Abstract
WRKY transcription factors (TFs) are of great importance in plant responses to different abiotic stresses. However, research on their roles in the regulation of thermotolerance remains limited. Here, we investigated the function of LlWRKY39 in the thermotolerance of lily (Lilium longiflorum ‘white heaven’). According to multiple alignment analyses, LlWRKY39 is in the WRKY IId subclass and contains a potential calmodulin (CaM)-binding domain. Further analysis has shown that LlCaM3 interacts with LlWRKY39 by binding to its CaM-binding domain, and this interaction depends on Ca2+. LlWRKY39 was induced by heat stress (HS), and the LlWRKY39-GFP fusion protein was detected in the nucleus. The thermotolerance of lily and Arabidopsis was increased with the ectopic overexpression of LlWRKY39. The expression of heat-related genes AtHSFA1, AtHSFA2, AtMBF1c, AtGolS1, AtDREB2A, AtWRKY39, and AtHSP101 was significantly elevated in transgenic Arabidopsis lines, which might have promoted an increase in thermotolerance. Then, the promoter of LlMBF1c was isolated from lily, and LlWRKY39 was found to bind to the conserved W-box element in its promoter to activate its activity, suggesting that LlWRKY39 is an upstream regulator of LlMBF1c. In addition, a dual-luciferase reporter assay showed that via protein interaction, LlCaM3 negatively affected LlWRKY39 in the transcriptional activation of LlMBF1c, which might be an important feedback regulation pathway to balance the LlWRKY39-mediated heat stress response (HSR). Collectively, these results imply that LlWRKY39 might participate in the HSR as an important regulator through Ca2+-CaM and multiprotein bridging factor pathways.
Subject terms: Plant sciences, Plant stress responses, Heat
Introduction
High temperature is one of the unfavorable factors affecting the growth of plants, generally impairing photosynthetic activity and negatively affecting cell division and growth1. Extreme high temperatures may result in a series of morphoanatomical and physiochemical changes in plant cells and even lead to severe economic losses in crops and other economically important plants2,3. Plants must produce various defense mechanisms against high temperature, including the accumulation of heat shock proteins (HSPs) and complex regulatory networks as established by transcription factors (TFs)4,5.
Lily (Lilium spp.) is one of the most popular cut flower products worldwide because of its attractive shape and color6. Lily adapts well to cool conditions but is sensitive to high temperatures (>30 °C), which not only reduces the quality of cut flowers but also leads to the degeneration of the bulb7. However, high temperatures will become an unavoidable environmental stress factor in the future because of the irreversible trend in global warming8,9. Therefore, an understanding of the HSR mechanisms of lily under HS is essential to improve the thermotolerance of lily.
TFs play major roles in increasing the stress tolerance of plants since they can regulate critical downstream genes by binding to cis-elements in gene promoters10,11. In the HSR, HS transcription factors (HSFs) can directly regulate the expression of downstream genes by binding to HS elements (HSEs; nGAAnnTTCn) in the promoters of downstream genes in response to HS12,13. Currently, most studies on thermotolerance in lily focus on HSFs. The overexpression of LlHSFA1 and LlHSFA2 from lily in Arabidopsis can enhance the thermotolerance of transgenic lines6,7,14. Two HSFA3 homologs of lily, LlHSFA3A and LlHSFA3B, increase the thermotolerance of transgenic Arabidopsis plants, possibly through a proline-mediated pathway15. In addition, LlHSFA3A and LlHSFA3B from lily can form a regulatory mechanism involving heat-inducible alternative splicing to sustain balance in the HSR16. Furthermore, lily LlDREB2B, a member of the DREB subfamily of the ERF/AP2 TF family, can increase the basal thermotolerance (BT) and acquired thermotolerance (AT) of transgenic Arabidopsis17.
Multiprotein bridging factor 1 (MBF1) is a highly conserved transcriptional coactivator with various forms involved in the regulation of diverse processes, such as oxidative stress, hormone-regulated seed germination, and translation18–20. In Arabidopsis, MBF1 cofactors are encoded by three genes: AtMBF1a (AT2G42680), AtMBF1b (AT3G58680), and AtMBF1c (AT3G24500)21. Among these genes, AtMBF1c is related to thermotolerance and functions upstream of salicylic acid (SA), ethylene, and trehalose signaling22,23. A previous study reported that AtMBF1c is regulated by AtHsfA1 cofactors because an hsfa1 quadruple mutant shows suppressed expression of AtMBF1c during HS24. Despite much information available indicating how plant MBF1c genes respond to HS, many questions about the relationship between MBF1c and other important TFs, e.g., WRKY TFs, remain.
WRKY TFs can participate in multiple adverse responses during plant growth and development25–27. The typical feature of WRKY TFs is the WRKY domain, which contains an invariant WRKYGQK sequence and a zinc finger motif (CX4–5CX22–23HXH or CX7CX23HXC)28,29. The WRKY superfamily is classified into three groups based on the number of conserved WRKY signatures (two WRKY sequences in group I and one WRKY sequence in groups II and III) and the composition of the zinc finger motif in which the zinc finger motif of groups I and II is CX4–5CX22–23HXH and the zinc finger motif of group III is CX7CX23HXC22. Group II is further classified into five subgroups (IIa to IIe) based on different conserved short motifs22,28. WRKY TFs can recognize and bind to W-box elements (TTGACC/T) in the promoters of resistance-related genes. The core sequence TGAC is essential for WRKY recognition, as indicated by different binding experiments30,31. This type of binding can regulate the expression of target stress genes and thus increase plant stress tolerance29,32.
In recent years, increasing evidence has shown that WRKYs are related to thermotolerance33–36. However, studies on WRKY TFs are primarily focused on crops or model plants such as rice, wheat, and Arabidopsis, and little research has been carried out on lily. In A. thaliana, the group II WRKY protein AtWRKY39 may play a positive role in thermotolerance by regulating the cooperation between the SA- and JA-activated signaling pathways34. Here, we identify a WRKY-IId factor in lily, LlWRKY39, which was induced by HS and can interact with LlCaM3 in a Ca2+-dependent manner. The thermotolerance of transgenic plants increased with the overexpression of LlWRKY39. Further analysis indicated that LlWRKY39 can bind to the promoter of LlMBF1c and activate its expression. However, the interaction between LlCaM3 and LlWRKY39 negatively affected the transactivation of LlMBF1c induced by LlWRKY39, which may imply feedback regulation of LlWRKY39 to maintain a balance during the HSR. The results of this study may help to reveal the biological function and mechanism of LlWRKY39 under HS and provide an important theoretical foundation for further perfecting the HS signal transduction network regulated by lily TFs.
Results
LlWRKY39 is a heat-inducible member of the WRKY group IId family of transcription factors
We searched AtWRKY39 from the Arabidopsis database with the online TAIR tool (https://www.arabidopsis.org/) and selected putative WRKY39 from the pollen transcriptome database of ‘little kiss’37. Then, we cloned putative WRKY39 from ‘white heaven’. Thus, the candidate was designated LlWRKY39. The full-length cDNA sequence of LlWRKY39 contains an 858-bp open reading frame (ORF) that encodes a 285-amino acid protein. The phylogenetic tree including all WRKY proteins in Arabidopsis showed that LlWRKY39 is closely related to AtWRKY39, AtWRKY74, and AtWRKY21 (Supplementary Fig. S1), which suggests that LlWRKY39 is a member of the WRKY group IId family. To identify the basic characteristics of LlWRKY39, amino acid sequence alignment of LlWRKY39, AtWRKY39, AtWRKY74, and AtWRKY21 was performed. The sequences exhibited a similar signature, which included a WRKYGQK sequence, one zinc-binding motif C–X5–C–X23–H–X1–H, one HARF (RTGHARFRR[A/G]P) motif, a nuclear localization site (NLS), and a potential calmodulin (CaM)-binding domain (CBD) (Fig. 1b). Then, we investigated the evolutionary relationship between LlWRKY39 and WRKY39 factors in different plants, including Arabidopsis, tomato, apple, wheat, brachypodium, date, oil palm, and pineapple. The phylogenetic results indicated that the closest relationship was established between LlWRKY39 and pineapple AcWRKY39 (Fig. 1c), which are both noncereal monocot species.
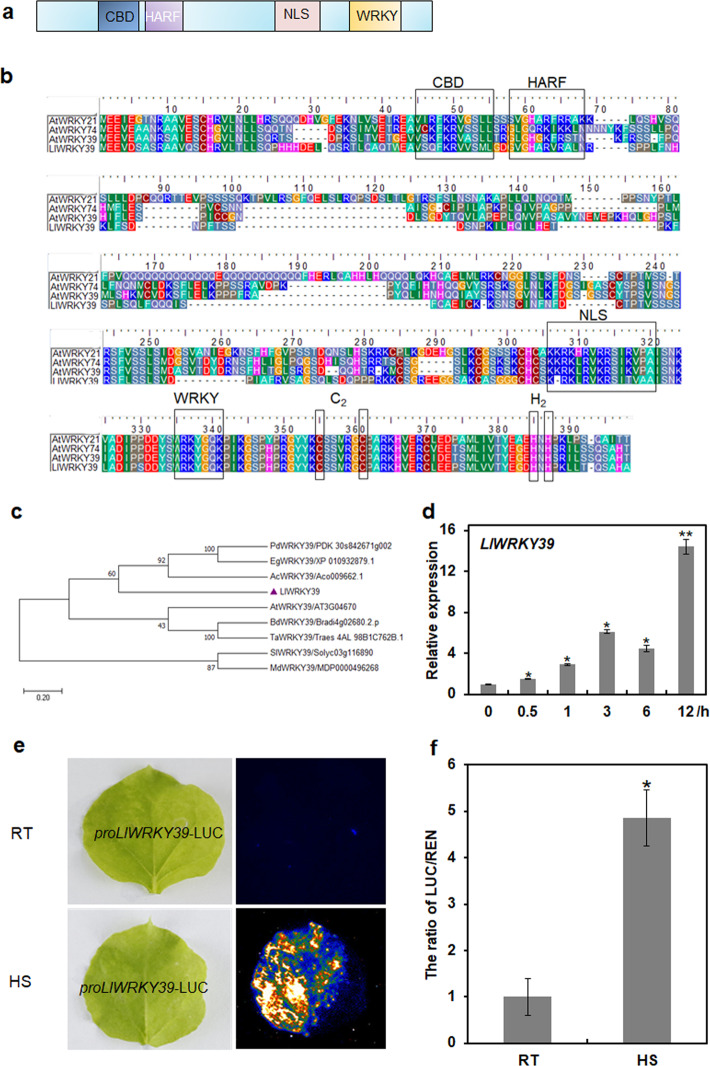
Fig. 1. LlWRKY39 is a heat-inducible member of the WRKY group IId of transcription factors.
a Structural diagram of the WRKY IId transcription factor. b Multiple alignments of LlWRKY39 with AtWRKY39 (AT3G04670), AtWRKY74 (AT5G28650), and AtWRKY21 (AT2G30590) using the Clustal W algorithm with default parameters. The approximate 60-amino acid WRKY domain, CaM-binding domain (CBD), HARF motif, and nuclear localization site (NLS) are indicated by black boxes. c Phylogenetic relationship of LlWRKY39 with other plant WRKY proteins was determined by the neighbor-joining method with 500 bootstrap replicates. LlWRKY39 is highlighted with a purple triangle. The following abbreviations are used to indicate the scientific names of plants with WRKY: AC ananas comosus, EG elaeis guineensis, PD phoenix dactylifera, BD brachypodium distachyon, TA triticum aestivum, AT arabidopsis thaliana, MD malus domestics, Sl solanum lycopersicum. d Relative expression of LlWRKY39 in lily leaves under HS for different lengths of time. Each bar indicates the mean ± SD of three repeated experiments (*P < 0.05 and **P < 0.01, Student’s t test). e Analysis of the promoter activity in N. benthamiana leaves under room temperature (RT) and HS (37 °C 2 h). f The relative value of LUC/REN. The ratio of LUC/REN at RT was set to 1 for normalization. All the values represent the mean ± SD of three repeated experiments (*P < 0.05, Student’s t test)
LlWRKY39 can be rapidly induced under HS at 37 °C for 0.5 h, and following prolonged HS for 12 h, the expression of LlWRKY39 is enhanced significantly (Fig. 1d). The expression results suggested that LlWRKY39 is induced by high temperature over a long period. To further examine the underlying mechanism of LlWRKY39 under HS, we isolated and analyzed the promoter of LlWRKY39. Different kinds of cis-elements were found in the promoter of LlWRKY39, such as light-responsive elements (I-BOX and GATA-BOX), an element for pollen-specific expression (POLLEN1), a binding site for WRKY TFs (W-box), CCAAT-BOX, and others, indicating that LlWRKY39 may be regulated by different types of TFs (Supplementary Table S1). Among these motifs, CCAAT-BOX is related to HS, and it can act cooperatively with HSEs to increase HS promoter activity38. To identify the promoter activity of LlWRKY39 under HS, we conducted a transient luciferase reporter assay with Nicotiana benthamiana leaves. The LUC signal increased profoundly after subjection to HS, although under normal conditions, the LUC signal was very low, which implied that HS activated the promoter activity of LlWRKY39 (Fig. 1e, f). Thus, these results indicate that LlWRKY39 is a heat-inducible WRKY group IId factor.
LlCaM3 interacts with LlWRKY39 by binding the CBD
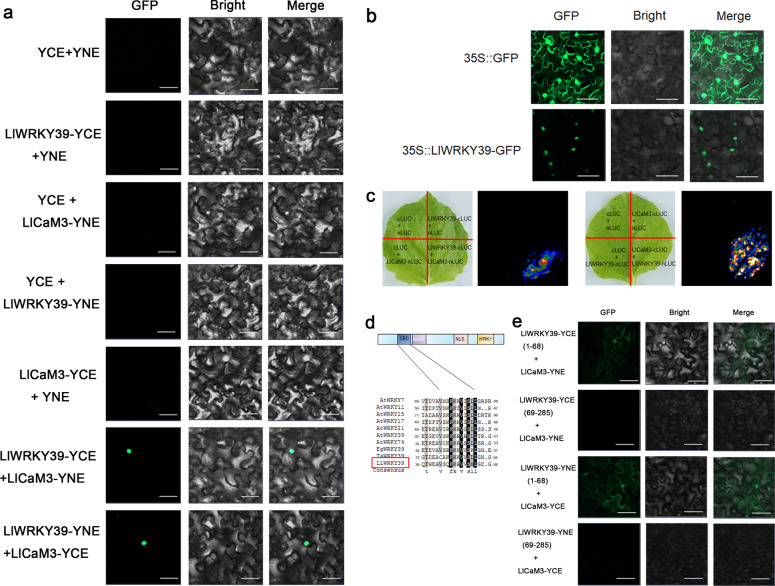
In the protein sequence assay, a potential CBD was found in LlWRKY39 (Fig. 1b), which suggests that LlWRKY39 is a CaM-binding protein (CBP). Therefore, we evaluated whether LlWRKY39 interacted with a heat-inducible CaM, LlCaM3, from lily (Supplementary Fig. S2). Bimolecular fluorescence complementation (BIFC) showed that the fluorescence generated upon LlWRKY39 interacting with LlCaM3 was emitted only in the nucleus (Fig. 2a). A previous study showed that LlCaM3 is a cytoplasmic and nuclear protein39. Here, we found that the fluorescence of the LlWRKY39-GFP fusion protein appeared only in the nucleus (Fig. 2b), which might explain why the fluorescence emitted by LlWRKY39 interacting with LlCaM3 appeared only in the nucleus. Similarly, firefly luciferase complementation imaging (FLC) assays also showed that LlWRKY39 interacted with LlCaM3 (Fig. 2c). However, compared with that of homologous genes, the CBD of LlWRKY39 is not highly conserved (Fig. 2d). Therefore, we sought to determine whether the interaction between LlWRKY39 and LlCaM3 depends on this domain. We truncated LlWRKY39 into two parts, one segment with the CBD and one without the CBD. In the BIFC assay, LlWRKY39 with the CBD interacted with LlCaM3, but LlWRKY39 lacking the CBD did not (Fig. 2e). Notably, the interaction signal was observed throughout the cytoplasm and nucleus, which may be explained by the deletion of the NLS of LlWRKY39 and LlCaM3 in the cytoplasm and nucleus30. In a split-ubiquitin assay with yeast cells, similar results were observed (Supplementary Fig. S3), which implies that the interaction between LlCaM3 and LlWRKY39 depends on the CBD region.
Fig. 2. Interaction between LlWRKY39 and LlCaM3 by binding the CaM-binding domain.
a BIFC assay. Fluorescence signals were observed using a confocal microscope. Bars = 50 μm. b Transient expression of LlWRKY39 in N. benthamiana leaves. Bar = 50 μm. c FLC assay. LUC signals were observed using a CCD camera. d Alignment of the CBD among WRKY group IId members in Arabidopsis, oil palm, wheat, and lily. Identical amino acids are shaded in black and gray. e Interaction of the CBD of LlWRKY39 and LlCaM3 as determined by BIFC. Bar = 50 μm
LlCaM3–LlWRKY39 interaction depends on Ca2+
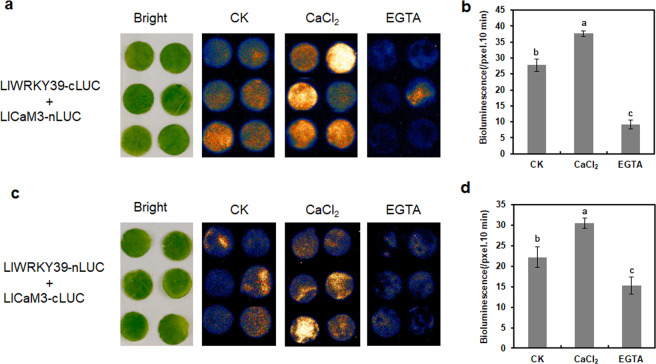
To determine whether the LlCaM3–LlWRKY39 interaction depended on Ca2+, we conducted a transient FLC assay with N. benthamiana leaves and CaCl2 and calcium ion chelator ethylene glycol tetraacetic acid (EGTA) treatments. The interaction signal in the leaves treated with CaCl2 was stronger than that in the leaves treated with water, but the interaction signal was significantly repressed in the leaves treated with EGTA, which implies that the LlCaM3–LlWRKY39 interaction might depend on Ca2+ (Fig. 3).
Fig. 3. Effects of CaCl2 and EGTA treatment on the interaction between LlWRKY39 and LlCaM3 in N. benthamiana leaves.
a LUC bioluminescence in N. benthamiana leaves coinfiltrated with mixed bacterial solutions of LlWRKY39-cLUC and LlCaM3-nLUC by different treatments. b, d LUC bioluminescence intensity was quantified using Andor Solis v15 software. c LUC bioluminescence in N. benthamiana leaves coinfiltrated with mixed bacterial solutions of LlWRKY39-nLUC and LlCaM3-cLUC by different treatments. Values represent the mean ± SD of three independent experiments. Significant differences are indicated by Student–Newman–Keuls test
Overexpression of LlWRKY39 increases the thermotolerance of transgenic plants
In a transient expression assay, the overexpression of LlWRKY39 in lily leaves did not affect their relative ion leakage under normal growth conditions. However, the relative ion leakage of ‘white heaven’ leaves in the control was significantly higher than that of leaves transformed with LlWRKY39 under HS, indicating that the transient expression of LlWRKY39 can protect the cells of lily from high temperature (Fig. 4a, b). Four homozygous T3 transgenic Arabidopsis lines were identified (Supplementary Fig. S4), and three transgenic Arabidopsis lines (OE-2, OE-4, and OE-5) were selected for BT and AT analyses. The survival rate of the WT plants was significantly lower than that of the three transgenic lines under both HS conditions (Fig. 4d, e), which showed that the overexpression of LlWRKY39 can increase the BT and AT of transgenic Arabidopsis. In addition, the expression levels of HS-inducible genes AtHSFA1, AtHSFA2, AtHSP101, AtDREB2A, AtMBF1c, AtGolS1, and AtWRKY39 increased significantly in transgenic Arabidopsis (Fig. 4f), which might facilitate the increase in thermotolerance of transgenic lines. Unexpectedly, the expression of AtHSFB2A, a transcriptional repressor40, was also significantly induced. In addition, the expression of AtHSP70, AtAPX1, AtAPX2, and AtHSFA3, which are the downstream genes of AtHSFA1, AtHSFA2, and AtDREB2A41–43, did not change significantly, suggesting that LlWRKY39 also participated in other pathways to control the expression of these genes. We also measured the expression level of genes such as LlHSFA1, LlHSFA2, and LlDREB2B in transiently overexpressed LlWRKY39 lily leaves. The qRT-PCR results showed that the expression levels of the LlHSFA1, LlHSFA2, and LlDREB2B genes were enhanced in lily leaves overexpressing LlWRKY39 (Supplementary Fig. S5).
Fig. 4. Thermotolerance assay in lily and transgenic Arabidopsis lines.
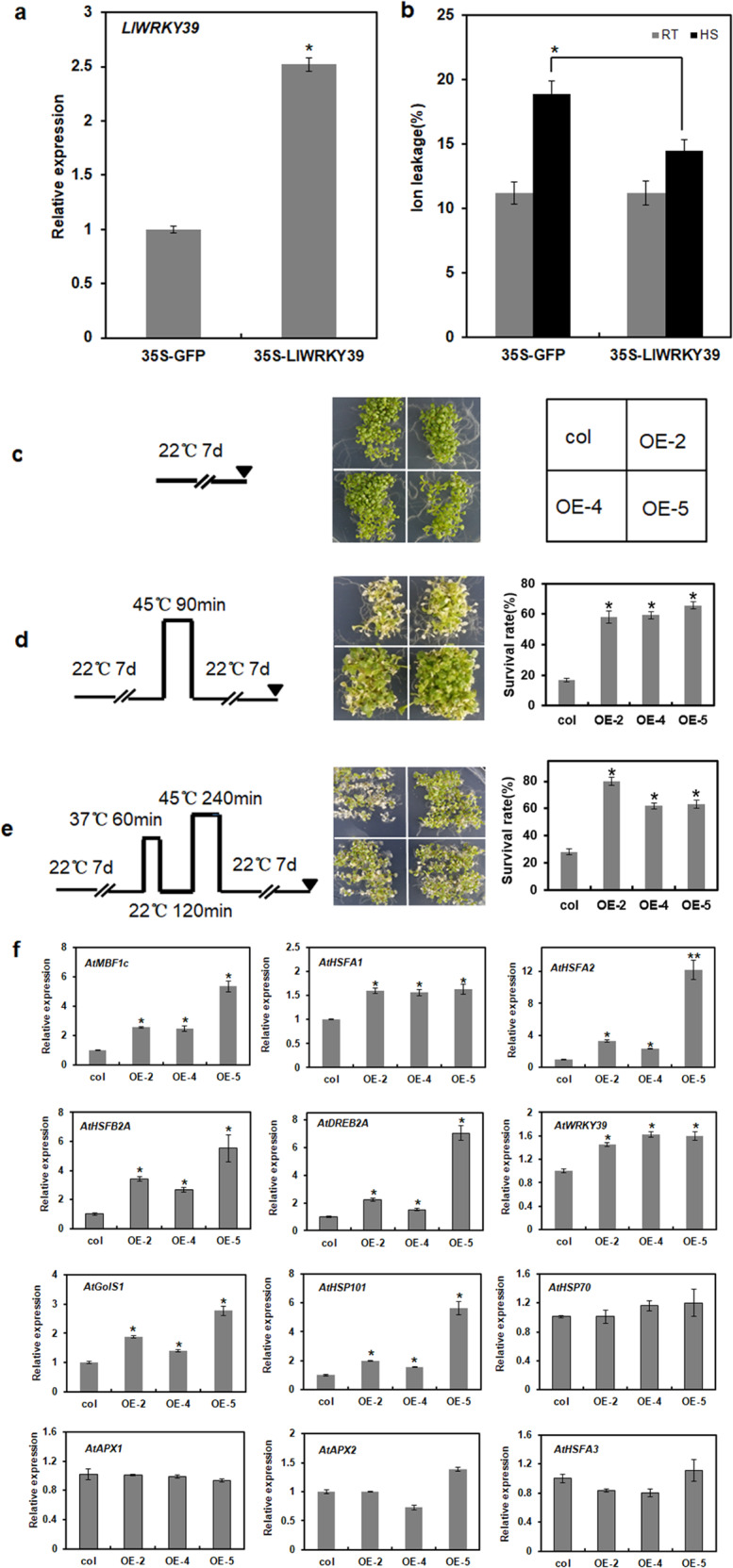
a The expression of LlWRKY39 in transient-overexpressing lily. b Relative ion leakage (%) of lily leaves under RT and HS (42 °C, 2 h). Each bar indicates the mean ± SD of three repeated experiments (t test, *P < 0.05). c Phenotypes of one-week-old seedlings before exposure to HS. d Phenotypes of one-week-old seedlings after the BT test. left: schematic representations of BT; right: survival rate of BT. e Phenotypes of one-week-old seedlings after the AT test. left: schematic representations of AT; right: survival rate of AT. A bar shows the mean of three independent experiments (*P < 0.05, t test). f Relative expression levels of HS-inducible genes in the transgenic plants. Each bar indicates the mean ± SD of three repeated experiments. Significant differences are indicated by t test (*P < 0.05 and **P < 0.01)
LlWRKY39 activates the expression of LlMBF1c
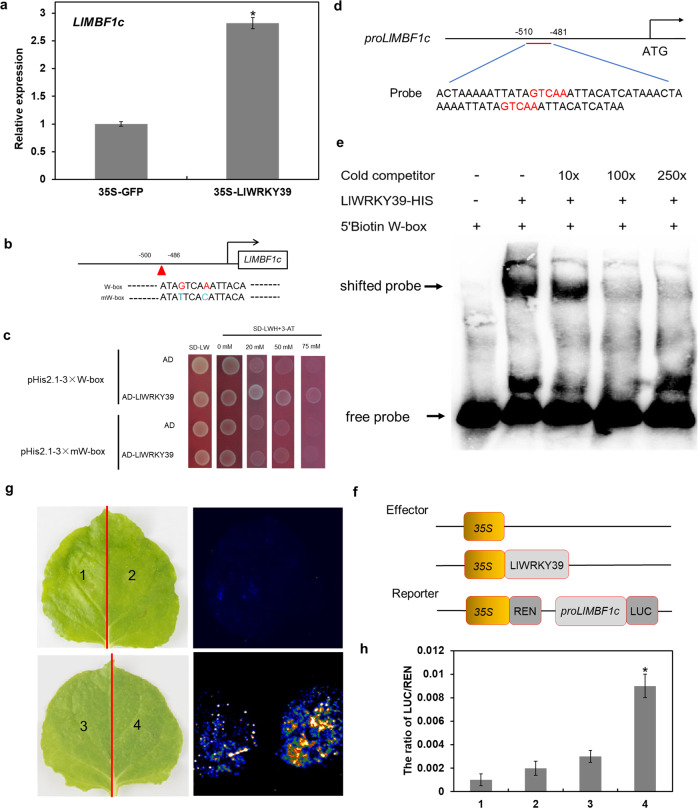
MBF1c is an extremely conserved transcriptional coactivator that plays an important role in the HSR22,23. Given that the overexpression of LlWRKY39 can increase the expression of AtMBF1c in Arabidopsis (Fig. 4f), Agrobacterium-mediated transient transformation was performed with lily leaves to verify whether the same regulation mode exists in lily. The overexpression of LlWRKY39 in lily activated the expression of LlMBF1c (Fig. 5a), although LlWRKY39 showed no transactivation activity in yeast cells (Supplementary Fig. S6). In addition, LlMBF1c and LlWRKY39 shared similar expression patterns under HS (Supplementary Fig. S7). To further identify the regulatory mechanism between LlWRKY39 and LlMBF1c, we isolated and analyzed the promoter of LlMBF1c. The LlMBF1c promoter contained various cis-elements, such as drought responsiveness elements (MYB2AT); light responsiveness elements (I-BOX and GATA-BOX); CCAAT-BOX, the binding site of the WRKY TF family (W-box); and others (Supplementary Table S2). Then, we conducted a yeast one-hybrid assay to confirm that LlWRKY39 binds to the W-box element in the promoter region (–500 to –486 bp) of LlMBF1c. The yeast cells cotransformed with LlWRKY39 and the W-box element of the LlMBF1c promoter survived on SD medium without Leu, Trp, and His (SD-LWH), even in the presence of 75 mM 3-amino-1,2,4-triazole (3-AT), which implies that LlWRKY39 has a high affinity for the W-box element (GTCAA). However, when the GTCAA sequence was mutated to TTCAC, LlWRKY39 failed to bind to it (Fig. 5c), implying that the G and A residues in GTCAA are essential for the recognition and combination of LlWRKY39. To further verify these results, we performed an electrophoretic mobility shift assay (EMSA). As shown in Fig. 5e, the LlWRKY39-HIS complex bound to the W-box element in the LlMBF1c promoter and produced a mobility shift, which implies that LlWRKY39 binds to the LlMBF1c promoter via the W-box to regulate its expression. The effector-reporter assay showed that the N. benthamiana leaves cotransformed with control combinations emitted a very low LUC signal, but the signal increased significantly after cotransformation with LlWRKY39 and proLlMBF1c-LUC, and the ratio of LUC/REN was also significantly higher than that of the control (Fig. 5g, h). Therefore, these results suggest that LlWRKY39 can activate the expression of LlMBF1c and that this activation may be achieved by directly binding the LlMBF1c promoter.
Fig. 5. LlWRKY39 activates the expression of LlMBF1c.
a Transient expression of LlWRKY39 in lily increased the expression of LlMBF1c. Each bar indicates the mean ± SD of three repeated experiments. Significant differences between the control and transient overexpression plants were determined by t test (*P < 0.05). b The sequence of the LlMBF1c promoter and its W-box mutated version; the red label indicates the W-box element. c Yeast one-hybrid assay. The interaction between LlWRKY39 and W-box elements was determined in SD-LWH medium with different concentrations of 3-AT. d Schematic diagram of the LlMBF1c promoter; the probe sequence is shown below the diagram. e LlWRKY39-HIS complex binds to the W-box in EMSA. The term 250× indicates the usage of excess unlabeled probe as a competitor, and “+” and “−” indicate its presence and absence, respectively. f The schematic diagram of the effector and reporter. The 836-bp fragment of the LlMBF1c promoter was used in this assay. g Bright field and dark field images of N. benthamiana leaves in the transient expression assays. h The ratio of LUC/REN. 1: mixed bacterial solutions of pGreenII-62-SK and pGreenII-0800-LUC (2:1); 2: mixed bacterial solutions of pGreenII-62-SK-LlWRKY39 and pGreenII-0800-LUC (2:1); 3: mixed bacterial solutions of pGreenII-62-SK and pGreenII-0800-proLlMBF1c-LUC (2:1); 4: mixed bacterial solutions of pGreenII-62-SK-LlWRKY39 and pGreenII-0800-proLlMBF1c-LUC (2:1). Each bar indicates the mean ± SD of three repeated experiments (*P < 0.05, t test)
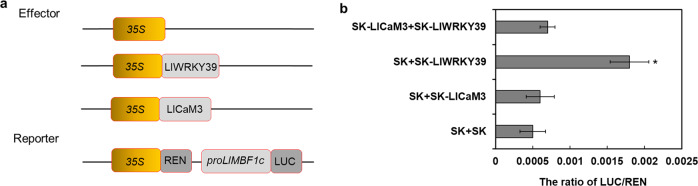
LlCaM3 negatively affects LlWRKY39 in the transactivation of LlMBF1c
To understand how the LlWRKY39–LlCaM3 interaction affects the function of LlWRKY39, we performed a dual-luciferase reporter assay. The ratio of LUC/REN when coexpressing LlWRKY39 together with LlCaM3 was significantly lower than that when only LlWRKY39 was expressed (Fig. 6b), which suggests that the LlWRKY39-LlCaM3 interaction represses the activation ability of LlWRKY39 for its target genes.
Fig. 6. Effect of LlCaM3 on the activation activity of LlWRKY39 at the promoter of LlMBF1c in N. benthamiana leaves according to a dual-luciferase reporter assay.
a The construction of the effect vectors and reporter vector. b Ratio of LUC/REN. Mixed bacterial solutions of effectors and reporter cultures (3:1). Each bar indicates the mean ± SD of three repeated experiments (*P < 0.05, t test)
Discussion
Calcium, a universal secondary messenger, is one of the main signal transducers and regulators in plant cells44–47. The transient changes in cytosolic Ca2+ concentration are called Ca2+ signatures, and Ca2+ sensors respond to these changes by activating or inactivating target proteins to participate in specific biochemical processes and regulate gene expression48. Most Ca2+ sensors are proteins with one or more EF-hands that have Ca2+-binding helix-turn-helix structures49. Ca2+ sensors with EF-hands are roughly classified into two groups: sensor responders and sensor relays50,51. CaMs are a group of well-characterized Ca2+ sensor relays that do not have any functional domains; however, when they bind to Ca2+, they can activate or inactivate interacting proteins by relaying the signal52. AtWRKY7 was the first WRKY TF to be reported to interact with CaM in a Ca2+-dependent manner by binding a short conserved motif (VSSFK[K/R]VISLL in the C-region), which is also called a CBD domain53. AtWRKY7 belongs to the WRKY group IId subfamily, and all members of this subfamily interact with Ca2+/CaM in Arabidopsis53. Here, we identified a new WRKY IId protein, LlWRKY39, from lily, which has a similar primary motif (HARF:RTGHARFRR[A/G]P) that is the distinctive characteristic of WRKY group IId TFs28. Additionally, LlCaM3, which is reportedly associated with HS in lily39, interacts with LlWRKY39 by binding the CBD in a Ca2+-dependent manner (Figs. 2 and 3). Ca2+-dependent CBD motifs are normally grouped into two categories, namely, motifs 1-5-10 and 1-8-14, the numbers of which imply the positions of conserved hydrophobic residues54. The CBD of AtWRKY7 does not belong to either of the two classical CBD motifs; however, AtWRKY7 is a Ca2+-dependent CBP53. LlWRKY39 may be similar to AtWRKY7, with a CBD that is not consistent with either of the two major classes (Fig. 2d). In this study, the interaction between LlWRKY39 and LlCaM3 depends on the CBD in the N-region of LlWRKY39, although it is not completely consistent with the CBD of the WRKY IId TFs in Arabidopsis (Fig. 2c, d), which suggests that the basic CBD can interact with the CaM.
In the present study, the induction of LlWRKY39 expression by HS implies that LlWRKY39 might participate in the regulation of HSR (Fig. 1d). Therefore, we identified the function of LlWRKY39 in the HSR process. The overexpression of LlWRKY39 increased the thermotolerance of lily and Arabidopsis (Fig. 4b, d, e). These results were consistent with those of previous studies on AtWRKY39 in Arabidopsis, in which the overexpressing plants showed greater thermotolerance during both seed germination and seedling growth than wrky39 mutants or wild-type plants under HS34. In the current study, some of the HS-responsive genes, namely, AtHSFA1, AtHSFA2, AtDREB2A, AtMBF1c, AtHSP101, and AtGolS1, were significantly upregulated in the transgenic lines (Fig. 4f). Their elevated expression may contribute to an increase in thermotolerance since all of these genes play positive roles in thermotolerance22,34,55–57. Notably, the basal expression of AtWRKY39 was also upregulated in the transgenic lines, possibly because of the self-activation of endogenous AtWRKY39. W-box elements in the promoters of stress-induced genes and WRKY genes indicate that WRKY TFs can also be self-regulated, e.g., WRKY33. The expression level of AtWRKY33 is usually low in healthy plants, but when plants are exposed to environmental stimuli, activated AtWRKY33 induces its own expression to generate a feedback mechanism for the rapid and strong induction of AtWRKY33 target genes that respond to biotic or abiotic stresses58–60. Nevertheless, HSFB2A, a transcriptional repressor40, is also induced, which is speculated to be a mechanism that maintains the balance in the HSR. In addition, although AtHSFA1, AtHSFA2, and AtDREB2A were induced in the transgenic plants, the expression of their target genes41,43, AtHSP70, AtAPX1, AtAPX2, and AtHSFA3, was not affected (Fig. 4f), which also suggests that another negative pathway exists in the LlWRKY39-mediated HSR to sustain the balance in the HSR. Among the detected genes, an important MBF1, AtMBF1c, was significantly induced in transgenic Arabidopsis. MBF1 proteins are highly conserved transcriptional coactivators that can build bridges between TFs and transcriptional regulation machinery22,61–63. MBF1c is a critical heat-response regulator in the HSR and functions upstream of SA, trehalose, and ethylene signaling to participate in the establishment of thermotolerance22,23. The homolog of MBF1c in lily, LlMBF1c, can be activated by LlWRKY39 by directly binding the W-box element in the prompter of LlMBF1c (Fig. 5), which suggested that LlWRKY39 might be an upstream regulator of LlMBF1c.
The proteins of the CaM-binding subgroup, AtWRKY7, AtWRKY11, and AtWRKY17, of the WRKY group IId proteins, play negative roles in the plant basal defense response64,65. wrky7 mutants exhibited increased resistance to Pseudomonas syringae 1, whereas plants overexpressing AtWRKY7 displayed a higher sensitivity to the pathogen by repressing the expression of SA-regulated defense genes or repressing weakly activated jasmonic acid (JA) signaling64. Similarly, wrky11 mutants showed increased resistance to P. syringae 1, and wrky11 and wrky17 double mutants exhibited a higher increase in resistance, which was due to the decreased JA levels and the downregulation of JA-responsive genes65. In contrast to previous studies, LlWRKY39 activated the expression of the downstream gene LlMBF1c. Additionally, the interaction between LlWRKY39 and LlCaM3 repressed the expression of LlMBF1c activated by LlWRKY39 (Fig. 6), suggesting that Ca2+/CaM played an important role in inhibiting excessive activation of LlWRKY39-mediated thermotolerance to sustain balance in the HSR. Additional examples show that MBF1 proteins can reduce tolerance to stresses, although MBF1 transcription cofactors often play positive roles against many stresses. For example, in pepper, CaMBF1 transcript levels are dramatically decreased in response to salt or cold stress, which suggests that MBF1 family factors have a negative effect on the stress response66. In addition, AtMBF1 genes can relieve abscisic acid (ABA)-dependent inhibition of germination, which implies that MBF1 proteins negatively regulate the ABA-dependent response to some extent19. ABA signaling can also be involved in the HSR67,68. In this study, the interaction between LlCaM3 and LlWRKY39 weakened the transcriptional activity of LlMBF1c activated by LlWRKY39 to a certain extent, suggesting that this feedback regulation balances the HSR to prevent the damage caused by excessive activation responses. There may be a similar regulatory mechanism in the downstream genes of LlMBF1c because some genes that negatively regulate thermotolerance in transgenic lines were upregulated, e.g., HSFB2A, and the expression of downstream HSP genes was not changed, e.g., HSP70, implying the existence of negative feedback regulation in the LlWRKY39-mediated HSR.
In conclusion, LlWRKY39 may act as a downstream component of the CaM-mediated calcium signaling pathway that lies upstream of LlMBF1c in the HSR. Considering these findings, we propose a simplified working model that may shed light on the mechanisms of the LlWRKY39-mediated HSR (Fig. 7). When lily is exposed to high temperature, LlWRKY39 is rapidly induced, which directly activates the expression of LlMBF1c and participates in the establishment of the HSR. Simultaneously, the feedback regulation mechanism also begins to respond, and Ca2+ enters the cell to activate LlCaM3, which upon interaction with LlWRKY39, can contribute to the prevention of the side effects caused by excessive activation to sustain balance in the HSR.
Fig. 7. A proposed working model of LlWRKY39-mediated HSR under HS.
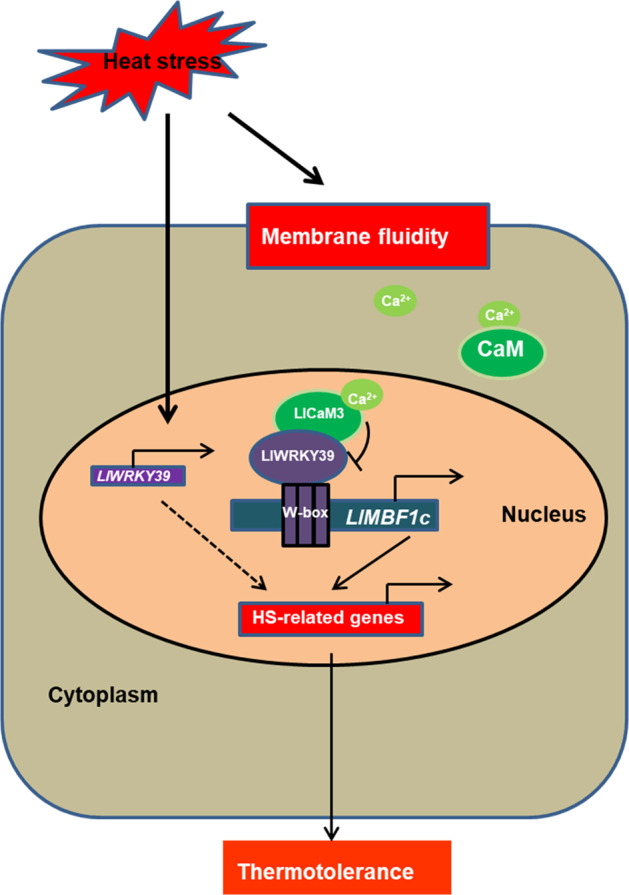
When lily suffers from high temperature, LlWRKY39 is rapidly induced, and its protein directly binds to the W-box element on the promoter of LlMBF1c to activate its expression. The feedback regulation mechanism also begins to respond, and Ca2+ flows into the cell to bind and activate LlCaM3; then, LlCaM3 interacts with LlWRKY39 to inhibit its activation-inducing function promoting the expression of downstream genes, e.g., LlMBF1c, which might contribute to the prevention of the side effects caused by excessive activation to sustain balance in the HSR
Materials and methods
Plant materials and growth conditions
The lily hybrid Lilium longiflorum ‘white heaven’ used in this experiment was cultured on Murashige and Skoog (MS) medium. Arabidopsis thaliana (Col-0) and N. benthamiana were grown in potting medium. All plant materials were cultured in a growth room at 22 °C with a 16-h photoperiod.
Molecular cloning and sequence analysis of LlWRKY39
Total RNA was extracted from the leaves of ‘white heaven’ with TRIzol reagent according to the manufacturer’s instructions (Invitrogen, USA). cDNA was biosynthesized by a reverse transcription system (TaKaRa, Japan). Primers for gene cloning are listed in Supplementary Table S3. The physicochemical properties of LlWRKY39 were estimated by EXPASY (http://web.expasy.org/compute_pi/, default setting).
Protein interaction assays
In the BIFC assay, the ORFs without stop codons of LlWRKY39 and LlCaM3 were amplified and inserted into pSPYCE (M) and pSPYNE173 vectors using a recombinant ligase (Vazyme, Nanjing, China). The reconstructed vectors and empty vectors were transformed into Agrobacterium tumefaciens strain GV3101. The combination of bacterial solutions (YCE:YNE:P19 = 3:3:1) was coinfiltrated into the leaves of N. benthamiana. After 48 h, a confocal laser-scanning microscope (LSM800, Zeiss, Germany) was used to observe the fluorescence signal. In the FLC assay, the ORFs without stop codons of LlWRKY39 and LlCaM3 were cloned into pCAMBIA1300-nLUC vectors, and the ORFs with stop codons of LlWRKY39 and LlCaM3 were cloned into pCAMBIA1300-cLUC vectors using recombinant ligase (Vazyme, Nanjing, China). The reconstructed vectors were transformed into A. tumefaciens strain GV3101. The corresponding combination of bacterial solutions (cLUC:nLUC = 1:1) was infiltrated into the leaves of N. benthamiana. After 48 h, the fluorescence signal was observed with a luminometer (PIXIS1024B, China). For a yeast two-hybrid assay, the split-ubiquitin vectors of pPR3-N and pBT3-STE were used. The ORF of LlCaM3 was inserted into pPR3-N, and the fragments (1 to 60; 1 to 45) of LlWRKY39 were cloned into pBT3-STE. The empty vectors pPR3-N and pBT3-STE were used as negative controls. The corresponding plasmids were cotransformed into yeast strain NMY51. The interaction was identified by spot assay on Leu-, Trp-, His-, and Ade-deficient SD medium with 3-AT (20 mM). All the primers used for vector construction are shown in Supplementary Table S3.
CaCl2 and EGTA treatment
The bacterial solutions (LlWRKY39-cLUC with LlCaM3-nLUC and LlWRKY39-nLUC with LlCaM3-cLUC) were collected by centrifugation and resuspended in buffer (10 mM MgCl2, 200 mM acetosyringone (AS), 10 mM 2-morpholino ethanesulfonic acid (MES), pH 5.6) to a final OD600 of 1.0. Then, the resuspended bacterial solutions were placed in the dark for 3 h. To induce transient expression, 1-cm diameter samples were cut from N. benthamiana leaves with a puncher. Then, the samples were placed into the bacterial suspension and infiltrated under vacuum for 10–15 min until the samples became transparent. After the vacuum was released, the samples were placed on medium (0.4% agar) in the dark. After 24 h, the samples were transferred to medium (0.4% agar) with CaCl2 (20 mM) or EGTA (20 mM) and incubated for 24 h. The samples on medium (0.4% agar) were used as controls. The fluorescence signal was observed with a luminometer (PIXIS1024B, China). The LUC activity measurement was performed using Andor Solis v15 software as described in a previous study69.
Subcellular localization of LlWRKY39
The ORF without the stop codon of LlWRKY39 was inserted into a pCAMBIA1300-GFP vector to generate a fusion protein (LlWRKY39-GFP). The primers used for vector construction are shown in Supplementary Table S3. The empty vector pCAMBIA1300-GFP was used as the control. The plasmids pCAMBIA1300-GFP and pCAMBIA1300-LlWRKY39-GFP were transformed into A. tumefaciens strain GV3101, and then, the bacterial solution was infiltrated into N. benthamiana. A confocal laser-scanning microscope (LSM800, Zeiss, Germany) was used to observe the fluorescence.
Transactivation activity assay of LlWRKY39 in yeast
The ORF of LlWRKY39 was inserted into the pGBKT7 vector using specific primers (Supplementary Table S3). The pGBKT7 vector, pGBKT7-GAL4, and pGBKT7-LlWRKY39 were transformed into yeast strain AH109. The transformed yeast cells were incubated on SD medium lacking Trp at 30 °C for 3 days. Positive clones were selected on SD medium (lacking Trp and His) containing 3-AT at 30 °C for 3 days.
Heat stress treatment of lily
To detect gene expression levels under HS, one-month-old ‘white heaven’ plants were exposed to 37 °C for 0, 0.5, 1, 3, 6, and 12 h. Heat treatment was applied in a temperature incubator (GZL-P80-A, Nanjing, China) without light. Samples of leaves were harvested for qRT-PCR analysis after heat treatment.
Transient transformation in lily leaves
The bacterial solutions of pCAMBIA1300-LlWRKY39 and pCAMBIA1300 (control) were collected by centrifugation and resuspended in the same buffer as described for the CaCl2 and EGTA treatment. The resuspended bacterial solutions were placed in the dark for 5 h and infiltrated into the leaves of ‘white heaven’. After 72 h, the infiltrated leaves were harvested for qRT-PCR analysis.
Gene expression analysis in lily
Total RNA was extracted using TRIzol as described above. A HiScript II Kit with gDNA Eraser (Vazyme, Nanjing, China) was used for cDNA biosynthesis with an oligo dT primer. qRT-PCR was performed using a 20-μL reaction system. Lily 18S rRNA was used as the endogenous gene. Three independent technical replicates were performed for each of three biological replicates. The relative expression level was calculated with the 2–∆∆Ct method70,71. The primers for qRT-PCR are shown in Supplementary Table S3.
Isolation and analysis of promoter sequences
The genomic DNA of ‘white heaven’ was extracted using a plant DNA extraction kit (Zomanbio, Beijing, China) following the manufacturer’s instructions. The promoter was isolated using hi-TAIL PCR72. PLACE databases (http://www.dna.affrc.go.jp/PLACE/) were used to analyze the cis-elements in the promoters.
Promoter activity analysis of LlWRKY39
The isolated 737-bp fragment of the LlWRKY39 promoter was fused to a pGreenII-0800-LUC vector using a recombinant ligase (Vazyme, Nanjing, China). The constructed vector and an empty vector (control) were transformed into A. tumefaciens strain GV3101 (pSoup), which was used to transform N. benthamiana leaves. After 48 h, one-half of the leaves with infiltrated material were treated at 37 °C for 2 h, and allowed to recover from HS at 22 °C for 12 h; then, the fluorescence signal was observed with a luminometer (PIXIS1024B, China). A luciferase reporter assay system (Promega, USA) was used to measure the activity of firefly luciferase (LUC) and Renilla luciferase (REN). The primers for vector construction are shown in Supplementary Table S3.
Plant transformation and generation of transgenic lines
It is difficult to obtain lily transgenic plants since a stable genetic transformation system of lily has not yet been established. Therefore, the model plant A. thaliana (Col-0) was used to verify the biological function of LlWRKY39 under HS. The bacterial solution of pCAMBIA1300-LlWRKY39 was collected by centrifugation and resuspended in a sucrose solution (5%). Arabidopsis plant genetic transformation was performed according to the floral dip method73. The harvested seeds were selected on MS medium containing 30 mg/L hygromycin until the T3 generation. One-week-old seedlings were collected to extract RNA for RT-PCR identification of the transgenic lines.
Phenotypic analysis and heat stress responsive gene expression in transgenic plants
Seeds of three transgenic lines (OE-2, OE-4, OE-5) and Col-0 were sown on the same MS medium, and after incubation, they were placed into the culture room at 4 °C for 3 days. To compare the thermotolerance of the transgenic line and wild-type (WT) plants, one-week-old seedlings were treated with two HS tests. One was a BT-test in which seedlings were directly treated at 45 °C (shown in Fig. 4d), whereas the other was an AT-test in which seedlings were first treated at 37 °C for 1 h, recovered for 2 h at 22 °C, and then treated again at 45 °C (shown in Fig. 4e). After these treatments, the plants were placed in the culture room for a 7-day recovery, and the phenotypes and survival rates of the seedlings were recorded. One-week-old seedlings of the transgenic and WT plants were collected to determine the expression levels of heat-related genes: AtHSFA1 (AT4G17750), AtHSFA2 (AT2G26150), AtHSFA3 (AT5G03720), AtHSFB2A (AT5G62020), AtDREB2A (AT2G40340), AtWRKY39 (AT3G04670), AtAPX2 (AT3G09640), AtAPX1 (AT1G07890), AtGolS1 (AT2G47180), AtMBF1c (AT3G24500), AtHSP70 (AT3G12580), and AtHSP101 (AT1G74310). AtActin2 (AT3G18780) was used as the endogenous gene. The primers used for qRT-PCR are shown in Supplementary Table S3.
Yeast one-hybrid assay
A fragment (-500 to -486) of the LlMBF1c promoter with three repeats was cloned into pHis2.1 to obtain the pHis2.1–3 × W-box; two sites of this fragment (Fig. 5b) were mutated and cloned into pHis2.1 to generate pHis2.1–3 × mW-box. LlWRKY39 was amplified and inserted into the pGADT7 vector. Different combinations of plasmids were cotransformed into yeast strain Y187 to identify positive clones. SD medium lacking Trp, Leu, and His was used to detect possible interactions. The primers used for vector construction are listed in Supplementary Table S3.
Luciferase reporter assay
The 836-bp fragment of the LlMBF1c promoter was cloned into pGreenII-0800-LUC to obtain the reporter vector. The ORF of LlWRKY39 was cloned into a pGreenII-62-SK vector to generate the effector vector. These vectors were transformed into A. tumefaciens strain GV3101 (pSoup). The mixed bacterial solution of the TF and promoter cultures (2:1) for the induction analysis was used to infiltrate N. benthamiana leaves. After 48 h, the LUC fluorescence of the infiltrated leaves was detected, and LUC and REN activities were measured using luciferase reporter assay reagents (Promega) as described by a previous study74. Three replicate experiments were used for statistical analysis by Student’s t test.
Electrophoretic mobility shift assay (EMSA)
An EMSA was performed using the Light Shift Chemiluminescent EMSA kit (Thermo Fisher, New York, USA) according to the manufacturer’s protocol. The biotinylated probes for the EMSAs were synthesized by TSINGKE Biological Technology (Nanjing). The recombinant proteins were purified as described in a previous study75. The samples were loaded onto a prerun native 4% polyacrylamide gel with TBE buffer as the electrolyte. After electroblotting onto a nylon membrane (Millipore, Darmstadt, Germany) and UV cross-linking for 2 min, the membrane was incubated in blocking buffer for 15 min and rinsed in washing buffer for 20 min. Finally, a CCD camera was used to visualize the signals in the membrane.
Measurement of relative ion leakage
Agrobacterium-mediated transformation was performed as described above, and then ‘white heaven’ plants were treated with HS at 42 °C for 2 h. The leaves were harvested to measure ion leakage (percentage) according to a previously described method6.
Dual-luciferase reporter assay
The ORFs of LlWRKY39 and LlCaM3 were cloned into pGreenII-62-SK76 for use as effector vectors. A fragment of the LlMBF1c promoter was inserted into pGreenII-0800-LUC76, which was used as the reporter vector. These vectors were transformed into A. tumefaciens strain GV3101 (pSoup). Mixed bacterial solutions of effector and reporter cultures (3:1) were used to infiltrate the leaves of N. benthamiana. The LUC and REN activity levels were measured as described above. The primers for the vector construction are shown in Supplementary Table S3.
Supplementary information
Acknowledgements
This work was supported by the National Key R&D Program of China (Grant No. 2019YFD1000400), the National Natural Science Foundation of China (31902055), the High Level Talent Project of the Top Six Talents in Jiangsu, China (NY-077), the Natural Science Foundation of Jiangsu Province, China (BK20190532), and the Fundamental Research Funds for the Central Universities (KJQN202032). We thank the Central Laboratory of the College of Horticulture of Nanjing Agricultural University, which is a large-scale instrument sharing platform.
Author contributions
N.T. conceived this project and designed all research with help from Z.W. L.D. and Z.W. conducted the experiments and processed the data. R.T. performed the EMSA. S.X., G.Y., X.C., and D.Z. provided technological assistances. X.C. provided the LlMBF1c and LlCaM3 sequences. L.D. and Z.W. wrote the first draft of the article, and all the authors read and revised the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Liping Ding, Ze Wu
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00473-7.
References
- 1.Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: An overview. Environ. Exp. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 3.Long SP, Ort DR. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010;13:241–248. doi: 10.1016/j.pbi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Fu W, et al. Molecular cloning and analysis of a cytosolic HSP70 gene from Enteromorpha prolifera (Ulvophyceae, Chlorophyta) Plant Mol. Biol. Rep. 2010;28:430–437. doi: 10.1007/s11105-009-0170-8. [DOI] [Google Scholar]
- 5.Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Xin H, et al. Cloning and characterization of HSFA2 from lily (Lilium longiflorum) Plant Cell Rep. 2010;29:875–885. doi: 10.1007/s00299-010-0873-1. [DOI] [PubMed] [Google Scholar]
- 7.Gong B, et al. LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 2014;33:1519–1533. doi: 10.1007/s00299-014-1635-2. [DOI] [PubMed] [Google Scholar]
- 8.Grover A, Mittal D, Negi M, Lavania D. Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci. 2013;205:38–47. doi: 10.1016/j.plantsci.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira EI, Fischer G, Van Velthuizen H, Walter C, Ewert F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013;170:206–215. doi: 10.1016/j.agrformet.2011.09.002. [DOI] [Google Scholar]
- 10.Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golldack D, Lueking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- 12.Nover L, et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Koskull-Doering P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Xin H, et al. Over-expression of LlHSFA2b, a lily heat shock transcription factor lacking trans-activation activity in yeast, can enhance tolerance to heat and oxidative stress in transgenic Arabidopsis seedlings. Plant Cell Tissue Organ Cult. 2017;130:617–629. doi: 10.1007/s11240-017-1251-2. [DOI] [Google Scholar]
- 15.Wu Z, et al. Overexpression of lily HSFA3s in Arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J. Exp. Bot. 2018;69:2005–2021. doi: 10.1093/jxb/ery035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z, et al. Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily. Plant Physiol. 2019;181:1651–1667. doi: 10.1104/pp.19.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, et al. A canonical DREB2-type transcription factor in lily is post-translationally regulated and mediates heat stress response. Front. Plant Sci. 2018;9:243. doi: 10.3389/fpls.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pamela Arce D, et al. The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J. Plant Physiol. 2010;167:194–200. doi: 10.1016/j.jplph.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Florencia Di Mauro M, et al. MBF1s regulate ABA-dependent germination of Arabidopsis seeds. Plant Signal. Behav. 2012;7:188–192. doi: 10.4161/psb.18843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blombach F, et al. Archaeal MBF1 binds to 30s and 70s ribosomes via its helix-turn-helix domain. Biochem. J. 2014;462:373–384. doi: 10.1042/BJ20131474. [DOI] [PubMed] [Google Scholar]
- 21.Tsuda K, Yamazaki K. Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim. Biophys. Acta. 2004;1680:1–10. doi: 10.1016/j.bbaexp.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 2008;283:9269–9275. doi: 10.1074/jbc.M709187200. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011;66:844–851. doi: 10.1111/j.1365-313X.2011.04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida T, et al. Arabidopsis HSFA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics. 2011;286:321–332. doi: 10.1007/s00438-011-0647-7. [DOI] [PubMed] [Google Scholar]
- 25.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, X. Y. et al. The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Horti. Res.10.1038/s41438-019-0147-1 (2019). [DOI] [PMC free article] [PubMed]
- 28.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 29.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Ulker B, Somssich IE. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012;15:431–437. doi: 10.1016/j.pbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Zhou X, Chen L, Huang W, Yu D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells. 2010;29:475–483. doi: 10.1007/s10059-010-0059-2. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 36.He GH, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16:116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, et al. Cytological and molecular characteristics of pollen abortion in lily with dysplastic tapetum. Hortic. Plant J. 2019;5:281–294. doi: 10.1016/j.hpj.2019.11.002. [DOI] [Google Scholar]
- 38.Rieping M, Schoffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimeric heat-shock genes in transgenic tobacco. Mol. Gen. Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- 39.Cao X, et al. Involvement of Ca2+ and CaM3 in regulation of thermotolerance in lily (Lilium longiflorum) Plant Mol. Biol. Rep. 2013;31:1293–1304. doi: 10.1007/s11105-013-0587-y. [DOI] [Google Scholar]
- 40.Ayako NY, et al. HSFA1d and HSFA1e involved in the transcriptional regulation of HSFA2 function as key regulators for the HSF signaling network in response to environmental stress. Plant Cell Physiol. 2011;52:933–945. doi: 10.1093/pcp/pcr045. [DOI] [PubMed] [Google Scholar]
- 41.Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013;163:276–290. doi: 10.1104/pp.113.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, et al. AtHSFA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China C Life Sci. 2005;48:540–550. doi: 10.1360/062005-119. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida T, et al. Functional analysis of an Arabidopsis heat-shock transcription factor HSFA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008;368:515–521. doi: 10.1016/j.bbrc.2008.01.134. [DOI] [PubMed] [Google Scholar]
- 44.Hepler PK. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17:2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mcainsh MR, Pittman JK. Shaping the calcium signature. New Phytol. 2009;181:275–294. doi: 10.1111/j.1469-8137.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- 46.Defalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 47.Hashimoto K, Kudla J. Calcium decoding mechanisms in plants. Biochimie. 2011;93:2054–2059. doi: 10.1016/j.biochi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 48.Luan S, Lan W, Lee SC. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 2009;12:339–346. doi: 10.1016/j.pbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Day IS, Reddy VS, Ali GS, Reddy ASN. Analysis of EF hand-containing proteins in Arabidopsis. Genome Biol. 2002;3:0056. doi: 10.1186/gb-2002-3-10-research0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park CY, et al. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005;579:1545–1550. doi: 10.1016/j.febslet.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 54.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HSFA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007;58:3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- 56.Charng YY, et al. A heat-inducible transcription factor, HSFA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34:738–751. doi: 10.1111/j.1365-3040.2011.02278.x. [DOI] [PubMed] [Google Scholar]
- 58.Zheng Z, Abu Qamar S, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 59.Lippok B, et al. Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 2007;20:420–429. doi: 10.1094/MPMI-20-4-0420. [DOI] [PubMed] [Google Scholar]
- 60.Mao G, et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim MJ, et al. Abiotic and biotic stress tolerance in Arabidopsis overexpressing the multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem. Biophys. Res. Commun. 2007;354:440–446. doi: 10.1016/j.bbrc.2006.12.212. [DOI] [PubMed] [Google Scholar]
- 62.Yan Q, et al. The grape VvMBF1 gene improves drought stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2014;118:571–582. doi: 10.1007/s11240-014-0508-2. [DOI] [Google Scholar]
- 63.Zhao Q, et al. Overexpression of a multiprotein bridging factor 1 gene DgMBF1 improves the salinity tolerance of chrysanthemum. Int. J. Mol. Sci. 2019;20:245. doi: 10.3390/ijms20020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim KC, Fan B, Chen Z. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas Syringae. Plant Physiol. 2006;142:1180–1192. doi: 10.1104/pp.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo WL, et al. Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. BMC Plant Biol. 2014;14:138. doi: 10.1186/1471-2229-14-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang YC, Niu CY, Yang CR, Jinn TL. The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016;172:1182–1199. doi: 10.1104/pp.16.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Wang X, Zhuang L, Gao Y, Huang B. Abscisic acid mediation of drought priming-enhanced heat tolerance in tall fescue (Festuca arundinacea) and Arabidopsis. Physiol. Plant. 2019;167:488–501. doi: 10.1111/ppl.12975. [DOI] [PubMed] [Google Scholar]
- 69.Lu J, Bai M, Ren H, Liu J, Wang C. An efficient transient expression system for gene function analysis in rose. Plant Methods. 2017;13:116. doi: 10.1186/s13007-017-0268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Willems E, Leyns L, Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 72.Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques. 2007;43:649–654. doi: 10.2144/000112601. [DOI] [PubMed] [Google Scholar]
- 73.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 74.Fan Y, et al. The RhHB1/RhLOX4 module affects the dehydration tolerance of rose flowers (Rosa hybrida) by fine-tuning jasmonic acid levels. Hortic. Res. 2020;7:74. doi: 10.1038/s41438-020-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W, et al. Hairy leaf 6, an AP2/ERF transcription factor, interacts with OsWOX3B and regulates trichome formation in rice. Mol. Plant. 2017;10:1417–1433. doi: 10.1016/j.molp.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 76.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.