Abstract
Plants in the genus Euphorbia produce a wide variety of pharmacologically active diterpenoids with anticancer, multidrug resistance reversal, and antiviral properties. Some are the primary industrial source of ingenol mebutate, which is approved for treatment of the precancerous skin condition actinic keratosis. Similar to other high value phytochemicals, Euphorbia diterpenoids accumulate at low concentrations in planta and chemical synthesis produces similarly low yields. We established genetically transformed root cultures of Euphorbia lathryis as a strategy to gain greater access to diterpenoids from this genus. Transformed roots produced via stem explant infection with Agrobacterium rhizogenes strain 15834 recapitulated the metabolite profiles of field-grown plant roots and aerial tissues. Several putative diterpenoids were present in transformed roots, including ingenol and closely related structures, indicating that root cultures are a promising approach to Euphorbia-specific diterpenoid production. Treatment with methyl jasmonate led to a significant, albeit transient increase in mRNA levels of early diterpenoid biosynthetic enzymes (farnesyl pyrophosphate synthase, geranylgeranyl pyrophosphate synthase, and casbene synthase), suggesting that elicitation could prove useful in future pathway characterization and metabolic engineering efforts. We also show the potential of transformed Euphorbia root cultures for natural product drug discovery applications by measuring their cytotoxic activities using a panel of human carcinoma cell lines derived from prostate, cervix, breast, and lung.
Keywords: Euphorbia, Agrobacterium, diterpene, anticancer, ingenol, specialized metabolism
1. INTRODUCTION
Diterpenoids from plants in the genus Euphorbia (family: Euphorbiaceae) are in the focus of natural product drug discovery due to their broad range of pharmacological activities. Over 650 unique structures have been reported (Vasas and Hohmann, 2014; Ernst et al. 2019), a number of which have shown potential as drug leads (Ernst et al., 2016; Saklani and Kutty, 2008). Multiple plant species within the genus Euphorbia contain the diterpenoid ingenol, which is generally present in conjugated forms and substituted by various carboxylic acids (Béres et al., 2018). Some naturally occurring ingenol conjugates possess anticancer and anti-HIV properties (Abreu et al., 2014; Vasas et al., 2012), whereas semi-synthetic derivatives show potential for increased efficacy (Silva et al., 2019). Ingenol mebutate, a naturally occurring ester derivative containing angelic acid, has gained significant attention due to its approval by the Food and Drug Administration in 2012 and the European Medicines Agency in 2013 for treatment of the precancerous skin condition, actinic keratosis (Braun et al., 2018; Martin and Swanson, 2013; Ogbourne and Parsons, 2014; Ortega Del Olmo and Salido-Vallejo, 2018). Originally isolated from Euphorbia peplus, ingenol mebutate has been extensively studied for its potent antitumor and antileukemic activities (Alchin, 2014; Doan et al., 2012). Current supply is limited because of its low-yield extraction from from E. peplus biomass (Hohmann et al., 2000), but it is also produced semi-synthetically starting from ingenol (Liang et al., 2012). An early total synthesis of ingenol involved 43 steps and yielded 0.007% (Winkler et al., 2002), whereas a recent 14-step semi-synthesis yields around 1% (Jørgensen et al., 2013). As such, the low yields via plant extraction and challenging synthetic routes have prompted investigation of alternative, economically viable sources of ingenol. One strategy for improving access to ingenol and other pharmaceutically relevant Euphorbia diterpenoids is to apply biotechnological approaches (Atanasov et al., 2015).
The precursor of all plant diterpenoids is geranylgeranyl pyrophosphate (GGPP), a C20 branched hydrocarbon chain bound to an ionizable diphosphate group (Cheng et al., 2007). Casbene synthase catalyzes the first step in taxonomically-restricted Euphorbiaceae diterpenoid biosynthesis via the cyclization of GGPP into casbene (Kirby et al., 2010). From casbene, the downstream biosynthetic steps involved in producing bioactive Euphorbia diterpenoids are poorly understood, but apparently proceed through intermediates such as jolkinol via cytochrome P450-catalyzed oxidations and a short-chain alcohol dehydrogenase (King et al., 2014; Luo et al., 2016; Zerbe et al., 2013). Engineering metabolic pathways or reconstructing them in heterologous hosts requires characterization of the biosynthetic steps that underlie metabolite production in the source plant (Gandhi et al., 2015; Kitaoka et al., 2015). Microbial hosts are attractive production platforms, however diterpenoid production in microbes is lagging behind other phytochemicals, with fewer studies and lower yields. Plant tissue cultures express biosynthetic pathways of the intact plant, are amenable to metabolic studies, and can generate uniform plant biomass for screening biological activities (Gandhi et al., 2015; la Parra and Quave, 2017; Ricigliano et al., 2015; Ricigliano et al., 2016; Yonekura-Sakakibara and Saito, 2009).
Euphorbia lathyris, a plant native to the Mediterranean region, accumulates cytotoxic and anticancer diterpenoids called Euphorbia factors. Some of these compounds inhibit p-glycoprotein transporters involved in multidrug resistant cancers (Jiao et al., 2009; Lee et al., 2018; Lin et al., 2017). Hydrolysis products of ingenol-based Euphorbia factors from E. lathyris seeds have been used in the semi-synthesis of ingenol mebutate (Liang et al., 2012). Transcriptomic and metabolomic analyses of E. lathyris seeds identified biosynthetic machinery involved in the conversion of casbene to jolkinol, but the full pathway leading to ingenol and structurally related diterpenoids remains to be elucidated (Luo et al., 2016). Nevertheless, E. lathyris cells and tissues possess the biosynthetic potential for a variety of Euphorbia diterpenoids, including ingenol (Adolf et al. 1984; Hou et al. 2011; Bicci et al. 2001; Jiao et al. 2013).
We established genetically transformed root cultures of E. lathyris to assess their diterpenoid biosynthetic capabilities and suitability for use in natural product drug screens. Transformed, or “hairy” root cultures have been established in plants that are susceptible to infection by Agrobacterium rhizogenes, a phytopathogen capable of horizontal genetic transfer of bacterial root-inducing DNA into the host plant genome (Nilsson and Olsson, 1997). Agrobacterium-derived roots exhibit many advantages compared to studying whole plants. Genetically transformed roots express the metabolite profiles of wild type plants, maintain high growth rates, and are amenable to functional genomic studies (Georgiev et al., 2007). Herein, transformed E. lathryis roots and field-grown plant tissues were analyzed by UPLC-PDA-HRMS (Ultra-Performance Liquid Chromatography-Photodiode Array-High Resolution Mass Spectrometry) to compare their metabolic equivalence. Culture treatment with the classical elicitor methyl jasmonate upregulated mRNA transcript levels of early diterpenoid biosynthetic enzymes, suggesting the utility of chemical elicitation to identify diterpenoid biosynthetic genes under similar regulation. We also tested the cytotoxic activities of transformed root extracts against a panel of human carcinoma cell lines derived from prostate, cervix, breast, and lung.
2. RESULTS
2.1. Genetic transformation and establishment of E. lathyris root cultures
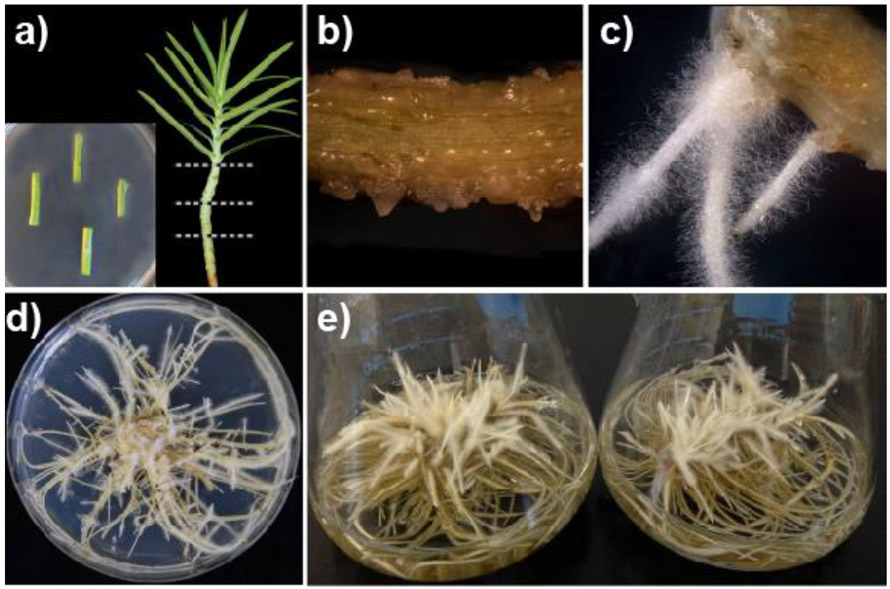
Transformed roots of E. lathyris were established via A. rhizogenes-mediated infection of greenhouse grown plants. Stem explants of E. lathyris were used as the target tissue for infection and root induction (Fig.1a). Root primordia were observed at the infection sites, emerging from callus as early as 15 days after inoculation (Fig.1b). Fully developed adventitious roots showed an abundance of root hairs along the axis that are characteristic of the “hairy root” phenotype (Fig.1c). An isogenic root line displaying vigorous growth was propagated on hormone-free solid media (Fig.1d), and then grown in quadruplicate liquid culture flasks on an orbital shaker to generate the experimental biomass used in subsequent experiments (Fig.1e).
Fig.1.
Establishment and culture maintenance of transformed E. lathyris roots. (a) Stem explants of 3-week-old greenhouse grown plants were used for co-culture with A. rhizogenes. (b) Roots emerged from callus at the site of infection after 2 to 3 weeks.(c) Adventitious roots displaying the characteristic of the “hairy root” phenotype. (d) Growth characteristics of the isogenic root line used in this study on agar media and in (e) liquid media.
2.2. Metabolite profiling
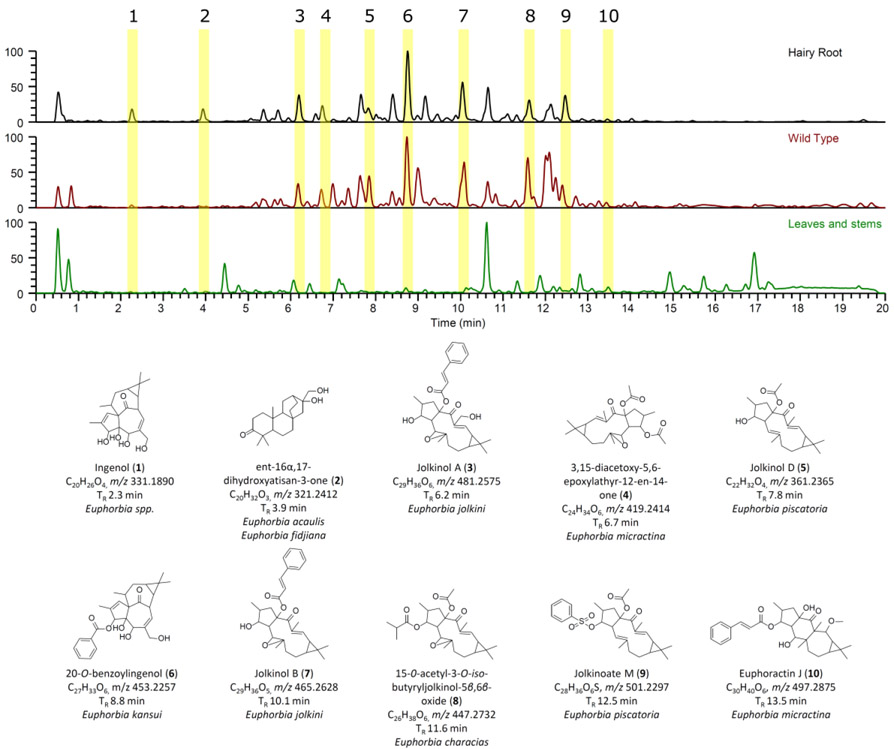
As a metabolic qualification of the transformed root cultures, chemical profiles of roots stably transformed with A. rhizogenes harboring Ri plasmid pRi15834 were compared to wild-type plant roots and aerial tissues (leaves and stems) from soil-grown E. lathyris plants. Samples were extracted and analyzed via a UPLC-PDA-HRMS system, including biological triplicates (Fig. S1); representative chromatograms are shown in Fig. 2. Chemical profiles of transformed roots closely resembled those of wild-type plant roots and shared some overlap with compounds detected in aerial tissues (Fig.2).
Fig.2.
Base peak chromatograms of transformed E. lathyris roots compared to wild-type plant roots and aerial parts with putatively assigned metabolites that are structurally related to ingenol.
Several putatively assigned diterpenoid compounds were present in transformed roots that are structurally related to ingenol (1). The structures were identified by comparing their accurate mass to metabolites that were isolated from Euphorbia spp. then analyzing the fragmentation patterns (MS/MS). This was carried out by searching the molecular formula identified from the mass spectra, identifying metabolites from Euphorbia spp. that have been reported in the literature, and then using the MS/MS data to assign the structures. Based on the UPLC-PDA-HRMS data, several compounds were putatively assigned. Several metabolites correlated to an identified accurate mass, and these metabolites were further narrowed down and putatively assigned based on the MS/MS fragmentation pattern as shown in Fig. 3. Based on these fragmentation patterns, nine compounds were putatively assigned to be structurally related to ingenol. These compounds are ent-16α,17-dihydroxyatisan-3-one (2), jolkinol A (3), 3,15-diacetoxy-5,6-epoxylathyr-12-en-14-one (4), jolkinol D (5), 3-O-benzoylingenol (6), jolkinol B (7), 15-O-acetyl-3-O-iso-butyryljolkinol-5β,6β-oxide (8), jolkinoate M (9), and euphoractin J (10) (Fig.2). The peak areas of compounds 1-10 revealed higher abundance in wild-type roots relative to aerial tissues. When peak areas from transformed roots were compared to wild type root and aerial tissues, relative diterpenoid abundances were between 1 and 4 times higher in transformed roots (Table 1).
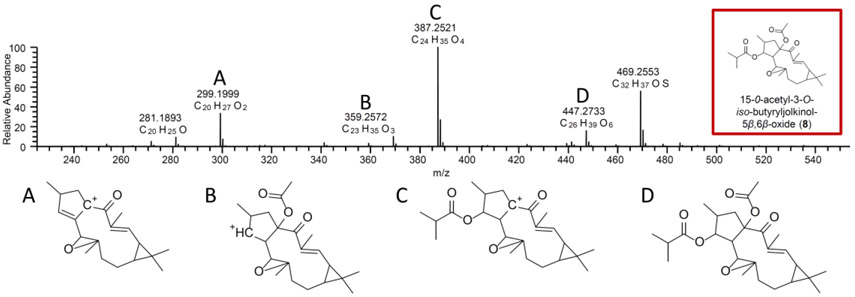
Fig.3.
The MS/MS (fragmentation) data of m/z 477.2732 aided in the putative assignment of compound 8 as 15-O-acetyl-3-O-iso-butyryljolkinol-5β,6β-oxide. The fragmentation structures and m/z values correspond to each other (i.e. structures and peaks A-D).
Table 1.
Peak areas of the putatively assigned diterpenes (1-10) in transformed roots, wild type, and leaves/stem. Peak area data were generated via an extracted ion chromatogram (i.e. XIC) followed by summing the area under the curves (i.e. AUC).
| m/z | Putative Molecular Formula |
TR (min) |
Putative ID | Peak Area (Transformed Root) |
Peak Area (Wild Type) |
Peak Area (Leaves and Stem) |
|---|---|---|---|---|---|---|
| 331.1890 | C20H26O4 | 2.3 | ingenol (1) | 1.27E+08 | 2.68E+07 | 4.76E+06 |
| 321.2412 | C20H32O3 | 3.9 | ent-16α,17-dihydroxyatisan-3-one (2) | 4.62E+07 | 1.71E+06 | ND* |
| 481.2575 | C29H36O6 | 6.2 | jolkinol A (3) | 1.72E+08 | 1.49E+08 | 3.41E+05 |
| 419.2414 | C24H34O6 | 6.7 | 3,15-diacetoxy-5,6-epoxylathyr-12-en-14-one (4) | 1.29E+08 | 1.46E+08 | ND* |
| 361.2365 | C22H32O4 | 7.8 | jolkinol D (5) | 6.16E+08 | 1.14E+08 | ND* |
| 453.2257 | C27H33O6 | 8.8 | 3-O-benzoylingenol (6) | 4.87E+08 | 4.66E+08 | 9.36E+06 |
| 465.2628 | C29H36O5 | 10.1 | jolkinol B (7) | 1.41E+09 | 6.56E+08 | 7.18E+05 |
| 447.2732 | C26H38O6 | 11.6 | 15-O-acetyl-3-O-iso-butyryljolkinol-5β,6β-oxide (8) | 1.52E+08 | 3.29E+08 | ND* |
| 501.2297 | C28H36O6S | 12.5 | jolkinoate M (9) | 9.11E+08 | ND* | 3.39E+04 |
| 497.2875 | C30H40O6 | 13.5 | euphoractin J (10) | 1.14E+08 | 1.23E+08 | 5.12E+05 |
ND Not detected.
2.3. Bioinformatic analysis and elicitation of early diterpenoid biosynthetic genes in transformed roots
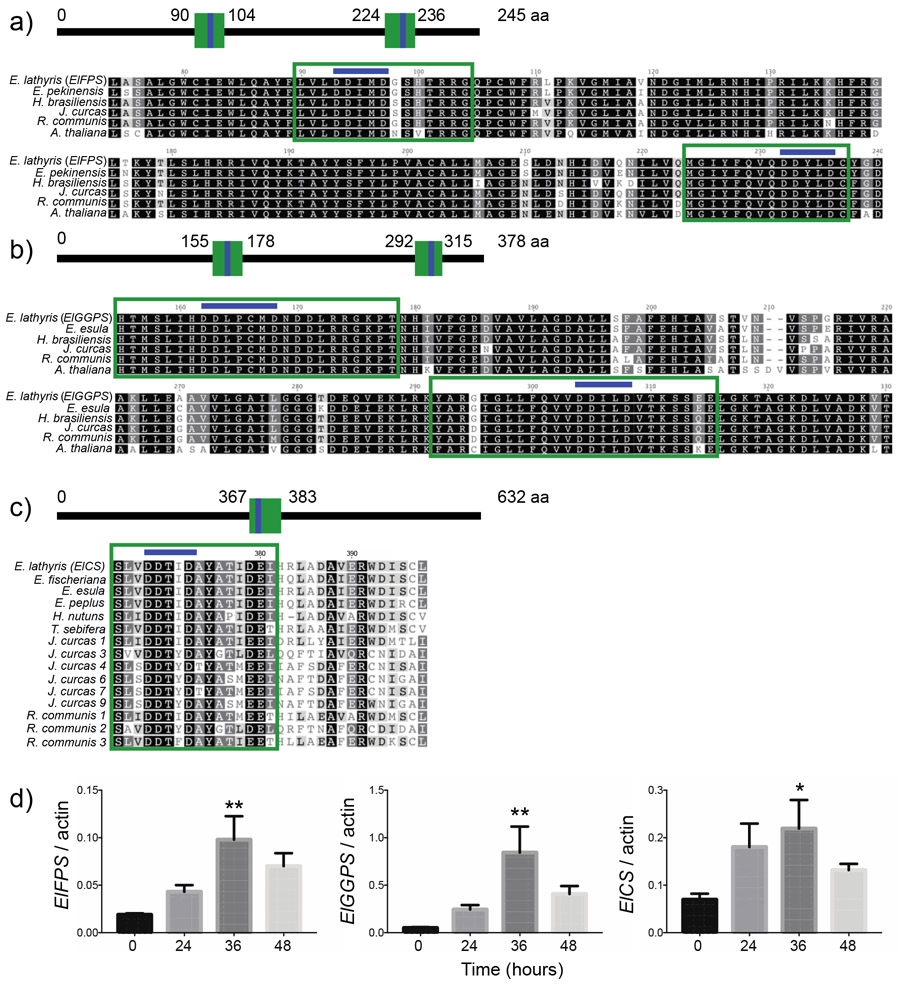
GGPP is derived from the activities of isoprenyl diphosphate synthases. These enzymes are classified according to the chain lengths of their end products, catalyzing the alkylation of one, two, or three units of isopentyl diphosphate (IPP; C5), with an allylic diphosphate such as dimethylallyl pyrophosphate (DMAPP; C5) to produce geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), and geranylgeranyl diphosphate (GGPP; C20). GGPP synthase (GGPS) alone or in regulated coordination with FPP synthase (FPS) may use DMAPP, GPP, or FPP as its allylic substrate to produce GGPP. In view of the key role of these enzymes in early diterpenoid biosynthesis, FPS and GGPS transcripts were amplified from E. lathyris transformed root mRNA for bioinformatic and gene expression analyses. The predicted amino acid sequences showed high similarity to other members of the Euphorbiaceae family and Arabidopsis. The deduced ElFPS sequence exhibited 98.9%, 93.1, 93.0%, 92.2%, and 83.4% identity to Euphorbia pekinensis (ACN63187), Hevea brasiliensis (ABR09548), Jatropha curcas (KDP35531), Ricinus communis (XP_002534338), and A. thaliana (U80605), respectively. The deduced ElGGPS sequence exhibited 89.7%, 88.5%, 84.2%, 83.4%, and 70.1% identity to Euphorbia esula (PRJNA411945), Hevea brasiliensis (BAF98302), Jatropha curcas (KDP31808), Ricinus communis (XP_002525096), A. thaliana (AAB67730), respectively. Two conserved aspartate-rich domains [DDxx(xx)D], FARM (First Aspartate Rich Motif) and SARM (Second Aspartate Rich Motif) are characteristic of prenyl diphosphate synthases (Gupta et al., 2011; Ohnuma et al., 1996). The FARM and SARM regions act as binding sites for allylic substrates and IPP, and are crucial to catalytic activity. Amino acid alignment of ElFPS with known FPP synthase accessions revealed the conserved FARM and SARM motifs at positions 93 – 97 aa and 232 – 235 aa, respectively (Fig.4a). Sequence alignment of ElGPS with known GGPP synthase accessions indicated the FARM and SARM motifs at positions 162 – 168 aa and 303 – 308 aa, respectively (Fig.4b).
Fig.4.
Bioinformatic analysis and chemical elicitation of early diterpenoid biosynthetic genes expressed in transformed E. lathyris roots. Comparison of the deduced amino acid sequences of (a) ElFPS and (b) ElGGPS highlighting two conserved aspartate-rich domains [DDxx(xx)D]. (c) Comparison of the deduced amino acid sequence of ElCS highlighting a conserved [DDxxD] motif that is essential to the cyclization functionalities of terpene synthases. (d) Time course of E. lathyris diterpenoid biosynthetic gene transcript levels in transformed root cultures treated with 100 μM methyl jasmonate. Asterisks indicate statistical significance in comparison to 0 h control assessed by one-way ANOVA (**,P< 0.01; *,P< 0.05)
Casbene synthase catalyzes the first committed step in the formation of diterpenoid backbones taxonomically restricted to the Euphorbiaceae family via the cyclization of GGPP into casbene. A casbene synthase transcript (ElCS) was amplified from transformed root mRNA. The deduced amino acid sequence of ElCS exhibited 90.8%, 89.7%, and 86.4% identity to Euphorbia fischeriana (JN862821), Euphorbia esula (GU332591), and Euphorbia peplus (AGN70884), respectively. Within the Euphorbiaceae family, ElCS shared the following pairwise amino acid identities: 76.4% with Triadica sebifera (GU332590), 67.9% with Homalanthus nutans (GU332592), 62.8 – 46.2% with J. curcas (BAJ53213, BAJ53216, BAJ53218, BAJ53219, BAJ53220, BAJ53221), and 73.9– 47.5% with R. communis (L32134, XM_002513323, XM_002513288). Protein alignment revealed a conserved [DDxxD] motif at positions 370 – 374 aa that is essential to the cyclization functionalities of terpene synthases, likely through association with Mg2+ (Aubourg et al., 2002). Taken together, these results suggest that ElFPS, ElGPS, and ElCS are catalytically active, possessing functional attributes that are comparable to homologous genes.
The transcript expression patterns of ElFPS, ElGPS, and ElCS were profiled in transformed roots treated with methyl jasmonate (MeJA), a classical elicitor of plant secondary metabolism. Gene expression was monitored over a 48-h period after addition of 100 μM MeJA. At 36 h, the ElFPS, ElGGPS, and ElCS mRNAs were transiently induced, 5.4-, 17.1-, and 3.2-fold, respectively (Fig.4c). These results indicate that ElFPS, ElGGPS, and ElCS are coordinately regulated at the transcriptional level in response to chemical elicitation.
2.4. Cytotoxicity bioassay
The cytotoxic properties of wild-type Euphorbia plant extracts and isolated diterpenoids are a topic of significant research interest, but studies are limited by low-level accumulation of medicinal compounds in planta. Tissue culture technologies enable the production of uniform plant biomass, thus improving access to these compounds for use in chemical and pharmacological studies. To test the antiproliferative effects of E. lathyris transformed roots in human cells, DU-145 (prostate), HeLa (cervix), MCF-7 (breast), MDA-MB-231 (breast), and H2347 (lung) carcinoma lines were treated with increasing concentrations of E. lathyris transformed root extract. After 48 h post-treatment, MCF-7 and MDA-MB-231 cells exhibited a significant reduction in viability in response to a concentration of 31.3 μg/mL extract (Fig. 5c-d). DU-145 cells exhibited a significant reduction in viability at a concentration of 62.5 μg/mL (Fig. 5a). HeLa cells showed a significant reduction in viability at a concentration of 125 μg/mL (Fig. 5b). In H2347 cells, a significant reduction in viability was observed at a concentration of 250 μg/mL (Fig. 5e). Relative to the carrier control, the 250 μg/mL concentration of root extract inhibited the different cell lines as follows: DU-145 (24.4%), HeLa (57.0%), MCF-7 (66.9%), MDA-MB-231 (38.1%), H2347 (70.6%). These results highlight the potential of E. lathyris transformed roots for natural product drug discovery applications.
Fig.5.
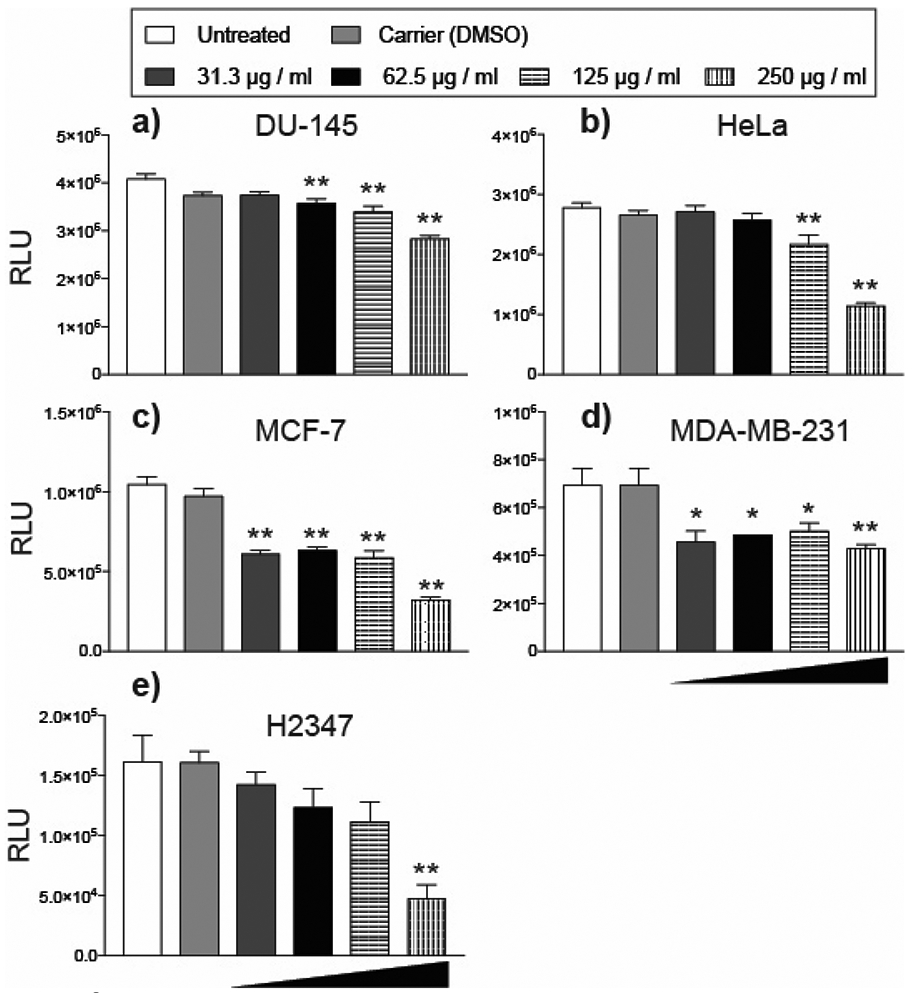
Cytotoxic activities of transformed E. lathyris root extract in human carcinoma and embryonic cell lines. a) DU-145 (prostate) b) HeLa (cervix) c) MCF-7 (breast) d) MDA-MB-231 (breast) e) and H2347 (lung) were treated with DMSO (carrier), or increasing concentrations of transformed root MeOH extracts (31.3 ug/ml, 52.5 ug/ml, 125 ug/ml, 250 ug/ml). Titer-Glo® (Promega) was used to count cell lines 48 h post-treatment. Each dose and timepoint was performed in triplicate. Asterisks indicate statistical significance in comparison to carrier (DMSO) control assessed by one-way ANOVA (**,P< 0.01; *,P< 0.05)
3. DISCUSSION
Currently, most Euphorbia and Euphorbiaceae family diterpenoids can only be obtained from the source plants. However, many species are slow-growing and accumulate small quantities of medicinal compounds. Our results indicate that E. lathyris transformed roots reproduce the metabolism of field-grown plants and are a promising source of Euphorbia diterpenoid structures. Additionally, transformed roots are a suitable platform to study Euphorbia diterpenoid biosynthesis and may prove useful in functional genomic screens of candidate metabolic genes. Since pharmacological studies are limited by low yields from plants, transformed root cultures could be a source of uniform Euphorbia biomass for natural product drug discovery applications
Alternative sources of ingenol have been a topic of significant research focus due to the interesting pharmacological properties exhibited by naturally occurring conjugates and semi-synthetic derivatives (Abreu et al., 2014; Silva et al., 2019; Vasas et al., 2012). Biotechnological production in microbes is a promising approach to increase the accessibility of high value diterpenoids (Zerbe et al., 2013). Some studies have used heterologous yeast expression systems to test the contribution of candidate Euphorbiaceae biosynthetic genes to diterpenoid production. This has proved to be useful in identifying casbene synthases responsible for the cyclization of GGPP into casbene (Kirby et al. 2010). From casbene, the routes to highly oxidized casbene derivatives, including ingenol, are less understood but likely proceed through intermediates such as jolkinol via cytochrome P450-catalyzed oxidations and a short-chain alcohol dehydrogenase (King et al., 2014; Luo et al., 2016; Zerbe et al., 2013). Recently, high-titer production of jolkinol by yeast was reported (Wong et al., 2018). Similar to other studies aimed at reconstituting complex oxidized terpenoid biosynthetic pathways in yeast, optimization of heterologous cytochrome P450 expression and precursor flux was a prerequisite (Paddon et al., 2013). Still, the primary bottleneck in heterologous ingenol production is complete pathway elucidation, which has proven difficult due to its structural complexity and the number of casbene modifiers encoded by Euphorbia genomes (Zerbe et al., 2013).
4. CONCLUDING REMARKS
Here, we found that transformed roots derived from A. rhizogenes-mediated transformation of E. lathryis possess the biosynthetic capacity for ingenol and related diterpenoid structures. A broad diversity of diterpenoids have been identified in aerial tissues and roots of E. lathryis, but most studies have focused on seed accumulation of ingenol-based Euphorbia factors. Indeed, E. lathyris seeds have been used as starting material to obtain ingenol hydrolysis products for the semi-synthesis of ingenol mebutate (Liang et al., 2012). Euphorbia spp. roots have been the focus of numerous studies in which diterpenoids were identified and assayed for biological activities (Lee et al., 2016; Liu et al., 2014; Shi et al., 2005; Ul-Haq et al., 2012). These findings are in agreement with our results that transformed roots are a viable approach to Euphorbia diterpenoid production in vitro. Induction of transformed root cultures in a variety of Euphorbiaceae species has potential to increase the current supply of diterpenoids. Other Euphorbiacae species contain diterpenoids of pharmacological significance that could potentially be produced via transformed roots. For example, prostratin from Homalanthus nutans (Kulkosky et al., 2001) and resiniferatoxin from E. resinifera (Kissin and Szallasi, 2011). Transformed roots are a genetically tractable platform that could be used to test the contributions of candidate biosynthetic genes to metabolite accumulation and pharmacological activities. Combinatorial biosynthesis employing non-native biosynthetic enzymes within the metabolic scaffold of select species could enable the future identification of novel metabolites with augmented pharmacological activities.
5. EXPERIMENTAL
5.1. Establishment and maintenance of E. lathyris tranformed root cultures
Seeds of Euphorbia lathryis were obtained from Plant World Seeds (Newton Abbot, UK) and used to produce the plant materials in this study. Relying on standard procedures described in the literature (Georgiev et al., 2007), stem segments from 6-week-old greenhouse grown E. lathyris plants were used for co-cultivation and root induction. A. rhizogenes strain 15834 harboring root-inducing plasmid pRi15834 was used to carry out the transformation. Explants were submerged in 1% sodium hypochlorite for 10 minutes, then rinsed three times with sterile H2O and 1 cm removed from each end with a sterile scalpel. The explants were submerged in Agrobacterium suspension, blotted dry, and incubated on hormone-free ½ MS medium (Murashige and Skoog, 1962) containing 3% sucrose and solidified with 1% agar for 25 ± 2 °C in the dark. Following 3 days of co-cultivation, the explants were subcultured to MS medium containing 150 mg/L timentin to kill the excess Agrobacterium. In approximately 2.5 weeks, hairy roots developed from the cut surfaces and along the axis of the explants. Single root segments were removed from different explants to avoid the unintended propagation of the same clone. For liquid cultures, approximately 200 mg of transformed root biomass was transferred to 30 mL of liquid MS medium and grown in darkness at 25 ± 2 °C on an orbital shaker at 100 rpm with subcultures performed every 3 weeks.
5.2. Metabolite extractions
Separately, biomass from transformed roots, wild-type roots, and wild-type leave/stems were ground using a mortar and pestle. MeOH was added to each sample of ground plant material to a concentration of 0.1g/mL. Then, the samples were shaken overnight (~16 h) at 100 rpm at room temperature. The samples were filtered using vacuum filtration, and the remaining residues were washed with MeOH. MeOH-H2O-hexanes (9:1:10) was added to the filtrate. The mixtures were stirred for 0.5 h, and then transferred into separatory funnels. The bottom layers were drawn off into round-bottom flasks, which were evaporated to dryness. The dried MeOH extracts were reconstituted in H2O-MeOH-chloroform (9:1:10). The biphasic solutions were shaken vigorously. The layers were separated and evaporated to dryness under vacuum.
5.3. Metabolite profiling via UPLC-PDA-HRMS
The QExactive Plus mass spectrometer operated over a range from m/z 130 to 1900 at a resolution of 70,000. The voltage for both positive and negative ionization modes were set to 3.7 kV, with a nitrogen sheath gas set to 25 arb and an auxiliary gas set to 5 arb. The S–Lens RF level was set to 50.0 with a capillary temperature at 350 °C. The flow rate of the UPLC was set to 0.3 mL/min using an Acquity UPLC HSS T3 column (2.1 × 50 mm × 1.7 μm) equilibrated at 40 °C. The mobile phase consisted of Fisher Optima LC–MS (Liquid Chromatography-Mass Spectrometry) grade acetonitrile–H2O (acidified with 0.1% formic acid), starting at 15% acetonitrile and increasing linearly to 100% acetonitrile over 17 min. It was held at 100% acetonitrile for 1.5 min before returning to starting conditions for re-equilibration. The PDA was set to acquire from 190 to 500 nm with 4 nm resolution.
5.4. Metabolite analyses
SciFinder (https://scifinder.cas.org/) was used to putatively assign the diterpenoid compounds structurally related to ingenol 1-10 (Fig. 2) based on mass spectral comparisons to previously reported structures isolated from Euphorbia spp. This was carried out by searching the molecular formula obtained from the mass spectra through the database, then identifying compounds that were previously reported from Euphorbia spp. Then by using the MS/MS data, the compounds were putatively identified. The peak areas for the compounds were determined from the area under the curve (AUC) of an Extracted-Ion Chromatogram (XIC) with a window of ± 5.0 ppm.
5.5. Sequence analyses of diterpenoid biosynthetic genes
The nucleotide and predicted amino acid sequences of early diterpenoid biosynthetic genes were analyzed using Geneious version 7.0 (Biomatters). Transcripts corresponding to farnesyl diphosphate synthase (FPS), geranylgeranyl diphosphate synthase (GGPS), and casbene synthase (CS) were amplified from a transformed root cDNA library using primers based on the consensus sequence of previously identified Euphorbiaceae orthologs. The nucleotide sequences of the E. lathryis biosynthetic genes amplified from transformed roots are deposited in EMBL/DDBJ/GenBank under the following accessions: ElFPS (MK512674), ElGGPS (MK512675), and ElCS (MK559432). GenBank accessions used in the ElFPS amino acid sequence comparison were as follows: Euphorbia pekinensis (ACN63187), Hevea brasiliensis (ABR09548), Jatropha curcas (KDP35531), Ricinus communis (XP_002534338), and A. thaliana (U80605). GenBank accessions used in the ElGGPS amino acid sequence comparison were as follows: Euphorbia esula (PRJNA411945), Hevea brasiliensis (BAF98302), Jatropha curcas (KDP31808), Ricinus communis (XP_002525096), A. thaliana (AAB67730). GenBank accessions used in the ElCS amino acid sequence comparison were as follows: Euphorbia fischeriana (JN862821), Euphorbia esula (GU332591), Euphorbia peplus (AGN70884), Triadica sebifera (GU332590), Homalanthus nutans (GU332592), J. curcas (BAJ53213, BAJ53216, BAJ53218, BAJ53219, BAJ53220, BAJ53221), and Ricinus communis (L32134, XM_002513323, XM_002513288).
5.6. Gene expression analyses
To test the effects of elicitation on biosynthetic gene expression, transformed roots were maintained in liquid ½ MS medium containing 3% sucrose in darkness on an orbital shaker at 25 ± 2 °C and 100 rpm. A 10 mM solution of MeJA was prepared in EtOH:H2O (1:1 v/v) and was added to 30 mL of liquid ½ MS to a final concentration of 100 μM. A solution of 1:1 v/v EtOH:H2O was added to the cultures as a control. Three-week-old vigorous cultures were sampled at 0, 24, 36, and 48 h time points following MeJA treatment. Total RNA was isolated using an RNeasy plant mini-kit (Qiagen) according to the manufacturer’s instructions.
Primers with annealing temperatures between 60 and 64 °C and amplicons of 185-292 bp were designed using Geneious version 7.0 (Biomatters) based on the sequence of biosynthetic gene transcripts amplified from E.lathyris transformed root cDNA. Primer specificity was confirmed by cloning and sequencing of the PCR products. Quantitative PCR was performed on an iCycler iQ™ Real-Time PCR system (Bio-Rad), using iScript™ One-Step RT-PCR Kit with SYBR® Green as per manufacturer instructions (Bio-Rad) with gene-specific primers. E.lathyris actin (GenBank: KC182753.1) was used as an internal reference for normalization. Three biological and three technical replicates were measured to confirm the accuracy of the results, and to determine standard error. Relative expression levels were determined using the 2 -ΔΔCt method (Livak and Schmittgen, 2001). Statistical analyses was performed using one-way ANOVA. Primers used in the qPCR expression analysis are as follows: actin (5’-AGCCGTTCTGTCATTGTATGCC-3’ and 5’-AAGTTTCTCCTTCATGTCACGG-3’), ElFPS (5’-AGACGTGGTCAGCCTTGTTGG-3’ and 5’-ATCATCTGTCCTGAAGCTGTTTGG-3’), ElGGPS (5’-TGCACATGGATGAACTCAAGCG-3’ and 5’-ACGATGACCTCCCGTGTATGG-3’), ElCS (5’-TCAAACAACACATAACGAACGCC-3’ and 5’-TTCGGCCATTCTGTCTCTTGCG-3’).
5.7. Human carcinoma cell culture and cytotoxicity bioassay
DU-145 (prostate), HeLa (cervical), MCF-7 (breast), MDA-MB-231 (breast), H2347 (lung) carcinoma cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cells were cultured in DMEM supplemented with FBS (5 % v/v), nonessential amino acids (100 mM), L-glutamine (5 mM), streptomycin (100 μg/mL), and penicillin (100 units/mL); all from BioWhittaker, Walkersville, MD. Cells were grown at 37 °C, in a 5% CO2, 95% air humidified tissue culture incubator.
For the transformed root extract used in the cytotoxicity bioassays, 3 g of air-dried transformed root biomass was ground using a mortar and pestle. MeOH was added to the ground plant material to a concentration of 0.1g/mL. The sample was shaken overnight at 100 rpm at room temperature. The sample were filtered using vacuum filtration and the MeOH was removed under vacuum to yield a yellow semi-solid that was dissolved in DMSO. To elucidate the antiproliferative activities of the root extract, carcinoma cell lines were treated with either DMSO (carrier), or increasing concentrations of E. lathyris root MeOH extracts (31.3 μg/mL, 52.5 μg/mL, 125 μg/mL, 250 μg/mL). Dilutions were prepared with cell culture medium in 1.5ml tubes, then vortexed thoroughly each time immediately prior to adding them to cell culture plates. After 48 h post-treatment, cells were trypsinized, harvested and vortexed to produce a single-cell suspension. Cells from each treatment condition were counted using CellTiter-Glo® Luminescent Cell Viability Assay (Promega). For each sample, a volume of CellTiter-Glo® Reagent equal to a volume of cell culture medium containing cells was placed into a standard 12mm luminescence tube and vortexed. Each tube was incubated for 10 minutes at room temperature to stabilize luminescent signal. Luminescence was determined using a Sirius single tube luminometer (Berthold). All CellTiter-Glo® luciferase results were reported as relative light units (RLU). Each dose and timepoint was performed in triplicate, and statistical analyses was performed using one-way ANOVA.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank personnel in the Ricigliano, Howarth, and Oberlies lab groups for their support, advice, and stimulating discussions. Sonja L. Knowles was supported in part by the National Institutes of Health via the National Center for Complementary and Integrative Health (F31 AT010558)
REFERENCES
- Abreu CM, Price SL, Shirk EN, Cunha RD, Pianowski LF, Clements JE, Tanuri A, Gama L, 2014. Dual role of novel ingenol derivatives from Euphorbia tirucalli in HIV replication: inhibition of de novo infection and activation of viral LTR. PLoS ONE 9, e97257–. doi: 10.1371/journal.pone.0097257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf W, Hecker E, & Becker H 1984. Macrocyclic lathyrane type diterpene esters (jolkinols) from callus cultures and roots of Euphorbia lathyris. Planta medica, 50(03), 259–261. [DOI] [PubMed] [Google Scholar]
- Alchin DR, 2014. Ingenol mebutate: a succinct review of a succinct therapy. Dermatol. Ther. (Heidelb) 4, 157–164. doi: 10.1007/s13555-014-0061-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov AG, Waltenberger B, Pferschy-Wenzig E-M, Linder T, Wawrosch C, Uhrin P, Temml V, Wang L, Schwaiger S, Heiss EH, Rollinger JM, Schuster D, Breuss JM, Bochkov V, Mihovilovic MD, Kopp B, Bauer R, Dirsch VM, Stuppner H, 2015. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv 33, 1582–1614. doi: 10.1016/j.biotechadv.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J, 2002. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745. doi: 10.1007/s00438-002-0709-y [DOI] [PubMed] [Google Scholar]
- Béres T, Dragull K, Pospíšil J, Tarkowská D, Dančák M, Bíba O, Tarkowski P, Doležal K, Strnad M, 2018. Quantitative analysis of ingenol in Euphorbia species via validated isotope dilution ultra-high performance liquid chromatography tandem mass spectrometry. Phytochem. Anal 29, 23–29. doi: 10.1002/pca.2711 [DOI] [PubMed] [Google Scholar]
- Bicchi C, Appendino G, Cordero C, Rubiolo P, Ortelli D, & Veuthey JL 2001. HPLC-UV and HPLC-positive-ESI-MS analysis of the diterpenoid fraction from caper spurge (Euphorbia lathyris) seed oil. Phytochem. Anal 12(4), 255–262. [DOI] [PubMed] [Google Scholar]
- Braun SA, Baran J, Schrumpf H, Buhren BA, Bölke E, Homey B, Gerber PA, 2018. Ingenol mebutate induces a tumor cell-directed inflammatory response and antimicrobial peptides thereby promoting rapid tumor destruction and wound healing. Eur. J. Med. Res 23, 45. doi: 10.1186/s40001-018-0343-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A-X, Lou Y-G, Mao Y-B, Lu S, Wang L-J, Chen X-Y, 2007. Plant terpenoids: biosynthesis and ecological functions. J. Integr. Plant Biol 49, 179–186. doi: 10.1111/j.1744-7909.2007.00395.x [DOI] [Google Scholar]
- Doan HQ, Gulati N, Levis WR, 2012. Ingenol mebutate: potential for further development of cancer immunotherapy. J Drugs Dermatol 11, 1156–1157. [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Saslis-Lagoudakis CH, Grace OM, Nilsson N, Simonsen HT, Horn JW, Rønsted N, 2016. Evolutionary prediction of medicinal properties in the genus Euphorbia L. Sci. Rep 6, 30531. doi: 10.1038/srep30531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Nothias LF, van der Hooft JJ, Da Silva RR, Saslis-Lagoudakis CH, Grace OM, Martinez-Swatson K, Hassemer G, Fuenz L, Simonsen HT and Medema MH, 2019. Assessing specialized metabolite diversity in the cosmopolitan plant genus Euphorbia L. Front. Plant Sci, 10, 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SG, Mahajan V, Bedi YS, 2015. Changing trends in biotechnology of secondary metabolism in medicinal and aromatic plants. Planta 241, 303–317. doi: 10.1007/s00425-014-2232-x [DOI] [PubMed] [Google Scholar]
- Georgiev MI, Pavlov AI, Bley T, 2007. Hairy root type plant in vitro systems as sources of bioactive substances. Appl. Microbiol. Biotechnol 74, 1175–1185. doi: 10.1007/s00253-007-0856-5 [DOI] [PubMed] [Google Scholar]
- Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK, 2011. Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul. 65, 93–100. doi: 10.1007/s10725-011-9578-x [DOI] [Google Scholar]
- Hohmann J, Evanics F, Berta L, Bartók T, 2000. Diterpenoids from Euphorbia peplus. Planta Medica 66, 291–294. doi: 10.1055/s-2000-8568 [DOI] [PubMed] [Google Scholar]
- Hou XR, Wan LL, Zhan ZJ, Li CP, & Shan WG 2011. Analysis and determination of diterpenoids in unprocessed and processed Euphorbia. lathyris seeds by HPLC–ESI-MS. J. Pharm. Anal 1(3), 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W, Dong W, Li Z, Deng M, Lu R, 2009. Lathyrane diterpenes from Euphorbia lathyris as modulators of multidrug resistance and their crystal structures. Bioorg. Med. Chem 17, 4786–4792. doi: 10.1016/j.bmc.2009.04.041 [DOI] [PubMed] [Google Scholar]
- Jiao W, Fang DM, Wu ZJ, Chen JZ, Shao HW, & Zhang GL 2013. Analysis of lathyrane diterpenes using electrospray ionization quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 27(1), 276–280. [DOI] [PubMed] [Google Scholar]
- Jørgensen L, McKerrall SJ, Kuttruff CA, Ungeheuer F, Felding J, Baran PS, 2013. 14-step synthesis of (+)-ingenol from (+)-3-carene. Science 341, 878–882. doi: 10.1126/science.1241606 [DOI] [PubMed] [Google Scholar]
- King AJ, Brown GD, Gilday AD, Larson TR, Graham IA, 2014. Production of bioactive diterpenoids in the euphorbiaceae depends on evolutionarily conserved gene clusters. The Plant Cell 26, 3286–3298. doi: 10.1105/tpc.114.129668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J, Nishimoto M, Park JG, Withers ST, Nowroozi F, Behrendt D, Rutledge EJG, Fortman JL, Johnson HE, Anderson JV, Keasling JD, 2010. Cloning of casbene and neocembrene synthases from Euphorbiaceae plants and expression in Saccharomyces cerevisiae. Phytochemistry 71, 1466–1473. doi: 10.1016/j.phytochem.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Kissin I, Szallasi A, 2011. Therapeutic targeting of TRPV1 by resiniferatoxin, from preclinical studies to clinical trials. Curr. Top. Med. Chem 11, 2159–2170. doi: 10.1101/128884 [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Lu X, Yang B, Peters RJ, 2015. The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol. Plant 8, 6–16. doi: 10.1016/j.molp.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, Boyd MR, Pomerantz RJ, 2001. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98, 3006–3015. [DOI] [PubMed] [Google Scholar]
- la Parra, de J, Quave CL, 2017. Ethnophytotechnology: harnessing the power of ethnobotany with biotechnology. Trends Biotechnol. 35, 802–806. doi: 10.1016/j.tibtech.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Lee JW, Jin Q, Jang H, Kim JG, Lee D, Kim Y, Hong JT, Lee MK, Hwang BY, 2018. Lathyrane-Type diterpenoids from the seeds of Euphorbia lathyris L. with Inhibitory Effects on NO production in RAW 264.7 cells. Chem. Biodivers 15, e1800144. doi: 10.1002/cbdv.201800144 [DOI] [PubMed] [Google Scholar]
- Lee JW, Lee C, Jin Q, Jang H, Lee D, Lee H-J, Shin JW, Han SB, Hong JT, Kim Y, Lee MK, Hwang BY, 2016. Diterpenoids from the roots of Euphorbia fischeriana with inhibitory effects on nitric oxide Production. J. Nat. Prod 79, 126–131. doi: 10.1021/acs.jnatprod.5b00789 [DOI] [PubMed] [Google Scholar]
- Liang X, Grue-Sørensen G, Petersen A, Högberg T, 2012. Semisynthesis of ingenol 3-Angelate (PEP005): efficient stereoconservative angeloylation of alcohols. Synlett 23, 2647–2652. doi: 10.1055/s-0032-1317415 [DOI] [Google Scholar]
- Lin M, Tang S, Zhang C, Chen H, Huang W, Liu Y, Zhang J, 2017. Euphorbia factor L2 induces apoptosis in A549 cells through the mitochondrial pathway. Acta Pharm Sin B 7, 59–64. doi: 10.1016/j.apsb.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-G, Li Z-L, Bai J, Meng D-L, Li N, Pei Y-H, Zhao F, Hua H-M, 2014. Anti-inflammatory diterpenoids from the roots of Euphorbia ebracteolata. J. Nat. Prod 77, 792–799. doi: 10.1021/np400873v [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo D, Callari R, Hamberger B, Wubshet SG, Nielsen MT, Andersen-Ranberg J, Hallström BM, Cozzi F, Heider H, Lindberg Møller B, Staerk D, Hamberger B, 2016. Oxidation and cyclization of casbene in the biosynthesis of Euphorbiafactors from mature seeds of Euphorbia lathyris L. Proc. Natl. Acad. Sci. U.S.A 113, E5082–E5089. doi: 10.1073/pnas.1607504113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Swanson N, 2013. Clinical findings using ingenol mebutate gel to treat actinic keratoses. J. Am. Acad. Dermatol 68, S39–S48. doi: 10.1016/j.jaad.2012.09.050 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F, 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nilsson O, Olsson O, 1997. Getting to the root: The role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiologia Plantarum 100, 463–473. doi: 10.1034/j.1399-3054.1997.1000307.x [DOI] [Google Scholar]
- Ogbourne SM, Parsons PG, 2014. The value of nature's natural product library for the discovery of New Chemical Entities: the discovery of ingenol mebutate. Fitoterapia 98, 36–44. doi: 10.1016/j.fitote.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Ohnuma SI, Narita K, Nakazawa T, Ishida C, Takeuchi Y, Ohto C, Nishino T, 1996. A role of the amino acid residue located on the fifth position before the first aspartate-rich motif of farnesyl diphosphate synthase on determination of the final product. J. Biol. Chem 271, 30748–30754. doi: 10.1074/jbc.271.48.30748 [DOI] [PubMed] [Google Scholar]
- Ortega Del Olmo R, Salido-Vallejo R, 2018. Ingenol mebutate for the treatment of actinic keratosis: effectiveness and safety in 246 patients treated in real-life clinical practice. J. Dermatolog. Treat 29, 393–399. doi: 10.1080/09546634.2017.1386272 [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD, 2013. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051 [DOI] [PubMed] [Google Scholar]
- Ricigliano V, Chitaman J, Tong J, Adamatzky A, Howarth DG, 2015. Plant hairy root cultures as plasmodium modulators of the slime mold emergent computing substrate Physarum polycephalum. Front. Microbiol 6, 375–10. doi: 10.3389/fmicb.2015.00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricigliano V, Kumar S, Kinison S, Brooks C, Nybo SE, Chappell J, & Howarth DG, 2016. Regulation of sesquiterpenoid metabolism in recombinant and elicited Valeriana officinalis hairy roots. Phytochemistry, 125, 43–53. 10.1016/j.phytochem.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Saklani A, Kutty SK, 2008. Plant-derived compounds in clinical trials. Drug Discov. Today 13, 161–171. doi: 10.1016/j.drudis.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Shi H-M, Williams ID, Sung HH-Y, Zhu H-X, Ip NY, Min Z-D, 2005. Cytotoxic diterpenoids from the roots of Euphorbia ebracteolata. Planta Medica 71, 349–354. doi: 10.1055/s-2005-864102 [DOI] [PubMed] [Google Scholar]
- Silva VAO, Rosa MN, Tansini A, Martinho O, Tanuri A, Evangelista AF, Cruvinel Carloni A, Lima JP, Pianowski LF, Reis RM, 2019. Semi-synthetic ingenol derivative from Euphorbia tirucalli inhibits protein kinase C isotypes and promotes autophagy and S-phase arrest on glioma cell lines. Molecules 24, 4265. doi: 10.3390/molecules24234265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul-Haq I, Ullah N, Bibi G, Kanwal S, Sheeraz Ahmad M, Mirza B, 2012. Antioxidant and cytotoxic activities and phytochemical analysis of Euphorbia wallichii root extract and its fractions. Iran J. Pharm. Res 11, 241–249. [PMC free article] [PubMed] [Google Scholar]
- Vasas A, Hohmann J, 2014. Euphorbia diterpenes: isolation, structure, biological activity, and synthesis (2008–2012). Chem. Rev 114, 8579–8612. doi: 10.1021/cr400541j [DOI] [PubMed] [Google Scholar]
- Vasas A, Rédei D, Csupor D, Molnár J, Hohmann J, 2012. Diterpenes from European Euphorbia species serving as prototypes for natural product-based drug discovery. Eur. J. Org. Chem 2012, 5115–5130. doi: 10.1002/ejoc.201200733 [DOI] [Google Scholar]
- Winkler JD, Rouse MB, Greaney MF, Harrison SJ, Jeon YT, 2002. The first total synthesis of (+/−)-ingenol. J. Am. Chem. Soc 124, 9726–9728. doi: 10.1371/journal.pgen.1006346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, de Rond T, d’Espaux L, van der Horst C, Dev I, Rios-Solis L, Kirby J, Scheller H, Keasling J, 2018. High-titer production of lathyrane diterpenoids from sugar by engineered Saccharomyces cerevisiae. Metab. Eng 45, 142–148. doi: 10.1016/j.ymben.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Saito K, 2009. Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep 26, 1466–1487. doi: 10.1039/b817077k [DOI] [PubMed] [Google Scholar]
- Zerbe P, Hamberger B, Yuen MMS, Chiang A, Sandhu HK, Madilao LL, Nguyen A, Hamberger B, Hamberger B, Bach SS, Bohlmann J, 2013. Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 162, 1073–1091. doi: 10.1104/pp.113.218347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.