Abstract
Streptococcus uberis is one of the most frequent mastitis-causing pathogens isolated from dairy cows. Further understanding of S. uberis genetics may help elucidate the disease pathogenesis. We compared the genomes of S. uberis isolates cultured from dairy cows located in distinctly different geographic regions of Australia. All isolates had novel multi locus sequence types (MLST) indicating a highly diverse population of S. uberis. Global clonal complexes (GCC) were more conserved. GCC ST86 and GCC ST143 represented 30% of the total isolates (n = 27) and were clustered within different geographic regions. Core genome phylogeny revealed low phylogenetic clustering by region, isolation source, and MLST. Identification of putative sortase (srtA) substrates and generation of a custom putative virulence factor database revealed genes which may explain the affinity of S. uberis for mammary tissue, evasion of antimicrobial efforts and disease pathogenesis. Of 27 isolates, four contained antibiotic resistance genes including an antimicrobial resistance cluster containing mel/mef(A), mrsE, vatD, lnuD, and transposon-mediated lnuC was also identified. These are novel genes for S. uberis, which suggests interspecies lateral gene transfer. The presence of resistance genes across the two geographic regions tested within one country supports the need for a careful, tailored, implementation and monitoring of antimicrobial stewardship.
Subject terms: Bioinformatics, Genomic analysis, Clinical microbiology, Pathogens, Epidemiology, Genetics research
Introduction
Mastitis poses a risk to public health, animal welfare, and farm profitabilty worldwide1–5. Increased costs related to clinical mastitis are the result of reduced milk production and price, discarded milk, animal culling, mortality, labour and herd veterinary costs5. Both contagious and environmental pathogens have been implicated in intramammary infections and bovine mastitis. Streptococcus uberis is one of the most frequently identified environmental pathogens responsible for mastitis in dairy herds, a trend which appears to be increasing worldwide4–6. Mastitis control programs have been effective in decreasing the prevalence of contagious pathogens in well managed dairy herds7, but S. uberis remains responsible for a significant proportion of both clinical and subclinical intramammary infections8. In Australia, S. uberis associated mastitis accounts for one of every three cases of clinical mastitis9. This pathogen can be found in a variety of sites in the dairy cow environment such as bedding materials and milking equipment, as well as on other items within the dairy cows’ daily living spaces10. However, S. uberis is not only an environmental risk, this pathogen has also been associated with cow-to-cow transmission consistent with a contagious pathogen role for S.uberis10–12 and indicative of the complexity of the bacteria, environment and dairy cow relationship.
A number of different DNA-based techniques have been used to investigate the genetic diversity of S. uberis isolated from cases of bovine mastitis, including MLST11, 13–16. While MLST has provided insights into the population diversity of S. uberis11, 16, 17, it is limited in its ability to delve deeply into genetic variation of isolates. The recent availability of low-cost Whole Genome Sequencing (WGS) technology and advances in computational biology has made it possible to compare the entirety of the S. uberis genome and completely characterise the levels of heterogeneity between strains18.
WGS analysis of the genomic sequence of S.uberis strains has revealed both a capacity for nutritional flexibility and a large variety of metabolic capabilities that promote bacterial survival in different environments18, potentially responsible for the high degree of genetic diversity of S. uberis within the dairy herd environments16, 19, 20. Variations in genetic profile among S. uberis strains were not only observed in isolates originating from different dairy herds, but also in the pathogens present within the same herd7, 21. This inherently high genetic diversity renders the attribution of a specific set of virulence factors (and associated pathogenesis) to a given S. uberis strain/s challenging12, 22, 23. The development of strategies to control this important mastitis-causing organism necessitates monitoring of the genetic diversity and determination of the significance of any detected diversity to the disease pathogenesis22, 24. Therefore, the main objectives of this study were to compare S. uberis isolates cultured from dairy cows with clinical mastitis in distinctly different geographic and climatic regions of Australia. We then identified antibiotic resistance genes, virulence factors, and mobile genetic elements which could then inform antimicrobial stewardship programs. A secondary objective was to examine the diversity of the isolates, in particular their specific antimicrobial resistance profiles from these different regions to identify geographic or climatic associations that may exist as a potential basis for development of more effective and sustainable mastitis treatments for dairy cows.
Results
S. uberis mastitis isolates, sequence types and phylogenetic relationships
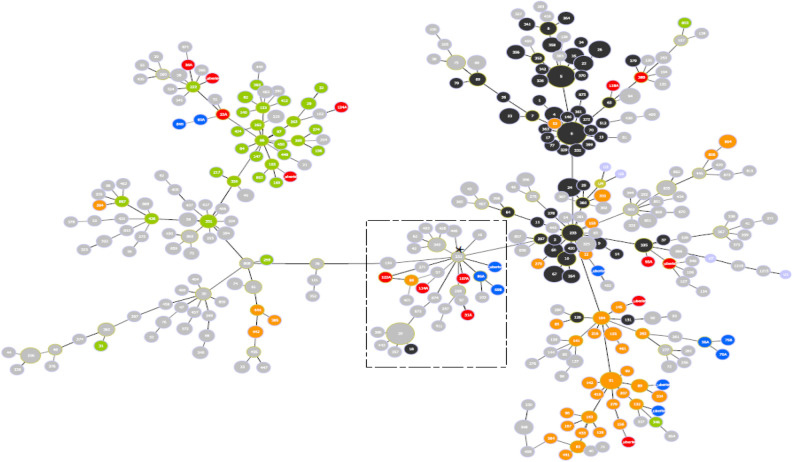
Eleven isolates from 10 locations in Victoria (VIC) and 16 isolates from 8 sites in Queensland (QLD) for a total of 27 isolates were collected and analysed in this study (Fig. 1). The 27 S. uberis genomes ranged in size from 1,832,555 to 2,019,389 bp (Table 1). There were 21 MLST16 sequence types (STs) represented within this collection. All sequence types (STs) were considered novel as they matched no known sequence types in the S. uberis MLST database (pubmlst.org/suberis)25. The closest ST matches were ST ~ 1223 (4/27) followed by ST ~ 222 (3/27). These were only present in QLD farms (Table 1). Analysis of GCC STs showed the most abundant STs to be GCC ST86 (8/27) and GCC ST143 (8/27) with GCC ST5 representing 4/27 isolates. All GCC ST86 isolates were from herds with high bulk milk somatic cell counts (270–400 × 103 cells/mL). GCC ST143 isolates were predominantly (p = 0.048) found in Victoria (75%) while GCC ST86 isolates were predominantly found in Queensland (88%). Seven isolates (27%) did not belong to a defined GCC. Based on the goeBURST analysis, a possible founder ST for those 7 isolates was ST251. The relationship between the STs of this study and those reported globally in the S. uberis MLST database (pubmlst.org/suberis) is depicted in Fig. 2.
Figure 1.
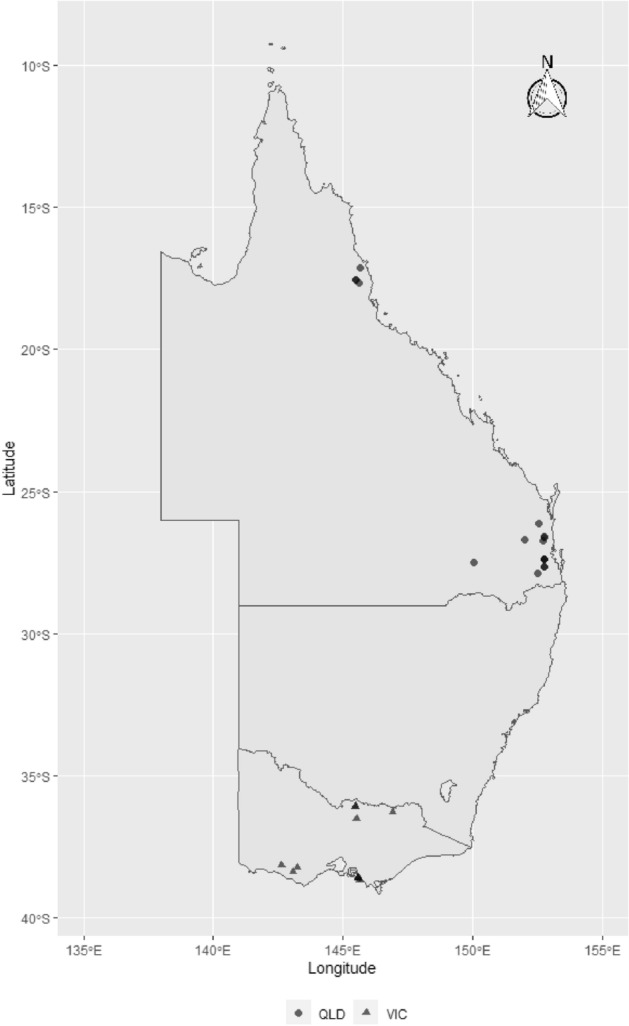
Map showing the geographical distribution of S. uberis isolates sourced from farms in climatically distinct regions across Australia: Queensland (QLD) farms (North QLD (circles), South QLD (circles)), and Victorian farms (triangles). The symbols represent the townships centroids were the farms are located. The map was generated using ozmap package version 0.3.6 [https://CRAN.R-project.org/package=ozmaps]99 implemented within R version 4.0.2 [https://www.R-project.org/]100).
Table 1.
Summary of S. uberis strains identified in this study, isolated from cows affected by bovine mastitis. MLST STs are also shown.
| Isolate | Genome size (bp) | Source | State | MLST ST | GCC ST |
|---|---|---|---|---|---|
| S. uberis 22A | 1,938,913 | QLD1 | Queensland | ~ 222 | 86 |
| S. uberis 31A | 1,900,760 | QLD2 | Queensland | ~ 97 | – |
| S. uberis 36A | 1,914,697 | QLD3 | Queensland | ~ 222 | 86 |
| S. uberis 38B | 1,958,229 | QLD3 | Queensland | ~ 253 | 5 |
| S. uberis 42 | 1,883,527 | VIC1 | Victoria | ~ 271 | 143 |
| S. uberis 43 | 1,925,541 | VIC2 | Victoria | ~ 402 | 5 |
| S. uberis 44 | 1,879,665 | VIC3 | Victoria | ~ 89 | 143 |
| S. uberis 45 | 1,961,274 | VIC4 | Victoria | ~ 190 | – |
| S. uberis 46 | 1,899,378 | VIC5 | Victoria | ~ 132 | 143 |
| S. uberis 47 | 1,971,757 | QLD4 | Queensland | ~ 27 | 143 |
| S. uberis 48 | 1,927,709 | QLD5 | Queensland | ~ 222 | 86 |
| S. uberis 49 | 2,019,389 | QLD6 | Queensland | ~ 145 | 143 |
| S. uberis 50 | 1,938,127 | QLD7 | Queensland | ~ 425 | 86 |
| S. uberis 51 | 1,984,097 | QLD8 | Queensland | ~ 56 | 86 |
| S. uberis 56A | 1,906,743 | VIC6 | Victoria | ~ 261 | 143 |
| S. uberis 60A | 1,967,409 | VIC6 | Victoria | ~ 412 | 86 |
| S. uberis 69B | 1,926,363 | VIC7 | Victoria | ~ 271 | – |
| S. uberis 70A | 1,919,457 | VIC8 | Victoria | ~ 256 | 143 |
| S. uberis 75B | 1,844,929 | VIC9 | Victoria | ~ 156 | 143 |
| S. uberis 86A | 1,977,432 | VIC10 | Victoria | ~ 90 | – |
| S. uberis 93A | 1,856,167 | QLD9 | Queensland | ~ 67 | 5 |
| S. uberis 106A | 1,832,555 | QLD6 | Queensland | ~ 1223 | 86 |
| S. uberis 107A | 1,832,592 | QLD6 | Queensland | ~ 1223 | – |
| S. uberis 114A | 1,927,632 | QLD7 | Queensland | ~ 1223 | – |
| S. uberis 118A | 1,894,094 | QLD8 | Queensland | ~ 850 | 5 |
| S. uberis 122A | 1,886,974 | QLD8 | Queensland | ~ 80 | – |
| S. uberis 124A | 1,909,330 | QLD8 | Queensland | ~ 1223 | 86 |
~ indicates closest ST. All STs were considered novel. The isolation sources have been given codes for privacy reasons.
Figure 2.
Multilocus sequence type (MLST)-based minimal spanning tree of 1220 Streptococcus uberis isolated from bovine mastitis milk as per NCBI database103 (as of 8 June 2020) to date. Coloured circles are Australian isolates from bovine origin (n = 240). The tree was calculated and generated using the goeBURST version 1.2.1 (http://www.phyloviz.net/goeburst/)113 full MST algorithm in Phyloviz version 2.0 (www.phyloviz.net)112. Node sizes reflect the number of isolates with specific MLST profile. Numbers within the nodes indicate the ST. Node colours refer to types of Global Clonal Complexes (GCC): GCC Strain Type (ST) 86 (green; left side of the plot), GCC ST143 (orange, right-bottom side of the plot), GCC ST5 (black, right-top side of the plot), isolates from Queensland are in red colour and Victorian isolates are in blue. Dashed rectangle show the location of the seven isolates to their possible ST founder ST251 (asterisk).
Pangenome analysis
Pangenome analysis showed a total gene pool of 3764 genes (including RNA encoding), with the core genome making up 1509 genes (Table 2). The soft-core genome, found in ≥ 26/27 isolates, was made up of 1542 genes. Of these, 1380 genes (89.49%) could be assigned a Gene Ontology (GO) identity and descriptors (Data S5). The most abundant class of genes, based on predicted localisation of the encoded proteins was as ‘integral member of the membrane’ followed by ‘cytoplasm location’. Most genes appeared to be housekeeping. In addition, 64 encoded proteins predicted to be associated with proteolysis, 5 associated with virulence, 4 with adhesion, 1 with capsule polysaccharide biosynthetic process, and 1 was pillus-associated. Of the 5 virulence-associated genes, 3 encoded proteins that were predicted to be secreted: pauA, LytR, and the YSIRK-type signal peptide-containing protein. Several proteins associated with nutrients commonly found in abundance within the mammary tissue of dairy cows were also identified including fibrinogen binding proteins (WP_046389274.1, WP_012658173.1) and lactose catabolism proteins (lacA, WP_000215993.1; lacB, WP_000686149.1).
Table 2.
Pangenome analysis results. The total number of genes and percent of total pangenome is shown.
| Pangenome breakdown | Number of isolatesa | Number of genes | Percentage of pangenome (%) |
|---|---|---|---|
| Core genes | 27 | 1509 | 40.09 |
| Soft core genes | 26 | 33 | 0.88 |
| Shell genes | 5–25 | 534 | 14.19 |
| Cloud genes | 1–4 | 1688 | 44.85 |
| Total genes | 1–27 | 3764 | 100 |
aNumber of isolates out of the total (n = 27).
Virulence factors
To address the lack of traditional S. uberis virulence factors detected from the Virulence Factor Database (VFDB), a custom S. uberis virulence database was constructed, named the S. uberis Putative Virulence Database; SuPVDB (Data S2). This was built based on three main sources. Firstly, identification of translated homologues from the VFDB using relaxed alignment statistics (> 70% query cover and > 60% similarity) (Data S3). Secondly, experimentally-confirmed virulence factors identified from a number of publications26–41. Finally, inclusion of genes with virulence-associated GO descriptors (Data S5). Despite HasA42, PauA43, and PauB39 being non-essential for bovine mastitis, these three were included in the search for the sake of completeness. MtsB and regulator ScaR were also included as potential virulence factors because they were associated with the MtuA operon. To identify homologous virulence factors in S. uberis, the Streptococcal VFDB amino acid sequences were screened against the translated pangenome. Any sequences with > 70% query cover and > 60% similarity were considered putative homologues. Twenty-eight previously undescribed putative virulence factors were identified in S. uberis based on homology to known virulence factors in other Streptococcal species (Data S3). This custom database consisted of 53 total sequences (Data S2), which were screened against each isolate, as well as all 40 S. uberis isolates present within GenBank (Data S6).
The GO virulence-associated genes encoded an enolase/phosphopyruvate hydratase (WP_012658173.1), LytR family regulatory protein (SQG46876.1), YSIRK-type signal peptide-containing protein (WP_154591130.1), PauA a plasminogen activator A (WP_046388868.1), and a HU family DNA-binding protein, HlpA (WP_012658741.1). 90 soft core proteins were predicted to be secreted including PauA, LytR, and the YSIRK-type signal peptide-containing protein were all predicted to be secreted, along with pneumococcal-type histidine triad protein (Pht).
The average number of virulence factors per isolate was 27.7 factors (± Standard deviation [σ] = 2.6) from the current study. When the publicly available S. uberis genomes were screened, an average of 27.86 ± 2.14 σ putative virulence factors were identified from isolates associated with bovine mastitis (Data S6). The 3 human mastitis isolates also contained a similar amount, 27.67 ± 4.04 σ. The aquatic S. uberis isolate CAIM 1894 (SAMN03093229) contained a total of 3 putative virulence factors.
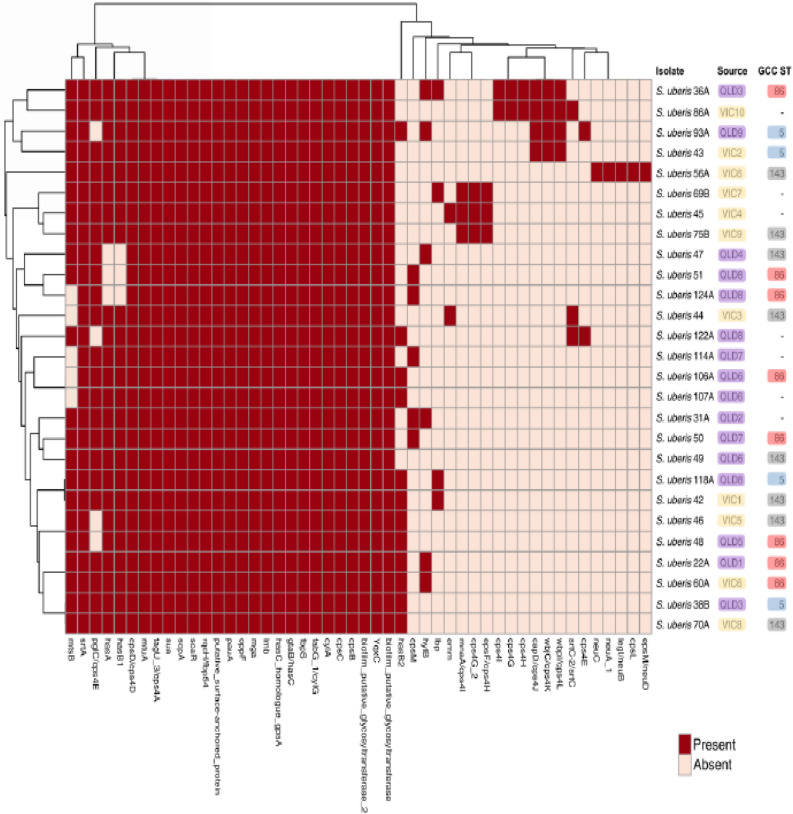
In the current study, the number of virulence factors per isolate ranged from 35 in S. uberis 36A to 24 in S. uberis 124A. Twenty-two putative virulence factors were found in every isolate. These factors included biofilm putative glycosyltransferase, biofilm putative glycosyltransferase 2, cpsB, cpsC, cpsD/cps4D, cylA, fabG 1/cylG, fbpS, gtaB/hasC, hasC homologue gpsA, lmb, mga, mtuA, oppF, pauA, putative surface-anchored protein, rqcH/fbp54, scaR, scpA, sua, tagU 3/cps4A and srtA (Fig. 4, Data S4). Moreover, there was a tendency for virulence factors to cluster by state (p = 0.09). No clustering was observed between isolation source and virulence factors. The accessory virulome was made up of putative virulence factors not present in every isolate. They consisted of 24 sequences (Fig. 4), with individual isolates containing the majority of them. S. uberis 56A was the only isolate to contain the putative virulence factors cpsL and neuABCD (epsM/neuD, legI/neuB, neuA_1, neuC).
Figure 4.
Heatmap showing the presence/absence of virulence and putative virulence genes (x-axis) within the S. uberis isolates identified in this study (y-axis). Presence of virulence genes shown in red, absence in light pink. Core virulence genes are found in every isolate (27/27), while accessory virulence genes are found in < 27 isolates. Victorian isolates are coloured in yellow, Queensland in purple. GCCs are coloured, GCC ST86 coloured red, GCC ST5 coloured blue and GCC ST143 coloured grey. Data generated using ABRicate version 1.0.1 (https://github.com/tseemann/abricate)121 with the VFDB122 and the custom SuPVDB (Data S2). Heatmap produced using ClustVis (https://biit.cs.ut.ee/clustvis/)124 and Inkscape version 0.92 (https://www.inkscape.org).
Putative sortase substrates, antimicrobial resistance and mobile genetic elements
Streptococcal sortase srtA has been shown to be essential for bovine mastitis26 and a previous study identified 9 proteins attached to the cell wall of S. uberis which were likely virulence-associated genes44. This enzyme cleaves and processes proteins containing particular motifs to allow external cell wall-anchoring. To confirm the importance of cell wall-anchored proteins, isolates were screened for the srtA gene, which had two variants with 85.9% similarity between them. 22/27 strains contained one srtA variant (WP_046390884.1) while 5/27 strains contained another (WP_037592200.1). This was the result of the 90% identity cut-off criteria used in the pangenome analysis. Within this constraint, sortase srtA was still considered a core gene.
To analyse SrtA substrates, core protein-coding sequences were screened for several motifs including LPXTG44, 45, LPXXXD44, LPXTA46, 47, QVPTGV and LPSTGE48. 12 protein-coding sequences were identified when excluding the more variable LPXXXD motif, and 102 including it. As only core genes were screened, there was an increased likelihood of finding essential virulence factors. Some putative sortase substrates of note included a transmembrane protein containing a xanthine permease domain (WP_046392124.1), 7 coding sequences (CDS) associated with the bacterial global response to DNA damage (SOS), peroxiredoxin ahpC (WP_015912043.1), and transmembrane branched-chain amino acid transport protein (WP_046393325.1) (Data S5).
Overall, antibiotic resistance genes were not commonly found within the genomes. Only S. uberis 45, 47, 48 and 51 contained resistance genes and most of these were predicted to encode resistance to lincosamides, due to presence of lnuC or lnuD (Table 3). Antibiotic resistance genes were found in both Victorian and Queensland isolates and in a number of STs. Screening NCBI for LnuC revealed it was not identified in any S. uberis isolates in the database. A multidrug antibiotic resistance cluster was identified in both S. uberis 48 and 51. The cluster consisted of mel/mef(A), mrsE, vatD and lnuD. Despite S. uberis 47 containing lnuD, it did not contain the cluster. The two isolates containing the cluster were found in different isolation sources within Queensland. They both belonged to GCC ST86. The gene cluster is associated with bacterial resistance to lincosamides, macrolides, oxazolidinones, phenicols, pleuromutilins, streptogramins and tetracyclines.
Table 3.
Table showing antibiotic resistance profiles of S. uberis isolates in this study.
| Isolate/resistance gene | Lincosamides | Streptogramins | Macrolides, oxazolidinones, phenicols, pleuromutilins, streptogramins, tetracyclines |
|---|---|---|---|
| S. uberis 45 | lnuD | ||
| S. uberis 47 | lnuC | ||
| S. uberis 48 | lnuD, mel, mrsE, | vatD | mel, mrsE |
| S. uberis 51 | lnuD, mel, mrsE, | vatD | mel, mrsE |
The antibiotic class is shown along the x-axis, and the isolate is shown along the y-axis. The gene responsible for resistance is shown.
One-third (9/27) of the isolates contained putatively intact bacteriophages (Table 4). S. uberis 38B contained two bacteriophages. The Streptococcus prophage 315.2 was found in isolates S. uberis 38B and S. uberis 47, both Queensland isolates. There were no virulence genes associated with any bacteriophages. Transposons and insertion sequences were identified in 21/27 isolates (77.78%) (Table 5). None were associated with virulence factors. No plasmids were found in the isolates.
Table 4.
Table of putatively intact bacteriophage presence within the S. uberis isolates identified in this study.
| Isolate | Region length (kb) | Closest bacteriophage hit | Accession of closest hit |
|---|---|---|---|
| S. uberis 38B | 36.8 | Streptococcus phage SMP | NC_008721 |
| S. uberis 38B | 42.2 | Streptococcus prophage 315.2 | NC_004585 |
| S. uberis 47 | 43.9 | Streptococcus prophage 315.2 | NC_004585 |
| S. uberis 49 | 40.8 | Enterococcus phage phiFL4A | NC_013644 |
| S. uberis 50 | 55.5 | Streptococcus phage 5093 | NC_012753 |
| S. uberis 60A | 59.5 | Streptococcus phage PH10 | NC_012756 |
| S. uberis 70A | 36.4 | Streptococcus phage T12 | NC_028700 |
| S. uberis 93A | 36.7 | Streptococcus phage 315.3 | NC_004586 |
| S. uberis 118A | 42.8 | Streptococcus phage P9 | NC_009819 |
| S. uberis 122A | 41.9 | Streptococcus phage T12 | NC_028700 |
The closest-related phage are also shown, along with the accession numbers.
Table 5.
Transposon and insertion sequences within the S. uberis isolates identified in this study.
| IS family | Isolates | Total |
|---|---|---|
| IS256 | S. uberis 46 | 1 |
| IS3 | S. uberis 106A; S. uberis 107A; S. uberis 114A; S. uberis 122A; S. uberis 122A; S. uberis 124A; S. uberis 22A; S. uberis 38B; S. uberis 43; S. uberis 44; S. uberis 47; S. uberis 49; S. uberis 51; S. uberis 56A; S. uberis 70A; S. uberis 75B; S. uberis 86A | 19 |
| IS30 | S. uberis 118A; S. uberis 42; S. uberis 44; S. uberis 56A; S. uberis 69B; S. uberis 70A; S. uberis 75B | 7 |
| IS6 | S. uberis 31A | 1 |
| ISL3 | S. uberis 86A | 1 |
| Novel | S. uberis 22A | 1 |
Isolates which contain IS families are separated by semicolons. S. uberis 122A contains two different IS3 regions and is included twice in this table.
Discussion
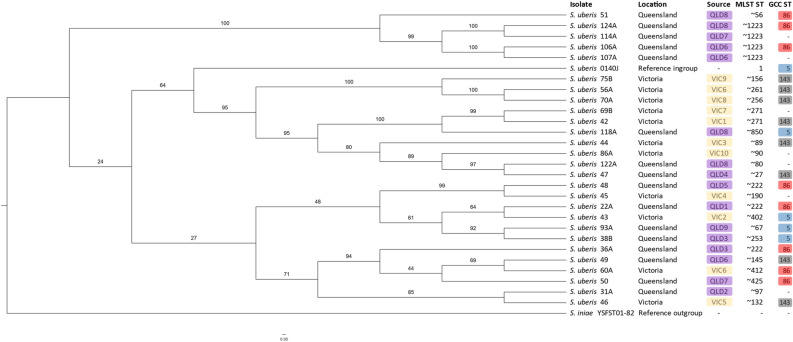
Twenty-nine herds located in three climatically distinct geographic regions of Australia were screened and from them 27 independent S. uberis isolates were obtained and their genetic relatedness was assessed. The diverse range of MLSTs observed in this study is consistent with previous surveys of bovine mastitis-associated S. uberis in that there was a large variety of STs identified49. In one study, 33 MLST STs were identified in their group of 46 isolates, the most common STs being MLST ST60 and ST15549. In the current study, none of the MLST STs in the isolates were identical, although some were similar such as MLST ST ~ 1223. However, GCC STs were more consistently represented (Table 1) with three GCC STs (ST86, ST143, GCC ST5) representing 20 of the 27 isolates. This finding is mostly consistent with the published literature that has identified GCC ST143 and GCC ST5 as the most common GCCs found in S. uberis isolates obtained from Australian cases of clinical mastitis11, 16, 17, 23. The observation that GCCs clustered by region was notable, given that GCC ST86 isolates were more prevalent in QLD compared to Victoria, while GCC ST143 isolates were more prevalent in Victoria. This study provides observational evidence that S. uberis GCC ST86 isolates, which had been previously thought to lack pathogenicity required to cause clinical mastitis49, can contribute to disease. Further investigations such as experimental animal trials will be required to confirm this observation. If GCC ST86 isolates were indeed responsible for clinical mastitis, it would also be consistent with the lack of strong phylogenetic clustering of GCC ST86 (Fig. 3); the putative virulence factor profiles which showed consistent differences between GCCs (Fig. 4). Isolation of S. uberis GCC ST86 strains from cows afflicted with bovine mastitis may be indicative of a shift in pathogenic lineages within the Australian S. uberis population. Alternatively, this provides evidence that sequence typing alone is not a sufficiently granular approach to associate with pathogenicity of S. uberis in the bovine mammary gland, neither the core genome-based phylogenetic clustering (Fig. 3) nor the putative virulence factor profiles (Fig. 4) align closely with GCC typing in parallel. Our results indicate that all isolates likely possess virulence factors promoting invasion of host tissue, survival in the host environment, evasion of the host immune response, and internalization in the mammary gland cells49.
Figure 3.
Maximum likelihood phylogenetic tree based on core genome alignment between the S. uberis isolates identified in this study. S. iniae YSFST01-82 (accession number: GCF_000831485.1) was used as an outgroup and S. uberis 0140J (accession number: AM946015) (MLST ST1, GCC ST5) as an ingroup. Bootstrap values are shown in numbers along branches. Purple labels show Queensland isolates. Yellow labels show Victorian isolates. MLST STs are shown, as well as GCC STs. GCCs are coloured, GCC ST86 coloured red, GCC ST 5 coloured blue, GCC ST143 coloured grey. Core genome alignment was performed in roary (version 3.13.0)115 and maximum likelihood tree was built in RAxML version 8.2.10 (raxmlHPC-PTHREADS-SSE3)120 with 1000 replicates. Tree visualised in FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). For figure clarity, the phylogenetic tree was visualised using a cladogram transformation. Data S1 shows the untransformed tree, along with an isolate-only tree which shows very high bootstrap support at almost every node (~ 100%) due to the increased number of genes in the core genome alignment.
These observations indicate that the relative importance of an environmental source and possible cow-to-cow transmission in the development and risk of mastitis also depend on geographical factors, and the transmissibility and disease potential of the particular strains circulating in the cow’s environment10, 20, 50. It could also impact on the relative success rate of treatment protocols51 and management practices and the apparent ‘success’ and ‘failure’ rates in different regions or on different properties. At present, attention to possible environmental sources of infection, good standards of milking hygiene, and proper treatment of S. uberis mastitis remain important measures in attempts to control transmission of S. uberis20, 23.
A maximum likelihood phylogeny of the core genome demonstrated the clustering of MLST STs and indicated that the scheme was, at times, suitable (Fig. 3) for S. uberis. For example, MLST ST ~ 1223 clustered well, however MLST ST ~ 222 did not. Given the amount of novel STs identified (100% of isolates), the usefulness of the ST was of limited value to describing the evolutionary lineage of this highly recombinant species52. While the GCCs clustered with other bovine mastitis-associated S. uberis (Fig. 2), the inconsistent phylogenetic clustering between GCC STs (Fig. 3) meant that these GCCs did not appear to accurately capture the assumed evolutionary lineages of the bovine mastitis-associated S. uberis isolates identified in this study. Given the reducing cost of whole genome sequencing and greater power for genomic and phylogenetic analysis compared with the multiple sanger sequencing reactions required for MLST and GCC ST analysis, it would be appropriate to rely more on higher resolution WGS in future diagnostics. There appeared to be two clades containing state-only isolates: a Queensland clade including S. uberis 51, 124A, 114A, 106A and 107A, which almost all shared the same MLST ST (~ 1223) and GCC ST (86); and a Victorian clade containing S. uberis 75B, 56A, and 70A, in which almost all sharing the same GCC ST (143). Within these two specific clades, it is likely that the isolates diverged from a common lineage within their respective states. Every other identified clade appeared to contain a mixture of Victorian and Queensland isolates. Phylogenetic clustering within a source point was not observed in many cases, such as with QLD7, QLD8, QLD9 and VIC6 isolates. This observed divergence was much greater than originally expected. The most likely explanation of this finding is environmental acquisition of novel S. uberis bovine mastitis-associated isolates23, 41, 50, rather than an acquisition of monophyletic descendants such as would be expected with a contagious pathogen. Given this observation, further analysis of environmental S. uberis reservoirs is recommended.
The pan and core genome genes were consistent with previous pangenome analyses, showing a core genome of 1550 genes using 13 clinical and sub-clinical bovine mastitis isolates27 and ~ 1530 genes using 21 bovine mastitis isolates52. Core genes associated with nutrients found in abundance within the mammary tissue of cows, including two fibrinogen binding proteins, were identified (WP_046389274.1, WP_012658173.1). This is a notable finding given the fibrinogen concentration in the milk of cows affected by mastitis is significantly higher than that of healthy cows53. Two genes associated with lactose catabolism were identified, lacA (WP_000215993.1) and lacB (WP_000686149.1), which is also notable given the abundance of lactose in bovine milk. This may explain the presence or adaptation of S. uberis to mammary tissue.
As strain specific pathogenicity has been observed with S. uberis54, this indicates bacterial pathogenic factors are involved, not host-specific factors. Notably, the isolates from this study shared similar numbers of putative virulence factors as the publicly available bovine mastitis isolates, as well as human mastitis isolates (Fig. 4, Data S4, Data S6). This indicates a common set of virulence determinants responsible for cross-host mastitis pathogenesis.
The core virulence factors identified in every isolate (Fig. 4, Data S4) may partially explain the pathogenic behaviour of S. uberis. There were several core genes of note including cpsBCD and cps4a which encode products involved in capsular polysaccharide formation40, as-well-as, two putative glycosyltransferases involved with biofilm formation38. cylAG were identified, which contribute to hemolytic activity55, corresponding to an ABC transporter and an enoyl reductase involved in fatty acid biosynthesis56. This analysis confirmed hasA as a non-essential virulence factor, as it was only found in 24/27 isolates, and also confirmed mtuA as an essential virulence factor33, as it was found in all isolates. rqcH/fbp54, a fibronectin-binding protein and putative fibronectin-binding protein fbpS were also found in all isolates. This has been shown to be important for adhesion and a key virulence factor in Streptococcus pyogenes57, as-well-as, in Streptococcus suis, but not essential for virulence58. Also found in every isolate was scpA, a C5a peptidase, which has been shown to delay accumulation of leukocytes attracted to S. pyogenes by cleaving the serum chemotaxin C5a, indicating it is an essential virulence factor59. hasC was also found in all isolates; it is essential for hyaluronic acid capsule production29. Upon loss of this gene, S. uberis becomes susceptible to phagocytosis by bovine neutrophils, indicating it is an important virulence factor29. lmb, encoding a laminin binding protein important for bacterial colonisation37 was also found in every isolate. Mga, a virulence regulator has been shown to be important for a number of phenotypes including biofilm formation, growth in whole human blood and soft tissue and phagocytosis resistance60, 61. This was also identified in all isolates of S. uberis in this study. sua, an adhesin, has been experimentally verified in S. uberis isolates from dairy cows and identified as a conserved gene62. Our study confirms this as it is conserved in all isolates. oppF, an oligopeptide permease, shown to be important for auxotrophic amino acid acquisition from bovine milk34, was also found in every isolate. scaR, another virulence regulator which upregulates during low levels of Mn2+63 was also found in every isolate. Given that manganese is present in cow’s milk at 0.02–0.05 µg/mL64, it is likely scaR upregulates virulence-associated genes in response to mastitis infection. Finally, a putative surface anchored protein was also found in all isolates. This was experimentally identified as a sortase substrate, anchored to the surface in S. uberis44. It contains a G5 domain, which is involved with N-acetylglucosamine binding and is found in a number of different enzymes, ranging from biofilm proteins to IgA-cleaving peptidases65. The neuABCD locus is involved in sialic acid synthesis and their presence in S. uberis 56A indicates it may produce a capsule to assist with immune system evasion66, 67.
Other core virulence genes included enolase/phosphopyruvate hydratase (WP_012658173.1), involved in plasminogen and adhesion in Streptococcal species68 and LytR family regulatory protein (SQG46876.1) essential for attaching capsules and teichoic acids to cell wall69. Also a YSIRK-type signal peptide-containing protein (WP_154591130.1), PauA a plasminogen activator A (WP_046388868.1) which has been already described as a non-essential virulence factor in S. uberis-related bovine mastitis43. Pht, a putatively secreted gene has been implicated in adhesion to epithelium in Streptococcus pneumonia70. Finally, a HU family DNA-binding protein, HlpA (WP_012658741.1) has been shown to bind to double-stranded DNA involved in repair and recombination71 in times of cellular stress72 and is involved in tissue inflammation of Streptococcal species73.
Scanning the core genome for putative sortase substrates revealed several genes which may also explain behaviour of S. uberis, including a transmembrane branched-chain amino acid transport protein (WP_046393325.1), which has been shown to transport leucine, isoleucine and valine via proton motive force74, all three of which are found in excess within bovine milk75. Furthermore, bacterial loads of Staphylococcus aureus, another bovine mastitis-causing organism have been correlated with these amino acids in milk76.
Another putative srtA substrate of note is a transmembrane protein containing a xanthine permease domain (WP_046392124.1), which allows uptake of xanthine. Xanthine and hypoxanthine are commonly found in bovine milk77, along with bovine enzyme xanthine oxidoreductase. Bovine xanthine oxidoreductase catalyses oxidation of hypoxanthine to hydrogen peroxide78 and xanthine, and of xanthine to uric acid, among many other substrates79. Of note, is the demonstrated antimicrobial activity against Gram-positive and negative bacteria in milk via production of reactive oxygen/nitrogen species, namely hydrogen peroxide and nitric oxide in both humans and cows78, 80. All S. uberis isolates identified in this study containing an externally-located protein which competes with this precursor antimicrobial compound may indicate a novel virulence factor which allows S. uberis to colonise and infect the udder, causing bovine mastitis. The core genome contained 7 genes associated with the bacterial SOS response (Data S5), which may be involved in combating damage caused to DNA by reactive oxygen/nitrogen species81. Also found in the core genome was peroxiredoxin ahpC (WP_015912043.1), which breaks down hydrogen peroxide and has been shown in Streptococcus agalactiae to bind heme82.
We propose the xanthine permease domain protein uptakes free hypoxanthine and xanthine found in bovine milk. This likely competes with bovine enzyme xanthine oxidoreductase which usually converts xanthine to the antimicrobial, reactive oxygen species, hydrogen peroxide. At least 7 genes associated with the bacterial SOS response are likely involved in combating damage caused to DNA by reactive oxygen/nitrogen species. The presence of core genes such as these demonstrate adaptation of S. uberis to the bovine host, and as one facet of S. uberis-associated bovine mastitis. It is possible the genes highlighted here demonstrate the Red Queen hypothesis83 between host and pathogen84, 85, which can be summarised as follows: organisms living within a dynamic environment (bovine host) require matching rates of evolution to maintain colonisation. Co-evolution between host and parasite is a widely recognised consequence of natural selection86 and continually develops as genetic arms-race.
There were a limited number of antibiotic resistance genes found in this study. This finding is consistent with some previous dairy herd surveys9, but inconsistent with others that detected a broad and diverse group of antibiotic resistance genes13, 87, 88. Four S. uberis isolates found across Victoria and Queensland herds contained a combination of InuC and a multidrug resistance cluster including mel/mef(A), mrsE, vatD and lnuD. The presence of an antimicrobial resistance gene cluster not associated with any known insertion sequences or mobilizable elements indicates this may have been acquired through uptake and integration of free DNA. The two isolates that contain this cluster, S. uberis 48 and 51 are not directly phylogenetically related in the context of other isolates, which may indicate this chromosomal integration occurred independently. Further evidence of this can be seen in the variable genomic context of this cluster, which is not consistently placed within the genome (data not shown). The mel/mef(A) gene has been reported in S. pneumoniae89 and S. pyogenes90 but not in S. uberis. S. uberis was then screened via NCBI for mel/mef(A). Only one protein sequence was identified (accession: WP_046391446.1) from S. uberis 6780 (accession: JATD00000000), also isolated from milk from a cow udder afflicted with bovine mastitis27. This provided further evidence that this observation is a rare occurrence not associated with known mobilizable elements. The mel/mef(A) gene encodes resistance to lincosamides, macrolides, oxazolidinones, phenicols, pleuromutilins, streptogramins and tetracyclines91. The msr family ABC-F type ribosomal protection gene, which is homologous to the coding sequence of MrsE (99% query coverage, 79% similarity), encodes resistance to streptogramins, phenicols, pleuromutilins, lincosamides, oxazolidinones, tetracyclines, macrolides and other antibiotics which target the peptidyl-transferase region of the ribosomal subunit92. The next gene in the cluster encodes a hypothetical protein with no known function or domains. This is followed by vatD, a streptogramin A O-acetyltransferase resistance gene93. The final gene in the cluster is lnuD, encoding a nucleotidyltransferase, which has been shown to provide lincosamides resistance. The lnuD and non-cluster lnuC genes encode O-nucleotidyltransferases and inactivate lincosamides by adenylylation94. The lnuD has been previously reported in S. uberis isolated from clinical bovine mastitis95, but the lnuC gene has only been identified in human S. agalactiae isolates96, 97, suggesting recent acquisition by bovine mastitis-associated S. uberis. As lnuC is a transposon-mediated resistance gene97, this may represent the first observed interspecies jump into bovine mastitis-associated S. uberis. Lincosamide (lincomycin, clindamycin, and pirlimycin) usage and resistance should be carefully monitored now this mobilizable gene is within bovine mastitis-associated S. uberis populations. It would be of value to determine if a similar pattern exists in other streptococcal species present in the dairy farm environment. Further work is needed to verify these findings and determine antimicrobial gene reservoirs and the mechanism of transfer. A limitation of this study was the small number of herds and limited number of isolates cultured from clinical mastitis cases. However, herd (farm) type, breed of dairy cow, and herd size were representative of Australian herds. Nevertheless, the presence of these genes in some of the isolates indicates that antibiotic usage should be carefully tracked and monitored to produce and maintain an up-to-date understanding of the level of antimicrobial resistance within dairy populations in the country.
Materials and methods
The study and all experimental procedures and protocols were approved by the Animal Ethics Committee at the University of Queensland (animal ethics approval numbers: SVS/ANRFA/540/18 and SVS/043/18/TERRAGEN), and all methods were performed in accordance with the relevant institutional guidelines and regulations. The methods described in the current study and reported results were compliant with ARRIVE guidelines for reporting animal research98.
Sample collection
This study was part of a larger project conducted between March and June 2019 based on 430 milk samples collected from 29 dairy herds located in Queensland [QLD] (North Queensland, n = 9 herds; Southeast QLD, n = 9 herds), and Victoria ([VIC], n = 11 herds) (Fig. 1; map figure was generated using ozmap package version 0.3.6 [https://CRAN.R-project.org/package=ozmaps]99 implemented within R version 4.0.2 [https://www.R-project.org/]100). A 2-week rolling average of herd bulk somatic cell counts (BMTSCCs) was used to arbitrarily divide the herds into three categories (≤ 150 × 1000 cells/mL, > 150 to 300 × 1000 cells/mL, and > 300 × 1000 cells/mL). BMTSCCs categories were used as an a priori assumption for the risk of mastitis in the herd. Herd selection was based on ease of access to the farm location and the cooperation of the dairy farm owners and their associated veterinary practices.
Detailed methodology is described elsewhere101. Briefly, milk samples were collected from eligible dairy cows with a new case of clinical mastitis. Chronic mastitis cases (apparently healthy cow with lumps palpable in the udder, and mild changes to milk) and subclinical mastitis cases were not eligible for study enrolment. An enrolment eligible clinical mastitis case was defined as a previously apparently healthy lactating dairy cow of any age, breed, or stage of lactation that was experiencing a new case of clinical mastitis, defined as either the first occurrence of a mastitis event in the current lactation or a mastitis event occurring at least 21 days following a previous mastitis event that has clinically resolved or achieved a clinical cure102. Eligible cases must have not received systemic or intramammary antimicrobials, anti-inflammatory medications, or topical treatments in the 2 weeks prior to becoming a new case, nor immediately prior to sample collection. Milk samples were collected aseptically from individual quarters into separate sterile tubes, immediately capped and placed in a − 20 °C freezer. Collected samples were transported and delivered frozen to the Veterinary Laboratory Services of the University of Queensland for bacterial culture.
Streptococcus uberis isolation
At the laboratory, milk samples were mixed thoroughly, 100 µL streaked onto Sheep Blood Agar (SBA, P2133 Sheep Blood Columbia Agar Plates, Thermofisher), and the SBA plate incubated aerobically at 37 °C for 18–24 h. S. uberis isolates were initially identified using conventional microbiology laboratory tests (Gram stain appearance and catalase production). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonik, Bremen, Germany) was used to confirm the identification. Individual colonies were sub-cultured on SBA plates and incubated aerobically at 37 °C for 18–24 h. Pure isolates were then incubated in 2 mL of Brain Heart Infusion (BHI) broth, subsequently mixed with 20% glycerol, and stored at − 80 °C.
DNA extraction
Stored S. uberis isolates were batch thawed and cultured in BHI broth (37 °C, orbital shaker at 300 rpm, 18–24 h). Genomic DNA was extracted using DNeasy PowerFood Microbial Kit (QIAGEN Chadstone, Victoria, Australia) with minor modifications. Eight mL of culture liquid was centrifuged (15 min, 4 °C, 20,000×g) to pellet the bacteria. The pellet was then resuspended in 450 µL of lysis buffer and incubated for 10 min at 65 °C. Proteinase K (Proteinase K, QIAGEN Chadstone, Victoria, Australia) (25 µL) was added and incubation continued for an additional 20 min at 65 °C. This Proteinase K modification to the DNeasy protocol increased the final quantity of extracted DNA (data not shown). Thereafter, the whole component was transferred to a Powerbead tube, secured horizontally to a vortex adapter, and vortexed at maximum speed for 10 min. After washing to remove protein and other inhibitors, purified DNA was eluted and the cconcentration and purity of the isolated genomic DNA determined using a NanoDrop ND-1000 spectrophotometer. A sample of DNA was considered acceptable if the A260/280 ratio was ~ 1.8.
Whole genome sequencing
The New England Biolabs NEBNext Ultra II FS DNA Library Prep Kit for Illumina for multiplexing was used according to the manufacturer’s instructions (New England Biolabs, Notting Hill, Victoria, Australia) to construct whole genome sequencing libraries. Libraries were sequenced (Illumina NextSeq 500 instrument, 300 cycle mid-output kit; 2 × 150 bp paired end) at the Centre of Health Genomics and Informatics, University of Calgary, Canada, to obtain an average coverage depth of > 100 and a read retention of > 98%. The draft genome sequences of the 27 S. uberis isolates have been deposited with GenBank103 under BioProject PRJNA669385.
Whole genome assembly and annotation
Primer sequences were removed and reads were quality trimmed using cutAdapt104. The Nullarbor pipeline105 was used to process the samples. SKESA assembler (version 2.3.0)106 was used for de-novo assembly. Assemblies were annotated using prokka (version 1.14.5)107. Details about the parameters used in different packages can be found at https://github.com/bananabenana/genomics_methods_S.uberis/blob/main/commands_run.
Multi-locus sequence typing
Assemblies were taxonomically assigned, analysed for MLST and were possible assigned a GCC, sequence types (STs)108, 109, and scored for genome distance using Type (Strain) Genome Server (TYGS) (https://tygs.dsmz.de)110 (date accessed: 19 May 2020). MLSTs were confirmed using mlst_check version 2.1.1706216111 and MLST databases accessed on 5 June 2020. Phyloviz version 2.0 (www.phyloviz.net)112 and the goeBURT algorithm113 were used to visualise the data. Pairwise comparison of genome sequences among the set of genomes were conducted by calculating precise distances using the Genome BLAST Distance Phylogeny approach (GBDP) under the algorithm 'coverage' and distance formula d5114. These distances were used to determine the genome similarities for each of the genome pairs. 100 distance replicates were calculated each. Digital DDH values and confidence intervals were calculated using the recommended settings of the GGDC 2.1114.
Pangenome analysis
Pangenome analysis was performed using roary (version 3.13.0)115, splitting paralogues and setting the identity threshold at 90%. This list was then annotated and characterised via screening sequences against NCBI and Gene Ontology116 using the PANNZER2 server117. Signal sequences were obtained from the core genome using SignalP 5.0 (Linux x86_64)118, 119.
Phylogenetic analysis
A core genome alignment generated by roary was performed, including an outgroup S. iniae YSFST01-82 (accession number: GCF_000831485.1) and in-group S. uberis 0140J (accession number: AM946015) ST1. RAxML (raxmlHPC-PTHREADS-SSE3 version 8.2.10)120 was used for maximum likelihood and rapid bootstrap analysis with 1000 replicates. The bipartitions output file was visualised in FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and bootstraps visualised. Inkscape version 0.92 (https://www.inkscape.org) was used to prepare the figure.
Identification of putative virulence factors
ABRicate version 1.0.1 (https://github.com/tseemann/abricate)121 was used with the VFDB122 and a custom gene list, the SuPVDB (Data S2). The custom gene list was based on sequences identified in the literature described to be S. uberis virulence factors with experimental or strong computational evidence26–41, as well identifying homologues of Streptococcal virulence factors. In addition to the isolates genomes reported in this current study, 40 additional publically available genomes (Data S6)27, 123 were retrieved from GenBank and screened for putative virulence factors. To identify homologous virulence factors in S. uberis, the Streptococcal VFDB amino acid sequences were screened against the translated pangenome. Any sequences with > 70% query cover and > 60% similarity were considered putative homologues (Data S3). The output was then visualised in ClustVis (https://biit.cs.ut.ee/clustvis/)123. No scaling was performed on the data and clustering was performed on rows and columns using the ‘Euclidean’ measure. To identify core, putative sortase A substrates, grep version 3.1 (https://git.savannah.gnu.org/cgit/grep.git/tree/AUTHORS) was used.
Antibiotic resistance determination
ABRicate version 1.0.1 (https://github.com/tseemann/abricate)121 was used with the CARD125, database accessed 4/5/2020.
Bacteriocin identification
Bacteriocins were identified by downloading the BAGEL4 database126 and converting into a multifasta file. The pangenome was then aligned against this using blastp version 2.9.0+127, 128 and results were filtered for ≥ 80% coverage and ≥ 50% similarity/positives (to account for different signal sequences).
Plasmid identification and analysis
Mob-suite version 3.0.0129 was used to identify plasmids using the mob recon function, then visualised in ClustVis (https://biit.cs.ut.ee/clustvis/)124. No scaling was performed on the data and clustering was performed on rows and columns using the ‘correlation’ distance measure. Putative mobilization of plasmids was determined using the mob_typer function.
Bacteriophage and prophages identification
The PHASTER (https://phaster.ca/)130 web server was used to identify bacteriophage and prophage regions using the API tool. ‘Questionable’ phages (score of 70–90) were grouped with ‘incomplete’.
Transposon/Insertion sequence identification
ISESCAN version 1.7.2 (https://github.com/xiezhq/ISEScan)131 was used to identify transposons and insertion sequences.
Statistical analysis
Fisher’s exact test was performed to identify significance in the proportion of strain types, virulence genes, and plasmids from isolates (unit of analysis) recovered from NQLD, SQLD and VICT. A two sided p-value obtained by Monte-Carlo simulation (n = 2000) of at least 0.05 was considered to be significant. Statistical analysis was conducted using stats package implement in R.
Supplementary Information
Acknowledgements
We would like to acknowledge Joanne Mollinger and Ameh James for their contributions to this work.
Author contributions
Conception and experimental design: J.I.A., T.O., M.S. Acquisition of data: H.A., H.R., J.I.A. Analysis and interpretation of data: B.V., J.I.A., R.M. Drafted the manuscript: B.V., J.I.A., H.A. All authors contributed to and revised the manuscript.
Competing interests
This work was funded by Science with Impact Scheme, the University of Queensland, Sub-tropical Dairy Board, Dairy Australia, and Terragen Biotech Ltd, Australia. The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ben Vezina and John I. Alawneh.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82357-3.
References
- 1.Rollin E, Dhuyvetter KC, Overton MW. The cost of clinical mastitis in the first 30 days of lactation: An economic modeling tool. Prev. Vet. Med. 2015;122:257–264. doi: 10.1016/j.prevetmed.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Huijps K, Lam TJGM, Hogeveen H. Costs of mastitis: Facts and perception. J. Dairy Res. 2008;75:113–120. doi: 10.1017/S0022029907002932. [DOI] [PubMed] [Google Scholar]
- 3.Hogeveen H, Huijps K, Lam TJGM. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011;59:16–23. doi: 10.1080/00480169.2011.547165. [DOI] [PubMed] [Google Scholar]
- 4.Shum LW, McConnel CS, Gunn AA, House JK. Environmental mastitis in intensive high-producing dairy herds in New South Wales. Aust. Vet. J. 2009;87:469–475. doi: 10.1111/j.1751-0813.2009.00523.x. [DOI] [PubMed] [Google Scholar]
- 5.Ruegg PL. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017;100:10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- 6.Verbeke J, Piepers S, Supre K, De Vliegher S. Pathogen-specific incidence rate of clinical mastitis in Flemish dairy herds, severity, and association with herd hygiene. J. Dairy Sci. 2014;97:6926–6934. doi: 10.3168/jds.2014-8173. [DOI] [PubMed] [Google Scholar]
- 7.Phuektes P, et al. Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J. Clin. Microbiol. 2001;39:1460–1466. doi: 10.1128/JCM.39.4.1460-1466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley A, Leach K, Breen J, Green L, Green M. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 2007;160:253–258. doi: 10.1136/vr.160.8.253. [DOI] [PubMed] [Google Scholar]
- 9.Charman N, Dyson R, Hodge A, Robertson N, Chaplin S. A Survey of Mastitis Pathogens in the South Eastern Australian Dairy Industry. Moncton: Dairy Focus; 2012. [Google Scholar]
- 10.Zadoks RN, et al. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol. Infect. 2003;130:335–349. doi: 10.1017/S0950268802008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pullinger GD, Coffey TJ, Maiden MC, Leigh JA. Multilocus-sequence typing analysis reveals similar populations of Streptococcus uberis are responsible for bovine intramammary infections of short and long duration. Vet. Microbiol. 2007;119:194–204. doi: 10.1016/j.vetmic.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Leelahapongsathon K, Schukken YH, Srithanasuwan A, Suriyasathaporn W. Molecular epidemiology of Streptococcus uberis intramammary infections: Persistent and transient patterns of infection in a dairy herd. J. Dairy Sci. 2020;103:3565–3576. doi: 10.3168/jds.2019-17281. [DOI] [PubMed] [Google Scholar]
- 13.Tomazi T, et al. Genotyping and antimicrobial resistance of Streptococcus uberis isolated from bovine clinical mastitis. PLoS One. 2019;14:e0223719. doi: 10.1371/journal.pone.0223719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan A, Khan I, Abdulmawjood A, Lämmler C. Evaluation of PCR methods for rapid identification and Differentiation of Streptococcus uberis and Streptococcus parauberis. J. Clin. Microbiol. 2001;39:1618–1621. doi: 10.1128/JCM.39.4.1618-1621.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg A, Nyman A, Unnerstad HE, Waller KP. Prevalence of bacterial genotypes and outcome of bovine clinical mastitis due to Streptococcus dysgalactiae and Streptococcus uberis. Acta Vet. Scand. 2014 doi: 10.1186/s13028-014-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffey TJ, et al. First insights into the evolution of Streptococcus uberis: A multilocus sequence typing scheme that enables investigation of its population biology. Appl. Environ. Microbiol. 2006;72:1420–1428. doi: 10.1128/AEM.72.2.1420-1428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas VL, Fenwick SG, Pfeiffer DU, Williamson NB, Holmes CW. Genomic typing of Streptococcus uberis isolates from cases of mastitis, in New Zealand dairy cows, using pulsed-field gel electrophoresis. Vet. Microbiol. 2000;75:27–41. doi: 10.1016/S0378-1135(00)00184-X. [DOI] [PubMed] [Google Scholar]
- 18.Ward PN, et al. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genom. 2009;10:54. doi: 10.1186/1471-2164-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pullinger GD, et al. Application of Streptococcus uberis multilocus sequence typing: Analysis of the population structure detected among environmental and bovine isolates from New Zealand and the United Kingdom. Appl. Environ. Microbiol. 2006;72:1429–1436. doi: 10.1128/AEM.72.2.1429-1436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundberg Å, et al. Udder infections with Staphylococcus aureus, Streptococcus dysgalactiae, and Streptococcus uberis at calving in dairy herds with suboptimal udder health. J. Dairy Sci. 2016;99:2102–2117. doi: 10.3168/jds.2015-9487. [DOI] [PubMed] [Google Scholar]
- 21.Reinoso EB, Lasagno MC, Odierno LM. Genetic patterns of Streptococcus uberis isolated from bovine mastitis. Rev. Argent. Microbiol. 2015;47:108–111. doi: 10.1016/j.ram.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 22.McDougall S, Parkinson TJ, Leyland M, Anniss FM, Fenwick SG. Duration of infection and strain variation in Streptococcus uberis isolated from cows' milk. J. Dairy Sci. 2004;87:2062–2072. doi: 10.3168/jds.S0022-0302(04)70024-7. [DOI] [PubMed] [Google Scholar]
- 23.Abureema S, Smooker P, Malmo J, Deighton M. Molecular epidemiology of recurrent clinical mastitis due to Streptococcus uberis: Evidence of both an environmental source and recurring infection with the same strain. J. Dairy Sci. 2014;97:285–290. doi: 10.3168/jds.2013-7074. [DOI] [PubMed] [Google Scholar]
- 24.Werner C, Sauerwald C, Sundrum A, El-Sayed A, Zschöck M. Genotyping of Streptococcus uberis isolates in healing process of bovine clinical mastitis. Int. J. Vet. Sci. Med. 2018;6:274–278. doi: 10.1016/j.ijvsm.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolley K, Bray J, Maiden M. Open-access bacterial population genomics: BIGSdb software, the PubMLST.rg website and their applications [version 1; peer review: 2 approved] Wellcome Open Res. 2018 doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leigh JA, Egan SA, Ward PN, Field TR, Coffey TJ. Sortase anchored proteins of Streptococcus uberis play major roles in the pathogenesis of bovine mastitis in dairy cattle. Vet. Res. 2010;41:63. doi: 10.1051/vetres/2010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hossain M, et al. Virulence related sequences; insights provided by comparative genomics of Streptococcus uberis of differing virulence. BMC Genom. 2015;16:334. doi: 10.1186/s12864-015-1512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan SA, Ward PN, Watson M, Field TR, Leigh JA. Vru (Sub0144) controls expression of proven and putative virulence determinants and alters the ability of Streptococcus uberis to cause disease in dairy cattle. Microbiology. 2012;158:1581–1592. doi: 10.1099/mic.0.055863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward PN, Field TR, Ditcham WGF, Maguin E, Leigh JA. Identification and disruption of two discrete loci encoding Hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect. Immun. 2001;69:392–399. doi: 10.1128/iai.69.1.392-399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver SP, Almeida RA, Calvinho LF. Virulence factors of Streptococcus uberis isolated from cows with mastitis. J. Vet. Med. B. 1998;45:461–471. doi: 10.1111/j.1439-0450.1998.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 31.Skalka B, Smola J. Lethal effect of CAMP-factor and UBERIS-factor—a new finding about diffusible exosubstances of Streptococcus agalactiae and Streptococcus uberis. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 1981;249:190–194. doi: 10.1016/S0174-3031(81)80073-X. [DOI] [PubMed] [Google Scholar]
- 32.Jiang M, Babiuk LA, Potter AA. Cloning, sequencing and expression of the CAMP factor gene of Streptococcus uberis. Microb. Pathog. 1996;20:297–307. doi: 10.1006/mpat.1996.0028. [DOI] [PubMed] [Google Scholar]
- 33.Smith AJ, et al. MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 2003;71:4842–4849. doi: 10.1128/iai.71.9.4842-4849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith AJ, Kitt AJ, Ward PN, Leigh JA. Isolation and characterization of a mutant strain of Streptococcus uberis, which fails to utilize a plasmin derived β-casein peptide for the acquisition of methionine. J. Appl. Microbiol. 2002;93:631–639. doi: 10.1046/j.1365-2672.2002.01723.x. [DOI] [PubMed] [Google Scholar]
- 35.Tian XY, et al. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019;131:33–39. doi: 10.1016/j.micpath.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 36.Lakshmi R. Understanding the regulation of Group B Streptococcal virulence factors. Future Microbiol. 2009;4:201–221. doi: 10.2217/17460913.4.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spellerberg B, et al. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human Laminin. Infect. Immun. 1999;67:871–878. doi: 10.1128/iai.67.2.871-878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czabanska A. Immunochemical Investigations of the Cell Envelope Components Isolated from Streptococcus uberis. Lübeck: University of Lübeck; 2013. [Google Scholar]
- 39.Ward PN, Leigh JA. Characterization of PauB, a novel broad-spectrum plasminogen activator from Streptococcus uberis. J. Bacteriol. 2002;184:119–125. doi: 10.1128/jb.184.1.119-125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morona JK, Miller DC, Morona R, Paton JC. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 2004;189:1905–1913. doi: 10.1086/383352. [DOI] [PubMed] [Google Scholar]
- 41.Zadoks RN, Schukken YH, Wiedmann M. Multilocus sequence typing of Streptococcus uberis provides sensitive and epidemiologically relevant subtype information and reveals positive selection in the virulence gene pauA. J. Clin. Microbiol. 2005;43:2407–2417. doi: 10.1128/JCM.43.5.2407-2417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field TR, Ward PN, Pedersen LH, Leigh JA. The Hyaluronic acid capsule of Streptococcus uberis is not required for the development of infection and clinical mastitis. Infect. Immun. 2003;71:132–139. doi: 10.1128/iai.71.1.132-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward PN, Field TR, Rapier CD, Leigh JA. The activation of bovine plasminogen by PauA is not required for virulence of Streptococcus uberis. Infect. Immun. 2003;71:7193–7196. doi: 10.1128/iai.71.12.7193-7196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egan SA, Kurian D, Ward PN, Hunt L, Leigh JA. Identification of sortase A (SrtA) substrates in Streptococcus uberis: Evidence for an additional hexapeptide (LPXXXD) sorting motif. J. Proteome Res. 2010;9:1088–1095. doi: 10.1021/pr901025w. [DOI] [PubMed] [Google Scholar]
- 45.Lalioui L, et al. The SrtA sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 2005;73:3342. doi: 10.1128/IAI.73.6.3342-3350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antos JM, et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuethrich I, et al. Site-specific chemoenzymatic labeling of aerolysin enables the identification of new aerolysin receptors. PLoS One. 2014;9:e109883. doi: 10.1371/journal.pone.0109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnett TC, Patel AR, Scott JR. A novel Sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV Motif to the cell wall. J. Bacteriol. 2004;186:5865–5875. doi: 10.1128/jb.186.17.5865-5875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomita T, et al. Identification of Streptococcus uberis multilocus sequence types highly associated with mastitis. Appl. Environ. Microbiol. 2008;74:114–124. doi: 10.1128/aem.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zadoks RN, Tikofsky LL, Boor KJ. Ribotyping of Streptococcus uberis from a dairy's environment, bovine feces and milk. Vet. Microbiol. 2005;109:257–265. doi: 10.1016/j.vetmic.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 51.AVPG. Australian Veterinary Prescribing Guidelines. https://vetantibiotics.fvas.unimelb.edu.au/bovine-medicine-guidelines/mastitis/. (2020).
- 52.Lang P, et al. Gene content differences across strains of Streptococcus uberis identified using oligonucleotide microarray comparative genomic hybridization. Infect. Genet. Evol. 2009;9:179–188. doi: 10.1016/j.meegid.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 53.Tabrizi AD, Batavani RA, Rezaei SA, Ahmadi M. Fibrinogen and ceruloplasmin in plasma and milk from dairy cows with subclinical and clinical mastitis. Pak. J. Biol. Sci. 2008;11:571–576. doi: 10.3923/pjbs.2008.571.576. [DOI] [PubMed] [Google Scholar]
- 54.Tassi R, et al. Strain-specific pathogenicity of putative host-adapted and nonadapted strains of Streptococcus uberis in dairy cattle. J. Dairy Sci. 2013;96:5129–5145. doi: 10.3168/jds.2013-6741. [DOI] [PubMed] [Google Scholar]
- 55.Spellerberg B, Martin S, Brandt C, Lütticken R. The cyl genes of Streptococcus agalactiae are involved in the production of pigment. FEMS Microbiol. Lett. 2000;188:125–128. doi: 10.1111/j.1574-6968.2000.tb09182.x. [DOI] [PubMed] [Google Scholar]
- 56.Spellerberg B, et al. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 1999;181:3212–3219. doi: 10.1128/jb.181.10.3212-3219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terao Y, et al. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 2001;42:75–86. doi: 10.1046/j.1365-2958.2001.02579.x. [DOI] [PubMed] [Google Scholar]
- 58.Li Q, et al. Fibronectin-/fibrinogen-binding protein (FBPS) is not a critical virulence factor for the Streptococcus suis serotype 2 strain ZY05719. Vet. Microbiol. 2017;208:38–46. doi: 10.1016/j.vetmic.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 59.O'Connor SP, Cleary PP. In Vivo Streptococcus pyogenes C5a peptidase activity: Analysis using transposon- and nitrosoguanidine-Induced mutants. J. Infect. Dis. 1987;156:495–504. doi: 10.1093/infdis/156.3.495. [DOI] [PubMed] [Google Scholar]
- 60.Le Breton Y, et al. Genome-wide discovery of novel M1T1 group A streptococcal determinants important for fitness and virulence during soft-tissue infection. PLoS Pathog. 2017;13:e1006584. doi: 10.1371/journal.ppat.1006584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 62.Luther DA, Almeida RA, Oliver SP. Elucidation of the DNA sequence of Streptococcus uberis adhesion molecule gene (sua) and detection of sua in strains of Streptococcus uberis isolated from geographically diverse locations. Vet. Microbiol. 2008;128:304–312. doi: 10.1016/j.vetmic.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 63.Jakubovics NS, Smith AW, Jenkinson HF. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 2000;38:140–153. doi: 10.1046/j.1365-2958.2000.02122.x. [DOI] [PubMed] [Google Scholar]
- 64.Lönnerdal B, Keen CL, Hurley LS. Iron, copper, zinc, and manganese in milk. Annu. Rev. Nutr. 1981;1:149–174. doi: 10.1146/annurev.nu.01.070181.001053. [DOI] [PubMed] [Google Scholar]
- 65.Bateman A, Holden MTG, Yeats C. The G5 domain: A potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2004;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 66.Feng Y, et al. Attenuation of Streptococcus suis virulence by the alteration of bacterial surface architecture. Sci. Rep. 2012;2:710. doi: 10.1038/srep00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auger J-P, Dolbec D, Roy D, Segura M, Gottschalk M. Role of the Streptococcus suis serotype 2 capsular polysaccharide in the interactions with dendritic cells is strain-dependent but remains critical for virulence. PLoS One. 2018;13:e0200453. doi: 10.1371/journal.pone.0200453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fulde M, Bergmann S. Moonlighting Proteins: Novel Virulence Factors in Bacterial Infections. New York: Wiley; 2017. pp. 245–268. [Google Scholar]
- 69.Ye W, et al. Pneumococcal LytR protein is required for the surface attachment of both capsular polysaccharide and teichoic acids: Essential for pneumococcal virulence. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kallio A, et al. Role of Pht proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect. Immun. 2014;82:1683–1691. doi: 10.1128/iai.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberto J, Nabti S, Jooste V, Mignot H, Rouviere-Yaniv J. The HU Regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One. 2009;4:e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stinson MW, McLaughlin R, Choi SH, Juarez ZE, Barnard J. Streptococcal histone-like protein: Primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect. Immun. 1998;66:259–265. doi: 10.1128/IAI.66.1.259-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stucky K, et al. Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbrdückii subsp. lactic DSM7290. Mol. Gen. Genet. 1995;249:682–690. doi: 10.1007/BF00418038. [DOI] [PubMed] [Google Scholar]
- 75.Wohlt JE, Clark JH, Derrig RG, Davis CL. Valine, leucine, and isoleucine metabolism by lactating bovine mammary tissue. J. Dairy Sci. 1977;60:1875–1882. doi: 10.3168/jds.S0022-0302(77)84118-0. [DOI] [PubMed] [Google Scholar]
- 76.Grispoldi L, et al. The relationship between S. aureus and branched-chain amino acids content in composite cow milk. Animals. 2019;9:20. doi: 10.3390/ani9110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tiemeyer W, Stohrer M, Giesecke D. Metabolites of nucleic acids in bovine milk. J. Dairy Sci. 1984;67:723–728. doi: 10.3168/jds.S0022-0302(84)81361-2. [DOI] [PubMed] [Google Scholar]
- 78.Ozturk G, et al. The antimicrobial activity of bovine milk xanthine oxidase. Int. Dairy J. 2020;102:104581. doi: 10.1016/j.idairyj.2019.104581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harrison R. Milk xanthine oxidase: Properties and physiological roles. Int. Dairy J. 2006;16:546–554. doi: 10.1016/j.idairyj.2005.08.016. [DOI] [Google Scholar]
- 80.Stevens CR, et al. Antibacterial properties of xanthine oxidase in human milk. Lancet. 2000;356:829–830. doi: 10.1016/S0140-6736(00)02660-X. [DOI] [PubMed] [Google Scholar]
- 81.Mendoza-Chamizo B, Løbner-Olesen A, Charbon G. Coping with reactive oxygen species to ensure genome stability in Escherichia coli. Genes. 2018;9:20. doi: 10.3390/genes9110565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lechardeur D, et al. The 2-cys peroxiredoxin alkyl hydroperoxide reductase C binds heme and participates in its intracellular availability in Streptococcus agalactiae. Int. J. Biol. Chem. 2010;285:16032–16041. doi: 10.1074/jbc.M109.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Valen L. A new evolutionary law. Evol. Theory. 1973;1:1–30. [Google Scholar]
- 84.Stockmann C, et al. Clinical and epidemiological evidence of the Red Queen hypothesis in pneumococcal serotype dynamics. Clin. Infect. Dis. 2016;63:619–626. doi: 10.1093/cid/ciw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonachela JA, Wortel MT, Stenseth NC. Eco-evolutionary Red Queen dynamics regulate biodiversity in a metabolite-driven microbial system. Sci. Rep. 2017;7:17655. doi: 10.1038/s41598-017-17774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schulte R, Makus C, Hasert B, Michels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl. Acad. Sci. USA. 2010;107:7359–7364. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Velez JR, et al. Whole-genome sequence analysis of antimicrobial resistance genes in Streptococcus uberis and Streptococcus dysgalactiae isolates from Canadian dairy herds. Front. Vet. Sci. 2017;4:63. doi: 10.3389/fvets.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kappeli N, et al. Sequence types and antimicrobial resistance profiles of Streptococcus uberis Isolated from bovine mastitis. Front. Vet. Sci. 2019;6:234. doi: 10.3389/fvets.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu X, et al. Distribution of serotypes, genotypes, and resistance determinants among macrolide-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 2010;54:1152–1159. doi: 10.1128/AAC.01268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clancy J, et al. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 91.MacAogáin M, et al. Metagenomics reveals a core macrolide resistome related to microbiota in chronic respiratory disease. Am. J. Respir. Crit. Care Med. 2020;202:433–447. doi: 10.1164/rccm.201911-2202OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su W, et al. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc. Natl. Acad. Sci. 2018;115:5157. doi: 10.1073/pnas.1803313115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hammerum AM, Flannagan SE, Clewell DB, Jensen LB. Indication of transposition of a mobile DNA element containing the vat(D) and erm(B) genes in Enterococcus faecium. Antimicrob. Agents Chemother. 2001;45:3223–3225. doi: 10.1128/AAC.45.11.3223-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bozdogan B, et al. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 1999;43:925–929. doi: 10.1128/AAC.49.7.2716-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petinaki E, et al. Lincomycin resistance gene inu (D) in Streptococcus uberis. Antimicrob. Agents Chemother. 2008;52:626–630. doi: 10.1128/AAC.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gravey F, et al. Lincosamide resistance mediated by inu (C)(L phenotype) in a Streptococcus anginosus clinical isolate. J. Antimicrob. Chemother. 2013;68:2464–2467. doi: 10.1093/jac/dkt255. [DOI] [PubMed] [Google Scholar]
- 97.Achard A, Villers C, Pichereau V, Leclercq R. New inu (C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 2005;49:2716–2719. doi: 10.1128/AAC.49.7.2716-2719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.ozmaps: Australia Maps. R package version 0.3.6. https://CRAN.R-project.org/package=ozmaps (2020).
- 100.R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2020).
- 101.Alawneh JI, et al. Survey and sequence characterisation of bovine mastitis-associated Escherichia coli in dairy herds. Front. Vet. Sci. 2020;7:1–16. doi: 10.3389/fvets.2020.582297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Santos J, Cerri R, Ballou M, Higginbotham G, Kirk J. Effect of timing of first clinical mastitis occurrence on lactational and reproductive performance of Holstein dairy cows. Anim. Reprod. Sci. 2004;80:31–45. doi: 10.1016/S0378-4320(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 103.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17(3):2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 105.Seemann, T. et al. Nullarbor. https://github.com/tseemann/nullarbor (2020).
- 106.Souvorov A, Agarwala R, Lipman DJ. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Z, et al. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. 2019;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jolley K, Bray J, Maiden M. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018 doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Page AJ, Taylor B, Keane JA. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J. Open Source Softw. 2016;8:118–119. doi: 10.21105/joss.00118. [DOI] [Google Scholar]
- 112.Nascimento M, et al. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 113.Francisco AP, Bugalho M, Ramirez M, Carrico JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009 doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Annon The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Törönen P, Medlar A, Holm L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018;46:W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 119.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seemann, T. ABRicate. https://github.com/tseemann/abricate.
- 122.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2015;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hill AW, Finch JM, Field TR, Leigh JA. Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow. FEMS Immunol. Med. Microbiol. 1994;8:109–117. doi: 10.1111/j.1574-695X.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 124.Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alcock BP, et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Heel AJ, et al. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Johnson M, et al. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robertson J, Nash JHE. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018 doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Arndt D, et al. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xie Z, Tang H. ISEScan: Automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics. 2017;33:3340–3347. doi: 10.1093/bioinformatics/btx433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.