Abstract
Background
Previous studies have demonstrated that various circular RNAs are involved in the malignant proliferation of cancers, such as liver cancer, lung cancer, breast cancer, and others. The potential role of circular RNAs in glioblastoma, however, is still uncertain. In this study, we aimed to study the potential role of hsa_circ_01844 in glioblastoma.
Methods
Using reverse transcription-polymerase chain reaction (RT-PCR) method, hsa_circ_01844 expression was measured in five glioblastoma samples and five normal brain samples. To evaluate the potential function of hsa_circ_01844 in glioblastoma, hsa_circ_01844 was overexpressed in glioblastoma cell lines (U251 and U87 cells). Using these two cell lines, in vitro experiments including the flow cytometry assay, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay, Transwell assay, and cell apoptosis assay were performed to investigate the role of hsa_circ_01844 in glioblastoma. Student t test and one-way analysis of variance were used for statistical analysis.
Results
The expression of circular RNA hsa_circ_01844 was lower in glioblastoma tissues when compared with the normal brain tissues by RT-PCR method (0.034 ± 0.036 vs. 1.630 ± 0.891, P < 0.001). Using two glioblastoma cell lines, we found that overexpression of hsa_circ_01844 in glioblastoma cells suppressed their proliferation, colony formation, migration, and increased the apoptotic rate compared with empty vector group and blank control group (all P < 0.05).
Conclusion
Hsa_circ_01844 shows decreased expression in glioblastoma and its overexpression induces apoptosis and inhibits proliferation, migration, and invasion of glioblastoma cells.
Keywords: Glioblastoma, Hsa_circ_01844, Apoptosis, Proliferation, Migration
Introduction
Glioma is the most common primary malignancy of the central nervous system. As the most biologically aggressive subtype, glioblastoma accounts for more than 80% of malignant glioma. Glioblastoma patients have a relatively short median survival of 14 to 17 months after surgery and chemoradiotherapy.[1–3] Only 3% to 5% of patients can survive more than 3 years.[4,5] The aggressive nature of glioblastoma lies in the diversity and complexity of its pathogenesis, which is also the main reason for its poor therapeutic effect.[6–8] Exploring the mechanisms of malignant proliferation in glioblastoma cells and how to inhibit their growth is a hot topic in the field of glioma research.
Circular RNAs (circRNAs) are a novel type of RNA molecule formed by a covalently closed loop. Many studies have indicated that circRNAs play key roles in various physiological and pathological processes.[9] Recent studies have demonstrated that circRNAs are expressed at variable levels in solid tumors such as esophageal cancer, liver cancer, colon cancer, and others.[10,11] Similar to long non-coding RNAs, circRNAs can also function as a microRNAs (miRNAs) sponge and interact with related miRNAs. For example, studies have shown that there were 70 miRNA-7 (miR-7) receptor binding sites on circRNA-7. The binding of miR-7 to circRNA-7 inhibits the expression of downstream targets of miR-7 and sequentially promotes the malignant proliferation of cervical cancer.[16] Some novel circRNAs have been identified to promote or inhibit the proliferation, migration, and invasion of glioma cells.[12–15] Over-expression of has_circ_067934 promotes proliferation and metastasis of malignant gliomas cells by up-regulating phosphatidylinositol 3 kinase/protein kinase B (PI3K-AKT) pathway.[17] Exploring the mechanisms of circRNAs in the etiology and pathogenesis of glioblastoma may provide new ideas for the treatment of this disease.[18]
A previous study using circRNA microarray analysis showed that a novel circRNA, hsa_circ_01844, had the lowest expression in glioblastoma tissues and its expression level was significantly different between glioblastoma and normal brain tissues.[19] In this study, we aimed to study the potential role of hsa_circ_01844 in glioblastoma.
Methods
Ethical approval and acquisition of tissue samples
This study was approved by the Ethics Committee of the 904th Hospital of Joint Logistic Support Force PLA (Wuxi Clinical College of Anhui Medical University) (LLSO20200846). All participants signed informed consent.
From May 2019 to November 2019, 5 patients with primary glioblastoma who received surgical treatment in the 904th Hospital of Joint Logistic Support Force PLA were enrolled for this study. After the glioblastoma tissue was removed from the patients, the specimens were frozen in liquid nitrogen and then stored in the refrigerator at −80°C immediately. Five normal brain tissues, which were taken from trauma patients in the 904th Hospital of Joint Logistic Support Force PLA were used as the control. In these control patients, partial resection of the normal brain was required as decompression treatment for their severe head injuries.
RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR)
Trizol reagent was added to the fresh glioblastoma tissues which were obtained intraoperatively, and the total RNA was extracted following the instructions of the reagent manufacturer. The first strand complementary DNA (cDNA) was synthesized by first strand cDNA synthesis kit (Invitrogen K1622, Carlsbad, California, USA). Quantitative real-time RT-PCR was performed using Roche480II Real-Time PCR System (Roche, Switzerland). Each specimen was tested in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. The primer sequences were: GAPDH forward primer, 5′-GCGAGATCCCTCCAAAATCAA-3′ and reverse primer, 5′-GTTCACACCCATGACGAACAT-3′; hsa_circ_01844 forward primer, 5′-AGCCACGTTGCTGTTAAACC-3′ and reverse primer, 5′-TAGTCGAGGGCCTTTTCAAG-3′.
Construction of lentivirus vector overexpressing hsa_circ_01844 in glioblastoma cell lines
The lentivirus vector was linearized by restriction enzyme digestion. Hsa_circ_01844 fragments were prepared by PCR amplification and inserted into the linearized lentivirus vector. The plasmids were transfected into bacteria and agar plates were used for bacterial culture and clone section. The positive clones were sequenced, analyzed, and then expanded to obtain high-quality plasmid for downstream virus packaging.
Cell lines and culture
Human glioblastoma cell lines (U87 and U251) were obtained from the American Type Culture Collection. Both U87 and U251 cells were cultured with 10% fetal bovine serum (FBS) and Dulbecco modified Eagle medium (DMEM) in a humidified atmosphere (5% CO2, 37°C).
Flow cytometric analysis
Logarithmic growth cells were inoculated into six-well plates (Corning, New York, USA) at a density of 5 × 105/mL (triplicate). After 48 h of culture, the cells were washed with phosphate-buffered saline (PBS) and fixed with 75% ethanol. Following the manufacturer's instructions, the cell cycle was measured by flow cytometry (BD Biosciences FACS Calibur, BD Biosciences, Piscataway, NJ, USA).
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
Cells in each group were cultured in DMEM with 10% FBS. Cells in logarithmic growth cycle were inoculated into 96-well plates (5000 cells/well) (triplicate) to grow for 1–4 days. Cell proliferation was analyzed following the protocol of MTT kit. The absorption value of each well was measured at 490 nm using a microplate reader (EL × 800, BIO-TEK, Vermont, USA). Data were collected from three independent assays.
Transwell assay
To measure cell migration, Transwell assay was performed. Transwell compartments with 24 holes and pore size of 8-μm (Corning) were coated with or without 150 μL Matrigel. Cell suspension was added in the upper chamber to postpone the logarithmic growth phase of the cells, and 600 μL medium containing 20% FBS was added in the lower chamber. After 24 h of culture, the disposed cells were fixed with 4% paraformaldehyde, stained with crystal violet solution (Sigma-Aldrich, Saint Louis, MO, USA), washed with PBS, and finally removed with cotton swabs.
Cell apoptosis analysis
Cells were cultured in Roswell Park Memorial Institute-1640 medium containing 10% FBS and 1% double antibody (penicillin-streptomycin mixture) for 24 h at a density of 300,000 cells/well. Cells were then collected 48 h after transfection. After washing with PBC, annexin V-fluorescein isothiocyanate apoptosis detection kit (BD Biosciences) was used to analyze cellular apoptosis by flow cytometry.
Statistical analysis
SPSS 20.0 software (IBM, Chicago, IL, USA) was used for statistical analysis. The data were obtained from at least three independent experiments, expressed as mean ± standard deviation. The Student's t test was used to analyze the differences between two groups. One-way analysis of variance was used to analyze the differences among three groups. A value of P < 0.05 was considered statistically significant.
Results
Hsa_circ_01844 was down-regulated in glioblastoma tissues
The expression level of hsa_circ_01844 was significantly lower in glioblastoma tissues when compared to normal control brain tissues (0.034 ± 0.036 vs. 1.630 ± 0.891, P < 0.001).
Overexpression of hsa_circ_01844 inhibited the cell cycle and proliferation of glioblastoma cells
As shown in Figure 1, the content of G2 phase in hsa_circ_01844 overexpression group (lentivirus group) was significantly lower than that in empty vector (EV) and blank control groups (all P < 0.05). This indicated that hsa_circ_01844 inhibited cell cycle progression and proliferation of U87 and U251 cells.
Figure 1.
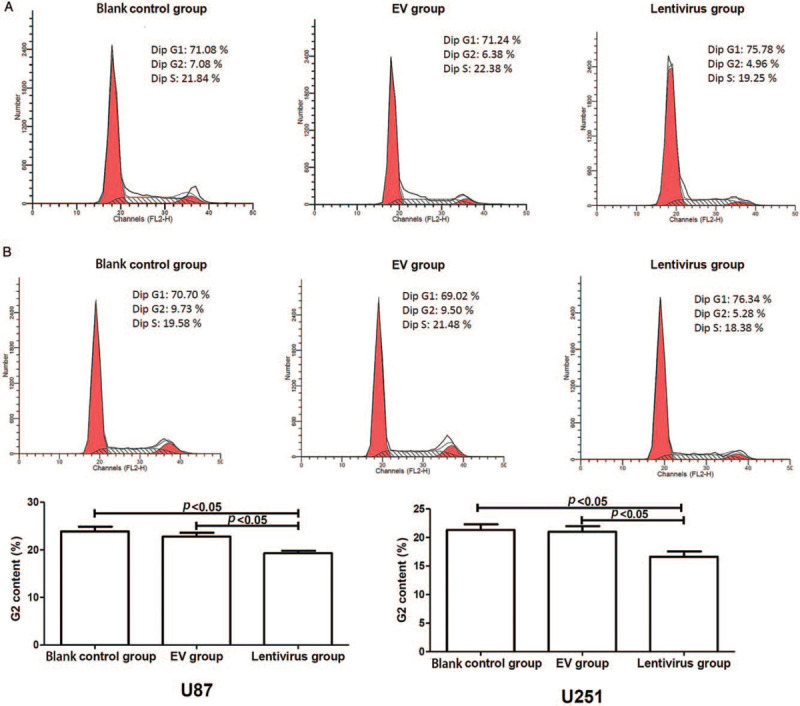
Flow cytometry was performed to monitor cell cycle in U87 and U251 cells. All data were collected from three independent assays. (A) U87 cells: the content of G2 phase in hsa_circ_01844 overexpression group (hsa_circ_01844 lentivirus) was significantly lower than that in EV and blank control (no transfection) groups. (B) U251 cells: the content of G2 phase in the hsa_circ_01844 overexpression group was significantly lower than that in EV and blank control groups. EV: Empty vector.
Overexpression of hsa_circ_01844 inhibited cell growth and reduced colony formation of glioblastoma cells
To determine the impact of hsa_circ_01844 on cell growth, MTT assay was performed using glioma cells transfected with hsa_circ_01844. Cells transfected with EVs and cells with no transfection were included as controls. As shown in Figure 2A and 2B, hsa_circ_01844 overexpression significantly inhibited cell growth (all P < 0.001). Next, we performed colony formation assay to examine the impact of hsa_circ_01844 overexpression on colony formation. As shown in Figure 2C and 2D, cells with hsa_circ_01844 overexpression showed significantly decreased numbers of colonies (all P < 0.01).
Figure 2.
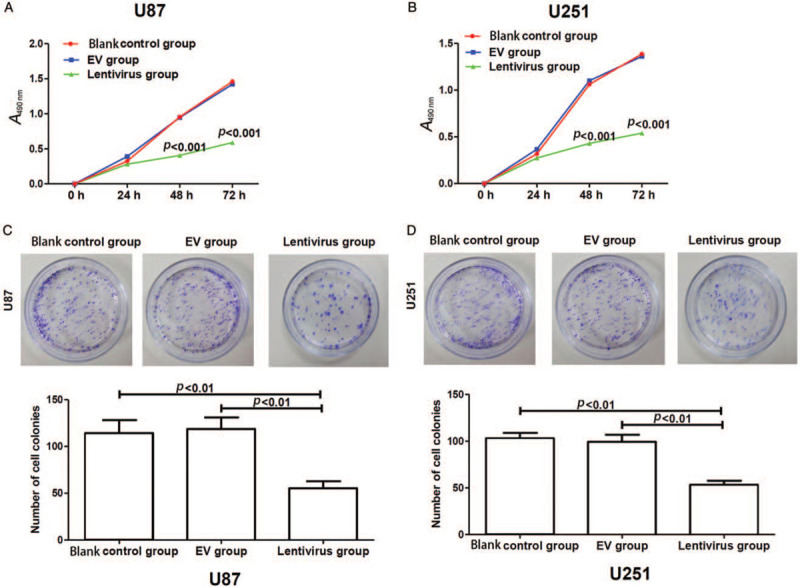
Overexpression of hsa_circ_01844 inhibited the growth and colony formation of U87 and U251 cells. All data were collected from 3 independent assays. (A and B) Cell growth was measured using MTT method. The overexpression of hsa_circ_01844 significantly inhibited the growth of glioma cells. (C and D) Colony formation was measured in hsa_circ_01844 overexpressed cells and control cells. The overexpression of hsa_circ_01844 significantly decreased the numbers of colonies. EV: Empty vector; MTT: 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide.
Up-regulation of hsa_circ_01844 inhibited the migration of glioblastoma cells
To determine whether hsa_circ_01844 has an impact on the migration of glioblastoma cells, transwell assays were conducted using U251 and U87 cells. As shown in Figure 3, overexpression of hsa_circ_01844 led to a significant decrease in the migration ability of glioblastoma cells when compared with control groups (all P < 0.001).
Figure 3.
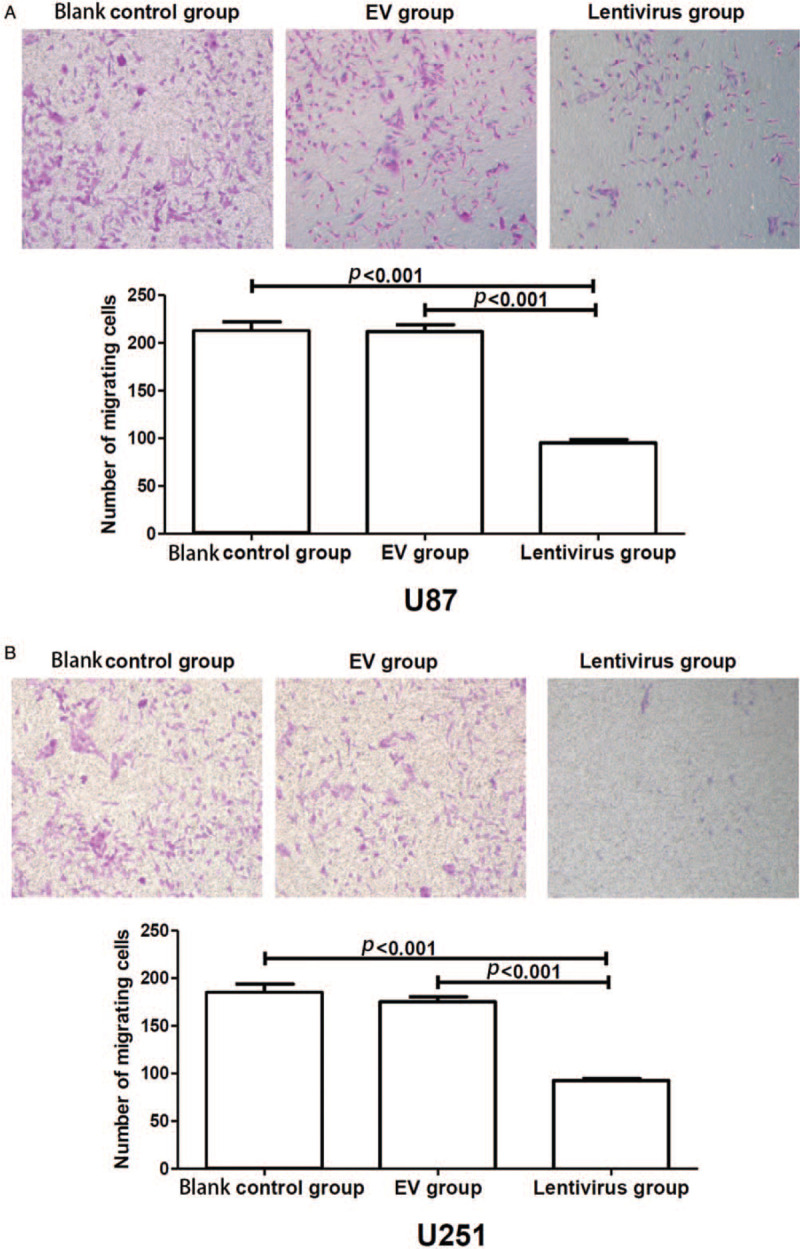
Hsa_circ_01844 inhibited cell migration in U87 and U251 cell lines in Traswell assay. All data were collected from three independent assays. (A) Overexpression of hsa_circ_01844 inhibited the U87 cells’ migration, and the number of migrating cells was significantly lower than that of control groups. (B) Overexpression of hsa_circ_01844 inhibited the U251 cells’ migration, and the number of migrating cells was significantly lower than that of control groups. Crystal violet staining. Original magnification, ×100.
Overexpression of hsa_circ_01844 promoted tumor cell apoptosis in glioblastoma cell lines
We next analyzed the apoptotic function of hsa_circ_01844 in glioblastoma cell lines by flow cytometry. As shown in Figure 4, cells with hsa_circ_01844 overexpression displayed a significant increased apoptotic rate when compared with control groups in both U87 and U251 cells (all P < 0.001). The result suggested that hsa_circ_01844 might accelerate the apoptosis in glioblastoma cell lines.
Figure 4.
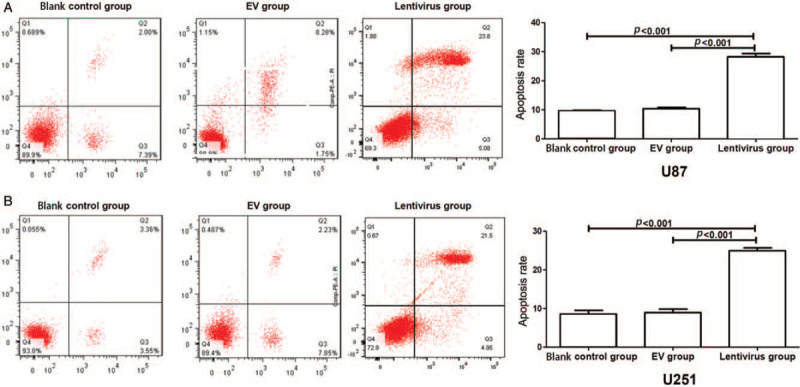
Overexpression of hsa_circ_01844 induced glioblastoma cells’ apoptosis. All data were collected from three independent assays. (A) Overexpression of hsa_circ_01844 promoted the apoptotic rates of U87 cells, as compared to the control groups. (B) Overexpression of hsa_circ_01844 promoted the apoptosis rate of U251 cells, as compared to the control groups. EV: Empty vector.
Discussion
With the rapid progress of high-throughput sequencing of non-coding RNAs, more attention turns to circRNAs, which are a special class of endogenous RNA, widely exist in human cells and take a great proportion of transcribing genetic information.[20,21] Recent studies have demonstrated that circRNA plays a crucial role in various physiological and pathological processes, including cancers pathogenesis.[10,11] Previous studies have shown that some novel circRNAs are involved in the proliferation of gliomas, such as circRNA ITCH, circRNA FBXW7, circRNA NFIX, and others.[22–24] In our study, we demonstrated that circRNA hsa_circ_01844 was down-regulated in glioblastoma tissues and it regulated the proliferation, migration, invasion, and apoptosis of glioblastoma cells. These findings have clinical implications for both diagnosis and potential therapeutic purposes.
The main mechanism of circRNAs in the role of tumor's pathogenesis may lie in the competitive adsorption of miRNA receptors or self-binding miRNA.[13] Compared with their linear isomers, circRNAs have higher expression levels and more miRNA binding sites, thus are more effective in sequestering miRNAs.[25–27] Therefore, the carcinogenic mechanisms driven by circRNAs may occur through their interaction with miRNAs to regulate subsequent pathways. Increasing evidence has supported this theory. Hsa_circ_005211 promotes the migration and invasion processes of breast cancer cells by acting as a sponge for mir-125a-5p.[28] Similarly, hsa_circ_0014359 acts as a miRNA-153 sponge in glioma cells and promotes the malignant proliferation of glioma cells by targeting miRNA-153/PI3K signaling pathway.[29]
In our study, we showed hsa_circ_01844 can potentially act as an inhibitor in glioblastomas by inducing apoptosis and inhibiting proliferation, migration of U251 and U87 cells. We found that the content of G2 phase in hsa-circ-01844 overexpression group (hsa_circ_01844 lentivirus) was significantly lower than that in control groups, which indicated that hsa_circ_01844 inhibit the U87 and U251 cells’ cell cycle progression. Furthermore, based on bioinformatics prediction, hsa_circ_01844 may interact with several miRNAs, such as miRNA-616 and miRNA-671-5p. MiRNA-616 might act as a tumor promoter and play oncogenic roles in glioma by regulating the Sry related HMG box-7 and wingless/integrated/β-catenin signaling,[30] and miR-671-5p can regulate complementarity determining region 1 pathway to inhibit the development of glioblastoma.[31] Accordingly, it was proposed that hsa_circ_01844 can sponge tumor promoter miRNAs and then inhibit the expression level of some targeted oncogenic genes, which can promote the glioblastoma progression.
Collectively, the present study investigates the expression level and function of has_circ_01844 in glioblastoma. Has_circ_01844 might serve as a novel therapeutic target for this malignant tumor in the future.
Conflicts of interest
None.
Footnotes
How to cite this article: Zhou JX, Chen KF, Hu S, Dong JR, Wang HX, Su X, Wang YH, Chu JS. Up-regulation of circular RNA hsa_circ_01844 induces apoptosis and suppresses proliferation and migration of glioblastoma cells. Chin Med J 2021;134:81–87. doi: 10.1097/CM9.0000000000000979
Jin-Xu Zhou and Ke-Fei Chen contributed equally to this work.
References
- 1.Cantrell JN, Waddle MR, Rotman M, Peterson JL, Ruiz-Garcia H, Heckman MG, et al. Progress toward long-term survivors of glioblastoma. Mayo Clin Proc 2019; 94:1278–1286. doi: 10.1016/j.mayocp.2018.11.031. [DOI] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Malkki H. Trial watch: glioblastoma vaccine therapy disappointment in phase III trial. Nat Rev Neurol 2016; 12:190.doi: 10.1038/nrneurol.2016.38. [DOI] [PubMed] [Google Scholar]
- 4.Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs 2016; 20:S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strobel H, Baisch T, Fitzel R, Schilberg K, Siegelin MD, Massler GK, et al. Temozolomide and other alkylating agents in glioblastoma therapy. Biomedicines 2019; 7:69.doi: 10.3390/biomedicines7030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alifieris C, Trafalis DT. Glioblastoma multiforme: pathogenesis and treatment. Pharmacol Ther 2015; 152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Polivka J, Polivka J, Holubec L, Kubikova T, Priban V, Hes O, et al. Advances in experimental targeted therapy and immunotherapy for patients with glioblastoma multiforme. Anticancer Res 2017; 37:21–33. doi: 10.21873/anticanres.11285. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh D, Nandi S, Bhattacharjee S. Combination therapy to checkmate glioblastoma: clinical challenges and advances. Clin Transl Med 2018; 7:33.doi: 10.1186/s40169-018-0211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee ECS, Elhassan SAM, Lim GPL, Kok WH, Tan SW, Leong EN, et al. The roles of circular RNAs in human development and diseases. Biomed Pharmacother 2019; 111:198–208. doi: 10.1016/j.biopha.2018.12.052. [DOI] [PubMed] [Google Scholar]
- 10.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 2019; 16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 12.Hao Z, Hu S, Liu Z, Song W, Zhao YY, Li MH. Circular RNAs: functions and prospects in glioma. J Mol Neurosci 2019; 67:72–81. doi: 10.1007/s12031-018-1211-2. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu JH, Li DP, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med 2019; 70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a largeclass of animal RNAs with regulatory potency. Nature 2013; 495:333–343. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Xin J, Zhang XY, Sun DK, Tian LQ, Xu P. Upregulated circular RNA hsa_circ_0067934 contributes to glioblastoma progression through activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2019; 23:3447–3454. doi: 10.26355/eurrev_201904_17709. [DOI] [PubMed] [Google Scholar]
- 18.Shea A, Harish V, Afzal Z, Chijioke J, Kedir H, Dusmatova S, et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med 2016; 5:1917–1946. doi: 10.1002/cam4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wang H, Chu J, Huang Q, Li G, Yan Y, et al. Circular RNA hsa_circ_0008344 regulates glioblastoma cell proliferation, migration, invasion, and apoptosis. J Clin Lab Anal 2018; 32:e22454.doi: 10.1002/jcla.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin S, Huang K, Zhu XG. Non-coding RNAs: regulators of glioma cell epithelial-mesenchymal transformation. Pathol Res Pract 2019; 215:152539.doi: 10.1016/j.prp.2019.152539. [DOI] [PubMed] [Google Scholar]
- 21.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018; 37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Ma K, Sun M, Shi S. Identication of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am J Transl Res 2018; 10:1373–1386. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Yang Y, Gao X, Zhang ML, Yan S, Sun CJ, Xiao FZ, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 2018; 110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci 2018; 225:1–19. doi: 10.3389/fnmol.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 2017; 242:1136–1141. doi: 10.1177/1535370217708978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Zhao K, Huang NN, Zhang N. Circular RNAs and human glioma. Cancer Biol Med 2019; 16:11–23. doi: 10.20892/j.issn.2095-3941.2018.0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to non-coding RNA. Science 2013; 340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhou SY, Zhu LP, et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother 2018; 107:1342–1353. doi: 10.1016/j.biopha.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 29.Jin P, Huang Y, Zhu P, Zou Y, Shao TT, Wang OY. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun 2018; 503:1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 30.Bai QL, Hu CW, Wang XR, Shang JX, Yin GF. MiR-616 promotes proliferation and inhibits apoptosis in glioma cells by suppressing expression of SOX7 via the Wnt signaling pathway. Eur Rev Med Pharmacol Sci 2017; 21:5630–5637. doi: 10.26355/eurrev_201712_14006. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol 2019; 234:1–13. doi: 10.1002/jcp.28061. [DOI] [PubMed] [Google Scholar]


