and visualization. The upper structures shows a LMNA homotetramer, with the mutated residues highlighted on one of the constituent homodimers, while the bottom structure is of a LMNA homodimer, as no higher-order structure is available for this region. (d) Lamin filaments are assembled in a hierarchical fashion. First, lamin proteins form dimers, which further assemble into head to tail polymers. These polymers then laterally assemble in an antiparallel fashion to form lamin protofilaments.
LMNB2 variants associated with microcephaly
Following identification of variants in LMNB1, we assessed whether variants in its close homolog LMNB2 were also associated with microcephaly. We identified four microcephalic individuals from the DDD cohort (P4, P5, P6, P7) and one individual from the 100kGP study (P12) with the same variant in LMNB2 (NM_032737.4: c.1192G>A, p.Glu398Lys) (Tables S1–S3). For cases P4, P5, and P12, the variant was established to be de novo, while in P7, it had been inherited from the proband’s mosaic, clinically unaffected, mother (Fig. S1). Finally, another case was identified with a distinct missense de novo variant (c.160A>C, p.Asn54His). These variants were absent from gnomAD (v2.1), at highly conserved residues (Fig. 1b) and predicted damaging (Table S4).
In total, in 13 microcephalic individuals we identified 5 separate variants absent from the general population, 9 of which were established to be de novo events, and 4 of which had occurred recurrently. Taken together, this provided strong genetic evidence for heterozygous variants in LMNB1 and LNMB2 as a cause of microcephaly.
Clinical phenotype of LMNB1/2 individuals
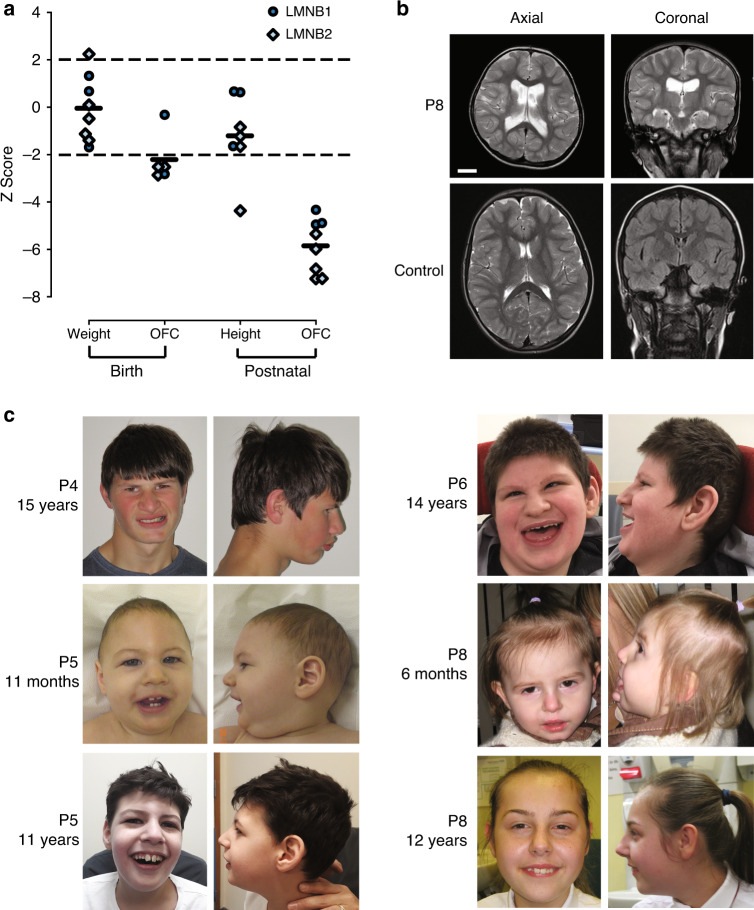
All LMNB1/2 cases had severe microcephaly (OFC −5.85 ± 1.14 SD, Fig. 2a, Table S5), evident from birth in all but one case. No intrauterine growth retardation was evident, with birth weights within normal range. Postnatally, height also remained within normal ranges for most cases. Global developmental delay of varying severity was evident and seizures present in four cases (Tables S1, S3). Major malformations were not reported. Neuroimaging demonstrated a structurally normal brain, without evidence of abnormal neuronal migration (Fig. 2b). In two cases gyral simplification was noted accompanied by global reduction in white matter volume alongside increased extra-axial spaces and enlarged ventricles. In P8, where serial scans were available, this was nonprogressive. Facially, aside from a sloping forehead in some individuals, a syndromic diagnosis was not evident, nor was a consistent facial gestalt observed (Fig. 2c).
Fig. 2. Pathogenic LMNB1 or LMNB2 variants in patients with primary microcephaly.
(a) Growth parameters of individuals with pathogenic LMNB1/2 variants plotted as Z-scores (standard deviations from the age and sex-matched mean). Black bars, mean Z-score. Dashed lines, 95% confidence interval for general population. (b) Neuroimaging demonstrates reduced cortical size with simplified gyri. Ventricular dilatation reflects accompanying global reduction in white matter. Comparison with age-matched control. Scale bar 2 cm. Axial images, T2-weighted. Coronal, T2 (P8), fluid-attenuated inversion recovery (FLAIR) (control). (c) Photographs of individuals P4, P5, P6, and P8. Written consent obtained from families for photography. OFC occipital frontal circumference.
Predicted consequences for lamin B1/B2 polymer assembly
While no LMNB1/2 protein structures are available that cover regions where the variants occur, all mutated residues are conserved with LMNA. Therefore, LMNA crystal structures could be used to model the substitutions. Each variant alters a residue in a coiled-coil segment of the α-helical rod domain of LMNB1/2 (Fig. 1a). LMNB1 Lys33 and LMNB2 Asn54 lie at the intradimer interface of coil 1A, LMNB1 Arg90 is at the interdimer interface of coil 1B, and LMNB2 Glu398 is at the intradimer interface of coil 2B. Substitution or deletion of these residues would therefore likely interfere with dimer or filament assembly. Notably, both p.Lys33Glu and p.Glu398Lys result in mutated residues with opposite electrostatic charges to the wild-type residue. LMNA Glu383, corresponding to LMNB2 Glu398, is established to separate the two lamin chains of the dimer at the end of the rod domain, where it repels Glu383 from the other chain.9 Therefore the heterozygous LMNB2 p.Glu398Lys substitution would result in a subset of mismatched dimers in which, instead, interaction of the two chains would be aberrantly stabilized, with a positively charged mutant lysine residue forming ionic interactions with the negatively charged wild-type glutamate. Molecular modeling with FoldX predicts this substitution to strongly stabilize the interdimer interaction (ΔΔG of −2.0 kcal/mol, Table S6), in keeping with a proposed stabilizing mechanism for pathogenic LMNA variants.10
Evidence for pathogenicity can also be drawn from the observation that all mutated codons are conserved in lamin A/C paralogs. Indeed, three of the LMNB variants (Lys33, Asn54, Arg90) correspond to residues in LMNA that are altered in lamin A/C associated disorders.11 As substitution of these sites in LMNA are disease causing, this would also be expected to be the case in LMNB1/B2. Furthermore, LMNB1 p.Lys33del has already been modeled in the ancestral C.elegans LMN (a protein most homologous to B-type lamins1) where it disrupts protofilament assembly in vitro and causes nuclear aggregates of lamin in vivo.12
Lastly, we examined the cellular distribution of GFP-lamin B fusion proteins containing microcephaly variants, in a similar manner to that done previously for LMNA variants.13 Cells expressing the LMNB1/B2 variants frequently contained nuclear aggregates and/or significantly altered nuclear morphology in comparison to cells expressing wild-type GFP-LMNB1/2 (Fig. S2, Tables S8 and S9). While GFP-LMNB1/2 proteins were expressed above endogenous levels, both aggregates and altered nuclear shape have previously been reported for pathogenic lamin A/C variants, both when overexpressed13 and at endogenous levels,12,14 leading us to conclude that lamin B variants may also disrupt lamin assembly in cells.
DISCUSSION
LMNB primary microcephaly: a new laminopathy
Here we identify recurrent and de novo heterozygous variants in LMNB1 and LMNB2 in eight microcephaly patients from the DDD cohort. The additional five cases ascertained from 100kGP on the basis of de novo or recurrent variants in LMNB1/2 also had microcephaly, confirming the phenotype–genotype link. A severe nonsyndromic microcephaly without other malformations was consistently present. In keeping with primary microcephaly, all but one case had a reduced OFC evident at birth, and neuroimaging demonstrated a structurally normal small brain with/without a simplified gyral pattern. Dilated ventricles and increased axial spaces have been previously documented,15,16 as have seizures in a minority of primary microcephaly cases. Primary microcephaly is usually accompanied by mild/moderate intellectual disability, although for LMNB1/2 global developmental delay varied substantially between cases, classified as severe in four of eight DDD cases, with speech not attained in three older children.
The LMNB1/B2 microcephaly phenotype is distinct from previously described lamin A/C laminopathies, pointing to different cellular and developmental roles for lamin B proteins, despite all lamins acting as major structural components of the nuclear envelope. The human microcephaly is in keeping with the previously reported mouse knockout model for Lmnb1 in which cerebral cortical size was markedly reduced.17 However, in both the Lmnb1−/− and Lmnb2−/− mice, (where brain size was not reduced at birth), there were neuronal migration defects resulting in abnormal layering of the cerebral cortex,18 suggesting potential discordance with the human phenotype and neuroimaging findings. This may be accounted for however by a different mechanism in human cases where variants are heterozygous rather than biallelic null. The pathogenic variants reported here are most likely dominant negative rather than haploinsufficient given that microcephaly is not present in the Lmnb1+/− mouse. The recurrent nature of the pathogenic variants at specific residues and absence of truncating variants favors gain of function or dominant negative mechanisms. Furthermore, substitutions in corresponding residues in lamin A/C do not destabilize the protein, rather disrupt lamin filament formation,19 a common scenario by which point variants in multimeric proteins can act in a dominant negative manner to perturb macromolecular complexes.20
LMNB1 and LMNB2 have overlapping but distinctive cellular and developmental roles.1 So it is striking that pathogenic variants in both cause primary microcephaly, suggesting an etiology in which a shared function is disrupted. Many primary microcephaly proteins are involved in mitosis, and often encode centrosomal or spindle components that when mutated result in altered spindle orientations disrupting symmetric cell division dynamics in neural progenitors.7 This reduces neuronal cell number generated during neurogenesis. B-type lamins associate with the mitotic spindle21 with defects in spindle orientation in neuronal precursor cells reported in Lmnb1 and Lmnb2 knockout mice,17 suggesting this could be the mechanism underlying microcephaly. Other mitotic processes may also be perturbed, given the involvement of LMNs in nuclear envelope reassembly. However, the potential for LMNB1/2 variants to alter the physical properties of lamin dimers and filaments (Fig. 1c) should also be considered, given that several of the variants are seen to impact on cellular assembly of nuclear lamina, and that the equivalent pathogenic LMNA variant to LMNB1 p.Lys33del results in fragile nuclei more prone to mechanical deformation.22 Such fragility in neuronal precursors would enhance susceptibility to mechanical stresses of interkinetic nuclear migration and neuronal migration and lead to increased cell death during brain development.23
Notably, several genes encoding components of the nuclear pore have also been implicated in microcephaly. While nuclear pores are also important components of the nuclear envelope, these are generally clinically distinct disorders, often associated with nephrotic syndrome (Galloway–Mowat syndrome24,25) or with a progressive microcephaly/encephalopathy.26,27 As well, nuclear–cytoplasmic transport has been demonstrated to be impaired,27 so the microcephaly in nuclear pore disorders could have a different mechanistic basis.
In conclusion, our findings establish heterozygous variants in LMNB1 and LMNB2 as causes of primary microcephaly, implicating the nuclear lamina in its etiology and defining a novel form of laminopathy. Future in vitro and in vivo studies will be important to provide further evidence for causality of the variants reported here, and to shed light on how they cause a laminopathy distinct from those previously described.
Supplementary information
Acknowledgements
We thank the families for their participation, Katya Lyulcheva-Bennett and Margaret MacDonald for assistance with clinical case characterization, Laura Murphy and IGMM imaging core for support with image analysis, and Andrea Robertson for technical assistance. Work was supported by the European Union’s Horizon 2020 research and innovation program European Research Council (ERC) Advanced Grant (grant agreement 788093); and by Medical Research Council (MRC) Unit core grant (U127580972). J.A.M., MRC Career Development Award (MR/M02122X/1) and Lister Institute Research Prize Fellowship; S.G.-H. Health Education England Genomics Education Programme research fellowship. The DDD Study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER (http://decipher.sanger.ac.uk), funded by Wellcome. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement. This research was also made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrew P. Jackson, Email: Andrew.Jackson@igmm.ed.ac.uk.
Genomics England Research Consortium:
J. C. Ambrose, P. Arumugam, E. L. Baple, M. Bleda, F. Boardman-Pretty, J. M. Boissiere, C. R. Boustred, H. Brittain, M. J. Caulfield, G. C. Chan, C. E. H. Craig, L. C. Daugherty, A. de Burca, A. Devereau, G. Elgar, R. E. Foulger, T. Fowler, P. Furió-Tarí, A. Giess, J. M. Hackett, D. Halai, A. Hamblin, S. Henderson, J. E. Holman, T. J. P. Hubbard, K. Ibáñez, R. Jackson, L. J. Jones, D. Kasperaviciute, M. Kayikci, A. Kousathanas, L. Lahnstein, K. Lawson, S. E. A. Leigh, I. U. S. Leong, F. J. Lopez, F. Maleady-Crowe, J. Mason, E. M. McDonagh, L. Moutsianas, M. Mueller, N. Murugaesu, A. C. Need, C. A. Odhams, A. Orioli, C. Patch, D. Perez-Gil, M. B. Pereira, D. Polychronopoulos, J. Pullinger, T. Rahim, A. Rendon, P. Riesgo-Ferreiro, T. Rogers, M. Ryten, K. Savage, K. Sawant, R. H. Scott, A. Siddiq, A. Sieghart, D. Smedley, K. R. Smith, S. C. Smith, A. Sosinsky, W. Spooner, H. E. Stevens, A. Stuckey, R. Sultana, M. Tanguy, E. R. A. Thomas, S. R. Thompson, C. Tregidgo, A. Tucci, E. Walsh, S. A. Watters, M. J. Welland, E. Williams, K. Witkowska, S. M. Wood, and M. Zarowiecki
Supplementary information
The online version of this article (10.1038/s41436-020-00980-3) contains supplementary material, which is available to authorized users.
References
- 1.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 2.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padiath QS, Saigoh K, Schiffmann R, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 4.Damiano JA, Afawi Z, Bahlo M, et al. Mutation of the nuclear lamin gene LMNB2 in progressive myoclonus epilepsy with early ataxia. Hum Mol Genet. 2015;24:4483–4490. doi: 10.1093/hmg/ddv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegele RA, Cao H, Liu DM, et al. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am J Hum Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verloes A, Drunat S, Gressens P, Passemard S. Primary autosomal recessive microcephalies and Seckel syndrome spectrum disorders—archived chapter, for historical reference only. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle: University of Washington; 1993. [PubMed]
- 7.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald TW, Gerety SS, Jones WD, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. Crystal structure of the human Lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J, Jo I, Kang S, et al. Structural basis for lamin assembly at the molecular level. Nat Commun. 2019;10:3757. doi: 10.1038/s41467-019-11684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer T, Misteli T. The lamin protein family. Genome Biol. 2011;12:1–14. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bank EM, Ben-Harush K, Wiesel-Motiuk N, et al. A laminopathic mutation disrupting lamin filament assembly causes disease-like phenotypes in Caenorhabditis elegans. Mol Biol Cell. 2011;22:2716–2728. doi: 10.1091/mbc.e11-01-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hübner S, Eam JE, Wagstaff KM, Jans DA. Quantitative analysis of localization and nuclear aggregate formation induced by GFP-lamin A mutant proteins in living HeLa cells. J Cell Biochem. 2006;98:810–826. doi: 10.1002/jcb.20791. [DOI] [PubMed] [Google Scholar]
- 14.Steele-Stallard HB, Pinton L, Sarcar S, et al. Modeling skeletal muscle laminopathies using human induced pluripotent stem cells carrying pathogenic LMNA mutations. Front Physiol. 2018;9:1332. doi: 10.3389/fphys.2018.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bond J, Roberts E, Mochida GH, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 16.Shaheen R, Maddirevula S, Ewida N. Genomic and phenotypic delineation of congenital microcephaly. Genet Med. 2019;21:545–552. doi: 10.1038/s41436-018-0140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, Sharov AA, McDole K, et al. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffinier C, Chang SY, Nobumori C, et al. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci U S A. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muchir A, Medioni J, Laluc M, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 20.Bergendahl LT, Gerasimavicius L, Miles J, et al. The role of protein complexes in human genetic disease. Protein Sci. 2019;28:1400–1411. doi: 10.1002/pro.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai M-Y, Wang S, Heidinger JM, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 22.Zwerger M, Jaalouk DE, Lombardi ML, et al. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet. 2013;22:2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen NY, Yang Y, Weston TA, et al. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc Natl Acad Sci U S A. 2019;116:25870–25879. doi: 10.1073/pnas.1917225116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosti RO, Sotak BN, Bielas SL, et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway–Mowat syndrome. J Med Genet. 2017;54:399–403. doi: 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- 25.Fujita A, Tsukaguchi H, Koshimizu E, et al. Homozygous splicing mutation in NUP133 causes Galloway–Mowat syndrome. Ann Neurol. 2018;84:814–828. doi: 10.1002/ana.25370. [DOI] [PubMed] [Google Scholar]
- 26.Shamseldin HE, Makhseed N, Ibrahim N, et al. NUP214 deficiency causes severe encephalopathy and microcephaly in humans. Hum Genet. 2019;138:221–229. doi: 10.1007/s00439-019-01979-w. [DOI] [PubMed] [Google Scholar]
- 27.Fichtman B, Harel T, Biran N, et al. Pathogenic variants in NUP214 cause “plugged” nuclear pore channels and acute febrile encephalopathy. Am J Hum Genet. 2019;105:48–64. doi: 10.1016/j.ajhg.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.