Abstract
Background
Musashi1 (MSI1) is an oncogenic protein with a crucial role in the proliferation and characteristics of the epithelial cells in breast cancer. The change in expression of MSI1 has a role in solid tumor progression. There are different factors that regulate MSI1 expression in various cancer tissues including microRNAs which are considered as one of the most important of these factors. The aim of our study is identification of the molecular cause of maximal expression of MSI1 in epithelial breast cancer cell lines.
Results
Among predicted microRNAs, miR-125b, miR-637 and miR-802 were able to significantly reduce the luciferase activity. In addition, the relative expression of these three miRNAs were measured in the cancerous cell lines that results showed a significant reduction in expression of all microRNAs. On the other hand, only the overexpression of miR-125b caused a change in the expression pattern of MSI1 in breast epithelial cancer cell lines. Accordingly, our results demonstrated that the exogenous expression of miR-125b decreased not only the MSI1 protein but also expression of epithelial markers in breast cancer cells.
Conclusions
The results of luciferase reporter assay showed that MSI1 is a direct target for miR-125b in epithelial breast cancer cells. Moreover, higher amount of MSI1 in those cell lines seems due to the reduced amount of miR-125b, which is responsible for epithelial features of those kinds of cancer cells. Therefore, the modulation of miR-125b may be a potential approach to help to combat against epithelial breast tumors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12860-021-00348-8.
Keywords: Breast cancer, Epithelial markers, microRNA, Musashi1
Background
Breast cancer has the highest incidence of female cancers worldwide. Analysis of global gene expression and DNA microarrays from cancer tissues asserted that a variety of genes are causative for this type of cancer. Accordingly, breast cancer is divided into five groups; a) HER2-positive, b) luminal A, c) luminal B, d) basal-like and e) normal-like subtypes [1].
In mammalian cells, the family of musashi (MSI) as RNA-binding proteins are highly conserved and have two members, musashi1 (MSI1) and musashi2 (MSI2). MSI1 has a main function in self renewal and maintenance of stem cells [2, 3]. MSI1 increases proliferation of mammary stem cells through triggering of two signaling pathways; Notch and Wnt [4]. Moreover, MSI1 is strongly expressed in multiple solid tumors such as brain [5–7], lung [8], ovary, squamous oral, colorectal and renal cancers [9–12]. On the other hand, MSI2 is reported to be upregulated in hematopoietic cancer cells [13]. The expression of MSI1 in HER2-positive breast cancer cell lines is correlated with HER2 and knock down of MSI1 reduces colony expansion of spheroid cultures in tumor cells [14]. Furthermore, MSI protein is essential for the preservation of epithelial property of breast cancer cells whereas overexpression of MSI2 regulates in vivo processing of epithelial-to-mesenchymal transition (EMT). Conversely, knocking down of MSI1 or MSI2 in the BT474 cell line enhances mesenchymal markers and decreases epithelial markers expression [15].
MicroRNAs belong to noncoding RNAs that control gene expression mostly through binding to 3’UTR of target genes. Generally, miRNAs regulate target gene expression via reduction of mRNA stability or translation [16–18]. MicroRNAs have diverse roles in various steps of breast cancer progression, which were indicated in recent studies [19–21]. 3`-untranslated region (3’UTR) of MSI1 has a length of 1800 nucleotides and is predicted to contain different sites for binding of endogenous miRNAs. The documentation shows that MSI1 has an oncogenic function in breast cancer progression, therefore any modulation in MSI1 expression could be considered as a possible therapeutic approach against breast cancer progression. Thus, the aim of our study was identification of putative miRNAs, which modulate MSI1 transcript in breast cancer cells. For this purpose, we first predicted the miRNAs that bind to 3’UTR of MSI1 using bioinformatic databases and then confirmed the direct binding of selected miRNAs to MSI1 3’UTR by luciferase assay. Finally, overexpression of miR-125b in human breast epithelial cancer cells modulated the expression of MSI1 as well as epithelial markers.
Results
Up-regulation of MSI1 was only detected in epithelial breast cancer cell
Analysis of transcript and protein of MSI1 were carried out in several breast cancer cell lines with different origins. The results demonstrated that MSI1 was highly expressed in T-47D and MCF-7 cells (epithelial cell line) but not in SKBR3 (HER2-positive cell line) and MBA-MD-231 cells (mesenchymal cell line) (Fig. 1).
Fig. 1.
MSI1 expression and western blot analyzing in breast cancer cell lines. a MSI1 expression was measured with real-time PCR. b MSI1 protein expression was analyzed using western blotting. Each expression was quantified and normalized with Gapdh. Represented value bars are the mean of triplicate independent experiments ± SEM
Bioinformatics prediction of microRNAs targeting MSI1
For prediction of those microRNAs which target MSI1, the following conditions were considered: the minimum folding free energy, context++ score, conservation, number and site seed type of miRNA binding to MSI1 3′-UTR [22]. Due to maximal expression of MSI1 in MCF-7 and T-47D cancer cell lines (epithelial cell lines) compared to MCF-10A (normal breast cell line), the levels of predicted miRNAs were assessed using a GEO dataset GSE70480, which was related to the expression profile of miRNAs in aforementioned cell lines. Based on extracted data, miR-125b-5p, miR-802, miR-490-3P, miR-338-3p and miR-637 were selected due to their effect on MSI1 downregulation (Fig. 2, supplementary Figure 1). On the other hand, miR-342-3p was excluded from the list as it had previously reported that its expression was high in MCF-7 similar to what reported for MSI1. Therefore, miR-342-3p could not be supposed to target MSI1 [23]. Also, the selection of miR-802 for further analysis was due to reduced expression in MCF-7 compared to normal breast cell line [24].
Fig. 2.

Schematic representation of bioinformatic sites for miRNAs prediction. For prediction of microRNA target sites, different scores in bioinformatic sites were used
Down-regulation of predicted microRNAs were verified in epithelial breast cancer cells
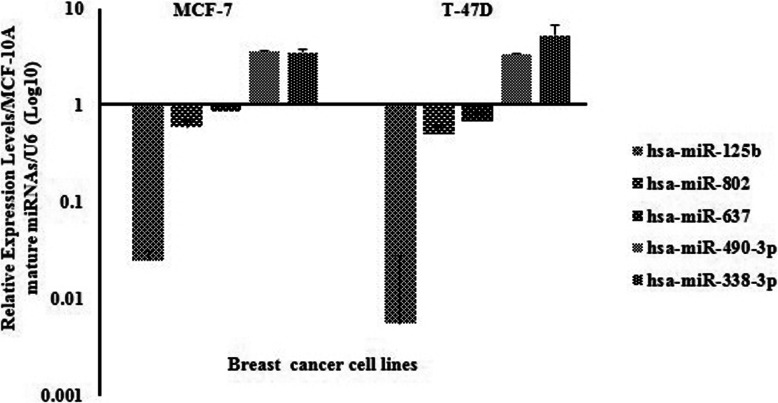
Since expression of MSI1 was high in both epithelial cancer cells (T47-D and MCF-7), the levels of predicted mature miRNAs were assessed in those cell lines (Fig. 3). Mature miRNAs expression were measured with MiRCURY LNA primer mix (Exicon, USA) and normalized with MiRCURY LNA U6 (Exicon, USA). However among them, only miR-125b-5p, miR-637 and miR-802 were selected for luciferase reporter assay because these microRNAs had lower expression values than others in both epithelial cancer cells compared to MCF-10A.
Fig. 3.
Expression of mature miRNAs in breast cancer cell lines in comparison of normal breast cell line, MCF-10A. Mature miRNAs expression were measured with real-time PCR. Expression of U6 was used for quantification and normalization of the results. Represented value bars are the mean of triplicate independent experiments ± SEM
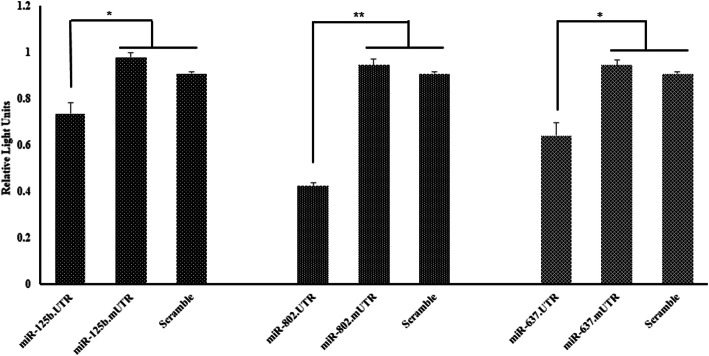
Confirmation of direct interaction of selected microRNAs with MSI1 transcript
To confirm direct binding of miR-125b-5p, miR-637 and miR-802 to 3′-UTR of MSI1 transcript (Supplementary Table 1), HEK293T cells were transfected with plasmids expressing precursors of both microRNAs as well as wild type or mutated MSI1 3’UTR. Transfected cells were harvested, lysed and the cell lysates were used for dual-luciferase reporter assay. Consistent with bioinformatics data, selected microRNAs significantly repressed luciferase activity. Elimination of microRNAs binding sites from 3’UTR of MSI1 resulted in significant increase in luminescent signals (Fig. 4). Therefore, direct targeting of MSI1 transcript by those microRNAs was verified.
Fig. 4.
Luciferase activity assay after overexpression of miRNA precursors. HEK393T cells were co-transfected with recombinant plasmids, which included MSI1 3’UTR downstream humanized Renilla luciferase reporter gene (hRluc) and miRNAs precursors. Also, HEK393T cells co-transfected with mutated Msi1 3’UTR for each miRNAs binding site downstream of hRluc luciferase reporter gene. For negative control analysis, HEK393Tcells co-transfected with a scrambled negative control miRNA (Control miRNA) and wild Msi1 3’UTR downstream hRluc. Then luminescence was measured. Error bars denote S.E.M. for three independent repeats in each experiment. Independent student t-test indicated differences between (*p < 0.05; **p < 0.01)
Elevated amount of miR-125b was able to inhibit MSI1 expression in cancer cells
To evaluate which one of microRNAs has a potential to negatively modulate the expression of MSI1 in epithelial cancer cells, MCF-7 and T-47D cells were transfected with plasmids expressing miRNA precursors. Expression of mature miRNAs in breast cancer cell lines after transfection were analyzed (Fig. 5). According to our data, ectopic expression of miR-125b reduced MSI1 transcript (Fig. 6 A, B) and its protein compared to negative control (Universal scramble) in both cell lines (Fig. 6 C, D). Surprisingly, in contrast to luciferase data, overexpression of miR-637 and miR-802 were not able to decrease the MSI1 expression in breast cancer cells. Consequently, we decided to use miR-125b for evaluating the effect of the microRNA on expression of epithelial markers in breast cancer cells.
Fig. 5.
Expression of mature miRNAs in breast cancer cell lines after transfection of recombinant plasmids [pBUD/precursor miRNAs expression plasmids] in MCF-7 and T-47D cell lines
Fig. 6.
MSI1 mRNA and protein expression analyzed in breast cancer cell lines. MSI1 expression analyzed in (a) MCF-7 and (b) T-47D breast cancer cells transfected with miRNA precursor plasmids and compared to cells transfected with a scrambled negative control miRNA. c MSI1 protein expression analyzed in MCF-7 and (d) T-47D breast cancer cells transfected with miRNA precursor plasmids as compared to cells transfected with a scrambled negative control miRNA (Control miRNA). Error bars denote S.E.M. for three independent repeats in each experiment. The expression of mRNA was analyzed using RT-qPCR, and normalized to GAPDH in comparison to untransfected cells. MSI1 protein expression was analyzed using western blotting Error bars signify S.E.M. for three independent repeats in each experiment. One-way ANOVA indicated differences (*p < 0.05)
Ectopic expression of miR-125b reduced epithelial markers
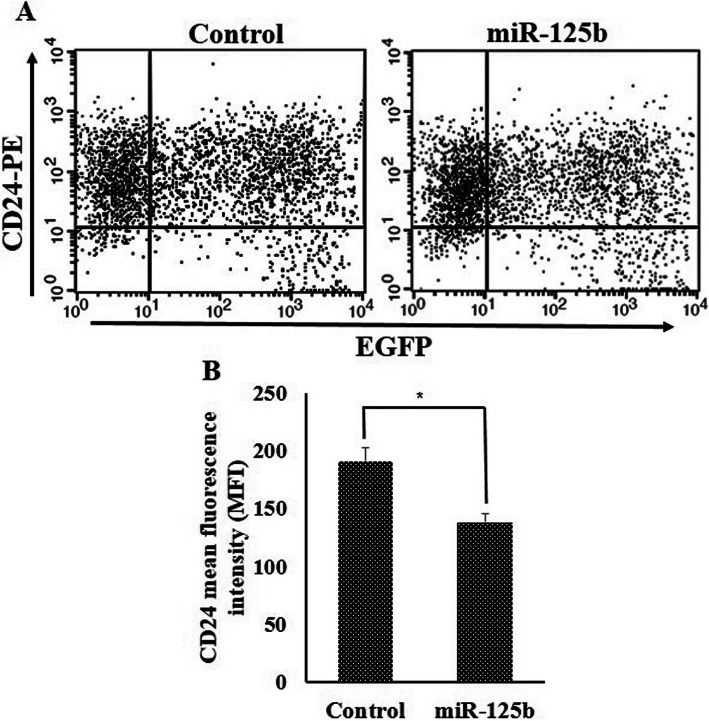
In previous studies, it was shown that knockdown of MSI1 in epithelial cells such as BT474 was responsible for reduction in epithelial markers including E-cadherin and epithelial cellular adhesion molecule (EpCAM) and CD24 [15]. Inversely, there was an increase in mesenchymal markerssuch as Fibronectin and jagged1 in similar circumstance. Our data were evident that ectopic expression of miR-125b in MCF-7 and T-47D cell lines gave rise to a reduction in E-cadherin, EpCAM and CD24 (Fig. 7a, c). However, increasing expression rate of miR-125 could not able to change the expression of mesenchymal markers (Fig. 7b, d). Moreover, Immunoblotting of E-cadherin in MCF-7 cells confirmed the reduction of epithelial markers followed by ectopic expression of miR-125b (Fig. 7e). Change of CD24 expression in EGFP positive population cells, which were transfected with miR-125b.pBUD and Usc. Pbud were evaluated by flow cytometry. A transfection efficiency of 30% was obtained in MCF7 cells (supplementary figure 2). Analysis of fluorescence intensity revealed that expression of CD24 was reduced after overexpression of miR-125b (Fig. 8). Reduction of MSI1 intensity in EGFP positive cell population in MCF-7 cells transfected with miR-125b.pBUD were shown in supplementary figure 3. Also, MCF-7 cells were transfected with EGFP.pBUD plasmid (USc.pBUD) as negative control. EGFP positive and negative cells have the same intensity for MSI1 (supplementary figure 4). Flow cytometry results were shown in supplementary figure 5. Eventually, according to the results, we concluded that down-regulation of the miR-125b is responsible for uprising of MSI1, which is crucial for maintenance of the epithelial phenotype in breast cancer cells (Fig. 9).
Fig. 7.
Analysis of epithelial and mesenchymal expression markers. a Epithelial marker expression in T-47D cells. b Expression of mesenchymal marker in T-47D cells. c Expression of Epithelial marker in MCF-7 cells. d Expression of mesenchymal marker in MCF-7 cells. e Western blot analysis for E-cadherin in MCF-7 cell lines. Error bars denote S.E.M. for three independent repeats in each experiment. Data were analyzed with the independent-samples t-test (*p < 0.05)
Fig. 8.
Analysis of CD24 expression through Flow cytometry. a Flow cytometry analysis of CD24 in EGFP positive cells showed that overexpression of mir-125b reduced the intensity of CD24 in MCF-7 cells. b Graphs of quantification of CD24 intensity by flow cytometry. Error bars denote S.E.M. for three independent repeats in each experiment. Data were analyzed with the independent-samples t-test (*p < 0.05)
Fig. 9.
Schematic representation of MSI1 regulation through overexpression of miR-125b in epithelial–luminal state of breast cancer cell lines. Overexpression of miR-125b in human breast epithelial cancer cells modulated the expression of MSI1 as well as epithelial markers
Discussion
RNA binding protein, MSI1, is highly expressed in different solid tumors [25, 26]. MSI1 has a crucial role in stem cell and cancer stem cell proliferation [27, 28]. MSI1 protein and notch signaling pathway are two key regulators for asymmetric division of the stem cell population in mammary epithelial cells [29]. In breast cancer, a positive correlation between expression of ErbB2 and MSI1 was observed. It has demonstrated that MSI1 expression is increased in spheroid culture derived from breast cancer lines such as T-47D and MCF-7 [14]. In breast cancer, MSI1 has also a critical role in EMT. Hence expression of MSI1 in T-47D and MCF-7 [epithelial cell lines] was significantly higher than MDA-MB-231 [mesenchymal cell line].
MicroRNAs regulate gene expression at post transcriptional levels. Recent data have shown that miR-802 as a tumor suppressor could control proliferation of MCF-7 cells through expression regulation of FOXM1 [24]. Similarly, miR-637 has also antitumor activity, such that its expression was reduced in hepatocellular carcinoma cell line (HCC) and clinical specimen of cancer patients (reference). The experiments revealed that overexpression of miR-637 in some hepatocellular carcinoma cell lines gave rise to a reduction in proliferation of cancer cells by regulation of signal transducer and activator of transcription 3 (STAT3) [30]. Furthermore, it was indicated that miR-637 has a tumor suppressor role, which prevents migration and invasion of cancer cells by suppression of AKT1 in human glioma [31]. Recently, it was reported that overexpression of miR-125b decreased the proliferation of breast cancer cells and promoted their apoptosis by regulation of G2/M phase. miR-125b controls cell cycle through regulation of a number of target genes such as multiple EGF-like-domains 9 (MEGF9), cyclin J (CCNJ), aminopeptidase A (ENPEP) and casein kinase 2-alpha (CK2-α) [32]. Likewise, it was reported that ectopic expression of miR-125b could prevent migration and proliferation of breast cancer cells through regulation of KIAA1522 [33].
The long 3’UTR fragment of MSI1 coding sequence has several potential binding sites for microRNA. In the present research, utilizing in silico analysis of a GEO dataset (GSE70480), we predicted several miRNAs that were able to bind 3’UTR of MSI1. Expression of the predicted miRNAs (miR-125b-5p, − 802, − 637, − 490-3p and − 338-3p) was assessed in some epithelial cancer cell lines in which MSI1 expression was high. Amongthem, three miRNAs (miR-125b-5p, − 802, and − 637) with low expression rate in tested breast cancer cells were chosen for further investigation. All of three selected miRNAs were shown to bind directly to MSI1 3’UTR as verified by luciferase reporter assay. Among the aforementioned miRs, miR-125b was able to regulate MSI1 expression whereas overexpression of miR-637 and -802 could not able to modulate transcript of MSI1 in MCF-7 and T-47D cells. Our further in silico analysis using RNA-binding protein-specific database (http://rbpdb.ccbr.utoronto.ca/) indicated that RNA binding motif protein X-Linked (RBMX) could attach to the same sequence of MSI1 3’UTR, which covers the miR-637 binding site. Interestingly, RBMX are actively expressed in breast cancer cells according to the human protein atlas data (https://www.proteinatlas.org) [34, 35]. In this respect, several independent studies have indicated that there is a counteraction between RNA binding protein and miRNAs for accessibility to 3’UTR of target genes [36, 37]. Therefore, it is more likely that RBMX compete with miR-637 for binding to its target sequence in MSI1 3’UTR. However to validate such hypothesis, further analyses are needed to be performed. On the other hand, following down regulation of MSI1 by miR-125b MCF-7 and T-47D cell lines, the expression of epithelial markers including E-cadherin, CD24 and EpCAM was also diminished. But conversely, transcript level of mesenchymal marker (Vimentin) was not affected in these cell lines, presumably due to the lack of enough expression of Vimentin as reported previously [38].
However, our data were in good agreement with the recent study that down regulation of MSI1 in epithelial cell line, BT474, caused a reduction in epithelial markers [15]. It seems that this effect is cell line dependent, as transfection of miR-125b inhibitor in triple negative breast cancer lines (TNBC) was responsible for reduction in cellular proliferation and process of EMT [39]. In these lines such as MDA-MB-468 and MDA-MB-231, the cells are negative for estrogen receptors, progesterone receptors, and excess HER2 protein, [39]. Similarly, in an agreement to our data, overexpression of miR-125 in Hs578T cells inhibited the EMT through a reduction in MAP 2 K7 [40].
Conclusions
Collectively, we indicated that MSI1 is a potential target of miR-125b. Hence, miR-125b could be considered as a potential factor for regulation of the progress of a number of breast cancer types and is potentially applicable for therapeutic purposes. However the exact mechanism underlying this phenomenon should be investigated in further studies as it would need to be critically looked at while comparing the same phenomenon with primary cells.
Methods
Bioinformatics sites for miRNAs prediction
In order to predict the miRNAs interact with MSI1 RNA, several bioinformatics database were implemented as Target Scan (targetscan.org), PicTar (pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi), miRWALK (mirwalk. Uni-hd. de/) and miRTar.human (http://mirtar.mbc.nctu.edu.tw/human).
Construction of expression plasmids
For construction of the plasmids expressing human miRs-802, − 637, −125b, miRNA precursors were PCR amplified by specific primers (Table 1, A) and using human genome as a template. Then, the desired sequences were cloned into SalI and XbaI sites of pBUD4.1 expression plasmid under CMV promoter. All restriction enzymes were obtained from Thermo Scientific, USA. To verify the expression of the constructs, the recombinant vectors were double digested with SalI and XbaI restriction enzymes. The recombinant plasmids were confirmed by restriction digestion (Supplementary figure 6). For negative control plasmid, universal scramble sequence was cloned into EGFP.pBUD plasmid (USc.pBUD) as reported previously [41]. Schematic representation of recombinant plasmids are displayed in supplementary figure 7. For Luciferase reporter assay, the multiple cloning site in pSICHECK2 plasmid (Promega, USA) was modified by cloning a synthetic DNA fragment into SalI and NotI sites in multiple cloning site and resulted vector was named as pSICHECK2.MCS. The synthetic DNA, which was constructed by annealing two oligonucleotides (Table 1, B) contained additional restriction sites (SalI-SacI-SpeI-AgeI-XhoI-NotI) for direct subcloning of target 3’UTRs under Renilla luciferase coding sequence. pSGG-MSI1 plasmid was gifted from Dr. Luiz O.F. Penalva (UT Health San Antonio, USA) and digested with NheI and XhoI restriction enzymes. The resulted wild type 3′-UTR fragment of MSII was ligated between SpeI and XhoI sites of pSICHECK2.MCS plasmid. Verification of final plasmid, pSICHECK2.MCS.MSI1, was performed through double digestion with XhoI and NheI enzymes and single digestion with XhoI. Restriction digestion patterns confirmed the plasmid size (Supplementary figure 8). Mutated type of MSI1 3’UTR for each miRNA binding site was constructed with site directed mutagenesis protocol [42] (Table 1, C) and these mutated fragments were cloned into pSICHECK2.MCS plasmid. Schematic representation of recombinant plasmids for luciferase assay were shown in Supplementary figure 9. The constructed vectors were named MSI13’UTR (miR-125b).pSICHECK2, MSI13’UTR (miR-637).pSICHECK2 and MSI13’UTR (miR-802).pSICHECK2. To confirm the sequence of the cloned fragments, final recombinant constructs were sent for sequencing by Metabion Company (Germany). Primers of Table 1, A and C were designed by oligo7.
Table 1.
List of primers used in this study
| A) Primers used for human miRNAs precursors amplification | ||||
| Genes | Forward Primer (5′-3′) | Reveres Primer (5′-3′) | ||
| miR-125b | GTCGACTGTGAAGGAAAGGAGC | TCTAGAACTTTAACAGAAATCCAGG | ||
| miR-637 | GTCGACACCCCAGGGATGATG | TCTAGA AGGCGGAGCACAGAAG | ||
| miR-802 | GTCGACCAGTTTGTTCCAGTGCC | TCTAGATAACACACACAGCCCCAG | ||
| Scramble | GTCGACCCGCTTGTTCGTTGGTAACTACATTCAAGAGATGTAGTTACC | GTCTAGAAAAAAGCTTGTTCGTTGGTAACTACATCTCTTGAATGTAGTTACC | ||
| B) Oligonucleotide sequences used for construction of pSICHECK2.MCS | ||||
| Name | (5′to3′) | |||
| OligoF | TCGACCGGAGCTCATACTAGTACACCGGTAACTCGAGTAGC | |||
| OligoR | GGCCGCTACTCGAGTTACCGGTGTACTAGTATGAGCTCCGG | |||
| C) Primers used for site-directed mutagenesis PCR | ||||
| Mutated site | Forward Primer (5′-3′) | Reveres Primer (5′-3′) | ||
| miR-125b |
F2:GCCCGCTCTCTCTTGGTTGGACCTGC F3:AAAGCAAAGGGCGTACCCCCACATTCTCTfC |
R2:GTCCAACCAAGGGCCTGAGAGAGCG R3:GAATGTGGGGGTACGCCCTTTGCTTTCC |
||
| miR-637 | F2:CTTGGCAGCCCCTCACCCCCCTTC | R2:GCCTCATAAAGAGGGACACACAGAAG | ||
| miR-802 |
F2:CCTCAGACACCGTAAGCTTGCAGGCCTCAG F3:GCAATAATCTTTGAGATGCGCGGCTGTTC |
R2:TTCACTCCAGCTATGCAC R3:GAACAGCCGCGCATCTCAAAGATTATTGCTTTGTG |
||
| Common primer | F1:GTCGACAGCGCCCCAGCCTGCA | R1:CTCGAGAACGTTTTCAAATAATTTATTAGG | ||
| D) Primers used for gene expression analysis by Q-PCR | ||||
| Genes | Forward Primer (5′-3′) | Reveres Primer (5′-3′) | TM (°C) | Accession No |
| MSI1 | TTGGCAGACTACGCAGGAAG | TGGTCCATGAAAGTGACGAAGC | 59 | NM_002442 |
| Gapdh | CCACTCCTCCACCTTTGACG | CCACCACCCTGTTGCTGTAG | 60 | NM_001357943 |
| E-cadherin | CTGCTGCTCTTGCTGTTTCTTC | CTTCTCCGCCTCCTTCTTCATC | 60 | NM_001317185 |
| EpCAM | CCATGTGCTGGTGTGTGAAC | CCTTCTGAAGTGGTCCGC | 60 | NM_002354 |
| CD24 | ATGTGGCAAGGAAAAACAGG | TTGGCATCCATCATCTAGTC | 60 | NM_001359084 |
| Vimentin | AAACTTAGGGGCGCTCTTGT | TGAGGGCTCCTAGCGGTTTA | 59 | NM_003380 |
| Snail | CCAGAGTTTACCTTCCAGCA | GATGAGCATTGGCAGCGA | 60 | NM_005985 |
Cell culture
HEK293T and breast cancer cells (T-47D, MCF-7, SK-BR-3 and MBA-MD-231) were purchased from the Pasteur Institute (Tehran, Iran). Cell culture was performed in 10% FBS supplemented DMEM high Glucose (Gibco, USA) including 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco, 15,070,063) as well as 2 mM L-glutamine (Gibco, 25,030–024), 1% non-essential amino acids (Gibco, 11,140–035). Normal breast cell line, MCF-10A, was cultured in DMEM/F12 (Gibco, 21,331–020) supplemented with 5% horse serum (ATOCEL, Aths-8100), insulin 10 μg/mL (Sigma, 16,334), cholera toxin (Sigma, c8052) 100 ng/mL, hydrocortisone (Sigma, H0888) 0.5 mg/mL, EGF (Sigma, 4127) 20 ng/mL, 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco, 15,070,063), 2 mM L-glutamine (Gibco, 25,030–024). Cell culture was performed under 5% CO2 at 37 °C.
Transcript analysis
The extraction of total RNA was achieved using TRI reagent (Thermo Scientific, 15,596–018) and cDNAs were synthesized using random hexamer primers and implementing MMLV reverse transcriptase according to the manufacturer’s protocol (TaKaRa, RR037A, Japan). To get rid of DNA contamination, DNase treatment was performed during cDNA synthesis. At the next step, 50 ng of cDNA was used for real-time PCR using SYBR green (TaKaRa, RR820Q, Japan) and specific primers on step one plus Real-Time PCR System (Applied Biosystems, USA). Primers were designed by the Beacon designer (Version 7.2, USA) and purchased from Metabion Company (Table 1, D). Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was used as reference gene for normalizing target gene expression and each experiment was repeated thrice. For microRNAs expression, cDNAs were synthesized using Universal cDNA synthesis kit II (Exicon, 203,301, Denmark). LNA™ primer mix for mature miRNAs and LNA™ Primer mix U6 (Exicon, Denmark) was used for Real time PCR. Finally, analysis of relative gene expression was performed using ΔΔCt method.
Immunoblot analysis
Proteins were extracted by means of TRI reagent (Thermo Scientific, 15,596–018) according to manufacturer instructions. Similar amount of protein for each sample, approximately 30 μg, was separated using SDS-PAGE and transferred to polyvinylidenedifluoride (PVDF; Bio Rad, 162–0176, USA) membranes. Blocking of the membranes was done in skim milk 10% (Millipore, 115,363) and then membranes were incubated with specific primary antibodies at 25 °C for 1.5 h. Primary antibodies were Anti-MSI1 antibody (Abcam, 52,865, USA; 1:4000), and mouse anti-Gapdh, clone 6C5 (Millipore, MAB374, USA; 1:5000). After this step, membranes were incubated at the same condition for the respective secondary antibodies, which were horseradish peroxidase (HRP)-conjugated a) goat anti-mouse IgG (Dako, P0447, USA; 1:5000) or b) goat anti-rabbit IgG (Santa Cruz, SC2301, USA; 1:16000). The HRP-conjugated IgG bound to each protein band was visualized by an Amersham ECL Advance Western Blotting Detection Kit (GE Healthcare, Germany).
Luciferase reporter assay
For Luciferase assay, HEK293T were seeded in 24-well plate to reach enough confluence for 24 h. Then, cells were co-transfected with pSICHECK2.MCS.MSI1 and each of precursor expressing plasmid. Besides, plasmid contained mutated type of MSI1 3’UTR for each miRNA binding site along with precursor expressing plasmid were co-transfected into HEK293T cells. Co-transfections were accomplished using Lipofectamine 2000 (Invitrogen, USA) according to its protocol. Approximately 2 days post-transfection, the cells were harvested and analyzed with dual-Luciferase reporter assay system (Promega, USA).
Immunofluorescence analysis
Approximately 48 h post-transfection, miR-125b.pBUD transfected MCF-7 cells were treated with 4% paraformaldehyde for 30 min at 25 °C. Permeabilization of the cells were achieved through incubating with 0.4% Triton X-100 for 20 min. After cell washing, primary antibody was diluted by blocking solution (BSA, 5 mg/mL) and used for cell staining. The cells were then incubated at 4 °C overnight. In the next step, the cells were stained with secondary antibody for 1 h at 37 °C and finally, they were stained with DAPI (D9542, Sigma) at final concentration of 1 μg/mL at room temperature for 5 min. Both first and secondary antibodies were Musashi-1 antibody (1:100, R&D: AF2628) and goat anti-rabbit IgG-FITC (1:100, Santa Cruz: Sc-2012). Images were captured under a fluorescent microscope (Olympus, Center Valley, PA, USA) using Olympus DP70 camera. CD24 expression was assayed using flow cytometry. Co-transfected MCF-7 cells with miR-125b.pBUD and pBUD.USc were detached using triple (Gibco, 12,604–012) after 48 h. Treated cells were stained with PE labeled mouse Anti-Human CD24 (BD Pharmingen™, 555,428, USA) for 30 min. After washing, the cells were evaluated by FACS Caliber flow cytometer (Becton Dickinson, USA and obtained results were analyzed with the ModFit LT™ v 4.0 program.
Statistical analysis
SPSS Statistical software (v. 20, IBM Corp., USA) was used to perform the data analysis and all data were reported as mean ± standard error of mean (SEM). Independent student t-test and one-way analysis of variance (ANOVA) were utilized to determine the statistical differences between groups, where p < 0.01 and p < 0.05 have been applied for indication of statistically significant differences.
Supplementary Information
Additional file 1: Figure S1. The results of GEO dataset GSE70480 for miRNAs selection. Related accession: GSM1782670 is for MCF-10A as normal breast cell line, GSM1782672 is for MCF-7 and GSM1782680 is for T-47D.
Additional file 2: Table S1. microRNA target sites in MSI1 3`UTR.
Additional file 3: Figure S2. MCF-7 cell line were transfected with miR-125b.pBUD and USc.pBUD expression plasmids. EGFP reporter gene indicated the accuracy of transfection.
Additional file 4: Figure S3. Reduction of MSI1 intensity in MCF-7 cell line which transfected with miR-125b.pBUD. The separated area demonstrated EGFP positive cells with decreased intensity of MSI1 as a result of overexpression of miR-125b.
Additional file 5: Figure S4. MSI1 intensity in MCF-7 cell line which transfected with (USc.pBUD) plasmid. Selected area shows EGFP positive and negative cells which have the same intensity for MSI1.
Additional file 6: Figure S5. Flow cytometry data confirmed reduction of MSI1 intensity in MCF-7 cell line after transfection with miR-125b.pBUD.
Additional file 7: Figure S6. Agarose electrophoresis of double digestion products of recombinant plasmids [pBUD/precursor miRNAs expression plasmids] which were digested with SalI and XbaI restriction enzymes. Double digestion with SalI and XbaI resulted in two distinct bands with approximate sizes of 5191 bp for plasmid pBUD and 378 bp for precursor of miR-637, 453 bp for precursor of miR-802 and 427 bp for precursor of miR-125b. M is 1kbp DNA ladder (Thermo Scientific, USA).
Additional file 8: Figure S7. Schematic representation of recombinant plasmids for ectopic expression of selected miRNAs which is involved miRNAs precursors and recombinant vector included scramble sequence as negative control.
Additional file 9: Figure S8. Agarose electrophoresis of double digestion products of recombinant plasmids [pSICHECK2.MCS.MSI1 plasmids] which were digested with XhoI and NheI restriction enzymes and single digestion with XhoI restriction enzyme. XhoI-linearized plasmid with a single band about 8053 bp and double digested plasmid fragments 5292 bp and 2761 bp. M is 1kbp DNA ladder (Thermo Scientific, USA).
Additional file 10: Figure S9. Schematic representation of pSICHECK2 plasmid which included MSI1 wild type and mutated MSI1 for direct binding of particular miRNAs.
Acknowledgments
Authors would like to convey their sincerest gratitude to Dr. Luiz O.F. Penalva (UT Health San Antonio, USA) for donation of pSGG-MSI1 plasmid.
Abbreviations
- CK2-α
Casein kinase 2-alpha
- EGF
Epidermal growth factor
- ENPEP
Amino peptidase A
- RT-qPCR
Real-time quantitative polymerase chain reaction
- CDS
Coding sequence
- CCNJ
Cyclin J
- JDMEM
Dulbecco’s modified Eagle medium
- EMT
Epithelial-mesenchymal transition
- FCS
Fetal calf serum
- MEGF9
Multiple EGF-like-domains
- M-MLV Reverse Transcriptase
Moloney Murine Leukemia Virus Reverse Transcriptase
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HER2
Human epidermal growth factor receptor 2
- HRP
Horseradish peroxidase
- Msi1
Musashi1
- PBS
Phosphate-buffered saline
- PVDF
Polyvinylidenedifluoride
- SEM
Standard error of mean
- STAT3
Signal transducer and activator of transcription 3
- TNBC
Triple negative breast cancer
- TRI
Trizol
- 3`-UTR
3`-untranslated region
Authors’ contributions
Study design was performed by M.F., K.D., and K.G. Data analysis and experiments were done by M.F., L.L. Interpretations of data were performed with M.F., K.D., and K.G. Manuscript writing was performed by M.F., K.G. and K.D. All authors have read and approved the manuscript.
Funding
This study was supported partly by a research budget from Research Department of University of Isfahan and Royan Institute for Biotechnology and partly by a grant-in-aid for the Ph. D students from the Iran National Science Foundation (NISF) [Ref. no. 97012028] awarded to K. G. (Supervisor) in support of M. F. for obtaining her Ph. D degree from the University of Isfahan.
Availability of data and materials
Supporting and raw data are available upon a reasonable request to the corresponding author.
Ethics approval and consent to participate
All protocols to use the Human Breast Cancer Cell lines were reviewed and confirmed by Ethics committee of Royan Institute (IR.ACECR.ROYAN.REC.1397.198).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Hossein Nasr-Esfahani, Email: mh.nasr-esfahani@royaninstitute.org.
Kamran Ghaedi, Email: kamranghaedi@sci.ui.ac.ir.
References
- 1.Perou CM, Sørlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115(7):1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 3.Chen H-Y, Wang M-L, Laurent B, Hsu C-H, Chen M-T, Lin L-T, Shen J, Chang W-C, Hsu J, Hung M-C. Musashi-1 promotes stress-induced tumor progression through recruitment of AGO2. Theranostics. 2020;10(1):201–217. doi: 10.7150/thno.35895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X-Y, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and notch pathways. Mol Cell Biol. 2008;28(11):3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muto J, Imai T, Ogawa D, Nishimoto Y, Okada Y, Mabuchi Y, Kawase T, Iwanami A, Mischel PS, Saya H. RNA-binding protein musashi1 modulates glioma cell growth through the post-transcriptional regulation of notch and PI 3 kinase/Akt signaling pathways. PLoS One. 2012;7(3):e33431. doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vo DT, Subramaniam D, Remke M, Burton TL, Uren PJ, Gelfond JA, de Sousa AR, Burns SC, Qiao M, Suresh U. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol. 2012;181(5):1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J-C, Tsai J-T, Chao T-Y, Ma H-I, Liu W-H. Musashi-1 enhances Glioblastoma migration by promoting ICAM1 translation. Neoplasia. 2019;21(5):459–468. doi: 10.1016/j.neo.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X-Y, Yu H, Linnpoila RI, Li L, Li D, Bo B, Okano H, Penalva LO, Glazer RI. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget. 2013;4(5):739–750. doi: 10.18632/oncotarget.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou G-Y, Yang T-W, Huang C-C, Tang C-Y, Yen J-Y, Tsai M-C, Chen H-Y, Fadhilah N, Lin C-C, Jong Y-J. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci Rep. 2017;7(1):2172. doi: 10.1038/s41598-017-02057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi C, Zhang Z. miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma. Tumor Biol. 2016;37(4):5437–5443. doi: 10.1007/s13277-015-4377-z. [DOI] [PubMed] [Google Scholar]
- 11.Xiao R, Yu Y, Shen S, Liu F, Kuang R. Musashi1 promotes tumor metastasis and is a prognostic marker for renal carcinoma. Int J Clin Exp Pathol. 2019;12(1):313–319. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CF, Zhang HC, Feng XM, Song XM, Wu YN. Knockdown of MSI1 inhibits the proliferation of human oral squamous cell carcinoma by inactivating STAT3 signaling. Int J Mol Med. 2019;44(1):115–124. doi: 10.3892/ijmm.2019.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X-Y, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9(1):221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz Y, Li F, Lambert NJ, Sokol ES, Tam W-L, Cheng AW, Airoldi EM, Lengner CJ, Gupta PB, Yu Z. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. Elife. 2014;3:e03915. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA–target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Jia Z, Liu Y, Gao Q, Han Y, Zhang G, Xu S, Cheng K. Zou W: miR-490-3p inhibits the growth and invasiveness in triple-negative breast cancer by repressing the expression of TNKS2. Gene. 2016;593(1):41–47. doi: 10.1016/j.gene.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Neel J-C, Lebrun J-J. Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal. 2013;25(7):1556–1566. doi: 10.1016/j.cellsig.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Tang H, Liu P, Yang L, Xie X, Ye F, Wu M, Liu X, Chen B, Zhang L, Xie X. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(12):3185–3197. doi: 10.1158/1535-7163.MCT-14-0243. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R, Tang JH. miR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Experimental and therapeutic medicine. 2013;5(3):813–818. doi: 10.3892/etm.2013.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan F, Wang W. MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep. 2015;12(3):4647–4651. doi: 10.3892/mmr.2015.3921. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Yang W-T, Zheng P-S. Msi1 promotes tumor growth and cell proliferation by targeting cell cycle checkpoint proteins p21, p27 and p53 in cervical carcinomas. Oncotarget. 2014;5(21):10870. doi: 10.18632/oncotarget.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uren PJ, Vo DT, de Araujo PR, Pötschke R, Burns SC, Bahrami-Samani E, Qiao M, de Sousa AR, Nakaya HI, Correa BR. RNA-binding protein Musashi1 is a central regulator of adhesion pathways in glioblastoma. Mol Cell Biol. 2015;35(17):2965–2978. doi: 10.1128/MCB.00410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNicol AM, Hardy LL, Spencer HJ, MacNicol MC. Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev Biol. 2015;15(1):15. doi: 10.1186/s12861-015-0064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and notch activation. J Cell Sci. 2010;123(19):3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 29.Clarke RB, Anderson E, Howell A, Potten CS. Regulation of human breast epithelial stem cells. Cell Prolif. 2003;36(s1):45–58. doi: 10.1046/j.1365-2184.36.s.1.5.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jf Z, He M, Wm F, Wang H, Lz C, Zhu X, Chen Y, Xie D, Lai P, Chen G. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54(6):2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 31.Que T, Song Y, Liu Z, Zheng S, Long H, Li Z, Liu Y, Wang G, Zhou J, Zhang X. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34(38):4952. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 32.Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F, Kondoh H. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One. 2013;8(10):e76247. doi: 10.1371/journal.pone.0076247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504(1):277–282. doi: 10.1016/j.bbrc.2018.08.172. [DOI] [PubMed] [Google Scholar]
- 34.Gómez-Esquer F, Agudo D, Martínez-Arribas F, Núñez-Villar M, Pollán M, Schneider J. The X-chromosome RBMX gene is expressed in mammary carcinoma. Cancer Genomics-Proteomics. 2004;1(1):39–44. [PubMed] [Google Scholar]
- 35.Martínez-Arribas F, Agudo D, Pollán M, Gómez-Esquer F, Díaz-Gil G, Lucas R, Schneider J. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97(6):1275–1282. doi: 10.1002/jcb.20725. [DOI] [PubMed] [Google Scholar]
- 36.Léveillé N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Vrielink JO, Le Sage C, Melo CA, Horlings HM, Wesseling J. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem. 2011;286(48):41442–41454. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang F, Zhang R, He Y, Zou M, Guo L, Xi T. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One. 2012;7(5):e35435. doi: 10.1371/journal.pone.0035435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nie J, Jiang H-C, Zhou Y-C, Jiang B, He W-J, Wang Y-F, Dong J. MiR-125b regulates the proliferation and metastasis of triple negative breast cancer cells via the Wnt/β-catenin pathway and EMT. Biosci Biotechnol Biochem. 2019;83(6):1062–1071. doi: 10.1080/09168451.2019.1584521. [DOI] [PubMed] [Google Scholar]
- 40.Hong L, Pan F, Jiang H, Zhang L, Liu Y, Cai C, Hua C, Luo X, Sun J. Chen Z: miR-125b inhibited epithelial–mesenchymal transition of triple-negative breast cancer by targeting MAP 2K7. OncoTargets and therapy. 2016;9:2639. doi: 10.2147/OTT.S99826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8(1):91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The results of GEO dataset GSE70480 for miRNAs selection. Related accession: GSM1782670 is for MCF-10A as normal breast cell line, GSM1782672 is for MCF-7 and GSM1782680 is for T-47D.
Additional file 2: Table S1. microRNA target sites in MSI1 3`UTR.
Additional file 3: Figure S2. MCF-7 cell line were transfected with miR-125b.pBUD and USc.pBUD expression plasmids. EGFP reporter gene indicated the accuracy of transfection.
Additional file 4: Figure S3. Reduction of MSI1 intensity in MCF-7 cell line which transfected with miR-125b.pBUD. The separated area demonstrated EGFP positive cells with decreased intensity of MSI1 as a result of overexpression of miR-125b.
Additional file 5: Figure S4. MSI1 intensity in MCF-7 cell line which transfected with (USc.pBUD) plasmid. Selected area shows EGFP positive and negative cells which have the same intensity for MSI1.
Additional file 6: Figure S5. Flow cytometry data confirmed reduction of MSI1 intensity in MCF-7 cell line after transfection with miR-125b.pBUD.
Additional file 7: Figure S6. Agarose electrophoresis of double digestion products of recombinant plasmids [pBUD/precursor miRNAs expression plasmids] which were digested with SalI and XbaI restriction enzymes. Double digestion with SalI and XbaI resulted in two distinct bands with approximate sizes of 5191 bp for plasmid pBUD and 378 bp for precursor of miR-637, 453 bp for precursor of miR-802 and 427 bp for precursor of miR-125b. M is 1kbp DNA ladder (Thermo Scientific, USA).
Additional file 8: Figure S7. Schematic representation of recombinant plasmids for ectopic expression of selected miRNAs which is involved miRNAs precursors and recombinant vector included scramble sequence as negative control.
Additional file 9: Figure S8. Agarose electrophoresis of double digestion products of recombinant plasmids [pSICHECK2.MCS.MSI1 plasmids] which were digested with XhoI and NheI restriction enzymes and single digestion with XhoI restriction enzyme. XhoI-linearized plasmid with a single band about 8053 bp and double digested plasmid fragments 5292 bp and 2761 bp. M is 1kbp DNA ladder (Thermo Scientific, USA).
Additional file 10: Figure S9. Schematic representation of pSICHECK2 plasmid which included MSI1 wild type and mutated MSI1 for direct binding of particular miRNAs.
Data Availability Statement
Supporting and raw data are available upon a reasonable request to the corresponding author.