Abstract
Previously we demonstrated that fluoride increased acetylated-p53 (Ac-p53) in LS8 cells that are derived from mouse enamel organ epithelia and in rodent ameloblasts. However, how p53 is acetylated by fluoride and how the p53 upstream molecular pathway responds to fluoride is not well characterized. Here we demonstrate that fluoride activates histone acetyltransferases (HATs) including CBP, p300, PCAF and Tip60 to acetylate p53. HAT activity is regulated by post-translational modifications such as acetylation and phosphorylation. HAT proteins and their post-translational modifications (p300, Acetyl-p300, CBP, Acetyl-CBP, Tip60 and phospho-Tip60) were analyzed by Western blots. p53-HAT binding was detected by co-immunoprecipitation (co-IP). Cell growth inhibition was analyzed by MTT assays. LS8 cells were treated with NaF with/without HAT inhibitors MG149 (Tip60 inhibitor) and Anacardic Acid (AA; inhibits p300/CBP and PCAF). MG149 or AA was added 1 h prior to NaF treatment. Co-IP results showed that NaF increased p53-CBP binding and p53-PCAF binding. NaF increased active Acetyl-p300, Acetyl-CBP and phosphor-Tip60 levels, suggesting that fluoride activates these HATs. Fluoride-induced phospho-Tip60 was decreased by MG149. MG149 or AA treatment reversed fluoride-induced cell growth inhibition at 24 h. MG149 or AA treatment decreased fluoride-induced p53 acetylation to inhibit caspase-3 cleavage, DNA damage marker γH2AX expression and cytochrome-c release into the cytosol. These results suggest that acetylation of p53 by HATs contributes, at least in part, to fluoride-induced toxicity in LS8 cells via cell growth inhibition, apoptosis, DNA damage and mitochondrial damage.Modulation of HAT activity may, therefore, be a potential therapeutic target to mitigate fluoride toxicity in ameloblasts.
Keywords: HAT, Tip60, CBP/p300, PCAF, Ameloblast, Dental Fluorosis
1. Introduction
Fluoride is a natural mineral that is used for prophylaxis of dental caries (Aoba and Fejerskov 2002). U.S. Public Health Service (PHS) recommends water fluoridation and fluoride concentration of 0.7 ppm to maintain caries prevention benefits and reduce the risk of dental fluorosis (Health and Human Services Federal Panel on Community Water 2015). However, fluoride is an environmental hazardous substance when large doses are taken acutely or when lower doses are taken chronically. There are high levels of fluoride in certain groundwaters worldwide, including in India, Africa and China. High fluoride concentrations in groundwater can lead to potential fluoride contamination in drinking water. Chronic excessive exposure to fluoride can cause enamel fluorosis and skeletal fluorosis (Asawa et al. 2015). A significant number of people in geographically high fluoride areas are affected by dental and skeletal fluorosis. In addition to mineralized tissues, fluoride can affect various organs, including kidney, gastrointestinal tract and central nervous system (Zuo et al. 2018). Recently it was reported that fluoride is possibly associated with neurotoxicity leading to low intelligence quotient (IQ) in children (Green et al. 2019) and autism spectrum disorder (Strunecka and Strunecky 2019).
Over 1.5 ppm of fluoride in drinking water during enamel development can lead to dental fluorosis. Dental fluorosis is a developmental hypomineralization of tooth enamel, which manifests as mottled or discolored enamel. The severity is positively correlated to the level of fluoride exposure. (Everett 2011). The USA National Health and Nutrition Examination Survey (1986 to 2012) showed that dental fluorosis severity and prevalence were increased in 2011–2012 compared with the previous surveys performed between 1986–1987 and 1999–2004 (Neurath et al. 2019). Other than the circumvention of exposure to high concentration of fluoride during enamel development, treatments to prevent dental fluorosis remain unknown.
Ameloblasts are derived from epithelial cells and are present adjacent to the forming enamel (Bao et al. 2016). Ameloblasts are responsible for enamel formation by secreting enamel matrix proteins and directing matrix mineralization (Lacruz et al. 2017). Enamel development progresses in stages. During the secretory stage, long thin crystallites grow out from the dentin. Later during the maturation stage these crystallites grow in width and thickness and fuse into enamel rods. It is during the maturation stage that most of the mineral precipitates and this releases numerous protons. Therefore, during the maturation stage, ameloblasts are in direct contact with the acidic (around pH 6.2) mineralizing enamel matrix (Smith et al. 1996). The low extracellular pH surrounding the maturation stage ameloblasts promotes the conversion of F− to HF and HF is extremely toxic. Approximately 25-fold more HF is formed at pH 6.0 as compared to pH 7.4. The low pH environment of maturation stage facilitates entry of HF into ameloblasts to enhance fluoride-induced cell stress (Sharma et al. 2010). This suggests that compared to the secretory stage (pH ~ 7.2), the low pH environment of the maturation stage reduces the threshold dose required to induce fluoride-mediated cytotoxicity in vivo. In contrast, the in vitro cell culture environment is neutral (pH ~ 7.3), which requires a higher fluoride dose than does a low pH environment to induce fluoride-mediated cytotoxicity. We showed that at low pH, lower doses of fluoride are required to activate stress-related proteins in LS8 cells in vitro (Sharma et al. 2010). This suggests that the neutral cell culture environment in vitro requires a higher dose of fluoride. Therefore, to assess fluoride toxic effects on LS8 cells (Chen et al. 1992) in vitro, we used NaF at 5 mM, which inhibits cell growth and induces apoptosis in LS8 cells.
Previously we reported that high concentrations of fluoride (5 mM) cause cell stress, ER stress (Kubota et al. 2005; Sharma et al. 2008) and oxidative stress followed by mitochondrial damage, DNA damage and apoptosis resulting in impairment of ameloblast function (Suzuki et al. 2015; Suzuki et al. 2014). Recently we demonstrated that fluoride induced acetylation of p53 (Ac-p53) in LS8 in vitro and rat ameloblasts in vivo (Suzuki et al. 2018). p53 is a transcription factor and responds to various genotoxic damage, cellular stress and affects many important cellular processes including proliferation, DNA repair, programmed cell death (apoptosis), autophagy, metabolism, and cell migration (Vousden and Prives 2009). p53 protein is regulated by post-translational modifications, including phosphorylation, acetylation, methylation and ubiquitination (Meek and Anderson 2009). Acetylation of p53 (Ac-p53) increases p53 protein half-life. The histone acetyltransferase (HAT) acetylates p53 to induce p53 downstream target genes, including p21. Ac-p53 is necessary for cell cycle checkpoint response to DNA damage and Ac-p53 regulates apoptosis. Acetylation of p53 is mediated by the HATs; CBP, p300, PCAF, MOZ, MOF and Tip60 (Reed and Quelle 2015). CREB-binding protein (CBP) and its homolog p300 are highly conserved and functionally related transcriptional co-activators and histone acetyltransferases. PCAF is CBP/p300 associate factor. CBP/p300 and PCAF promote a p53 open conformation to acetylate p53 and enhance p53 transcriptional activity, leading to growth arrest and/or apoptosis (Reed and Quelle 2015). Acetylation of CBP (Ac-CBP) at Lys1535 and acetylation of p300 (Ac-p300) at Lys1499 are known to enhance their HAT activity, thereby Ac-CBP/Ac-p300 can increase Ac-p53 levels (Ghazi et al. 2017). CBP/p300 and PCAF HAT activity is inhibited by Anacardic acid (AA) from cashew nut shell liquid (Balasubramanyam et al. 2003). In response to DNA damage, phosphorylated-Tip60 (p-Tip60) [Ser86] promotes its HAT activity, thereby inducing p53 acetylation (Reed and Quelle 2015). Tip60 and MOF-mediated acetylation of p53 selectively increases p53 binding to apoptotic gene promoters, including Bax and PUMA to induce cell death (Li et al. 2009; Sykes et al. 2006; Tang et al. 2006). The 6-alkylsalicylate (MG149) is a potent HAT inhibitor for Tip60 and MOF (van den Bosch et al. 2017). Recently we characterized the Ac-p53 downstream pathway in fluoride toxicity. Fluoride-mediated Ac-p53 increased expression of downstream targets p21 and Mdm2 and the p53/Mdm2/p21 pathway promoted fluoride toxicity (Deng et al. 2019). However, how p53 is acetylated by fluoride and how the p53 upstream molecular pathway responds to fluoride toxicity is not well characterized. Here we demonstrate that fluoride activates HATs including CBP/p300, PCAF and Tip60 that then acetylate p53 to promote fluoride toxicity in LS8 cells.
2. Materials and Methods
2.1. Cell Culture
LS8 cells derived from the mouse enamel organ epithelia (Chen et al. 1992) were maintained in alpha minimal essential medium with GlutaMAX (Thermo Fisher Scientific, Waltham, MA) supplemented with fetal bovine serum (10%) and sodium pyruvate (1 mM). Cells were treated with sodium fluoride (NaF; Thermo Fisher Scientific) with/without Anacardic Acid (AA; inhibitor of CBP/p300 and PCAF) and MG149 (Inhibitor of Tip60) as indicated (Selleck Chemicals, Houston, TX).
2.2. Cell proliferation assay
MTT assays were performed to quantify cell proliferation. Cells were cultured in 96-well plates overnight. Cells were treated with indicated concentrations of NaF with/without Anacardic Acid or MG149. After 24 h, MTT reagent was added into each well to incubate for 3 h followed by taking pictures of cells. After that, media was removed and DMSO was added to lyse cells with shaking on orbital shaker for 15–30 min and OD was measured at 590 nm.
2.3. Western blot analysis
After LS8 cells were lysed, total proteins were extracted with RIPA buffer (Thermo Fisher Scientific) with Thermo Scientific Halt protease inhibitor cocktail. Mitochondrial and cytosolic fractions were extracted using mitochondrial isolation kit for cultured cells (Thermo Fisher Scientific). Protein concentration was measured by BCA protein assay kit (Thermo Fisher Scientific). Equal amounts of protein sample were loaded into Mini-Protean TGX gels (Bio-Rad, Hercules, CA) and transferred to nitrocellulose filter membranes followed by blocking in nonfat dry milk (5%) or BSA (5%) for 1 h at RT. Membranes were incubated with the primary antibodies overnight at 4 °C. The following primary antibodies were used; rabbit anti-acetylated p53 (Lys379), rabbit anti-cleaved caspase 3, rabbit anti-γH2AX, rabbit anti-PCAF, rabbit anti-acetylated P300/CBP, rabbit anti-Tip60, rabbit anti-beta actin (Cell Signaling Technology, Boston, MA), and rabbit anti-phosphorylated Tip60 (Ser86) (Thermo Fisher Scientific). After antibody treatment, the membranes were incubated with the HRP-conjugated anti-rabbit IgG secondary antibodies (Bio-Rad) for 1 h at RT. Enhanced chemiluminescence was performed with SuperSignal West Pico (Thermo Fisher Scientific). Signal was detected by myECL imager (Thermo Fisher Scientific). Bands were quantified by MyImage analysis software (Thermo Fisher Scientific). Representative images are shown in the Results section. Each experiment was performed at least three times and Relative protein expression and statistical significance were analyzed by one-way analysis of variance (ANOVA) with Fisher’s least significant difference (LSD) post-hoc test using the SPSS statistics 20 software (version 20). Statistical analyses of relative protein expression are shown in Supplementary figures 1–6.
2.4. Real time quantitative PCR (qPCR) analysis
Total RNA was extracted from LS8 cells (Direct-zol RNA Mini Prep, Zymo Research Corp, Irvine, CA) to synthesize cDNA using iScript (Bio-Rad). Real-time qPCR amplification was performed on QuantStudio 3 (Thermo Fisher Scientific). Primers were; Bax (NM 007527.3), forward: 5’-AGCTGCCACCCGGAAGAAGACCT-3’, reverse: 5’-CCGGCGAATTGGAGATGAACTG-3’; Bcl-2 (NM 009741.5), forward: 5’- TCAGGCTGGAAGGAGAAGATG-3’, reverse: 5’- TGTCACAGAGGGGCTACGAGT-3’; Gapdh (NM 001289726), forward: 5’- GCAAAGTGGAGATTGTTGCCAT-3’, reverse: 5’-CCTTGACTGTGCCGTTGAATTT-3’.
The qPCR data were analyzed using the 2-ΔΔCT method (Pfaffl 2001). At least three biological replicates were analyzed for each experiment.
2.5. Immunoprecipitation
The co-IP assay was performed using the Pierce™ Co-immunoprecipitation Kit (Thermo Fisher Scientific). Cells were lysed and protein were extracted using ice-cold IP Lysis/Wash Buffer. Protein concentration was determined by BCA protein assay kit (Thermo Fisher Scientific). The samples were then split. 20 μg protein was used for general tryptic digestion to determine the amounts of total p53, CBP and PCAF. 1 mg of total protein was used for co-IP. Proteins were incubated with mouse anti-p53, rabbit anti-CBP, or negative control mouse- or rabbit-IgG (Cell Signaling Technology), Immunocomplexes were immunoprecipitated using protein G-agarose beads (Thermo Fisher Scientific). The immunocomplexes were analyzed by Western blot and probed with antibodies; rabbit anti-p53, rabbit anti-PCAF, mouse anti-p53 or mouse anti-CBP. At least three biological replicates for each experiment were performed and representative images are shown.
2.6. Statistical Analysis
Data were analyzed by one-way analysis of variance (ANOVA) with Fisher’s least significant difference (LSD) post-hoc test using the SPSS statistics 20 software (version 20). Significance was assessed at P < 0.05.
3. Results
3.1. Fluoride increased CBP-p53 and PCAF-p53 binding and Anacardic Acid (AA) inhibited fluoride-induced p53 acetylation in LS8 cells.
CBP/p300 and PCAF can directly bind to p53 and induce p53 acetylation (Reed and Quelle 2015). Acetylated-(Ac)-CBP (Lys1535) and Ac-p300 (Lys1499) are known to enhance HAT activity, and thereby increase Ac-p53 levels (Ghazi et al. 2017). We asked if fluoride increases acetylation of CBP/p300 and PCAF in LS8 cells. Western blot results show that fluoride (5 mM) treatment for 2–6 h significantly increased Ac-CBP/p300 levels (Fig. 1A) and PCAF protein levels (Fig. 1B), suggesting that fluoride increased HAT activity of CBP/p300 and PCAF in LS8 cells. Lower concentrations of fluoride (1 mM and 3 mM) showed increasing Ac-CBP/p300 and PCAF levels with fluoride dose. Fluoride (3 mM) treatment for 2 h significantly increased Ac-CBP/p300 (P < 0.05), but there were no significant differences in Ac-CBP/p300 nor PCAF between control and 1mM or 3 mM fluoride treatments (Supplementary Fig. 2). Fluoride treatment (5 mM) for 6 h increased CBP-p53 binding (Fig. 1C) and PCAF-p53 binding (Fig. 1D). The CBP/p300 and PCAF inhibitor Anacardic Acid (AA) inhibited the fluoride-induced acetylation of p53 at 6 h (Fig. 1 E), indicating that CBP/p300 and PCAF play critical roles in fluoride-induced p53 acetylation.
Figure 1. Fluoride increased HAT activity and acetylation of p53, which was inhibited by Anacardic Acid (AA).
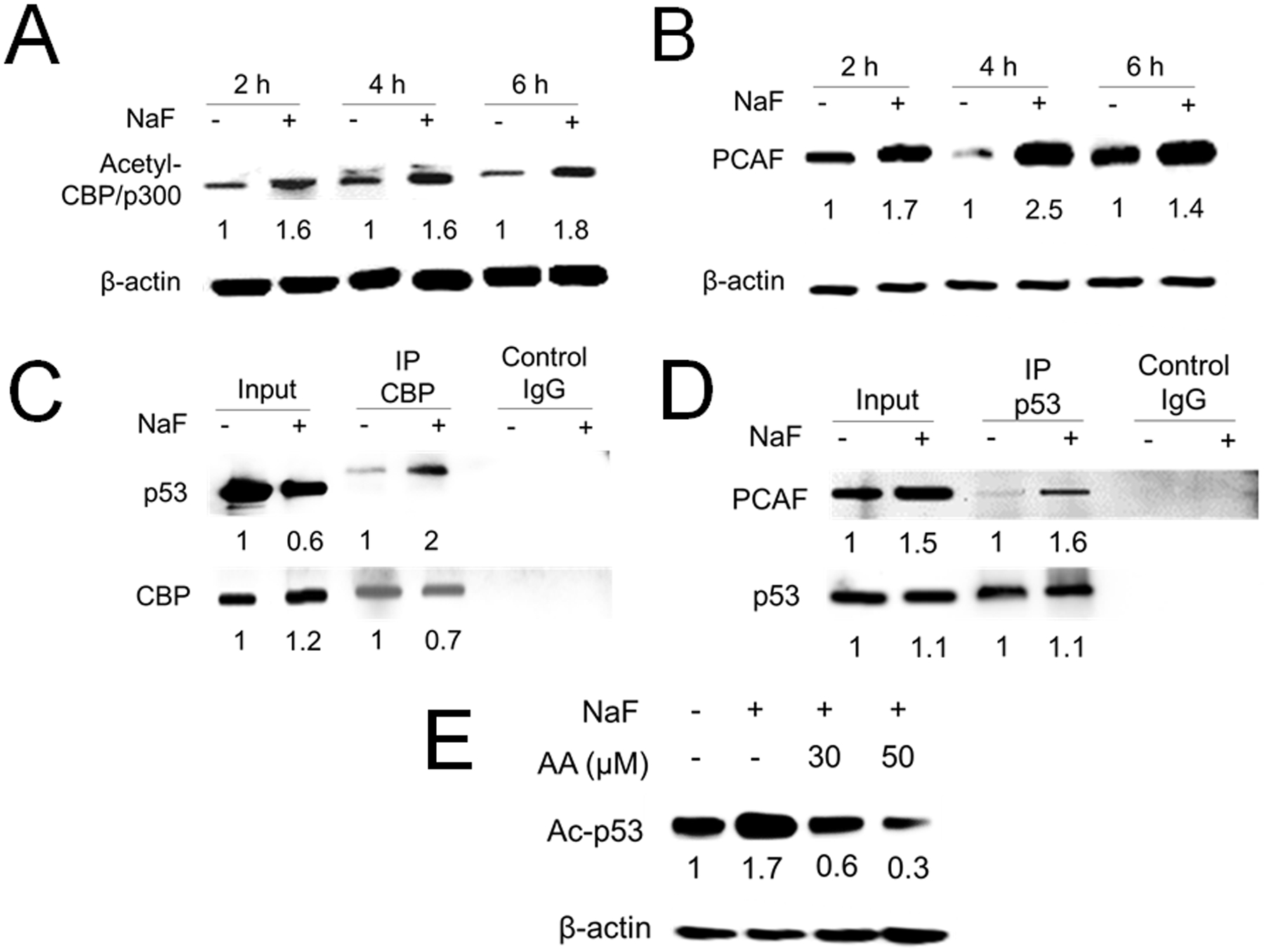
LS8 cells were treated with NaF (5 mM) for indicated times. (A) Acetyl-CBP/p300 (300 kDa) protein was quantified by western blot. Fluoride treatment significantly induced Acetyl-CBP/p300 at 2, 4 and 6 h. (B) PCAF (93 kDa) protein was quantified by western blot. Fluoride (5 mM) treatment for 4 h significantly increased PCAF protein. The numbers show the relative expression normalized by the β-actin loading control for each treatment group (44 kDa). (C) LS8 cells were treated with NaF (5 mM) for 6 h. Protein was immunoprecipitated using anti-CBP antibody and CBP-p53 binding was detected by the anti-p53 antibody. Fluoride treatment increased CBP-p53 binding. (D) LS8 cells were treated with NaF (5 mM) for 6 h. Protein was immunoprecipitated using anti-p53 antibody and PCAF-p53 binding was detected by the anti-PCAF antibody. Fluoride treatment increased PCAF-p53 binding. Control IgG was used as a negative control. The numbers show relative protein expression vs Control (0 mM NaF) for Input and separately for IP lanes. (E) LS8 cells were treated with NaF (5 mM) with/without a CBP/p300/PCAF inhibitor Anacardic Acid (AA) (30 and 50 μM) for 6 h. Acetyl-p53 (53 kDa) was detected by western blot. AA inhibited fluoride-induced acetylation of p53. The numbers show the relative expression normalized by the β-actin loading control (44 kDa). Statistical analyses of relative protein expression are shown in Supplementary Fig. 1
3.2. Fluoride-induced Tip60 phosphorylation was attenuated by MG149, which decreased fluoride-induced p53 acetylation.
Phosphorylation of Tip60 promotes its acetyltransferase activity. GSK-3 kinase (glycogen synthase kinase 3) phosphorylates Tip60 at Ser86 in response to DNA damage, thereby inducing p53 acetylation (Reed and Quelle 2015). LS8 cells were treated with NaF (5 mM) for indicated times, and phospho-(p)-Tip60 protein levels were significantly increased by NaF at 2–24 h (Fig. 2A). Treatments with lower doses of fluoride (1 mM or 3 mM) for 4 h and 18 h did not significantly increase p-Tip60 levels compared to control conditions without NaF (Supplementary Fig. 4). Tip60 inhibitor MG149 attenuated fluoride-induced p-Tip60 levels at 6–24 h (Fig. 2B). MG149 treatment significantly decreased fluoride-induced Ac-p53 at 6 h (Fig. 2C). These results suggest that p-Tip60 contributes to acetylation of p53 after fluoride treatment.
Figure 2. Fluoride induced Tip60 phosphorylation and MG149 attenuated this and attenuated acetylation of p53.
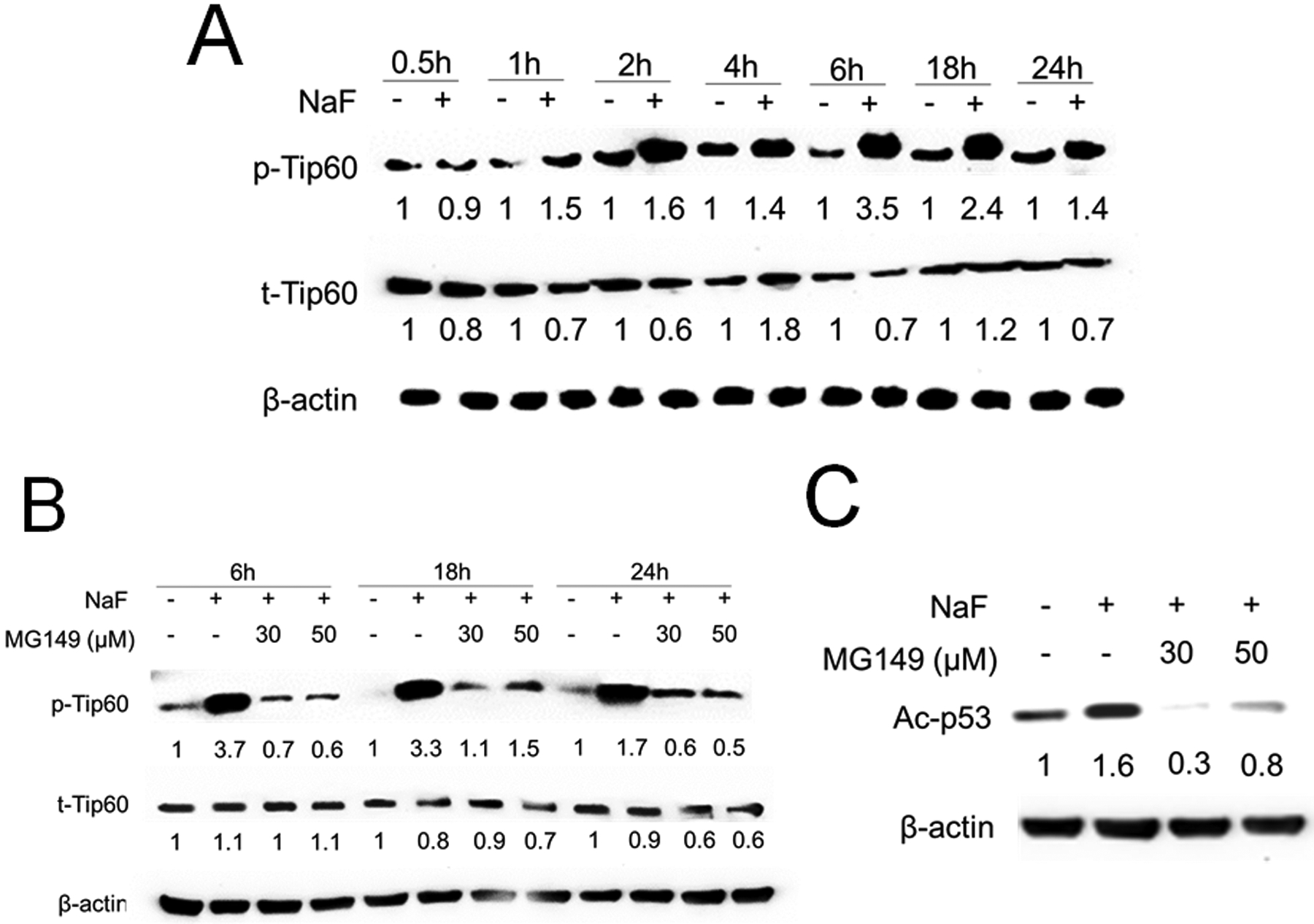
(A) LS8 cells were treated with NaF (5 mM) for indicated times. phospho-(p)-Tip60 (75 kDa) and Tip60 (60 kDa) were detected by western blot. Fluoride induced phosphorylation of Tip60 from 1 h to 24 h. (B) LS8 cells were treated with NaF (5 mM) with/without a Tip60 inhibitor; MG149 (30 and 50 μM) for 6 h, 18 h and 24 h. MG149 inhibited fluoride-induced p-Tip60 levels from 6 h to 24 h. (C) Cells were treated with NaF (5 mM) with/without MG149 for 6 h, Ac-p53 (53 kDa) was detected by western blot. MG149 inhibited fluoride-induced Ac-p53. The numbers show relative expression normalized by the β-actin loading control for each treatment group (44 kDa). Statistical analyses of relative protein expression are shown in Supplementary Fig. 3
3.3. Anacardic Acid (AA) and MG149 mitigated fluoride-induced cell growth inhibition, apoptosis and mitochondrial damage in LS8 cells.
Previously we demonstrated that fluoride induced Ac-p53 levels to increase apoptosis, DNA damage and mitochondrial damage. Decrease of Ac-p53 levels by SIRT1 overexpression protected cells from fluoride toxicity (Suzuki et al. 2018). Here, we assessed the effects of Ac-p53 inhibition by HAT inhibitors; AA and MG149 on fluoride toxicity in LS8 cells. Cells were treated with NaF (5 mM) with/without AA (10 – 50 μM) or MG149 (10 – 50 μM) for 24 h. MTT assay results show that fluoride significantly inhibited cell growth compared to control (P < 0.01). Addition of AA (at 30 μM; P < 0.01 and at 50 μM; P < 0.05) (Fig. 3A) or MG149 (at 30 μM and 50 μM; P < 0.05) (Fig. 3B) significantly increased cell growth compared to NaF alone.
Figure 3. Anacardic Acid (AA) and MG149 increased cell growth compared to NaF alone in LS8 cells.
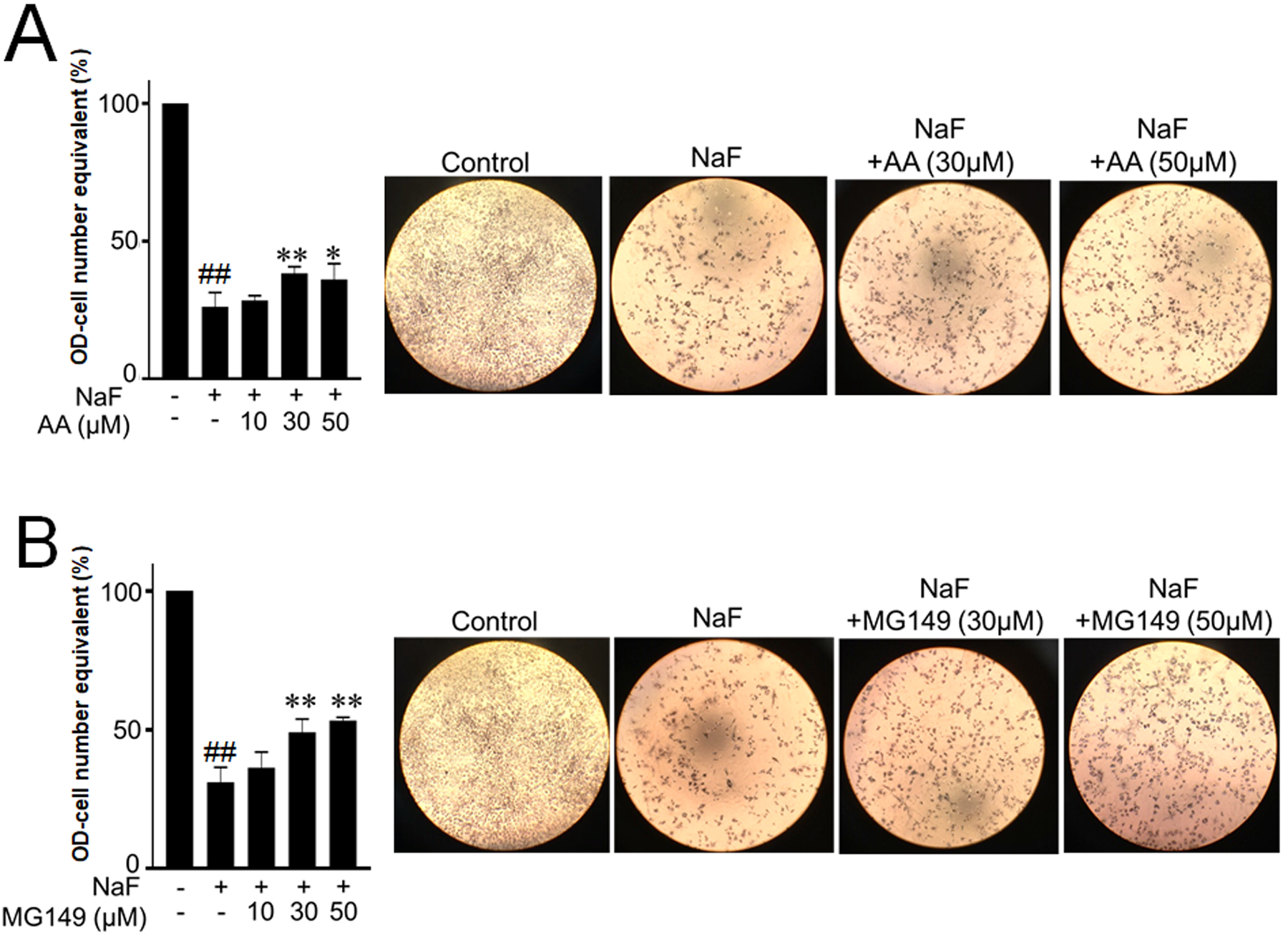
Cells were treated with NaF (5 mM) with/without AA (10–50 μM) or MG149 (10–50 μM) for 24 h. Cell proliferation was detected by MTT assays. Pictures show representative images of cells. (A) AA significantly increased cell growth at 30 μM and 50 μM compared to NaF alone. (B) MG149 increased cell growth at 30 μM and 50 μM compared to NaF alone. Data are presented as means ± SD. ##; P < 0.01 vs control, *; P < 0.05 vs NaF alone, **; P < 0.01 vs NaF alone.
Figure 4 shows the effects of AA and MG149 on fluoride-induced apoptosis and DNA damage in LS8 cells. Western blot results show that NaF (5 mM) treatment for 18 h and 24 h significantly increased cleaved-caspase-3 and DNA damage marker γH2AX. This was suppressed by AA (30 μM and 50 μM) (Fig. 4A) and by MG149 (30 μM and 50 μM) (Fig. 4B). The Bax and Bcl-2 mRNA ratio was assessed by real-time qPCR. NaF treatment significantly increased the Bax/Bcl-2 mRNA ratio compared to control at 24 h (P < 0.01). The Fluoride-induced Bax/Bcl-2 mRNA ratio was significantly suppressed by AA (30 μM and 50 μM) (Fig. 4C) or MG149 (30 μM and 50 μM) (P < 0.01) (Fig. 4D).
Fig. 4. Anacardic Acid (AA) and MG149 attenuated fluoride-induced apoptosis in LS8 cells.
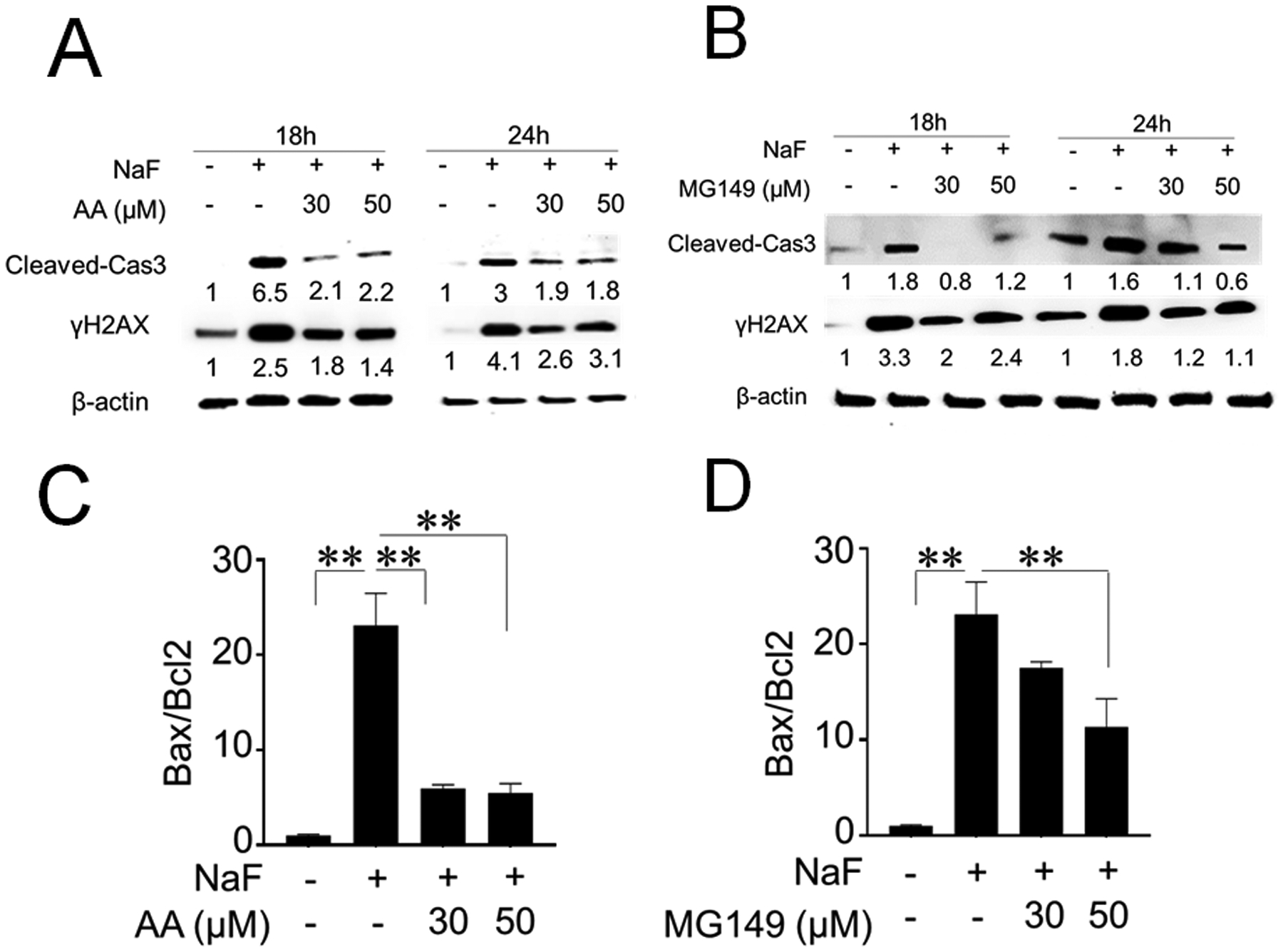
Cells were treated with NaF (5 mM) with/without AA (30 μM and 50 μM) or MG149 (30 μM and 50 μM) for 18 h and 24 h. Cleaved caspase-3 (17 kDa) and DNA damage marker γH2AX (15 kDa) expression were detected by western blot. (A) AA attenuated fluoride-induced cleavage of caspase-3 and γH2AX expression at 18 h and 24 h. (B) MG149 inhibited fluoride-induced cleavage of caspase-3 and γH2AX expression at 18 h and 24 h. The numbers show relative protein expression normalized by the β-actin loading control for each treatment group (44 kDa). Statistical analyses of relative protein expression are shown in Supplementary Fig. 5. (C) qPCR results showed that AA (30 μM and 50 μM) significantly decreased the Bax/Bcl-2 mRNA ratio compared to NaF treatment alone (P < 0.01). (D) MG149 (50 μM) significantly decreased the Bax/Bcl-2 mRNA ratio compared with NaF treatment alone (P < 0.01). Data are presented as means ± SD. **; P < 0.01.
Cytochrome-c plays an important role in mitochondrial function. Mitochondrial damage increases Cytochrome-c release into the cytosol. Fluoride treatment for 6 h significantly increased cytochrome-c release into the cytosol, while cytochrome-c levels in mitochondria were reduced. This cytochrome-c release into cytosol was attenuated by AA (Fig. 5A) or MG149 (Fig. 5B). These results suggest that inhibition of Ac-p53 by HAT inhibitors (AA or MG149) mitigates fluoride-induced cell growth inhibition, apoptosis, DNA damage and mitochondrial damage in LS8 cells.
Fig. 5. Anacardic Acid (AA) and MG149 inhibited fluoride-induced cytochrome-c release in LS8 cells.
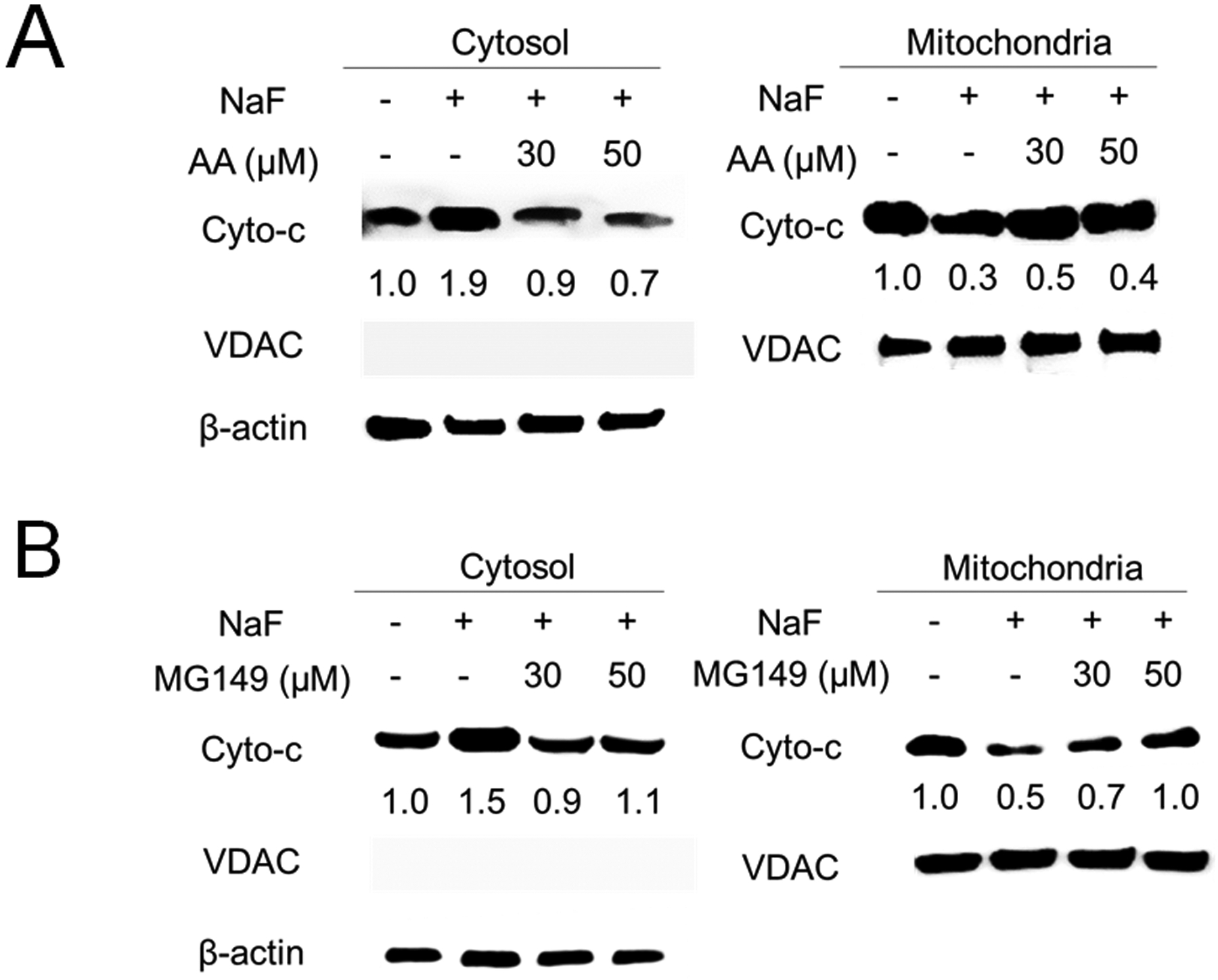
Cells were treated with NaF (5 mM) with/without AA or MG149 for 6 h. Cytochrome-c (14 kDa) in the cytosol and in the mitochondria were detected by western blot. (A) AA (30 μM and 50 μM) attenuated fluoride-induced cytochrome-c release in the cytosol. (B) MG149 (30 μM and 50 μM) inhibited fluoride-induced cytochrome-c release in the cytosol. β-actin (44 kDa) was used as a cytosol loading control. VDAC/Porin (35 kDa) was used as a mitochondrial loading control. The numbers show relative protein expression normalized by the loading control (β-actin or VDAC/Porin). Statistical analyses of relative protein expression are shown in Supplementary Fig. 6.
4. Discussion
Fluoride toxic effects and its mechanisms are reported in a variety of cell types and tissues reviewed in (Zuo et al. 2018). Fluoride-mediated cell stress including ER stress (Kubota et al. 2005) and oxidative stress (Suzuki et al. 2015) are considered as important mediators of fluoride-induced apoptosis. The p53 tumor suppressor protein exerts growth arrest and apoptosis in response to various types of cellular stress, including DNA damage (Vousden and Prives 2009). Fluoride induces apoptosis and/or cell cycle arrest via the p53 pathway in various cell types, including ameloblasts, lymphocytes, hepatocytes, lung cells and renal cells (Jiang et al. 2019a; Lin et al. 2018; Wen et al. 2019; Ying et al. 2017). Following DNA damage, p53 is acetylated at Lys379 to promote apoptosis (Sakaguchi et al. 1998). Fluoride-mediated acetylation of p53 has been reported in ameloblasts (Suzuki et al. 2018), human neuroblastoma (Tu et al. 2018) and osteoblasts (Gu et al. 2019). Deacetylation of p53 may play an important role in down-regulating p53 activity and promoting cell survival following a cell stress response. SIRT1 is a histone deacetylase and deacetylates p53 to promote cell survival (Adedara et al. 2016). Activation of SIRT1 by resveratrol or overexpression of SIRT1 decreased fluoride-induced Ac-p53 (Suzuki et al. 2018) to mitigate fluoride-induced apoptosis in LS8 cells (Suzuki and Bartlett 2014) and in neuroblastoma SH-SY5Y cells (Tu et al. 2018). These studies suggest that acetylation of p53 plays a critical role in promoting fluoride toxicity. However, the molecular mechanism of how fluoride acetylates p53 has not been well characterized. Here, we characterized the p53 upstream molecular pathway and factors responsible for fluoride-induced acetylation of p53. Our results show that HATs, including CBP/p300, PCAF and Tip60 contribute to fluoride-mediated p53 acetylation to promote fluoride toxicity (cell growth inhibition, apoptosis, DNA damage and mitochondrial damage) in LS8 cells (Summarized in Fig. 6).
Fig. 6. Schematic summary depicting fluoride-mediated HAT activation and acetylation of p53 to promote fluoride toxicity in LS8 cells.
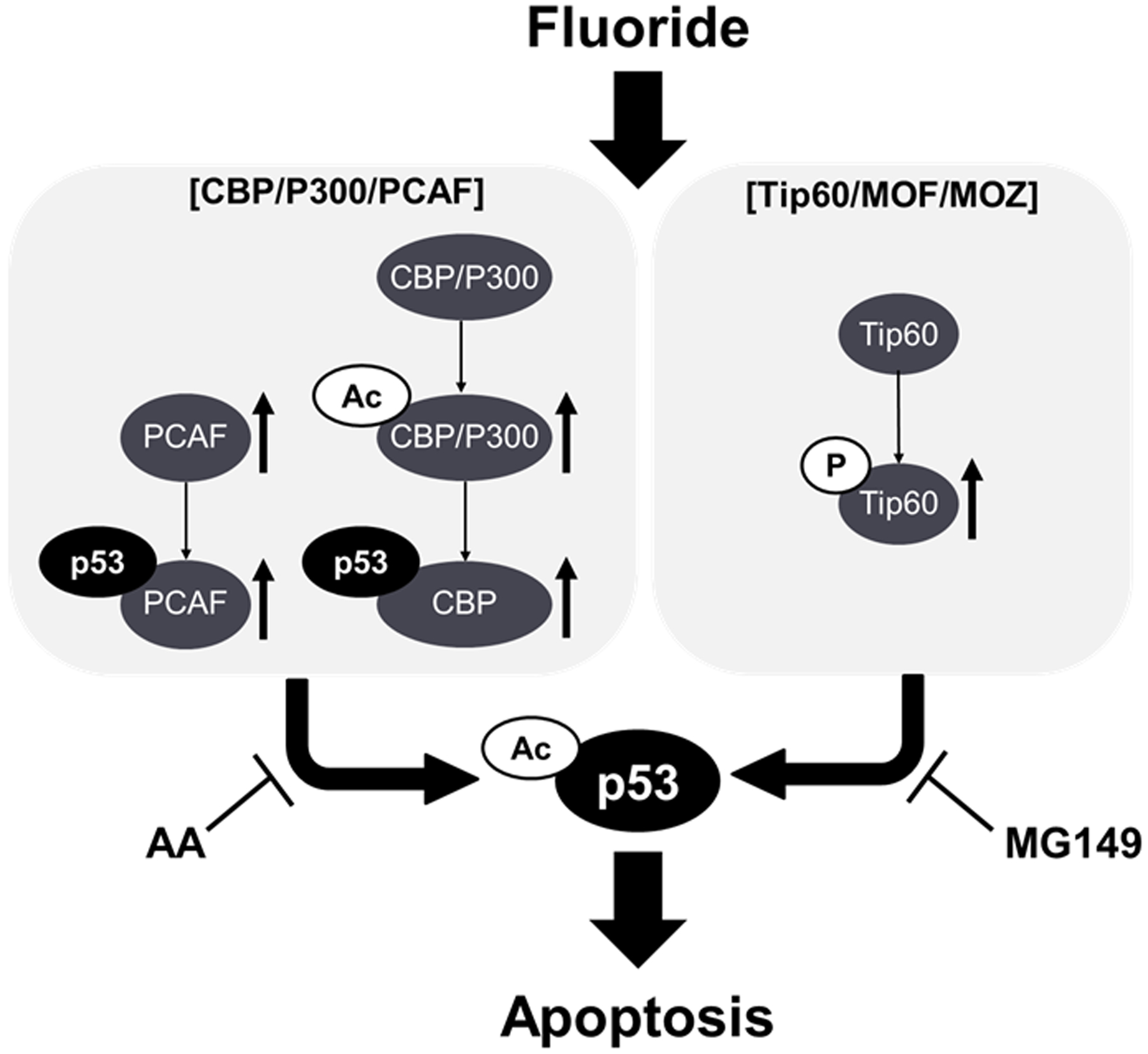
NaF activated HATs increasing Acetyl-CBP/P300 (Ac-CBP/P300), PCAF and phosho-Tip60 (p-Tip60). NaF increased p53-CBP binding and p53-PCAF binding to increase acetyl-p53 (Ac-p53). Anacradic Acid; AA (CBP/P300 and PCAF inhibitor) or MG149 (Tip60 inhibitor) mitigated fluoride toxicity including cell growth inhibition and apoptosis in LS8 cells.
Two different groups of HAT (CBP/p300/PCAF and Tip60/MOF/MOZ) acetylate p53 (Reed and Quelle 2015). Posttranslational modifications regulate CBP/p300 and Tip60 HAT activity. The acetylation of CBP [K1535] and p300 [K1499] (Ac-CBP/p300) is known to enhance CBP/p300 HAT activity and CBP/p300 can auto acetylate (Thompson et al. 2004). Phosphorylation enhances Tip60 HAT activity. Our results show that fluoride induced Ac-CBP/p300 and p-Tip60 in LS8 cells, suggesting that fluoride activates both HAT groups (CBP/p300/PCAF and Tip60) to acetylate p53. To assess fluoride toxic effects on LS8 cells, we used NaF at 5 mM to inhibit cell growth and induce apoptosis. Compared to NaF (5 mM) treatment, lower doses of NaF (1 and 3 mM), which does not induce apoptosis in LS8 cells, did not significantly increase Ac-CBP/p300 and p-Tip60 compared to control (0 mM of NaF). These results suggest that fluoride-mediated Ac-CBP/p300 and p-Tip60 participate in fluoride toxicity, including apoptosis.
Intriguingly, there were differences in response to fluoride (5 mM) between CBP/p300/PCAF and Tip60. Fluoride increased PCAF and Ac-CBP/p300 at an early phase (2 h to 6 h) and maximum effects of fluoride on PCAF and Ac-CBP/p300 were observed at 4 h and 6 h respectively. Fluoride-mediated up-regulation of PCAF and Ac-CBP/p300 was diminished at the late phase (18 h to 24 h) (data not shown). On the other hand, fluoride (5 mM) increased p-Tip60 at 1 h and maximum effect was observed at 6 h. Fluoride-mediated p-Tip60 lasted until late phase (18 h and 24 h). These results suggest that the upstream signaling response to fluoride that initiates HAT activity may differ between the two HAT groups (CBP/p300/PCAF and Tip60/MOF/MOZ).
GSK-3 kinase (glycogen synthase kinase 3) phosphorylates Tip60 at S86 in response to DNA damage, thereby inducing p53 acetylation and H4 acetylation at the PUMA promoter to increase PUMA expression, which promotes apoptosis (Charvet et al. 2011). GSK-3β plays an important role in regulating ameloblast differentiation via Wnt and TGF-β pathways (Yang et al. 2018). A recent study showed that fluoride activates GSK-3β/β-catenin signaling in rodent brain to induce fluoride-mediated neurotoxicity (Jiang et al. 2019a). Although fluoride effects on GSK-3β during enamel development remain to be elucidated, GSK-3 signaling may participate in upstream signaling of fluoride-mediated Tip60 phosphorylation and enamel malformation during amelogenesis.
Since inhibition of p53 acetylation by overexpressing SIRT1 (deacetylase) mitigated fluoride-induced toxicity (Suzuki et al. 2018), we assessed the effect of HAT inhibitors (AA and MG149) on fluoride-induced cell growth inhibition, apoptosis, DNA damage and mitochondrial damage in LS8 cells. Our results showed that AA inhibited p53 acetylation, reversed fluoride-induced cell growth inhibition, reduced cleaved-caspase 3 and DNA damage marker γH2AX levels, decreased Bax/Bcl-2 mRNA ratio, and suppressed cytochrome-c release into the cytosol. These results are in concordance with previous studies showing the effects of AA on other types of cells. Genistein-induced p53 acetylation and apoptosis were significantly diminished by AA in human non-small cell lung carcinoma (NSCLC) cell lines, A549 and H640 (Wu et al. 2016). AA down-regulated UV-enhanced Acetyl-H3 and γH2AX levels in human dermal fibroblasts (Kim et al. 2009). Recent animal studies suggest that AA might be a potent therapeutic agent to treat pathophysiological diseases, such as epilepsy (Luiz Gomes et al. 2018), cancer (Wu et al. 2011) and oxidative stress (Jiang et al. 2019b). These studies indicate that AA may be a promising agent to mitigate fluoride toxicity. MG149 (Tip60 inhibitor) can inhibit the p53 and NF-kappa B signaling pathways in myeloma cell lines (Legartová et al. 2013) and can reduce the expression of pro-inflammatory factors in mice (van den Bosch et al. 2017). Our results showed that MG149 treatment significantly inhibited fluoride-induced p-Tip60 levels and attenuated Ac-p53 to suppress fluoride toxicity in LS8 cells. These results indicate that inhibition of Tip60 may ameliorate fluoride toxicity through suppression of p53. However, AA and/or MG149 can suppress HAT activity to inhibit acetylation of both histone and non-histone proteins. This may alter subsequent signaling pathways, which could cause adverse effects. Therefore, optimization of treatment regimen and more studies are required to confirm the promising results in order to minimize untoward effects prior to therapeutic applications.
5. Conclusion
We characterized the p53 up-stream molecular pathway in LS8 cells exposed to 5 mM sodium fluoride. Results suggest that histone acetyltransferases (HATs), including CBP/p300, PCAF and Tip60 contribute, at least in part, to fluoride-mediated p53 acetylation to promote fluoride toxicity (cell growth inhibition, apoptosis, DNA damage and mitochondrial damage) in LS8 cells. Pharmacological modulation of HAT activity may be a potential therapeutic target to mitigate fluoride toxicity.
Supplementary Material
Fluoride activates histone acetyltransferase (HAT) in enamel organ-derived LS8 cells
HAT inhibitors suppressed fluoride-mediated acetylation of p53 and cell toxicity
Modulation of HAT activity may be a potential target to mitigate fluoride toxicity
Acknowledgments:
We thank Dr. Malcolm L. Snead for generously providing us with LS8 cells.
Funding: Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award number R01DE018106 (J.D.B.), R01DE027648 (M.S.) and was supported by a Seed Grant from The Ohio State University, College of Dentistry under award number 21-100300 (M.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no conflict of interest.
Declaration of competing financial interests: The authors declare they have no actual or potential competing financial interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Adedara IA, Abolaji AO, Idris UF, Olabiyi BF, Onibiyo EM, Ojuade TD, Farombi EO. 2016. Neuroprotective influence of taurine on fluoride-induced biochemical and behavioral deficits in rats. Chem Biol Interact. 261:1–10, doi: 10.1016/j.cbi.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Aoba T, Fejerskov O. 2002. Dental fluorosis: Chemistry and biology. Crit Rev Oral Biol Med. 13(2):155–170. [DOI] [PubMed] [Google Scholar]
- Asawa K, Singh A, Bhat N, Tak M, Shinde K, Jain S. 2015. Association of temporomandibular joint signs & symptoms with dental fluorosis & skeletal manifestations in endemic fluoride areas of dungarpur district, rajasthan, india. Journal of clinical and diagnostic research: JCDR. 9(12):ZC18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. 2003. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 278(21):19134–19140, doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- Bao L, Diao H, Dong N, Su X, Wang B, Mo Q, Yu H, Wang X, Chen C. 2016. Histone deacetylase inhibitor induces cell apoptosis and cycle arrest in lung cancer cells via mitochondrial injury and p53 up-acetylation. Cell Biol Toxicol. doi: 10.1007/s10565-016-9347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, Mellert H, Brandenburg M, Lindner SE, Breit B et al. 2011. Phosphorylation of tip60 by gsk-3 determines the induction of puma and apoptosis by p53. Mol Cell. 42(5):584–596, doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Couwenhoven RI, Hsu D, Luo W, Snead ML. 1992. Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch Oral Biol. 37(10):771–778, doi: 10.1016/0003-9969(92)90110-t. [DOI] [PubMed] [Google Scholar]
- Deng H, Ikeda A, Cui H, Bartlett JD, Suzuki M. 2019. Mdm2-mediated p21 proteasomal degradation promotes fluoride toxicity in ameloblasts. Cells. 8(5)doi: 10.3390/cells8050436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett ET. 2011. Fluoride’s effects on the formation of teeth and bones, and the influence of genetics. J Dent Res. 90(5):552–560, doi: 10.1177/0022034510384626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi T, Nagiah S, Tiloke C, Sheik Abdul N, Chuturgoon AA. 2017. Fusaric acid induces DNA damage and post-translational modifications of p53 in human hepatocellular carcinoma (hepg2) cells. J Cell Biochem. 118(11):3866–3874, doi: 10.1002/jcb.26037. [DOI] [PubMed] [Google Scholar]
- Green R, Lanphear B, Hornung R, Flora D, Martinez-Mier EA, Neufeld R, Ayotte P, Muckle G, Till C. 2019. Association between maternal fluoride exposure during pregnancy and iq scores in offspring in canada. JAMA Pediatr. doi: 10.1001/jamapediatrics.2019.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Z, Gao J, Han D, Zhang L, Chen P, Luo G, Han B. 2019. Sirt1 suppresses p53-dependent apoptosis by modulation of p21 in osteoblast-like mc3t3-e1 cells exposed to fluoride. Toxicol In Vitro. 57:28–38, doi: 10.1016/j.tiv.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Health USDo, Human Services Federal Panel on Community Water F. 2015. U.S. Public health service recommendation for fluoride concentration in drinking water for the prevention of dental caries. Public Health Rep. 130(4):318–331, doi: 10.1177/003335491513000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Li G, Zhou X, Wang C, Qiao Y, Liao D, Shi D. 2019a. Chronic fluoride exposure induces neuronal apoptosis and impairs neurogenesis and synaptic plasticity: Role of gsk-3beta/beta-catenin pathway. Chemosphere. 214:430–435, doi: 10.1016/j.chemosphere.2018.09.095. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhang H, Mehmood K, Li K, Zhang L, Yao W, Tong X, Li A, Wang Y, Jiang J et al. 2019b. Effect of anacardic acid against thiram induced tibial dyschondroplasia in chickens via regulation of wnt4 expression. Animals (Basel). 9(3)doi: 10.3390/ani9030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Shin JM, Eun HC, Chung JH. 2009. The role of p300 histone acetyltransferase in uv-induced histone modifications and mmp-1 gene transcription. PLoS One. 4(3):e4864, doi: 10.1371/journal.pone.0004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Lee DH, Tsuchiya M, Young CS, Everett ET, Martinez-Mier EA, Snead ML, Nguyen L, Urano F, Bartlett JD. 2005. Fluoride induces endoplasmic reticulum stress in ameloblasts responsible for dental enamel formation. J Biol Chem. 280(24):23194–23202, doi: 10.1074/jbc.M503288200. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Habelitz S, Wright JT, Paine ML. 2017. Dental enamel formation and implications for oral health and disease. Physiol Rev. 97(3):939–993, doi: 10.1152/physrev.00030.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legartová S, Stixová L, Strnad H, Kozubek S, Martinet N, Dekker FJ, Franek M, Bártová E. 2013. Basic nuclear processes affected by histone acetyltransferases and histone deacetylase inhibitors. Epigenomics. 5(4):379–396. [DOI] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CAS, Kunkel S, Dou Y. 2009. Two mammalian mof complexes regulate transcription activation by distinct mechanisms. Molecular cell. 36(2):290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zheng L, Fan L, Kuang W, Guo R, Lin J, Wu J, Tan J. 2018. The epigenetic regulation in tooth development and regeneration. Curr Stem Cell Res Ther. 13(1):4–15, doi: 10.2174/1574888X11666161129142525. [DOI] [PubMed] [Google Scholar]
- Luiz Gomes AJ, Dimitrova Tchekalarova J, Atanasova M, da Conceicao Machado K, de Sousa Rios MA, Paz M, Gaman MA, Gaman AM, Yele S, Shill MC et al. 2018. Anticonvulsant effect of anacardic acid in murine models: Putative role of gabaergic and antioxidant mechanisms. Biomed Pharmacother. 106:1686–1695, doi: 10.1016/j.biopha.2018.07.121. [DOI] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. 2009. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol. 1(6):a000950, doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath C, Limeback H, Osmunson B, Connett M, Kanter V, Wells CR. 2019. Dental fluorosis trends in us oral health surveys: 1986 to 2012. JDR Clin Trans Res. 4(4):298–308, doi: 10.1177/2380084419830957. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time rt-pcr. Nucleic Acids Res. 29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S, Quelle D. 2015. P53 acetylation: Regulation and consequences. Cancers. 7(1):30–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12(18):2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Tsuchiya M, Bartlett JD. 2008. Fluoride induces endoplasmic reticulum stress and inhibits protein synthesis and secretion. Environ Health Perspect. 116(9):1142–1146, doi: 10.1289/ehp.11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Tsuchiya M, Skobe Z, Tannous BA, Bartlett JD. 2010. The acid test of fluoride: How ph modulates toxicity. PLoS One. 5(5):e10895, doi: 10.1371/journal.pone.0010895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Issid M, Margolis HC, Moreno EC. 1996. Developmental changes in the ph of enamel fluid and its effects on matrix-resident proteinases. Adv Dent Res. 10(2):159–169. [DOI] [PubMed] [Google Scholar]
- Strunecka A, Strunecky O. 2019. Chronic fluoride exposure and the risk of autism spectrum disorder. Int J Environ Res Public Health. 16(18)doi: 10.3390/ijerph16183431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Bandoski C, Bartlett JD. 2015. Fluoride induces oxidative damage and sirt1/autophagy through ros-mediated jnk signaling. Free Radic Biol Med. 89:369–378, doi: 10.1016/j.freeradbiomed.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Bartlett JD. 2014. Sirtuin1 and autophagy protect cells from fluoride-induced cell stress. Biochim Biophys Acta. 1842(2):245–255, doi: 10.1016/j.bbadis.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ikeda A, Bartlett JD. 2018. Sirt1 overexpression suppresses fluoride-induced p53 acetylation to alleviate fluoride toxicity in ameloblasts responsible for enamel formation. Arch Toxicol. 92(3):1283–1293, doi: 10.1007/s00204-017-2135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Sierant ML, Antone JV, Everett ET, Whitford GM, Bartlett JD. 2014. Uncoupling protein-2 is an antioxidant that is up-regulated in the enamel organ of fluoride-treated rats. Connect Tissue Res. 55 Suppl 1:25–28, doi: 10.3109/03008207.2014.923854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Molecular cell. 24(6):841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Molecular cell. 24(6):827–839. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J et al. 2004. Regulation of the p300 hat domain via a novel activation loop. Nat Struct Mol Biol. 11(4):308–315, doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Tu W, Zhang Q, Liu Y, Han L, Wang Q, Chen P, Zhang S, Wang A, Zhou X. 2018. Fluoride induces apoptosis via inhibiting sirt1 activity to activate mitochondrial p53 pathway in human neuroblastoma sh-sy5y cells. Toxicol Appl Pharmacol. 347:60–69, doi: 10.1016/j.taap.2018.03.030. [DOI] [PubMed] [Google Scholar]
- van den Bosch T, Leus NG, Wapenaar H, Boichenko A, Hermans J, Bischoff R, Haisma HJ, Dekker FJ. 2017. A 6-alkylsalicylate histone acetyltransferase inhibitor inhibits histone acetylation and pro-inflammatory gene expression in murine precision-cut lung slices. Pulmonary pharmacology & therapeutics. 44:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: The growing complexity of p53. Cell. 137(3):413–431. [DOI] [PubMed] [Google Scholar]
- Wen P, Wei X, Liang G, Wang Y, Yang Y, Qin L, Pang W, Qin G, Li H, Jiang Y et al. 2019. Long-term exposure to low level of fluoride induces apoptosis via p53 pathway in lymphocytes of aluminum smelter workers. Environ Sci Pollut Res Int. 26(3):2671–2680, doi: 10.1007/s11356-018-3726-z. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lin YC, Chen HL, Huang PR, Liu SY, Yeh SL. 2016. The enhancing effect of genistein on apoptosis induced by trichostatin a in lung cancer cells with wild type p53 genes is associated with upregulation of histone acetyltransferase. Toxicol Appl Pharmacol. 292:94–102, doi: 10.1016/j.taap.2015.12.028. [DOI] [PubMed] [Google Scholar]
- Wu Y, He L, Zhang L, Chen J, Yi Z, Zhang J, Liu M, Pang X. 2011. Anacardic acid (6-pentadecylsalicylic acid) inhibits tumor angiogenesis by targeting src/fak/rho gtpases signaling pathway. J Pharmacol Exp Ther. 339(2):403–411, doi: 10.1124/jpet.111.181891. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li Z, Chen G, Li J, Li H, Yu M, Zhang W, Guo W, Tian W. 2018. Gsk3beta regulates ameloblast differentiation via wnt and tgf-beta pathways. J Cell Physiol. 233(7):5322–5333, doi: 10.1002/jcp.26344. [DOI] [PubMed] [Google Scholar]
- Ying J, Xu J, Shen L, Mao Z, Liang J, Lin S, Yu X, Pan R, Yan C, Li S et al. 2017. The effect of sodium fluoride on cell apoptosis and the mechanism of human lung beas-2b cells in vitro. Biol Trace Elem Res. 179(1):59–69, doi: 10.1007/s12011-017-0937-y. [DOI] [PubMed] [Google Scholar]
- Zuo H, Chen L, Kong M, Qiu L, Lu P, Wu P, Yang Y, Chen K. 2018. Toxic effects of fluoride on organisms. Life Sci. 198:18–24, doi: 10.1016/j.lfs.2018.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


