Abstract
Introduction:
Dysfunctional serotonin signaling has been linked to the pathogenesis of autism, obsessive compulsive disorder, mood disorders and schizophrenia. While the hypo-activity of serotonin signaling is involved in the pathogenesis of depression, anxiety and obsessive compulsive disorder; LSD, an agonist of serotonin type 2 receptor (5-HTR2A) induces psychosis. Therefore, anxiety and depressive disorders are treated by SSRIs which inhibit serotonin transporter (5-HTT) while psychotic disorders are controlled by drugs that block serotonin and/or dopamine receptors. Since genetic polymorphisms and epigenetic dysregulation of 5-HTT are involved in the pathogenesis of mental diseases, we analyzed DNA methylation of 5-HTT promoter in post-mortem brains and saliva samples of patients with schizophrenia (SCZ) and bipolar disorder (BD) to evaluate its potential application as a diagnostic and/or therapeutic biomarker in SCZ and BD.
Methods:
Whole genome DNA methylation profiling was performed for a total of 24 samples (including two saliva samples) using the Illumina 450K DNA methylation array platform, followed by bisulfite sequencing to identify candidate CpGs for further analysis. Quantitative methylation specific PCR (qMSP) was used to assess the degree of CpG methylation of 5-HTT promoter in 105 post-mortem brains (35 controls, 35 SCZ and 35 BD) and 100 saliva samples (30 controls, 30 SCZ, 20 BD and 20 first degree relatives of SCZ or BD). The U133 2.0 Plus Human Transcriptome array for a total of 30 post-mortem brain samples (each group 10) followed by quantitative real-time PCR was used to study 5-HTT expression in 105 post-mortem brain samples.
Results:
The qMSP analysis for 5-HTT promoter region showed DNA hypermethylation in post-mortem brain samples of SCZ patients (~30%), particularly in drug free patients (~60%, p=0.04). Similarly, there was a trend for DNA hypermethylation in antipsychotic free BD patients (~50%, p=0.066). qMSP analysis of DNA extracted from the saliva samples also exhibited hypermethylation of 5-HTT promoter in patients with SCZ (~30%, p=0.039), which was more significant in drug naïve SCZ patients (>50%, p= 0.0025). However, the difference was not significant between the controls and unaffected first degree relatives of patients with SCZ (p=0.37) versus patients using antipsychotic drugs (p=0.2). The whole genome transcriptome analysis of post mortem brain samples showed reduced expression of 5-HTT in SCZ compared to the control subjects (~50%, p=0.008), confirmed by quantitative real-time PCR analysis (~40%, p=0.035) which was more significant in drug free SCZ patients (~70%, p=0.022).
Conclusion:
A correlation between reduction in 5-HTT expression and DNA hypermethylation of the 5-HTT promoter in drug naïve SCZ patients suggest that an epigenetically defined hypo-activity of 5-HTT may be linked to SCZ pathogenesis. Furthermore, this epigenetic mark in DNA extracted from saliva can be considered as one of the key determinants in a panel of diagnostic and/or therapeutic biomarkers for SCZ.
Keywords: Serotonin transporter, DNA methylation, Brain, Saliva, Schizophrenia
Introduction
It is well documented that serotonin has major roles in the brain development, memory and synapse formation, mood and cognitive state (Broman and Fletcher 1999; Kandel 2001). Dysfunctional serotonin signaling has been implicated in the pathogenesis of several mental diseases such as autism, obsessive compulsive disorder, mood disorders and schizophrenia (Sadock et al., 2009; Werner and Coveñas 2010). While the hypo-activity of serotonin signaling has been linked to the pathogenesis of non-psychotic mental diseases such as depression, anxiety and obsessive compulsive disorder, the use of LSD which resembles serotonin and a partial agonist for serotonin type 2 receptor (5-HTR2A) may induce psychotic phenotype. These lines of experimental evidence have been instrumental in the establishment of current therapeutics in psychiatry. While, on one hand, the majority of anxiety and depressive disorders are treated by serotoninergic drugs such as inhibitors of serotonin transporter (5-HTT) which reuptakes the secreted serotonin from the synaptic cleft, on the other hand, most of the psychotic disorders are controlled by drugs that block serotonin and/or dopamine receptors (Sadock et al., 2009).
In animal studies, 5-HTT knockout mouse elicit depressive-like behavior associated with a 50% decrease in the number of serotoninergic cells and a substantial decrease in firing rate in the dorsal raphe nucleus (Lira et al., 2003). Interestingly, the brains of 5-HTT -null mice utilize less glucose compared to the wild type mice both in resting and stimulated conditions (Esaki et al., 2005) linking the behavioral phenotype to metabolic anomalies reported in major psychiatric disorders (e.g. Spelman et al 2007; Fernandez-Egea et al., 2008). A transient inhibition of 5-HTT in post natal period by serotonin specific reuptake inhibitors also causes a persistent emotional abnormality in adult mice (Ansorge et al., 2008) underlining the importance of 5-HTT in normal development.
Genetic variations of serotonin receptors and serotonin transporter have been linked to the pathogenesis of anxiety and mood disorders as well as schizophrenia. For example the C allele of T102C promoter polymorphism of HTR2A is linked to schizophrenia which has been confirmed by two meta-analysis (Williams et al., 1999; Abdolmaleky et al 2004). Additionally, the STin2 VNTR polymorphism of 5-HTT is linked to schizophrenia pathogenesis (Fan and Sklar 2005). Furthermore, based on the meta-analyses of published studies, the short allele of 5-HTT promoter polymorphism with a reduced 5-HTT expression has been linked to neuroticism (Sen et al. 2004), alcohol dependence (Feinn et al., 2005), depressive disorders (Kiyohara and Yoshimasu 2010), bipolar disorder (Lasky-Su et al., 2005) but not schizophrenia (Fan and Sklar 2005). The short allele of 5-HTT promoter polymorphism has been implicated with a poor response to SSRI treatment as well (Murphy et al., 2004). A summary of the known and engineered 5-HTT alterations and their corresponding behavioral effects are listed in Table 1.
Table 1.
Behavioral effects of altered functionality of 5-HTT gene
| Study type | Modification | Change of function | Linked to | Ref |
|---|---|---|---|---|
| Genetic (human) | STin2 VNTR polymorphism | unknown | SCZ | Fan and Sklar 2005 |
| Promoter polymorphism (short allele) | Reduced expression | BD, neuroticism, alcoholism, depression, poor response to SSRIs | ||
| Animal studies (mice) | Knocked out of 5-HTT | Reduced expression | depressive-like behavior, reduced brain glucose use | |
| Transient inhibition of 5-HTT by SSRIs | Inhibition | persistent emotional abnormality in adult mice | ||
| Animal study (rats) | Knocked out of 5-HTT | Reduced expression | Altered characteristics of the raphe neurons and their outgrowing neurites to medial prefrontal cortex and altered identity of the cortical neurons | |
| Human | SSRIs use in pregnancy | Inhibition | Autism spectrum disorder in offspring | |
| Childhood bullying victimization | Promoter DNA hypermethylation | blunted cortisol response to stress | ||
| Childhood sexual abuse | Promoter DNA hypermethylation | antisocial behavior in adulthood | ||
| Female nurses working in stressful environment | Promoter DNA hypomethylation | - |
The small effect size of known genetic variants in disease pathogenesis and to inconsistency in research findings (Hamshere et al., 2012), led us to hypothesize that epigenetic fine tuning may be a buffering mechanism to compensate for genetic defects. We have previously sought to define the potential contribution of epigenetic variations of mono-aminergic genes by examining both the DNA derived from post-mortem brains and saliva of patients with schizophrenia and bipolar disorder. We found that epigenetic dysregulation of MB-COMT and HTR2A in the brain of patients with SCZ and BD, associated with an early age of disease onset, was attenuated with anti-psychotic drugs (Abdolmaleky et al., 2006 and 2011). We also observed the retention of some of these epigenetic modifications in DNA extracted from saliva samples of patients with SCZ and BD (Nohesara at al., 2011; Ghadiri et al., 2011). Other investigators have also reported age dependent epigenetic variations of other mono-aminergic genes such as DRD4, MAOA and 5-HTT in the blood of dizygotic as well as monozygotic twins (Wong et al., 2010). The latter studies also uncovered that bullying victimization is associated with an increased 5-HTT promoter DNA methylation and a blunted cortisol response to stress in affected twins versus non-bullied twins as examined at age 10 (Ouellet-Morin et al., 2012). However, other studies on adults found hypomethylation of 5-HTT promoter in the blood cells of female nurses working in highly stressed environment (Alasaari et al., 2012). Additionally, it has also been reported that higher degree of 5-HTT promoter methylation may be protective against PTSD in individuals exposed to traumatic events (Koenen et al., 2011). In non-human primates, while DNA methylation of 5-HTT was not different in female bonnet macaques exposed to early life stress versus those raised in control condition, a greater degree of 5-HTT methylation “was associated with higher behavioral stress reactivity” (Kinnally et al., 2011).
Recent reports also indicated that, while the expression of 5-HTT is affected by methylation of specific CpGs of 5-HTT promoter, the methylation of these CpGs are affected by genotype as well as the incidence of childhood sexual abuse (Vijayendran et al., 2012). Hypermethylation of 5-HTT promoter DNA has also been reported in the blood as well as post-mortem brain samples of patients with bipolar disorder (Sugawara et al., 2011). Collectively, these findings suggest an important role for epigenetic dysregulation of 5-HTT in the pathogenesis of major mental diseases including SCZ.
Here, we describe the promoter DNA methylation analysis for the 5-HTT gene in post mortem brain as well as saliva samples to examine if similar epigenetic aberrations could be observed in the brain and saliva samples of patients with SCZ.
Materials and Methods
Post-mortem Brain Samples and Statistical Analysis:
We obtained 105 DNA and RNA samples extracted from the frontal lobe dissects of patients with SCZ, BD and matched control subjects (Table 2) from the Stanley Medical Research Institute (S. Additionally, a total of 100 saliva samples from the patients with SCZ, BD and control subjects (consisting of 30 controls, 30 SCZ, 20 BD and 20 first degree relatives of SCZ or BD patients) were obtained from the saliva bank of Tehran Psychiatric Institute. The saliva samples were collected according to the regulations of the local institutional review board and donated to us without identifiers for genetic/epigenetic analysis. Before the sample collection the study subjects were informed of the purpose of study and upon their informed consent they were referred to the Tehran psychiatric Institute for confirmatory diagnostic evaluations by two psychiatrists based on the Structured Clinical Interview for DSM VI-R. Data related to demographics, family history, age of disease onset, duration of disease, and drug use was also recorded. The patients with substance dependency (with the exception of cigarette smoking), mental retardation, neurological diseases and active medical conditions were excluded from the study. Following the consenting process, 2 ml saliva samples were collected using Oragene Saliva Collection Kit (DNAgenotek, Ottawa, Canada). An additional 20 saliva samples, matched for age and other demographics, were also collected from unaffected first degree relatives of patients with SCZ (13 samples) and BD (7 samples) for the same epigenetic analysis. Matched normal controls were also interviewed and subjects with a history or family history of mental diseases, substance dependency (with the exception of cigarette smoking) and active medical conditions were excluded.
Table 2.
Post-mortem brain sample characteristics.
| Sample source | Diagnosis | Number of cases | Sex M/F | Age mean (SD) | Laterality L/R | Smoker | Severely alcoholic | Committed suicide | Drug free |
|---|---|---|---|---|---|---|---|---|---|
| SMRI | SCZ | 35 | 26/9 | 42.5 (8.47) | 17/18 | 23 | 9 | 7 | 3 |
| BD | 35 | 8/17 | 45.2 (10.5) | 20/15 | 16 | 7 | 15 | 8 | |
| Control | 35 | 26/9 | 44.2 (7.63) | 16/19 | 9 | 0 | 0 | N/A |
Determination of methylation status and mapping differentially methylated CpG islands of 5HTT promoter:
The whole genome DNA methylation profiling was initially performed for a total of 24 samples (including two saliva samples) using the Illumina 27K (for 12 samples) and 450K (for another 12 samples) DNA methylation array platforms, followed by bisulfite sequencing to identify candidate CpGs for subsequent target gene analysis in the total samples. Quantitative methylation specific PCR (qMSP) was employed to assess the degree of CpG methylation of 5-HTT promoter using previously described methods (Abdolmaleky et al., 2008) and primers shown in Table 3. In brief, DNA was extracted from the brain sample using TRIzol, and from the saliva samples according to the instruction of the manufacturer (DNAgenotek, Ottawa, Canada). Next, 1μg of genomic DNA was chemically modified with sodium bisulfite using Qiagen bisulfite modification kit (EpiTec Bisulfite Kit, Cat#59104) to convert the unmethylated cytosines to thymine. Several primer pairs were used (Table 3 and Figure 1) to amplify 5-HTT promoter region for bisulfite sequencing as well as to perform MSP to detect methylated and unmethylated DNA in separate PCR reactions (Figure 2, A & B). PCR products were run in a 6% Acrylamide gel to ascertain that the primers amplify a single targeted product (Figure 2, B & C). Melting curve analysis was also used in subsequent real time PCR based qMSP using SYBR green to confirm the presence of a single PCR product.
Table 3.
primers for bisulfite sequencing and qMSP analysis of 5-HTT promoter region
| Primers/application | Forward | Reverse | Ann. Temp. |
|---|---|---|---|
| Bisulfite sequencing F1R1 (Site A) | TAAGGGTTTTTAAGTTGAGTTTATATTT | AAAATCCTAACTTTCCTACTCTTTAACTTTA | 55º C |
| Bisulfite sequencing nested F2R2 (Site A) | TTAGGTTTTAGGAAGAAAGAGAGAGTAGT | AAAAAAAAAAAACTACACAAAAAAACAAATATA | 57º C |
| Bisulfite sequencing F3R3 (Site B) | GGAGGYGTATATTTGTTTTTTTGTGTAGTT | AACCCTCACATAATCTAATCTCTAAATAA | 55º C |
| MSP F1R1 M | CGGGCGCGTATTTCGTTTCGTAGC | GCCAAAAAACTCTTAAAAAATTTTTACG | 57º C |
| MSP F1R1 U | TGGGTGTGTATTTTGTTTTGTAGT | ACCAAAAAACTCTTAAAAAATTTTTACA | 57º C |
| qMSP F1R1 M (Site A) | TCGAGGTTAAGAGAAAGCGGTAC | AAAAAAAAAAAACTACACAAAAAAACAAATATACG | 57º C |
| qMSP F1R1 U (Site A) | TTGAGGTTAAGAGAAAGTGGTAT | AAAAAAAAAAAACTACACAAAAAAACAAATATACG | 57º C |
| qMSP M F2R2 (Site B) | CGCGGAGTCGCGGTCGC | CCCTCACATAATCTAATCTCTAAATAACCG | 57º C |
| qMSP U F2R2 (Site B) | TGTGGAGTTGTGGTTGT | CCCTCACATAATCTAATCTCTAAATAACCA | 57º C |
| β-Actin promoter for qMSP analysis | GGTGGGTTTAGATTTAGGTTGTGTA | TAAAACTACCTACTTTTAAAAATAACAATCAC | 57º C |
| 5-HTT expression | GCATCCCCACATATATAGCTTATCGG | GGAATTTCTGTTGGTGTTTCTGG | 60º C |
| β-Actin expression | CGAGCACAGAGCCTCGCCTTTGCC | TGTCGACGACGAGCGCGGCGATAT | 60º C |
M: methylated primer, U: unmethylated primer
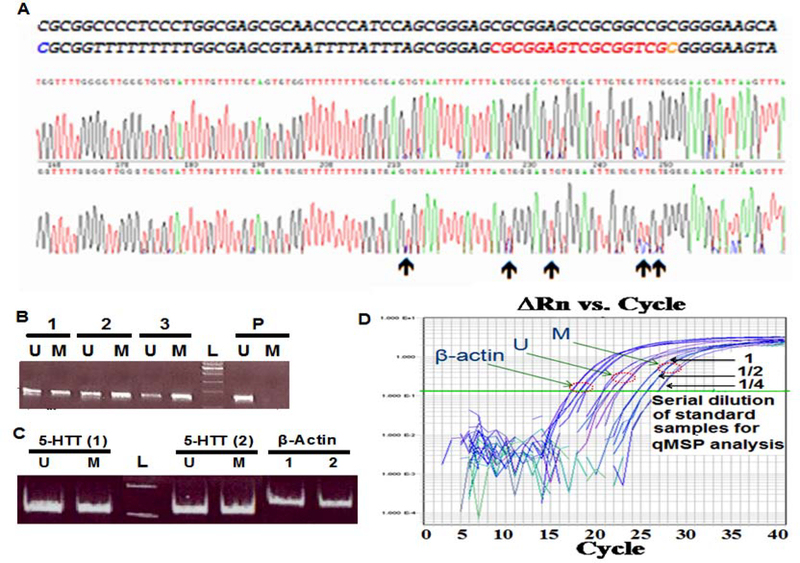
Figure 1. The 5-HTT promoter sequence and the location of primers.
The upper lines show original DNA sequence and the lower lines bisulfite modified DNA sequence assuming that all of cytosines (C) followed by guanine (G) are methylated. The candidate CpGs for methylation are in bold and gray. DNA sequences flanking the MSP and qMSP primer sites amplifying methylated (M) and unmethylated (U) products are underlined. The site corresponding to the Probe # CG26741280 of the Illumina DNA methylation array is underlined and bolded in original DNA sequence. There are two SP1 binding sites in 5-HTT promoter region which are marked with yellow color (GGGCGG and CCGCCC).
Figure 2. Bisulfite sequencing of the 5-HTT promoter region and MSP analysis.
A. Bisulfite sequencing of 5-HTT promoter region in representative samples for identification overall methylation pattern and partially methylated sites (indicated by arrows) for subsequent MSP and qMSP analysis. The original DNA sequence (top line) and bisulfite modified DNA sequence (bottom line) are placed at the top of sequence traces. The red segment of the bottom line shows a representative differentially methylated site. B. MSP analysis of a representative control (1), SCZ (2) and BD (3) using post-mortem brain samples. Placental DNA (P) was used as a negative control for methylation. C. MSP analysis of saliva DNA of a representative control subject (1) and a SCZ patient (2) using F1R1 qMSP primers. The β-Actin promoter amplified with primers designed from a CpG free region was used for the normalization of unmethylated (U) and methylated (M) products during qMSP analysis. L indicates a 100bp DNA ladder. D. Serial dilution of standard samples to optimize conditions for qMSP analysis in the test trials.
Optimizing the conditions for qMSP analysis using SYBR green:
The optimal conditions to obtain a reliable standard curves was achieved using different concentrations of primers as well as serially diluted standard samples, including unmethylated placental DNA and in vitro methylated DNA in the test trials (Figure 2C). SYBR green based relative qMSP analysis was performed to evaluate the degree of promoter DNA methylation using the ΔΔCT method of quantification normalized with the PCR product of β-Actin promoter as well as the 5-HTT promoter (using F2R2 sequencing primers) amplified with primers designed from a CpG free region (Table 3 and Figure 1). Brain and saliva samples were analyzed by qPCR using the ABI 7900 and ABI 7500 instruments, respectively. Similar to other qPCR analyses and based on the instruction of ABI, the fold changes were calculated as 2-ΔΔCT for relative quantification of methylated and methylated products. In brief, first the CT (cycle threshold) of unmethylated or methylated PCR product of each sample was subtracted from the CT of β-Actin PCR product (amplified with primers designed from CpG free region of β-Actin promoter) to obtain the ΔCT of methylated and unmethylated PCR product for each sample. Then the values for methylated and unmethylated products were subtracted from the ΔCT of in vitro methylated and unmethylated placental DNA, respectively to obtain ΔΔCT of methylated and unmethylated PCR products for each sample. Next using the 2-ΔΔCT formula, the value for unmethylated and methylated PCR products were obtain, separately. In order to calculate the degree of DNA methylation, we used M/M+U equation, where M and U represent the values for methylated and unmethylated products, respectively.
Total RNA isolation and 5-HTT expression analysis:
RNA was extracted from the brain dissects using TRIzol and mRNA was reverse transcribed to synthesize cDNA. In test trials the cDNA was amplified using Platinum Taq DNA polymerase (Invitrogen) and gene specific primers as previously described (Abdolmaleky et al., 2006). The expression of 5-HTT was normalized to β -Actin expression during qRT-PCR analysis using SYBR green methodology and fold changes were calculated as 2- ΔΔCT. The t test was used for the statistical analysis of quantitative PCR data for the comparison of different study groups. Additionally, we performed whole transcriptome analysis for 30 postmortem brain samples dissected from the dorso-lateral frontal cortex of patients with SCZ and BD and control subjects (group of 10 each) using Affymetrix GeneChip Human Genome U133 Plus 2.0 array platform. The array data was analyzed according the standard methods (Gower et al., 2011). The data from the entire sets of expression and DNA methylation analyses will be presented elsewhere depicting comprehensive gene connections (manuscript in preparation). However, here we use all the relevant expression/methylation data pertaining to 5-HTT to aid in the current study.
Results
Higher 5-HTT promoter DNA methylation in post-mortem brains and saliva of drug naïve SCZ patients:
The Illumina whole genome DNA methylation profiling along with bisulfite DNA sequencing of the 5-HTT promoter region (Figure 2, panel A) in representative samples defined partially methylated CpGs of the 5-HTT promoter in DNA extracted from the human post mortem brain tissues affected by SCZ and BD as well as from representative saliva samples. Bisulfite sequencing and MSP analysis of 5-HTT promoter region showed both unmethylated and methylated product in SCZ and BD patients as well as in controls (Figure 2, panel A–C).
The qMSP analysis targeted partially methylated CpGs of the Illumina probe # CG26741280 (Figure 1, site A showing an average of ~20% methylation based on the DNA array data) exhibited higher degree of methylation only in three drug free SCZ patients compared to the controls and other SCZ patients (Almost doubled, p=0.04 and p=0.038, respectively) in post mortem brain samples. However, the difference between controls and those patients who were under antipsychotic treatment was not statistically significant (data not shown).
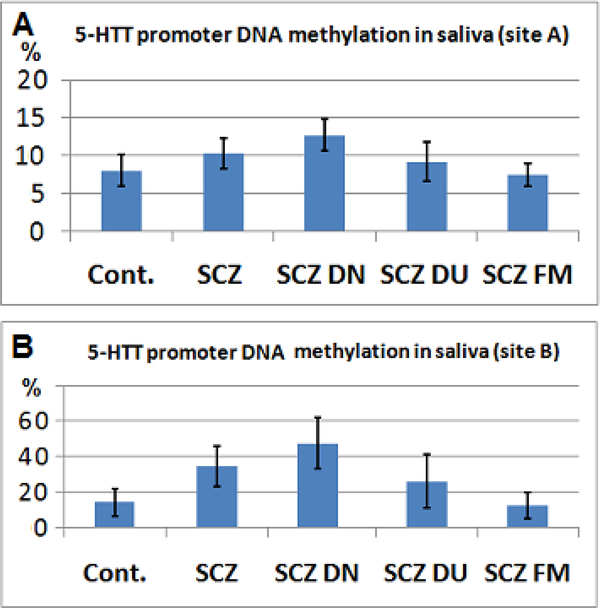
This site also exhibited a significant hypermethylation of 5-HTT promoter in DNA extracted from the saliva of patients with SCZ (~30%, p=0.039, Figure 3A), which was much more significant in ten drug naïve SCZ patients compared to controls (>50%, p= 0.0025, two tailed t test). Similar to the post-mortem brain samples, there was no statistically significant difference between the controls and those patients who were under antipsychotic drugs (Figure 3A). Additionally, the difference between the controls and unaffected first degree relatives of patients with SCZ was not statistically significant (p=0.37).
Figure 3. Promoter DNA methylation status of 5-HTT in SCZ versus control subjects.
A. There was higher promoter DNA methylation in SCZ patients compared to the controls and first degree relatives of SCZ patients (SCZ FM) at site A. Notably, SCZ patients and drug naïve SCZ patients (DN SCZ) exhibit higher degree of DNA methylation. Drug use in SCZ patients (DU SCZ) reduced 5-HTT promoter DNA hypermethylation. B. Similar to site A, the 5-HTT promoter DNA methylation in SCZ was higher than controls at site B, particularly in drug naïve patients.
The qMSP analysis targeted site B (Figure 1) showed that, the degree of methylation in DNA extracted from saliva at this site was more than the brain tissues and significantly higher in SCZ compared to controls (~two times, p=0.038), particularly in drug naïve SCZ patients (~ three times, 0.0035, Figure 3B). However, there was no significant DNA methylation differences between controls and the first degree relatives of SCZ patients (p=0.34) as well as in controls and BD patients (p=0.4) or controls and the SCZ patients who were under antipsychotic treatment (p=0.3).
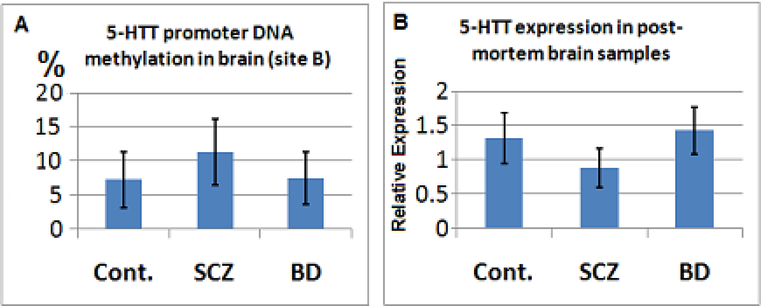
The qMSP analysis of targeted CpGs surrounding site B for the post-mortem brain samples showed a trend for DNA hypermethylation of 5-HTT promoter in SCZ patients (~35%, Figure 4A), particularly in drug free SCZ patients (~60%, p=0.061). Similarly in antipsychotic free BD patients, there was a trend for higher DNA methylation compared to other BD patients (~50%, p=0.066). Nevertheless, the overall DNA methylation levels at this site was trivial (~10%) in post mortem brain samples. There was also a trend towards reduced 5-HTT promoter methylation with older age in the brains of control subjects (data not shown), but not in SCZ or BD patients that could be a reflection of drug use. Furthermore, a longer duration of antipsychotic use (particularly atypical antipsychotics) was associated with reduced 5-HTT promoter DNA methylation (data not shown).
Figure 4. Promoter DNA methylation and expression and analysis for 5-HTT gene in post-mortem brains of patients with SCZ and BD versus the control subjects.
A: The Y axis indicates promoter DNA methylation of 5-HTT (site B) normalized with the PCR product of β-Actin promoter in SCZ and BD versus controls. B: The Y axis indicates relative expression of 5-HTT normalized with β-Actin expression. As shown, in SCZ the expression of 5-HTT is significantly less than controls as well as BD patients.
Lower expression of 5-HTT in post mortem brain tissues of SCZ patients compared to control subjects and BD patients:
The whole genome transcriptome analysis of post mortem brain samples from patients with SCZ and BD versus control samples (each group 10) revealed a highly significant reduction in the expression of 5-HTT in SCZ patients (~50%, p=0.008). Real-time PCR analysis of the whole post mortem brain samples (each group 35 samples) also found statistically significant decrease in the 5-HTT expression in SCZ (~40%, p=0.035, student t test) but not in BD (p=0.3) versus the control subjects. In fact, in BD the expression of 5-HTT was significantly more than SCZ patients (p=0.014, two tailed t test; Figure 4B). In three drug free SCZ patients the expression of 5-HTT in post-mortem brains was half compared to other SCZ patients (p=0.072) and 1/3 compared to the controls (p=0.022, two tailed t test). The expression of 5-HTT in post-mortem brain samples was inversely correlated with the promoter DNA methylation of site B, in particular (Figure 4A vs. 4B).In BD patients antipsychotic use was associated with 15% increase in 5-HTT expression though non-significant. There was a trend for high 5-HTT Expression in smokers (data not shown). In general and in each study group. Therefore, considering higher rate of smoking in SCZ patients (Table 2), the magnitude of reduced 5-HTT expression might be partially overcome due to smoking of patients. However, we found no conclusive pattern of altered DNA methylation as a result of smoking.
Discussion
We found that there was reduction in the expression of 5-HTT in postmortem brain tissues of patients with SCZ (but not in BD) that was associated with corresponding increase in the promoter DNA methylation. The magnitude of the reduced expression was much larger in drug free SCZ patients. Follow up studies using DNA extracted from the saliva of patients with SCZ and BD also exhibited DNA hypermethylation of 5-HTT promoter in drug naïve SCZ patients (but not in BD). In fact, the difference between SCZ patients who were under drug treatment compared to the control subjects was not statistically significant, suggesting that antipsychotic drugs reverse epigenetic dysregulation of 5-HTT in SCZ patients. This also indicate that an increased serotonin signaling due to epigenetically determined hypo-activity of 5-HTT resulting in decreased reuptake of the released serotonin from the synaptic cleft, may contribute to SCZ pathogenesis. Consistent with this hypothesis, while LSD, a partial agonist of serotonin type-2 receptors (HTR2A) induces psychosis, the most effective antipsychotic drugs block HTR2A to inhibit the transmission of serotonin signaling to postsynaptic neurons. Therefore, it has been suggested that while nor-epinephrine and dopamine reuptake is responsible for euphoria and the development of dependency, serotonin reuptake inhibition is linked to the development of psychosis (Rothman et al., 2001).
Although each serotoninergic neuron modulate ~500,000 target neurons and each cortical neuron receives almost 200 inputs from the serotoninergic neurons, and an appropriate amount of serotonin is re-quired for neuronal growth and synaptogenesis (Kandel, 2001; Sadock et al., 2009), an excess amount of serotonin may lead to improper syn-apse formations due to recruitment of cell adhesion molecules (Broman and Fletcher, 1999). Based on animal studies knockdown of 5-HTT results in extensive developmental alterations in the characteristics of the raphe neurons, projection of their outgrowing neurites to the frontal lobe and the identity of medial prefrontal cortex neurons (Witteveen et al., 2013). In humans the uses of serotonin specific reuptake inhibitors during pregnancy, particularly in the first trimester has been linked to higher frequency of autism spectrum disorders in offsprings (Croen et al., 2011) supporting that an epigenetic down-regulation of 5-HTT may have similar impacts on the brain architecture and disease development.
From the analysis of the post-mortem brains, we found that atypical antipsychotic drugs were much more efficient in reducing 5-HTT promoter DNA methylation as well as increasing the gene expression compared to classical antipsychotics both in SCZ and BD patients. Interestingly, our studies revealed that the degree of DNA methylation of 5-HTT promoter region in antipsychotic free SCZ and BD patients were almost doubled compared to the patients who were under antipsychotic treatment both in post-mortem brains as well as saliva samples. In fact, SCZ and BD patients who were under atypical antipsychotic drugs, exhibited a ~50% decrease in the degree of 5-HTT promoter DNA methylation with an associated approximately 40% and 20% increase in the gene expression, respectively, compared to the other patients. While animal studies also showed that antipsychotic drugs can change the expression levels of many other genes involved in the pathogenesis and/or treatment of SCZ (Fatemi et al., 2012), several studies have shown that epigenetic alterations mediate a large part of these expression changes (e.g. Li et al., 2004, Grayson 2010; Abdolmaleky et al., 2006 and 2011; Akbarian 2012). Interestingly, antipsychotic drugs have also been shown to decrease 5-HTT promoter DNA methylation in other studies (Vijayendran et al., 2012) associated with increased 5-HTT expression. The results of our studies indicate that, in addition to the blockade of 5-HT2A receptors, antipsychotic drugs may exert their effects through epigenetic up-regulation of 5-HTT and thus a more efficient serotonin reuptake from the synaptic cleft.
More importantly, our data suggest that 5-HTT promoter methylation status of DNA extracted from saliva (or blood) could serve as a disease biomarker to select the best antipsychotic drug for the treatment of the affected individuals, and to perform subsequent 5-HTT promoter methylation analysis to monitor the efficacy of the drug during the course of SCZ treatment. These findings gains additional support from the fact that; the identified aberrant promoter DNA methylation of 5-HTT is not present in the first degree relative of SCZ patients as well as patients with bipolar disorder. Hence, it can be considered as a disease specific epigenetic biomarker responsive to drug treatment. In this line, a recent in vivo positron emission tomography (PET) analysis also provided evidence that serotonin synthesis in the orbitofrontal cortex of human brain is correlated to the 5-HTT promoter methylation status of the blood cells (Wang et al., 2012).
In conclusion, despite there has been recent progress in identifying the potential utilities of individual epigenetic biomarkers and therapeutic targets, it is also important to note that, complex diseases such as schizophrenia and bipolar disorder are polygenic and multi-factorial in origin and hence, a panel of diagnostic or therapeutic biomarkers, and an understanding of their network interactions could provide more accurate and comprehensive approach to the disease management.
ACKNOWLEDGMENT
Postmortem DNA and RNA samples were donated by The Stanley Brain Collection courtesy of Drs. Michael B. Knable, E. Fuller Torrey, Maree J. Webster and Robert H. Yolken. Saliva samples for DNA extraction were donated by the Tehran Psychiatric Institute, Tehran University of Medical Sciences (TUMS). The authors express their gratitude to the Stanley Medical Research Institute and TUMS for providing DNA and RNA samples. This work was supported by a grant from Mental Health Research Center, TUMS, NARSAD Independent Investigator Award to Dr. Sam Thiagalingam and CTSI, Boston University (NIH CTSA, UL1-TR00157).
References:
- Abdolmaleky HM, Faraone SV, Glatt SJ and Tsuang MT., 2004. Meta-analysis of Association Between the T102C Polymorphism of the 5HT2a Receptor Gene and Schizophrenia. Schizophrenia Res, 67(1) 53–62. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aleali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S., 2006. Hypomethylation of MB-COMT Promoter is a Major Risk Factor for Schizophrenia and Bipolar Disorder, Hum Mol Genet. 15(21) 3132–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Smith CL, Zhou JR, Thiagalingam S (2008). Epigenetic alterations of the dopaminergic system in major psychiatric disorders. Methods Mol Biol. 448:187–212. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Yaqubi S, Papageorgis P, Lambert AW, Ozturk S, Sivaraman V, Thiagalingam S., 2011. Epigenetic dysregulation of HTR2A in the brain of patients with schizophrenia and bipolar disorder. Schizophr Res. 129(2–3) 183–90.21550210 [Google Scholar]
- Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimäki M, Vahtera J, Kronholm E, Härmä M, Puttonen S, Paunio T., 2012. Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS One. 7(9) e45813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, 2010. Epigenetics of schizophrenia. Curr Top Behav Neurosci. 4:611–28. [DOI] [PubMed] [Google Scholar]
- Ansorge MS, Morelli E, Gingrich JA., 2008. Inhibition of serotonin but not norepinephrine transport during development produces delayed, persistent perturbations of emotional behaviors in mice. J Neurosci. 28(1) 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman SH. and Fletcher JM. 1999: The changing nervous system. Neurobehavioral consequences of early brain disorder. New York: P3–93. [Google Scholar]
- Croen LA, Grether JK, Yoshida CK, Odouli R, Hendrick V (2011). Antidepressant use during pregnancy and childhood autism spectrum disorders. Arch. Gen. Psychiatry, 68(11):1104–1112. [DOI] [PubMed] [Google Scholar]
- Esaki T, Cook M, Shimoji K, Murphy DL, Sokoloff L, Holmes A., 2005. Developmental disruption of serotonin transporter function impairs cerebral responses to whisker stimulation in mice. Proc Natl Acad Sci U S A. 102(15) 5582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JB, Sklar P., 2005. Meta-analysis reveals association between serotonin transporter gene STin2 VNTR polymorphism and schizophrenia. Mol Psychiatry. 10(10) 928–938. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Reutiman TJ, Novak J, Engel RH., 2012. Comparative gene expression study of the chronic exposure to clozapine and haloperidol in rat frontal cortex. Schizophr Res. 2012 Feb;134(2–3):211–8. [DOI] [PubMed] [Google Scholar]
- Feinn R, Nellissery M, Kranzler HR., 2005. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 133B(1) 79–84. [DOI] [PubMed] [Google Scholar]
- Fernandez-Egea E, Miller B, Bernardo M, Donner T, Kirkpatrick B., 2008. Parental history of type 2 diabetes in patients with nonaffective psychosis. Schizophr Res. 98(1–3) 302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadirivasfi M, Nohesara S, Ahmadkhaniha HR, Eskandari MR, Mostafavi S, Thiagalingam S, and Abdolmaleky HM., 2011. Hypomethylation of the serotonin receptor type-2A gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder, Am J Med Genet B Neuropsychiatr Genet. 156(5) 536–45. [DOI] [PubMed] [Google Scholar]
- Gower AC, Steiling K, Brothers JF 2nd, Lenburg ME, Spira A, 2011. Transcriptomic studies of the airway field of injury associated with smoking-related lung disease. Proc Am Thorac Soc. 8(2) 173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR. 2010. Schizophrenia and the epigenetic hypothesis. Epigenomics. 2(3) 341–4. [DOI] [PubMed] [Google Scholar]
- Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, Jones I, Forty L, Jones L, Gordon-Smith K, Riley B, O’Neill T, Kendler KS, Sklar P, Purcell S, Kranz J., 2012. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 18(6) 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER., (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294:1030–8. [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Feinberg C, Kim D, Ferguson K, Leibel R, Coplan JD, John Mann J., 2011. DNA methylation as a risk factor in the effects of early life stress. Brain Behav Immun. 25(8) 1548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara C, Yoshimasu K.. 2010. Association between major depressive disorder and a functional polymorphism of the 5-hydroxytryptamine (serotonin) transporter gene: a meta-analysis. Psychiatr Genet. 20(2) 49–58. [DOI] [PubMed] [Google Scholar]
- Koenen KC, Uddin M, Chang SC, Aiello AE, Wildman DE, Goldmann E, Galea S., 2011. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depress Anxiety. 28(8) 639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su JA, Faraone SV, Glatt SJ, Tsuang MT., 2005. Meta-analysis of the association between two polymorphisms in the serotonin transporter gene and affective disorders. Am J Med Genet B Neuropsychiatr Genet. 133B(1) 110–115. [DOI] [PubMed] [Google Scholar]
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, Ferris C, Konradi C, Akbarian S., 2004. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem. 90(5) 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira A, Zhou M, Castanon N, Ansorge MS, Gordon JA, Francis JH, Bradley-Moore M, Lira J, Underwood MD, Arango V, Kung HF, Hofer MA, Hen R, Gingrich JA. 2003. Altered depression-related behaviors and functional changes in the dorsal raphe nucleus of serotonin transporter-deficient mice. Biol Psychiatry. 54(10) 960–71. [DOI] [PubMed] [Google Scholar]
- Murphy GM Jr, Hollander SB, Rodrigues HE, Kremer C, Schatzberg AF., 2004. Effects of the serotonin transporter gene promoter polymorphism on mirtazapine and paroxetine efficacy and adverse events in geriatric major depression. Arch Gen Psychiatry. 61(11) 1163–9. [DOI] [PubMed] [Google Scholar]
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM., 2011. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 45(11) 1432–8. [DOI] [PubMed] [Google Scholar]
- Park BY, Lee BC, Jung KH, Jung MH, Park BL, Chai YG, Choi IG., 2011. Epigenetic changes of serotonin transporter in the patients with alcohol dependence: methylation of an serotonin transporter promoter CpG island. Psychiatry Investig. 8(2) 130–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet-Morin I, Wong CC, Danese A, Pariante CM, Papadopoulos AS, Mill J, Arseneault L., 2012. Increased serotonin transporter gene (SERT) DNA methylation is associated with bullying victimization and blunted cortisol response to stress in childhood: a longitudinal study of discordant monozygotic twins. Psychol Med. 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadock BJ, Sadock VA, Ruiz P., 2009. Kaplan and Sadock’s Comprehensive Textbook of Psychiatry, 9 ed, Vol 1 Philadelphia, PA: Lippincott Williams & Wilkins. [Google Scholar]
- Sen S, Burmeister M, Ghosh D., 2004. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet. 127B(1) 85–9. [DOI] [PubMed] [Google Scholar]
- Spelman LM, Walsh PI, Sharifi N, Collins P, Thakore JH., 2007. Impaired glucose tolerance in first-episode drug-naive patients with schizophrenia. Diabet. Med., 24, pp. 481–485. [DOI] [PubMed] [Google Scholar]
- Sugawara H, Iwamoto K, Bundo M, Ueda J, Miyauchi T, Komori A, Kazuno A, Adati N, Kusumi I, Okazaki Y, Ishigooka J, Kojima T, Kato T., 2011. Hypermethylation of serotonin transporter gene in bipolar disorder detected by epigenome analysis of discordant monozygotic twins. Transl Psychiatry. 1 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendran M, Beach SR, Plume JM, Brody GH, Philibert RA., 2012. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Front Psychiatry. 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Szyf M, Benkelfat C, Provençal N, Turecki G, Caramaschi D, Côté SM, Vitaro F, Tremblay RE, Booij L., 2012. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS One. 7(6) e39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner FM, Coveñas R., 2010. Classical neurotransmitters and neuropeptides involved in major depression: a review. Int J Neurosci. 120(7) 455–470. [DOI] [PubMed] [Google Scholar]
- Williams J, McGuffin P, Nothen M, Owen MJ, 1997. Meta-analysis of association between the 5-HT2a receptor T102C polymorphism and schizophrenia. EMASS Collaborative Group. European Multicentre Association Study of Schizophrenia. Lancet. 26,349(9060)1221. [DOI] [PubMed] [Google Scholar]
- Witteveen JS, Middelman A, van Hulten JA, Martens GJ, Homberg JR, Kolk SM., 2013. Lack of serotonin reuptake during brain development alters rostral raphe-prefrontal network formation. Front Cell Neurosci. 7: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J., 2010. A longitudinal study of epigenetic variation in twins. Epigenetics. 5(6) 516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]