Dong et al. show that the epigenetic regulator Brd4 forms a complex with transcription factors IRF8/PU.1 to activate the transcription of NLRC4 inflammasome component Naips in macrophages. Brd4-dependent inflammasome activation protects mice from Salmonella infection via cytokine release and pyroptosis.
Abstract
NLRC4 inflammasome activation and the subsequent maturation of IL-1β and IL-18 are critical for protection against infection by bacterial pathogens. The epigenetic regulator Brd4 has emerged as a key player in inflammation by regulating the expression of inflammatory cytokines. However, whether Brd4 has any role in inflammasome activation remains undetermined. Here, we demonstrated that Brd4 is an important regulator of NLRC4 inflammasome activation in response to Salmonella typhimurium infection. Brd4-deficient bone marrow–derived macrophages (BMDMs) displayed impaired caspase-1 activation, ASC oligomerization, IL-1β maturation, gasdermin-D cleavage, and pyroptosis in response to S. typhimurium infection. RNA sequencing and RT-PCR results revealed that the transcription of Naips was decreased in Brd4-deficient BMDMs. Brd4 formed a complex with IRF8/PU.1 and bound to the IRF8 and PU.1 binding motifs on the promoters of Naips to maintain the expression of Naips. Furthermore, myeloid lineage–specific Brd4 conditional knockout mice were more susceptible to S. typhimurium infection with increased mortality, bacterial loads, and tissue damage; impaired inflammasome-dependent cytokine production; and pyroptosis. Our studies identify a novel function of Brd4 in innate immunity by controlling inflammasome-mediated cytokine release and pyroptosis to effectively battle S. typhimurium infection.
Introduction
Inflammasomes modulate cellular responses to pathogen- or danger-associated molecular patterns and are essential for the innate immune response against pathogen infection. The inflammasome complex is a cytosolic multiprotein complex consisting of a sensor of nucleotide-binding domain and leucine-rich repeat receptors (NLRs) or absent in melanoma 2 (AIM2)-like receptors (ALRs), the adaptor molecule apoptosis-associated speck-like protein containing a CARD (ASC), and the effector subunit pro–caspase-1 (Guo et al., 2015). Upon recognizing specific substances produced during infection or tissue damage, the corresponding NLRs, including NLR family pyrin domain containing 1 (NLRP1), NLRP3, or NLR family CARD domain-containing protein 4 (NLRC4), nucleate adaptor ASC to form ASC speck structures around the nucleus. This leads to subsequent clustering of pro–caspase-1, resulting in the autocleavage of pro–caspase-1 into activated caspase-1, which catalyzes proteolytic cleavage of pro–IL-1β and pro–IL-18 to mature IL-1β and IL-18 (Lu et al., 2014). In addition, activated caspase-1 cleaves gasdermin-D (GSDMD), and the resulting N-terminal fragment of GSDMD forms pores on the cell membrane to initiate a form of inflammatory cell death, termed “pyroptosis” (Latz et al., 2013; Rathinam et al., 2012).
In response to infection or cellular stress, host cells activate distinct inflammasomes to trigger an inflammatory response (Karki and Kanneganti, 2019). For example, the NLRP3 inflammasome is activated by a variety of substances, including ATP, bacterial toxins, microbial products, viral RNAs, and particulate matter (He et al., 2016). Activation of the NLRP3 inflammasome requires the priming signal that induces NLRP3 expression through nuclear factor-κB (NF-κB) in response to microbial or endogenous molecules (He et al., 2016). The AIM2 inflammasome is activated by cytosolic foreign DNA from invading pathogens or mislocalized self-DNA when the nucleus loses its integrity (Lugrin and Martinon, 2018). The NLRC4 inflammasome is activated in response to bacterial flagellin and type III secretion system (T3SS) apparatus proteins (rod and needle) detected by nucleotide-binding domain and NLR family apoptosis inhibitory proteins (Naips; Miao et al., 2010; Zhao and Shao, 2015). Specifically, Naip1, the orthologue of human NAIP, interacts with the needle component of the T3SS; Naip2 detects the inner rod component of T3SS; and Naip5 and Naip6 both serve as cytoplasmic sensors for bacterial flagellin (Miao et al., 2010; Zhao and Shao, 2015). By binding to Naips, NLRC4 associates with ASC, leading to its oligomerization and the interaction with inactive pro–caspase-1 to facilitate activation of caspase-1 (Duncan et al., 2007). The NLRC4 inflammasome is critically involved in host defense against enteric pathogens, including Salmonella typhimurium (Kofoed and Vance, 2011; Zhao et al., 2011). Naip2-, Naip5-, or Nlrc4-knockout mice were more susceptible to S. typhimurium infection, highlighting the essential role of the NAIP-NLRC4 inflammasome in the induction of host defenses following bacterial infection (Carvalho et al., 2012; Franchi et al., 2012; Zhao et al., 2016). While a great deal is known about how different Naips sense flagellin and T3SS components for NLRC4 inflammasome activation, little is known about how these NLRC4 inflammasome components are regulated.
Due to the essential role of inflammasomes in the innate immune responses to infection and their involvement in the development of inflammatory diseases, the activation of inflammasomes must be tightly controlled to provide appropriate protection against infection while avoiding host tissue damage (Man and Kanneganti, 2015). Various components of the inflammasome can be regulated transcriptionally by different transcription factors, preparing the cells for optimal inflammasome activation (Man and Kanneganti, 2015). For instance, NF-κB–dependent transcriptional priming of Nlrp3 is a prerequisite for NLRP3 inflammasome activation (Bauernfeind et al., 2009). Meanwhile, the transcription of ASC is controlled by IFN-activated gene Ifi205 (Ghosh et al., 2017). Furthermore, the transcription of Gsdmd is regulated by IFN regulatory factor 2 (IRF2) via its direct binding to the promoter (Kayagaki et al., 2019). A recent study also indicates that IRF8 and lineage-determining factor PU.1 are responsible for the transcription of basal levels of Naips and Nlrc4 in resting macrophages (Karki et al., 2018). Irf8-knockout mice have a higher susceptibility to S. typhimurium infection, and Irf8-deficient BMDMs show reduced transcription of Naips and Nlrc4 and impaired NLRC4 inflammasome activation (Karki et al., 2018). IRF8 has relatively weak DNA binding activity, and its dimerization with PU.1 enhances its binding to DNA and chromatin, facilitating IRF8 target gene expression in unstimulated macrophages (Langlais et al., 2016; Laricchia-Robbio et al., 2005; Mancino et al., 2015; Salem et al., 2020). IRF8 and PU.1 bind to the Ets/IRF composite element or IRF-Ets composite sequence (IECS) within the promoters to maintain the basal expression of many genes essential for macrophage functions and differentiation (Mancino et al., 2015; Tamura et al., 2005). Nevertheless, how IRF8 and PU.1 regulate the basal levels of pathogen recognition proteins such as Naips in macrophages remains elusive.
Bromodomain-containing protein 4 (Brd4) has emerged as a key epigenetic regulator for the expression of genes involved in inflammation. Brd4 regulates gene transcription by binding to acetylated histones or nonhistone proteins via its two bromodomains at promoters or enhancers, activating CDK9 of P-TEFb (positive transcription elongation factor b) to stimulate RNA polymerase II (RNAPII)-dependent transcription elongation (Hargreaves et al., 2009; Huang et al., 2009; Jang et al., 2005; Yang et al., 2005). Deletion of Brd4 in myeloid lineage cells or inhibition of Brd4 by small molecules suppresses inflammatory gene expression in macrophages and lipopolysaccharide (LPS)-induced sepsis (Bao et al., 2017; Chen et al., 2016; Dey et al., 2019; Nicodeme et al., 2010). In addition to transcription elongation, Brd4 also regulates inflammatory gene expression via stimulation of enhancer RNAs (Chen et al., 2016; Hah et al., 2015; Xiao et al., 2018). Furthermore, Brd4 could regulate inflammatory gene expression by affecting protein translation (Bao et al., 2017). Brd4 modulates inflammatory gene expression through Mnk2-eIF4E pathway-dependent translational control of IκBα resynthesis (Bao et al., 2017). While these studies underline the critical role of Brd4 in the inflammatory response by its ability to regulate inflammatory gene expression, whether Brd4 has any role in inflammasome-mediated production of IL-1β and IL-18 remains undetermined.
Here, we demonstrate that Brd4 is essential for the optimal expression of the NLRC4 inflammasome components. Brd4 forms a complex with IRF8 and PU.1 and binds to IRF8 and PU.1 binding motifs on the promoters of Naips to facilitate IRF8/PU.1-mediated Naip transcription. These studies identify a novel function of Brd4 in the innate immune response for its ability to regulate expression of Naips and NLRC4 inflammasome activation to control infection from bacteria, including S.typhimurium.
Results
Brd4 is indispensable for optimal NLRC4 inflammasome activation
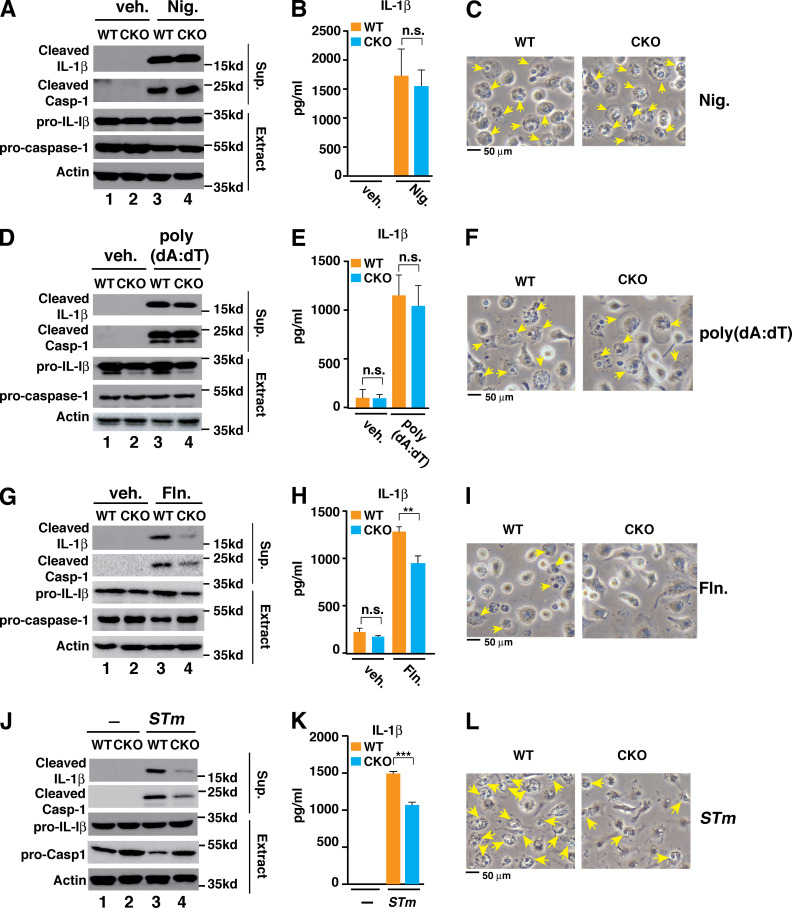
To determine the potential role of Brd4 in inflammasome activation, we challenged the WT and Brd4-deficient BMDMs with various stimuli to activate different inflammasomes, including NLRP3, AIM2, and NLRC4. When nigericin was used to stimulate LPS-primed WT BMDMs, inflammasome activity was apparent as measured by IL-1β processing and secretion, caspase-1 activation, and pyroptotic cell death (Fig. 1, A–C). These inflammasome activation–associated phenotypes were similarly observed in LPS-primed Brd4-deficient BMDMs after nigericin treatment (Fig. 1, A–C), indicating that Brd4 is not involved in nigericin-induced NLRP3 inflammasome activation. We next investigated the involvement of Brd4 in poly(deoxyadenylic-deoxythymidylic) acid sodium salt (poly(dA:dT))-induced AIM2 inflammasome activation. Similar to nigericin-treated macrophages, there were no significant differences in the IL-1β processing and secretion, caspase-1 activation, and pyroptotic cell death between poly(dA:dT)-stimulated WT and Brd4-deficient BMDMs (Fig. 1, D–F). These data suggest that Brd4 is also dispensable for poly(dA:dT)-induced AIM2 inflammasome activation.
Figure 1.
Brd4 is critical for optimal NLRC4 inflammasome activation. (A–C) BMDMs were primed with LPS (0.5 µg/ml) for 4 h followed by treatment with vehicle (veh.) or 10 µM nigericin (Nig.) for 30 min. Culture supernatant (Sup.) and cell lysates (Extract) were collected and immunoblotted for the indicated proteins (A). Levels of IL-1β in the culture media were measured by ELISA (B). (C) Representative images of cells from A. Pyroptotic cells are indicated by yellow arrows. (D–F) BMDMs were primed with LPS (0.5 µg/ml) for 4 h followed by vehicle or 2 µg/ml poly(dA:dT) treatment for 4 h. Immunoblots (D), ELISA for IL-1β (E), and cell images of pyroptosis (F) were assessed the same as in A–C. (G–I) BMDMs were primed with LPS (0.5 µg/ml) for 4 h followed by treatment of vehicle or Salmonella flagellin (Fln.; 2 µg/ml) for 2 h. Immunoblots (D), ELISA for IL-1β (E), and cell images of pyroptosis (F) were assessed the same as in A–C. (J–L) WT and Brd4-deficient BMDMs were infected with S. typhimurium (STm) for 1 h (MOI, 10). Immunoblots (D), ELISA for IL-1β (E), and cell images of pyroptosis (F) were assessed the same as in A–C. Results are presented as mean ± SD of three independent experiments. **, P < 0.01; and ***, P < 0.005 (Student’s t test). n.s., not significant.
We further explored Brd4’s involvement in flagellin-mediated NLRC4 inflammasome activation. As expected, flagellin activated the NLRC4 inflammasome in WT BMDMs with robust IL-1β processing, caspase-1 activation, IL-1β secretion, and pyroptotic cell death (Fig. 1, G–I). In contrast, flagellin-induced inflammasome activation was dramatically impaired in Brd4-deficient BMDMs (Fig. 1 G–I), indicating that Brd4 is required for flagellin-induced NLRC4 inflammasome activation. To further confirm Brd4’s involvement in NLRC4 inflammasome activation, we infected WT and Brd4-deficient BMDMs with log-phase S. typhimurium, which is a potent NLRC4 inflammasome activator (Miao et al., 2010), and found that infection-associated inflammasome activation was decreased in Brd4-deficient BMDMs compared with WT BMDMs (Fig. 1, J–L). Collectively, these data demonstrate that Brd4 is actively involved in the activation of NLRC4 inflammasome but is not required for nigericin-triggered NLRP3 or poly(dA:dT)-activated AIM2 inflammasome activation in macrophages.
Brd4 regulates S. typhimurium Salmonella pathogenicity island 1 (SPI-1)–dependent NLRC4 inflammasome activation
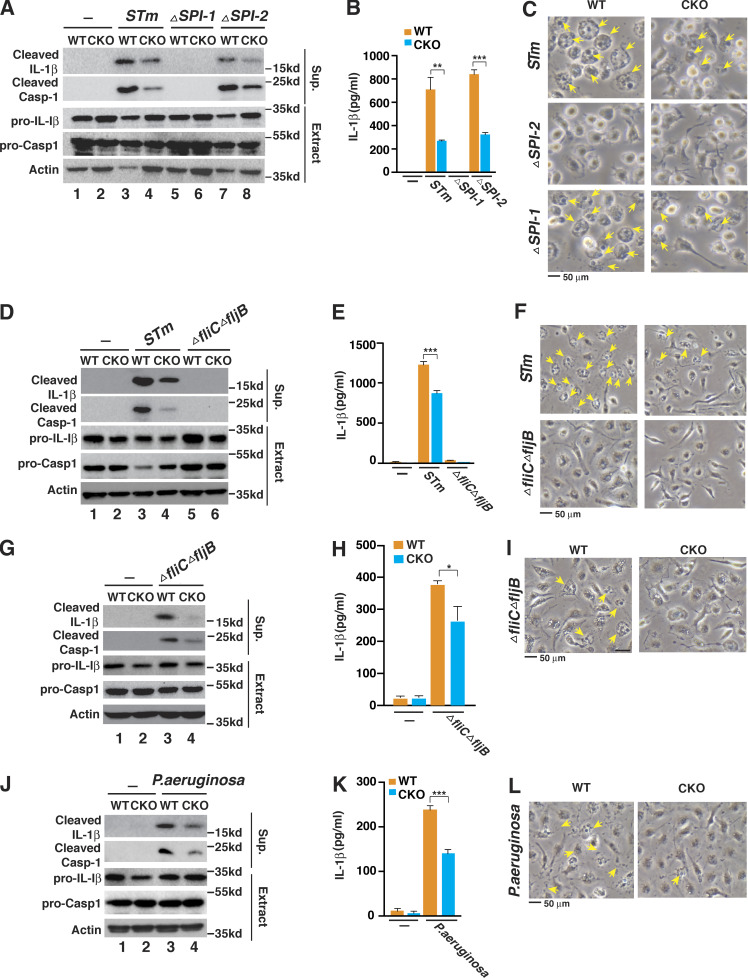
The two different T3SSs, SPI-1 and SPI-2, play different roles in the pathogenicity of S. typhimurium. The SPI-1 T3SS plays a role in early bacterial invasion and inflammasome activation, whereas the SPI-2 T3SS is required for intracellular survival and replication at later time points in infection (Figueira and Holden, 2012; LaRock et al., 2015). To address the role of the T3SSs in Brd4-mediated inflammasome activation, we infected WT and Brd4-deficient BMDMs with WT and isogenic S. typhimurium mutants deleted for SPI-1 (ΔSPI-1) or SPI-2 (ΔSPI-2). Compared with WT S. typhimurium, the ΔSPI-1 mutant failed to activate the NLRC4 inflammasome, while the ΔSPI-2 mutant retained the ability to activate the NLRC4 inflammasome in BMDMs (Fig. 2, A–C). These results are consistent with the notion that SPI-1 but not SPI-2 is critically involved in S. typhimurium–induced NLRC4 inflammasome activation (Zhao and Shao, 2015). In Brd4-deficient BMDMs, we also observed reduced inflammasome activation upon infection with the ΔSPI-2 mutant (lanes 7 and 8 in Fig. 2, A–C), supporting a role of Brd4 in SPI-1–dependent NLRC4 inflammasome activation.
Figure 2.
Brd4 is essential for the SPI-1–dependent NLRC4 inflammasome. (A–C) WT or Brd4-deficient BMDMs were infected with S. typhimurium (STm) or ΔSPI-1 or ΔSPI-2 isogenic mutant for 1 h (MOI, 10). Culture supernatant (Sup.) and cell lysate (Extract) were collected and immunoblotted for the indicated proteins (A). Levels of IL-1β in the culture media were measured by ELISA (B). (C) Representative images of cells from A. Pyroptotic cells are indicated by yellow arrows. (D–F) WT or Brd4-deficient BMDMs were infected with S. typhimurium (STm) or ΔfliCΔfljB isogenic mutant for 1 h (MOI, 10). Immunoblots (D), ELISA for IL-1β (E), and cell images of pyroptosis (F) were assessed the same as in A–C. (G–I) WT or Brd4-deficient BMDMs were infected with S. typhimurium or ΔfliCΔfljB isogenic mutant for 4 h (MOI, 10). Immunoblots (G), ELISA for IL-1β (H), and cell images of pyroptosis (I) were assessed the same as in A–C. (J–L) WT or Brd4-deficient BMDMs were infected with P. aeruginosa for 4 h (MOI, 20). Immunoblots (J), ELISA for IL-1β (K), and cell images of pyroptosis (L) were assessed the same as in A–C. Results are presented as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.005 (Student’s t test).
To further evaluate the role of flagellin in Brd4-mediated NLRC4 inflammasome activation in response to S. typhimurium, we infected WT and Brd4-deficient BMDMs with WT or isogenic flagellin deletion mutant (ΔfliCΔfljB) of S. typhimurium. Notably, ΔfliCΔfljB mutant resembled a ΔSPI-1 mutant and was unable to activate inflammasome at the early time point after S. typhimurium infection (1 h; MOI, 10; Fig. 2, D–F). The ΔfliCΔfljB mutant of S. typhimurium has been shown to activate the inflammasome at a later time point via Naip2 (Zhao et al., 2016). When we infected the BMDMs with the ΔfliCΔfljB mutant for a longer period of time (4 h), we observed activation of caspase-1 and IL-1β release in WT BMDMs, but the activation was drastically reduced in Brd4-deficient cells (Fig. 2, G–I), indicating that Brd4 is essential for Naip2-mediated inflammasome activation. Collectively, these data suggest that Brd4 is engaged in SPI-1– and flagellin-dependent NLRC4 inflammasome activation in response to S. typhimurium infection.
In addition to S. typhimurium, other pathogens, including Pseudomonas aeruginosa, Shigella flexneri, and Burkholderia pseudomallei, can also activate the NLRC4 inflammasome via T3SS components (Miao et al., 2010; Sutterwala et al., 2007). For example, P. aeruginosa infection triggers the NLRC4 inflammasome in an ice protease-activating factor–dependent but flagellin-independent manner (Sutterwala et al., 2007). We further infected the WT and Brd4-deficient BMDMs with P. aeruginosa and found that Brd4 deficiency resulted in lower IL-1β processing, caspase-1 activation, IL-1β secretion, and pyroptotic cell death, as in S. typhimurium infection (Fig. 2, J–L). These data reveal that Brd4 is important for P. aeruginosa infection–induced NLRC4 inflammasome activation, suggesting that Brd4-mediated NLRC4 inflammasome activation could be a more general mechanism applied not only to enteric pathogens but also to nonenteric pathogens.
Brd4 deficiency reduces inflammasome assembly and GSDMD-mediated pyroptosis
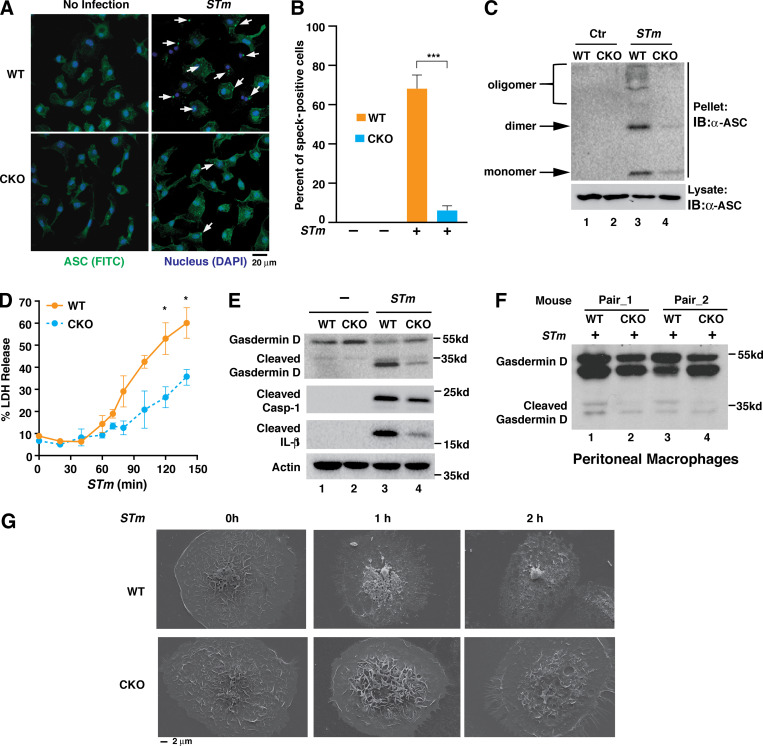
ASC specks serve as platforms for pro–caspase-1 recruitment during inflammasome activation (Stutz et al., 2013). We next investigated whether Brd4 deficiency would affect ASC speck formation, the hallmark of inflammasome assembly (Latz et al., 2013), upon S. typhimurium infection. In response to infection, ASC aggregated and formed a speck in many of WT BMDMs (Fig. 3 A). In contrast, ASC aggregation and speck formation were dramatically reduced in Brd4-deficient BMDMs after S. typhimurium infection (Fig. 3, A and B). ASC speck formation results from its oligomerization (Masumoto et al., 1999). As expected, S. typhimurium stimulated the oligomerization of ASC in WT BMDMs (Fig. 3 C). However, S. typhimurium barely induced the oligomerization of ASC in Brd4-deficient BMDMs (Fig. 3 C).
Figure 3.
Brd4 is required for ASC speck formation and GSDMD cleavage. (A) Confocal microscopy of WT and Brd4-deficient BMDMs infected with S. typhimurium (STm) for 1 h (MOI, 10) staining for ASC (FITC) and nucleus (DAPI). ASC specks are marked with arrows. (B) Percentage of infected cells from A containing ASC specks. A total of at least 100 cells were counted from 5 different fields. Quantification represents mean ASC speck numbers from two independent experiments. ***, P < 0.005 (Student’s t test). (C) WT and Brd4-deficient BMDMs were infected with S. typhimurium (STm) or control (Ctr) for 1 h (MOI, 10). Triton X-100 soluble (lysate) and insoluble (pellet) fractions were immunoblotted with ASC antibody. IB, immunoblot. (D) The levels of released LDH were measured from WT and Brd4-deficient BMDMs infected with S. typhimurium (STm) for the indicated time points (MOI, 10). Results are presented as mean ± SD of two independent experiments. *, P < 0.05 (Student’s t test). (E) WT and Brd4-deficient BMDMs were infected with S. typhimurium (STm) for 1 h (MOI, 10). Levels of GSDMD and caspase-1 were immunoblotted as indicated. (F) WT and Brd4-CKO mice were infected with S. typhimurium (STm; 103 CFU) intraperitoneally for 2 h. Levels of GSDMD in peritoneal macrophages from infected mice were immunoblotted as indicated. (G) Scanning EM of WT and Brd4-deficient BMDMs infected with S. typhimurium (MOI, 10) with indicated times.
Pyroptosis is another important feature associated with inflammasome activation. We next investigated the role of Brd4 in pyroptosis. Lactate dehydrogenase (LDH) release to the extracellular space is considered a hallmark of pyroptosis due to pyroptosis-associated pore formation in the plasma membrane, cell swelling, and plasma membrane disruption (Rayamajhi et al., 2013). We first measured the levels of released LDH activity from WT and Brd4-deficient BMDMs after S. typhimurium infection. The levels of released LDH were increased from WT BMDMs after infection with S. typhimurium (Fig. 3 D). However, the levels of released LDH were significantly lower in Brd4-deficient BMDMs (Fig. 3 D). These data suggest that Brd4 regulates the pyroptosis in S. typhimurium–infected macrophages.
GSDMD is the executor of pyroptosis and is cleaved by caspase-1 as a result of the inflammasome activation (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). The cleaved form of GSDMD, the gasdermin-N domain, perforates the plasma membrane to induce cell swelling and osmotic lysis (Shi et al., 2015). S. typhimurium infection induced the cleavage of GSDMD associated with activated caspase-1 and IL-1β processing in WT BMDMs (Fig. 3 E). However, the cleavage of GSDMD was reduced in infected Brd4-deficient BMDMs, as were caspase-1 activation and IL-1β processing (Fig. 3 E). Impaired cleavage of GSDMD could also be observed in peritoneal macrophages isolated from Brd4–conditional knockout (Brd4-CKO) mice intraperitoneally infected with S. typhimurium (Fig. 3 F). These data indicate Brd4 is important for caspase-1–dependent cleavage of GSDMD in vitro and in vivo after S. typhimurium infection.
We next investigated pyroptosis by observing the morphologic changes of BMDMs with scanning EM. WT BMDMs showed protruding nuclei and cell membrane rupture when infected with S. typhimurium (Fig. 3 G). In contrast, Brd4-deficient BMDMs maintained the regular cell morphology with intact cell membranes (Fig. 3 G). Collectively, these results demonstrate that Brd4 is essential for ASC speck formation, caspase-1–dependent cleavage of GSDMD, and GSDMD-mediated pyroptosis during S. typhimurium infection, consistent with a defect in NLRC4 inflammasome activation.
Brd4 regulates the transcription of NLRC4 inflammasome components
Because of the nature of Brd4 as a transcription regulator, we reasoned that Brd4 would most likely regulate NLRC4 inflammasome activation in a transcription-dependent manner. To profile the global effect of Brd4 deficiency on gene transcription, we performed RNA sequencing (RNA-seq) in WT and Brd4-deficient BMDMs with or without S. typhimurium infection. S. typhimurium infection altered gene expression in both WT and Brd4-deficient BMDMs (principal component 1 [PC1]). However, a significantly altered expression was observed between Brd4-deficient and WT BMDMs independent of S. typhimurium infection (PC2; Fig. 4 A). These results indicate that Brd4 is critical for maintaining the basal levels of transcription for many genes in macrophages. When we analyzed the genes altered in the absence of infection (WT 0 h vs. CKO 0 h) and in the presence of infection (WT 2 h vs. CKO 2 h), we identified a total of 617 overlapping genes (fold change, ≥1.8; false discovery rate [FDR], ≤0.1%; Fig. 4 B). Gene Ontology (GO) enrichment analysis indicated that many of these overlapped genes were involved in innate immune responses (Fig. 4 D), supporting the critical role of Brd4 in inflammatory gene expression in innate immunity (Bao et al., 2017; Dey et al., 2019). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis unveiled that genes classified in the NLR family were significantly altered (Fig. 4 C). Notably, NLR signaling is one of the primary pattern recognition receptors that senses intracellular microbe- or damage-derived molecules (Takeuchi and Akira, 2010). Several NLRs, including NLRP3, NLRC4, pyrin, and Naips, have been well characterized for their involvement in the activation of inflammasomes (Karki and Kanneganti, 2019). Hence, we plotted the heat map for genes altered in the NLR signaling pathway and found that expression of several genes encoding NLRC4 inflammasome components, including Naip5, Naip6, and Nlrc4, was down-regulated in Brd4-deficient BMDMs (Fig. 4 E), These RNA-seq data support a potential role of Brd4 in the transcriptional regulation of NLRC4 inflammasome components.
Figure 4.
Depletion of Brd4 alters the transcriptional landscape in macrophages. (A) PC analysis of WT and Brd4-deficient BMDMs with or without S. typhimurium infection (2 h; MOI, 10). (B) Venn diagram indicates the number of genes with significantly altered expression (fold change, ≥1.8; FDR, ≤0.1%) in WT and Brd4-deficient BMDMs with or without infection. (C and D) A list of 617 overlapped genes from the Venn diagram in B was classified with KEGG pathway enrichment (C) and GO analysis (D). (E) Heat map representation of 25 genes in NLR signaling from C, color coded by Z-score. (F) WT and Brd4-deficient BMDMs infected with S. typhimurium (STm) for the indicated time points (MOI, 10). Expression of indicated genes was measured by quantitative RT-PCR. Results are presented as mean ± SD in three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.005. AGE, advanced glycation end products; n.s., not significant; PI3K, phosphatidylinositol 3-kinase; RAGE, receptor for advanced glycation end products.
We next performed quantitative RT-PCR to confirm the RNA-seq results. In agreement with the RNA-seq data, we found that expression of all of Naip1, Naip2, Naip5, Naip6, and Nlrc4 was decreased at the basal level in Brd4-deficient BMDMs (Fig. 4 F). Upon S. typhimurium infection, Naips and Nlrc4 were down-regulated in both WT and Brd4-deficient BMDMs, with a faster decrease in WT cells than in Brd4-deficient BMDMs (Fig. 4 F), likely reflecting one of the mechanisms by which S. typhimurium inhibits inflammasome activation to promote its persistence (Perez-Lopez et al., 2013). Interestingly, S. typhimurium infection significantly increased the expression of Nlrp3 (Fig. 4 F), likely via LPS. However, the transcriptional up-regulation appeared to be Brd4 independent, because Brd4 deficiency had no effect on the induced transcription of Nlrp3 (Fig. 4 F).
Brd4 cooperates with IRF8 and PU.1 to regulate the transcription of Naips
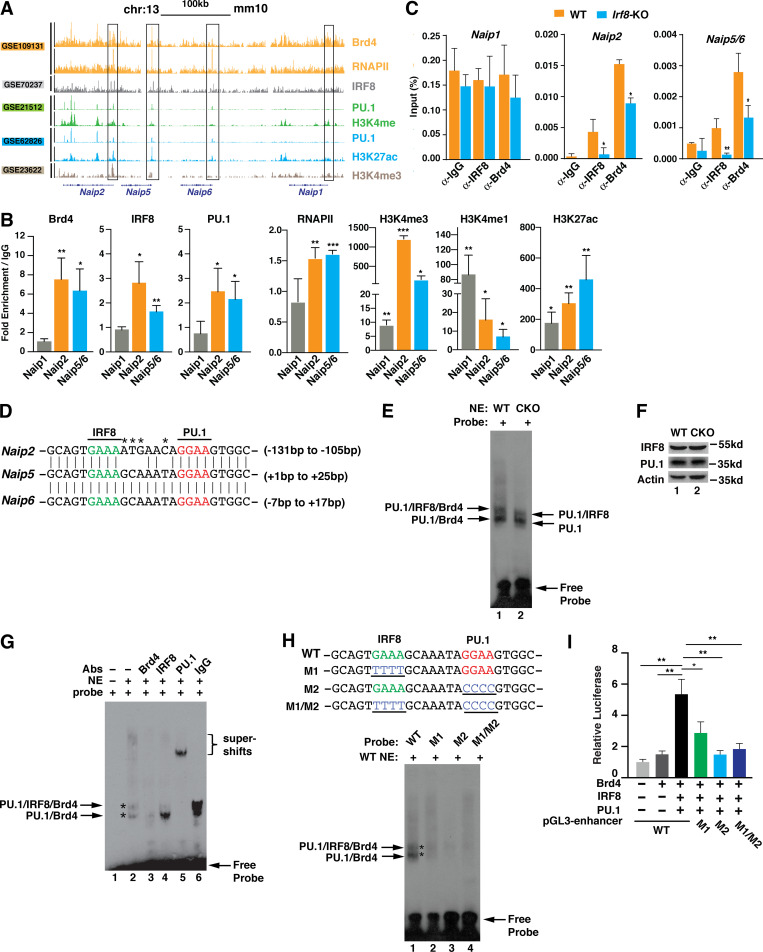
To determine the mechanism by which Brd4 regulates the transcription of Naips, we sought to identify the key transcription factors that are involved in Naips expression. A recent study indicated that transcription of Naips, including Naip1, Naip2, Naip5,and Naip6, was regulated by IRF8 and PU.1 (Karki et al., 2018). We hypothesized that Brd4 might cooperate with IRF8/PU.1 to regulate the transcription of Naips. To test this hypothesis, we first analyzed the available chromatin immunoprecipitation–sequencing (ChIP-seq) datasets from BMDMs to determine the enrichment of Brd4, IRF8, PU.1, RNAPII, and certain histone marks on the promoters or enhancers of Naips (Fig. 5 A). Mouse Naip1, Naip2, Naip5, and Naip6 are located in proximity on chromosome 13 (Fig. 5 A). For Naip1, there was no enrichment for Brd4, PU.1, IRF8, or RNAPII at the promoter region (Fig. 5 A). This could possibly explain the lower basal levels of Naip1 in BMDMs (Yang et al., 2013). Different from Naip1, there were clear enrichments of Brd4, IRF8, PU.1, and RNAPII at the promoter regions of Naip2, Naip5, and Naip6, which also had high H3K4m3/H3H4me1 ratios and H3K27ac signals (Fig. 5 A), suggesting active transcriptional initiation at these promoters in these unstimulated conditions. This ChIP-seq data analysis suggests that IRF8, PU.1, and Brd4 might function together to regulate the basal transcription of Naip2, Naip5, and Naip6 via binding to the promoters.
Figure 5.
Brd4 forms a complex with IRF8 and PU.1 to regulate the expression of Naips. (A) Naips gene tracks of ChIP-seq peaks for Brd4, IRF8, PU.1, RNAPII, and the indicated histone modifications on chromosome 13, mm10. Tracks from the same study are coded with the same color. The y axis indicates normalized ChIP-seq signals. (B) ChIP assays were performed using antibodies against Brd4, IRF8, PU.1, RNAPII, and the indicated histone modifications and probed for the promoters of indicated Naips in BMDMs. Results were analyzed by qPCR and are shown as mean (fold change over IgG) ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.005. (C) ChIP assays were performed using antibodies against Brd4 and IRF8 and probed for the promoters of indicated Naips in WT and Irf8-KO iBMDMs. Results were analyzed by qPCR and are shown as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.005. (D) Sequence alignment of the promoter regions of Naip2, Naip5, and Naip6 containing the IRF8 and PU.1 binding motifs. Asterisks mark the different nucleotides of IR8F/PU.1 binding sites between Naip2 and Naip5/6 promoters. (E) EMSA was performed using nuclear extracts (NE; 5 µg) of WT or Brd4-deficient BMDMs with Naip5/6 probe (2 pmol). Protein complexes are indicated by arrows. (F) Protein expression levels of IRF8 and PU.1 in WT and Brd4-deficient BMDMs. (G) EMSA was performed using nuclear extracts (NE; 5 µg) of WT BMDMs with Naip5/6 probe (2 pmol) with or without the addition of the indicated antibodiesbs) or IgG (1 μg). Asterisks1 µg. Asterisks mark two complexes formed as (H) EMSA was performed using nuclear extracts (NE; 5 µg) of BMDMs with Naip5/6 probe or probes with the indicated mutations in the IRF8 or PU.1 binding motifs. Asterisks mark two complexes formed as indicated. (I) The pGL3-enhancer reporter plasmids (0.1 µg) containing the WT sequence or the indicated mutant sequences were cotransfected with expression vectors for Brd4 (0.1 µg), IRF8 (0.05 µg), and PU.1 (0.05 µg) into 293T cells. Luciferase activity was measured 48 h after transfection. Data represent the mean ± SD of triplicates from two independent experiments. *, P < 0.05; **, P < 0.01 (Student’s t test).
We next validated the ChIP-seq analysis with ChIP–quantitative PCR (qPCR) on the promoters of these Naip genes. Interestingly, Naip5 and Naip6 share a 97% sequence identity in their promoter regions around the transcription start sites from −1,000 to +2,000 bps, indicating that the regulation of Naip5 and Naip6 transcription is likely the same. We therefore referred to the promoters of Naip5 and Naip6 as Naip5/6. Consistent with the ChIP-seq data, Brd4, IRF8, PU.1, and RNAPII were found to be enriched at the promoters of Naip2 and Naip5/6, which was associated with higher levels of H3K4me3/H3K4me1 ratio and H3K27ac signal (Fig. 5 B). There were much lower enrichment signals on the promoter of Naip1 (Fig. 5 B). These data further support the notion that Brd4 might cooperate with IRF8/PU.1 to regulate the transcription of Naip2 and Naip5/6 via binding to their promoters.
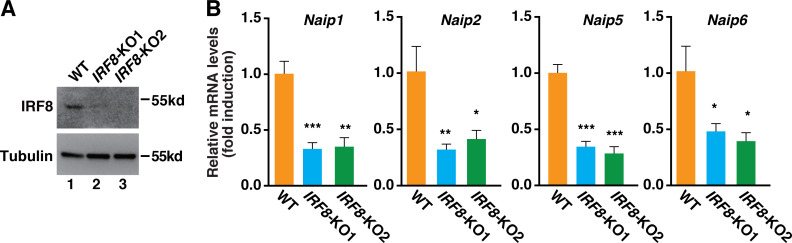
Because IRF8 regulates the transcription of Naips (Karki et al., 2018), we suspected that IRF8 might be involved in the recruitment of Brd4. To test this hypothesis, we generated IRF8-deficient immortalized BMDMs (iBMDMs) using CRISPR-Cas9 technology (Fig. S1 A). Similar to IRF8-deficient BMDMs (Karki et al., 2018), deletion of IRF8 in iBMDMs reduced the basal transcription of Naips, including Naip1, Naip2, and Naip5/6 (Fig. S1 B). When we performed ChIPs in these IRF8-deficient iBMDMs, we found that the recruitment of Brd4 to the promoters of Naip2 and Naip5/6 but not Naip1 was decreased (Fig. 5 C), suggesting that IRF8 is involved in the recruitment of Brd4 to stimulate the transcription of Naip2 and Naip5/6.
Figure S1.
Depletion of Irf8 in iBMDMs using the CRISPR/Cas9 system reduced the expression of Naips. (A) The levels of IRF8 and tubulin in the whole-cell lysates were detected by immunoblotting in Irf8-depleted iBMDMs. (B) mRNA levels of Naip1, Naip2, Naip5, and Naip6 were measured by real-time PCR in WT or Irf8-KO iBMDMs. Results are presented as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.00; and ***, P < 0.005 (Student’s t test).
Examining the promoter sequences of Naip2, Naip5, and Naip6, we observed that they all contained IRF8 and PU.1 binding motifs (Fig. 5 D). These promoter sequences were highly conserved between Naip5/6 and Naip2, with only four nucleotide differences (Fig. 5 D). Because PU.1 is required for a macrophage-specific open chromatin landscape at promoters and enhancers and the dimerization of IRF8 with PU.1 significantly enhanced IRF8’s accessibility to DNA (Heinz et al., 2010; Langlais et al., 2016), it is possible that IRF8 and PU.1 bind to these motifs on the promoters and form a complex with Brd4 to stimulate the transcription of Naips. To test this possibility, we employed an electrophoretic mobility shift assay (EMSA) using a biotin-labeled probe containing IRF8 and PU.1 binding motifs from the promoters of Naip5/6 (Fig. 5 D). Incubation of nuclear extracts from WT BMDMs with an Naip5/6 promoter probe resulted in slower migrations of the biotin-labeled probe (Fig. 5 E), reflecting complex formation on the promoters. There were two complexes formed on the biotin-labeled probe, and both complexes migrated faster when using nuclear extracts from Brd4-deficient BMDMs (Fig. 5 E), where expression of IRF8 and PU.1 was not affected by Brd4 depletion (Fig. 5 F), suggesting that these two complexes might contain Brd4. Supporting this, addition of anti-Brd4 antibodies to the nuclear extracts abolished both complexes (Fig. 5 G). Addition of anti-PU.1 antibodies also abolished both complexes, while addition of anti-IRF8 antibodies only removed the slower-migrating complex (Fig. 5 G). There were not obvious supershifts with the addition of anti-Brd4 or anti-IRF8 antibodies (Fig. 5 G), likely due to the failure of the large antibody-bound complexes entering the gels. These data suggest that Brd4, IRF8, and PU.1 form two complexes on the promoters of Naip2 and Naip5/6 with a slower-migrating complex containing Brd4, IRF8, and PU.1 and a faster-migrating complex containing PU.1 and Brd4 but not IRF8.
IRF8 and PU.1 are transcription factors that can form dimers on their DNA binding motifs to regulate gene expression (Langlais et al., 2016). To further determine the roles of IRF8 and PU.1 on the Brd4-containing complex formation, we mutated the IRF8 or PU.1 binding motif individually (M1 or M2) or in combination (M1/M2) by altering four nucleotides within each motif (Fig. 5 H, upper panel). Mutation of IRF8 or PU.1 binding motif alone or in combination abolished complex formation (Fig. 5 H). Together, these data suggest that binding of IRF8 and PU.1 dimers to these motifs is essential for the recruitment of Brd4 to the promoters to facilitate Naips transcription. Supporting this, we found that coexpression of IRF8, PU.1, and Brd4 activated the pGL3-enhancer luciferase reporter containing the WT sequence but not the reporter containing M1 or M2 alone or in combination (Fig. 5 I).
Brd4-CKO mice are more susceptible to S. typhimurium infection
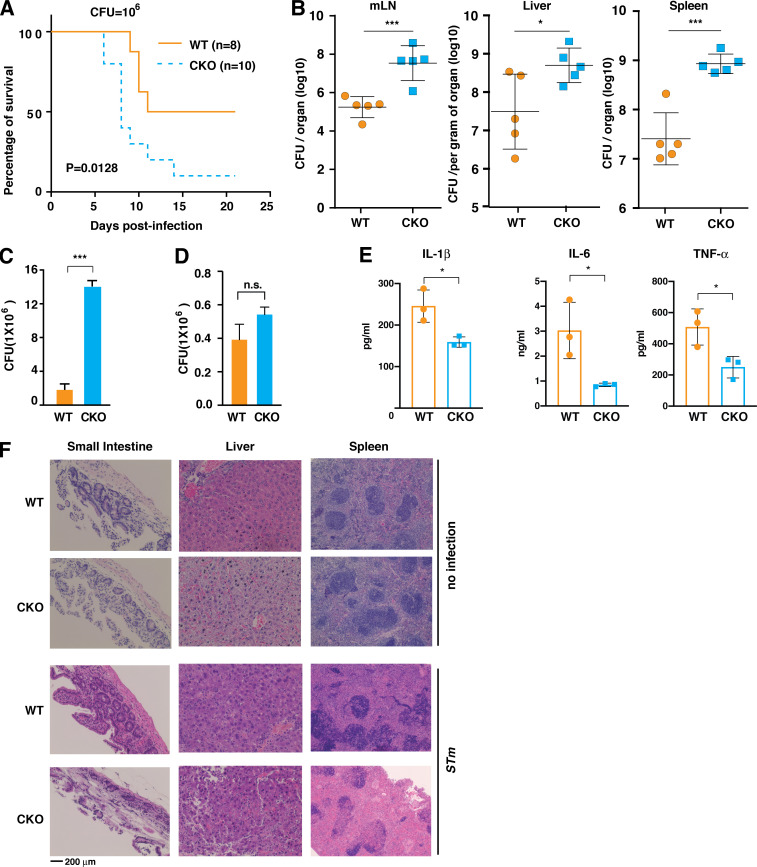
Having identified the critical role of Brd4 in activating the NLRC4 inflammasome by stimulating the expression of Naips in macrophages, we next sought to examine the in vivo relevance of Brd4 during S. typhimurium infection. We first orogastrically challenged the WT and Brd4-CKO mice with S. typhimurium and examined the response from these mice. Brd4-CKO mice died gradually during the course of 3 wk, while WT mice died more slowly with 50% survival (Fig. 6 A). The increased vulnerability of Brd4-CKO mice to S. typhimurium infection might be due to the failure of Brd4-CKO mice in the clearance of bacteria because the NLRC4 inflammasome is essential for restricting S. typhimurium propagation in animals (Sellin et al., 2014). Therefore, we measured the bacterial burden in various tissues of these infected mice and found that Brd4-CKO mice had significantly higher bacterial loads in mesenchymal lymph nodes, liver, and spleen (Fig. 6 B). In line with this increased bacterial burden in Brd4-CKO mice, we observed a significant increase in survival/replication of intracellular S. typhimurium in Brd4-deficient BMDMs (Fig. 6 C). This increased intracellular S. typhimurium was not due to the change in phagocytosis, because both WT and Brd4-deficient BMDMs displayed similar phagocytosis potential (Fig. 6 D). We also found decreased production of cytokines, including IL-1β, IL-6, and TNF-α, in the plasma of Brd4-CKO mice (Fig. 6 E), consistent with a compromised inflammatory response to S. typhimurium. Histological analysis also revealed more severe tissue damage to the small intestine, liver, and spleen in Brd4-CKO mice than in WT mice after S. typhimurium (Fig. 6 F). Collectively, these data suggest that Brd4-mediated NLRC4 inflammasome activation is essential for protecting the host against S. typhimurium infection.
Figure 6.
Brd4-CKO mice are more susceptible to S. typhimurium infection. (A) Kaplan-Meier survival plots of WT and Brd4-CKO mice infected orogastrically with S. typhimurium at 106 CFU. The statistical significance was determined by using the log-rank test. (B) Bacterial burden of WT and Brd4-CKO mice (n = 5) in the mesenchymal lymph node, liver, and spleen 6 d after orogastric infection with S. typhimurium (106 CFU). (C) Intracellular S. typhimurium numbers in WT or Brd4-deficient BMDMs after 30-min infection (MOI, 10), followed by culturing in media containing gentamicin for 24 h to remove extracellular bacteria. (D) Intracellular S. typhimurium numbers in WT or Brd4-deficient BMDMs after 30-min infection (MOI, 10), followed by culturing in media containing gentamicin for 1.5 h to remove extracellular bacteria. Results are presented as mean ± SD of two independent experiments. n.s., not significant; *, P < 0.05; ***, P < 0.005 (Student’s t test). (E) Plasma levels of IL-1β, IL-6, and TNF-α were measured 6 d after orogastric infection (n = 3). (F) WT and Brd4-CKO mice were orogastrically infected with S. typhimurium (STm; 106 CFU) for 6 d, and the small intestine, liver, and spleen were assessed by hematoxylin and eosin staining.
Discussion
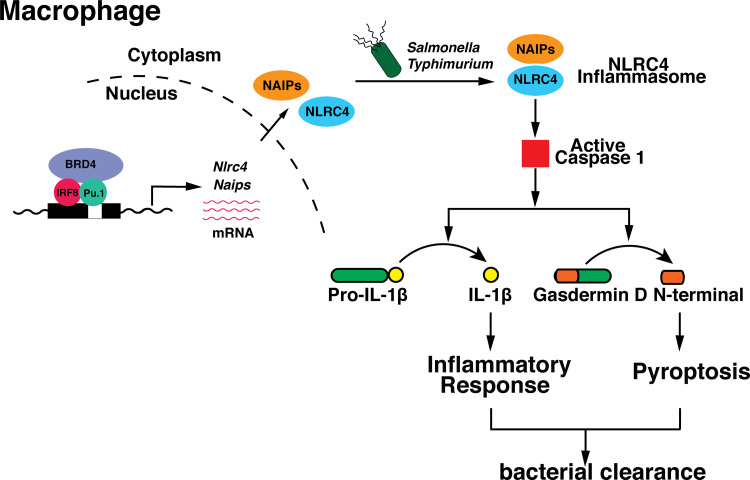
Brd4 has emerged as a critical regulator of inflammatory cytokine production in innate immune response (Bao et al., 2017; Dey et al., 2019). In this study, we identified a novel mechanism by which Brd4 regulates inflammatory cytokine production via inflammasome activation. Brd4 forms a complex with IRF8 and PU.1 on the promoters of Naips to maintain their basal expression in macrophages. These basal levels of Naips and Nlrc4 are essential for the activation of caspase-1 and the subsequent cleavage of pro–IL-1β and GSDMD upon S. typhimurium infection. The IL-1β secretion and GSDMD-mediated pyroptosis provide the protective immune response against bacterial infection (Fig. 7).
Figure 7.
Schematic representation of Brd4-medaited NLRC4 inflammasome during Salmonella infection. Brd4, together with IRF8 and PU.1, maintains the basal transcription of Naips, which sense the different components of S. typhimurium, leading to the activation of caspase-1 and the subsequent cleavage of pro–IL-1β and GSDMD, contributing to the infection-mediated inflammatory response and pyroptosis.
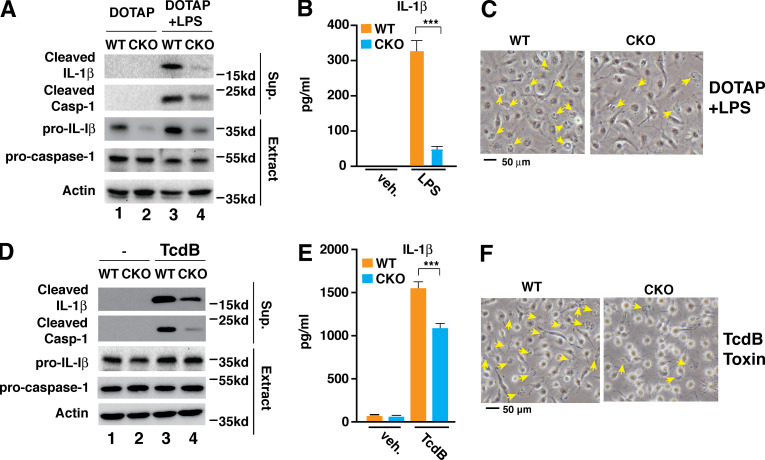
Deficiency of Brd4 impaired flagellin and S. typhimurium–induced NLRC4 inflammasome activation but had little effect on nigericin-triggered NLRP3 inflammasome or double-stranded DNA–induced AIM2 inflammasome activation (Fig. 1). NLRP3 inflammasome activation requires NF-κB–dependent transcriptional activation of Nlrp3 as a priming signal (Bauernfeind et al., 2009). Although Brd4 is a known NF-κB coactivator (Huang et al., 2009), we did not observe much difference in nigericin-induced NLRP3 inflammasome activation between LPS-primed WT and Brd4-deficient BMDMs (Fig. 1, A–C). In addition, there was not any difference of induced Nlrp3 expression in S. typhimurium–infected WT and Brd4-deficient BMDMs (Fig. 4 F). Therefore, Brd4 might not be involved in LPS-induced transcriptional activation of Nlrp3, likely reflecting the ability of Brd4 to selectively activate a subset of NF-κB target genes (Huang et al., 2009). While Brd4 is not involved in the activation of NLRP3 inflammation, Brd4 regulated caspase-11–mediated noncanonical NLRP3 inflammasome activation because intracellular LPS-activated cleavage of pro–caspase-1 and pro–IL-1β, secretion of IL-1 β, and pyroptosis were decreased in Brd4-deficient BMDMs (Fig. S2, A–C). Additionally, we found that Brd4 was also involved in pyrin inflammasome activation in response to Clostridium difficile toxin B (TcdB; Fig. S2, D–F). In addition to the NLRC4 inflammasome, Brd4 appears to regulate the activation of different forms of inflammasome, and the detailed mechanisms warrant further investigation.
Figure S2.
Brd4 was involved in the activation of the noncanonical NLRP3 inflammasome and the pyrin inflammasome. (A–C) WT or Brd4-deficient BMDMs were primed with LPS (0.5 µg/ml) for 4 h followed by DOTAP or DOTP + LPS transfection for 4 h. Culture supernatant (Sup.) and cell lysates (Extract) were collected and immunoblotted for the indicated proteins (A). Levels of IL-1β in the culture media were measured by ELISA (B). (C) Representative images of cells from A. Pyroptotic cells are indicated by yellow arrows. (D–F) WT or Brd4-deficient BMDMs were primed with LPS (0.5 µg/ml) for 4 h followed by vehicle (veh.) or TcdB toxin treatment (1 µg/ml) for 4 h. Culture supernatant (Sup.) and cell lysates (Extract) were collected and immunoblotted for the indicated proteins (D). Levels of IL-1β in the culture media were measured by ELISA (E). (F) Representative images of cells from D. Results are presented as mean ± SD of three independent experiments. ***, P < 0.005 (Student’s t test).
S. typhimurium injects flagellin and other effector proteins via SPI-1–encoded T3SS, which could be sensed by different Naips to activate the NLRC4 inflammasome (Duncan and Canna, 2018). In mice, Naip1 or Naip2 recognizes the T3SS needle protein PrgI or inner rod protein PrgJ, respectively, while both Naip5 and Naip6 recognize flagellin (Duncan and Canna, 2018). Importantly, all these NLRC4 inflammasome sensor proteins were attenuated in Brd4-deficient BMDMs (Fig. 4 E). Flagellin deletion ΔfliCΔfljB mutant failed to activate the NLRC4 inflammasome (Fig. 2, D–F), and flagellin-induced NLRC4 inflammasome activation was impaired in Brd4-deficient BMDMs (Fig. 1, G–I), suggesting that Brd4-mediated expression of Naip5 and Naip6 is critical for NLRC4 inflammasome activation. Because SPI-1 T3SS plays a dominant role in the activation of the NLRC4 inflammasome during the early phase of Salmonella infection (Figueira and Holden, 2012), ΔSPI-1 but not the ΔSPI-2 isogenic mutant failed to activate Brd4-mediated NLRC4 inflammasome activation after 1-h infection (Fig. 2 A). After infection for a longer period of time, S. typhimurium could activate inflammasome independent of flagellin, but largely relying on the Naip2-mediated sensing of SPI-1 inner rod proteins (Zhao et al., 2016). When WT or Brd4-deficient BMDMs were infected with ΔfliCΔfljB mutant for a longer period of time (4 h), we observed that flagellin-independent activation of the inflammasome was decreased in Brd4-deficient BMDMs (Fig. 2, G–I). These data suggest that Brd4-mediated Naip2 expression is also involved in NLRC4 inflammasome activation. As such, the impaired NLRC4 inflammasome activation in Brd4-deficient BMDMs and in Brd4-CKO mice would represent the combined defects from sensing of flagellin by Naip5/6 and T3SS inner rod protein by Naip2.
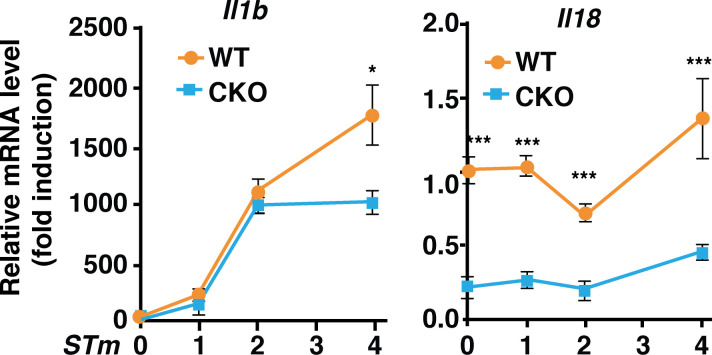
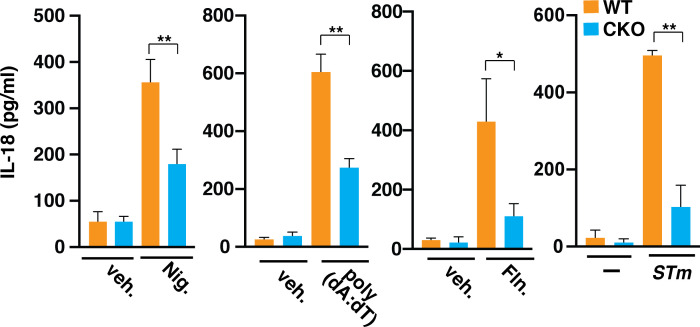
Inflammasome activation leads to the secretion of mature IL-1β and IL-18 (Latz et al., 2013). Deficiency of Brd4 in macrophages resulted in a compromised NLRC4 inflammasome activation with reduced IL-1β secretion after S. typhimurium infection (Fig. 1). Brd4 has been shown to be involved in the transcription of Il1b in gastric epithelial cells in response to Helicobacter pylori infection (Chen et al., 2016). However, Brd4 does not seem to be involved in the induced transcription of Il1b during the early time point of S. typhimurium infection (within 2 h), because there was no significant difference in the infection-induced Il1b transcription in WT and Brd4-deficient BMDMs (Fig. S3). However, on the one hand, IL-1β secretion during the early time point (within 2 h) decreased in Brd4-deficient BMDMs when challenged with flagellin or S. typhimurium (Fig. 1, G, H, J, and K), supporting Brd4’s role in NLRC4 inflammasome activation and IL-1β maturation independent of its ability to activate transcription. On the other hand, the transcription of Il18, which was constitutively activated and was not induced by infection, was significantly decreased in Brd4-deficient BMDMs (Fig. S3). Consistently, we also observed reduced IL-18 secretion in Brd4-defcient BMDMs in response to different inflammasome activators (Fig. S4). Overall, these data indicate the essential role of Brd4 in regulating IL-1β and IL-18 production through both transcriptional regulation and inflammasome-mediated post-transcriptional processing.
Figure S3.
Brd4 differentially regulated the transcription of Il1b and Il18. WT and Brd4-deficient BMDMs infected with S. typhimurium (STm) for the indicated time points (MOI, 10). Expression of Il1b and Il18 was measured by quantitative RT-PCR. Results are presented as mean ± SD in three independent experiments. *, P < 0.05; ***, P < 0.005.
Figure S4.
IL-18 secretion was decreased in Brd4-defcient BMDMs. WT or Brd4-deficient BMDMs were stimulated with vehicle (veh.) or different inflammasome activators, including Nigericin (Nig.), poly(dA:dT), Flagellin (Fln.), and S. typhimurium (STm), as indicated. Levels of IL-18 in the culture media were measured by ELISA. Results are presented as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01 (Student’s t test).
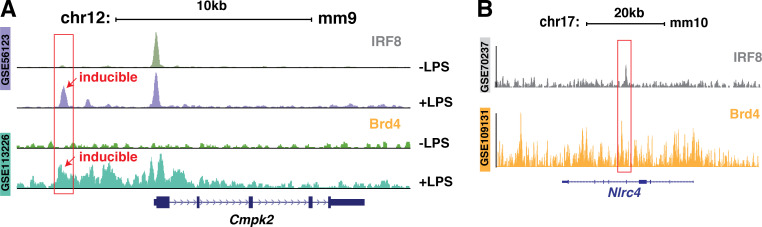
IRF8 plays a prominent role in maintaining the steady-state epigenetic and transcriptional level of critical genes in macrophages (Langlais et al., 2016). At the molecular level, IRF8 is known to form ternary complexes with other transcription factors, including IRF1 or IRF2, AP1, and PU.1 (Salem et al., 2020). Specifically, IRF8 and PU.1 bind to Ets/IRF composite elements or IECS to stimulate basal gene expression in resting macrophages (Salem et al., 2020; Tamura et al., 2005). IRF8 and PU.1 have been indicated in the transcription of Naips and Nlrc4 in BMDMs (Karki et al., 2018), likely through their binding to the promoters of Naip2, Naip5, and Naip6. Via ChIPs, we found that IRF8 and PU.1 were enriched on the promoter regions containing the unique IRF8 and PU.1 binding motifs (Fig. 5 B). Brd4 was also enriched in the same regions and formed a complex with IRF8 and PU.1 on these motifs, and the enrichment of Brd4 on these regions appears to be IRF8 dependent (Fig. 5, B and C). These binding motifs resemble the IECS with six nucleotides rather than two or three nucleotides between IRF and Ets binding motifs (Tamura et al., 2005), likely representing a nontypical IECS (Fig. 5 D). Both IRF8 and PU.1 binding sites are essential for the binding of the IRF8–PU.1–Brd4 complex to the DNA because mutation of either binding motif completely abolished the complex formation (Fig. 5 H). These results define the critical role of IRF8 and PU.1 in the recruitment of Brd4 to the promoters of Naips to maintain their steady-state expression levels. In addition to the constitutive binding to DNA with PU.1 to regulate the basal transcription of genes responsible for macrophage functions, IRF8 is also actively involved in the inducible inflammatory gene expression in response to LPS (Mancino et al., 2015). Brd4 is known to be a critical regulator of inflammatory gene expression in LPS-treated macrophages (Bao et al., 2017; Dey et al., 2019; Nicodeme et al., 2010), and it is possible that Brd4 could also regulate IRF8-dependent inducible gene expression. Supporting this, we found that LPS stimulated the recruitment of Brd4 and IRF8 to the same regulatory region of some IRF8-dependent inducible genes, such as Cmpk2 (Mancino et al., 2015; Fig. S5 A).
Figure S5.
ChIP-seq analysis of recruitment of IRF8 and Brd4 on Cmpk2 and Nlrc4. (A) Gene tracks of ChIP-seq peak for IRF8 (GEO accession no. GSE56123) and Brd4 (GEO accession no. GSE113226) to a representative genomic region containing the LPS-inducible gene Cmpk2. The y axis indicates normalized ChIP-seq signals. (B) Gene tracks of ChIP-seq peaks for IRF8 (GEO accession no. GSE70237) and Brd4 (GEO accession no. GSE109131) on the Nlrc4 gene. Tracks on the y axis indicate normalized ChIP-seq signals.
In addition to Naips, the basal transcription of Nlrc4 was also decreased in Brd4-deficient BMDMs (Fig. 4 F), which could also contribute to the impaired inflammasome activation in response to S. typhimurium infection. While our study demonstrates how Brd4 regulates the expression of Naips via forming a complex with IRF8 and PU.1 on their promoters, how Brd4 regulates the expression of Nlrc4 remains unclear because there is no IRF8 binding motif on the promoter of Nlrc4, although its transcription is decreased in Irf8-deficient BMDMs (Karki et al., 2018). An intronic region of Nlrc4 has been found to be enriched with IRF8 and is likely involved in the transcription of Nlrc4 (Karki et al., 2018). Of interest, we found that Brd4 was also enriched in the same intronic region as IRF8 (Fig. S5 B). Whether and how Brd4 might cooperate with IRF8 to regulate the transcription of Nlrc4 through this unique intronic region remains an interesting question and warrants further investigation.
Failure to control the propagation of S. typhimurium due to the defect in NLRC4 inflammasome signaling contributes to the accelerated mortality in infected Nlrc4-knockout mice (Franchi et al., 2012). The Brd4-CKO mice resembled Nlrc4-knockout mice with the enhanced bacterial loads in various tissues and increased mortality upon S. typhimurium infection (Fig. 6). Brd4-CKO mice displayed a defect in NLRC4 inflammasome activation with reduced GSDMD cleavage in macrophages and reduced serum level of IL-1β after S. typhimurium infection (Figs. 3 F and 6 D). The increased bacterial loads from Brd4-CKO mice likely reflect the compromised NLRC4 inflammasome–mediated bacterial killing in macrophages (Jorgensen et al., 2017) because Brd4-deficient BMDMs exhibited reduced killing activity against intracellular S. typhimurium without affecting the phagocytosis potential (Fig. 6, C and D). These data support the critical role of Brd4-mediated optimal NLRC4 inflammasome activation in host survival. It has to be noted that the serum levels of proinflammatory cytokine TNF-α and IL-6 were decreased in S. typhimurium–infected Brd4-CKO mice compared with infected WT mice (Fig. 6 E). Therefore, the reduced expression of TNF-α and IL-6 might also contribute to the increased susceptibility of Brd4-CKO mice to S. typhimurium infection.
Brd4 plays a critical role in inflammatory response by stimulating the expression of inflammatory genes via its binding to promoters and enhancers for the synthesis of mRNA and enhancer RNAs (Shi and Vakoc, 2014). Brd4 binds to acetylated NF-κB or histones to facilitate NF-κB–dependent inflammatory gene expression (Brown et al., 2014; Huang et al., 2009). Here, we identified IRF8 and PU.1 as novel transcription partners of Brd4 in inflammatory cytokine production. Brd4 cooperates with the IRF8–PU.1 complex to stimulate the inflammasome-mediated processing of IL-1β. It appears that Brd4 serves as a key regulator of inflammatory response by its ability to cooperate with various transcription factors to orchestrate the optimal inflammatory response for host defense against infection. Future studies should define how Brd4 is directed to different transcription factors to dictate both steady-state and signal-dependent inflammatory gene expression.
Materials and methods
Mice
WT (Brd4Flox/Flox) and Brd4-CKO (Brd4Flox/Flox-LyzMCre-cre) mice have been previously described (Bao et al., 2017). Mice were kept under specific pathogen–free conditions at the animal facilities of University of Illinois at Urbana-Champaign (UIUC). For all experiments, sex- and age-matched mice were used. All animal experiments were approved by the UIUC Institutional Animal Care and Use Committee.
Reagents and antibodies
LPS (Escherichia coli O111:B4, L2630), nigericin (N7143) and N-(2,3-dioleoyloxy-1-propyl)trimethylammonium methyl sulfate (DOTAP) liposomal transfection reagent (144189-73-1) were from Sigma-Aldrich. Ultrapure flagellin from S. typhimurium (tlrl-epstfla-5) and poly(dA:dT; tlrl-patn) were purchased from InvivoGen. TcdB from List Labs (155). Lipofectamine RNAiMAX Transfection Reagent (13778150) was from Invitrogen. Mouse TNF-α (88-7324-88), IL-1β (88-7013-88), IL-18 (BMS618-3), and IL-6 (88-7064-22) ELISA kits were from Invitrogen. The LightShift Chemiluminescent EMSA Kit (20148) was from Thermo Fisher Scientific Inc. Anti–IL-1β (AF-401-NA) was from R&D Systems. Anti–caspase-1 (AG-20B-0042) and anti-ASC (AG-25B-0006-C100) were from AdipoGen. Anti-GSDMD (ab209845) and anti-H3K27ac (ab4729) were from Abcam. Antiactin (sc-47778), anti-IRF8 (sc-365042), anti-RNAPII (sc-47701), and anti-PU.1 (sc-390405) were from Santa Cruz Biotechnology, Inc. Anti-Brd4 (A301-985A) was from Bethyl Laboratories. Anti-H3K4me1 (07-436) and anti-H3K4me3 (05-745R) were from EMD Millipore.
Preparation of BMDMs
BMDMs were prepared as previously described (Bao et al., 2017). Briefly, bone marrow was isolated from tibias and femurs using an aseptic technique. To differentiate bone marrow cells into macrophages, cells were cultured in DMEM/F12 with 10% FBS, L-glutamine (2 mM), penicillin/streptomycin (1:100), Hepes buffer (10 mM), and 20% conditioned medium of L929 cells in sterile plastic Petri dishes. After 7 d, BMDMs were ready for further simulation or infection.
Bacterial infection
The WT S. typhimurium (14028; American Type Culture Collection) strain and all the S. typhimurium isogenic mutants were grown in Lysogeny broth (LB) medium at 37°C and 10% CO2 for overnight culture. Bacteria were diluted (1:100) and subcultured for an extra 6 h before infection. For in vivo infection, sex- and age-matched WT and Brd4-CKO mice were subjected to either orogastric injection (106 colony-forming units [CFU]) or intraperitoneal injection (102 CFU) at a total volume of 0.2 ml per mouse. Survival was monitored upon infection. For in vitro infection, WT S. typhimurium and the isogenic mutants were added to BMDMs at an MOI of 10 for the indicated time points. The P. aeruginosa PAO1 strain (a gift from Dr. G. Lau, UIUC) was grown in LB medium at 37°C and 10% CO2 for overnight culture. The bacteria were subcultured for an extra 4 h before infection at an MOI of 25.
Inflammasome activation in BMDMs
To induce inflammasome activation, 1.0 × 106 BMDMs were plated in six-well plates overnight. The next day, cells were primed with LPS (0.5 µg/ml) for 4 h before stimulation. For NLRP3 inflammasome activation, BMDMs were treated with 10 µM nigericin for 30 min. For AIM2 inflammasome activation, BMDMs were transfected with 2 µg/ml poly(dA:dT) for 4 h using Lipofectamine RNAiMAX according to the manufacturer’s instruction (Invitrogen). For NLRC4 inflammasome activation, BMDMs cells were either infected with S. typhimurium for 1 h (MOI, 10) or P. aeruginosa for 2 h (MOI, 25) or transfected with ultrapure flagellin (2 µg/ml) for 2 h using DOTAP following the manufacturer’s protocol (Roche). For noncanonical NLRP3, LPS were transfected into BMDMs for 4 h using DOTAP following the manufacturer’s protocol (Roche). For pyrin inflammasome activation, BMDMs were treated with TcdB (1 µg/ml) for 4 h. Cells and supernatant were collected for immunoblotting and ELISA.
Environmental scanning EM
BMDMs growing on cover slides after S. typhimurium infection were fixed with a solution containing both glutaraldehyde (2.5%) and PFA (2.0%) in a 0.1 µM sodium-cacodylate buffer. BMDMs were rinsed with 0.1 µM sodium cacodylate and dehydrated with a graded series of ethanol solutions (37%, 63%, 95%, and 100%). Samples were transferred from absolute ethanol to a Tousimis 931 critical point dryer and mounted on a stub with double-stick carbon tape before coating with gold-palladium. After coating, cells were visualized with an FEI Quanta FEG 450 environmental scanning electron microscope.
ChIP-seq analysis
ChIP-seq raw data (.srx files) were downloaded from the Sequence Read Archive and converted to FASTQ files using the Sequence Read Archive toolkit. FASTQ files were uploaded to Biocluster2 (High-Performance Biological Computing, Carl R. Woese Institute for Genomic Biology, UIUC) and mapped to the GRCm38/mm10 genome using Bowtie2. Bedgraph files were generated using HOMER and visualized on the University of California, Santa Cruz Genome Browser (https://genome.ucsc.edu/). Peaks were called and further identified when passing the HOMER threshold (P < 0.005; FDR, <0.1%). In this study, we reanalyzed ChIP-seq raw data corresponding to IRF8 (Gene Expression Omnibus [GEO] accession no. GSE70237); Brd4 and Pol II (GEO accession no. GSE109131); H3K4me and PU.1 (GEO accession no. GSE21512); H3K4me3 (GEO accession no. GSE23622); and H3K27ac, H3K4me2, and PU.1 (GEO accession no. GSE62826).
RNA-seq
BMDMs were infected with S. typhimurium (MOI, 10) for 2 h, and total RNA was prepared using the Aurum Total RNA Mini Kit (7326820; Bio-Rad Laboratories). A quality check of RNA samples (three biological replicates) was performed using the Agilent 2100 BioAnalyzer. cDNA library construction and sequencing were performed by BGI Genomics with the BGISEQ-500 platform. Clean reads were mapped to the GRCm38/mm10 genome using STAR (Spliced Transcripts Alignment to a Reference), and differentially expressed genes were identified with the DEGseq package. Genes with fold change ≥1.8 and FDR ≤0.1% were considered to be statistically significant. GO enrichment and KEGG pathway enrichment were further analyzed using the R function phyper.
ELISA analysis of cytokine levels
Levels of TNF-α, IL-1β, IL-18, and IL-6 from cell culture supernatant and serum were measured using mouse ELISA kits (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time PCR
BMDMs were infected with S. typhimurium (MOI, 10) for different time points. Total RNA was isolated with Aurum Total RNA Mini Kit (7326820; Bio-Rad Laboratories) and reverse transcribed with the iScript cDNA Synthesis Kit (170-8891; Bio-Rad Laboratories) according to the manufacturer’s instructions. Real-time PCR was performed on ABI 7300 instruments using iTaq Universal SYBR Green Supermix (172-5124; Bio-Rad Laboratories) and appropriate primers. Primer sequences used for RT-PCR were as follows (F, forward; R, reverse): Naip1 (F, 5′-CAACCACGATGATCCAGCAG-3′; R, 5′-TCATGTGGCGAAAAGTGGCTT-3′), Naip2 (F, 5′-AGCTTGGTGTCTGTTCTCTGT-3′; R, 5′-GCGGAAAGTAGCTTTGGTGTAG-3′), Naip5 (F, 5′-TGCCAAACCTACAAGAGCTGA-3′; R, 5′-CAAGCGTTTAGACTGGGGATG-3′), Naip6 (F, 5′-TAACAGGCCAAGCACAGGTC-3′; R, 5′-GGGGCCAGTCCTTAAACGTG-3′), Nlrc4 (F, 5′-TTGAAGGCGAGTCTGGCAAAG-3′; R, 5′-GGCGCTTCTCAGGTGGATG-3′), Nlrp3 (F, 5′-ATCAACAGGCGAGACCTCTG-3′; R, 5′-GTCCTCCTGGCATACCATAGA-3′), Il1b (F, 5′-GCAACTGTTCCTGAACTCAACT-3′; R, 5′-ATCTTTTGGGGTCCGTCAACT-3′), Il18 (F, 5′-GACTCTTGCGTCAACTTCAAGG-3′; R, 5′-CAGGCTGTCTTTTGTCAACGA-3′), and Actin (F, 5′-GTGACGTTGACATCCGTAAAGA-3′; R, 5′-GCCGGACTCATCGTACTCC-3′).
ChIP-qPCR
ChIP assays were performed as described previously (Bao et al., 2017). Briefly, BMDMs (∼2 × 107 cells) were cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-link was quenched by 125 mM glycine, and cells were sonicated with a Diagenode Bioruptor 300 to reach the desired genomic fragment length (∼300–700 bp). Immunoprecipitation was performed overnight at 4°C by mixing cell lysates, antibodies of interest, and Protein A Magnetic Dynabeads (10001D; Thermo Fisher Scientific). Protein–DNA complexes were reverse cross-linked at 65°C overnight with the addition of proteinase K (0.2 mg/ml). DNAs were further purified with a Qiagen quick spin column, and real-time PCR was performed as described above. Primer sequences used for the ChIP-PCR were as follows: Naip1 (forward, 5′-ATAGCCTGGCCCAATTCTTT-3′; reverse, 5′-GGCTTGGCAGCTTTGATTAG-3′), Naip2 (forward, 5′-CAGCAAGGGGGCAGAGAAAAT-3′; reverse, 5′-ACAGGCAGCTCATGGTTTGAG-3′), and Naip5/Naip6 (forward, 5′-TATAGCCTGGTGCCACTTCC-3′; reverse, 5′-AACCCTGACAAAAGCAGTTCA-3′).
Colony enumeration assay
Mice were orally administered an S. typhimurium PBS suspension (107 CFU) at a volume of 0.2 ml. After 7 d, mice were euthanized, and their organs were collected and homogenized. 10-fold serial dilutions of each organ were plated on LB agar plates overnight at 37°C. Bacterial CFUs were counted the next day.
Hematoxylin and eosin staining
Paraffin tissue section slides from the small intestine, liver, and spleen were prepared by the Department of Animal Sciences, College of Agricultural, Consumer and Environmental Sciences, UIUC. Tissue sections were deparaffinized and rehydrated by submerging them into xylene for 10 min, followed by a gradient of ethanol washes (100%, 95%, and 80% H2O). Tissue section slides were stained with hematoxylin for 3 min and eosin for 45 s. After staining, slides were dehydrated with 95% ethanol, 100% ethanol, and xylene. Images were captured using an EVOS XL Core microscope (Life Technologies).
Immunofluorescence staining
BMDMs grown on coverslips (∼5 × 105) were infected with S. typhimurium (MOI, 10) for 1 h. Cells were fixed and permeabilized with 100% methanol (prechilled at −20°C) at room temperature for 5 min. After being washed with PBS three times, cells were incubated with blocking buffer (1% BSA, 22.52 mg/ml glycine in PBS with Tween 20) for 30 min, followed by overnight ASC primary antibody incubation (2 µg/ml). The next day, cells were incubated with goat antirabbit FITC secondary antibody (2 µg/ml, F2765; Thermo Fisher Scientific) for 1 h in the dark. DAPI staining solution (1 µg/ml) was added to the coverslips for 5 min. Coverslips were mounted with mounting medium (0.3 µg/ml DAPI, 10% Mowiol 4-88, 1% 1,4-diazabicyclo[2.2. 2]octane, 25% glycerol, and 0.1 M Tris, pH 8.5) and images were captured using a Zeiss LSM 510 Meta confocal microscope with Plan-Neofluar 40×/1.3 NA oil objective lenses and a Zeiss AxioCam HR camera. Images were acquired using ZEN lite software (Zeiss) and processed by using ImageJ software.
Bacterial killing and phagocytosis assays
BMDMs were infected with S. typhimurium (MOI, 10) for 30 min. Medium was replaced with fresh medium supplemented with 50 µg/ml gentamicin. For the bacterial killing assay, after 1.5 h, the medium was further replaced with fresh medium containing 10 µg/ml gentamicin for 24 h. BMDMs were lysed, and supernatant was plated on LB agar plates using a 10-fold serial dilution technique. Colony numbers were counted 24 h later. For the phagocytosis assay, after 1.5 h, BMDMs were lysed, and supernatant was plated on LB agar plates using a 10-fold serial dilution technique. Colony numbers were counted 24 h later.
EMSAs
All the biotinylated probes were purchased from Integrated DNA Technologies. 5 µg of nuclear extracts isolated from BMDMs with hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5 mM DTT, 1 mM PMSF, and protease inhibitor cocktail) and cell extraction buffer (10 mM Tris-HCl, pH 7.4, 300 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM EGTA, 0.5 mM DTT, 1 mM PMSF, protease inhibitor cocktail) were incubated with various biotinylated probes following the protocol of the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). Sequences of the WT probes were as follows: forward, 5′-GCAGTGAAAGCAAATAGGAAGTGGC-3′; reverse, 5′-GCCACTTCCTATTTGCTTTCACTGC-3′.
Generation of Irf8 CRISPR/Cas9 knockout cell lines
A CRISPR/Cas9 plasmid targeting mouse Irf8 was purchased from Santa Cruz Biotechnology (sc-421016) and transfected into iBMDMs (a kind gift from Dr. D. Wang, Duke University) using UltraCruz Transfection Reagent (sc-395739) according to the manufacturer’s instruction. GFP-positive cells were sorted into single clones via flow cytometry (BD FACSAria III). Genomic DNA was purified from individual clones and PCR amplified using primers spanning the designed knockout region. Knockout was confirmed by sequencing the PCR product and immunoblotting.
LDH release
The LDH in the culture medium was determined with a LDH cytotoxicity assay kit (Promega) according to the manufacturer's instructions.
ASC oligomerization
WT and Brd4-deficient BMDMs (106 cells) were infected with S. typhimurium (MOI, 10) for 1 h. Cells were collected and lysed with TBS buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, EDTA-free protease inhibitor cocktail) for 30 min at 4°C. After centrifugation, Triton X-100 soluble fractions were collected as lysates, and insoluble pallets were cross-linked with disuccinimidyl suberate (4 mM) at 37°C for 30 min. Lysates and pallets were dissolved in 2× Laemmli sample buffer and subjected to immunoblotting.
Statistical analysis
GraphPad Prism 8.0 software was used for all data analysis. Data were presented as mean ± SD. Student’s t test was used to compute P values and determine significance in group comparisons. Log-rank tests were used to compute P values for survival curves.
Online supplemental material
Fig. S1 shows that depletion of Irf8 in iBMDMs using the CRISPR/Cas9 system reduced the expression of Naips. Fig. S2 shows that Brd4 was involved in the activation of the noncanonical NLRP3 inflammasome and the pyrin inflammasome. Fig. S3 shows how Brd4 differentially regulated the transcription of Il1b and Il18. Fig. S4 shows that IL-18 secretion was decreased in Brd4-defcient BMDMs. Fig. S5 shows the results of ChIP-seq analysis of recruitment of IRF8 and Brd4 on Cmpk2 and Nlrc4.
Acknowledgments
We thank Dr. Gee Lau (University of Illinois at Urbana-Champaign, Urbana, IL) for providing the P. aeruginosa strain, Dr. Gang Huang (Cincinnati Children's Hospital Medical Center, Cincinnati, OH) for PU.1 expression vector, and Dr. Donghai Wang (Duke University School of Medicine, Durham, NC) for iBMDMs. We also thank all members in the Chen laboratory for discussion.
X. Dong was a recipient of the Herbert E. Carter Fellowship of the Department of Biochemistry, University of Illinois at Urbana-Champaign. This work is supported in part by funds provided by the University of Illinois at Urbana-Champaign to L.-F. Chen (RB18061) and funds provided by Fujian Medical University to X. Hu (XRCZX2019008).
The authors declare no competing financial interests.
Author contributions. X. Dong, X. Hu, Y. Bao, X.-d. Yang, and L.-F. Chen designed the experiment; X. Dong, X. Hu, G. Li, and Y. Bao performed the experiments; J.M. Slauch provided critical reagents; X. Dong, X. Hu, Y. Bao, and L.-F. Chen analyzed the data; L.-F. Chen supervised the research; X. Dong, J.M. Slauch, and L.-F. Chen wrote the manuscript.
References
- Bao, Y., Wu X., Chen J., Hu X., Zeng F., Cheng J., Jin H., Lin X., and Chen L.F.. 2017. Brd4 modulates the innate immune response through Mnk2-eIF4E pathway-dependent translational control of IκBα. Proc. Natl. Acad. Sci. USA. 114:E3993–E4001. 10.1073/pnas.1700109114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind, F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B.G., Fitzgerald K.A., et al. 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183:787–791. 10.4049/jimmunol.0901363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., Bair S., Newton G., Lichtman A., Kung A., et al. 2014. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 56:219–231. 10.1016/j.molcel.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, F.A., Nalbantoglu I., Aitken J.D., Uchiyama R., Su Y., Doho G.H., Vijay-Kumar M., and Gewirtz A.T.. 2012. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol. 5:288–298. 10.1038/mi.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Wang Z., Hu X., Chen R., Romero-Gallo J., Peek R.M. Jr., and Chen L.F.. 2016. BET inhibition attenuates Helicobacter pylori-induced inflammatory response by suppressing inflammatory gene transcription and enhancer activation. J. Immunol. 196:4132–4142. 10.4049/jimmunol.1502261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, A., Yang W., Gegonne A., Nishiyama A., Pan R., Yagi R., Grinberg A., Finkelman F.D., Pfeifer K., Zhu J., et al. 2019. BRD4 directs hematopoietic stem cell development and modulates macrophage inflammatory responses. EMBO J. 38:e100293. 10.15252/embj.2018100293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, J.A., and Canna S.W.. 2018. The NLRC4 inflammasome. Immunol. Rev. 281:115–123. 10.1111/imr.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., and Sul H.S.. 2007. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27:79–101. 10.1146/annurev.nutr.27.061406.093734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira, R., and Holden D.W.. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology (Reading). 158:1147–1161. 10.1099/mic.0.058115-0 [DOI] [PubMed] [Google Scholar]
- Franchi, L., Kamada N., Nakamura Y., Burberry A., Kuffa P., Suzuki S., Shaw M.H., Kim Y.G., and Núñez G.. 2012. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 13:449–456. 10.1038/ni.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Wallerath C., Covarrubias S., Hornung V., Carpenter S., and Fitzgerald K.A.. 2017. The PYHIN protein p205 regulates the inflammasome by controlling Asc expression. J. Immunol. 199:3249–3260. 10.4049/jimmunol.1700823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Callaway J.B., and Ting J.P.. 2015. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21:677–687. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah, N., Benner C., Chong L.W., Yu R.T., Downes M., and Evans R.M.. 2015. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc. Natl. Acad. Sci. USA. 112:E297–E302. 10.1073/pnas.1424028112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves, D.C., Horng T., and Medzhitov R.. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 138:129–145. 10.1016/j.cell.2009.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, W.T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.H., Zhong C.Q., and Han J.. 2015. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25:1285–1298. 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Hara H., and Núñez G.. 2016. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 41:1012–1021. 10.1016/j.tibs.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Yang X.D., Zhou M.M., Ozato K., and Chen L.F.. 2009. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29:1375–1387. 10.1128/MCB.01365-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., and Ozato K.. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 19:523–534. 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Jorgensen, I., Rayamajhi M., and Miao E.A.. 2017. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17:151–164. 10.1038/nri.2016.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, R., and Kanneganti T.D.. 2019. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer. 19:197–214. 10.1038/s41568-019-0123-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, R., Lee E., Place D., Samir P., Mavuluri J., Sharma B.R., Balakrishnan A., Malireddi R.K.S., Geiger R., Zhu Q., et al. 2018. IRF8 regulates transcription of Naips for NLRC4 inflammasome activation. Cell. 173:920–933.e13. 10.1016/j.cell.2018.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki, N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kayagaki, N., Lee B.L., Stowe I.B., Kornfeld O.S., O’Rourke K., Mirrashidi K.M., Haley B., Watanabe C., Roose-Girma M., Modrusan Z., et al. 2019. IRF2 transcriptionally induces GSDMD expression for pyroptosis. Sci. Signal. 12:eaax4917. 10.1126/scisignal.aax4917 [DOI] [PubMed] [Google Scholar]
- Kofoed, E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais, D., Barreiro L.B., and Gros P.. 2016. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J. Exp. Med. 213:585–603. 10.1084/jem.20151764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laricchia-Robbio, L., Tamura T., Karpova T., Sprague B.L., McNally J.G., and Ozato K.. 2005. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc. Natl. Acad. Sci. USA. 102:14368–14373. 10.1073/pnas.0504014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock, D.L., Chaudhary A., and Miller S.I.. 2015. Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13:191–205. 10.1038/nrmicro3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz, E., Xiao T.S., and Stutz A.. 2013. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13:397–411. 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., Schröder G.F., Fitzgerald K.A., Wu H., and Egelman E.H.. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 156:1193–1206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin, J., and Martinon F.. 2018. The AIM2 inflammasome: sensor of pathogens and cellular perturbations. Immunol. Rev. 281:99–114. 10.1111/imr.12618 [DOI] [PubMed] [Google Scholar]
- Man, S.M., and Kanneganti T.D.. 2015. Regulation of inflammasome activation. Immunol. Rev. 265:6–21. 10.1111/imr.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino, A., Termanini A., Barozzi I., Ghisletti S., Ostuni R., Prosperini E., Ozato K., and Natoli G.. 2015. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29:394–408. 10.1101/gad.257592.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto, J., Taniguchi S., Ayukawa K., Sarvotham H., Kishino T., Niikawa N., Hidaka E., Katsuyama T., Higuchi T., and Sagara J.. 1999. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 274:33835–33838. 10.1074/jbc.274.48.33835 [DOI] [PubMed] [Google Scholar]
- Miao, E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., and Aderem A.. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme, E., Jeffrey K.L., Schaefer U., Beinke S., Dewell S., Chung C.W., Chandwani R., Marazzi I., Wilson P., Coste H., et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature. 468:1119–1123. 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lopez, A., Rosales-Reyes R., Alpuche-Aranda C.M., and Ortiz-Navarrete V.. 2013. Salmonella downregulates Nod-like receptor family CARD domain containing protein 4 expression to promote its survival in B cells by preventing inflammasome activation and cell death. J. Immunol. 190:1201–1209. 10.4049/jimmunol.1200415 [DOI] [PubMed] [Google Scholar]
- Rathinam, V.A., Vanaja S.K., and Fitzgerald K.A.. 2012. Regulation of inflammasome signaling. Nat. Immunol. 13:333–342. 10.1038/ni.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi, M., Zhang Y., and Miao E.A.. 2013. Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol. Biol. 1040:85–90. 10.1007/978-1-62703-523-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, S., Salem D., and Gros P.. 2020. Role of IRF8 in immune cells functions, protection against infections, and susceptibility to inflammatory diseases. Hum. Genet. 139:707–721. 10.1007/s00439-020-02154-2 [DOI] [PubMed] [Google Scholar]
- Sellin, M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shi, J., and Vakoc C.R.. 2014. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 54:728–736. 10.1016/j.molcel.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Stutz, A., Horvath G.L., Monks B.G., and Latz E.. 2013. ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040:91–101. 10.1007/978-1-62703-523-1_8 [DOI] [PubMed] [Google Scholar]
- Sutterwala, F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., and Flavell R.A.. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245. 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O., and Akira S.. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tamura, T., Thotakura P., Tanaka T.S., Ko M.S., and Ozato K.. 2005. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 106:1938–1947. 10.1182/blood-2005-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Fan Y., Li J., Zhang X., Lou X., Dou Y., Shi X., Lan P., Xiao Y., Minze L., et al. 2018. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J. Exp. Med. 215:559–574. 10.1084/jem.20170928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., and Zhou Q.. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 19:535–545. 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- Yang, J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., and Shao F.. 2015. The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol. Rev. 265:85–102. 10.1111/imr.12293 [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Shi J., Shi X., Wang Y., Wang F., and Shao F.. 2016. Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. J. Exp. Med. 213:647–656. 10.1084/jem.20160006 [DOI] [PMC free article] [PubMed] [Google Scholar]