Abstract
Background
Non‐small cell lung cancer (NSCLC) is one of the most malignant cancers worldwide and its pathogenesis is not completely clear. In this study, we explored the functions and mechanisms of exosomes transferring miR‐3180‐3p in NSCLC progression.
Methods
The expression levels of miR‐3180‐3p in NSCLC tissues and paracarcinoma tissues was obtained from the GEO database (GEO: GSE53882). Exosomes derived from A549 cells were identified. Proliferation, migration and invasion were measured after treatment with exosomal miR‐3180‐3p or transfection using miR‐3180‐3p mimics. The relationship between miR‐3180‐3p and forkhead box P4 (FOXP4) was predicted using a bioinformatic tool and measured using a dual‐luciferase reporter gene assay and western blotting. Finally, a mouse xenograft model of NSCLC cells was established to verify the function of exosomal miR‐3180‐3p in vivo.
Results
We found that miR‐3180‐3p decreased in both NSCLC cell lines and patient tissues. Overexpression of miR‐3180‐3p or treatment with exosomal miR‐3180‐3p significantly suppressed cell proliferation and metastasis in NSCLC cell lines. Subsequently, we found miR‐3180‐3p downregulated FOXP4 protein expression levels. Furthermore, the volumes and weights of nude mouse tumors expressing exosomal miR‐3180‐3p were significantly reduced.
Conclusions
Exosomal miR‐3180‐3p suppresses NSCLC progression by downregulating FOXP4 expression.
Key points
Significant findings of the study
We found that exosomal miR‐3180‐3p suppressed NSCLC progression and also identified a miR‐3180‐3p target gene. These findings provide a foundation to determine innovative therapeutic strategies.
What this study adds
This study contributes to research investigating exosomal containing miRNAs.
Keywords: Exosomes, FOXP4, miR‐3180‐3p, non‐small cell lung cancer
We found that exosomal miR‐3180‐3p suppressed NSCLC progression and also identified a miR‐3180‐3p target gene.
Introduction
Lung cancer is one of the most common tumors with the highest mortality rate. Non‐small cell lung cancer (NSCLC) accounts for 85% of all lung cancer cases. 1 Despite progress in developing treatments for NSCLC, the five‐year survival rate is still less than 15%. 2 , 3 It is a priority to better understand the mechanisms behind NSCLC pathogenesis and find potential prognostic biomarkers for this disease.
MicroRNAs (miRNAs) are single‐stranded non‐coding RNAs that are 19–24 nt in length and have the ability to regulate gene expression at the post‐transcriptional level by targeting the 3′‐untraslated region (3′‐UTR) of a mRNA. 4 Aberrant expression of miRNAs is closely related to the tumor microenvironment. 5 MiR‐421 was reported to contribute to NSCLC cell migration, invasion and inhibit cell apoptosis by mediating the β‐catenin‐miR‐421‐KEAP1 axis. 6 MiR‐330‐3p promotes brain metastasis and epithelial‐mesenchymal transition (EMT) by mediating the GRIA3‐TGF‐β1 interaction. 7
Exosomes are extracellular vesicles containing a lipid bilayer‐enclosed structure that are smaller than 150 nm. They contain proteins and nucleic acids that can be released from many cell types. 8 , 9 Exosomes often contain RNA molecules or proteins, which provide a mechanism for these bioactive compounds to transfer into neighboring cells, thus mediating intercellular communication. 10 , 11
RNAs concentrated in exosomes include miRNAs, mRNAs, long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs). 12 , 13 , 14 Increasing evidence has shown that miRNAs enriched in exosomes can be shuttled to neighboring cells and regulate their biological functions. 15 , 16 Exosomes derived from tumor cells have been found to be associated with tumor angiogenesis, invasion and chemoresistance. 17 , 18 , 19 For example, human mesenchymal stem cell (MSC) derived exosomes enriched miR‐143 transfer to prostate cancer cells and inhibited PC3 cell proliferation and metastasis by suppressing TFF3 protein expression. 20 Exosomes with low miR‐34c‐3p expression levels promote migration and invasion of NSCLC by upregulating integrin α2β1. 21 With increasing attention on the function of miRNAs enriched in exosomes, we focused on the mechanisms behind miR‐3180‐3p in exosomes in this study.
Methods
Cell culture
All NSCLC cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China). Cell lines were cultured in RPMI‐1640 (Sigma) supplemented with 10% FBS (Thermo Fisher Scientific), 100 units/mL penicillin and 100 μg/mL streptomycin at 37°C in 5% CO2. To remove exosomes, FBS used in cell culture was ultracentrifuged at 100 000 g for 10 hours.
Isolation and characterization of exosomes
After culturing cells for 48 hours in exosome‐free FBS, medium was collected and filtrated using a 0.22 μm filter. Exosomes were obtained using ultracentrifugation‐based isolation techniques. 22 Briefly, the medium was centrifuged at 300 × g for 10 minutes, 2000 × g for 15 minutes, 5000 × g at 15 minutes and 12 000 × g for 30 minutes, then the supernatant was ultracentrifuged at 100 000 × g for 70 minutes. The supernatant was discarded and PBS used to wash the exosomes. Exosomes were pelleted using ultracentrifugation at 100 000 × g for 70 minutes. All ultracentrifugation steps were performed at 4°C in a Beckman ultracentrifuge (TL‐100). Exosomes were assessed using nanoparticle tracking analysis (NTA; ZetaView, Particle Metrix, DE), western blotting and transmission electron microscopy (TEM; JEM‐1400, JEOL, Japan).
For exosome‐tracking experiments, exosomes from A549 cells were labeled using PKH67 membrane dye (Sigma‐Aldrich) and labeled exosomes were washed twice with medium that did not include FBS. A549 cells were seeded into 24‐well chambers and labeled exosomes were added and incubated for three hours. Cells were fixed using 4% paraformaldehyde for 10 minutes. Cells were then washed three times with PBS and blocked with 0.1% Triton‐X 100 for one hour, then incubated with an actin antibody at 4°C overnight. The next day, cells were stained using 20 μg/mL Alexa Fluor 594 IgG (H + L) (Life Technologies, Grand Island, NY) for 10 minutes at room temperature. Cellular nuclei were stained using DAPI. Merged images were captured using an electron microscope (LSM 710, Carl Zeiss).
Dual‐luciferase reporter assay
The FOXP4 3′UTR nucleotide sequence predicted miR‐3180‐3p target sites and mutation sequences were synthesized (Genewiz, Beijing, China), digested with XhoI and NotI and cloned into the psiCHECK‐2 dual luciferase vector (Promega, Madison, WI, USA) to generate the FOXP4‐3′UTR wt and FOXP4‐3′UTR mut. Subsequently, NSCLC cells were plated in a 24‐well plate and co‐transfected with 50 ng FOXP4‐3′UTR wt or FOXP4‐3′UTR mut and 20 nM of miR‐3180‐3p mimics (5′‐UGGGGCGGAGCUUCCGGAG‐3′) or miR‐NC (5′‐UUCUCCGAACGUGUCACGUTT‐3′). Approximately 48 hours after transfection, cells were collected and luciferase activities were measured using a Dual‐Luciferase Reporter assay kit (Promega) on a TD‐20/20 Luminometer (Turner Designs, Sunnyvale, CA, USA).
Western blotting
Cells or exosome lysates were collected using RIPA lysis buffer (Beyotime, Shanghai, China) and separated by 10% SDS‐PAGE. Proteins were transferred onto PVDF membranes and blocked with 5% solution of nonfat milk for one hour. Membranes were treated with primary antibodies recognizing FOXP4 (ab251688, Abcam) and GAPDH (#5174, CST) overnight at 4°C. Subsequently, membranes were incubated with a secondary antibody for 1 hour at room temperature. Protein bands were detected using ECL substrate (Tanon Science and Technology, Shanghai, China). GAPDH was used as an endogenous loading control.
qRT‐PCR
Total RNA was extracted using TRIzol (Invitrogen, CA, USA) according to the manufacturer's instructions. cDNA synthesis with reverse transcriptase (RT) was performed using a M‐MLV First Strand kit (Life Technologies). A MiRNA First‐Strand cDNA Synthesis Kit (Thermo Fisher Scientific) was used for miRNA reverse transcriptase. The miScript SYBR Green PCR Kit (Qiagen) was used for RT‐PCR reactions. Relative expression levels of miRNAs or mRNAs were calculated using the ∆∆Ct method. GAPDH and U6 small nuclear RNA (U6 snRNA) were used for normalization of mRNA and miRNA. Primer sequences are presented in Table 1.
Table 1.
Primer sequences used in qRT‐PCR
| Gene | Primer sequence (5′–3′) |
|---|---|
| miR‐3180‐3p | F: GCGTGGGGCGGAGCTT |
| R: AGTGCAGGGTCCGAGGTATT | |
| U6 | F: AGAGCCTGTGGTGTCCG |
| R: CATCTTCAAAGCACTTCCCT | |
| GAPDH | F: CCTTCCGTGTCCCCACT |
| R: GCCTGCTTCACCACCTTC | |
| FOXP4 | F: AGGACACGGAGGGTTCAG |
| R: TGTGGGGAGAGCTGGTG |
F, forward; FOXP4, forkhead box P4; miR‐3180‐3p, microRNA‐3180‐3p; R, reverse.
Cell proliferation and transwell assay
A transwell assay was performed using 24‐well chambers containing membrane filter inserts. Then, 48 hours after transfection, A549 and H460 cell lines were collected and resuspended in serum‐free RPMI1640 medium at a cell density of 1 × 105 cells/mL. Chambers were used for migration assays and chambers coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were used for invasion assays. A total of 200 μL cell suspension was added to the upper chambers and 600 μL of RPMI1640 medium with 20% FBS was added to the lower chambers. After 24 hours, the chambers were fixed with 4% paraformaldehyde for 30 minutes, stained with 0.5% crystal violet solution for 10 minutes and washed three times with PBS. Cells in the upper chambers were removed using a cotton swab. Transmembrane cells were counted in three random fields. The experiment was repeated three times.
Tumor xenograft and tail vein injection
A total of 24 nude mice that were four weeks old were divided into two groups where five mice in each group were randomly chosen for tumor growth observation. A total of 1 × 107 A549 cells were resuspended with 50% Matrigel and 0.2 mL of cells were subcutaneously injected into mice. To observe the effects of exosomes on tumor growth in vivo, 100 μg of exosomes from transfected A549 cells also transfected with miR‐3180‐3p mimics or a negative control in 100 μL PBS were injected into mice by the tail vein every three days based on previous reports. 23 Tumor size was measured every seven days. After five weeks all nude mice were sacrificed and tumors were removed and weighed. This study was approved and conducted by the Institutional Animal Care and Use Committee of the Soochow University.
Statistical analysis
Significance tests and correlation analyses were performed using GraphPad Prism 5.0 (GraphPad Software). The Student's t‐test was used in statistical analysis. A P‐value less than 0.05 was considered statistically significant.
Results
miR‐3180‐3p was expressed at low levels in NSCLC patients
NSCLC expression profile data was extracted from the GEO database (GEO: GSE53882). We found that the miR‐3180‐3p was significantly lower than that compared with paired para‐carcinoma tissues (Fig 1a). Furthermore, we detected miR‐3180‐3p expression in NSCLC cell lines and found that miR‐3180‐3p levels were significantly lower in A549 and H460 cell lines compared to the normal cell lines HBE and BEAS‐2B (Fig 1b).
Figure 1.
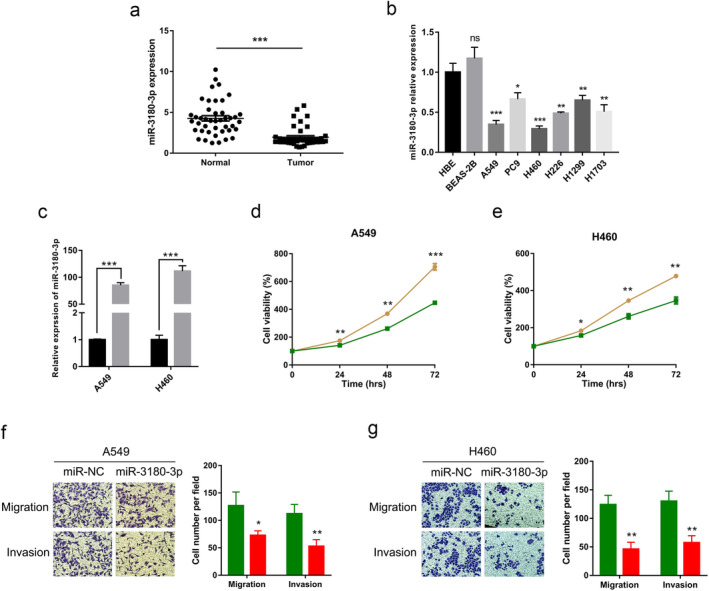
Overexpression of miR‐3180‐3p inhibits proliferation, migration and invasion of in NSCLC cells. (a) The expression of miR‐3180‐3p in NSCLC and paracarcinoma tissues obtained from the GEO database (GEO: GSE53882). (b) The expression of miR‐3180‐3p in six NSCLC cell lines and two normal cell lines. (c) The expression of miR‐3180‐3p after overexpression miR‐3180‐3p in A549 and H460 cell lines  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. (d, e) A CCK‐8 assay was performed in A549 and H460 cells transfected with miR‐3180‐3p or miR‐NC. A549:
, miR‐3180‐3p. (d, e) A CCK‐8 assay was performed in A549 and H460 cells transfected with miR‐3180‐3p or miR‐NC. A549:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. H460:
, miR‐3180‐3p. H460:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. (f, g) The migratory and invasive abilities of A549 and H460 cells transfected with miR‐3180‐3p and miR‐NC was measured using a transwell assay. A549:
, miR‐3180‐3p. (f, g) The migratory and invasive abilities of A549 and H460 cells transfected with miR‐3180‐3p and miR‐NC was measured using a transwell assay. A549:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. H460:
, miR‐3180‐3p. H460:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. *P < 0.05, **P < 0.01, ***P < 0.001, and n ≥ 3.
, miR‐3180‐3p. *P < 0.05, **P < 0.01, ***P < 0.001, and n ≥ 3.
Overexpression of miR‐3180‐3p suppresses proliferation, migration, and invasion of NSCLC cells
To explore the functions of miR‐3180‐3p in NSCLC cells, A549 and H460 cell lines were transfected with miR‐NC and miR‐3180‐3p mimics. The proliferation, migration and invasion of NSCLC cells were examined. First, the transfection efficiency was confirmed using qRT‐PCR. Results revealed that the expression of miR‐3180‐3p was strikingly increased (Fig 1c). Compared with the control group, the proliferation, migration and invasion abilities were significantly decreased in the miR‐3180‐3p mimic group (P < 0.05) (Fig 1d–g).
FOXP4 is a target gene of miR‐3180‐3p
To determine miR‐3180‐3p target genes, we used TargetScan software, which revealed FOXP4 as a possible target gene (Fig 2a). To validate its accuracy, a dual luciferase assay was performed. A549 and H460 cells were cotransfected with miR‐NC or miR‐3180‐3p mimics and FOXP4‐3′UTR wild‐type (WT) or FOXP4‐3′UTR mutant (MUT) luciferase reporter vectors. As shown in Fig 2b,c, compared with the control group, miR‐3180‐3p mimics significantly decreased luciferase activity in the WT group (P < 0.05), but had no significant effects on the MUT group (P > 0.05).
Figure 2.
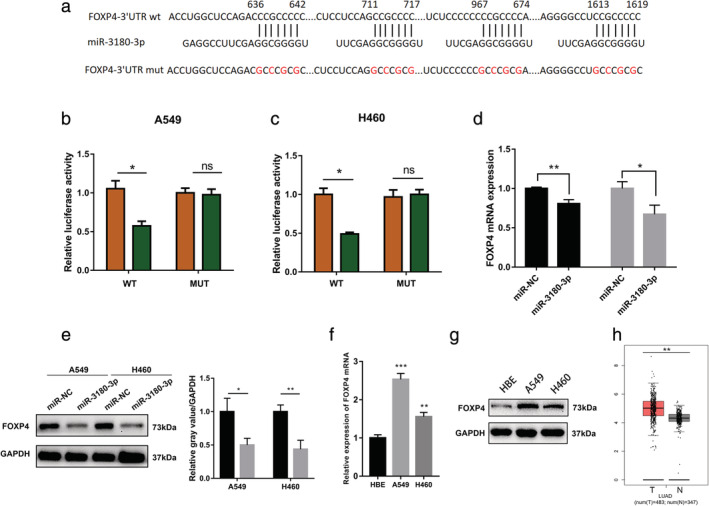
Identification of FOXP4 as a miR‐3180‐3p target gene. (a) The wild‐type and mutated binding site between miR‐3180‐3p and FOXP4. (b, c) A549 and H460 cell lines were cotransfected with miR‐NC and miR‐3180‐3p with a WT and MUT 3′UTR. Luciferase activity was measured after 24 hours cotransfection. A549:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. H460:
, miR‐3180‐3p. H460:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. (d, e) A549 and H460 cells were transfected with miR‐NC or miR‐3180‐3p mimics for 48 hours. The mRNA and protein levels of FOXP4 were analyzed using qRT‐PCR and western blotting
, miR‐3180‐3p. (d, e) A549 and H460 cells were transfected with miR‐NC or miR‐3180‐3p mimics for 48 hours. The mRNA and protein levels of FOXP4 were analyzed using qRT‐PCR and western blotting  , A549;
, A549;  , H460;
, H460;  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p. (f, g) The mRNA and protein levels of FOXP4 were analyzed using qRT‐PCR and western blotting in HBE, A549 and H460 cells. (h) FOXP4 mRNA expression levels were extracted from The Cancer Genome Atlas (TCGA). *P < 0.05, **P < 0.01, and n ≥ 3.
, miR‐3180‐3p. (f, g) The mRNA and protein levels of FOXP4 were analyzed using qRT‐PCR and western blotting in HBE, A549 and H460 cells. (h) FOXP4 mRNA expression levels were extracted from The Cancer Genome Atlas (TCGA). *P < 0.05, **P < 0.01, and n ≥ 3.
To further demonstrate FOXP4 as a target gene of miR‐3180‐3p, we detected FOXP4 mRNA and protein expression after transfecting miR‐3180‐3p mimics. The results showed that both FOXP4 mRNA and protein expression were significantly decreased (Fig 2d,e). The FOXP4 mRNA and protein levels were higher in A549 and H460 cells compared with HBE cells (Fig 2f,g). The FOXP4 mRNA expression was verified using the TCGA database (Fig 2h). These results suggested that FOXP4 as the target gene of miR‐3180‐3p.
FOXP4 knockdown and miR‐3180‐3p upregulation show the same phenotypes in NSCLC cells
To explore the effects of FOXP4 on NSCLC, FOXP4‐siRNAs were used to suppress FOXP4 in A549 and H460 cells. The protein expression levels of FOXP4 were significantly reduced in the two siRNA groups compared with the control group (Fig 3a). To further clarify the effects of FOXP4‐siRNAs on cancer cell proliferation and metastasis, CCK‐8 and transwell assays were used to determine the effects of FOXP4 on NSCLC cells. The CCK‐8 assay revealed that the growth rate of A549 and H460 cells transfected with siRNAs were significantly lower than control group cells (Fig 3b). Transwell assay revealed that FOXP4 knockdown remarkably inhibited the migration and invasion of NSCLC cells (Fig 3c,d).
Figure 3.
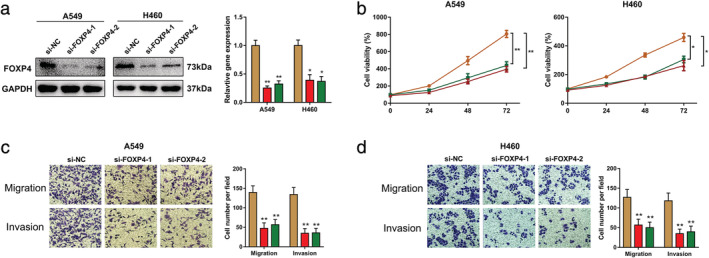
Knockdown of FOXP4 inhibits proliferation, migration and invasion in NSCLC cells. (a) The protein levels of FOXP4 in A549 and H460 cells were analyzed by western blotting after transfection with siRNA targeting FOXP4  , si‐NC;
, si‐NC;  , si‐FOXP4‐1;
, si‐FOXP4‐1;  , si‐FOXP4‐2. (b) A CCK‐8 assay was performed in A549 and H460 cells after suppression of FOXP4 using siRNAs. A549:
, si‐FOXP4‐2. (b) A CCK‐8 assay was performed in A549 and H460 cells after suppression of FOXP4 using siRNAs. A549:  , si‐NC;
, si‐NC;  , si‐FOXP4‐1;
, si‐FOXP4‐1;  , si‐FOXP4‐2. H460:
, si‐FOXP4‐2. H460:  , si‐NC;
, si‐NC;  , si‐FOXP4‐1;
, si‐FOXP4‐1;  , si‐FOXP4‐2. (c, d) The migratory and invasive ability of A549 and H460 cells were measured using a transwell assay after knockdown of FOXP4 by siRNAs. A549:
, si‐FOXP4‐2. (c, d) The migratory and invasive ability of A549 and H460 cells were measured using a transwell assay after knockdown of FOXP4 by siRNAs. A549:  , si‐NC;
, si‐NC;  , si‐FOXP4‐1;
, si‐FOXP4‐1;  , si‐FOXP4‐2. H460:
, si‐FOXP4‐2. H460:  , si‐NC;
, si‐NC;  , si‐FOXP4‐1;
, si‐FOXP4‐1;  , si‐FOXP4‐2. *P < 0.05, **P < 0.01, and n ≥ 3.
, si‐FOXP4‐2. *P < 0.05, **P < 0.01, and n ≥ 3.
Overexpression of FOXP4 reverses the effects of miR‐3180‐3p on NSCLC cells
Since our data suggested that miR‐3180‐3p inhibited cell growth and metastasis by downregulating FOXP4, we explored whether FOXP4 is a direct functional mediator of the inhibitory effects of miR‐3180‐3p. As expected, we observed that exogenous miR‐3180‐3p significantly suppressed the expression of FOXP4 (Fig 4a,b). CCK‐8 assays showed that miR‐3180‐3p suppressed the proliferation of NSCLC cells and that this suppression could be blocked by overexpressing FOXP4 (Fig 4c,d). Furthermore, transwell migration and invasion assays showed that miR‐3180‐3p inhibited metastasis of NSCLC cells and that inhibition could be blocked by increased FOXP4 (Fig 4e,f). Collectively, these observations suggested that miR‐3180‐3p inhibited the growth and metastasis of NSCLC, at least partially, through the miR‐3180‐3p‐FOXP4 pathway.
Figure 4.
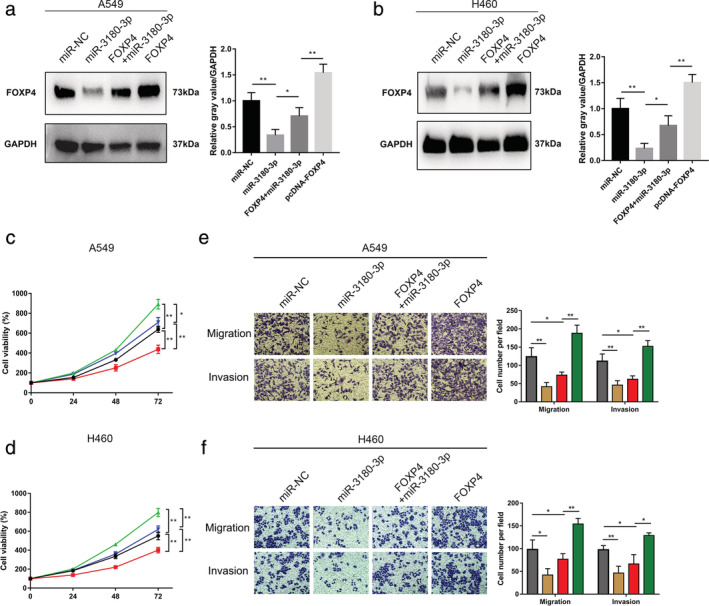
Partial reversal of the inhibitory effects of miR‐3180‐3p on NSCLC cell phenotypes through FOXP4. A549 and H460 cells were transfected with miR‐NC, miR‐3180‐3p, miR‐3180‐3p + FOXP4 or FOXP4. (a, b) Western blots were performed to detect FOXP4 protein expression levels. (c, d) A CCK‐8 assay was performed. A549:  , NC;
, NC;  , miR‐3180‐3p;
, miR‐3180‐3p;  , FOXP4;
, FOXP4;  , FOXP4+miR‐3180‐3p. H460:
, FOXP4+miR‐3180‐3p. H460:  , NC;
, NC;  , miR‐3180‐3p;
, miR‐3180‐3p;  , FOXP4;
, FOXP4;  , FOXP4+miR‐3180‐3p. (e, f) The migratory and invasive ability of A549 and H460 cells were measured using a transwell assay. A549:
, FOXP4+miR‐3180‐3p. (e, f) The migratory and invasive ability of A549 and H460 cells were measured using a transwell assay. A549:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p;
, miR‐3180‐3p;  , FOXP4+miR‐3180‐3p;
, FOXP4+miR‐3180‐3p;  , FOXP4. H460:
, FOXP4. H460:  , miR‐NC;
, miR‐NC;  , miR‐3180‐3p;
, miR‐3180‐3p;  , FOXP4+miR‐3180‐3p;
, FOXP4+miR‐3180‐3p;  , FOXP4. *P < 0.05, **P < 0.01, and n ≥ 3.
, FOXP4. *P < 0.05, **P < 0.01, and n ≥ 3.
Exosomal miR‐3180‐3p inhibits proliferation, migration, and invasion of NSCLC cells
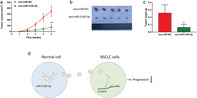
It is known that exosomes play an important role in cell‐to‐cell communication and that they can change the physiological functions of recipient cells through biological active factors. 13 From Exocarta we know that miR‐3180‐3p exists in exosomes. To further verify that miR‐3180‐3p was enriched in exosomes, we extracted exosomes after transfecting miR‐3180‐3p mimics into A549 cells. The canonical morphology of exosomes was captured using TEM which confirmed the presence of extracted exosomes (Fig 5a). The size of these exosomes was measured using particle size distribution with approximately 30–100 nm diameter (Fig 5b). The expression of exosome markers such as Annexin V, Flotillin‐1 and CD63 were examined using a western blot, which further confirmed successful isolation of exosomes from supernatants obtained from NSCLC cells (Fig 5c).
Figure 5.
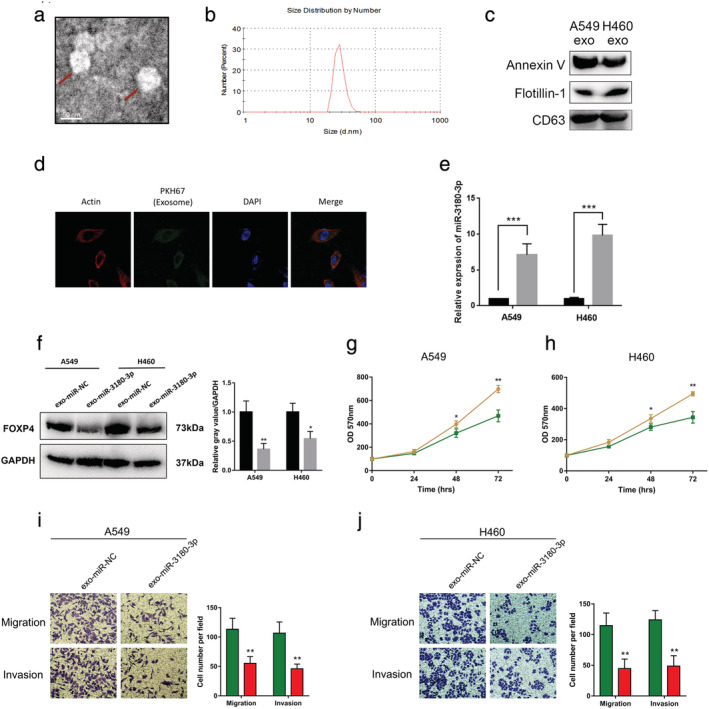
Exosomal miR‐3180‐3p derived from A549 and H460 cells suppresses proliferation and metastasis of NSCLC cells. (a) Observation of the morphology of exosomes under TEM (scale bar, 50 nm). (b) Size distribution of exosomes. (c) Expression of exosomal markers Annexin V, Flotillin‐1 and CD63. (d) A representative immunofluorescence image shows the internalization of PKH67‐labeled A549‐derived exosomes (green) by macrophages. Blue represents nuclear DNA staining by DAPI. Red represents cytoplasm incubation with a β‐Actin antibody. (e) MiR‐3180‐3p levels in exo‐A549 and exo‐H460 were measured using RT‐PCR  , miR‐NC exo;
, miR‐NC exo;  , miR‐3180‐3p exo. (f) Western blots were performed
, miR‐3180‐3p exo. (f) Western blots were performed  , exo‐miR‐NC;
, exo‐miR‐NC;  , exo‐miR‐3180‐3p. (g, h) A CCK‐8 assay was performed. A549:
, exo‐miR‐3180‐3p. (g, h) A CCK‐8 assay was performed. A549:  , exo‐miR‐NC;
, exo‐miR‐NC;  , exo‐miR‐3180‐3p. H460:
, exo‐miR‐3180‐3p. H460:  , exo‐miR‐NC;
, exo‐miR‐NC;  , exo‐miR‐3180‐3p. (i, j) The migratory and invasive ability of A549 and H460 cells were measured using transwell assay. A549:
, exo‐miR‐3180‐3p. (i, j) The migratory and invasive ability of A549 and H460 cells were measured using transwell assay. A549:  , miR‐NC exo;
, miR‐NC exo;  , miR‐3180‐3p exo. H460:
, miR‐3180‐3p exo. H460:  , miR‐NC exo;
, miR‐NC exo;  , miR‐3180‐3p exo. *P < 0.05, **P < 0.01, ***P < 0.001 and n ≥ 3.
, miR‐3180‐3p exo. *P < 0.05, **P < 0.01, ***P < 0.001 and n ≥ 3.
We next sought to determine whether purified exosomes could enter cells and perform functions. Exosomes derived from A549 cells were labeled with a green fluorescent marker, PKH67. The results showed that PKH67 was localized in the cytoplasm of recipient cells (Fig 5d). QPCR determined the effective functions of exosomal miR‐3180‐3p. The results showed that the expression of miR‐3180‐3p was significantly increased in recipient NSCLC cells treated with Exo‐miR‐3180‐3p (Fig 5e).
Following the observation that miR‐3180‐3p is enriched in exosomes, we speculated that miR‐3180‐3p shuttled by exosomes could inhibit proliferation, migration and invasion of NSCLC cells. To access the role of miR‐3180‐3p in exosomes, NSCLC cells were treated with Exo‐miR‐NC or Exo‐miR‐3180‐3p. The results showed that FOXP4 protein levels in A549 and H460 cells exposed to miR‐3180‐3p exosomes were significantly decreased compared to miR‐NC exosomes (Fig 5f). In addition, proliferation was inhibited by miR‐3180‐3p exosomes (Fig 5g,h) and the migratory and invasive abilities showed similar findings (Fig 5i,j).
Effects of exosomal miR‐3180‐3p on tumors in vivo
To further explore the role of exosomal miR‐3180‐3p in NSCLC in vivo, A549 cells were subcutaneously injected into nude mice. Exosomes extracted from A549 cells transfected with miR‐3180‐3p or miR‐NC mimics were injected into mice through the tail vein. In line with our analysis in vitro, tumor tissues of nude mice injected with miR‐3180‐3p exosomes attenuated tumor size and weight (Fig 6a–c).
Figure 6.
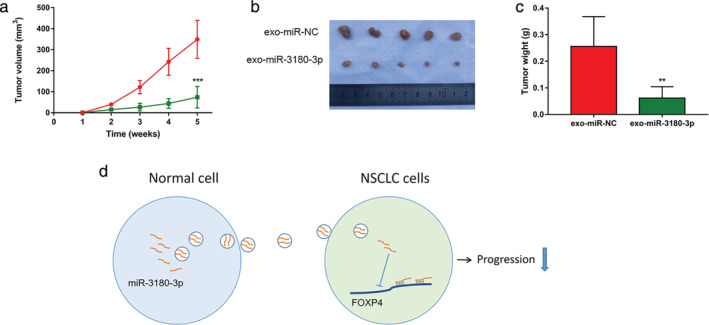
Exosomes enriched with miR‐3180‐3p suppress tumor growth in vivo. (a) Statistical analysis of tumor volume growth curves in mice  , exo‐miR‐NC;
, exo‐miR‐NC;  , exo‐miR‐3180‐3p. (b, c) Photographs of tumor and curves of tumor weight. (d) Schematic model of exosomal miR‐3180‐3p showing inhibition of NSCLC progression. **P < 0.01, ***P < 0.001 and n > 3.
, exo‐miR‐3180‐3p. (b, c) Photographs of tumor and curves of tumor weight. (d) Schematic model of exosomal miR‐3180‐3p showing inhibition of NSCLC progression. **P < 0.01, ***P < 0.001 and n > 3.
Discussion
With the rapid development of technology, many diseases can be treated or even cured in the clinic. However, there is no effective treatment for lung cancer. Many studies have reported that miRNAs play important roles in cancers. There is considerable evidence indicates that miRNAs are involved in the progression of NSCLC. This evidence suggests that miRNAs are a promising approach to treat cancer. However, the mechanisms behind miR‐3180‐3p in the regulation of NSCLC remains unclear.
In this study, we investigated the role of miR‐3180‐3p in NSCLC. We found that miR‐3180‐3p levels were downregulated in both NSCLC cell lines and human samples. An upregulation of miR‐3180‐3p suppressed NSCLC cell proliferation and metastasis. We further determined that the tumor oncogene FOXP4 was a direct target of miR‐3180‐3p.
FOXP4 belongs to the human forkhead‐box (FOX) gene family. Forkhead proteins play key roles in cell cycle regulation and oncogenesis. Yin et al. reported that microRNA‐491‐5p suppresses osteosarcoma cell proliferation and promotes apoptosis by targeting FOXP4. 24 Restoration of FOXP4 expression significantly promotes breast cancer cell proliferation, migration and invasion. 25 Yang et al. found the mRNA and protein levels of FOXP4 were higher in NSCLC tissues than in normal tissues. 26 We found that downregulated FOXP4 prominently suppressed NSCLC cell proliferation, migration and invasion.
Exosomes are secreted by cells, circulate in the blood and induce a series of cellular reactions when being captured by recipient cells. Recent studies have suggested that tumor‐derived exosomes may play an important role in tumor progression. 27 , 28 , 29 HCC cells release exosomes containing both SMAD3 protein and mRNA and these exosomes enhance recipient HCC cell SMAD3 signaling and increase their adhesive ability. 30 Wei et al. Showed that miRNAs within exosomes play important roles in cancer. For example, exosomal miR‐222–3p promotes proliferation, gemcitabine resistance, migration and invasion of NSCLC cells by targeting the promoter of SOCS3. 31 We isolated exosomes derived from the two NSCLC cell lines A549 and H460 by overexpressing miR‐3180‐3p. We found that miR‐3180‐3p can be enriched in exosomes. The overexpression of exosomal miR‐3180‐3p and silencing of FOXP4 both suppressed NSCLC cell proliferation, migration and invasion.
In summary, to the best of our knowledge, this is the first time that exosomal miR‐3180‐3p has been demonstrated to inhibit NSCLC proliferation and metastasis by targeting FOXP4 (Fig 6d). These results revealed a new regulatory network involving miR‐3180‐3p and FOXP4, which may be applicable to the therapeutic treatment of NSCLC in the future.
In conclusion, in this study we demonstrated that miR‐3180‐3p carried by exosomes could suppress NSCLC cell proliferation and metastasis by downregulating FOXP4. These findings may provide a foundation for innovative therapeutic strategies.
Disclosure
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81672934 and 81873417).
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: From targets to treatments. Cancer Treat Rev 2015; 41: 361–75. [DOI] [PubMed] [Google Scholar]
- 3. Wu X, Ruan L, Yang Y, Mei Q. Analysis of gene expression changes associated with human carcinoma‐associated fibroblasts in non‐small cell lung carcinoma. Biol Res 2017; 50: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bayraktar R, Van Roosbroeck K, Calin GA. Cell‐to‐cell communication: microRNAs as hormones. Mol Oncol 2017; 11: 1673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rupaimoole R, Calin GA, Lopez‐Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov 2016; 6: 235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duan FG, Wang MF, Cao YB et al MicroRNA‐421 confers paclitaxel resistance by binding to the KEAP1 3'UTR and predicts poor survival in non‐small cell lung cancer. Cell Death Dis 2019; 10: 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei C, Zhang R, Cai Q et al MicroRNA‐330‐3p promotes brain metastasis and epithelial‐mesenchymal transition via GRIA3 in non‐small cell lung cancer. Aging 2019; 11: 6734–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med 2013; 91: 431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest 2016; 126: 1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol 2013; 200: 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Silva N, Samblas M, Martinez JA, Milagro FI. Effects of exosomes from LPS‐activated macrophages on adipocyte gene expression, differentiation, and insulin‐dependent glucose uptake. J Physiol Biochem 2018; 74: 559–68. [DOI] [PubMed] [Google Scholar]
- 12. Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion‐infected neuronal cells. Nucleic Acids Res 2012; 40: 10937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang X, Yuan T, Tschannen M et al Characterization of human plasma‐derived exosomal RNAs by deep sequencing. BMC Genomics 2013; 14: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y, Zheng Q, Bao C et al Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res 2015; 25: 981–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–9. [DOI] [PubMed] [Google Scholar]
- 16. Cha DJ, Franklin JL, Dou Y et al KRAS‐dependent sorting of miRNA to exosomes. Elife 2015; 4: e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of Exosomal circRNAs in cancer. Mol Ther Nucl Acids 2020; 21: 367–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR‐135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor‐inhibiting HIF‐1. Blood 2014; 124: 3748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramteke A, Ting H, Agarwal C et al Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol Carcinog 2015; 54: 554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Che Y, Shi X, Shi Y et al Exosomes derived from miR‐143‐overexpressing MSCs inhibit cell migration and invasion in human prostate cancer by downregulating TFF3. Mol Ther Nucl Acids 2019; 18: 232–44. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Huang W, Yan Y, Liu Y et al Exosomes with low miR‐34c‐3p expression promote invasion and migration of non‐small cell lung cancer by upregulating integrin alpha2beta1. Signal Transduct Target Ther 2020; 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics 2017; 7: 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen G, Huang AC, Zhang W et al Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature 2018; 560: 382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin Z, Ding H, He E, Chen J, Li M. Up‐regulation of microRNA‐491‐5p suppresses cell proliferation and promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell Prolif 2017; 50 (1): e12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang N, Gu Y, Li L et al Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR‐4316. Arch Biochem Biophys 2018; 653: 63–70. [DOI] [PubMed] [Google Scholar]
- 26. Yang T, Li H, Thakur A et al FOXP4 modulates tumor growth and independently associates with miR‐138 in non‐small cell lung cancer cells. Tumor Biol 2015; 36 (10): 8185–91. [DOI] [PubMed] [Google Scholar]
- 27. Zeng Z, Li Y, Pan Y et al Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 2018; 9: 5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo D, Lui GYL, Lai SL et al RAB27A promotes melanoma cell invasion and metastasis via regulation of pro‐invasive exosomes. Int J Cancer 2019; 144: 3070–85. [DOI] [PubMed] [Google Scholar]
- 29. Faict S, Muller J, De Veirman K et al Exosomes play a role in multiple myeloma bone disease and tumor development by targeting osteoclasts and osteoblasts. Blood Cancer J 2018; 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu Q, Zhang Q, Lou Y et al Primary tumor‐derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene 2018; 37: 6105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei F, Ma C, Zhou T et al Exosomes derived from gemcitabine‐resistant cells transfer malignant phenotypic traits via delivery of miRNA‐222‐3p. Mol Cancer 2017; 16: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]


