Abstract
STAT1 is a master regulator that orchestrates type 1 and 2 interferon (IFN)-induced IFN-stimulated gene (ISG) expression. The mechanisms by which STAT1 is phosphorylated and activated upon IFN signaling remain elusive. Our work demonstrated that ubiquitination of STAT1 mediated by the E3 ligase RNF220 contributed significantly to STAT1 activation and innate immune responses. Rnf220 gene deficiency resulted in the downregulation of IFN signaling and decreased expression of ISGs in response to type 1 and 2 IFNs stimulation and Acinetobacter baumannii and HSV-1 infection. Mechanistically, RNF220 interacted with STAT1 and mediated the K63-linked polyubiquitination of STAT1 at residue K110, which promoted the interaction between STAT1 and the kinase JAK1. The expression of RNF220 was induced by pathogenic infection and IFN signaling. RNF220 promoted STAT1 ubiquitination and phosphorylation through a positive feedback loop. RNF220 haploinsufficiency impaired IFN signaling, and RNF220-defective mice were more susceptible to A. baumannii and HSV-1 infection than WT mice. Our work offers novel insights into the mechanisms of STAT1 modulation and provides potential therapeutic targets against bacterial and viral infection and inflammatory diseases.
Subject terms: Antimicrobial responses, Cell death and immune response, Interferons, Infectious diseases, Signal transduction
Introduction
Type 1 interferon (IFN) signaling is crucial for the host defense against pathogenic infection and autoimmune diseases; thus, the production and downstream events of type 1 IFN are tightly regulated [1]. Type 1 IFN is produced in three major pathways: the TLR3- and TLR4-mediated TIR domain-containing adapter-inducing IFN-β (TRIF)-dependent pathway, the intracellular RNA-induced mitochondrial antiviral signaling (MAVS)-dependent pathway, the intracellular DNA-induced cytosolic sensors cyclic GMP-AMP synthase (cGAS)-stimulator of IFN genes (STING) axis-dependent pathway [2–4]. The produced type 1 IFN further activates on IFN-α receptor (IFNAR) and induces the downstream JAK kinase-mediated phosphorylation of STAT proteins, such as phosphorylated STAT1, STAT2, and IRF9, which form the heterotrimeric IFN-stimulated gene factor 3 (ISGF3) complex. The ISGF3 binds to IFN-stimulated response elements (ISREs), and phosphorylated STAT1 forms homodimers that bind to the IFN-γ activation sequence (GAS) to initiate transcription [1, 5].
STAT1 regulates various biological effects downstream of type 1 and type 2 IFN receptors and many other cytokine receptors in multiple cell types [6]. The cooperative binding of STAT1 to DNA was reported to be essential for the promoter recruitment of STAT1 homodimers but not that of ISGF3, which distinguishes type 1 from type 2 IFN signaling [7]. Canonical STAT1 activation is regulated by the JAK-mediated phosphorylation of STAT1 at tyrosine reside 701 (Y701). Phosphorylation-defective mutant STAT1Y701F was also demonstrated to stimulate the expression of a subset of interferon-inducible genes that contribute to the antibacterial immune responses [8, 9]. In addition to its phosphorylation, STAT1 is also regulated by the other posttranslational modifications, such as ubiquitination and SUMOylation. STAT1 SUMOylation was indicated to have an inhibitory effect on STAT1 activation through decreasing STAT1 tyrosine phosphorylation and DNA-binding activity [10, 11]. Ubiquitination of STAT1 was initially illustrated to destabilize IFN-γ-activated STAT1 by using a proteasome inhibitor, and the ubiquitin-proteasome pathway was shown to negatively regulate IFN-γ induced STAT1 activation [12]. To date, a number of ubiquitinating E3 ligases and deubiquitinases have been shown to contribute to STAT1 ubiquitination and physiological activity. For instance, the SMURF1 protein negatively regulates interferon-γ signaling through promoting STAT1 protein ubiquitination and degradation [13]. In contrast, the deubiquitinases ubiquitin-specific protease 13 (USP13) and USP2a remove the K48-linked polyubiquitin from total and phosphorylated STAT1 and regulate IFN signaling and antiviral activity by stabilizing STAT1 [14, 15]. The NKLAM-mediated K63-linked polyubiquitination of STAT1 promotes the interaction between STAT1 and JAK1 in the nucleus and positively regulates STAT1 transcriptional activity [16]. Recently, the nuclear RNF2-mediated K33-linked polyubiquitination STAT1 was demonstrated to promote STAT1 disassociation from DNA [17]. STAT1 activity is tightly regulated at multiple levels in response to different kinds of pathogenic infections.
RNF220 is a RING domain E3 ubiquitin ligase that was demonstrated to mediate the ubiquitination of multiple targets and is involved in various developmental processes and disease progression through previous investigations by our and other groups. Our previous work showed that RNF220 interacted with Sin3B for Sin3B ubiquitination and proteasomal degradation, which was recently implicated in the development of the sperm head in human patients [18, 19]. We also reported that RNF220 interacted with the deubiquitinase USP7 to mediate the deubiquitination of β-catenin and promoted canonical Wnt signaling [20]. Our previous work also revealed that the RNF220-mediated K63-linked polyubiquitination of Gli and its nuclear export contributed to the formation of a Gli gradient and neural patterning in neural development [21–23]. Furthermore, Kim et al. recently reported that RNF220 interacted with ZC4H2 to mediate the ubiquitination and proteasomal degradation of Dbx1/2 and Nkx2.2 to specify spinal progenitor domains [24]. Despite advances in understanding RNF220 function in the development and signaling pathways, the role of RNF220 in the innate immune responses during pathogenic infections remains unknown.
Here, we showed that the expression of Rnf220 was upregulated during bacterial and viral infection. RNF220 mediated the K63-linked polyubiquitination of STAT1, which sustained the STAT1 activation through regulating the interaction between JAK1 and STAT1. RNF220 deficiency impaired type 1 and 2 IFN signaling and increased host susceptibility to A. baumannii and HSV-1 infection.
Results
Rnf220 expression is induced during infection and stimulation
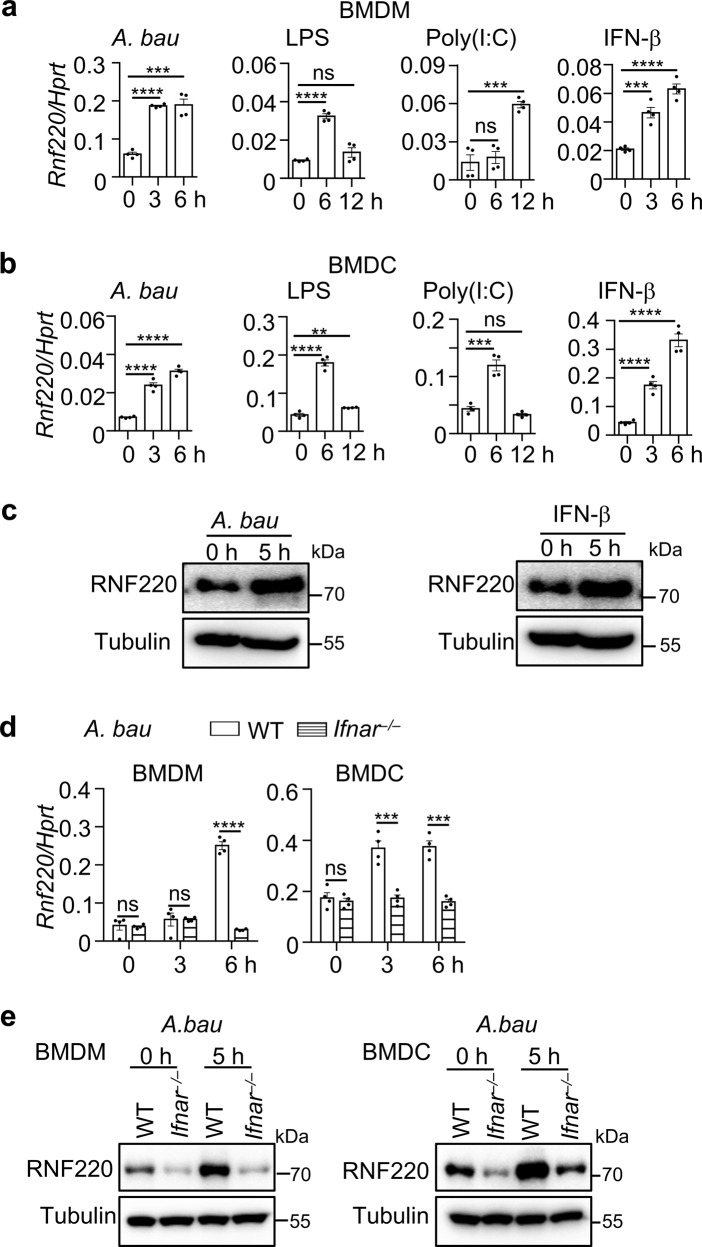
To examine the role of RNF220 in host immune responses, we investigated the expression of Rnf220 during pathogenic infection and ligand stimulation. The expression of Rnf220 was substantially increased in primary bone-marrow-derived macrophages (BMDMs) and bone-marrow-derived dendritic cells (BMDCs) in response to A. baumannii infection, and treatments with LPS, Poly (I:C), and IFN-β (Fig. 1a, b). The increased expression of RNF220 induced by A. baumannii infection and IFN-β treatment was also confirmed by western blot analysis (Fig. 1c). We identified several ISRE and GAS elements within the promoter region of Rnf220 gene (Supplementary Fig. 1a). To determine whether type 1 IFN signaling is crucial for the expression of Rnf220, WT and Ifnar–/– BMDMs and BMDCs were infected with A. baumannii. Notably, the expression of Rnf220 induced by A. baumannii infection was significantly reduced in Ifnar–/– BMDMs and BMDCs compared with that in WT compartments (Fig. 1d, e).
Fig. 1. RNF220 expression is induced in response to A. baumannii infection and ligand stimulation.
a, b Gene expression analysis of Rnf220 in WT BMDMs (a) and BMDCs (b) in response to A. baumannii infection (50 MOI), LPS (200 ng/ml), Poly(I:C) (40 μg/ml), and IFN-β (2 ng/ml) treatment for indicated times. c Immunoblot analysis of RNF220 in WT BMDMs in response to A. baumannii infection and IFN-β treatment for indicated times. d Gene expression analysis of Rnf220 in WT and Ifnar–/– BMDMs and BMDCs in response to A. baumannii infection for indicated times. e Immunoblot analysis of RNF220 in WT and Ifnar–/– BMDMs and BMDCs in response to A. baumannii infection for indicated times. Data represent three independent experiments and are presented as mean ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant.
RNF220 regulates the expression of type 1 IFN-stimulated genes
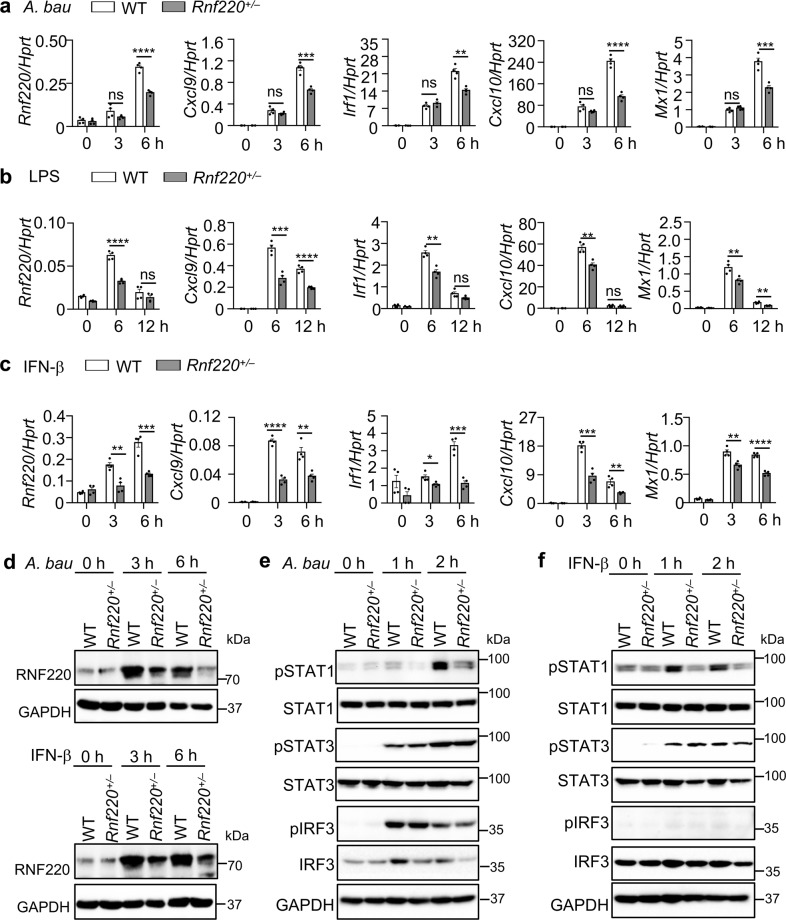
Rnf220 homozygous knockout mice have an embryonic lethal phenotype [22]. The expression of Rnf220 in Rnf220 heterozygous (Rnf220+/–) BMDMs was partially reduced compared with that in WT cells (Fig. 2a). We infected BMDMs from WT and Rnf220+/– mice with A. baumannii and examined the inflammatory cytokine expression and type 1 IFN signaling. Interestingly, the expression of Cxcl9, Cxcl10, Irf1, and Mx1, which are downstream molecules of type 1 IFN, was substantially reduced in Rnf220+/– BMDMs compared with that in WT BMDMs (Fig. 2a). However, the expression of Tnf, Il1a, and Il6 was comparable between WT and Rnf220+/– BMDMs (Supplementary Fig. 1b). The expression of type 1 IFN-inducible genes but not inflammatory cytokine genes was downregulated in Rnf220+/– BMDMs compared with that in WT BMDMs upon LPS and IFN-β treatments (Fig. 2b, c and Supplementary Fig. 1c, d). Notably, the protein level of RNF220 was substantially induced by A. baumannii and IFN-β in WT BMDMs, but not Rnf220+/– BMDMs (Fig. 2d). The reduced production of CXCL9 and TNF in Rnf220+/– BMDMs compared with WT BMDMs triggered by A. baumannii and IFN-β were confirmed by ELISA analysis (Supplementary Fig. 1e). Consistently, the phosphorylation of STAT1 induced by A. baumannii infection and IFN-β treatment was substantially reduced in Rnf220+/– BMDMs compared with that in WT BMDMs, whereas the phosphorylation of IRF3 and STAT3 was not affected in Rnf220+/– BMDMs (Fig. 2e, f). The phosphorylation of ERK and IκBα was comparable between WT and Rnf220+/– BMDMs during A. baumannii infection, while IFN-β mainly activates the IFNAR downstream signaling, and has a slight effect on the phosphorylation of IRF3, ERK, and IκBα (Fig. 2f and Supplementary Fig. 1f, g).
Fig. 2. Type 1 IFN signaling is impaired in Rnf220+/– BMDMs.
a–c Gene expression analysis of Rnf220, Cxcl9, Irf1, Cxcl10, and Mx1 in WT and Rnf220+/– BMDMs in response to A. baumannii infection (50 MOI, a), LPS (200 ng/ml, b), and IFN-β treatment (2 ng/ml, c) for indicated times. d Immunoblot analysis of RNF220 in WT and Rnf220+/– BMDMs in response to A. baumannii infection and IFN-β treatment for indicated times. e, f Immunoblot analysis of phosphorylation of STAT1, STAT3, and IRF3 in WT and Rnf220+/– BMDMs in response to A. baumannii infection (e) and IFN-β treatment (f) for indicated times. Data represent three independent experiments and are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant.
RNF220 haploinsufficiency impairs Type 1 IFN signaling
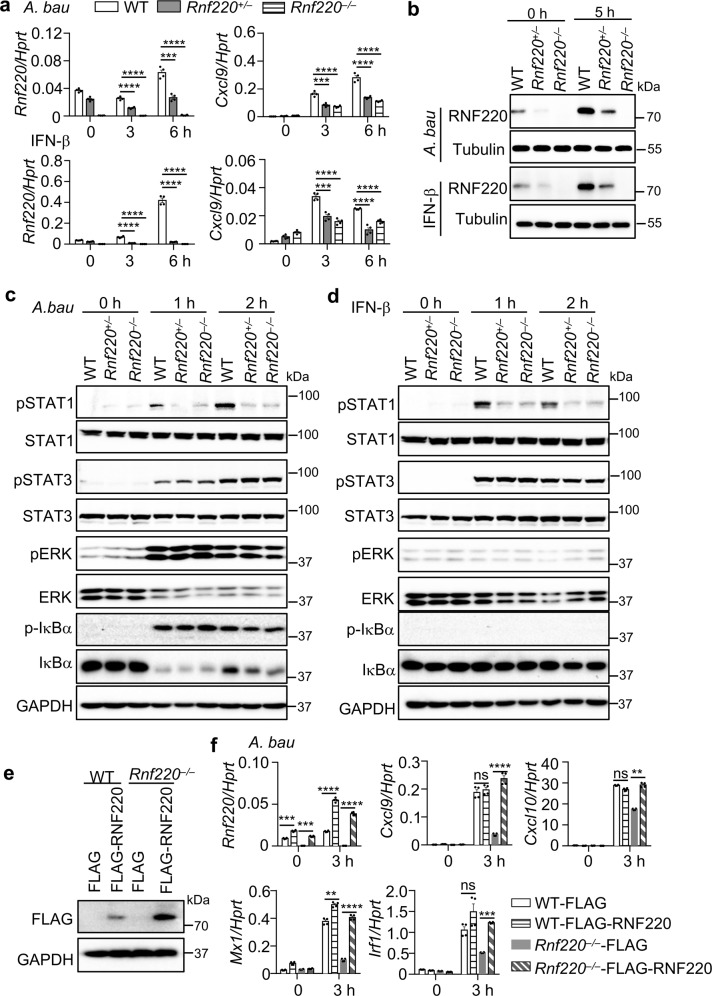
To further investigate type 1 IFN signaling in Rnf220 homozygous knockout cells, we developed macrophages from embryonic liver stem cells and found that the expression of Rnf220 was partially impaired in Rnf220+/– cells and abolished in Rnf220–/– cells (Fig. 3a). The protein expression of RNF220 in WT, Rnf220+/– and Rnf220–/– cells in response to A. baumannii infection and IFN-β treatment were confirmed by western blot analysis (Fig. 3b). The expression of Cxcl9 triggered by A. baumannii infection and IFN-β treatment was similarly reduced in Rnf220+/– and Rnf220–/– cells compared with WT cells, whereas the expression of Il6 and Il1a induced by A. baumannii infection was not reduced in Rnf220+/– or Rnf220–/– cells compared with WT cells (Fig. 3a and Supplementary Fig. 2). However, the expression of Il6 and Il1a was not induced by IFN-β (Supplementary Fig. 2). Consistent with the gene expression data, the phosphorylation of STAT1, but not STAT3 in A. baumannii-infected and IFN-β-treated macrophages was equally reduced in Rnf220+/– and Rnf220–/– cells compared with WT cells (Fig. 3c, d). ERK and IκBα activation induced by A. baumannii infection was not affected in Rnf220+/– or Rnf220–/– cells (Fig. 3c).
Fig. 3. RNF220 haploinsufficiency impairs type 1 IFN signaling.
a Gene expression analysis of Rnf220 and Cxcl9 in WT, Rnf220+/–, and Rnf220–/– macrophages derived from embryonic liver stem cells in response to A. baumannii infection (50 MOI) and IFN-β treatment (2 ng/ml) for indicated times. b Immunoblot analysis of RNF220 in WT, Rnf220+/–, and Rnf220–/– macrophages of a in response to A. baumannii infection and IFN-β treatment for indicated times. c, d Immunoblot analysis of phosphorylation of STAT1, STAT3, ERK, and IκBα in WT, Rnf220+/–, and Rnf220–/– embryonic stem cell-derived macrophages in response to A. baumannii infection (c) and IFN-β treatment (d) for indicated times. e Immunoblot analysis of RNF220 in immortalized WT and Rnf220–/– macrophages derived from embryonic stem cell and transfected with RNF220-expressing plasmid (FLAG-RNF220) and vector control (FLAG) by lentivirus. f Gene expression analysis of Rnf220, Cxcl9, Cxcl10, Mx1, and Irf1 in uninfected and A. baumannii-infected immortalized WT and Rnf220–/– macrophages transfected with RNF220-expressing plasmid and vector control by lentivirus of (e). Data represent three independent experiments and are presented as mean ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant.
To confirm the critical role of RNF220 in type 1 IFN signaling, we performed an RNF220 rescue experiment in Rnf220–/– immortalized macrophages. The transfection of WT RNF220 in Rnf220–/– cells restored the decreased expression of Cxcl9, Cxcl10, Mx1, and Irf1 induced by A. baumannii infection (Fig. 3e, f). Furthermore, the transfection of WT RNF220 in Rnf220+/– mouse skin fibroblasts (MSFs) also rescued the decreased expression of Cxcl9, Cxcl10, Mx1, and Irf1 compared with their expression in WT cells in response to A. baumannii infection (Supplementary Fig. 3a, b). Taken together, these results indicate that RNF220 regulates type 1 IFN signaling and it is haploinsufficient (one copy of the gene is not sufficient to support the gene’s biological functions).
RNF220 interacts with STAT1
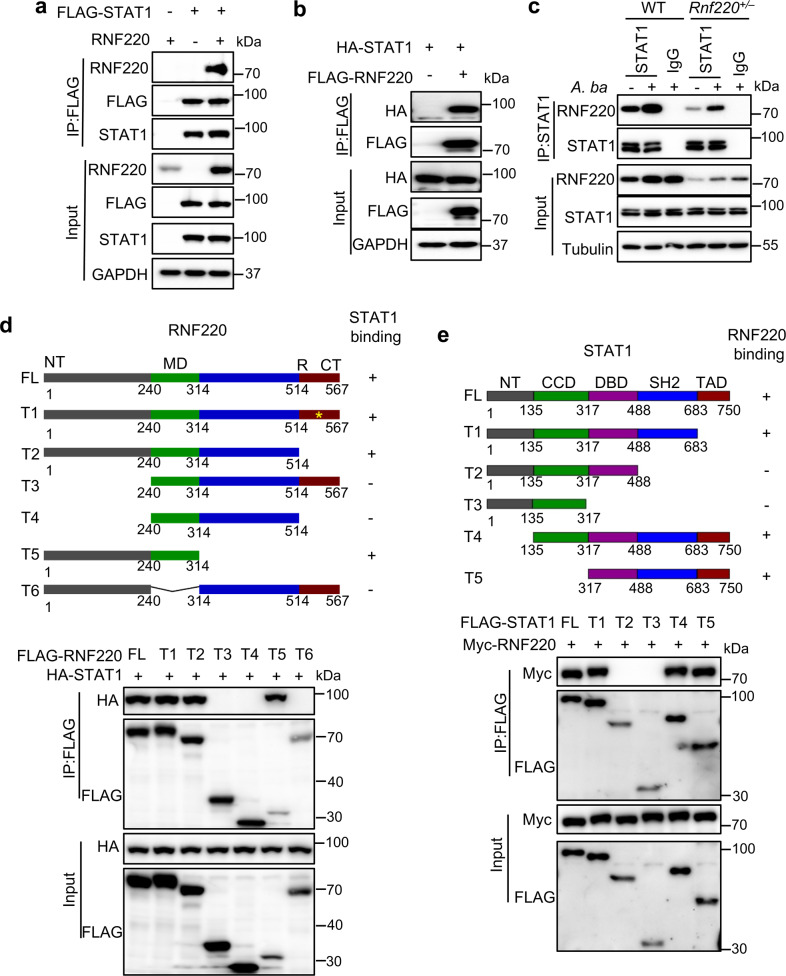
To define the targets regulated by RNF220, we performed co-immunoprecipitation (co-IP) experiments to immunoprecipitate RNF220 with STAT1, STAT2, and STAT3, which are critical downstream molecules of IFNAR [1]. Notably, RNF220 indeed interacted with STAT1, but not STAT2 or STAT3, in transfected HEK293T cells (Fig. 4a and Supplementary Fig. 4a, b). The interaction between RNF220 and STAT1 was also confirmed by reverse IP experiment (Fig. 4b). To investigate the interaction between endogenous RNF220 and STAT1, we performed co-IP analysis with RNF220 and STAT1 in WT and Rnf220+/– BMDMs. Indeed, RNF220 was immunoprecipitated by STAT1 in WT BMDMs, and the interaction was substantially reduced in Rnf220+/– BMDMs under normal condition. The A. baumannii infection increased the interaction between RNF220 and STAT1 in both WT and Rnf220+/– BMDMs (Fig. 4c). To further define the domain of RNF220 that interacts with STAT1, truncated RNF220 and RNF220 with site mutations underwent co-IP analysis with STAT1. Both the N-terminal domain (1–240 aa) and middle domain (240–314 aa) of RNF220 were essential for STAT1 binding (Fig. 4d). Neither the RING domain deletion nor the catalytic site was required for RNF220 binding with STAT1 (Fig. 4d). Furthermore, we performed co-IP analysis of RNF220 with truncated STAT1, and found that the SH2 domain of STAT1 was required for its interaction with RNF220 (Fig. 4e).
Fig. 4. RNF220 interacts with STAT1.
a Immunoblot analysis of RNF220 that coimmunoprecipitated with FLAG-STAT1 from lysates of HEK293T cells transfected with plasmids as indicated. b Immunoblot analysis of STAT1 that coimmunoprecipitated with FLAG-RNF220 from lysates of HEK293T cells transfected with plasmids as indicated. c Co-IP analysis of endogenous of STAT1 and RNF220 in WT and Rnf220+/– BMDMs with or without A. baumannii infection for 2.5 h. d Immunoblot analysis of STAT1 that coimmunoprecipitated with full length and truncated FLAG-RNF220 from lysates of HEK293T cells transfected with plasmids as indicated. e Immunoblot analysis of RNF220 that coimmunoprecipitated with full length and truncated FLAG-STAT1 from lysates of HEK293T cells transfected with plasmids as indicated. Data represent three independent experiments.
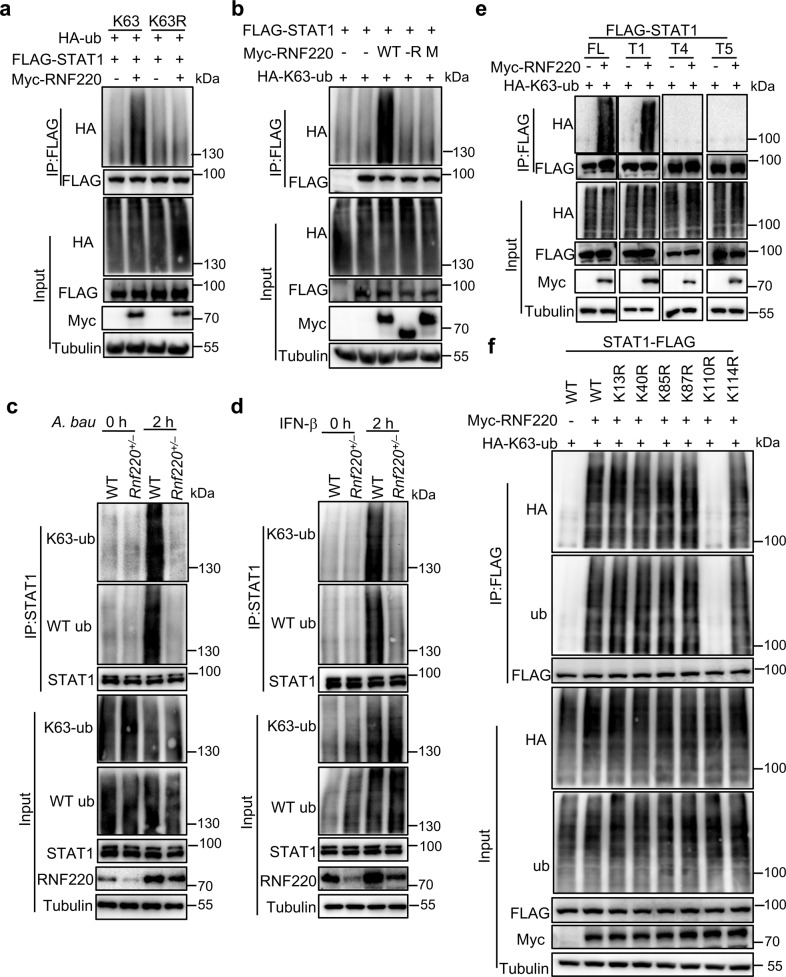
RNF220 mediates the K63-linked polyubiquitination of STAT1
RNF220 was reported to mediate the ubiquitination or recruitment of deubiquitination enzymes [18, 20]. To investigate whether RNF220 mediates STAT1 ubiquitination, we transfected HEK293T cells with RNF220; STAT1; HA-tagged WT ubiquitin; and K6-, K11-, K27-, K29-, K33-, K48-, or K63-specific ubiquitin. RNF220 specifically promoted the polyubiquitination of STAT1 in the presence of HA-tagged K63-specific ubiquitin, but not HA-tagged WT or other linkage-specific ubiquitin (Fig. 5a and Supplementary Fig. 5a). However, RNF220 did not catalyze STAT1 polyubiquitination in the presence of K63R mutant ubiquitin, which harbored a lysine-to-arginine substitution at position 63 (Fig. 5a). Furthermore, the RNF220 catalytic site mutant (M) and RING domain deletion (-R) construct failed to promote STAT1 polyubiquitination (Fig. 5b). To confirm the endogenous RNF220-mediated ubiquitination of STAT1, we analyzed endogenous ubiquitination in WT and Rnf220+/– BMDMs in response to A. baumannii infection and IFN-β treatment. Strikingly, the total and K63-linked ubiquitination of endogenous STAT1 induced by A. baumannii infection and IFN-β treatment was reduced in Rnf220+/– BMDMs compared with that in WT BMDMs (Fig. 5c, d).
Fig. 5. RNF220 mediates the K63-linked polyubiquitination of STAT1.
a Co-IP analysis of WT K63- and K63 mutant (K63R)-linked polyubiquitination of STAT1 mediated by RNF220 in HEK293T cells transfected with plasmids as indicated. b Co-IP analysis of the K63-linked polyubiquitination of STAT1 mediated by WT RNF220, RING domain deletion mutant (-R), and catalytic site mutant (M) in HEK293T cells transfected with plasmids as indicated. c, d Co-IP analysis WT and the K63-linked polyubiquitination of endogenous STAT1 in WT and Rnf220+/– BMDMs in response to A. baumannii infection (30 MOI, c) and INF-β treatment (2 ng/ml, d) for indicated times. e Co-IP analysis of the K63-linked polyubiquitination of full length and truncated STAT1 mediated by RNF220 in HEK293T cells transfected with plasmids as indicated. f Co-IP analysis of the K63-linked polyubiquitination of WT STAT1 and STAT1 mutants (K13R, K40R, K85R, K87R, K110R, and K114R) mediated by RNF220 in HEK293T cells transfected with plasmids as indicated. Data represent two independent experiments.
To determine which lysine residues of STAT1 are essential for the RNF220- mediated K63-linked polyubiquitination of STAT1, we first investigated the ubiquitination of the STAT1 truncated mutants T1, T4, and T5 (Fig. 4e), which interacted with RNF220 and spanned the full length of STAT1. RNF220 remarkably mediated the ubiquitination of full-length STAT1 and the T1 truncation construct, but not the T4 or T5 truncation constructs (Fig. 5e), suggesting that the residues within the N-terminal domain are critical for RNF220-mediated STAT1 ubiquitination. To pinpoint the residues required for STAT1 ubiquitination, we generated 6 N-terminal site-directed STAT1 mutants (K13R, K40R, K85R, K87R, K110R, and K114R) in which lysine residues were replaced with arginine residues at positions 13, 40, 85, 87, 110, and 114, respectively. The interaction between RNF220 and STAT1 was not altered in the presence of the site-directed STAT1 mutants compared with the full-length WT STAT1 (Supplementary Fig. 5b). Strikingly, the K63-linked polyubiquitination of K110R STAT1 was diminished, but not that of other STAT1 mutants (Fig. 5f). Our data collectively indicate that RNF220 mediates the K63-linked polyubiquitination of STAT1 at K110.
RNF220 regulates the interaction between JAK1 and STAT1
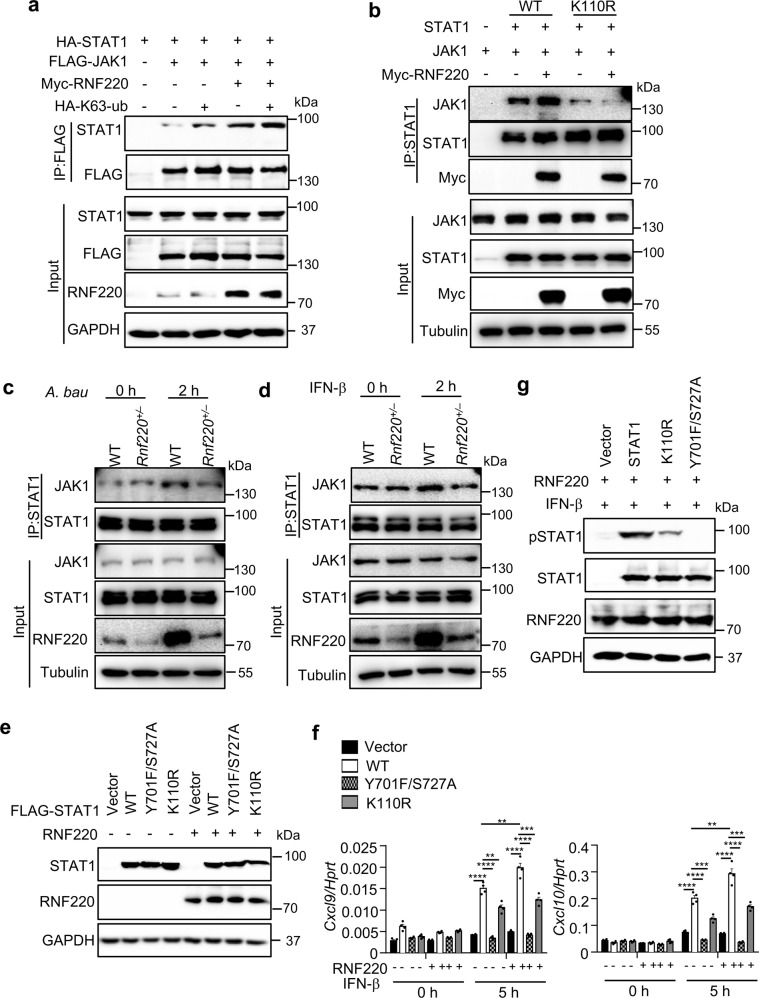
The K63-linked and K33-linked polyubiquitination of STAT1 were previously demonstrated to be important for STAT1 phosphorylation and its DNA-binding activity [16, 17]. To investigate whether the K63-linked polyubiquitination of STAT1 mediated by RNF220 is critical for STAT1 phosphorylation and activation, we performed co-IP experiments in HEK293T cells transfected with RNF220, STAT1, and the kinase JAK1. RNF220 indeed promoted the interaction between STAT1 and JAK1, and K63-ubiquitin further increased their interaction (Fig. 6a). The interaction of STAT1 and JAK1 in the presence or absence of RNF220 was substantially reduced when STAT1 was mutated at K110 (Fig. 6b). Furthermore, the endogenous interaction of STAT1 and JAK1 was reduced in Rnf220+/– BMDMs compared with that in WT compartments during A. baumannii infection and IFN-β treatment (Fig. 6c, d). These data indicate that the K63-linked polyubiquitination of STAT1 mediated by RNF220 is essential for the interaction between STAT1 and JAK1, which contributes to STAT1 phosphorylation.
Fig. 6. RNF220 promotes the interaction between STAT1 and JAK1.
a Immunoblot analysis of STAT1 that coimmunoprecipitated with FLAG-JAK1 in the presence and absence of RNF220 and K63-ubiquitin from lysates of HEK293T cells transfected with plasmids as indicated. (b) Immunoblot analysis of JAK1 that coimmunoprecipitated with WT STAT1 and STAT1 mutant (K110R) in the presence and absence of RNF220 from lysates of HEK293T cells transfected with plasmids as indicated. c, d Immunoblot analysis of JAK1 that coimmunoprecipitated with STAT1 in WT and Rnf220+/– BMDMs in response to A. baumannii infection (c) and IFN-β treatment (d) for indicated times. e Immunoblot analysis of STAT1 and RNF220 in Stat1–/– L929 cells transfected with WT and mutated STAT1 plasmids (K110R, Y701F/S727A), RNF220 plasmid and vector control by lentivirus as indicated. f Gene expression analysis of Cxcl9 and Cxcl10 in untreated and IFN-β treated cells in e. g Immunoblot analysis of phosphorylation of STAT1 in Stat1–/– L929 cells transfected with WT and mutated STAT1 plasmids (K110R, Y701F/S727A), RNF220 and vector control in e with IFN-β treatment. Data represent two independent experiments and are presented as mean ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001.
To define the critical role of K110 site for STAT1 activity, WT STAT1, STAT1 phosphorylation sites mutant Y701F/S727A, and K110 site mutant K110R were transfected into Stat1–/– L929 cells [17], followed by IFN-β treatment. Notably, the expression of Cxcl9 and Cxcl10 was rescued by WT STAT1, but not Y701F/S727A STAT1 mutant. K110R STAT1 mutant only partially rescued the expression of Cxcl9 and Cxcl10 induced by IFN-β (Fig. 6e, f). Consistently, the phosphorylation of STAT1 induced by IFN-β was also reduced in the presence of K110R mutant (Fig. 6g). Taken together, our data demonstrate that RNF220 promotes STAT1 ubiquitination and phosphorylation, which sustains STAT1 activation.
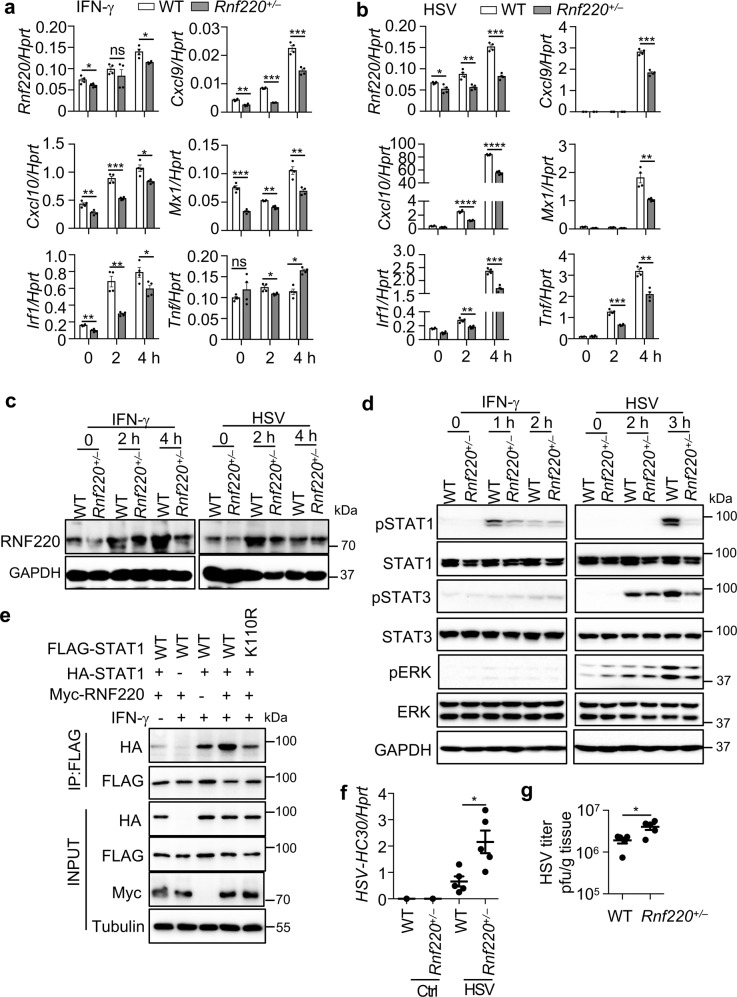
RNF220 is critical for type 2 IFN signaling and HSV-1 infection
The engagement of type 2 IFN (IFN-γ) and IFNGR also triggers STAT1 phosphorylation and the formation of STAT1 homodimer that in turn binds to GAS elements to induce gene expression [6]. To determine whether RNF220 is also involved in the type 2 IFN signaling, we treated WT and Rnf220+/– BMDMs with IFN-γ. As hypothesized, the expression of Cxcl9, Cxcl10, Mx1, and Irf1 induced by IFN-γ was reduced in Rnf220+/– BMDMs compared with that in WT BMDMs (Fig. 7a). To further examine the role of RNF220 in the immune responses upon viral infection, WT and Rnf220+/– BMDMs were infected with herpes simplex virus type 1 (HSV-1). Notably, the expression of the both IFN- and NF-κB-activated genes Cxcl9, Cxcl10, Mx1, Irf1, and Tnf induced by HSV-1 was impaired in Rnf220+/– BMDMs compared with that in WT BMDMs (Fig. 7b).
Fig. 7. RNF220 mediates type 2 IFN and HSV-1 infection.
a, b Gene expression analysis of Rnf220, Cxcl9, Cxcl10, Mx1, Irf1, and Tnf in WT and Rnf220+/– BMDMs in response to IFN-γ treatment (10 ng/ml, a) and HSV-1 infection (3 MOI, b) for indicated times. c Immunoblot analysis of RNF220 in WT and Rnf220+/– BMDMs in response to IFN-γ treatment (10 ng/ml) and HSV-1 infection (3 MOI) for indicated times. d Immunoblot analysis of phosphorylation of STAT1, STAT3, and ERK in WT and Rnf220+/– BMDMs in response to IFN-γ treatment and HSV-1 infection for indicated times. e Immunoblot analysis of STAT1 dimerization in Stat1–/– L929 cells transfected with WT FLAG-STAT1, mutated FLAG-STAT1 (K110R), WT HA-STAT1, and RNF220 plasmids as indicated with or without IFN-γ treatment. f, g WT and Rnf220+/– mice were intranasally infected with HSV-1 (5.0 × 106 pfu per mouse) for 2 days and pulmonary viral burden was analyzed by qRT-PCR (f) and plaque assay (g). Each symbol indicates an individual mouse for f and g. Data represent two independent experiments and are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant.
The increased expression of Rnf220 induced by both IFN-γ treatment and HSV-1 infection was confirmed by western blot analysis (Fig. 7c). In line with the qRT-PCR data, the production of CXCL9 induced by IFN-γ and HSV-1 was reduced in Rnf220+/– BMDMs compared with that in WT BMDMs, and the production of TNF was reduced in Rnf220+/– BMDMs upon HSV-1 infection (Supplementary Fig. 6). Consistent with these data, the phosphorylation of STAT1 induced by IFN-γ and HSV-1 were reduced in Rnf220+/– BMDMs (Fig. 7d). The phosphorylation of STAT3 and ERK were slightly inhibited in Rnf220+/– BMDMs compared with that in WT BMDMs upon HSV-1 infection (Fig. 7d). We transfected both HA-STAT1 and FLAG-STAT1 into Stat1–/– L929 cells to analysis the effect of RNF220 and K110R mutant on the dimerization of STAT1 induced by IFN-γ treatment. Notably, the dimerization of STAT1 induced by IFN-γ treatment was indeed increased by RNF220, and the K110R mutation inhibited the dimerization of STAT1 (Fig. 7e).
To investigate whether RNF220 plays roles in host defense against HSV-1 infection, WT and Rnf220+/– mice were infected with HSV-1, and the viral burden at day 2 was measured in vivo. Remarkably, the HSV-1 viral burden in the lungs of WT mice was significantly lower than that in the lungs of Rnf220+/– mice (Fig. 7f, g), indicating that RNF220 plays a protective role in host defense against HSV-1 infection.
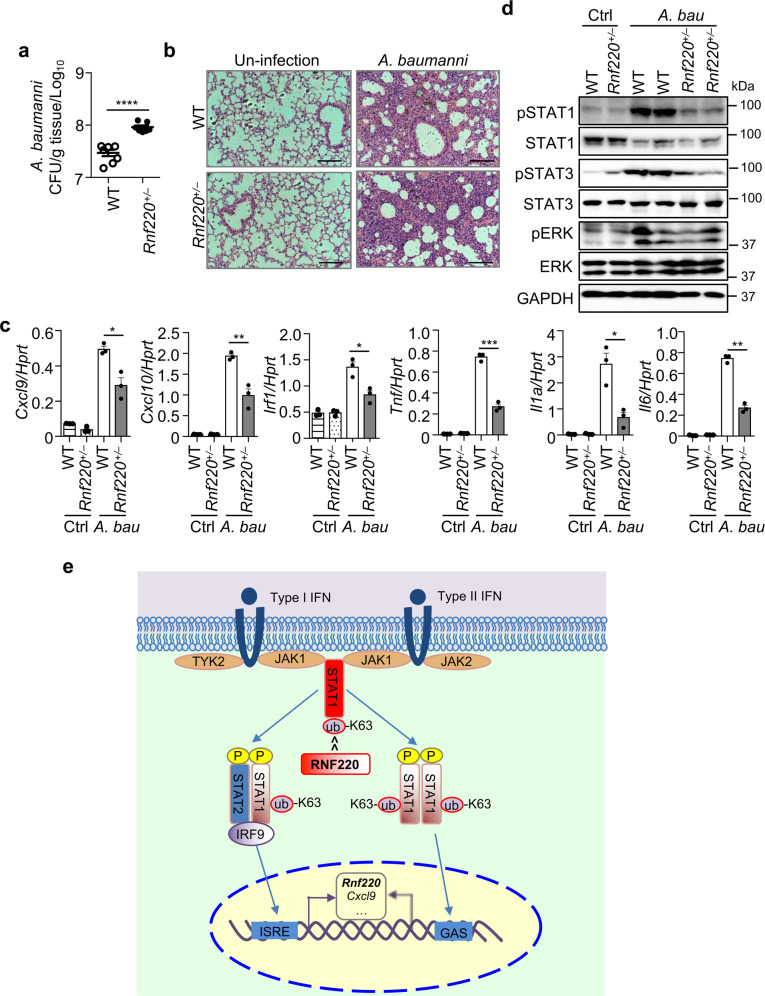
RNF220 promotes host defense against A. baumannii infection
To define the role of RNF220 in host defense against A. baumannii infection, WT and Rnf220+/– mice were intranasally infected with A. baumannii, and the bacterial burden in the lung was enumerated 2 days after infection. Bacterial loads in the lungs of Rnf220+/– mice were significantly higher than those in WT mice (Fig. 8a). Hematoxylin and Eosin (H&E) staining and cytokine gene expression analysis revealed that the inflammatory responses in the lung induced by A. baumannii infection were reduced in Rnf220+/– mice (Fig. 8b, c), which is consistent with our previous finding that the Ifnar–/– mice were susceptible to A. baumannii infection due to an impaired inflammatory responses [25]. Furthermore, the phosphorylation of STAT1, STAT3, and ERK in the lung triggered by A. baumannii infection were reduced in Rnf220+/– mice compared with those in WT mice (Fig. 8d). Overall, our data reveal that RNF220 contributes to increase type 1 and 2 IFN signaling responses by a positive feedback loop through mediating the K63-linked polyubiquitination of STAT1, which promotes host defense against A. baumannii and HSV-1 infection (Fig. 8e).
Fig. 8. RNF220 increases host defense against A. baumannii infection.
a WT and Rnf220+/– mice were intranasally infected 4.0 × 108 CFU of A. baumannii per mouse. Bacterial burden in the lungs on day 2 after infection was measured. b H&E staining of lung sections from WT and Rnf220+/– mice on day 2 after A. baumannii infection and control mice. c Gene expression analysis of Cxcl9, Cxcl10, Irf1, Tnf, Il1a, and Il6 in the lungs of WT and Rnf220+/– mice on day 2 after A. baumannii infection and control mice. d Immunoblot analysis of phosphorylation of STAT1, STAT3, and ERK in the lungs of WT and Rnf220+/– mice on day 2 after A. baumannii infection and control mice. e A model of the RNF220-mediated STAT1 ubiquitination in response to type 1 and 2 IFNs. Upon IFN treatment, STAT1 is phosphorylated and interacts with phosphorylated STAT2 and IRF9 to form ISGF3, or phosphorylated STAT1 forms homodimer, which bind to DNA elements for activating ISRE- and GAS-driven gene expression. RNF220 mediates the K63-linked polyubiquitination of STAT1, which promotes STAT1 phosphorylation and activation. The expression of Rnf220 is induced by type 1 and 2 IFNs signaling. RNF220 contributes to a positive feedback regulation of STAT1 between the ubiquitination and phosphorylation, thus leading to increased host defense against A. baumannii and HSV-1 infection. Each symbol indicates an individual mouse for a and c. Scale bars, 100 μm for b. Data represent two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
STAT1 is a pivotal transcription factor that orchestrates interferon signaling—triggered, ISRE—and GAS-driven gene transcription and host immune responses [1]. The phosphorylation of STAT1 is tightly regulated to ensure nucleocytoplasmic STAT1 shuttling, DNA binding and transcriptional activation. The mechanism by which STAT1 is phosphorylated and activated upon IFN signaling remains elusive. Our data demonstrate that the K63-linked polyubiquitination of STAT1 mediated by RNF220 contributes significantly to STAT1 activation and innate immune responses.
The phosphorylation of STAT1 at tyrosine 701 (Y701) and serine 727 (S727) leads to the formation of STAT1 homodimers and STAT1/2 heterodimers associated with IRF9 and their subsequent nuclear translocation in response to interferon signaling [26]. Increasing evidence indicates that STAT1 modifications, such as ubiquitination, SUMOylation, and acetylation, contribute significantly to the STAT1 activation. Both the SUMOylation and acetylation of STAT1 were proposed to inhibit STAT1 activity through decreasing STAT1 phosphorylation and DNA binding [11, 27, 28]. Ubiquitination of STAT1 exerted distinct effects on STAT1 activation depending on the ubiquitin linkage type. Both the K48- and K33-linked polyubiquitination of STAT1 negatively regulate its activity through mediating STAT1 proteasomal degradation and its disassociation from DNA, respectively [14, 15, 17]. Our work reveals that the K63-linked polyubiquitination of STAT1 promotes its activation, which is consistent with previous observations that the K63-linked ubiquitination of STAT1 mediated by NKLAM promotes STAT1 nuclear retardation and transcriptional activity in response to IFN-γ signaling, although the site of NKLAM-regulated STAT1 ubiquitination was not identified [16]. To the best of our knowledge, our data demonstrate that the K63-linked polyubiquitination of STAT1 at site K110 is essential for STAT1-mediated gene transcription in response to both type 1 and type 2 interferons for the first time, and describe a new layer of physiologically relevant STAT1 regulation. Our results define RNF220 as master regulator for maintaining STAT1 activation through a positive feedback loop. Our previous study revealed that the K63-linked polyubiquitination of TRAF3 is critical for type 1 IFN production during bacterial and viral infections [4]. Collectively, our results indicate the effect of K63-linked polyubiquitination on multiple targets for the regulation of type 1 IFN signaling.
RNF220 was previously reported to directly mediate K48-linked polyubiquitination for proteasomal degradation and the recruitment of deubiquitinase [18–20, 24]. Our recent findings revealed that RNF220 also mediates the K63-linked polyubiquitination of Gli transcription factors and their cellular localization during neural patterning [21, 22]. In this study, the K63-linked polyubiquitination of STAT1 mediated by RNF220 was shown to contribute to type 1 and 2 IFN signaling and the host defense against A. baumannii and HSV-1 infection, which is consistent with our previous finding of the protective role of type 1 IFN in the host defense against A. baumannii infection [25]. RNF220 interacted with STAT1, but not STAT2 or STAT3, highlighting its specific roles in STAT1-dependent signaling. The expression of Rnf220 induced by infection and ligand stimulation suggests that RNF220 is a critical host factor evolved for defense against pathogenic infection. The RNF220 haploinsufficiency also indicates the level of RNF220 and that induced RNF220 expression is required for its functional activity, which renders RNF220 as a potential drug target for treating dysregulated IFN signaling responses. How RNF220 activation and cellular localization are regulated upon infection and stimulation and the posttranslational regulation of RNF220 need to be further examined.
Unphosphorylated STAT1 was shown to be a functional transcriptional factor independent of its phosphorylation. It prolongs the immune response by promoting the late gene expression induced by interferons [29]. Instead, full biological activation of STAT1 requires its continuous phosphorylation at Y701, nuclear translocation, chromatin binding and phosphorylation at S727 [26]. S727 was demonstrated to be essential for the SUMOylation of STAT1, which contributed to the reduced phosphorylation of STAT1 and diminished transcription of IFN-responsive genes [11, 30]. The acetylation of STAT1 was also shown to catalyze STAT1 dephosphorylation and counteract IFN-induced immune responses [28]. In contrast, our findings reveal a positive feedback between the ubiquitination and phosphorylation of STAT1 mediated by RNF220, which promotes STAT1-mediated immune responses and host defense. Our study highlights a deeper understanding of the regulation of STAT1 activity and provides potential therapeutic targets against bacterial and viral infection and inflammatory diseases.
Methods
Mice
Rnf220 gene knockout mice were generated as previously described [22]. All mice were kept in specific pathogen-free conditions within the Animal Resource Center at Kunming Institute of Zoology, Chinese Academy of Sciences. All animal experiments were conducted in accordance with the guidelines and were approved by the Kunming Institute of Zoology, Chinese Academy of Sciences Animal Care and Use Committee, Kunming, China.
Bacterial and viral culture and infection of mice
The bacterial strain A. baumannii was growing as previously described [25]. HSV-1 strain was provided J. Zhou (Kunming Institute of Zoology) and propagated as described previously [4].
Eight- to ten-week-old Rnf220+/– mice and WT controls were infected intranasally with A. baumannii (4.0 × 108 CFUs per mouse) and HSV-1 (5.0 × 106 pfu per mouse) as indicated. Mice were weighted and monitored daily over time. Mice were euthanized at days as indicated after infection and lung was harvested to determine the bacterial and viral burden as described previously [31].
Preparation of BMDM, BMDC, and MSF for infection
To generate bone-marrow-derived macrophages (BMDMs), bone marrow (BM) cells were cultured in L929 cell-conditioned Dulbecco’s Modified Eagle Medium and Nutrient Mixture F-12 (DMEM/F-12) supplemented with 10% FBS, 1% non-essential amino acids and 1% Penicillin-streptomycin for 5 days. BMDMs were seeded in 12-well plates (1 million cells per well) and cultured overnight. Next day, cells were washed and supplied with fresh media without antibiotics. For BMDC generation, BM cells were cultured in RPMI1640 media supplemented with GM-CSF (20 ng/ml), 10% FBS, 1% non-essential amino acids, and 1% penicillin-streptomycin for 7 days. MSFs were prepared as previously described [32]. Briefly, pinnae of adult wild type mice were minced and digested with 20 mg/ml collagenase type I (C0130, Sigma–Aldrich) for 3 h, followed by filtration through 70-μm strainers. Cells were cultured in 50% FBS in DMEM supplemented with HEPES, 1% penicillin and streptomycin, L-glutamine, sodium pyruvate, non-essential amino acids, and β-mercaptoethanol for the first 3–4 days. Cells were subcultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. Cells were infected with bacteria or virus for the indicated times. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with protease inhibitor and phosphatase inhibitors (Roche) for immunoblot analysis.
Preparation of tissue sample for histopathologic staining and immunoblot analysis
Lung tissues were fixed in 10% formalin and embedded in paraffin for sections (5-μm thickness), which were stained with hematoxylin and eosin and examined. Tissues were homogenized in RIPA buffer with protease and phosphatase inhibitors for immunoblot analysis. Protein concentration was determined using bicinchoninic acid assay (BCA) kit (Pierce) according to manufacturer’s instructions.
Immunoblot analysis and antibodies
Samples were separated by 12% SDS-PAGE and followed by electrophoretic transfer onto PVDF membranes and blocked with 5% non-fat milk and further incubated overnight in primary antibody at 4 °C as described previously [31]. The following primary antibodies were used: anti-RNF220 (Sigma, HPA027578), anti-JAK1 (CST, 3344), anti-IκBα (CST, 9242), anti-phosphorylated (p)-IκBα (CST, 2859), anti-ERK (Santa Cruz, SC-94), anti-p-ERK (Santa Cruz, SC-7383), anti-p-IRF3 (CST, 4947), anti-IRF3 (CST, 4302), anti-p-STAT1 (CST, 7649), anti-STAT1 (CST, 14994 s), anti-p-STAT3 (CST, 9131), anti-STAT3 (CST, 9139), anti-HA (Santa Cruz, SC-805), anti-FLAG (Sigma, F3165), anti-Ubiquitin (Millipore 04–263), anti-K63-ubiquitin (CST, 5621), anti-Myc (CST, 2278), anti-Tubulin (Proteintech, 66031–1-Ig), and anti-GAPDH (CST, 5174). HRP-labeled anti-rabbit, anti-mouse or anti-goat (CST) was used as the secondary antibody.
Plasmids transfection and Co-immunoprecipitation
The plasmids expressing WT, truncated, catalytic site-mutated, and RING domain-deleted RNF220 were previously described [22]. STAT1 was amplified from cDNA library and cloned into pCDH vector, truncated STAT1 sequence was amplified from full-length of STAT1 and cloned into pCDH vector. Site-directed mutations were generated using the QuikChange site-directed mutagenesis kits. All plasmids were confirmed by DNA sequencing. Lipofectamine 2000 reagents (Invitrogen) were used for transient transfection of plasmids into HEK293T cells.
For immunoprecipitation (IP), whole HEK293T or L929 cells collected 36 h post transfection and WT and Rnf220+/– BMDMs with or without A. baumannii infection or IFN-β (R&D Systems, 8234-MB-010) treatment were lysed in IP buffer composed of 1.0% Triton X-100, 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, and protease/phosphatase inhibitor cocktails (Biotool, USA). After centrifugation, supernatants were collected and incubated with protein A/G Plus–Agrose (Santa Cruz, sc-2003) and 5 μg of the corresponding antibodies for 6 h at 4 °C, followed by washing five times with IP buffer. Immunoprecipitated components were eluted by boiling in the 1% (wt/vol) SDS buffer (60 mM Tris-HCl (pH 6.8), 1% (wt/vol) SDS, 5% (vol/vol) glycerol, 0.005% (wt/vol) bromophenol blue and 1% (vol/vol) 2-mercaptoethanol) for 10 min. For immunoblot analysis, immunoprecipitates and input lysates were separated by SDS-PAGE, followed by transferring onto PVDF membranes and detected by specific antibodies.
Ubiquitination analysis
For polyubiquitination analysis of STAT1 in HEK293T cells, HEK293T cells were transfected with plasmids expressing WT RNF220 or RNF220 mutant, HA–ubiquitin (WT) or HA–ubiquitin (K48) or HA–ubiquitin (K63) or HA–ubiquitin mutant (K63R) or HA–ubiquitin (K6) or HA–ubiquitin (K11) or HA–ubiquitin (K27) or HA–ubiquitin (K29) or HA–ubiquitin (K33) with MG132, and FLAG-STAT1 (WT and mutants). For ubiquitination analysis of STAT1 in BMDMs, WT and Rnf220+/– BMDMs were treated with IFN-β or A. baumannii infection for indicated times. Transfected HEK293T cells and BMDMs were lysed in lysis buffer (50 mM Tris-HCl (pH 6.8), 1.5% SDS) and boiling for 15 min, the denatured samples were immunoprecipitated with the anti-FLAG and anti-STAT1 antibodies, and the IP products were analyzed by immunoblot with anti-HA, anti-ubiquitin and specific antibodies.
Lentivirus production and infection
RNF220 and STAT1 were cloned into the lentiviral expression vector pCDH-CMV-MCS-EF1-Puro. The viral particles were prepared by transfecting HEK293T cells with RNF220- and STAT1-expressing, or control plasmids in combination with packaging vectors. Twelve hours later, media was replaced with fresh complete DMEM. Viral supernatant was harvested and passed through 0.45-μm syringe filter at 48 and 72 h after transfection. To establish stably infected cells, WT and knockout cells were infected with lentivirus as indicated in the presence of polybrene (8 μg/ml) for three times. The infected cells were cultured in fresh media for at least 3 days and then performed with A. baumannii infection for gene expression analysis.
Real-time quantitative PCR
Total RNA was isolated from tissues or cells using Trizol (Invitrogen) and purified by RNeasy Mini Kit (QIAGEN), cDNA was reverse transcribed by using Superscript III (Invitrogen). Real-time qPCR was performed on the Bio-Rad CFX-96 Touch Real-Time Detection System. Primer sequences are listed in Supplementary Table 1.
ELISA
Cell culture supernatant from media and treated BMDMs were analyzed for cytokine and chemokine release using ELISA MAX™ Standard (BioLegend) following the manufacturer’s instructions.
Statistical analysis
Data are given as mean ± SEM. Statistical analyses were performed using one-way ANOVA with multiple comparisons, two-tailed Student t-test, and LogRank test. P values ≤0.05 were considered significant.
Supplementary information
Acknowledgements
We thank C. Gao (Shandong University School of Basic Medical Sciences, Jinan) for mutated Stat1 plasmids; X. Cao (Nankai University, Tianjin) for Stat1–/– L929 cells; C. Chen (Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming) for ubiquitin plasmids; J. Zhou (Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming) for HSV-1; S.-Z. Duan (Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai) for L929 cells. This work was supported by the National Key Research and Development Program of China (2017YFD0500300), the Chinese Academy of Sciences (XDB13000000 and HXDTZX-2019-1), the National Natural Science Foundation of China (31970896, 31871483, 31671521, 31500847, 31701134, and 81701578) and Yunnan Province (2019FJ008, 2018FA038, 2018FB127, 2018FB131, HXDT-2019-2, and AMHD-2018-2).
Author contributions
XQ and BM designed the study; XG, PM, YL, YY, CW, TX, HW, CL, BM, and XQ performed experiments and analyzed the data; XQ wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaomin Guo, Pengcheng Ma
Contributor Information
Bingyu Mao, Email: mao@mail.kiz.ac.cn.
Xiaopeng Qi, Email: qixiaopeng@mail.kiz.ac.cn.
Supplementary information
The online version of this article (10.1038/s41418-020-00609-7) contains supplementary material, which is available to authorized users.
References
- 1.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K, Liu J, Cao X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J Autoimmun. 2017;83:1–11. doi: 10.1016/j.jaut.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Wang L, Berman M, Kong Y-Y, Dorf Martin E. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–40. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F, Li Y, Liang H, Xu T, Kong Y, Huang M, et al. HECTD3 mediates TRAF3 polyubiquitination and type I interferon induction during bacterial infection. J Clin Investig. 2018;128:4148–62. doi: 10.1172/JCI120406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–35. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–50. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, et al. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat Immunol. 2014;15:168. doi: 10.1038/ni.2794. [DOI] [PubMed] [Google Scholar]
- 8.Majoros A, Platanitis E, Kernbauer-Hölzl E, Rosebrock F, Müller M, Decker T. Canonical and non-canonical aspects of JAK–STAT signaling: lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majoros A, Platanitis E, Szappanos D, Cheon H, Vogl C, Shukla P, et al. Response to interferons and antibacterial innate immunity in the absence of tyrosine-phosphorylated STAT1. EMBO Rep. 2016;17:367–82. doi: 10.15252/embr.201540726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grönholm J, Vanhatupa S, Ungureanu D, Väliaho J, Laitinen T, Valjakka J, et al. Structure-function analysis indicates that sumoylation modulates DNA-binding activity of STAT1. BMC Biochem. 2012;13:20. doi: 10.1186/1471-2091-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begitt A, Droescher M, Knobeloch K-P, Vinkemeier U. SUMO conjugation of STAT1 protects cells from hyperresponsiveness to IFNγ. Blood. 2011;118:1002–7. doi: 10.1182/blood-2011-04-347930. [DOI] [PubMed] [Google Scholar]
- 12.Kim TK, Maniatis T. Regulation of interferon-γ-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–9. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 13.Yuan C, Qi J, Zhao X, Gao C. Smurf1 protein negatively regulates interferon-γ signaling through promoting STAT1 protein ubiquitination and degradation. J Biol Chem. 2012;287:17006–15. doi: 10.1074/jbc.M112.341198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh H-M, Yu C-Y, Yang H-C, Ko S-H, Liao C-L, Lin Y-L. Ubiquitin-specific protease 13 regulates IFN signaling by stabilizing STAT1. J Immunol. 2013;191:3328–36. doi: 10.4049/jimmunol.1300225. [DOI] [PubMed] [Google Scholar]
- 15.Ren Y, Zhao P, Liu J, Yuan Y, Cheng Q, Zuo Y, et al. Deubiquitinase USP2a sustains interferons antiviral activity by restricting ubiquitination of activated STAT1 in the nucleus. PLOS Pathog. 2016;12:e1005764. doi: 10.1371/journal.ppat.1005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence DW, Kornbluth J. E3 ubiquitin ligase NKLAM ubiquitinates STAT1 and positively regulates STAT1-mediated transcriptional activity. Cell Signal. 2016;28:1833–41. doi: 10.1016/j.cellsig.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Jiang M, Wang W, Liu W, Song X, Ma Z, et al. Nuclear RNF2 inhibits interferon function by promoting K33-linked STAT1 disassociation from DNA. Nat Immunol. 2018;19:41–52. doi: 10.1038/s41590-017-0003-0. [DOI] [PubMed] [Google Scholar]
- 18.Kong Q, Zeng W, Wu J, Hu W, Li C, Mao B. RNF220, an E3 ubiquitin ligase that targets Sin3B for ubiquitination. Biochemical Biophysical Res Commun. 2010;393:708–13. doi: 10.1016/j.bbrc.2010.02.066. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X, Wang X, Zhang X, Xiao Z, Zhang C, Liu X, et al. A homozygous RNF220 mutation leads to male infertility with small-headed sperm. Gene. 2019;688:13–18. doi: 10.1016/j.gene.2018.11.074. [DOI] [PubMed] [Google Scholar]
- 20.Ma P, Yang X, Kong Q, Li C, Yang S, Li Y, et al. The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of β-catenin. Mol Cell Biol. 2014;34:4355–66. doi: 10.1128/MCB.00731-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma P, Song N, Xu Y, Gao X, Ding Y, Mao B. RNF220 mediated K-63 linked ubiquitination induces sequestration of Gli to pattern the ventral neural tube. Mechanisms Dev. 2017;145:S100–S101. doi: 10.1016/j.mod.2017.04.261. [DOI] [Google Scholar]
- 22.Ma P, Song N-N, Li Y, Zhang Q, Zhang L, Zhang L, et al. Fine-tuning of Shh/Gli signaling gradient by non-proteolytic ubiquitination during neural patterning. Cell Rep. 2019;28:541–e544. doi: 10.1016/j.celrep.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Ma P, Song N-N, Cheng X, Zhu L, Zhang Q, Zhang L, et al. ZC4H2 stabilizes RNF220 to pattern ventral spinal cord through modulating Shh/Gli signaling. J Mol Cell Biol. 2019;12:337–44. doi: 10.1093/jmcb/mjz087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Choi T-I, Park S, Kim MH, Kim C-H, Lee S. Rnf220 cooperates with Zc4h2 to specify spinal progenitor domains. Development. 2018;145:dev165340. doi: 10.1242/dev.165340. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Guo X, Hu C, Du Y, Guo C, Di W, et al. Type I IFN operates pyroptosis and necroptosis during multidrug-resistant A. baumannii infection. Cell Death Differ. 2018;25:1304–18. doi: 10.1038/s41418-017-0041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadzak I, Schiff M, Gattermeier I, Glinitzer R, Sauer I, Saalmüller A, et al. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci USA. 2008;105:8944–9. doi: 10.1073/pnas.0801794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ungureanu D, Vanhatupa S, Grönholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–6. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 28.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Gührs K-H, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–35. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci USA. 2009;106:9373–8. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhatupa S, Ungureanu D, Paakkunainen M, Silvennoinen O. MAPK-induced Ser727 phosphorylation promotes SUMOylation of STAT1. Biochem J. 2008;409:179–85. doi: 10.1042/BJ20070620. [DOI] [PubMed] [Google Scholar]
- 31.Qi X, Man SM, Malireddi RKS, Karki R, Lupfer C, Gurung P, et al. Cathepsin B modulates lysosomal biogenesis and host defense against Francisella novicida infection. J Exp Med. 2016;213:2081–97. doi: 10.1084/jem.20151938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L, Guo X, Shen C, Hao X, Sun P, Li P, et al. Salivary factor LTRIN from Aedes aegypti facilitates the transmission of Zika virus by interfering with the lymphotoxin-β receptor. Nat Immunol. 2018;19:342–53. doi: 10.1038/s41590-018-0063-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.