Abstract
Experimental evidence points to the role of Zinc fingers and homeoboxes protein 1 and 2 (ZHX1 and ZHX2) in the development and progression of several types of cancer, including hematological malignancies. Here, we determined whether the altered expression of ZHX1 and ZHX2 has clinical implications in patients with CLL. Interestingly, CLL patients with low expression ZHX1 and ZHX2 presented higher WBC counts. Importantly, our data showed that CLL patients with cytogenetic alterations presented reduced transcriptional levels of ZHX1 and ZHX2 in comparison with patients with normal karyotype. Moreover, when stratifying CLL patients according to the karyotype prognosis value, we observed that the expression of ZHX1 and ZHX2 was significantly reduced in CLL patients presenting adverse karyotypes. Finally, we stratified patients according to the number of chromosomal aberrations and observed a negative association between ZHX1 and ZHX2 expression and the accumulation of chromosomal abnormalities in CLL patients. Our data showed that the low expression of ZHX1 and ZHX2 is associated with a worse prognosis in CLL, followed by a greater number of leukemic cells and unfavorable cytogenetics findings in the diagnosis. Further studies will be important to confirm the prognostic value of ZHX1 and ZHX2 in independent CLL cohorts.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40364-021-00263-2.
Keywords: ZHX1, ZHX2, Chronic lymphocytic leukemia, Karyotype, Prognosis
To the Editor,
Zinc fingers and homeoboxes protein 1 and 2 (ZHX1 and ZHX2) are two members of the zinc-finger and homeobox families, which act as transcriptional repressors. Experimental evidence points to the role of such ZHX members in the development and progression of several types of cancer, including hematological malignancies [1]. Nevertheless, the expression of ZHX members on CLL remains unknown.
In this article, we determined whether the altered expression of ZHX1 and ZHX2 has clinical implications in patients with CLL. Initially we accessed the BloodSpot database by the International Microarray Innovations in Leukemia (MILE) study group [2], which was performed in a cohort of 73 healthy bone marrow samples and 448 CLL patients. Additionally, ZHX1 and ZHX2 expression was determined by quantitative real-time PCR in peripheral blood mononuclear cells (PBMC) obtained from 51 CLL patients, diagnosed according to Matute’s score, and treated at the University Hospital of the Medical School of Ribeirão Preto (University of São Paulo, Brazil). For comparison purposes, 8 samples of peripheral blood B-sorted cells from age-matched healthy volunteers were included. Flow cytometry analysis of ZAP70 expression was performed, following the identification of the CD19+ CD5+ population. Patients presenting at least 20% of leukemic cells expressing ZAP-70 were designated as “ZAP-70 positive”. Cytogenetic analysis was performed by G-banding and according to the results, the prognostic classification of this cohort was determined as favorable, intermediate, and adverse [3, 4]. The statistical analysis is described in the supplementary information file.
Data from the MILE study group showed that ZHX1 mRNA levels were increased in CLL (p ≤ .0001), regardless of the probe analyzed. Using the probe 203556_at, no differences regarding the expression of the ZHX2 gene between CLL and healthy bone marrow samples were found (p > 0.05). Conversely, the probe 1557706_at revealed an increased expression of this transcript in CLL samples (p = 0.002). In order to validate these data and explored possible prognostic implications in CLL, we decided to investigate the levels of expression of ZHX1 and ZHX2 in 51 patients with CLL and 8 B-cell samples obtained from healthy donors. The median age of patients was 65 years (range: 43–85), and 21 of them were female (41%). Overall, 35 patients were classified as Binet A, followed by 11 Binet B, and 5 Binet C. The expression of the ZHX1 (p > 0.05) and ZHX2 (p > 0.05) genes were similar between CLL and control samples (supplementary Fig. 1). Although we have not explored the functions of ZHX family members at a functional level, we speculate that such contrasting data between in silico analysis and our validation can be explained by the differences between control samples. While the bloodspot database uses bone marrow cells, the primary samples from our court were previously purified B-cell samples.
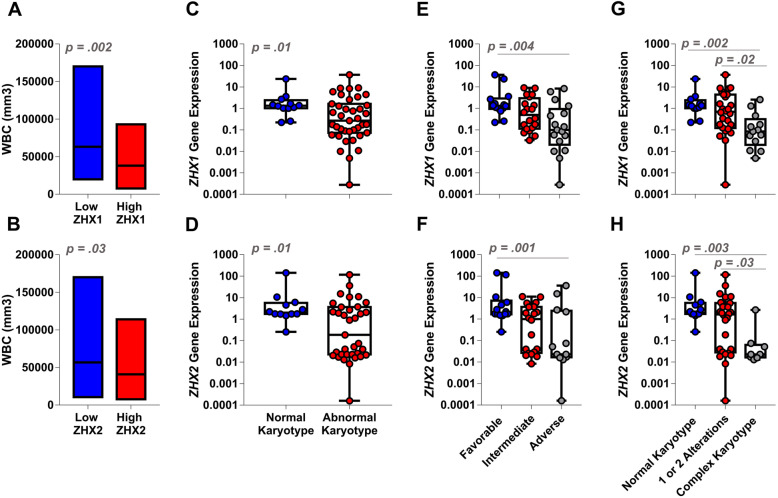
In order to compare the clinical and laboratorial features of CLL patients grouped according to ZHX1 and ZHX2 transcript levels, we adopted the median value of ZHX1 and ZHX2 expression as the cut-off level. Clinical and baseline characteristics revealed that patients with low ZHX1 expression had higher leukocyte (WBC) counts (p = 0.03) and more frequent karyotype alterations (p = 0.007) indicatives of intermediate and adverse prognosis (p = 0.002). Patients with low ZHX2 expression also showed karyotype changes (p = 0.04) categorized into intermediate and adverse risk groups (p = 0.09). Furthermore, female patients tended to have lower ZHX2 expression (p = 0.03) (Table 1). Differential expression of ZHX1 and ZHX2 had no impact on Binet stage, platelet number, and expression of ZAP-70 protein (p > 0.05). However, when patients with CLL were dichotomized according to the level of expression of ZHX1 and ZHX2, patients with low expression ZHX1 and ZHX2 presented higher WBC counts (p = 0.002 and p = 0.03, respectively). Importantly, our data showed that CLL patients with cytogenetic alterations presented reduced transcriptional levels of ZHX1 and ZHX2 in comparison with patients with normal karyotype (p = 0.01). Moreover, when stratifying CLL patients according to the karyotype prognosis value (favorable, intermediate, and adverse), we observed that the expression of ZHX1 and ZHX2 was significantly reduced in CLL patients presenting adverse karyotypes (p = 0.004 and p = 0.01, respectively). Finally, we stratified patients according to the number of chromosomal aberrations (0 aberrations: normal karyotype, 1–2 alterations, and ≥ 3 aberrations: complex karyotype) and observed a negative association between ZHX1 and ZHX2 expression and the accumulation of chromosomal abnormalities in CLL patients (p = 0.002 and p = 0.003, respectively) (Fig. 1).
Table 1.
Clinical and Baseline characteristics
| Characteristics | All Patients (%) | ZHX 1 | p | ZHX2 | p | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
|
Age, y Median (range) |
65 (43, 85) | 65.5 (54, 80) | 59 (43, 85) | .44 | 66.5 (54, 80) | 59 (43, 85) | .15 |
| Sex | .24 | .01 | |||||
| Female | 21 (41.1) | 9 (34.6) | 12 (48) | 5 (20.8) | 13 (56.5) | ||
| Male | 30 (58.9) | 17 (65.4) | 13 (52) | 19 (79.2) | 10 (43.5) | ||
| Binet | .26 | .60 | |||||
| A | 35 (68.7) | 17 (68) | 18 (69.2) | 16 (48.5) | 17 (51.5) | ||
| B | 11 (21.5) | 4 (16) | 7 (27) | 5 (50) | 5 (50) | ||
| C | 5 (9.8) | 4 (16) | 1 (3.8) | 3 (75) | 1 (25) | ||
| Platelets | .16 | .55 | |||||
| median (range), X109 /L | 142.5 (11, 312) | 137.5 (15, 261) | 147 (11, 312) | 141.5 (15, 312) | 140 (11, 284) | ||
| WBC | .03 | .15 | |||||
| median (range), X109 /L | 45.2 (7.8, 170.1) | 61.1 (29, 170.1) | 23.4 (7.8, 93.2) | 45.3 (10.5, 170.1) | 30.1 (7.8, 93.2) | ||
| ZAP-70a | .18 | .37 | |||||
| Negative | 16 (32) | 6 (24) | 10 (40) | 7 (30.4) | 9 (39.1) | ||
| Positive | 34 (68) | 19 (76) | 15 (60) | 16 (69.6) | 14 (60.9) | ||
| Karyotype | .007 | .04 | |||||
| Normal | 12 (23.5) | 2 (7.7) | 10 (40) | 3 (12.5) | 9 (39.1) | ||
| Abnormal | 39 (76.5) | 24 (92.3) | 15 (50) | 21 (87.5) | 14 (60.1) | ||
| Cytogenetic risk | .002 | .09 | |||||
| Favorable | 14 (27.5) | 2 (7.7) | 12 (48) | 4 (16.7) | 10 (43.5) | ||
| Intermediate | 20 (39.2) | 11 (42.3) | 9 (36) | 11 (45.8) | 9 (39.1) | ||
| Adverse | 17 (33.3) | 13 (50) | 4 (16) | 9 (37.5) | 4 (17.4) | ||
| Number of karyotypic changes | .003 | .01 | |||||
| 0 | 12 | 2 (7.7) | 10 (40) | 3 (12.5) | 9 (39.1) | ||
| 1 or 2 | 26 | 13 (50) | 13 (52) | 13 (54.2) | 13 (56.5) | ||
| 3 or more | 13 | 11 (42.3) | 2 (8) | 8 (33.3) | 1 (4.4) | ||
Values represent number (percentage), or median (range) when indicated in the row headings
a Missing values were excluded for statistical analysis
Statistically significant differences are in bold
Fig. 1.
ZHX1 and ZHX2 expression in CLL and association with WBC and cytogenetic findings. CLL patients with low expression of ZHX1 (a) and ZHX2 (b) showed higher WBC counts. Low expression of ZHX1 and ZHX2 was significantly associated with abnormal (c, d), adverse (e, f), and complex (g, h) karyotype. Mann–Whitney test was employed to compare differences between two groups. Kruskal–Wallis followed by the multiple comparison test (Dunn’s) was used to compare differences between three groups
Our data showed that the low expression of ZHX1 and ZHX2 is associated with a worse prognosis in CLL, followed by a greater number of leukemic cells and unfavorable cytogenetics findings in the diagnosis [5]. Recently, several studies have been conducted to unravel the influence of members of the ZHX family on cancer. Interestingly, ZHX1 was found to be down-regulated in renal cell carcinoma, and the mRNA reduction of this gene was associated with lower survival rates [6]. ZHX1 is also downregulated in gastric cancer, and the reduced expression of this gene has been associated with other clinical parameters that are indicative of aggressive disease [7]. ZXH2 was found to have reduced expression in hepatocellular carcinoma, and the decreased levels of this gene were associated with the low survival of patients [8]. Moreover, the impact of ZHX2 on hematological neoplasms has been investigated. It was demonstrated that patients with multiple myeloma who have reduced ZHX2 expression tend to have a worse prognosis, with resistance to chemotherapy and unfavorable karyotype [9]. Besides, this gene acts as a tumor suppressor in Hodgkin lymphoma [10]. Considering our findings, we hypothesize that ZHX1 and ZHX2 may also play a role as tumor suppressor in CLL, the aberrant expression of these genes possibly contributing to an increase in the number of malignant cells and chromosomal instability.
To the best of our knowledge, our study represents the first step to demonstrate the prognostic impact of ZHX members in CLL. However, it is important to highlight that in our cohort, only 14% of the patients in the favorable prognosis group have del (13q), while in the adverse prognosis group, del (17p) and 11q23 is found in 29% of cases. We cannot rule out that some classic chromosomal changes may have been underestimated in this study. The fact that the samples were not analyzed using FISH method to identify these chromosomal alterations represents a limitation of this work. Further studies will be important to characterize the functional role of ZHX1 and ZHX2 in this leukemia and also to confirm the prognostic value of both genes in independent CLL cohorts.
Supplementary Information
Additional file 1: Fig. S1. Gene expression of ZHX1 and ZHX2 in CLL. ZHX1 gene expression was analyzed in CLL patients by accessing the probes 223213_s_at (A) and 223214_s_at (B) available in the Blood Spot database. (C) ZHX1 gene expression also was determined in CLL samples by real-time quantitative PCR. ZHX2 gene expression was analyzed in CLL patients, by accessing the probes 203556_at (D) and 1557706_at (E) available in the Blood Spot database. (F) ZHX2 gene expression also was determined in CLL samples by real-time quantitative PCR. Horizontal bars represent the median expression of the genes. Mann–Whitney test was employed to compare differences between the groups.
Acknowledgments
We are very grateful to all participants in this study.
Abbreviations
- CLL
Chronic lymphocytic leukemia
- MILE
Microarray innovations in leukemia
- PBMC
peripheral blood mononuclear cells
- ZHX 1
Zinc fingers and homeoboxes protein 1
- ZHX2
Zinc fingers and homeoboxes protein 2
Authors’ contributions
N.I.G.M and L.D.C.F.B. performed experiments, collected, analyzed, and interpreted data, and drafted the manuscript; F.A.R.N. interpreted the data, and drafted the manuscript; E.M.R. and A.R.L.A. obtained patient samples, updated the clinical data, analyzed, and interpreted data, and drafted the manuscript; F.S.A. conceived and designed the study, interpreted the data, and reviewed the manuscript. The author (s) read and approved the final manuscript.
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ).
Availability of data and materials
All data underlying the findings are fully available.
Ethics approval and consent to participate
The present study was approved by the local Ethical Committee. Written informed consent was obtained from all subjects included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu Y, Ma D, Ji C. Zinc fingers and homeoboxes family in human diseases. Cancer Gene Ther. 2015;22:223–226. doi: 10.1038/cgt.2015.16. [DOI] [PubMed] [Google Scholar]
- 2.Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sønderby CK, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44:D917–D924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rigolin GM, Cavallari M, Quaglia FM, Formigaro L, Lista E, Urso A, et al. In CLL, comorbidities and the complex karyotype are associated with an inferior outcome independently of CLL-IPI. Blood. 2017;129:3495–3498. doi: 10.1182/blood-2017-03-772285. [DOI] [PubMed] [Google Scholar]
- 4.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JA, Bomben R, Pozzo F, Tissino E, Härzschel A, Hartmann TN, et al. An updated perspective on current prognostic and predictive biomarkers in chronic lymphocytic leukemia in the context of chemoimmunotherapy and novel targeted therapy. Cancers. 2020;12. Available from: http://dx.doi.org/10.3390/cancers12040894 [DOI] [PMC free article] [PubMed]
- 6.Kwon R-J, Kim YH, Jeong DC, Han M-E, Kim J-Y, Liu L, et al. Expression and prognostic significance of zinc fingers and homeoboxes family members in renal cell carcinoma. PLoS One. 2017;12:e0171036. doi: 10.1371/journal.pone.0171036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7:6–8. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue X, Zhang Z, Liang X, Gao L, Zhang X, Zhao D, et al. Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E. Gastroenterology. 2012;142:1559–70.e2. doi: 10.1053/j.gastro.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armellini A, Sarasquete ME, García-Sanz R, Chillón MC, Balanzategui A, Alcoceba M, et al. Low expression of ZHX2, but not RCBTB2 or RAN, is associated with poor outcome in multiple myeloma. Br J Haematol. 2008;141:212–215. doi: 10.1111/j.1365-2141.2007.06956.x. [DOI] [PubMed] [Google Scholar]
- 10.Nagel S, Schneider B, Meyer C, Kaufmann M, Drexler HG, Macleod RAF. Transcriptional deregulation of homeobox gene ZHX2 in Hodgkin lymphoma. Leuk Res. 2012;36:646–655. doi: 10.1016/j.leukres.2011.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Gene expression of ZHX1 and ZHX2 in CLL. ZHX1 gene expression was analyzed in CLL patients by accessing the probes 223213_s_at (A) and 223214_s_at (B) available in the Blood Spot database. (C) ZHX1 gene expression also was determined in CLL samples by real-time quantitative PCR. ZHX2 gene expression was analyzed in CLL patients, by accessing the probes 203556_at (D) and 1557706_at (E) available in the Blood Spot database. (F) ZHX2 gene expression also was determined in CLL samples by real-time quantitative PCR. Horizontal bars represent the median expression of the genes. Mann–Whitney test was employed to compare differences between the groups.
Data Availability Statement
All data underlying the findings are fully available.