Abstract
Background
Reverse transcription polymerase chain reaction (RT-PCR) is the gold standard for detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Previously, the accuracy of the quantitative LUMIPULSE SARS-CoV-2 antigen test was demonstrated using samples collected retrospectively. In this study, the LUMIPULSE antigen test was clinically validated using prospective samples.
Methods
In total, 1033 nasopharyngeal swab samples were collected from 1033 individuals, and an additional 275 follow-up samples were collected from 43 patients who subsequently tested positive for coronavirus disease 2019 (COVID-19). All 1308 samples were subjected to quantitative RT-PCR (RT-qPCR) and the antigen test. The antibody response was investigated for patients with discordant results to clarify if seroconversion had occurred.
Results
RT-qPCR identified 990 samples as negative and 43 as positive, while the antigen test identified 992 samples as negative, 37 as positive and four as inconclusive. The overall concordance rate was 99.7% (1026/1029). Sensitivity, specificity, positive predictive value and negative predictive value of the antigen test were 92.5% (37/40), 100% (989/989), 100% (37/37) and 99.7% (989/992), respectively, after exclusion of the four inconclusive results. The kappa coefficient was 0.960 (95% confidence interval 0.892–0.960), suggesting excellent agreement between the two tests. Seropositivity in five of seven patients with discordant results suggested that the discrepancy was caused by samples collected during the late phase of infection. Using follow-up samples, correlation was observed between the antigen level and the viral load or cycle threshold value. The concordance rate between these test results tended to be high among samples collected 0–9 days after symptom onset, but this decreased gradually in samples collected thereafter.
Conclusions
This prospective study demonstrated that the LUMIPULSE antigen test is a highly accurate diagnostic test for SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Antigen, RT-qPCR, LUMIPULSE
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has spread rapidly around the world. Approximately 80% of infected patients with coronavirus disease 2019 (COVID-19) develop mild symptoms and recover without specific treatment (Matheson and Lehner, 2020). However, 20% of patients deteriorate rapidly within 7–10 days of symptom onset, and 25% of these patients will face mechanical ventilation and high mortality rates (Matheson and Lehner, 2020).
Reverse transcription polymerase chain reaction (RT-PCR) is a sensitive and specific assay that is considered the gold standard for SARS-CoV-2 testing (Corman et al., 2020, Hirotsu et al., 2020a). However, RT-PCR requires specialized equipment and skilled technicians, and is time-consuming and expensive to perform. As an alternative, the antigen test has been approved for the diagnosis of patients with COVID-19 (US Food and Drug Administration, n.d.).
Most antigen tests are developed for use as a rapid diagnostic at the point of care. These tests are based on paper assays or lateral flow immunochromatography and provide qualitative detection of SARS-CoV-2. The simple-to-use format of the rapid antigen test does not require special equipment or operator skills. In general, these rapid kits have high specificity but low sensitivity, which can yield false-negative results for samples with low viral loads (Albert et al., 2020, Cerutti et al., 2020, Linares et al., 2020, Mak et al., 2020, Scohy et al., 2020). For instance, sensitivity is 30.2% (32/106) for the COVID-19 Ag Respi-Strip (Coris BioConcept, Wallonia, Belgium) (Scohy et al., 2020), 45.7% (16/35) for the Biocredit Covid-19 Ag Detection Kit (BioVendor, Brno, Czech Republic) (Mak et al., 2020), 70.6% (77/109) for the STANDARD Q COVID-19 Ag Test (SD Biosensor, Suwon, Republic of Korea) (Cerutti et al., 2020), and 73.3–79.6% (44/60 and 43/55) for the Panbio COVID-19 Ag Rapid Test (Abbott, Abbott Park, IL, USA) (Albert et al., 2020, Linares et al., 2020).
The World Health Organization recommends that rapid diagnostic tests should be used for symptomatic individuals within the first 5–7 days following symptom onset, but should not be used for individuals without any symptoms (World Health Organization, 2020). There is growing demand for the clinical utility and high accuracy of the quantitative antigen test to be demonstrated. However, few prospective validation studies have been reported for large cohorts.
The authors previously evaluated the accuracy of the LUMIPULSE SARS-CoV-2 antigen test, which is a fully automated system based on the chemiluminescent enzyme immunoassay principle (Hirotsu et al., 2020b). This antigen test can measure the antigen level of SARS-CoV-2 nucleocapsid protein quantitatively. The antigen test has been approved in Japan and is widely used in hospitals, clinical laboratory centres and airport quarantines. Fujirebio Europe acquired CE marking in August 2020, and have started to supply the antigen test globally (e.g. at German airports) (Fujirebio et al., 2020a, Fujirebio et al., 2020b).
This article reports the findings from a prospective validation study of the LUMIPULSE antigen test using a total of 1308 nasopharyngeal swab samples. Of these, 1033 were initial samples collected prospectively from 1033 individuals. In addition, 275 follow-up samples were collected longitudinally from 42 confirmed cases of COVID-19 in this cohort. The antigen test and quantitative RT-PCR (RT-qPCR) were conducted for each sample. In addition, the antibody response in seven patients with discordant results was examined to clarify their seroconversion status.
Materials and methods
Patients and samples
In total, 1308 nasopharyngeal swab samples were collected from 1033 individuals. First, 1033samples from 1033 individuals–including symptomatic individuals (i.e. those with a fever, cough, sore throat, fatigue and/or headache), asymptomatic individuals who had been in contact with an infected patient, and returnees from abroad–were analysed prospectively. Among these 1033 individuals, 43 were confirmed as PCR-positive individuals (hereafter termed ‘patients with COVID-19’), including 36 symptomatic and seven asymptomatic patients. Second, 275 follow-up samples were collected longitudinally from the 43 patients with COVID-19. Thus, in total, 318 samples (43 initial and 275 follow-up samples) were collected from the 43 patients with COVID-19 (average of 7.4 samples per patient, range 1–27 samples). All 1308 samples were subjected to RT-qPCR and the LUMIPULSE antigen test.
The Institutional Review Board of the Clinical Research and Genome Research Committee at Yamanashi Central Hospital approved this study and the use of an opt-out consent method (Approval Nos. C2019-30 and C2020-9). The requirement for written informed consent was waived as this was an observational study and due to the urgent need to collect COVID-19 data. Participation in the study by patients was optional. All methods were performed in accordance with the relevant guidelines and regulations, and the Declaration of Helsinki.
Sample collection and processing
All nasopharyngeal swab samples were collected using cotton swabs and placed in 3 mL of viral transport media (VTM) obtained from Copan Diagnostics (Murrieta, CA, USA); 700 μL of VTM was used for the antigen test immediately after sample collection. The residual VTM was stored temporarily at 4 °C, and 200 μL of VTM was used for nucleic acid extraction within 2 h of sample collection.
SARS-CoV-2 antigen test (LUMIPULSE)
The sample antigen levels were determined quantitatively using the LUMIPULSE SARS-CoV-2 antigen test (Fujirebio, Inc., Tokyo, Japan) in accordance with the manufacturer’s instructions (Hirotsu et al., 2020b, Hirotsu et al., 2021). In brief, 700 μL of the VTM samples was vortexed, transferred into a sterile tube, and centrifuged at 2000×g for 5 min. Aliquots (100 μL) of the supernatant were used for testing on the LUMIPULSE G600II automated system (Fujirebio). For samples with an antigen level >5000 pg/mL, the samples were diluted with the kit diluent and retested, and the antigen level was calculated taking the dilution factor into account. Samples with an antigen level ≥10 pg/mL were considered positive, samples with an antigen level ≥1.0 pg/mL and <10.0 pg/mL were considered inconclusive, and samples with an antigen level <1.0 pg/mL were considered negative, in accordance with the manufacturer’s guidelines.
Viral nucleic acid extraction
Total nucleic acid was isolated from the samples using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) on the KingFisher Duo Prime System (Thermo Fisher Scientific), as described previously (Hirotsu et al., 2020c, Hirotsu et al., 2020d). Briefly, 200 μL of VTM, 5 μL of proteinase K, 265 μL of binding solution, 10 μL of total nucleic acid binding beads, 0.5 mL of wash buffer, and 0.5–1 mL of 80% ethanol was added to each well of a 96-well plate. The nucleic acids were eluted with 70 μL of elution buffer. The total nucleic acids were subjected to RT-qPCR immediately.
RT-qPCR
According to the protocol developed by the National Institute of Infectious Diseases in Japan (Hirotsu et al., 2020a, Hirotsu et al., 2020c, Shirato et al., 2020), one-step RT-qPCR was performed to detect SARS-CoV-2. This PCR amplifies the nucleocapsid (N) gene of SARS-CoV-2 (NC_045512.2). The reaction mixture comprises 5 μL of 4 × TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific), 1.0 μL of 10 μM forward primer (5′-AAATTTTGGGGACCAGGAAC-3′), 1.4 μL of 10 μM reverse primer (5′-TGGCAGCTGTGTAGGTCAAC-3′), 0.8 μL of 5 μM probe (5′-FAM-ATGTCGCGCATTGGCATGGA-TAMRA-3′), 6.8 μL of nuclease-free water, and 5 μL of nucleic acid sample in a 20-μL total volume. The expected amplicon size is 158 bp. The human ribonuclease P protein subunit p30 (RPP30) gene was used (Integrated DNA Technologies, Coralville, IA, USA) as the internal positive control (Hirotsu et al., 2020a).
RT-qPCR assays were conducted on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) with the following cycling conditions: 50 °C for 5 min for reverse transcription, 95 °C for 20 s, and 45 cycles at 95 °C for 3 s and 60 °C for 30 s. The threshold was set at 0.2.
A cycle threshold (Ct) value was assigned to each PCR reaction, and the amplification curve was assessed visually. According to the national protocol (Version 2.9.1) (Shirato et al., 2020), a sample was considered positive when a visible amplification plot was observed, and a sample was considered negative when no amplification was observed.
The absolute copy number of viral loads was determined using serial diluted DNA control targeting the N gene of SARS-CoV-2 (Integrated DNA Technologies), as described previously (Hirotsu et al., 2020a). The limit of detection of RT-qPCR using the primer/probe was considered as two copies in accordance with the previous report (Shirato et al., 2020).
SARS-CoV-2 antibody test
To clarify the discordant results observed between RT-qPCR and the antigen test in seven individuals, the pan-immunoglobulin level was measured against the full length of SARS-CoV-2 nucleocapsid recombinant protein expressed in Escherichia coli. The serum samples were subjected to the Elecsys Anti-SARS-CoV-2 test (Roche Diagnostics, Basel, Switzerland) on the cobas 8000 automated platform (Roche Diagnostics) (Muench et al., 2020). This assay uses the electrochemiluminescence immunoassay principle. Samples with a cut-off index (COI; electrochemiluminescent signal of the test sample/cut-off value of the calibration sample) <1.0 were considered negative, while those with COI ≥ 1.0 were considered positive.
Statistical analysis
Sensitivity, specificity, positive predictive value and negative predictive value were calculated, using the results of RT-qPCR as the reference, and with exclusion of samples with an inconclusive result on antigen testing. Student’s t-test was calculated to determine significant differences between the groups, and P < 0.05 was considered to indicate statistical significance. Cohen’s kappa coefficient of results between the two tests with 95% confidence intervals (CI) was calculated using R Version 3.1.2 (http://www.r-project.org/). Cohen’s kappa values >0.81 were interpreted to indicate near-perfect agreement (Landis and Koch, 1977).
Results
Comparison of the antigen test and RT-qPCR results
In total, 1308 nasopharyngeal swab samples (1033 initial samples and 275 follow-up samples) were collected from 1033 individuals. Each sample was subjected to both RT-qPCR and the LUMIPULSE SARS-CoV-2 antigen test.
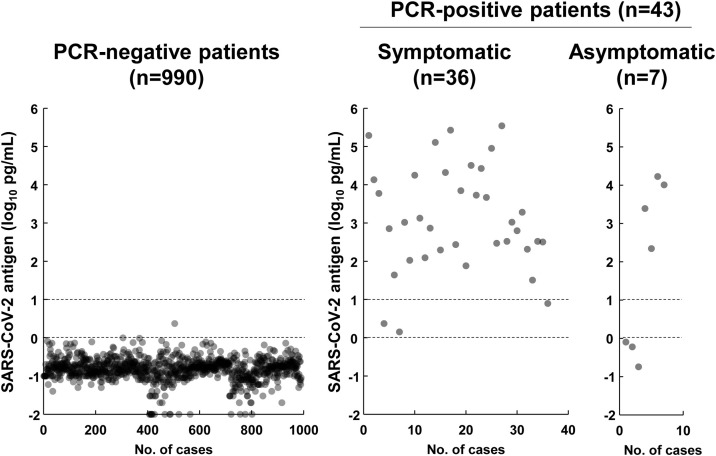
Among the 1033 initial samples, RT-qPCR identified 990 as negative and 43 as positive (symptomatic, n = 36; asymptomatic, n = 7; Figure 1 ). The antigen level of RT-qPCR-positive patients was significantly higher than that of RT-qPCR-negative patients (P = 0.78 × 10−31, Student's t-test) (Figure 1). The mean antigen level was −0.7 log10 pg/mL (range −2.0 to 0.4 log10 pg/mL) for RT-qPCR-negative patients and 4.4 log10 pg/mL (range 0.7–5.5 log10 pg/mL) for RT-qPCR-positive patients (Figure 1). There was no significant difference in antigen levels between symptomatic and asymptomatic patients (P = 0.36, Student's t-test). To determine the accuracy of the antigen test, the results from RT-qPCR were compared with those from the antigen test for each sample. The antigen test identified 992 samples as negative, 37 as positive, and four as inconclusive (Table 1 ). Thus, the rate of inconclusive results with the antigen test was 0.4% (4/1033).
Figure 1.
Prospective study of the antigen levels in 1033 initial samples collected from 1033 individuals. The dot plots show the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) antigen levels in the samples identified by polymerase chain reaction (PCR) as negative (n = 990) or positive (n = 43). Among the 43 PCR-positive individuals, 36 were symptomatic and seven were asymptomatic. The two dashed lines indicate the decision threshold for the LUMIPULSE antigen test. The lower dashed line indicates 0 log10 pg/mL (1 pg/mL) and the upper dashed line indicates 1 log10 pg/mL (10 pg/mL).
Table 1.
Comparison of LUMIPULSE antigen test and quantitative reverse transcription polymerase chain reaction (RT-qPCR) results.
| Antigen test | RT-qPCR | Number of samples (%) | Mean antigen level (range) (log10 pg/mL) | Mean viral load (range) (log10 copies/μL) | Ct value, mean (range) |
|---|---|---|---|---|---|
| Negative | Negative | 989 (95.7%) | −0.74 (−2 to −0.0044) | NA | NA |
| Negative | Positive | 3 (0.3%) | −0.28 (−0.74 to −0.97) | 1.3 (1.0–1.9) | 36.7 (35–39) |
| Positive | Negative | 0 (0%) | NA | NA | NA |
| Positive | Positive | 37 (3.6%) | 4.5 (1.4–5.6) | 5.4 (1.4–7.7) | 21.9 (14–35) |
| Inconclusive | Negative | 1 (0.1%) | 0.37 | NA | NA |
| Inconclusive | Positive | 3 (0.3%) | 0.59 (0.15–0.89) | 1.7 (1.3–2.2) | 34.3 (33–36) |
| Total | 1033 (100%) |
Ct, threshold cycle; NA, not available.
Antigen levels were determined by the LUMIPULSE antigen test. Viral load and Ct value were determined by RT-qPCR. There were significant differences in viral load (P = 0.88 × 10−5) and Ct value (P = 0.13 × 10−5) between the antigen-test-negative and RT-qPCR-positive samples, and samples that were positive on both tests.
The mean viral load (log10 copies/μL) was 1.3 [median ± standard deviation (SD) 1.1 ± 0.5] in the antigen-test-negative and RT-qPCR-positive samples, and 5.4 (median ± SD 5.6 ± 1.3) in the samples judged to be positive by both tests (P = 0.88 × 10−5, Student's t-test) (Table 1). The mean Ct value of RT-qPCR was 36.7 (median ± SD 36.0 ± 2.1) in the antigen-test-negative and RT-qPCR-positive samples, and 21.9 (median ± SD 21.0 ± 4.4) in the samples judged to be positive by both tests (P = 0.13 × 10−5, Student's t-test). The Ct value of samples that were on the borderline between positive and negative based on the antigen test results was approximately 35.
The overall concordance rate between the two tests was 99.7% (1026/1029). The antigen test exhibited sensitivity of 92.5% (37/40), specificity of 100% (989/989), positive predictive value of 100% (37/37) and negative predictive value of 99.7% (989/992). The kappa coefficient was 0.960 (95% CI 0.892–0.960), suggesting excellent agreement between the two tests. These results suggest that the antigen test can judge the positive or negative status of almost all samples with high accuracy compared with RT-qPCR.
Discrepant results attributable to samples with low viral load collected from seropositive patients
There were seven discordant results (0.7%, 7/1033) between RT-qPCR and the antigen test (Table 2 ). Of these, three were negative on the antigen test and positive on RT-qPCR (Cases #1–3), three were inconclusive on the antigen test and positive on RT-qPCR (Cases #4–6), and one was inconclusive on the antigen test and negative on RT-qPCR (Case #7).
Table 2.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR), LUMIPULSE antigen test, and antibody test results for the seven discordant cases.
| Case # | Residence | Antigen test | Antigen level (pg/mL) | RT-qPCR | Viral load (log10 copies/μL) | Antibody test | COI at admission |
|---|---|---|---|---|---|---|---|
| #1 | Brazil | Negative | 0.8 | Positive | 1.1 | Positive | 69.9 |
| #2 | Brazil | Negative | 0.6 | Positive | 1.9 | Positive | 84.3 |
| #3 | India | Negative | 0.18 | Positive | 1.0 | Positive | 1.0 |
| #4 | Japan | Inconclusive | 2.35 | Positive | 2.2 | Positive | 2.6 |
| #5 | Japan | Inconclusive | 1.42 | Positive | 1.3 | Positive | 3.2 |
| #6 | Japan | Inconclusive | 7.85 | Positive | 1.7 | Negative | 0.3 |
| #7 | Japan | Inconclusive | 2.36 | Negative | NA | Negative | 0.09 |
COI, cut-off index; NA, not available.
Cases #1–3 had returned from overseas (two from Brazil and one from India) and tested positive on RT-PCR at airport quarantine in Japan. Cases #4–7 lived in the district and were admitted to the study hospital. Case #7 was found in cardiopulmonary arrest at home and was an emergency admission to the hospital but passed away shortly afterwards.
At the beginning of hospitalization, the antibody levels were positive in Cases #1–5 but the viral loads were low (range 1.0–2.2 log10 copies/μL), indicating that these patients were seropositive for SARS-CoV-2 at a later phase of infection (Table 2). Conversely, the antibody response had likely only just commenced in Case #6, as reported previously (Omata et al., 2021) (Table 2).
Correlation between antigen level and viral load of SARS-CoV-2
In this prospective study, 318 nasopharyngeal swab samples were collected from 43 patients with COVID-19. These 318 samples included 43 initial samples and an additional 275 follow-up samples collected during hospitalization. Of 318 samples, 43 initial samples had been analysed previously (Figure 1), and these data were used for the following analyses.
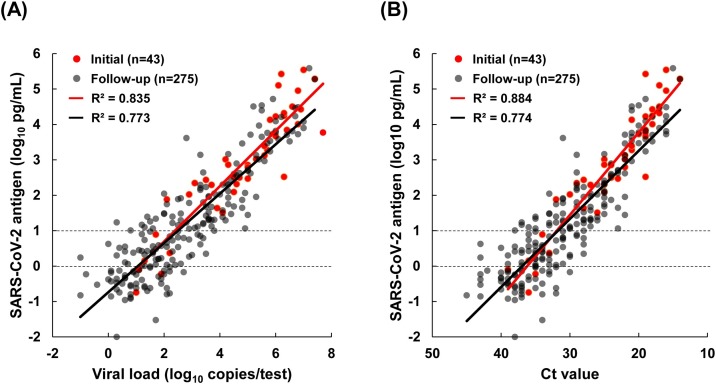
Consistent with the previous report (Hirotsu et al., 2020b), the viral load determined by RT-qPCR and the antigen level were correlated for both the initial samples (R 2 = 0.835) and the follow-up samples (R 2 = 0.773) (Figure 2 A). Similarly, there was correlation between the Ct value and the antigen level in both the initial samples (R 2 = 0.884) and the follow-up samples (R 2 = 0.774) (Figure 2B). The coefficient of determination for the initial samples was slightly higher than that for the follow-up samples. These results suggest that variability increased in follow-up samples because these samples included lower viral load samples collected from hospitalized patients who were in a late phase of infection or recovery.
Figure 2.
Correlation between the antigen level and viral load or threshold cycle (Ct) value. In the study cohort, 43 patients with coronavirus disease 2019 were identified. A total of 318 samples were collected from these patients, including 43 initial and 275 follow-up samples. The dot plots show the correlation between the antigen level as determined in the LUMIPULSE antigen test and the viral load (A) or Ct value (B) as determined on quantitative reverse transcription polymerase chain reaction. SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
Relationship between test results and timing of sample collection from symptom onset
Of 318 samples collected from 43 patients with COVID-19, 250 nasopharyngeal swab samples were collected from 36 symptomatic patients and 68 samples were collected from seven symptomatic patients. To investigate the relationship between time since symptom onset and test results, these 250 samples from symptomatic patients were examined up to 30 days after symptom onset.
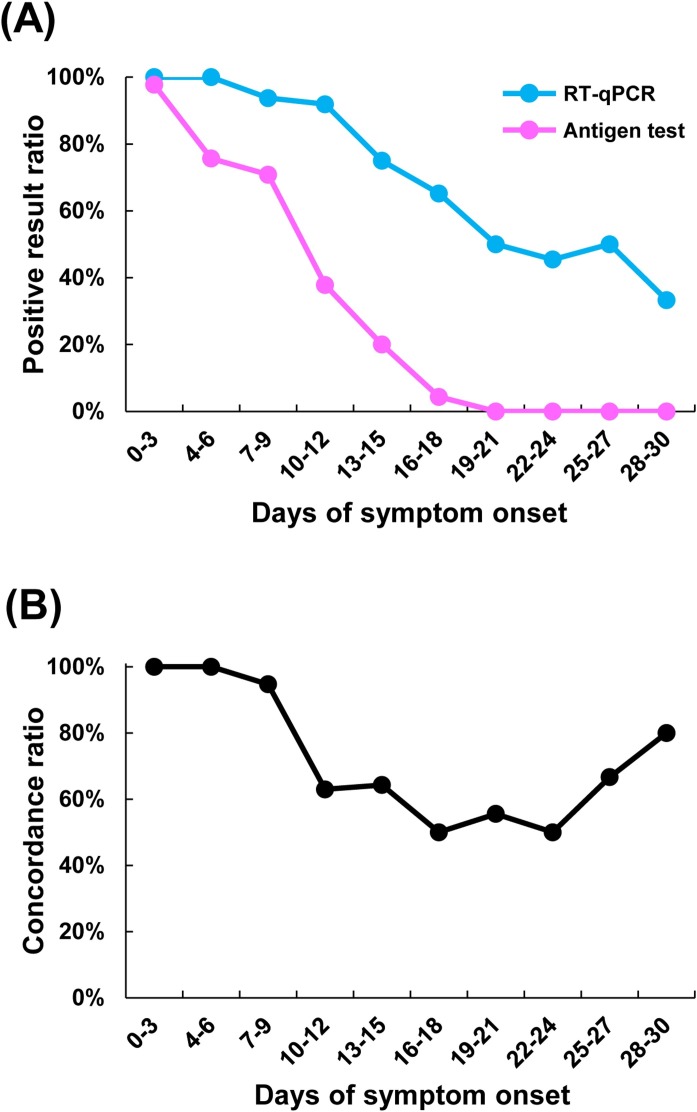
Positive results on both the antigen test (71%–98%) and RT-qPCR (94–100%) were observed in a high proportion of samples collected within 0–9 days of symptom onset (Figure 3 A). However, the positive result ratio of the antigen test declined over 10–30 days after symptom onset (Figure 3A). Overall, the positive result ratio of RT-qPCR was higher than that for the antigen test over the entire observation period (Figure 3A). The concordance ratio between RT-qPCR and the antigen test was high (95–100%) for samples collected within 0–9 days of onset, low (50–64%) for samples collected within 10–24 days of symptom onset, and slightly increased (67–80%) for samples collected within 25–30 days of symptom onset (Figure 3B).
Figure 3.
Effect of the timing of sample collection since symptom onset on test results. In total, 250 samples were collected from 36 symptomatic patients with coronavirus disease 2019 for 30 days after symptom onset, and these samples were subjected to both the LUMIPULSE antigen test and quantitative reverse transcription polymerase chain reaction (RT-qPCR). (A) Positive result ratio was calculated by the number of positive samples on RT-qPCR or antigen test during the period since symptom onset divided by the total number of samples tested in that period. (B) The concordance ratio indicates the percentage of samples with equivalent results from the antigen test and RT-qPCR when excluding samples with inconclusive antigen test results.
Detection rate of antigen test decrease after 10 days of symptom onset
Among the 250 follow-up samples from 36 symptomatic patients, the overall concordance rate was 80.6% (162/201), sensitivity was 76.1% (124/163), and specificity was 100% (38/38) (Table 3 ). Sensitivity was high (94.4–100%) for the samples collected within 0–9 days of symptom onset; however, this declined gradually for samples collected ≥10 days after symptom onset. Notably, specificity was 100% throughout the observation period (Table 3).
Table 3.
Relationship between timing of sample collection following symptom onset and LUMIPULSE antigen test and quantitative reverse transcription polymerase chain reaction (RT-qPCR) results.
| Days since symptom onset | Number of samples | Concordant (positive on both tests) | Concordant (negative on both tests) | Discordant (antigen test: negative/RT-qPCR: positive) | Discordant (antigen test: positive/RT-qPCR: negative) | Inconclusive on antigen test | Overall agreementa | Sensitivitya | Specificitya |
|---|---|---|---|---|---|---|---|---|---|
| 0–3 | 44 | 43 | 0 | 0 | 0 | 1 | 100% | 100% | NA |
| 4–6 | 37 | 28 | 0 | 0 | 0 | 9 | 100% | 100% | NA |
| 7–9 | 48 | 34 | 2 | 2 | 0 | 10 | 94.7% | 94.4% | 100% |
| 10–12 | 37 | 14 | 3 | 10 | 0 | 10 | 63.0% | 58.3% | 100% |
| 13–15 | 20 | 4 | 5 | 5 | 0 | 6 | 64.3% | 44.4% | 100% |
| 16–18 | 23 | 1 | 8 | 9 | 0 | 5 | 50.0% | 10.0% | 100% |
| 19–21 | 12 | 0 | 5 | 4 | 0 | 3 | 55.6% | 0% | 100% |
| 22–24 | 11 | 0 | 5 | 5 | 0 | 1 | 50.0% | 0% | 100% |
| 25–27 | 12 | 0 | 6 | 3 | 0 | 3 | 66.7% | 0% | 100% |
| 28–30 | 6 | 0 | 4 | 1 | 0 | 1 | 80.0% | 0% | 100% |
| Total | 250 | 124 | 38 | 39 | 0 | 49 | 80.6% | 76.1% | 100% |
NA, not available.
These data were calculated after excluding the 49 inconclusive samples from the total of 250 samples.
Discussion
This study prospectively validated the performance of the LUMIPULSE SARS-CoV-2 antigen test. Compared with RT-qPCR, the accuracy of the antigen test was high when the test was conducted using initial samples from 1033 patients who visited the hospital. The sensitivity of the LUMIPULSE antigen test (92.5%) was higher than that of a conventional rapid antigen test (approximately 30–70%) (Albert et al., 2020, Cerutti et al., 2020, Linares et al., 2020, Mak et al., 2020, Scohy et al., 2020). The positive detection rate of the antigen test decreased gradually in follow-up samples collected from patients hospitalized with COVID-19. To the authors’ knowledge, this is the first longitudinal, prospective study to provide real-world data illustrating the clinical validity of antigen tests for COVID-19 screening.
In 1033 individuals, three (0.3%) were judged as positive on RT-qPCR and negative on the antigen test (Cases #1–3 in Table 2). This study found that these false-negative results were attributed to the samples with very low viral load obtained from seropositive patients. Furthermore, in hospitalized patients, the antigen test tended to judge negative for samples collected ≥10 days after symptom onset, but RT-qPCR often remained positive (Figure 3). In a previous study, the authors consistently observed discordant results in samples collected from a persistent viral-shedding patient (Hirotsu et al., 2021). These results suggest that the viral load was low and protein translation was likely to be attenuated in host cells. It is believed that assessing the immune response with an antibody test is useful to interpret the discrepant results (Omata et al., 2021). In antigen-test-negative and RT-PCR-positive samples, the two possibilities need to be examined carefully: namely, the sample was collected from a patient in a late or recovery phase of infection, or the sample was collected from a patient in a very early phase of infection.
False-positive results are a burden to both patients and healthcare workers, necessitating patient quarantine and surveys of up to 100 individuals who had close contact with these patients. To prevent misleading false-positive results, a highly specific test is needed. Previously, a case report showed a false-positive result with the LUMIPULSE antigen test (Ogawa et al., 2020). However, the percentage of false-positive results is 0.3% (1/301) according to the data on the kit's package insert, suggesting that the LUMIPULSE antigen test yields a robust result. In the present study, specificity was 100% for both the initial samples (989/989 samples) and follow-up samples (38/38 samples). The authors encountered fluctuating results when using viscous samples in a preliminary study, but resolved this issue by centrifuging the samples sufficiently and using the supernatants for the antigen test (data not shown). It is hoped that further scrutiny of false-positive samples will lead to increased accuracy regardless of the nature of the sample.
Data on SARS-CoV-2 infectivity are accumulating (Rhee et al., 2020). RT-PCR yields a positive result for a long duration (6–7 weeks following infection), even in the presence of an extremely low viral load (Sun et al., 2020, Hirotsu et al., 2021). However, RT-PCR cannot directly indicate the presence of viable and infectious SARS-CoV-2. In-vitro studies have revealed that the infectivity of SARS-CoV-2 is only maintained in clinical samples for approximately 8–10 days after symptom onset (Bullard et al., 2020, La Scola et al., 2020, Million et al., 2020, Perera et al., 2020, van Kampen et al., 2020, Wolfel et al., 2020). Therefore, RT-PCR-positive results can reflect the presence of non-infectious viral ‘debris’ in samples collected several weeks after symptom onset or recovery. Notably, this study observed that the rate of positive results with the antigen test declined rapidly at approximately 9 days after symptom onset. Based on in-vitro studies, the timing of the decrease in antigen levels appears to mark the point when the levels of infectious virus particles diminish (Bullard et al., 2020, La Scola et al., 2020, Million et al., 2020, Perera et al., 2020, van Kampen et al., 2020, Wolfel et al., 2020). Further studies using cell-based models and non-human primate models are required to clarify the relationship between antigen levels and virus infectivity (Munster et al., 2020, Wolfel et al., 2020), and to investigate whether monitoring the antigen level will help to determine the length of quarantine needed, likely response to treatment and timing of hospital discharge.
A possible limitation of this study is the portability of the LUMIPULSE antigen test. Although it is easier to conduct than RT-qPCR, it requires specific equipment and centrifugation. Therefore, it is difficult to test outside of the clinical laboratory. There is also a serious concern about how to manage this test in low-income countries. There is a need to develop tests that can be applied to a wide range of circumstances by making the equipment more accessible and improving the protocol.
In conclusion, the LUMIPULSE SARS-CoV-2 antigen test system can be applied easily in the majority of hospitals that are caring for patients with COVID-19, and it offers good sensitivity and high specificity as a clinical tool. The antigen level, viral load and Ct value determined with the antigen test and RT-qPCR provide meaningful information regarding the stage of infection in patients with COVID-19.
Conflict of interest
None declared.
Funding
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), a Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from Takeda Science Foundation (to Y.H.).
Contributions
YH contributed to study design, data collection, data analysis and writing (review and editing). MM, MS, KA, YN, KH and HS were involved in sample preparation and data collection, and data analysis. MH and HM supervised the research. TT, YK and YM provided resources. MO supervised the research, and was also involved in writing (review and editing).
Acknowledgements
The authors wish to thank Shintaro Yagi and Satoshi Kojima (Fujirebio) for technical discussion, and all of the medical and ancillary hospital staff and patients for consenting to participate. The authors also wish to thank Natasha Beeton-Kempen, PhD, from Edanz Group (https://en-author-services.edanz.com/) for editing a draft of this manuscript.
References
- Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M.Á. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centers. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.11.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujirebio . Fujirebio; Ghent: 2020. Fujirebio Europe and CENTOGENE Enter Partnership to Provide Rapid And High-Quality Preventive SARS-CoV-2 Antigen Testing. Available at: https://www.fujirebio.com/en/news-events/fujirebio-europe-and-centogene-enter-partnership-to-provide-rapid-and-highquality [Accessed 15 February 2021] [Google Scholar]
- Fujirebio . Fujirebio; Ghent: 2020. Fujirebio Europe Announces CE Marking of the Fully Automated Lumipulse® G SARS-CoV-2 Antigen Assay and the Rapid Antigen Device ESPLINE® SARS-CoV-2. Available at: https://www.fujirebio.com/en/news-events/fujirebio-europe-announces-ce-marking-of-the-fully-automated-lumipulser-g-sarscov2 [Accessed 15 February 2021] [Google Scholar]
- Hirotsu Y., Mochizuki H., Omata M. Double-quencher probes improve detection sensitivity toward severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020;284:113926. doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci Rep. 2020;10:18899. doi: 10.1038/s41598-020-76043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Nakajima M., Mochizuki H., Omata M. Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: a patient from the Diamond Princess cruise ship. Infect Control Hosp Epidemiol. 2020;41:1105–1106. doi: 10.1017/ice.2020.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K. Analysis of a persistent viral shedding patient infected with SARS-CoV-2 by RT-qPCR, FilmArray Respiratory Panel v2.1, and antigen detection. J Infect Chemother. 2021;27:406–409. doi: 10.1016/j.jiac.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Linares M., Pérez-Tanoira R., Carrero A., Romanyk J., Pérez-García F., Gómez-Herruz P. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson N.J., Lehner P.J. How does SARS-CoV-2 cause COVID-19? Science. 2020;369:510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- Million M., Lagier J.C., Gautret P., Colson P., Fournier P.E., Amrane S. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: a retrospective analysis of 1061 cases in Marseille, France. Travel Med Infect Dis. 2020;35:101738. doi: 10.1016/j.tmaid.2020.101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench P., Jochum S., Wenderoth V., Ofenloch-Haehnle B., Hombach M., Strobl M. Development and validation of the Elecsys Anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01694-20. e01694-e01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Fukumori T., Nishihara Y., Sekine T., Okuda N., Nishimura T. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. J Clin Virol. 2020;131:104612. doi: 10.1016/j.jcv.2020.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata M., Hirotsu Y., Sugiura H., Maejima M., Nagakubo Y., Amemiya K. The dynamic change of antibody index against Covid-19 is a powerful diagnostic tool for the early phase of the infection and salvage PCR assay errors. J Microbiol Immunol Infect. 2021 doi: 10.1016/j.jmii.2020.12.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C., Kanjilal S., Baker M., Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1249. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Sun J., Xiao J., Sun R., Tang X., Liang C., Lin H. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg Infect Dis. 2020;26:1834–1838. doi: 10.3201/eid2608.201097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration. Emergency Use Authorization. White Oak, MD; FDA; n.d. Available at: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization [Accessed 15 February 2021].
- van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020 doi: 10.1038/s41467-020-20568-4. 2020.2006.2008.20125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . WHO; Geneva: 2020. Antigen Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays. Available at: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays [Accessed 25 January 2021] [Google Scholar]