Abstract
Microbial-root associations are important to help plants cope with abiotic and biotic stressors. Managing these interactions offers an opportunity for improving the efficiency and sustainability of agricultural production. By characterizing the bacterial and archaeal community (via 16S rRNA sequencing) associated with bulk and rhizosphere soil of sixteen strawberry cultivars in two controlled field studies, we explored the relationships between the soil microbiome and plant resistance to two soil-borne fungal pathogens (Verticillium dahliae and Macrophomina phaseolina). Overall, the plants had a distinctive and genotype-dependent rhizosphere microbiome with higher abundances of known beneficial bacteria such as Pseudomonads and Rhizobium. The rhizosphere microbiome played a significant role in the resistance to the two soil-borne pathogens as shown by the differences in microbiome between high and low resistance cultivars. Resistant cultivars were characterized by higher abundances of known biocontrol microorganisms including actinobacteria (Arthrobacter, Nocardioides and Gaiella) and unclassified acidobacteria (Gp6, Gp16 and Gp4), in both pathogen trials. Additionally, cultivars that were resistant to V. dahliae had higher rhizosphere abundances of Burkholderia and cultivars resistant to M. phaseolina had higher abundances of Pseudomonas. The mechanisms involved in these beneficial plant-microbial interactions and their plasticity in different environments should be studied further for the design of low-input disease management strategies.
Subject terms: Agroecology, Applied microbiology, Microbial ecology, Microbiome
Introduction
Soil microorganisms support key soil processes such as decomposition, mineralization, aggregate formation and biocontrol of plant diseases, all highly relevant to agricultural production1–3. However, understanding the complexity and drivers of the soil microbiome, remains a challenge, and the practical application of ecologically based agricultural management strategies not feasible4. Soil bacterial diversity and abundance is primarily regulated by small- and large-scale changes in abiotic factors such as soil pH, oxygen concentration, and the availability as well as quality of carbon (C)1. Large variations in these factors exist in the macro (i.e. cm scale) and microenvironments (i.e. mm to μm scale) within a soil profile, producing hotspots of microbial diversity and activity. The rhizosphere, the volume of soil that is directly influenced by plant roots and which spans usually 1 mm from the root surface, can contain up to 30,000 bacterial and archaeal species5. Furthermore, the rhizosphere typically has higher microbial biomass and activity when compared to the surrounding bulk soil6. This microbial hotspot within soil habitats is driven by plant root presence, nutrient and water uptake as well as the continuous release of carbon compounds through root rhizodeposits7,8. Differential bioavailability and assimilation of carbon compounds by soil microorganisms results in the selective promotion of certain taxa and inhibition of others9.
Root-associated microorganisms, either symbiotic or free living are key for plant nutrition and health and, because of this, the rhizosphere is frequently compared with the human gut10. Soil bacteria such as Pseudomonas, Bacillus and Rhizobium can increase the concentration of plant available phosphorus (P) in soil by releasing phosphatase enzymes or organic chelates11. Root-exudate production and the release of C compounds to usually C-deprived soil microorganisms, is known to trigger microbial activity and the production of exoenzymes which in turn further accelerates the decomposition of organic molecules in the rhizosphere and nutrient release12,13. As a result, rhizosphere soil typically has larger N mineralization rates and higher concentrations of bioavailable nitrogen (N) and P when compared to bulk soil, regardless of the plant species considered6. Additionally, certain strains of Bacillus, Pseudomonas, Burkholderia and Acinetobacter are thought to increase the availability and uptake of micronutrients such as Zn and Fe for mung bean, rice and corn14–16. Increases in plant growth have also been related to the induction of changes in plant root architecture and increase in root absorptive surface through the bacterially secreted plant hormones like indole-3-acetic acid, gibberellins and cytokinins17. Plants with adequate nutrition may be more resilient to biotic stress; in addition, rhizosphere microorganisms are known to have also direct impacts in plant health by triggering plant defenses10. Induction of systemic resistance in tomato plants by the bacterium Azospirillum brasilense was attributed to the capacity of this microbial strain to produce ethylene, a hormone involved in plant defense17. In tomato plants rhizosphere, bacteria from Bacillus, Pseudomonas, and Azotobacter triggered the production of antioxidant peroxidase and polyphenol oxidase enzymes, reducing the incidence of early blight disease by the pathogenic fungus Alternaria solani18. Finally, numerous bacteria have been identified that are antagonists of filamentous fungal pathogens such as Verticillium dahliae and are therefore able to directly suppress the pathogen via competition, production of antibiotic substances, siderophores, protein degrading enzymes and volatile compounds19.
Cultivated strawberry (Fragaria x ananassa Duchesne ex Rozier) is a fruit crop highly appreciated throughout the world, and therefore of major economic importance20. World production of this crop increased sharply over the last decades reaching over 13 million tons in 201721. Currently, the largest producer is China, followed by the USA, Mexico, Egypt, Turkey and Spain21. Within the USA, California strawberry production accounted for 88% of the U.S. market in 2017, representing the 4th most valuable agricultural commodity in the state with a total revenue of 3.10 billion dollars22. Since the late 1950s, the California strawberry industry relied on the use of preplant soil fumigation with methyl bromide to control soil-borne diseases23. However, since the complete phase out of methyl bromide in 2016 under the Montreal Protocol due to its stratospheric ozone depleting nature, strawberry growers are facing the challenge of maintaining their productivity while using alternative disease management strategies24. At the same time, new diseases such as the soil-borne fungus Macrophomina phaseolina, are starting to emerge threatening the strawberry industry even further25. Other fumigants such as chloropicrin are currently in use, although their efficacy is substantially lower than methyl bromide26. Furthermore, urban development and encroachment in the areas with the most intensive strawberry production, severely limits the application of pesticides. The above-mentioned restrictions in fumigant use have shifted the interest of the industry towards integrated strategies combining cultural techniques such as anaerobic soil disinfestation, cover crops, or crop rotations to improve soil health, as well as the use of resistant cultivars23. To date, strawberry cultivars have been identified that are tolerant to soil-borne pathogens like Verticillium dahliae27,28 and Fusarium oxysporum f. sp. fragariae. Resistance to these soil-borne pathogens seems to be achieved through the expression of multiple genes29 thought to regulate the production of substances with antifungal properties such as catechin, caffeic acid, or citric acid30. Cultivars with different degrees of resistance to Macrophomina crown rot have also been identified, yet little is known on the mechanisms involved despite its recently increasing incidence in California and other regions of the world31–33. The interplay between roots and soil microorganisms is likely involved in the resistance to these soil-borne fungal pathogens, as it has been previously observed for cucumber, common bean and tomato34–36. Yet, to date no information is available on how the rhizosphere microbiome influences the resistance of strawberry cultivars to these pathogens. This knowledge is instrumental in the selection of strawberry cultivars that will allow the strawberry industry to maintain its high yields and competitiveness, while reducing the need for agrochemicals with the associated reduction in production costs and environmental impacts.
Here, we used two ongoing field trials established to determine the host resistance to M. phaseolina and V. dahliae of strawberry cultivars and elite breeding lines, to explore potential resistance mechanisms by assessing the role of the rhizosphere microbiome in the resistance to these soil-borne fungal pathogens in a subset of 16 cultivars. More precisely we aimed to: (i) characterize the rhizosphere microbiome in strawberry plants under commercial field conditions; (ii) identify changes in the rhizosphere microbiome in the presence of two soil-borne fungal pathogens (V. dahliae and M. phaseolina) and potential increases in the presence of beneficial rhizosphere microorganisms and (iii) evaluate the relationships between the rhizosphere microbiome and plant traits such as resistance against soil-borne pathogens, plant growth and nutrition. We hypothesized that strawberry plants have the capacity to recruit beneficial microorganisms and establish a rhizosphere microbiome different from bulk soil. Further, variations in plant traits derived from decades of breeding cultivars would also have an impact on belowground root traits and therefore different strawberry cultivars would show different microbial communities. Finally, plants with higher resistance will exhibit a different microbiome from plants with low resistance having a higher abundance of beneficial bacteria and archaea; resistance to soil-borne pathogens, growth and nutrient uptake, would be corelated with the composition of the rhizosphere microbiome.
Results
Rhizosphere microbiome of the strawberry cultivars
A total of 3,939,353 high quality bacterial and archaeal sequences were generated across both the bulk soil and rhizosphere soil samples. After filtering chloroplast and unassigned reads, 3,786,463 sequences were clustered into 5,600 OTUs representing 33 Archaeal and 5567 Bacterial OTUs from 353 genera.
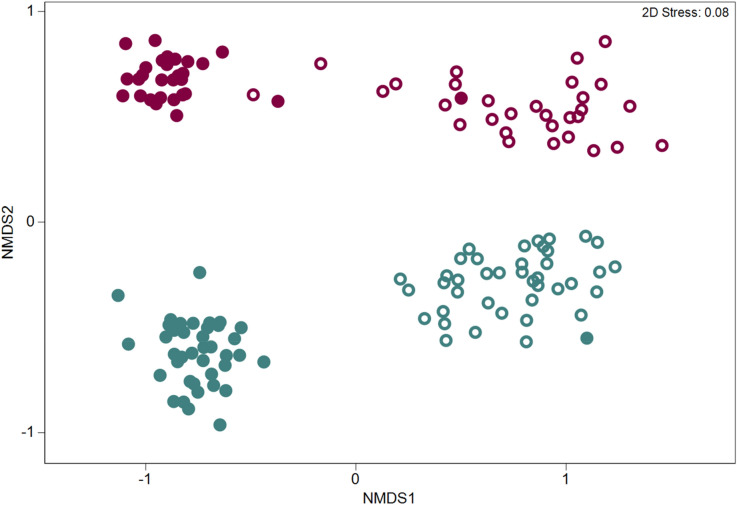
Differences in OTU community structure between sample types (rhizosphere and bulk soils) in the two pathogen trials were examined via non-metric multidimensional scaling (NMDS) using a Bray–Curtis dissimilarity of the standardized and transformed data (Fig. 1).
Figure 1.
Non-metric multidimensional scaling (NMDS) ordinations based on the Bray–Curtis similarity of the OTU-based bacterial and archaeal community structure in the rhizosphere (open symbols) and bulk soil (filled symbols) in two pathogen trials inoculated or infested with Macrophomina phaseolina (blue) and Verticillium dahliae (red).
We observed significant differences in the soil microbiome between pathogen trials although the magnitude of the differences depended on the sample type (PERMANOVA results for pathogen trial x sample type: Pseudo-F = 9.17, p = 0.001) (Fig. 1). A pairwise comparison via PERMANOVA between rhizosphere soils in each pathogen trial showed significant differences in their microbiomes (t = 4.14, p = 0.001).
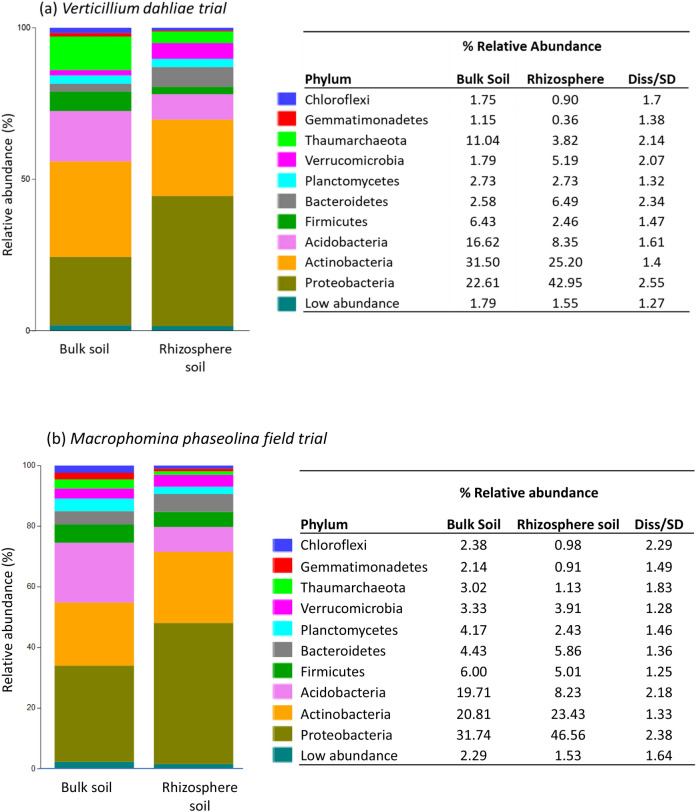
SIMPER analysis (performed with taxa grouped at the phylum level) revealed that the rhizosphere and bulk soil samples differed by 17.87% and 13.56% in the V. dahliae and M. phaseolina trial, respectively. In the V. dahliae trial, the bacterial phyla contributing most to these differences included the archaeal phylum Thaumarchaeota, Firmicutes and Acidobacteria, which exhibited higher abundances in the bulk soil relative to the rhizosphere soil (Fig. 2a). Additionally, the phyla Verrucomicrobia, Bacteroidetes, and Proteobacteria exhibited higher abundances in the rhizosphere soil relative the bulk soil (Fig. 2a). In the M. phaseolina trial, the archaeal phylum Acidobacteria contributed the most to differences between sample types, followed by the archaeal phylum Thaumarchaeota and the bacterial phyla Chloroflexi, Gemmatimonadetes, Planctomycetes and Firmicutes, all of which exhibited higher abundances in the bulk soil relative to rhizosphere soil (Fig. 2b). The phyla Proteobacteria, Verrucomicrobia, Bacteroidetes and Actinobacteria exhibited a greater relative abundance in the rhizosphere soil compared with the bulk soil (Fig. 2b).
Figure 2.
Differences in the average relative abundance of the main archaeal and bacterial phyla in the bulk and rhizosphere soil samples collected from the pathogen trials infested with Verticillium dahliae (a) and inoculated with Macrophomina phaseolina (b). Phyla representing < 1% of the average relative abundance were grouped into ‘low abundance’.
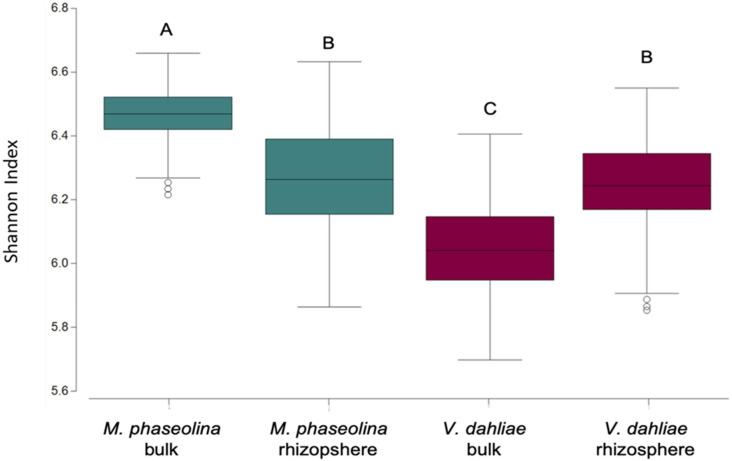
Diversity of OTUs, determined using the Shannon diversity index (H) was significantly higher in the bulk soil of the M. phaseolina trial than in the V. dahliae trial but remained similar in the rhizosphere soil across trials (pathogen trial x sample type: F = 40.75, p < 0.0001; Fig. 3).
Figure 3.
Diversity of the microbial community in the bulk and rhizosphere soil from the pathogen trials inoculated with Macrophomina phaseolina and infested with Verticillium dahliae. Different letters denote significant differences at p < 0.005.
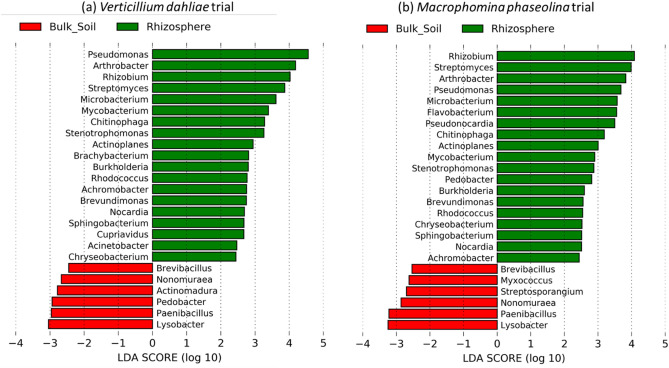
We used linear discriminant effect size analysis (LEfSe) to assess the differences in the relative abundance of the different microorganisms at the genus level, specifically focusing on a subset of well-known antagonists of filamentous fungal plant pathogens19. The LEfSe analysis performed on samples from within in the V. dahliae trial data revealed 191 taxonomic clades that exhibited greater relative abundance in the bulk soil relative to the rhizosphere (Table S1), 6 of which are known fungal antagonists (Fig. 4a). This analysis also revealed 231 taxonomic clades that exhibited greater relative abundances in the rhizosphere, 19 of which are genera known to include fungal antagonistic species (Fig. 4a). On samples from within the M. phaseolina trial, LefSe analysis revealed 219 taxonomic clades that exhibited greater relative abundance in the bulk soil relative to the rhizosphere soil, 6 of which are known fungal antagonists (Table S1). Additionally, there were 220 taxonomic clades that exhibited greater relative abundances in the rhizosphere, 19 of which are known fungal antagonists (Fig. 4b).
Figure 4.
Linear discriminant analysis (LDA) effect size (LEfSe) of genera known to include species antagonistic to filamentous fungal pathogen in the rhizosphere and bulk soil of strawberry grown in fields infested with Verticilium dahliae and inoculated with Macrophomina phaseolina. Genera listed were both significantly different (α = 0.05) and had an LEfSe score of > 2.0. A positive LDA score represents depletion in bulk soil and enrichment in rhizosphere (green) and a negative LDA score represents the opposite (red).
Effects of the soil-borne fungal pathogens on the rhizosphere microbiome
The effect of pathogen presence on the rhizosphere microbiome of strawberry plants was determined by comparing the rhizosphere microbiomes in the non-infested controls with the experimental plots that were inoculated (M. phaseolina trial) or grown in soil with a high concentration of the fungal pathogen (V. dahliae trial). To compare control vs inoculated plants, we aggregated all the plots, regardless of the cultivar. Thus, the control had a total of 10 replicates (plots), while inoculated plants had 40 replicates (plots) in each trial. The rhizosphere microbial community composition of plants exposed to V. dahliae was significantly different from control plants (ANOSIM: R = 0.3, p = 0.004). An average dissimilarity of 27.11% in OTU relative abundances was found between infested and control plants based on SIMPER analysis (performed with taxa grouped at the genus level) in the V. dahliae trial. The genera found in higher relative abundance in the control plants include Arthrobacter, Steroidobacter, Pseudomonas, Azotobacter, Luteolibacter, Nakamutella, Saccharibacteria (genera incertae sedis), WPS-1 (genera incertae sedis), Micromonospora, Arenimonas. On the other hand, the archaeal genus Nitrosphaera and bacterial genera Gaiella, Hydrocarboniphaga, Rubrobacter, Streptomyces, Mucilaginibacter, Rhizobium, Pseudonocardia, Niastella and Aminobacter exhibited greater relative abundances in V. dahliae infested plants (Table 1).
Table 1.
Average percent relative abundances of bacterial genera contributing to at least 1% to the observed variability between the rhizosphere of infested vs. control plants via SIMPER analysis.
| Genus | Infested | Control | Diss/SD | Contribution (%) |
|---|---|---|---|---|
| Verticilium dahliae field trial | ||||
| Nitrososphaera | 7.96 | 2.84 | 1.43 | 2.66 |
| Arthrobacter | 3.26 | 6.35 | 1.72 | 1.99 |
| Steroidobacter | 2.05 | 3.55 | 1.42 | 1.9 |
| Gaiella | 4.05 | 1.73 | 1.38 | 1.82 |
| Pseudomonas | 1.35 | 2.04 | 1.23 | 1.64 |
| Hydrocarboniphaga | 0.97 | 0.64 | 1.25 | 1.56 |
| Azotobacter | 0.26 | 1.43 | 0.7 | 1.54 |
| Rubrobacter | 1.21 | 1.11 | 0.88 | 1.51 |
| Streptomyces | 3.24 | 2.87 | 1.35 | 1.48 |
| Mucilaginibacter | 2.61 | 1.64 | 1.38 | 1.44 |
| Luteolibacter | 0.74 | 1.52 | 1.64 | 1.37 |
| Nakamurella | 0.55 | 1.16 | 1.09 | 1.33 |
| Saccharibacteria_genera_incertae_sedis | 0.95 | 1.8 | 1.54 | 1.31 |
| Rhizobium | 3 | 2.19 | 1.32 | 1.25 |
| Methylobacterium | 1.1 | 1.11 | 1.32 | 1.25 |
| WPS-1_genera_incertae_sedis | 1.06 | 1.74 | 1.48 | 1.24 |
| Micromonospora | 0.16 | 0.72 | 2.22 | 1.22 |
| Pseudonocardia | 2.36 | 1.38 | 1.69 | 1.14 |
| Niastella | 0.76 | 0.59 | 1.02 | 1.13 |
| Aminobacter | 1.24 | 1.12 | 1.33 | 1.09 |
| Arenimonas | 0.57 | 0.74 | 1.33 | 1.02 |
| Macrophomina phaseolina field trial | ||||
| Pseudomonas | 9.1 | 4.12 | 1.57 | 2.49 |
| Nakamurella | 0.73 | 2.53 | 1.48 | 2.17 |
| Gemmatimonas | 0.32 | 1.33 | 0.92 | 1.6 |
| Isoptericola | 0.68 | 0.63 | 1.13 | 1.45 |
| Arthrobacter | 5.75 | 6.61 | 1.35 | 1.42 |
| Xanthomonas | 0.89 | 0.68 | 1.29 | 1.39 |
| Flavobacterium | 1.08 | 1.69 | 1.02 | 1.36 |
| Phycicoccus | 0.97 | 1.92 | 1.41 | 1.33 |
| Saccharibacteria genera incertae sedis | 1.18 | 1.89 | 1.45 | 1.29 |
| Steroidobacter | 2.09 | 1.59 | 1.35 | 1.19 |
| Streptomyces | 2.34 | 1.62 | 1.38 | 1.16 |
| Burkholderia | 0.15 | 0.58 | 1.07 | 1.16 |
| Mucilaginibacter | 1.64 | 0.91 | 1.36 | 1.13 |
| Bradyrhizobium | 0.74 | 1.06 | 1.27 | 1.09 |
| Gaiella | 1.78 | 1.16 | 1.43 | 1.09 |
| Novosphingobium | 4.2 | 4.38 | 1.34 | 1.07 |
| Nitrososphaera | 2.46 | 1.77 | 1.45 | 1.07 |
| Luteolibacter | 0.67 | 0.92 | 1.35 | 1.06 |
| Massilia | 0.67 | 0.53 | 1.2 | 1.03 |
| Arenimonas | 0.55 | 0.81 | 1.46 | 1.02 |
| Sphingomonas | 4.32 | 5.26 | 1.37 | 1.01 |
Diss/SD values represent the average dissimilarity divided by the standard deviation.
The rhizosphere microbiome was also significantly different between inoculated and control plants in the M. phaseolina trial (R = 0.51, p < 0.001). SIMPER analysis (performed with taxa grouped at the genus level) revealed an average dissimilarity of 27.07% between infested and non-infested treatments. The bacterial genera Nakamurella, Gemmatimonas, Arthrobacter, Flavobacterium, Phycicoccus, Saccharibacteria (genera incertae sedis), Burkholderia, Bradyrhizobium, Novosphingobium, Luteolibacter, Arenimonas and Sphingobium exhibited higher relative abundances in the control relative to the inoculated plants and contributed at least 1% to the total dissimilarity between groups. The bacteria in the genera Pseudomonas, Isoptericola, Xanthomonas, Steroidobacter, Streptomyces, Mucilaginibacter, Gaiella and Massilia exhibited higher relative abundances in the rhizosphere of plants inoculated with M. phaseolina and contributed at least 1% to the total dissimilarity between groups (Table 1).
Cultivar-dependent selection of rhizosphere microorganisms
Within the fungal-inoculated plots, significant differences were observed in the rhizosphere microbiome of the 10 strawberry cultivars grown in the V. dahliae trial (ANOSIMS R = 0.43, p < 0.001), showing the cultivar-dependent selection of microorganisms. Similar results were observed for the 10 cultivars grown in the M. phaseolina trial (ANOSIMS R = 0.24, p < 0.001). Furthermore, Shannon diversity in the rhizosphere microbiome was also found to be significantly different between cultivars in the V. dahliae trial (ANOVA: F = 2.41, p = 0.05, Table 2), but not in the M. phaseolina trial (ANOVA: F = 0.58, p = 0.8, Table 3).
Table 2.
Aboveground traits (biomass and leaf nutrient contents), rhizosphere microbial diversity (Shannon Index) and mortality of the strawberry cultivars grown in the presence of the soil-borne fungal pathogen Verticillium dahliae.
| Mortality (%) | Resistance | Plant biomass (g) | Leaf N (%) | Leaf P (%) | Leaf K (%) | Leaf Ca (%) | Leaf Mg (%) | Leaf Ca:Mg | Shannon Index | |
|---|---|---|---|---|---|---|---|---|---|---|
| UC | 2.2 ± 2.2c | High | 96.44 ± 5.64abc | 1.79 ± 0.09bc | 0.30 ± 0.01a | 1.16 ± 0.06ab | 1.20 ± 0.12ab | 0.52 ± 0.03ab | 2.29 ± 0.10ab | 6.17 ± 0.16 |
| CR | 4.5 ± 2.2c | High | 60.95 ± 11.25cde | 1.94 ± 0.08bc | 0.33 ± 0.01a | 1.00 ± 0.04b | 1.26 ± 0.08ab | 0.54 ± 0.01ab | 2.32 ± 0.11ab | 6.25 ± 0.02 |
| MS | 4.5 ± 2.2c | High | 117.32 ± 11.08a | 1.80 ± 0.05bc | 0.28 ± 0.01ab | 1.24 ± 0.01ab | 1.19 ± 0.11ab | 0.47 ± 0.02b | 2.51 ± 0.13a | 6.16 ± 0.04 |
| SA | 6.7 ± 3.9c | High | 43.07 ± 2.27de | 2.01 ± 0.05bc | 0.21 ± 0.01b | 1.05 ± 0.09b | 1.53 ± 0.08a | 0.63 ± 0.03a | 2.42 ± 0.08ab | 6.33 ± 0.02 |
| PA | 6.7 ± 6.7c | High | 51.71 ± 4.92de | 1.93 ± 0.05bc | 0.27 ± 0.01ab | 1.08 ± 0.05b | 1.30 ± 0.02a | 0.62 ± 0.01a | 2.09 ± 0.07ab | 6.39 ± 0.03 |
| OA | 57.1 ± 25.7ab | Intermediate | 23.70 ± 4.92e | 2.06 ± 0.01ab | 0.31 ± 0.01a | 1.45 ± 0.004a | 1.16 ± 0.27ab | 0.50 ± 0.07ab | 2.31 ± 0.24ab | 6.29 ± 0.06 |
| BG1 | 60.0 ± 13.3b | Intermediate | 94.65 ± 7.71abc | 1.73 ± 0.02c | 0.31 ± 0.01a | 1.19 ± 0.1ab | 1.27 ± 0.06ab | 0.47 ± 0.03b | 2.69 ± 0.03a | 6.01 ± 0.09 |
| FEL | 60.0 ± 20.4ab | Intermediate | 79.06 ± 2.21bcd | 2.06 ± 0.05ab | 0.31 ± 0.02a | 1.19 ± 0.05ab | 1.35 ± 0.07a | 0.54 ± 0.02ab | 2.51 ± 0.20a | 6.41 ± 0.09 |
| BG4 | 75.6 ± 4.4a | Low | 109.64 ± 9.51ab | 1.92 ± 0.09bc | 0.27 ± 0.02ab | 1.17 ± 0.04ab | 1.18 ± 0.15ab | 0.48 ± 0.02b | 2.44 ± 0.20ab | 6.13 ± 0.02 |
| BA | 93.3 ± 6.7a | Low | 40.67 ± 8.65e | 2.32 ± 0.06a | 0.25 ± 0.03ab | 1.16 ± 0.02ab | 0.77 ± 0.04b | 0.43 ± 0.03b | 1.82 ± 0.09b | 6.13 ± 0.11 |
| P-Value | < 0.0001 | < 0.0001 | < 0.0001 | 0.0023 | 0.008 | 0.009 | < 0.0001 | 0.0085 | 0.05 | |
| F Ratio | 9 | 18.33 | 8.09 | 4.64 | 3.71 | 3.57 | 6.1 | 3.64 | 2.41 |
Values are means of three replicates ± standard error.
Different letters within the same column indicate significant differences at p < 0.005.
Table 3.
Aboveground traits (biomass and leaf nutrient contents), rhizosphere microbial diversity (Shannon Index) and mortality of strawberry cultivars inoculated with Macrophomina phaseolina.
| Resistance | Mortality (%) | Plant biomass (g) | Leaf N (%) | Leaf P (%) | Leaf K (%) | Leaf Ca (%) | Leaf Mg (%) | Leaf Ca:Mg | Shannon Index | |
|---|---|---|---|---|---|---|---|---|---|---|
| GNA | High | 10.7 ± 4.6e | 62.13 ± 5.02c | 2.02 ± 0.06ab | 0.23 ± 0.01 | 1.12 ± 0.04abc | 1.15 ± 0.1bcd | 0.50 ± 0.02bc | 2.30 ± 0.14bc | 6.29 ± 0.08 |
| MS | High | 11.3 ± 3.1e | 101.12 ± 11.15ab | 1.98 ± 0.10ab | 0.26 ± 0.02 | 1.24 ± 0.07a | 1.09 ± 0.06d | 0.45 ± 0.01c | 2.43 ± 0.11abc | 6.33 ± 0.04 |
| PA | Intermediate | 25.14 ± 7.3de | 73.03 ± 4.67bc | 2.08 ± 0.09ab | 0.26 ± 0.03 | 1.06 ± 0.11abc | 1.38 ± 0.08abcd | 0.62 ± 0.02ab | 2.24 ± 0.12c | 6.16 ± 0.11 |
| BG4 | Intermediate | 30.6 ± 5.2cde | 101.66 ± 3.00ab | 1.99 ± 0.10ab | 0.21 ± 0.01 | 0.96 ± 0.04abc | 1.24 ± 0.05bcd | 0.51 ± 0.02bc | 2.41 ± 0.07abc | 6.18 ± 0.07 |
| DR | Intermediate | 36.3 ± 2.5cde | 53.57 ± 2.50c | 2.14 ± 0.07ab | 0.30 ± 0.02 | 1.11 ± 0.01abc | 1.55 ± 0.03abc | 0.57 ± 0.01abc | 2.71 ± 0.08abc | 6.33 ± 0.10 |
| ABN | Intermediate | 55.0 ± 4.4abc | 57.42 ± 6.14c | 2.06 ± 0.07ab | 0.26 ± 0.01 | 0.90 ± 0.04bc | 1.57 ± 0.05ab | 0.57 ± 0.03abc | 2.75 ± 0.06ab | 6.26 ± 0.12 |
| ELD | Intermediate | 68.8 ± 2.6 bcd | 120.85 ± 12.22a | 1.77 ± 0.07b | 0.28 ± 0.03 | 1.26 ± 0.03a | 1.37 ± 0.11abcd | 0.48 ± 0.02c | 2.81 ± 0.15a | 6.23 ± 0.06 |
| MOY | Intermediate | 69.1 ± 8.9bcd | 59.40 ± 6.19c | 1.89 ± 0.01ab | 0.26 ± 0.02 | 0.83 ± 0.05c | 1.76 ± 0.08a | 0.65 ± 0.02a | 2.71 ± 0.06abc | 6.15 ± 0.13 |
| FEL | Low | 81.6 ± 7.4ab | 69.49 ± 6.87bc | 2.14 ± 0.04ab | 0.25 ± 0.01 | 1.16 ± 0.03ab | 1.13 ± 0.08 cd | 0.48 ± 0.03c | 2.37 ± 0.1abc | 6.22 ± 0.07 |
| UCJ | Low | 92.86 ± 5.1a | 79.19 ± 12.47bc | 2.20 ± 0.15a | 0.26 ± 0.03 | 1.00 ± 0.15abc | 1.43 ± 0.19abcd | 0.55 ± 0.06abc | 2.58 ± 0.11abc | 6.13 ± 0.09 |
| P-Value | < 0.0001 | < 0.0001 | 0.036 | 0.14 | 0.0013 | 0.00016 | < 0.0001 | 0.0027 | 0.8 | |
| F Ratio | 25.65 | 8.4 | 2.38 | 1.67 | 4.26 | 5.59 | 6.16 | 3.81 | 0.58 |
Analysis of variance was used to compare groups.
Different letters within the same column indicate significant differences at p < 0.005.
Four of the 16 cultivars were represented in both pathogen trials to test the stability of rhizosphere plant-microbial interactions and whether the selection of microorganisms would lead to similar rhizosphere microbial communities, regardless of the environmental conditions and bulk soil microbial community. Significant differences in the microbiome were observed among cultivars (Pseudo-F = 1.83, p = 0.001, Fig. S1), and pathogen trial (Pseudo-F = 9.6, p = 0.001, Fig. S1), but no significant interaction was found between cultivar and pathogen trial (Pseudo-F = 1.11, p = 0.257), indicating the selection of microbes by cultivar in the rhizosphere depended strongly on the environmental conditions and the microbiome of the surrounding bulk soil.
Relationship between the rhizosphere microbiome and health of the strawberry cultivars
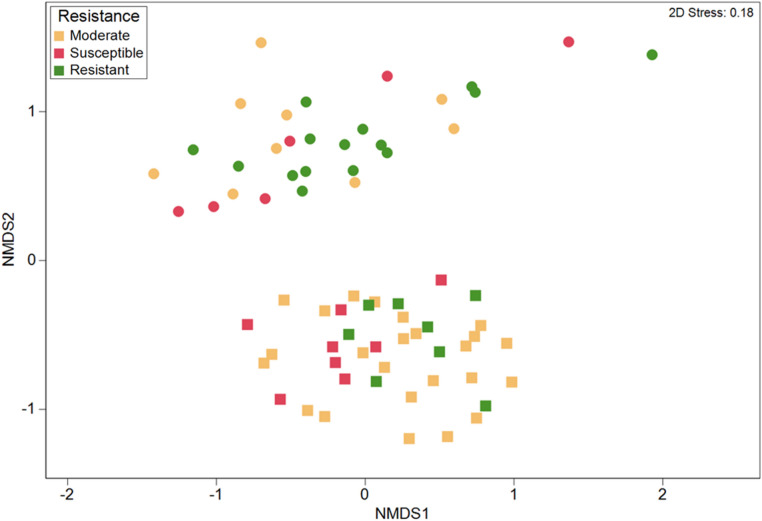
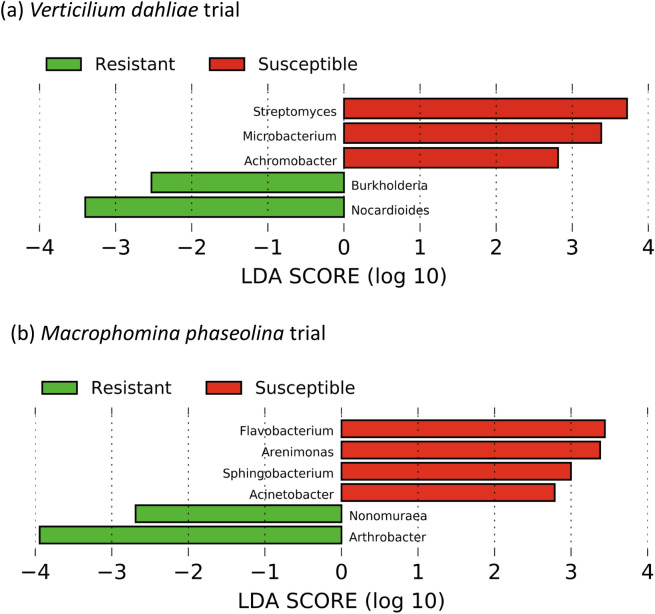
The ten cultivars selected within each pathogen trial, showed remarkable differences in their mortality or resistance to the soil-borne fungal pathogens, aboveground biomass and nutrient uptake. Three distinct disease resistance groups were observed in the V. dahliae trial, with five cultivars showing mortality lower than 10% (resistant), three cultivars showing resistance between 50–70% (moderate) and two cultivars showing more than 70% mortality (susceptible) (Table 2). These differences in plant resistance were matched with differences in the rhizosphere microbiome between the three groups (ANOSIM, R = 0.189, p = 0.015, Fig. 5). We further explored the relationship among the rhizosphere microbiome and disease resistance by comparing the microbiome between the two susceptible and the five resistant cultivars. A SIMPER analysis conducted at the genus level, showed that the resistant and susceptible cultivars in the V. dahliae field trial differed in 31% of the OTUs, and 23 genera contributing to at least 1% of the differences (Table 4). Twelve genera were more abundant in the resistant cultivars including Gaiella, unclassified Acidobacteria Gp4, Gp6, Gp16, Novosphingobium, Arthrobacter, Sphingomonas, Nocardioides, Rubrobacter, Variovorax, Phycioccus and Pseudonocardia (Table 4). The archaeal genera Nitrososphaera, Streptomyces, Rhizobium, Pseudomonas or Flavobacterium were more abundant in the susceptible cultivars (Table 4). The LEfSe analysis showed a total of 10 clades that were significantly more abundant in the resistant cultivars as compared to the susceptible (Table S3), including Burkholderia and Nocardioides, two known fungal antagonists (Fig. 6a).
Figure 5.
Non-metric multidimensional scaling (NMDS) ordinations based on the Bray–Curtis similarity of bacterial and archaeal OTU community structure among strawberry cultivars exhibiting different resistance against Verticillium dahliae (squares) and Macrophomina phaseolina (circles).
Table 4.
SIMPER analysis showing the average percent relative abundances of bacterial genera contributing to at least 1% to the observed variability in the rhizosphere microbiome between cultivars with high and low resistance to Verticilium dahliae and Macrophomina phaseolina.
| Genus | Resistant | Susceptible | Diss/SD | Contribution (%) |
|---|---|---|---|---|
| Verticilium dahliae field trial | ||||
| Nitrososphaera | 7.62 | 8.24 | 1.1 | 9.85 |
| Gp6 | 7.24 | 6.13 | 1.1 | 5.59 |
| Gaiella | 3.87 | 3.37 | 1.25 | 3.66 |
| Novosphingobium | 4.66 | 4.33 | 1.09 | 2.99 |
| Gp16 | 2.59 | 2.37 | 1.55 | 2.54 |
| Arthrobacter* | 3.3 | 2.46 | 1.28 | 2.38 |
| Streptomyces* | 2.31 | 3.46 | 1.38 | 2.35 |
| Rhizobium* | 2.4 | 3.08 | 1.36 | 2.28 |
| Pseudomonas* | 1.05 | 1.91 | 1.31 | 2.23 |
| Sphingomonas* | 4.48 | 4.3 | 1.24 | 2.18 |
| Gp4 | 2.53 | 1.63 | 1.29 | 2.15 |
| Mucilaginibacter | 2.07 | 2.58 | 1.33 | 2.13 |
| Phenylobacterium | 2.82 | 2.88 | 1.41 | 1.71 |
| Hydrocarboniphaga | 0.8 | 0.96 | 1.24 | 1.67 |
| Steroidobacter | 1.49 | 1.73 | 1.42 | 1.56 |
| Aminobacter | 1.09 | 1.27 | 1.09 | 1.43 |
| Nocardioides* | 2.44 | 1.86 | 1.48 | 1.29 |
| Flavobacterium* | 0.81 | 1.35 | 0.73 | 1.26 |
| Rubrobacter | 1.1 | 0.55 | 0.69 | 1.21 |
| Variovorax | 1.48 | 1.38 | 1.26 | 1.14 |
| Phycicoccus | 1.21 | 1.16 | 1.3 | 1.05 |
| Neorhizobium | 1.21 | 1.64 | 1.45 | 1.04 |
| Pseudonocardia | 2.21 | 1.96 | 1.37 | 1.04 |
| Macrophomina phaseolina field trial | ||||
| Pseudomonas* | 8.12 | 7.65 | 0.91 | 8.77 |
| Gp6 | 8.05 | 5.29 | 1.33 | 6.02 |
| Arthrobacter* | 5.27 | 3.67 | 1.4 | 4.26 |
| Sphingomonas* | 4.11 | 4.42 | 1.23 | 3.09 |
| Novosphingobium | 4.15 | 4 | 1.24 | 2.8 |
| Gp16 | 2.95 | 2.51 | 1.32 | 2.7 |
| Phenylobacterium | 2.88 | 3.11 | 1.36 | 2.19 |
| Steroidobacter | 1.63 | 2.06 | 1.32 | 2.11 |
| Xanthomonas | 0.41 | 1.34 | 1.18 | 1.91 |
| Nitrososphaera | 2.65 | 1.82 | 1.63 | 1.8 |
| Gp4 | 2.04 | 1.28 | 1.34 | 1.78 |
| Flavobacterium* | 0.53 | 1.23 | 0.96 | 1.41 |
| Nocardioides* | 2.13 | 2.1 | 1.35 | 1.32 |
| Mucilaginibacter | 1.38 | 1.74 | 1.43 | 1.28 |
| Aminobacter | 0.76 | 1.34 | 1.09 | 1.23 |
| Streptomyces* | 2.32 | 2.08 | 1.04 | 1.2 |
| Microvirga | 2.05 | 1.83 | 1.2 | 1.2 |
| Rhizobium* | 2.3 | 2.55 | 1.45 | 1.12 |
| Gaiella | 1.73 | 1.41 | 1.57 | 1.07 |
| Isoptericola | 0.42 | 0.61 | 1.11 | 1.04 |
Diss/SD values represent the average dissimilarity divided by the standard deviation.
*Potential fungal antagonists19.
Figure 6.
Linear discriminant analysis (LDA) effect size (LEfSe) showing the differential abundance of genera known to include species antagonistic to filamentous fungal pathogen in the rhizosphere of strawberry plants with high and low resistance against Verticillium dahliae (a) and Macrophomina phaseolina (b). Genera listed were significantly different (α = 0.05) and had an LEfSe score of > 2.0. A negative LDA score represents depletion in the rhizosphere of low resistance cultivars and enrichment in the rhizosphere of resistant cultivars (green) and a positive LDA score represents the opposite (red).
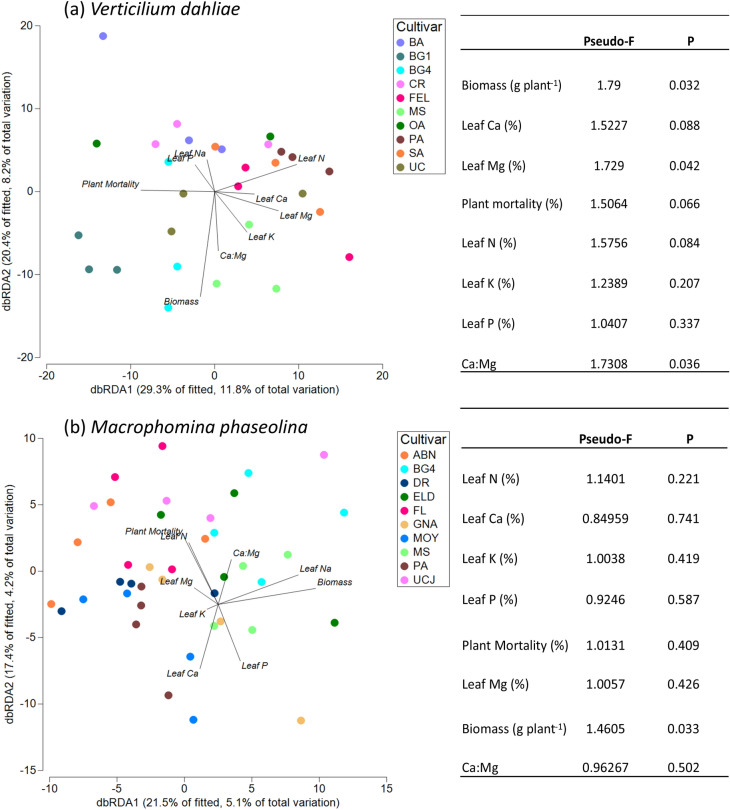
No significant correlation was found between the diversity of the rhizosphere microbiome and plant mortality in response to V. dahliae (p = 0.06), although the Shannon diversity index was significantly positively correlated to the contents of Ca (p = 0.04) and Mg in the leaves of the strawberry plants (p = 0.004; Table S5). We further explored the relationship between the rhizosphere bacterial and archaeal community, plant mortality and other aboveground plant traits by using distance-based redundancy analysis (db-RDA). We observed that microbial community composition in the rhizosphere of the strawberry plants grown in the presence of V. dahliae, was significantly associated with aboveground plant biomass, the leaf Mg content as well as the leaf Ca:Mg ratio of the plants (Fig. 7a).
Figure 7.
Distance-based redundancy analysis (dbRDA) on rhizosphere bacterial and archaeal communities at OTU level and selected plant traits in the Verticillium dahliae (a) and Macrophomina phaseolina (b) trials.
Differences in plant mortality among the cultivars in the M. phaseolina trial were significant but not as clear as in the V. dahliae trial, with a susceptible (more than 80% mortality), a moderate (20–70% mortality), and a resistant group (< 20% mortality) (Table 3). No differences among these resistance groups were found in their rhizosphere microbiome (R = − 0.048, p > 0.05, Fig. 5). We further explored the relationship among the rhizosphere microbiome and disease resistance by comparing the microbiome between the two cultivars with the highest mortality rates (FL, UCJ) and the two cultivars with the lowest mortality (GNA, MS). We found significant differences in the overall OTU community between susceptible and high resistant cultivars (R = 0.2, p = 0.009). A SIMPER analysis on these cultivars (conducted with taxa grouped at the genus level) found an average dissimilarity of 28.8% between susceptible and resistant cultivars. A total of 20 genera contributed to at least 1% of the differences between resistant and susceptible cultivars (Table 4). Eleven genera were more abundant in the resistant cultivars, including Pseudomonas, Gp6 and Arthobacter, which contributed to the largest differences, as well as Novosphingobium, Gp16, Nitrososphaera, Gp4, Nocardioides, Streptomyces, Microvirga and Gaiella. On the other hand, the genera Sphingomonas, Phenylobacterium, Xanthomonas, Flavobacterium, Mucilaginibacter, Aminobacter, Rhizobium and Isoptericola were all more abundant in the rhizosphere of the cultivars susceptible to M. phaseolina (Table 4). LEfSe analysis revealed that resistant cultivars had 34 clades that were significantly more abundant as compared to susceptible cultivars (Table S4), including the fungal antagonists Arthobacter and Nonomuraena (Fig. 6b). The fungal antagonistic bacteria from the genus Flavobacterium, Arenimonas, Sphingobacterium and Acinetobacter were more abundant in the rhizosphere of susceptible plants.
Plant mortality was not found to be significantly correlated to plant aboveground biomass, leaf nutrient content or to the microbial diversity in the rhizosphere of the M. phaseolina trial (Table S6). Plant biomass was significantly positively correlated to leaf K content (p < 0.001), and some other significant correlations were found between leaf K and leaf Ca, Mg and P, although none of these leaf nutrients were correlated to the rhizosphere microbial diversity (Shannon index). The dbRDA analysis revealed that plant biomass was significantly associated with the composition of the microbiome in the rhizosphere, but not with the plant leaf nutrient contents (Fig. 7b).
Discussion
Understanding plant-microbial interactions is instrumental for the development of low input management strategies in high value crops. Here we studied the role of the rhizosphere bacterial and archaeal community in the growth and health of strawberry plants subjected to the biotic stress imposed by two different soil-borne fungal pathogens. To do this, we used 16 cultivars with remarkably different phenotypes including aboveground biomass, nutrient uptake and resistance to soil-borne diseases and we grew them under commercial field conditions. As we had initially hypothesized, the rhizosphere microbiome of the strawberry plants was clearly different from the soil microbiome in the two pathogen trials, showing that the plants are able to select from the pool of microorganisms available in the soil. This has been observed in many other plant species such as rice, corn, tomatoes, beans, barley and Arabidopsis34,37–42; to the best of our knowledge, the research presented here constitutes the first evidence of a distinct rhizosphere microbiome in different strawberry cultivars and environmental conditions. The differences between bulk and rhizosphere soil were observed consistently in the two pathogen trials which had a different soil management history and a different soil microbiome. Interestingly, the structure and diversity of the rhizosphere microbiome of the strawberry cultivars in the two soils were more similar to each other than to their respective bulk soils. This suggests that plant roots, even though they can only select from the pool of microbes available in their surrounding environment, exhibit strong and consistent selection regardless of environmental conditions. This rhizosphere filter has been attributed to the change in pH, redox conditions and production of rhizodeposits including the release of root exudates of different nature7,8,43,44.
The relative abundance of most bacterial and archaeal phyla was lower in the rhizosphere than in the surrounding bulk soil with the exception of Proteobacteria, as has been previously observed in other plant species42,45, as well as Verrucomicrobia and Bacteroidetes in the V. dahliae trial. The phyla Proteobacteria and Bacteroidetes include several bacteria that are known to promote plant growth and/or to be antagonists of soil-borne fungal pathogens, such as Rhizobium, Pseudomonas, Burkholderia and Flavobacterium19,46. All of these taxa, along with other known fungal antagonists, were found in higher abundances in the rhizosphere of the strawberry plants. This capacity of the plants to recruit beneficial microorganisms that help them cope with biotic and abiotic stress has been reported previously for other crops5,47,48. The genus Pseudomonas is known to include several plant growth promoting species and strains capable of producing plant hormones such as auxins, induce systemic resistance, and produce cell wall degrading enzymes and siderophores that chelate iron18,19,49. Similarly, rhizosphere free-living species from the genus Rhizobium have been reported to produce antibiotic substances, chelates and induce systemic resistance in a wide range of crops50,51. Interestingly, the rhizosphere of the strawberry plants was enriched with bacteria that are known to be able to fix atmospheric N, such as Rhizobium and Burkholderia. Rhizobium includes N-fixing bacteria known to establish symbiotic interactions in legumes, however recent evidence shows that Rhizobium could also be naturally present in the rhizosphere of non-legume crops such as sunflower, corn, barley or rice, and can be successfully inoculated in the rhizosphere of vegetables such as tomato, pepper, lettuce and radish, increasing their growth significantly52,53. The genus Burkholderia includes several diazotrophic species, which have been recently found to play a significant role in nitrogen fixation in tomatoes, corn and sugarcane54–56.
In addition to the differences in the overall structure of the microbiome, microbial diversity was different in the rhizosphere of the strawberry plants compared to the bulk soil. Consistent with previous studies that show a decrease in microbial diversity closer to the roots39,45, a lower microbial diversity was observed in the rhizosphere of the plants grown in the M. phaseolina trial. The opposite trend was observed in the V. dahliae trial. Despite this, no significant differences were found in rhizosphere-associated microbial diversity between the two pathogen trials, again showing the homogenizing effect of the microbial filter of the rhizosphere. Similar results were observed by Mendes et al.34 when growing common bean in two different soils.
We assessed whether the presence of the soil-borne fungal pathogens increased the abundance of beneficial bacteria in the rhizosphere of the strawberry plants. The presence of V. dahliae has been shown to induce a change in the fungal microbial community of strawberry in a previous field study57. In our study, the rhizosphere microbiome was significantly modified in the treatment versus the control plots with increases in the relative abundance of the actinobacteria Pseudonocardia and Gaiella as well as the ammonia-oxidizing archaea Nitrososphaera in the plots with V. dahliae; and increases in the relative abundance of Povalibacter, a saprophytic proteobacterium, and Pseudomonas in the plots inoculated with M. phaseolina. These changes could be attributed to a change in the root exudation patterns in the presence of the soil-borne pathogens, a higher prevalence of dead roots, and microbial competition7,10,58,59. Nevertheless, it is also possible that the different management (i.e. fumigation) in the case V. dahliae controls versus the inoculated plots, contributed to the observed differences in this field.
Differences in rhizosphere microbiome between the cultivars showed genotype-dependent selection of the rhizosphere microorganisms as has been observed in other plant species39,41. However, comparison of the same cultivars grown parallelly in the two pathogen trials, showed that their microbial communities changed depending on the field where they were grown, suggesting strong genotype by environment interactions. In fact, these same cultivars showed very different leaf nutrient concentrations and mortality rates, that could be driven by the different soil-borne pathogens, initial soil fertility and microbiome in both pathogen trials60.
Previous studies have shown that, cultivar-specific rhizosphere microbiomes are associated with increased resistance to soil-borne diseases. Resistance can be due to the presence of biocontrol taxa that directly or indirectly suppress the disease (specific disease suppression), and plant growth promoting bacteria that induce systemic disease resistance in the plant. However it is also possible that disease suppression is the result of the coordinated action of all the microorganisms present in the rhizosphere (general disease suppression)61. Mendes et al.34 studied the rhizosphere microbiome of different common bean cultivars with different degrees of resistance to Fusarium oxysporum, and observed that a resistant cultivar had a higher abundance of certain bacterial families such as Pseudomonadaceae, Bacillaceae, Solibacteraceae and Cytophagaceae. Using a combination of 16S rRNA sequencing, shotgun metagenomics, and plate cultures, Kwak et al.36 were able to determine that the resistance of the tomato cultivar Hawaii 7996 to the soil-borne pathogen Ralstonia solanacearum was conferred by the presence of a Flavobacterium strain. In our study, comparisons in the rhizosphere microbiome between cultivars with different levels of resistance against the two fungal pathogens showed significant differences. Correlation analysis carried out through db-RDA showed no relationship between the whole community of the rhizosphere and plant mortality suggesting that resistance could be associated to the presence of biocontrol taxa but not general suppression of the soil-borne diseases. Taxa found in higher abundance in the cultivars resistant to V. dahliae included, Burkholderia, Sphingomonas and Novosphingonium which have been found to suppress this fungal pathogen in previous studies19,62. Additionally, higher abundances of unclassified Acidobacteria (Gp6, Gp16 and Gp4) and Actinobacteria (Arthrobacter, Nocardioides and Gaiella) were found in resistant strawberry cultivars. Acidobacteria have been found to correlate well with apple tree growth before63 and to be negatively correlated to the abundance of Fusarium oxysporum in banana64. Arthrobacter is a known antagonist to V. dahliae through the production of cell wall degrading enzymes19,65, whereas Nocardioides and Gaiella have recently shown to be essential to the resistance of strawberry plants against F. oxysporum66. A recent study performed with soils collected at the continental scale, found that acidobacteria and actinobacteria are key players in the suppression of the fungal pathogen Fusarium oxysporum67. Results of this study show that both acidobacteria and actinobacteria are also key taxa in the resistance of strawberry cultivars against V. dahliae.
In the M. phaseolina trial, resistant cultivars were characterized by greater abundances of Pseudomonas, actinobacteria (Nonomuraea, Arthrobacter, Streptomyces, Nocardioides and Gaiella), which are well-known to be fungal antagonists and plant growth promoters19,48,68. Additionally, the rhizosphere of resistant plants was also characterized by higher abundances of unclassified Acidobacteria (Gp6, Gp16 and Gp4). Interestingly, the same taxa seemed to be in higher abundances in the resistant cultivars in the two field trials, showing a clear role of the rhizosphere microbiome and suggesting a common mechanism in the resistance to soil-borne diseases in the strawberry cultivars.
In addition to playing a role in plant resistance to biotic stress, the rhizosphere microbiome was significantly correlated to other plant traits including plant biomass and leaf Mg and Ca:Mg ratio in the V. dahliae field trial suggesting a role in the response to abiotic stress. The soil at the experimental fields had a high concentration of Mg and low Ca: Mg ratio that could create nutrient stress in the plants as shown by the significant correlations between leaf Mg, Ca, Ca:Mg with plant mortality. Together, these findings show a role of the rhizosphere microbiome in regulating the response of the plant to biotic and abiotic (nutritional) stress. The specific mechanisms and plasticity of these beneficial plant-microbial interactions needs to be further investigated. In particular, given the strong genotype by environment interactions observed when cultivars were compared across the two fields, it is necessary to determine what are the optimal soil conditions and management strategies that can promote beneficial plant-microbial interactions.
Conclusions
Strawberry plants can recruit microorganisms to their rhizosphere from the pool of microbes available in the soil, increasing the relative abundance of beneficial microorganisms involved in nutrient uptake and resistance to soil-borne diseases. Results of two field trials using 16 cultivars show a higher abundance of diazotrophic bacteria in the rhizosphere of strawberry plants which should be further explored as a strategy to reduce inputs of N fertilizers. Furthermore, cultivars resistant to the two soil-borne pathogens consistently showed higher abundances of biocontrol microorganisms such as Pseudomonas, and actinobacteria such as Arthrobacter, Nocardioides and Gaiella. Selection of rhizosphere microorganisms is strongly associated to the different genotypes and therefore seems to be a genetic trait that could be selected through breeding; nevertheless, the microbiome selected by a specific cultivar depends largely on the environmental conditions and microbiome of the surrounding soil. Thus, this suggests that soil health may be important in the establishment of these beneficial plant-microbial interactions.
Material and methods
Plant material
Strawberry cultivars from different breeding programs and representing a range of resistance to soil-borne fungal diseases, were selected from two large ongoing field trials, evaluating 90 cultivars and elite selections for their resistance to Macrophomina phaseolina and Verticillium dahliae. These trials were established in October through July in 2015–2016, 2016–2017 and 2017–2018. Plant material for the current study was selected from the two pathogen trials established in October 2016. We selected 10 cultivars exhibiting a range of resistance, from high to low, to Macrophomina phaseolina and Verticillium dahliae. Out of the 10 cultivars, 6 were unique to each trial and 4 were grown in both trials, for a total of 16 different cultivars evaluate in this study. The names and breeding programs of the cultivars selected from each field trial are shown in Table S7.
Field trials
Two parallel (located 0.4 km apart) replicated pathogen trials were established on the California Polytechnic State University campus in San Luis Obispo, California (USA) to evaluate the 90 strawberry cultivars and elite selections for their resistance to the soil-borne fungal pathogens. The soil at both field sites was classified as Pachic Haploxeroll on a 0–2% slope with a clay loam texture. The soil in the V. dahliae trial had a pH of 7, electrical conductivity (EC) of 1.2, cation exchange capacity (CEC) of 25.5, and 3% organic matter. The soil in the M. phaseolina trial had a pH of 6.8, EC of 1.6, CEC of 17.4, and 3% organic matter.
The trials followed a complete randomized block design with four blocks. Briefly, in each trial, strawberry plants were grown in 1.6 × 2.5 m plots established in raised beds (0.3 m high). Plots contained 20 plants of the same cultivar and constituted the experimental unit. Ninety plots (one per cultivar) were arranged randomly within each block and 4 replicates (blocks) were established (Fig. S1). This design resulted in a total of 360 plots per field trial (90 cultivars × 4 replicates or beds).
Differences in pathogen history between the two field sites prompted distinct methods for ensuring host infection. The V. dahliae trial relied on high levels of microsclerotia found naturally in the field soil (20 CFU g−1 soil). The Andersen sampler technique69 was used to estimate V. dahliae populations in field soil with the following methods. Eight soil cores (2.5 by 30 cm) were collected from each replicated plot, bulked, and thoroughly mixed for analysis. Soil aggregates from each sample were broken up and thoroughly mixed for 5 min. After air drying the soil at room temperature for 1 week, each sample was ground into a fine powder with a mortar and pestle. An Andersen air sampler was used to distribute the soil on the surface of NP-10 medium70. Three replicates of 5 total plates were processed for each sample. After 10 days of incubation in the dark at 20 °C, the surface of the agar medium was gently washed under a stream of water to remove soil particles, and microsclerotial colonies of V. dahliae were counted with the aid of a dissecting microscope. The estimate of soil inoculum density was expressed as an average number of CFU of V. dahliae g−1 of dry soil.
Macrophomina phaseolina was not naturally present in the soil at the field site and had to be inoculated. Prior to the inoculation, the field was fumigated (392 kg ha-1 of a 50% chloropicrin + 50% methyl bromide solution) to ensure the removal of all other soil-borne fungal pathogens. Fumigation was carried out in 2015, two years before the start of this trial, since the effects last usually 2–3 years. At transplanting, 5 g of M. phaseolina inoculum (20,500–25,200 CFU g-1) was applied to the soil-crown interface. Macrophomina inoculum was produced from field isolates collected from diseased plants in 2014 and 2015 following procedure by Mihail71. Briefly, a homogenized 1:0.4:0.4 sand: cornmeal: deionized water mixture was autoclaved for one hour on two separate days in separate 250 mL containers (Nalgene, Rochester, NY). After cooling to room temperature, the containers were inoculated with plugs of the pathogen. Inoculated cornmeal-sand containers were then incubated at 30 °C for two weeks and shaken vigorously by hand daily to aid rapid colonization. After incubation the colonized cornmeal-sand inoculum was spread over a metal tray to air dry for five days. Once dry, this inoculum was stored in the dark at room temperature for two weeks before being applied in the field. The density of viable microsclerotia within the cornmeal-sand inoculum was enumerated with direct plating of 10–3 to 10–4 serial dilutions onto NP-10 medium70. Ten replicate plates of each dilution were used. The plates were incubated in the dark at 30 °C for 7 days before counting colony forming units (CFU).
Additionally, in each pathogen trial, one block was established as a non-inoculated control where the soil-borne pathogens were eliminated through chemical fumigation prior to planting (V. dahliae trial) or not introduced artificially (M. phaseolina trial). The control block contained one 20-plant plot per cultivar for a total of 90 plots (90 cultivars). Due to the lack of replication, no comparisons were made between inoculated and control plots at the cultivar level. Nevertheless, control plots were used to assess the overall effects of pathogen presence in the rhizosphere microbiome of the plants.
Bare-root strawberry seedlings were transplanted to the control and experimental plots in mid-October 2016 and grown until the end of July 2017. Fertilization was carried out through subsurface drip irrigation (fertigation) during the growing season and preplant slow release fertilizer banded in the bed for a total input of 112 kg N ha−1.
Soil and plant sample collection
During fruit set, we determined plant mortality in the 90 cultivars visually by assessing the percentage of dead plants in each plot. Additionally, presence of the pathogens in symptomatic plants was confirmed by plating tissue pieces of internal crown tissue on selective media. Tissue isolations were conducted on 10 symptomatic plants in each replication in the host resistance trial on a biweekly basis. Subsequently, we selected ten representative cultivars exhibiting a range of high to low resistance within each pathogen trial for further study of their rhizosphere microbiome. A summary of the cultivars used in this study with their respective breeding programs is shown in Table S7.
At harvest (July 2017), we performed another visual evaluation to determine actual plant mortality per plot at the moment of harvest (% of dead plants). Plants were classified as susceptible (more than 70% mortality), moderate (20–70% mortality), and resistant (< 20% mortality). One shoot, soil and rhizosphere composite sample was collected subsequently from each plot for each of the selected cultivars (Fig. S2). Briefly, within each plot, four plants were randomly selected while avoiding dead or dying plants to reduce the confounding effects of the dead root tissue, typical of symptomatic plants, in the soil microbiome. The above and belowground components of each plant were separated and then consolidated separately by plot to obtain one root and one shoots sample per plot. Fresh shoot biomass was recorded in the field after removing fruit. Subsamples of the shoot biomass were then collected in each plot for oven drying and the determination of aboveground plant biomass and leaf nutrient content. A preliminary sampling showed that the highest concentration of primary and secondary roots was found in a radius of 5 cm from the crown and 10 cm from the soil surface. Root samples were collected using 5-cm diameter cores centered around the strawberry crown to a depth of approximately 10 cm. A 750 cm3 bulk soil sample (without the presence of roots) from each plot was also collected from the center of each strawberry bed (50 cm from the nearest plant) at a depth of 10 cm. Rhizosphere soil was collected by dry sieving roots on sterilized 500 µm sieves. Samples were lyophilized and stored at − 80 °C prior to DNA extraction.
Leaf tissue analysis
Tissue analysis was performed using leaf and petiole samples dried for 24 h at 55 °C. Dry samples were crushed using a mortar and pestle, followed by two 2-min cycles of grinding in a Bel-Art Micro-Mill grinder (Wayne, NJ, USA). Tissue carbon and nitrogen concentrations were analyzed from a 300 mg subsample using a vario MAX CNS Macro Elemental Analyzer (Elementar Analysensysteme GmbH, Hanau, Germany). Quality control was ensured by analyzing duplicate samples and NIST SRM 1573A tomato leaf standards every ten samples. Concentrations of leaf P, K, Ca and Mg were determined using a modified USEPA72 acid digestion method in the Ultima 2 ICP-OES (Horiba Jobin Yvon S.A.S., Logjumeau, France).
DNA isolation and extraction from rhizosphere and bulk soils
To study bacterial and archaeal diversity in the soil and rhizosphere samples, DNA was extracted from a 250 mg subsample using the DNeasy PowerSoil Kit (QIAGEN, Venlo, Netherlands) following the manufacturer’s recommendations. DNA yield was measured on a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Samples with yield below 5 ng/µL were extracted again prior to amplification. The V4 region of the 16S rRNA was subsequently sequenced using bacterial/archaeal universal primers 515F (5′ -GTG YCA GCM GCC GCG GTA A- 3′) and 806R (3′ -GGA CTA CNV GGG TWT CTA AT-3′)73. PCR reagents included 25 µL of DDH20, 2.5 µL of each primer, 12.5 µL Phusion Hot start flex 2 × Master Mix (Thermo Fisher Scientific), and 50 ng of template DNA. The thermal cycling scheme consisted of 30 s at 98 °C, 10 s at 98 °C, 35 cycles of 54 °C for 30 s, 45 s at 72 °C and 10 min at 72 °C. After the first round of PCR, sequencing adapters and barcodes were added for further amplification. PCR amplification products were visualized by 2% agarose gel electrophoresis. The target fragments were recovered using the AxyPrep PCR Cleanup Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions. The PCR product was further purified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific). The library was quantified on the QuantiFluor fluorescence quantification system (Promega Corporation, Madison, WI, USA). Each qualified sequencing library (i.e., had concentrations above 2 mM) was diluted and pooled/multiplexed. The pooled library was loaded on Illumina MiSeq and the MiSeq Reagent Kit v2, 500 cycles (Illumina) was used for paired-end sequencing (2 × 250 bp). Raw sequence data has been deposited in GenBank (accession number PRJNA666409).
Sequence data processing
Paired-end sequences obtained on the MiSeq platform were de-multiplexed, followed by raw data filtering and processing utilizing the UPARSE pipeline (v11.0.667, www.drive5.com/usearch). Paired end reads were merged with an allowance of 10 bp max differences and with a maximum allowance of 10 bp differences and > 80% identification. Sequences with less than 50 bp were filtered and all sequences trimmed to 280 bp. Low quality sequences were filtered using a maximum expected error threshold of 1.0. Sequences were clustered into denovo Operational Taxonomic Units (OTUs) based on 97% similarity and singletons were discarded. Taxonomy was assigned to representative OTU sequences using the Naïve Bayesian classifier74 based on the Ribosomal Database Project37,75. Chloroplast and unassigned sequences were removed from OTU tables prior to statistical analyses. Samples were rarefied to match the sequences of the lowest count sequence of 6,666 for analyses using taxonomic rank abundance.
Data analysis
Resemblance matrices for the OTU tables were generated using Bray–Curtis similarity of the standardized and Log (x + 1) transformed data76. Interpretations of multivariate distances were conducted using nonmetric multi-dimensional scaling (NMDS) ordinations based on the resemblance matrices using a maximum of 50 restarts and the lowest stress solutions. The differences between sample types (rhizosphere or bulk soil), pathogen trials (V. dahliae or M. phaseolina), plant genotypes and resistance to the soil-borne diseases in the soil microbial community composition within each pathogen trial was assessed through analysis of similarities (ANOSIMS) or PERMANOVA (for bifactorial analyses) using Spearman rank correlation with 999 max permutations of the resemblance matrices were used to determine significance levels between groups. Similarity percentage (SIMPER) analyses were conducted using the Bray–Curtis similarity matrix. Correlations between rhizosphere OTU relative abundances and plant variables (leaf nutrient contents and mortality) were made using the distance-based linear model (DistLM) with 999 permutations. All microbial community data were analyzed using PRIMER-e version 7 with the PERMANOVA + addon (Quest Research Limited, Auckland, NZ).
The effect of ‘sample type’ (bulk vs. rhizosphere soil), ‘cultivar’ and ‘cultivar resistance’ to the fungal pathogens on the microbial diversity (Shannon diversity index), plant mortality, and plant nutrient comparisons among groups were conducted using general linear models and Tukey HSD post hoc tests in JMP Pro (SAS, Cary, NC); Pearson’s correlations were carried out among the different plant traits analyzed using JMP Pro 13. A list of bacterial genera known to include species antagonistic to filamentous fungal plant pathogens was used to compare treatment groups (Inderbitzin et al., 2017). Linear discriminant analysis (LDA) effect size (LEfSe) comparing taxonomic features between groups was conducted online (www.huttenhower.sph.harvard.edu/galaxy) with an LDA threshold of 2.0 and an alpha value of 0.05.
Supplementary Information
Acknowledgements
This work was supported by the California Agriculture Research Institute of the California State University (CSU ARI 19-03-008), and the California Strawberry Commission.
Author contributions
C.L., K.I. and G.H. designed the study and obtained research funding. E.B., C.L., K.I., conducted the experiments. E.B., A.P., S.H. and C.L. analyzed the data, C.L. and E.B. prepared figures and tables. C.L. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82768-2.
References
- 1.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 2.Bardgett RD, van der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515:505–511. doi: 10.1038/nature13855. [DOI] [PubMed] [Google Scholar]
- 3.van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 4.Sergaki C, Lagunas B, Lidbury I, Gifford ML, Schäfer P. Challenges and approaches in microbiome research: from fundamental to applied. Front. Plant Sci. 2018;9:1205. doi: 10.3389/fpls.2018.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 6.Finzi AC, et al. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Change Biol. 2015;21:2082–2094. doi: 10.1111/gcb.12816. [DOI] [PubMed] [Google Scholar]
- 7.Haichar FZ, Santaella C, Heulin T, Achouak W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014;77:69–80. doi: 10.1016/j.soilbio.2014.06.017. [DOI] [Google Scholar]
- 8.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. doi: 10.1007/s11104-009-9925-0. [DOI] [Google Scholar]
- 9.Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- 10.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Richardson AE, Simpson RJ. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 2011;156:989–996. doi: 10.1104/pp.111.175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu B, et al. Rhizosphere priming effects on soil carbon and nitrogen mineralization. Soil Biol. Biochem. 2014;76:183–192. doi: 10.1016/j.soilbio.2014.04.033. [DOI] [Google Scholar]
- 13.Moreau D, Bardgett RD, Finlay RD, Jones DL, Philippot L. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019;33:540–552. doi: 10.1111/1365-2435.13303. [DOI] [Google Scholar]
- 14.Sharma A, Johri BN, Sharma AK, Glick BR. Plant growth-promoting bacterium Pseudomonas sp. strain GRP3 influences iron acquisition in mung bean (Vigna radiata L. Wilzeck) Soil Biol. Biochem. 2003;35:887–894. doi: 10.1016/S0038-0717(03)00119-6. [DOI] [Google Scholar]
- 15.Vaid, S. K., Kumar, B., Sharma, A., Shukla, A. K. & Srivastava, P. C. Effect of zinc solubilizing bacteria on growth promotion and zinc nutrition of rice. J. Soil Sci. Plant Nutr. 22 (2014).
- 16.Goteti PK, Emmanuel LDA, Desai S, Shaik MHA. Prospective zinc solubilising bacteria for enhanced nutrient uptake and growth promotion in maize (Zea mays L.) Int. J. Microbiol. 2013;2013:1–7. doi: 10.1155/2013/869697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacheron, J. et al. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci.4 (2013). [DOI] [PMC free article] [PubMed]
- 18.Narendra Babu A, Jogaiah S, Ito S, Kestur Nagaraj A, Tran L-SP. Improvement of growth, fruit weight and early blight disease protection of tomato plants by rhizosphere bacteria is correlated with their beneficial traits and induced biosynthesis of antioxidant peroxidase and polyphenol oxidase. Plant Sci. 2015;231:62–73. doi: 10.1016/j.plantsci.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Inderbitzin P, et al. Soil Microbiomes associated with verticillium wilt-suppressive broccoli and chitin amendments are enriched with potential biocontrol agents. Phytopathology. 2018;108:31–43. doi: 10.1094/PHYTO-07-17-0242-R. [DOI] [PubMed] [Google Scholar]
- 20.Simpson D. The Economic Importance of Strawberry Crops. In: Hytönen T, Graham J, Harrison R, editors. The Genomes of Rosaceous Berries and Their Wild Relatives. Berlin: Springer; 2018. pp. 1–7. [Google Scholar]
- 21.Food and Agriculture Organization of the United Nations (FAO). FAO Global Statistical Yearbook. (2018).
- 22.California Department of Food and Agriculture. California Agricultural Statistics Review 2017–2018. (2018).
- 23.Holmes, G. J., Mansouripour, S. M. & Hewavitharana, S. Strawberries at the Crossroads: Management of Soilborne Diseases in California without Methyl Bromide. Phytopathology PHYTO-11-19-0406-IA (2020) 10.1094/PHYTO-11-19-0406-IA. [DOI] [PubMed]
- 24.Lloyd M. Growing for the future: Collective action, land stewardship and soilborne pathogens in California strawberry production. Calif. Agric. 2016;70:101–103. doi: 10.3733/ca.2016a0009. [DOI] [Google Scholar]
- 25.Koike, S. T. Crown Rot of Strawberry Caused by Macrophomina phaseolina in California. Plant Dis.92 (2018). [DOI] [PubMed]
- 26.Guthman J. Land access and costs may drive strawberry growers’ increased use of fumigation. Calif. Agric. 2017;71:184–191. doi: 10.3733/ca.2017a0017. [DOI] [Google Scholar]
- 27.Shaw DV, Gubler D, Hansen J. Field resistance of California strawberries to Verticilium dahliae at three conidial inoculum concentrations. HortScience. 1997;32:711–713. doi: 10.21273/HORTSCI.32.4.711. [DOI] [Google Scholar]
- 28.Shaw DV, Gordon TR, Hansen J, Kirkpatrick SC. Relationship between the extent of colonization by Verticillium dahliae and symptom expression in strawberry (Fragaria × ananassa ) genotypes resistant to verticillium wilt. Plant Pathol. 2010;59:376–381. doi: 10.1111/j.1365-3059.2009.02203.x. [DOI] [Google Scholar]
- 29.Antanaviciute L, et al. Mapping QTL associated with Verticillium dahliae resistance in the cultivated strawberry (Fragaria × ananassa) Hortic. Res. 2015;2:15009. doi: 10.1038/hortres.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besbes F, Habegger R, Schwab W. Induction of PR-10 genes and metabolites in strawberry plants in response to Verticillium dahliae infection. BMC Plant Biol. 2019;19:128. doi: 10.1186/s12870-019-1718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez S, Henríquez JL, Urcola LA, Scott A, Gambardella M. Susceptibility of strawberry cultivars to root and crown rot caused by Macrophomina phaseolina. J. Berry Res. 2016;6:345–354. doi: 10.3233/JBR-150114. [DOI] [Google Scholar]
- 32.Pastrana AM, Basallote-Ureba MJ, Aguado A, Capote N. Potential inoculum sources and incidence of strawberry soilborne pathogens in Spain. Plant Dis. 2017;101:751–760. doi: 10.1094/PDIS-08-16-1177-RE. [DOI] [PubMed] [Google Scholar]
- 33.Viejobueno, J., Ramallo, A. C., Kirschbaum, D. S., Baino, O. M. & Salazar, S. M. Severe outbreaks of strawberry crown and root charcoal rot caused by Macrophomina phaseolina in Tucumán, Argentina. 5 (2017).
- 34.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018;12:212–224. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao H, Wu F. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt: Soil microbial community structure in cucumber rhizosphere. FEMS Microbiol. Ecol. 2010;72:456–463. doi: 10.1111/j.1574-6941.2010.00859.x. [DOI] [PubMed] [Google Scholar]
- 36.Kwak M-J, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018;36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 37.Lee SA, et al. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 2019;9:9300. doi: 10.1038/s41598-019-45660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards KR, Kaštovská E, Borovec J, Šantrůčková H, Picek T. Species effects and seasonal trends on plant efflux quantity and quality in a spruce swamp forest. Plant Soil. 2018;426:179–196. doi: 10.1007/s11104-018-3610-0. [DOI] [Google Scholar]
- 39.Brisson VL, Schmidt JE, Northen TR, Vogel JP, Gaudin ACM. Impacts of maize domestication and breeding on rhizosphere microbial community recruitment from a nutrient depleted agricultural soil. Sci. Rep. 2019;9:15611. doi: 10.1038/s41598-019-52148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulgarelli D, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walters WA, et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. 2018;115:7368–7373. doi: 10.1073/pnas.1800918115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? Root exudates and rhizosphere bacteria. FEMS Microbiol. Ecol. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- 44.Hinsinger P, Plassard C, Tang C, Jaillard B. Origins of root-mediated pH changes in the rhizosphere and their responses to environmental constraints: a review. Plant Soil. 2003;248:43–59. doi: 10.1023/A:1022371130939. [DOI] [Google Scholar]
- 45.Peiffer JA, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Youseif SH. Genetic diversity of plant growth promoting rhizobacteria and their effects on the growth of maize plants under greenhouse conditions. Ann. Agric. Sci. 2018;63:25–35. doi: 10.1016/j.aoas.2018.04.002. [DOI] [Google Scholar]
- 47.Rolli E, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait: Root bacteria protect plants from drought. Environ. Microbiol. 2015;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- 48.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 49.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 50.Chakraborty U, Chakraborty BN. Interaction of Rhizobium leguminosarum and Fusarium solani f.sp. pisi on pea affecting disease development and phytoalexin production. Can. J. Bot. 1989;67:1698–1701. doi: 10.1139/b89-214. [DOI] [Google Scholar]
- 51.Tonelli ML, Figueredo MS, Rodríguez J, Fabra A, Ibañez F. Induced systemic resistance -like responses elicited by rhizobia. Plant Soil. 2020 doi: 10.1007/s11104-020-04423-5. [DOI] [Google Scholar]
- 52.Alami Y, Achouak W, Marol C, Heulin T. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 2000;66:3393–3398. doi: 10.1128/AEM.66.8.3393-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Fraile P, et al. Rhizobium promotes non-legumes growth and quality in several production steps: towards a biofertilization of edible raw vegetables healthy for humans. PLoS ONE. 2012;7:e38122. doi: 10.1371/journal.pone.0038122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perin L, et al. Diazotrophic burkholderia species associated with field-grown maize and sugarcane. Appl. Environ. Microbiol. 2006;72:3103–3110. doi: 10.1128/AEM.72.5.3103-3110.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martinez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007;73:5308–5319. doi: 10.1128/AEM.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Deynze A, et al. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biol. 2018;16:e2006352. doi: 10.1371/journal.pbio.2006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nallanchakravarthula S, Mahmood S, Alström S, Finlay RD. Influence of soil type, cultivar and Verticillium dahliae on the structure of the root and rhizosphere soil fungal microbiome of strawberry. PLoS ONE. 2014;9:e111455. doi: 10.1371/journal.pone.0111455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Y, et al. Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils. 2016;52:997–1005. doi: 10.1007/s00374-016-1136-2. [DOI] [Google Scholar]
- 59.Dudenhöffer J, Scheu S, Jousset A. Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J. Ecol. 2016;104:1566–1575. doi: 10.1111/1365-2745.12626. [DOI] [Google Scholar]
- 60.Wei Z, et al. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019;5:eaaw0759. doi: 10.1126/sciadv.aaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 62.Snelders, N. C. et al. A Plant Pathogen Utilizes Effector Proteins for Microbiome Manipulation. (2020). 10.1101/2020.01.30.926725.
- 63.Franke-Whittle IH, Manici LM, Insam H, Stres B. Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil. 2015;395:317–333. doi: 10.1007/s11104-015-2562-x. [DOI] [Google Scholar]
- 64.Fu L, et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol. Biochem. 2017;104:39–48. doi: 10.1016/j.soilbio.2016.10.008. [DOI] [Google Scholar]
- 65.Morrissey RF, Dugan EP, Koths JS. Chitinase production by an Arthrobacter sp. lysing cells of Fusarium roseum. Soil Biol. Biochem. 1976;8:23–28. doi: 10.1016/0038-0717(76)90016-X. [DOI] [Google Scholar]
- 66.Zhao F, et al. Vermicompost can suppress Fusarium oxysporum f. sp. lycopersici via generation of beneficial bacteria in a long-term tomato monoculture soil. Plant Soil. 2019;440:491–505. doi: 10.1007/s11104-019-04104-y. [DOI] [Google Scholar]
- 67.Trivedi P, et al. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol. Biochem. 2017;111:10–14. doi: 10.1016/j.soilbio.2017.03.013. [DOI] [Google Scholar]
- 68.Cha J-Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butterfield EJ. Reassessment of soil assays for Verticillium dahliae. Phytopathology. 1977;77:1073. doi: 10.1094/Phyto-67-1073. [DOI] [Google Scholar]
- 70.Kabir Z, Bhat RG, Subbarao KV. Comparison of media for recovery of Verticillium dahliae from soil. Plant Dis. 2004;88:49–55. doi: 10.1094/PDIS.2004.88.1.49. [DOI] [PubMed] [Google Scholar]
- 71.Mihail, J. D. Macrophomina. In Methods for Research on Soilborne Phytopathogenic Fungi 134–136 (American Phytopathological Society, Saint Paul).
- 72.U.S. EPA. Method 3050B: Acid Digestion of Sediments, Sludges, and Soils (1996).
- 73.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole JR, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucl. Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clarke KR, Warwik RM. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. Plymouth: PRIMER-e Ltd; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.