Abstract
CARM1 is a protein arginine methyltransferase (PRMT) that acts as a coactivator in a number of transcriptional programs. CARM1 orchestrates this coactivator activity in part by depositing the H3R17me2a histone mark in the vicinity of gene promoters that it regulates. However, the gross levels of H3R17me2a in CARM1 KO mice did not significantly decrease, indicating that other PRMT(s) may compensate for this loss. We thus performed a screen of type I PRMTs, which revealed that PRMT6 can also deposit the H3R17me2a mark in vitro. CARM1 knockout mice are perinatally lethal and display a reduced fetal size, whereas PRMT6 null mice are viable, which permits the generation of double knockouts. Embryos that are null for both CARM1 and PRMT6 are noticeably smaller than CARM1 null embryos, providing in vivo evidence of redundancy. Mouse embryonic fibroblasts (MEFs) from the double knockout embryos display an absence of the H3R17me2a mark during mitosis and increased signs of DNA damage. Moreover, using the combination of CARM1 and PRMT6 inhibitors suppresses the cell proliferation of WT MEFs, suggesting a synergistic effect between CARM1 and PRMT6 inhibitions. These studies provide direct evidence that PRMT6 also deposits the H3R17me2a mark and acts redundantly with CARM1.
Keywords: arginine methylation, PRMT6, CARM1, transcriptional coactivator, epigenetics, histone methylation, post-translational modification (PTM), post-transcriptional regulation
Growing evidence shows that protein arginine methylation plays a significant role in a number of cellular processes, including gene regulation, mRNA splicing, translation, DNA damage response, and signal transduction (1). In mammalian cells, three types of methylarginine are catalyzed by nine protein arginine methyltransferases (PRMTs): Type I methyltransferases generating asymmetric dimethylarginine (ADMA), Type II methyltransferases generating symmetric dimethylarginine (SDMA), and Type III methyltransferases generating monomethylarginine (MMA) (2). CARM1 (also called PRMT4) is recognized as a transcriptional activator and belongs to the Type I methyltransferase class. CARM1 deposits methylation marks on both histone tails (3) and a number of nonhistone proteins (4–6). CARM1 executes its coactivator function by modifying a diverse substrate repertoire that often shares a splicing and/or transcriptional theme (7–10) and includes the H3R17 histone code mark as one of the major regulatory nodes (8, 11–13). CARM1 can methylate histone H3 on both Arg-17 and Arg-26 residues but has a preference for Arg-17 over Arg-26 (14), at least in vitro. It is the only mammalian PRMT known to methylate H3R17 (15); however, interestingly, ablation of CARM1 in mouse embryonic fibroblasts (MEFs) does not result in the total loss of the H3R17me2a mark (9), indicating that another PRMT(s) may methylate this site when CARM1 is absent.
PRMT6 is also a Type I arginine methyltransferase that catalyzes the H3R2me2a mark, which in turn blocks enzymes from depositing the adjacent H3K4me3 and impedes the binding of the effectors of this transcriptional activation mark (16–18). Thus, in this setting, PRMT6 functions as a transcriptional corepressor. Most of the PRMTs methylate glycine- and arginine-rich (GAR) motifs within their substrates, but CARM1 displays unique substrate specificity and cannot methylate GAR motifs (19, 20). Similar to other PRMTs, PRMT6 typically methylates GAR motifs in substrates like HMGA1 and fibrillarin (21, 22), but it also methylates non-GAR motifs in the HIV-1 Tat protein and on PRMT6 itself (an automethylation site) (23, 24). PRMT6 methylates multiple sites on histones apart from H3R2, including H2A/H4R3 (17), H2AR29 (25), and H3R42 (26), all of which are non-GAR motifs. Interestingly, both CARM1 and PRMT6 can methylate histone H3 at Arg-42 (26), suggesting that PRMT6 and CARM1 can share substrates in some scenarios. Moreover, an in vitro methylation assay performed on nearly 200 putative CARM1 substrates revealed that PRMT6 could also methylate some of these substrates, but PRMT1 and PRMT5 could not (27). Supporting this possible redundant role for PRMT6 and CARM1 is the evidence that both of these PRMTs function as coactivators in the context of nuclear receptor–mediated transcriptional activation (28).
In this study, we performed in vitro methylation assays using a panel of recombinant type I PRMTs, showing that PRMT6 can efficiently methylate the histone H3R17 site. The CARM1 knockout mice died at birth and showed a reduced fetal size (9). In contrast, the PRMT6 knockout mice were viable (29), allowing us to generate CARM1 knockout embryos on a PRMT6 knockout background. We found that embryos that are null for both CARM1 and PRMT6 are smaller than littermates that are only null for CARM1. Importantly, MEFs generated from these double knockout embryos displayed a reduction in the H3R17me2a mark. We also detected elevated levels of DNA damage in these double knockout cells. Using two recently developed small molecular compounds that specifically inhibit CARM1 (30) and PRMT6 (31), we show that there is a synergistic effect between CARM1 and PRMT6 inhibition on cell proliferation. Thus, these data unmask partially redundant functions for CARM1 and PRMT6.
Results
PRMT6 generates the H3R17me2a mark in vitro
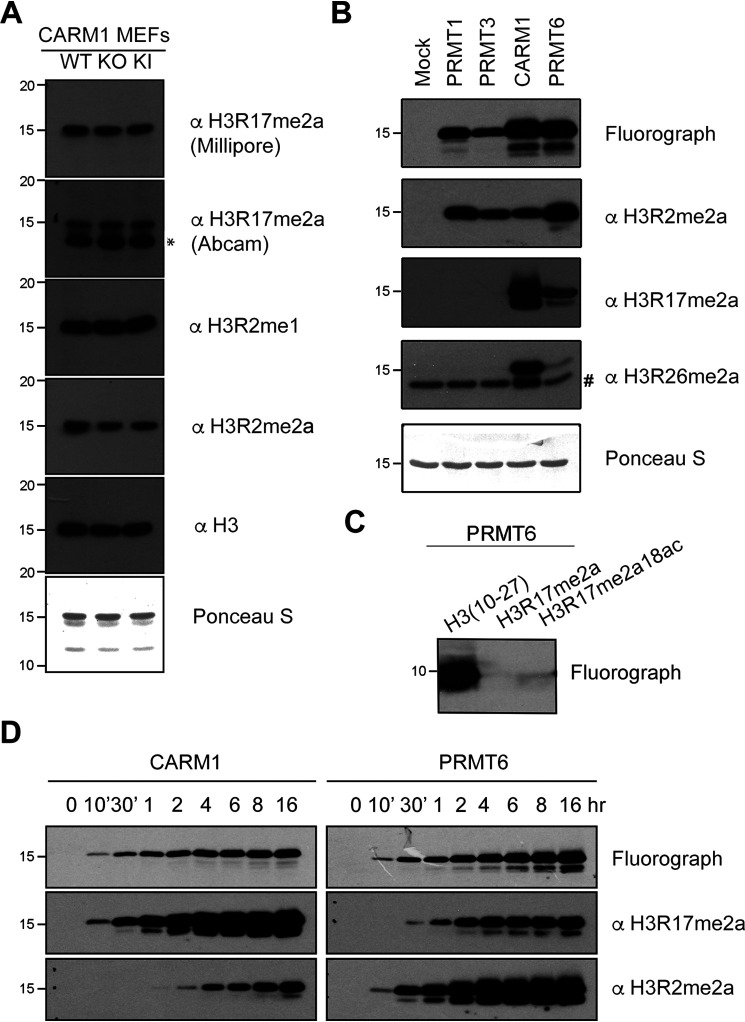
Previously, we showed that global levels of H3R17me2a are not impacted by the loss of CARM1 (9), suggesting that other PRMT(s) may also be able to methylate this site. To test this hypothesis, we purified core histones from the aforementioned CARM1 knockout MEFs and CARM1 enzyme-dead knock-in MEFs (32) (Fig. S1A). Using two different H3R17me2a-specific antibodies, we observed no decrease in H3R17me2a levels in either the CARM1 knockout or the enzyme-dead knock-in MEF lines (Fig. 1A). We confirmed the specificity of these antibodies by using a panel of arginine-methylated peptides (Fig. S1B). The Millipore antibody for H3R17me2a was used in the following studies because there is an additional nonspecific band (Fig. 1A) using the Abcam antibody. To search for the PRMTs that generate the H3R17me2a mark, we performed an in vitro methylation assay with a few recombinant type I PRMTs: PRMT1, PRMT3, PRMT6, and CARM1. All four PRMTs can methylate histone H3, to varying degrees, to generate the H3R2me2a mark; however, only CARM1 and PRMT6 can generate the H3R17me2a mark (Fig. 1B). Like CARM1, PRMT6 can generate the H3R26me2a mark, albeit weakly (Fig. 1B). In addition, recombinant PRMT6 can methylate a histone H3 peptide (amino acids 10–27) that harbors the H3R17 motif; however, when the H3R17 site is masked with a dimethyl mark or mutated to alanine, it can no longer be methylated by PRMT6 (Fig. 1C and Fig. S1C). Finally, we performed a time course in vitro methylation assay and observed a steady increase in the methylation of both H3R17 and H3R2 sites by PRMT6 (Fig. 1D). These findings suggest that both CARM1 and PRMT6 deposit an ADMA mark on H3R17, at least in vitro.
Figure 1.
PRMT6 methylates histone H3 at the Arg-17 site in vitro. A, acid-extracted core histones from CARM1 WT, KO, and enzyme-dead KI MEFs were immunoblotted using the indicated methyl-specific antibodies. Ponceau staining served as an additional loading control. B, in vitro methylation reactions were performed using recombinant histone H3 (2 µg) with the indicated GST-tagged PRMTs (0.5 µg) in the presence of [3H]SAM. Reactions were separated on SDS-PAGE, transferred to PVDF membranes for fluorograph, and immunoblotted using the indicated arginine methyl-specific antibodies to histone H3. C, unmethylated histone H3 peptide and Arg-17–methylated peptides were incubated with GST-PRMT6 and [3H]SAM. Methylation signal by PRMT6 was detected by fluorography. D, in vitro methylation reactions were performed using recombinant histone H3 with the indicated recombinant GST-fused PRMT in the presence of [3H]SAM and incubated at the indicated time. Reactions were separated on an SDS-polyacrylamide gel, transferred to PVDF membranes for fluorograph, and immunoblotted using the indicated methyl-specific antibodies. *, nonspecific band detected by the Abcam H3R17me2a antibody. #, nonspecific band detected by the H3R26me2a antibody.
PRMT6 overexpression results in elevated levels of the H3R17me2a mark in HeLa cells
To determine whether PRMT6 can methylate the H3R17 site in mammalian cells, PRMT6 and CARM1 were overexpressed in HeLa cells, and the levels of H3R17me2a were tested by Western blotting. Increased levels of H3R2me2a by PRMT6 overexpression were observed as reported previously (18). In addition, the levels of H3R17me2a were also elevated when PRMT6 was ectopically expressed (Fig. 2A). In contrast, the overexpression of CARM1 in HeLa cells did not cause an increase in the H3R17me2a mark, likely because the ectopically expressed CARM1 is predominantly cytoplasmic (Fig. S2A) and does not engage its nuclear substrates. We also note that the GFP-CARM1 protein level is lower than the endogenous CARM1 levels (Fig. 2A), whereas ectopic PRMT6 expression is much higher than its endogenous counterpart. When CARM1 is forced to express in the nucleus (using a GFP expression vector with a nuclear localization signal), a substantial increase in the levels of H3R17me2a was observed as expected (Fig. S2, B and C). Next, we analyzed a HeLa cell line that stably expresses a TAP-tagged form of PRMT6. The protein G domains in the TAP tag allow tight binding by IgG; thus, the ectopic expression of PRMT6 can be visualized by any IgG-type antibody (33). Again, increased H3R17me2a levels were observed in the PRMT6-overexpressing lines (Fig. 2B). We also generated a cell line that stably expresses an ER-PRMT6 fusion protein, which was also linked to a FLAG tag (34). Upon treatment with tamoxifen, the fusion protein was stabilized and translocated into the nucleus. In this experiment, we mixed ER-PRMT6 cells with the same number of WT cells, to have PRMT6-overexpressing cells and control cells adjacent to each other in the same field of view. This mixture culture was treated with tamoxifen and then prepared for immunofluorescence with anti-FLAG and anti-H3R17me2a antibodies (Fig. 2C). FLAG-tagged ER-PRMT6–positive cells showed significantly higher H3R17me2a signals. Even though the H3R17me2a antibody can recognize additional CARM1 substrates, the predominant signal generated by this antibody in ER-PRMT6–positive cells is due to an increase of the H3R17me2a histone mark (Fig. S2D). Together, these findings indicate that PRMT6 can methylate the H3R17 site both in vitro and in cells.
Figure 2.
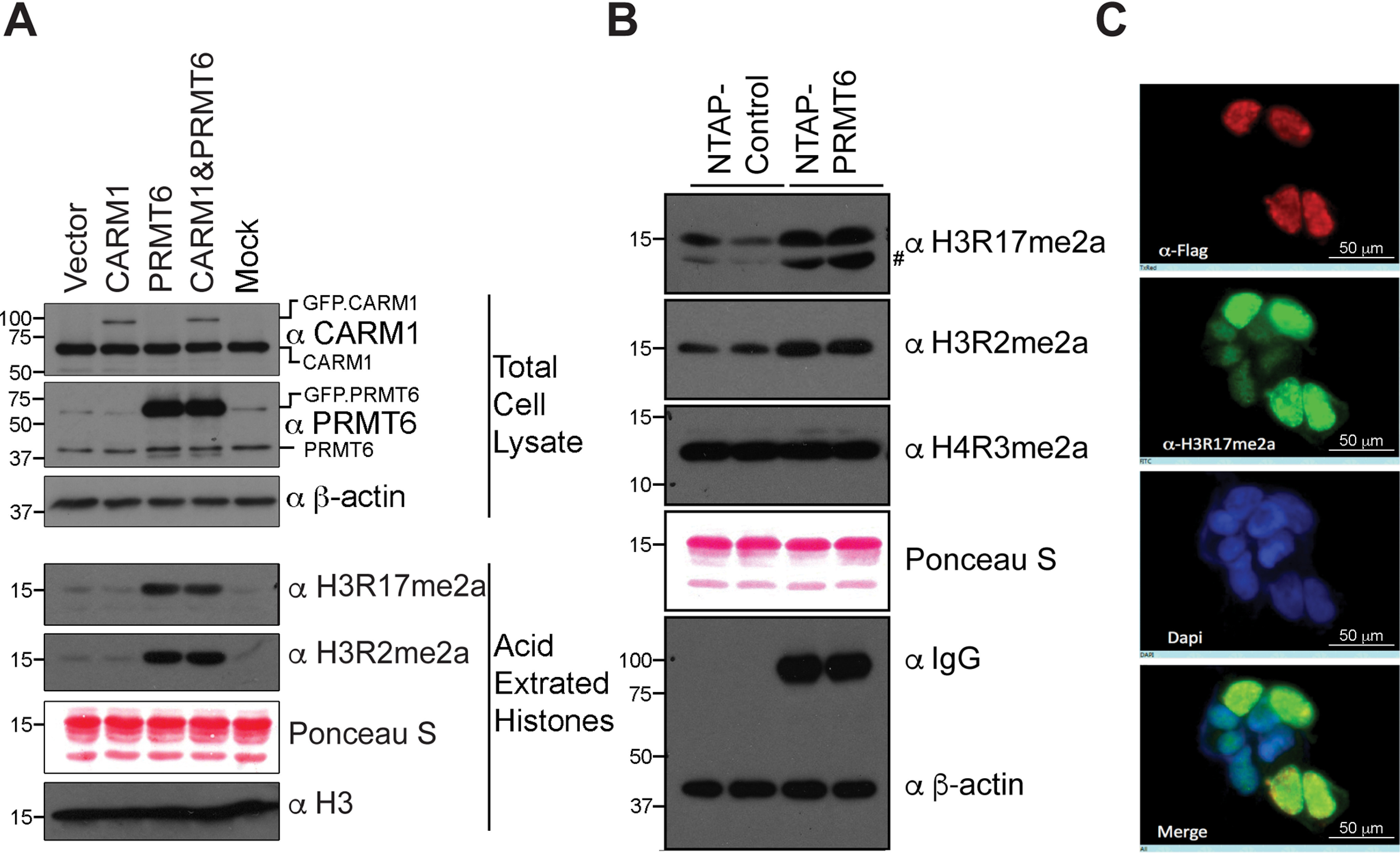
PRMT6 methylates histone H3 at Arg-17 site in cells. A, total cell lysates and core histones were extracted from HeLa cells transfected with CARM1 alone and PRMT6 alone or together. The levels of CARM1, PRMT6, H3R17me2a, and H3R2me2a are detected by immunoblots using methyl-specific antibodies. B, core histones were extracted from cell lines stably expressing NTAP or NTAP-tagged PRMT6. The levels of indicated histone arginine methylation were detected by Western blotting. Ectopic expression of PRMT6 was monitored using an IgG-type antibody. C, HEK293 cells stably expressing ER*-FLAG-PRMT6 were mixed with WT HEK293 cells with a 1:1 ratio in culture. The mixture was then treated with 4-hydroxytamoxifen for 48 h. The levels of FLAG-PRMT6 and H3R17me2a were visualized by immunostaining with the indicated antibodies. #, nonspecific band.
Double knockout mice reveal cross-talk between CARM1 and PRMT6
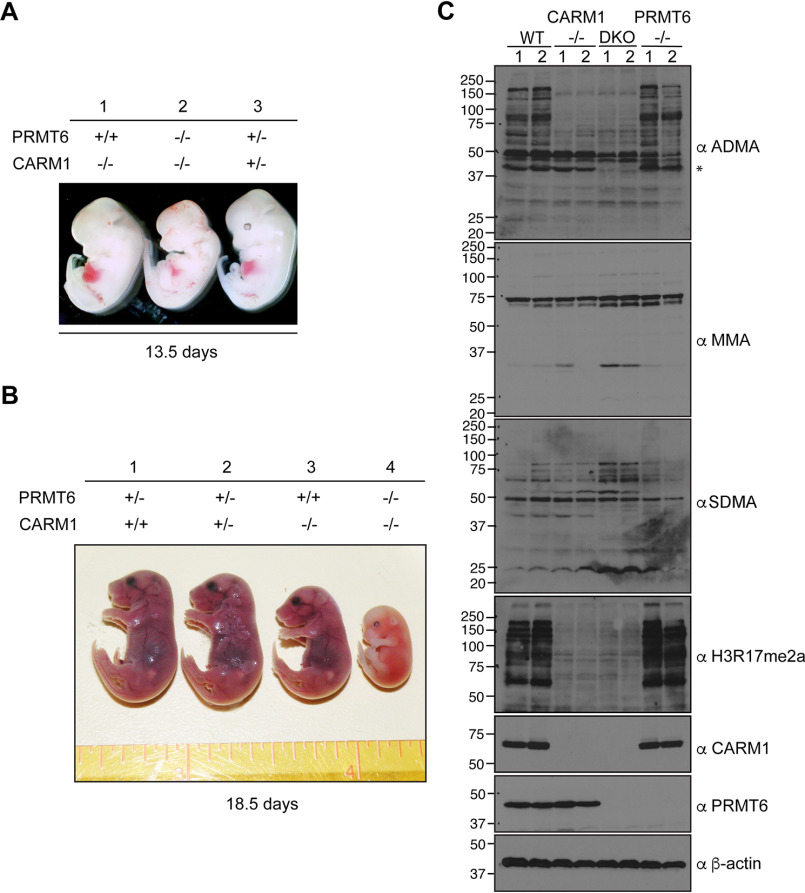
Because both CARM1 and PRMT6 have the ability to deposit the H3R17me2a mark, it would be important to gauge the degree of redundancy between these two PRMTs in mice. Previously, we made CARM1 knockout mice and showed that CARM1 knockout embryos are similar in size to their WT littermates at E12.5 but are significantly smaller than their littermates at E18.5 (9). Whereas CARM1 knockout mice died at birth, PRMT6 knockout mice survived to adulthood and did not show any signs of stunted growth (29). Importantly, no PRMT double knockouts have yet been performed. First, we intercrossed PRMT6+/− CARM1+/− double heterozygous mice to generate the single and double knockout embryos in the same litter. At E13.5, both the CARM1 knockout and heterozygous embryos were the same size, and the CARM1/PRMT6 double knockout embryos were distinctly smaller than the CARM1 knockout embryos (Fig. 3A). At E18.5, CARM1 knockout embryos were smaller than CARM1 WT embryos, as we previously reported (9), and the CARM1/PRMT6 double knockout embryos were even smaller (Fig. 3B). Second, we crossed CARM1+/− mice onto a PRMT6 null background, which is fertile, and then intercrossed these mice. Again, CARM1/PRMT6 double knockout embryos were smaller than single knockout embryos at E13.5 (Fig. S3A). Genotyping was performed by PCR (Fig. S3B), and Western blot analysis confirmed the loss of PRMT6 and CARM1 in both embryo extracts and the corresponding cell lines derived from those embryos (Fig. S2C). Next, we used two independent sets of primary MEF lines derived from WT, CARM1−/−, PRMT6−/−, and CARM1/PRMT6 double knockout embryos and tested the level of MMA, ADMA, and SDMA, using GAR motif antibodies, as well as the H3R17me2a antibody (which recognizes a cadre of CARM1 substrates (9)) (Fig. 3C). Interestingly, we observed that the depletion of CARM1 and PRMT6 together led to the disappearance of a strong ADMA signal of approximately 42 kDa (Fig. 3C), which could represent an as yet uncharacterized shared substrate for these two PRMTs. In addition, the extracts from CARM1/PRMT6 double knockout embryos display an increase in both MMA and SDMA signals, suggesting that PRMT5 may methylate these substrates when they are no longer blocked with ADMA marks. The phenomenon of substrate scavenging by PRMTs has previously been reported in both PRMT1 knockout cells and CARM1 knockout cells (27, 35). These data, particularly the reduced size of the CARM1/PRMT6 double knockout embryos, provide genetic evidence for a degree of redundancy between PRMT6 and CARM1. It should be noted that a very small subset of CARM1 substrates are likely methylated by PRMT6, because the H3R17me2a antibody, which cross-reacts with a large number of CARM1 substrates, does not lose any immunoreactivity on whole-cell lysates from PRMT6 knockout lines. We next focused on the histone H3R17me2a mark itself, using acid-purified core histones from these mutant MEF lines.
Figure 3.
CARM1/PRMT6 double knockout embryos are smaller compared with single knockout. CARM1/PRMT6 double knockout embryos and their littermates at age E13.5 (A) and E18.5 (B) were obtained from cross-breeding CARM1+/−PRMT6+/− mice. C, primary MEFs were generated from the WT, CARM1 KO, PRMT6 KO, and DKO embryos. The gross levels of ADMA, MMA, SDMA, and CARM1 substrates (indicated by H3R17me2a antibody) are shown. *, highlighted ADMA signal only lost in CARM1/PRMT6 double knockout MEFs.
Loss of CARM1 and PRMT6 leads to reduced H3R17me2a levels in mitosis
Previous studies performed by Sakabe and Hart indicate that the H3R17me2a mark is increased during mitosis (36). To validate these published findings, we performed an expanded cell cycle analysis of the H3R17me2a mark, along with known cell cycle markers like cyclin B1 levels and histone H3S10 phosphorylation states (37). We synchronized HeLa cells with thymidine and nocodazole and tested the levels of H3R17me2a and H4R3me2a at the indicated time points after the cells were released from cell cycle arrest. The intensity of the H3R17me2a mark mirrors the H3Ser10ph signal, clearly supporting the regulation of this mark during the cell cycle (Fig. 4A). The H4R3me2a mark was not altered during this time course (Fig. 4A). Next, we thus focused on studying the impact of CARM1 and PRMT6 loss on the H3R17me2a mark during mitosis, when the level of this mark peaks. WT, CARM1−/−, PRMT6−/−, and CARM1/PRMT6 double knockout MEFs were arrested in mitosis, and the levels of H3R17me2a were tested by Western blotting. During mitosis, H3R17me2a levels were elevated in WT, CARM1 knockout, and PRMT6 knockout MEFs. However, elevated levels of H3R17me2a were not seen in the double knockout MEFs (Fig. 4B). Asynchronous CARM1/PRMT6 double knockout MEFs only showed a slightly lower level of H3R17me2a than other asynchronous MEF cells, suggesting that other methyltransferases may help maintain the basal level of H3R17me2a in the double knockout MEFs or that this antibody weakly recognizes the unmethylated histone. H3Ser10ph is widely known to be a biomarker during mitosis, and elevated levels of it are found in the mitotic cells of all four genotypes tested (Fig. 4B). These findings indicate that both CARM1 and PRMT6 are required for the deposition of the H3R17me2a mark during mitosis.
Figure 4.
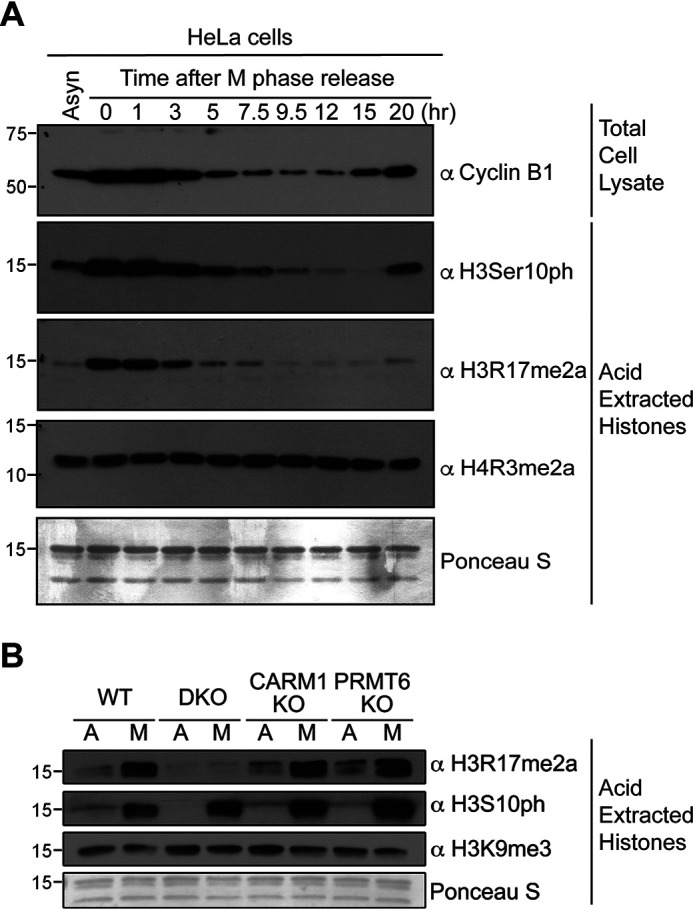
CARM1 and PRMT6 are both required for the elevation of H3R17me2a in mitosis. A, HeLa cells were arrested in mitosis by treatment with thymidine followed by nocodazole, and the cells were harvested after the cells were released from cell cycle arrest at the indicated time points. Then total cell lysates and core histones were prepared and probed with the indicated antibodies. Core histones and total cell lysates from asynchronized (Asyn) HeLa cells were loaded as controls. B, core histones from asynchronous (A) and mitotic (M) WT, CARM1 KO, PRMT6 KO, and DKO primary MEFs were immunoblotted using the indicated antibodies.
CARM1/PRMT6 double knockout MEFs experience increased double-strand DNA damage
CARM1 is reported to methylate p300, which promotes the recruitment of BRCA1 to the promoter of p21 and is required for the induction of p21 and GADD45 expression (38). Also, p21 levels are up-regulated in PRMT6-deficient MEFs (29), whereas GADD45a expression is suppressed by PRMT6 in the early development of zebrafish (39). PRMT6 methylates DNA polymerase β to promote efficient base excision repair (40). CARM1 deposits marks that are read by the TDRD3 effector protein, which in turn recruits TOP3B to resolve R-loops and suppress DNA damage (41). These findings implicate both CARM1 and PRMT6 in the DNA damage response. Thus, we were interested to examine the impact of CARM1/PRMT6 loss on the DNA damage response. PRMT6 knockout MEFs showed increased staining for γ-H2AX, as compared with WT or CARM1 knockout MEFs, whereas the double knockout MEFs showed the strongest signal of this biomarker for DNA damage (Fig. 5A). Western blotting analysis further confirmed that there is an increase in γ-H2AX in double knockout MEFs (Fig. 5B). In summary, we showed that there is significantly increased DNA damage response in the CARM1/PRMT6 double knockout MEFs compared with the single knockout lines.
Figure 5.
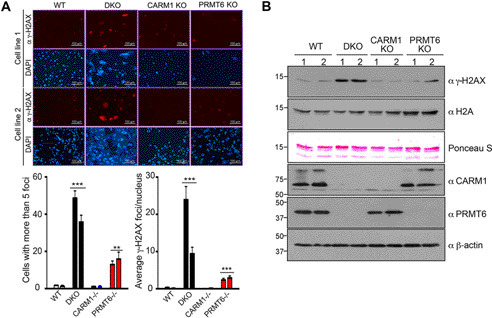
Increased DNA damage in CARM1/PRMT6 double knockout primary MEFs. A, two individual MEF lines generated from WT, CARM1 KO, PRMT6 KO, and DKO embryos were immunostained with anti-γ-H2AX antibody. In a certain field of view, cells with more than five γ-H2AX foci were counted. Also, the average number of γ-H2AX foci per nucleus was calculated and plotted. B, acid-extracted core histones as well as total cellular lysates from WT, CARM1 KO, PRMT6 KO, and DKO MEFs were separated on an SDS-polyacrylamide gel and subsequently immunoblotted with the indicated antibodies.
Synergistic effect between CARM1 and PRMT6 small molecule inhibitions
Recent advances in small molecular compounds allow us to specifically inhibit the enzyme activities of certain PRMTs (42). TP-064 is a selective and noncompetitive inhibitor to CARM1 (30), and EPZ020411 is a potent and selective inhibitor to PRMT6 (31). By using these compounds, we can avoid the variation between different cell lines. We thus hypothesize that the loss of activity of both CARM1 and PRMT6 will result in reduced cell proliferation, as suggested from the genetic experiments (Fig. 3 (A and B) and Fig. S3A). To test this hypothesis, we combined increasing concentrations of CARM1 (TP-064) and PRMT6 (EPZ020411) inhibitors to treat WT MEFs, and we tested the relative proliferation of the cells after 6 days (Fig. 6A). As individual agents, 3–10 μm EPZ020411 mildly inhibited the cell growth of WT MEFs, whereas 3–10 μm of TP-064 slowed the proliferation rates. When combined, lower concentrations of TP-064 and EPZ020411 (0.3–3 μm) inhibited the proliferation rates of WT MEF cells (Fig. 6A), suggesting that there is a synergistic effect between the two compounds. After 4 days of the combined use of a 3 μm concentration of the CARM1 inhibitor and 3 μm PRMT6 inhibitor, a clear inhibition of cell growth was observed with combination treatment over single inhibitor treatment (Fig. S4). Next, we analyzed the combined drug effect values shown in Fig. 6A by using the combination index theorem of Chou and Talalay (43), which is based on the mass-action law principle. A combinatorial index (CI) of <1 indicates synergism, whereas a CI of >1 demonstrates antagonism. Effect-oriented mapping of the combination data demonstrated synergism between the CARM1 inhibitor and the PRMT6 inhibitor (Fig. 6B). These results show that the loss of enzyme activity of both CARM1 and PRMT6 has dramatic effects on cell proliferation rates and cell viability of WT MEFs.
Figure 6.
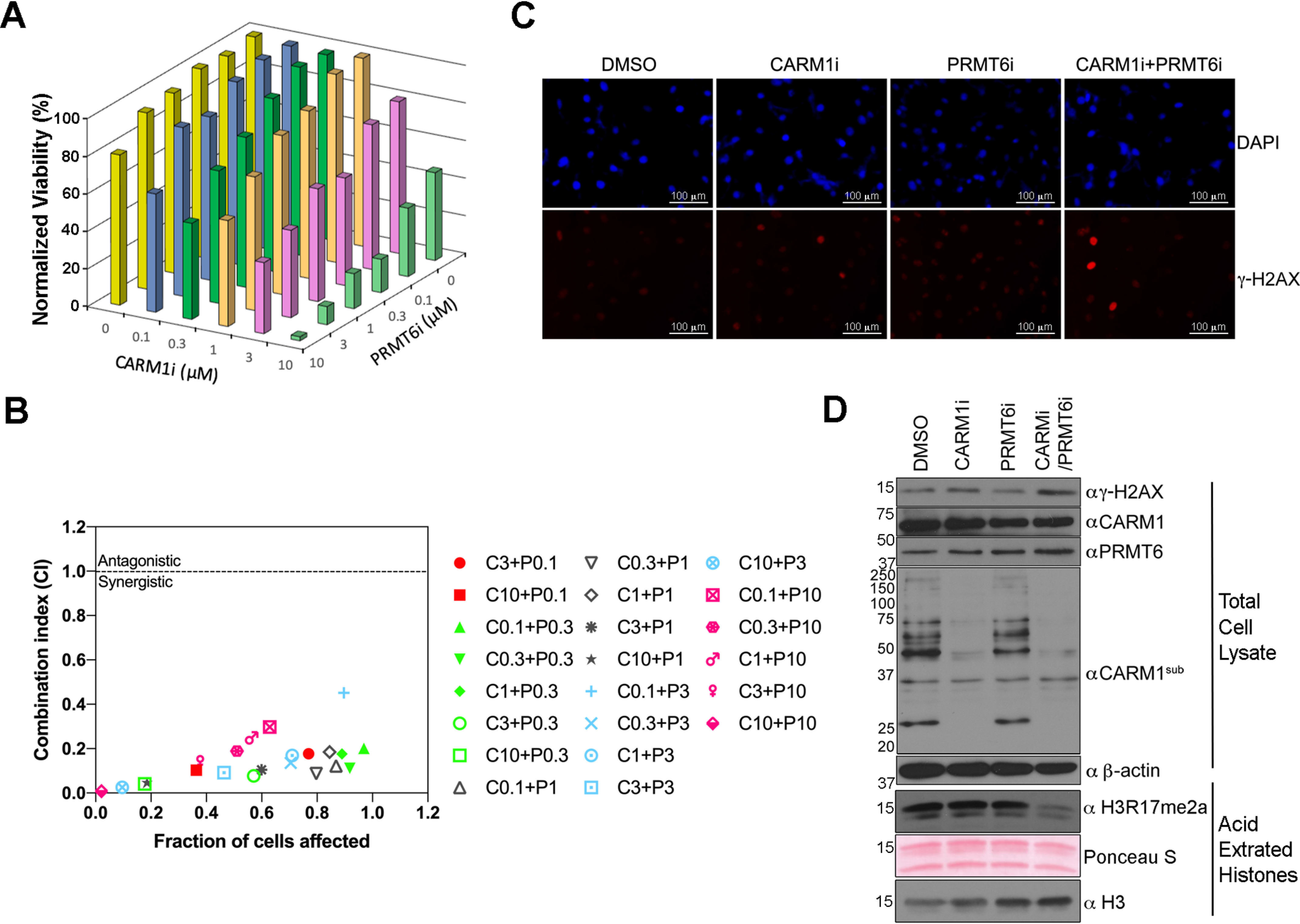
Synergistic effect of CARM1 and PRMT6 inhibitors on the cell proliferation of WT MEFs. A, WT MEFs were treated with a combination of the indicated concentration of CARM1 inhibitor (CARM1i) and PRMT6 inhibitor (PRMT6i) for a total of 6 days. At the end of the culture, cell counting was performed with the CellTiterGlo® kit, and the viabilities of different drug treatment groups were normalized by the group with no CARM1 inhibitor or PRMT6 inhibitor (which was set as 100%). B, normalized cell viabilities lower than 100% in A were allowed to be used to calculate the CI values, with CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA). Synergistic interactions are implied by values of <1, whereas values of >1 indicate antagonistic interactions. C and D, WT MEFs were treated with 3 μm CARM1 inhibitor alone, 3 μm PRMT6 inhibitor alone, or the inhibitors combined for 6 days. The treated cells were stained with anti-γ-H2AX or DAPI to visualize the DNA damage foci in the nucleus (C) or lysed for Western blotting to detect the levels of γ-H2AX and H3R17me2a (D).
CARM1 and PRMT6 enzyme activities are required for normal DNA damage response
Compared with the MEFs deficient in CARM1 or PRMT6 only, MEFs with both CARM1 and PRMT6 knocked out displayed increased γ-H2AX levels (Fig. 5). To make sure this effect on DNA damage response was not due to the idiosyncrasies related to the establishment of different primary MEF lines, we applied the CARM1 inhibitor and the PRMT6 inhibitor to WT MEFs and tested the γ-H2AX level by immunostaining (Fig. 6C) and Western blotting (Fig. 6D). The effect of the CARM1 inhibitor was monitored by a pan-CARM1 substrate antibody (αCARM1sub) that we generated in-house (Fig. 6D). The specificity of this antibody was demonstrated by Western blotting with lysates from HEK293T cells treated with different PRMT inhibitors, including the inhibitors to type I PRMT (MS023), CARM1 (TP-064), and PRMT6 (EPZ020411). The signals generated by this antibody appreciably decreased when CARM1 was inhibited (Fig. S5). With the combination of CARM1/PRMT6 inhibitor treatment, a decrease in H3R17me2a levels is observed (Fig. 6D), and a significant signal increase in the DNA damage biomarker in WT MEFs was seen in both assays (Fig. 6, C and D). These data further suggest that CARM1 and PRMT6 work together to ensure and maintain DNA integrity and that the enzyme activities of both are required for this regulation to occur.
Discussion
In this study, we discovered that PRMT6, like CARM1, has the ability to methylate histone H3 at Arg-17 both in vitro and in vivo (Figs. 1 and 2). To date, PRMT6 has been shown to deposit methylation marks on histone H3R2 (16), H3R17, H3R42 (26), H2A/H4R3 (17), and H2AR29 (25). Among these sites, H3R2me2a and H2AR29me2a are associated with repressed gene expression (16, 17, 25), whereas H3R17me2a, H4R3me2a, and H3R42me2a are generally associated with transcriptional activation (26, 44). Histone H3R2 was thought to be the major histone target site of PRMT6 in cells, and PRMT6 was widely considered a transcriptional repressor (29, 44–46); however, in a few other cases, PRMT6 was reported to act as a transcriptional coactivator (34, 47). Moreover, recent RNA-Seq data from PRMT6 knockout cells showed almost equal amounts of up-regulated and down-regulated genes (48). Interestingly, ∼25% of deregulated genes have promoters that are associated with H3R2me2a peaks, and 70% of these H3R2me2a-marked genes are down-regulated in the absence of PRMT6 (48). These data suggest that PRMT6 functions as both a transcriptional repressor and activator, depending on the context. It is unclear how these opposite functions of PRMT6 are regulated, but it could be due to its presence in different transcriptional complexes, which may direct the enzyme activity of this PRMT to different sites on histone tails. Our data revealed that PRMT6 can methylate H3R17 and that it can impinge on the CARM1 pathway.
Most PRMTs, including PRMT6, prefer a GAR motif in their substrates, whereas CARM1 has a preference for a proline residue near the methylated arginine (4, 19). In a few cases, PRMT6 was shown to methylate non-GAR motifs in HIV Tat proteins and in PRMT6 itself (23, 24). Another study showed that both CARM1 and PRMT6 can methylate histone H3 at Arg-42, stimulating the activation of p53-dependent transcription in vitro (26). In this study, we showed that both CARM1 and PRMT6 can methylate H3R2, H3R17, and H3R26 in vitro (Fig. 1, B and D), with varying degrees of efficiency. In cells, we validated that PRMT6 has the ability to methylate H3R17 (Fig. 2), and we showed the loss of an unknown methylated protein in CARM1 and PRMT6 double knockout MEFs (Fig. 3). Our data further support the hypothesis that CARM1 and PRMT6 share additional common substrates (Fig. 3C), apart from H3R17 and H3R42.
The best example of PRMTs collaborating during certain cellular processes is PRMT1 and PRMT5. Together, they regulate hypoxia and ischemia-induced apoptosis (49), the formation of stress granules (50), cell proliferation (51), and RNA splicing (52). Besides the collaboration between PRMT1 and PRMT5, PRMT1 and CARM1 work together to synergistically regulate p53-dependent and p53-independent genes (53–56). Also, knocking down PRMT1 or CARM1 leads to an increased RNA-binding ability of hnRNPUL1 (57). Even though PRMT6 and PRMT1 in vitro share similar substrates, such as H4R3 (58), Npl3 (21), and TOP3B (59), not much is known about their collaboration in vivo. In the case of CARM1 and PRMT6, they are known to work together to positively regulate hormone-dependent transcription (28, 60). Knockdown of both CARM1 and PRMT6 significantly inhibits the estrogen-stimulated proliferation of breast cancer cells, but it does not inhibit cell proliferation in the absence of estrogen (28). In a recent study, MCF-7 cells were treated with a small molecular inhibitor of both CARM1 and PRMT6, and the estrogen-dependent transcription of GREB1 significantly increased, whereas the loss of CARM1 alone only partially mimicked this effect (61). Finally, in early mouse embryonic development, CARM1 methylation of the H3R26 site seems to be critical for driving cell fate decisions (62, 63), but it remains unclear whether PRMT6, which can weakly methylate the H3R26 site (Fig. 1, B and D), functions redundantly in this setting. In summary, previous reports showed that the combinatorial effect of CARM1 and PRMT6 regulates cell proliferation in a hormone-dependent manner (28). Here, we provide a mechanistic explanation for the reported synergy between CARM1 and PRMT6, which relies on the ability of these two PRMTs to methylate the same motifs on histone tails and perhaps also on nonhistone substrates.
Materials and methods
Antibodies
Two H3R17me2a antibodies used in this study were from Millipore (#07-214) and Abcam (#ab8284), mostly from Millipore (if not indicated). Other antibodies were anti-H3R2me1 (Abcam, #ab15584), anti-H3R2me2a (Millipore, #07-585), anti-H3R26me2a (Millipore, #07-215), anti-H4R3me2a (Active Motif, #39705), anti-histone H3 (Abcam, #ab18521), anti-FLAG (Sigma, #F1804), anti-β-actin (Sigma, #A1978), anti-CARM1 (Bethyl, #A300-421A), anti-PRMT6 (Bethyl, #A300-929A), anti-H3Ser10ph (Active Motif, #61623), anti-cyclin B1 (BD Biosciences, #554177), anti-H3K9me3 (Active Motif, #39161), anti-SDMA (collaboration with Cell Signaling Technologies), anti-ADMA (collaboration with Cell Signaling Technologies), anti-MMA (collaboration with Cell Signaling Technologies), pan-CARM1 substrate antibody (made in-house), anti-γ-H2AX (Millipore, #05-636), and anti-histone H2A (Millipore, #ABE327).
Knockout mice and genotyping
CARM1+/− mice (9) were bred with PRMT6−/− mice (29) to generate CARM1+/− PRMT6+/− double heterozygous mice. CARM1+/− PRMT6+/− mice were further intercrossed to produce WT, CARM1−/−, PRMT6−/−, and CARM1−/−PRMT6−/− embryos. For timed pregnancies, CARM1+/−PRMT6+/− mice were mated overnight. Females were inspected for vaginal plugs the following morning, and the following noon was taken as day 0.5 of gestation (E0.5). Genomic DNA was isolated from liver biopsies and analyzed by PCR using the following primers: 5′-AGT CCA TGC TGA GCT CCG T-3′ and 5′-TCC ATG CAG CTC ATA TCC A-3′ for PRMT6 WT allele; 5′-AAG GTC ACT GGA AGA AGG-3′ and 5′-ACT CTC AGA ATT GCC TAG-3′ for PRMT6 knockout allele; 5′-CCC ACT TCT GTT ACC TCC TTT G-3′ and 5′-TAA CTA AAA GAA AAT GGA ATG G-3′ for CARM1 genotyping (both WT allele and KO allele). All mouse experiments were reviewed and approved by an institutional animal care and use committee at MD Anderson Cancer Center.
Plasmids and cell culture
GST-tagged PRMTs were expressed from constructs based on the vector pGEX-6P-1 (Amersham Biosciences). Constructs expressing GST-PRMT1, GST-PRMT3, GST-CARM1, and GST-PRMT6 have been described previously (21). NTAP-PRMT6 was generated by cloning PRMT6 cDNA into pCeMM NTAP(GS) vector (EUROSCARF). CARM1 and PRMT6 cDNAs were inserted into peGFP-C1 vector to generate constructs expressing GFP-tagged CARM1 and PRMT6 in mammalian cells. GFP-NLS-CARM1 was generated by inserting mouse CARM1 coding sequence into the BglII site in pAcGFP1-Nuc (Takara). CARM1 WT, KO, and KI MEFs have been described previously (32). HeLa and HEK293 cell lines were obtained from ATCC. HeLa cells stably expressing NTAP or NTAP-PRMT6 were generated by transfecting NTAP or NTAP-PRMT6 construct into WT HeLa cells, followed by selection of single clones using G418. Stable ER*-FLAG-PRMT6 HEK293 cell line has been described previously (34). Individual embryos (WT, CARM1 KO, PRMT6 KO, CARM1, and PRMT6 double KO (DKO)) from CARM1+/−PRMT6+/− inbred at E13.5 were sheared and placed into culture to generate primary MEF cells as described previously (32). All cell lines were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum.
Histone purifications
Core histones were purified from cell pellets using a Histone Purification Mini kit (Active Motif) according to the manufacturer's instructions.
In vitro methylation
GST-tagged PRMT1, PRMT3, CARM1, and PRMT6 were purified and used as an enzyme source. Recombinant histones or peptides were incubated with GST-tagged PRMTs and [3H]SAM at 30 °C for 1 h (or the indicated time course). SDS loading buffer was added to the reactions, and the samples were boiled for 5 min. Samples were then applied to electrophoresis on an SDS-polyacrylamide gel and transferred to a PVDF membrane. Then the membrane was dried, and the radioactive signals were recorded with X-ray films.
Immunostaining
Cells were seeded on coverslips. At the end of cell culture, the indicated cells were rinsed with PBS and subsequently fixed and permeabilized in buffer containing 2% paraformaldehyde and 0.5% Triton X-100 in PBS at 4 °C for 30 min. Then the cells were blocked in 20% fetal bovine serum and incubated with the indicated primary antibody for 1 h at room temperature. Cells were then washed and incubated with Alexa Fluor 647–conjugated secondary antibody (Invitrogen) for 30 min. Finally, they were washed again and stained with DAPI for 5 min at room temperature.
Western blotting
Cells were harvested and lysed, and the cell lysates were applied to Western blot analysis. In brief, cells were harvested and washed three times with cold PBS, and then radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, 5 mm EDTA, supplemented with proteinase inhibitor mixture) was added to obtain the cell lysates. Cell debris was pelleted and discarded, whereas the supernatant was kept. Protein samples were added with SDS loading buffer and boiled for 5 min, followed by SDS-PAGE. Then the proteins were transferred to a PVDF membrane. The membrane was blocked with 5% fat-free milk for 1 h at room temperature and then incubated with primary antibodies at 4 °C overnight. The blot was then washed three times with PBST and incubated with secondary antibodies for 1 h at room temperature. After washing three times with PBST, the membrane was incubated with ECL reagent, and the signals were detected on X-ray film.
Synchronization
HeLa cells were cultured to 40% confluence and treated with 2 mm thymidine for 24 h. Then thymidine was removed by washing with PBS. Fresh medium was then added, and the cells were incubated for 3 h. Afterward, nocodazole was added at a concentration of 100 ng/ml for 12 h to arrest cells at mitosis.
PRMT inhibitor treatment and cell counting
TP-064 (Cayman Chemical, #20256) and EPZ020411 (Cayman Chemical, #19160) were dissolved in DMSO. Mouse embryonic fibroblasts were treated with these drugs for a total of 6 days, with the medium and drugs changed every other day. At the end of the culture, CellTiter-Glo luminescent kit (Promega) was used to measure the cell viability. Briefly, cells were trypsinized and diluted with fresh Dulbecco's modified Eagle's medium to the culture volume. Then 100 μl of prewarmed reagent was directly added to 100 μl of the harvested cell culture. The plates were shaken on a horizontal shaker at room temperature for 10 min and applied to a luminescence plate reader. Cell viabilities were normalized to the untreated group.
Data availability
The data sets supporting the conclusions of this article are included within the article and the supporting information.
Supplementary Material
Acknowledgments
We thank Rebecca Deen for editing the manuscript.
This article contains supporting information.
Author contributions—D. C., G. G., M. O. H., and M. T. B. conceptualization; D. C. and G. G. data curation; D. C. and G. G. formal analysis; D. C., G. G., A. D. L., S. J., and M. T. B. investigation; D. C., G. G., and M. T. B. methodology; D. C., G. G., and M. T. B. writing-original draft; A. D. L., S. J., M. O. H., and S. R. writing-review and editing; S. R. and M. T. B. resources; M. T. B. funding acquisition; M. T. B. project administration.
Funding and additional information—M. T. B. is supported by Cancer Prevention and Research Institute of Texas (CPRIT) Grant RP180804 and National Institutes of Health Grant GM126421. S. R. is funded by Canadian Institutes of Health Research Grant FDN-154303. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—M. T. B. is a co-founder of EpiCypher.
- PRMT
- protein arginine methyltransferase
- CARM
- coactivator-associated arginine methyltransferase
- MMA
- mono methylarginine
- SDMA
- symmetric dimethylarginine
- ADMA
- asymmetric dimethylarginine
- GAR
- glycine- and arginine-rich
- γ-H2AX
- phosphorylated histone H2AX
- DAPI
- 4′,6-diamidino-2-phenylindole
- MEF
- mouse embryonic fibroblast
- E
- embryonic day
- CI
- combinatorial index
- KO
- knockout
- KI
- knock-in
- DKO
- double knockout
- PVDF
- polyvinylidene difluoride.
References
- 1. Guccione, E., and Richard, S. (2019) The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657 10.1038/s41580-019-0155-x [DOI] [PubMed] [Google Scholar]
- 2. Jarrold, J., and Davies, C. C. (2019) PRMTs and arginine methylation: cancer's best-kept secret? Trends Mol. Med. 25, 993–1009 10.1016/j.molmed.2019.05.007 [DOI] [PubMed] [Google Scholar]
- 3. Schurter, B. T., Koh, S. S., Chen, D., Bunick, G. J., Harp, J. M., Hanson, B. L., Henschen-Edman, A., Mackay, D. R., Stallcup, M. R., and Aswad, D. W. (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40, 5747–5756 10.1021/bi002631b [DOI] [PubMed] [Google Scholar]
- 4. Cheng, D., Côté, J., Shaaban, S., and Bedford, M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25, 71–83 10.1016/j.molcel.2006.11.019 [DOI] [PubMed] [Google Scholar]
- 5. Cheng, D., Vemulapalli, V., Lu, Y., Shen, J., Aoyagi, S., Fry, C. J., Yang, Y., Foulds, C. E., Stossi, F., Treviño, L. S., Mancini, M. A., O'Malley, B. W., Walker, C. L., Boyer, T. G., and Bedford, M. T. (2018) CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci. Alliance 1, e201800117 10.26508/lsa.201800117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abeywardana, T., Oh, M., Jiang, L., Yang, Y., Kong, M., Song, J., and Yang, Y. (2018) CARM1 suppresses de novo serine synthesis by promoting PKM2 activity. J. Biol. Chem. 293, 15290–15303 10.1074/jbc.RA118.004512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frietze, S., Lupien, M., Silver, P. A., and Brown, M. (2008) CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 68, 301–306 10.1158/0008-5472.CAN-07-1983 [DOI] [PubMed] [Google Scholar]
- 8. El Messaoudi, S., Fabbrizio, E., Rodriguez, C., Chuchana, P., Fauquier, L., Cheng, D., Theillet, C., Vandel, L., Bedford, M. T., and Sardet, C. (2006) Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the cyclin E1 gene. Proc. Natl. Acad. Sci. U. S. A. 103, 13351–13356 10.1073/pnas.0605692103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yadav, N., Lee, J., Kim, J., Shen, J., Hu, M. C., Aldaz, C. M., and Bedford, M. T. (2003) Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 100, 6464–6468 10.1073/pnas.1232272100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daujat, S., Bauer, U. M., Shah, V., Turner, B., Berger, S., and Kouzarides, T. (2002) Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12, 2090–2097 10.1016/S0960-9822(02)01387-8 [DOI] [PubMed] [Google Scholar]
- 11. Miao, F., Li, S., Chavez, V., Lanting, L., and Natarajan, R. (2006) Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-κB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol. Endocrinol. 20, 1562–1573 10.1210/me.2005-0365 [DOI] [PubMed] [Google Scholar]
- 12. Kim, D. I., Park, M. J., Lim, S. K., Choi, J. H., Kim, J. C., Han, H. J., Kundu, T. K., Park, J. I., Yoon, K. C., Park, S. W., Park, J. S., Heo, Y. R., and Park, S. H. (2014) High-glucose-induced CARM1 expression regulates apoptosis of human retinal pigment epithelial cells via histone 3 arginine 17 dimethylation: role in diabetic retinopathy. Arch. Biochem. Biophys. 560, 36–43 10.1016/j.abb.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 13. Yang, G., Zhou, C., Wang, R., Huang, S., Wei, Y., Yang, X., Liu, Y., Li, J., Lu, Z., Ying, W., Li, X., Jing, N., Huang, X., Yang, H., and Qiao, Y. (2019) Base-editing-mediated R17H substitution in histone H3 reveals methylation-dependent regulation of Yap signaling and early mouse embryo development. Cell Rep. 26, 302–312.e4 10.1016/j.celrep.2018.12.046 [DOI] [PubMed] [Google Scholar]
- 14. Jacques, S. L., Aquino, K. P., Gureasko, J., Boriack-Sjodin, P. A., Porter Scott, M., Copeland, R. A., and Riera, T. V. (2016) CARM1 preferentially methylates H3R17 over H3R26 through a random kinetic mechanism. Biochemistry 55, 1635–1644 10.1021/acs.biochem.5b01071 [DOI] [PubMed] [Google Scholar]
- 15. Gayatri, S., and Bedford, M. T. (2014) Readers of histone methylarginine marks. Biochim. Biophys. Acta 1839, 702–710 10.1016/j.bbagrm.2014.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guccione, E., Bassi, C., Casadio, F., Martinato, F., Cesaroni, M., Schuchlautz, H., Lüscher, B., and Amati, B. (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 10.1038/nature06166 [DOI] [PubMed] [Google Scholar]
- 17. Hyllus, D., Stein, C., Schnabel, K., Schiltz, E., Imhof, A., Dou, Y., Hsieh, J., and Bauer, U. M. (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21, 3369–3380 10.1101/gad.447007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iberg, A. N., Espejo, A., Cheng, D., Kim, D., Michaud-Levesque, J., Richard, S., and Bedford, M. T. (2008) Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283, 3006–3010 10.1074/jbc.C700192200 [DOI] [PubMed] [Google Scholar]
- 19. Gayatri, S., Cowles, M. W., Vemulapalli, V., Cheng, D., Sun, Z. W., and Bedford, M. T. (2016) Using oriented peptide array libraries to evaluate methylarginine-specific antibodies and arginine methyltransferase substrate motifs. Sci. Rep. 6, 28718 10.1038/srep28718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee, J., and Bedford, M. T. (2002) PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 3, 268–273 10.1093/embo-reports/kvf052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frankel, A., Yadav, N., Lee, J., Branscombe, T. L., Clarke, S., and Bedford, M. T. (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277, 3537–3543 10.1074/jbc.M108786200 [DOI] [PubMed] [Google Scholar]
- 22. Miranda, T. B., Webb, K. J., Edberg, D. D., Reeves, R., and Clarke, S. (2005) Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem. Biophys. Res. Commun. 336, 831–835 10.1016/j.bbrc.2005.08.179 [DOI] [PubMed] [Google Scholar]
- 23. Boulanger, M. C., Liang, C., Russell, R. S., Lin, R., Bedford, M. T., Wainberg, M. A., and Richard, S. (2005) Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79, 124–131 10.1128/JVI.79.1.124-131.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singhroy, D. N., Mesplède, T., Sabbah, A., Quashie, P. K., Falgueyret, J. P., and Wainberg, M. A. (2013) Automethylation of protein arginine methyltransferase 6 (PRMT6) regulates its stability and its anti-HIV-1 activity. Retrovirology 10, 73 10.1186/1742-4690-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waldmann, T., Izzo, A., Kamieniarz, K., Richter, F., Vogler, C., Sarg, B., Lindner, H., Young, N. L., Mittler, G., Garcia, B. A., and Schneider, R. (2011) Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11 10.1186/1756-8935-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casadio, F., Lu, X., Pollock, S. B., Leroy, G., Garcia, B. A., Muir, T. W., Roeder, R. G., and Allis, C. D. (2013) H3R42me2a is a histone modification with positive transcriptional effects. Proc. Natl. Acad. Sci. U. S. A. 110, 14894–14899 10.1073/pnas.1312925110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shishkova, E., Zeng, H., Liu, F., Kwiecien, N. W., Hebert, A. S., Coon, J. J., and Xu, W. (2017) Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat. Commun. 8, 15571 10.1038/ncomms15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harrison, M. J., Tang, Y. H., and Dowhan, D. H. (2010) Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 38, 2201–2216 10.1093/nar/gkp1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neault, M., Mallette, F. A., Vogel, G., Michaud-Levesque, J., and Richard, S. (2012) Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 40, 9513–9521 10.1093/nar/gks764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakayama, K., Szewczyk, M. M., Dela Sena, C., Wu, H., Dong, A., Zeng, H., Li, F., de Freitas, R. F., Eram, M. S., Schapira, M., Baba, Y., Kunitomo, M., Cary, D. R., Tawada, M., Ohashi, A., et al. (2018) TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget 9, 18480–18493 10.18632/oncotarget.24883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitchell, L. H., Drew, A. E., Ribich, S. A., Rioux, N., Swinger, K. K., Jacques, S. L., Lingaraj, T., Boriack-Sjodin, P. A., Waters, N. J., Wigle, T. J., Moradei, O., Jin, L., Riera, T., Porter-Scott, M., Moyer, M. P., et al. (2015) Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med. Chem. Lett. 6, 655–659 10.1021/acsmedchemlett.5b00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim, D., Lee, J., Cheng, D., Li, J., Carter, C., Richie, E., and Bedford, M. T. (2010) Enzymatic activity is required for the in vivo functions of CARM1. J. Biol. Chem. 285, 1147–1152 10.1074/jbc.M109.035865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bürckstümmer, T., Bennett, K. L., Preradovic, A., Schütze, G., Hantschel, O., Superti-Furga, G., and Bauch, A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 10.1038/nmeth968 [DOI] [PubMed] [Google Scholar]
- 34. Di Lorenzo, A., Yang, Y., Macaluso, M., and Bedford, M. T. (2014) A gain-of-function mouse model identifies PRMT6 as a NF-κB coactivator. Nucleic Acids Res. 42, 8297–8309 10.1093/nar/gku530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dhar, S., Vemulapalli, V., Patananan, A. N., Huang, G. L., Di Lorenzo, A., Richard, S., Comb, M. J., Guo, A., Clarke, S. G., and Bedford, M. T. (2013) Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311 10.1038/srep01311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakabe, K., and Hart, G. W. (2010) O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 10.1074/jbc.M110.158170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johansen, K. M., and Johansen, J. (2006) Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 14, 393–404 10.1007/s10577-006-1063-4 [DOI] [PubMed] [Google Scholar]
- 38. Lee, Y. H., Bedford, M. T., and Stallcup, M. R. (2011) Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 25, 176–188 10.1101/gad.1975811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao, X. X., Zhang, Y. B., Ni, P. L., Wu, Z. L., Yan, Y. C., and Li, Y. P. (2016) Protein arginine methyltransferase 6 (Prmt6) is essential for early zebrafish development through the direct suppression of gadd45αa stress sensor gene. J. Biol. Chem. 291, 402–412 10.1074/jbc.M115.666347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. El-Andaloussi, N., Valovka, T., Toueille, M., Steinacher, R., Focke, F., Gehrig, P., Covic, M., Hassa, P. O., Schär, P., Hübscher, U., and Hottiger, M. O. (2006) Arginine methylation regulates DNA polymerase β. Mol. Cell 22, 51–62 10.1016/j.molcel.2006.02.013 [DOI] [PubMed] [Google Scholar]
- 41. Yang, Y., McBride, K. M., Hensley, S., Lu, Y., Chedin, F., and Bedford, M. T. (2014) Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 10.1016/j.molcel.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li, A. S. M., Li, F., Eram, M. S., Bolotokova, A., Dela Sena, C. C., and Vedadi, M. (2020) Chemical probes for protein arginine methyltransferases. Methods 175, 30–43 10.1016/j.ymeth.2019.11.017 [DOI] [PubMed] [Google Scholar]
- 43. Chou, T. C. (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70, 440–446 10.1158/0008-5472.CAN-09-1947 [DOI] [PubMed] [Google Scholar]
- 44. Blanc, R. S., and Richard, S. (2017) Arginine methylation: the coming of age. Mol. Cell 65, 8–24 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 45. Stein, C., Riedl, S., Rüthnick, D., Nötzold, R. R., and Bauer, U. M. (2012) The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 40, 9522–9533 10.1093/nar/gks767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stein, C., Nötzold, R. R., Riedl, S., Bouchard, C., and Bauer, U. M. (2016) The arginine methyltransferase PRMT6 cooperates with Polycomb proteins in regulating HOXA gene expression. PLoS ONE 11, e0148892 10.1371/journal.pone.0148892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scaramuzzino, C., Casci, I., Parodi, S., Lievens, P. M. J., Polanco, M. J., Milioto, C., Chivet, M., Monaghan, J., Mishra, A., Badders, N., Aggarwal, T., Grunseich, C., Sambataro, F., Basso, M., Fackelmayer, F. O., et al. (2015) Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85, 88–100 10.1016/j.neuron.2014.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bouchard, C., Sahu, P., Meixner, M., Nötzold, R. R., Rust, M. B., Kremmer, E., Feederle, R., Hart-Smith, G., Finkernagel, F., Bartkuhn, M., Savai Pullamsetti, S., Nist, A., Stiewe, T., Philipsen, S., and Bauer, U. M. (2018) Genomic location of PRMT6-dependent H3R2 methylation is linked to the transcriptional outcome of associated genes. Cell Rep. 24, 3339–3352 10.1016/j.celrep.2018.08.052 [DOI] [PubMed] [Google Scholar]
- 49. Lim, S. K., Jeong, Y. W., Kim, D. I., Park, M. J., Choi, J. H., Kim, S. U., Kang, S. S., Han, H. J., and Park, S. H. (2013) Activation of PRMT1 and PRMT5 mediates hypoxia- and ischemia-induced apoptosis in human lung epithelial cells and the lung of miniature pigs: the role of p38 and JNK mitogen-activated protein kinases. Biochem. Biophys. Res. Commun. 440, 707–713 10.1016/j.bbrc.2013.09.136 [DOI] [PubMed] [Google Scholar]
- 50. Tsai, W. C., Gayatri, S., Reineke, L. C., Sbardella, G., Bedford, M. T., and Lloyd, R. E. (2016) Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 291, 22671–22685 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao, G., Zhang, L., Villarreal, O. D., He, W., Su, D., Bedford, E., Moh, P., Shen, J., Shi, X., Bedford, M. T., and Xu, H. (2019) PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res. 47, 5038–5048 10.1093/nar/gkz200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fong, J. Y., Pignata, L., Goy, P. A., Kawabata, K. C., Lee, S. C., Koh, C. M., Musiani, D., Massignani, E., Kotini, A. G., Penson, A., Wun, C. M., Shen, Y., Schwarz, M., Low, D. H., Rialdi, A., et al. (2019) Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209.e9 10.1016/j.ccell.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. An, W., Kim, J., and Roeder, R. G. (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 10.1016/j.cell.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 54. Kleinschmidt, M. A., Streubel, G., Samans, B., Krause, M., and Bauer, U. M. (2008) The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 36, 3202–3213 10.1093/nar/gkn166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hassa, P. O., Covic, M., Bedford, M. T., and Hottiger, M. O. (2008) Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 377, 668–678 10.1016/j.jmb.2008.01.044 [DOI] [PubMed] [Google Scholar]
- 56. Koh, S. S., Chen, D., Lee, Y. H., and Stallcup, M. R. (2001) Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276, 1089–1098 10.1074/jbc.M004228200 [DOI] [PubMed] [Google Scholar]
- 57. Larsen, S. C., Sylvestersen, K. B., Mund, A., Lyon, D., Mullari, M., Madsen, M. V., Daniel, J. A., Jensen, L. J., and Nielsen, M. L. (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 9, rs9 10.1126/scisignal.aaf7329 [DOI] [PubMed] [Google Scholar]
- 58. Zhang, Y., van Haren, M. J., and Martin, N. I. (2019) Peptidic transition state analogues as PRMT inhibitors. Methods 175, 24–29 10.1016/j.ymeth.2019.08.003 [DOI] [PubMed] [Google Scholar]
- 59. Huang, L., Wang, Z., Narayanan, N., and Yang, Y. (2018) Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 46, 3061–3074 10.1093/nar/gky103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen, D., Huang, S. M., and Stallcup, M. R. (2000) Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275, 40810–40816 10.1074/jbc.M005459200 [DOI] [PubMed] [Google Scholar]
- 61. Stossi, F., Dandekar, R. D., Mancini, M. G., Gu, G., Fuqua, S. A. W., Nardone, A., De Angelis, C., Fu, X., Schiff, R., Bedford, M. T., Xu, W., Johansson, H. E., Stephan, C. C., and Mancini, M. A. (2020) Estrogen-induced transcription at individual alleles is independent of receptor level and active conformation but can be modulated by coactivators activity. Nucleic Acids Res. 48, 1800–1810 10.1093/nar/gkz1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hupalowska, A., Jedrusik, A., Zhu, M., Bedford, M. T., Glover, D. M., and Zernicka-Goetz, M. (2018) CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175, 1902–1916.e13 10.1016/j.cell.2018.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang, J., Wang, L., Feng, G., Wang, Y., Li, Y., Li, X., Liu, C., Jiao, G., Huang, C., Shi, J., Zhou, T., Chen, Q., Liu, Z., Li, W., and Zhou, Q. (2018) Asymmetric Expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175, 1887–1901.e18 10.1016/j.cell.2018.11.039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this article are included within the article and the supporting information.