Abstract
Dilated cardiomyopathy (DCM) is associated with mutations in cardiomyocyte sarcomeric proteins, including α-tropomyosin. In conjunction with troponin, tropomyosin shifts to regulate actomyosin interactions. Tropomyosin molecules overlap via tropomyosin–tropomyosin head-to-tail associations, forming a continuous strand along the thin filament. These associations are critical for propagation of tropomyosin's reconfiguration along the thin filament and key for the cooperative switching between heart muscle contraction and relaxation. Here, we tested perturbations in tropomyosin structure, biochemistry, and function caused by the DCM-linked mutation, M8R, which is located at the overlap junction. Localized and nonlocalized structural effects of the mutation were found in tropomyosin that ultimately perturb its thin filament regulatory function. Comparison of mutant and WT α-tropomyosin was carried out using in vitro motility assays, CD, actin co-sedimentation, and molecular dynamics simulations. Regulated thin filament velocity measurements showed that the presence of M8R tropomyosin decreased calcium sensitivity and thin filament cooperativity. The co-sedimentation of actin and tropomyosin showed weakening of actin-mutant tropomyosin binding. The binding of troponin T's N terminus to the actin-mutant tropomyosin complex was also weakened. CD and molecular dynamics indicate that the M8R mutation disrupts the four-helix bundle at the head-to-tail junction, leading to weaker tropomyosin–tropomyosin binding and weaker tropomyosin–actin binding. Molecular dynamics revealed that altered end-to-end bond formation has effects extending toward the central region of the tropomyosin molecule, which alter the azimuthal position of tropomyosin, likely disrupting the mutant thin filament response to calcium. These results demonstrate that mutation-induced alterations in tropomyosin–thin filament interactions underlie the altered regulatory phenotype and ultimately the pathogenesis of DCM.
Keywords: muscle, tropomyosin, cardiomyopathy, cardiac muscle, in vitro motility, molecular dynamics, DCM, M8R
Cardiac muscle contraction, generated by the ATP-dependent attachment and detachment of myosin and actin, is regulated by a variety of proteins along the thick and thin filaments of the sarcomere. Sarcomeric protein mutations have been shown to have profound effects on cross-bridge cycling and often interfere with the proper regulation of contraction (1). Inherited, autosomal dominant mutations are related to more than 80% of clinical cases of dilated cardiomyopathy (DCM); these mutations frequently occur in sarcomeric protein genes including the thin filament proteins actin, all three of the troponin subunits, and tropomyosin (2). Understanding the effects of these mutations on protein–protein interaction is needed to inform future therapeutic design and personalized drugs of choice (3).
Tropomyosin molecules twist together as a parallel dimer that forms an α-helical coiled-coil. The tightly wound nature of tropomyosin provides little protection from local mutation-induced structural changes having long-range effects (4, 5) that can act a hundred or more angstroms away from the site of the mutation (4, 6). Each tropomyosin dimer binds seven consecutive actin subunits and, when bound end-to-end with adjacent tropomyosin dimers, creates a continuous cable that wraps around the actin filament. The overlap of ∼11 residues of each terminus of neighboring tropomyosin dimers form a four-helix bundle (7). The N terminus of the troponin subunit T forms a complex with this head-to-tail four-helix bundle of tropomyosin (8, 9), offering additional stabilization (10, 11), which slows myosin–ATPase activity (12) when assessed in vitro. The C terminus of troponin T and the troponin complex subunits I and C bind both to the central portion of the tropomyosin dimer and to actin (9, 13, 14).
Each tropomyosin dimer interacts weakly with actin primarily via electrostatic interactions (15–17); however, the observed high affinity of tropomyosin for actin results from the end-to-end bonds between adjacent tropomyosin dimers, which stabilize the tropomyosin cable along the long-pitch helix of actin, preventing tropomyosin detachment. This binding arrangement allows for tropomyosin to saturate the thin filament while also allowing movement between three regulatory states (18, 19). In the absence of calcium, the regulatory complex consisting of troponin and tropomyosin blocks the myosin-binding sites along actin, preventing the myosin-actin strong-binding state. At higher calcium levels, the troponin C subunit binds calcium, resulting in a cascade of conformational changes that lead to a shift in tropomyosin from the “blocked” state, favoring “closed” and “open” states, each of which has a structural correlate: the thin filament “B-state,” “C-state,” and “M-state” positions. This process allows myosin heads to interact with actin and undergo cross-bridge cycling to generate force (11, 15, 18).
Strong tropomyosin head–tail bonds allow the azimuthal shift of tropomyosin to be propagated between adjacent tropomyosin dimers along the thin filament, thus playing a role in cooperative activation of actin–myosin interactions. Therefore, the stiffness of the tropomyosin dimer, the affinity of tropomyosin for actin and troponin, and the strength of the tropomyosin–tropomyosin bond all will influence the transition between regulatory states and ultimately control muscle contraction.
Mutations in the tropomyosin–tropomyosin overlap region can cause devastating and progressive dysfunction of the heart (20–22). For example the methionine at position 8 is part of the four-helix bundle's hydrophobic core in the tropomyosin overlap complex (7, 23) and when mutated is connected to DCM or to nemaline myopathy (21, 24). Here, the mutation to a positively charged arginine in α-tropomyosin is expected to disrupt the four-helix bundle structure and perturb biochemical and functional properties.
To test the effects of the M8R mutation in tropomyosin on actomyosin contractile activity, we expressed tropomyosin containing the Met-to-Arg substitution in Escherichia coli. Modeling the junctional tropomyosin–tropomyosin interactions following molecular dynamics (MD) simulations showed that the solvent-accessible surface area of the four-helix bundle was much greater in the M8R tropomyosin, consistent with the observed alterations in tropomyosin structure and reduced tropomyosin–tropomyosin end-to-end bond strength. Furthermore, MD simulations of mutant tropomyosin on actin indicate structural alterations that correspond to disrupted regulatory transitions of tropomyosin on the actin filament. Measurements of co-sedimentation in the presence and absence of a troponin T N-terminal fragment suggested that the structural change in the four-helix bundle diminished troponin T's ability to bind to actin–tropomyosin. In addition, the in vitro motility assay was used to assess the ability of the mutant tropomyosin to properly confer calcium regulation of actin propulsion by myosin and shows that the cooperativity and calcium sensitivity of the troponin–tropomyosin regulatory complex were reduced. Taken together, these data demonstrate that both local and long-range perturbations in tropomyosin structure underlie DCM mutation-induced changes in actin–tropomyosin interactions that contribute to disease development.
Results
M8R–tropomyosin overlap structure is distorted
CD and chemical denaturant-induced unfolding
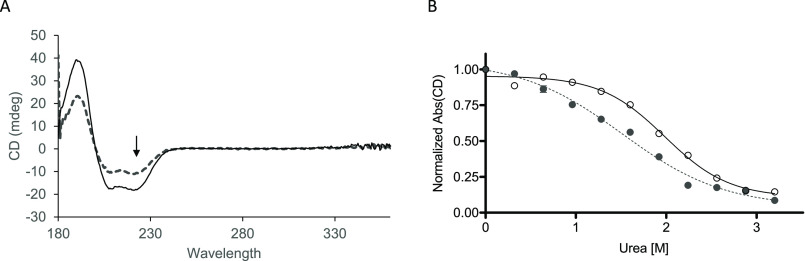
The N terminus of tropomyosin is a critical determinant of tropomyosin's ability to interact with actin and to perform its regulatory functions in striated muscle (14). Using CD, we determined that both WT and M8R mutant tropomyosin exhibit the characteristic pattern for an α-helical twist, with a peak at 222 nm that is typically used to quantify the presence of the α-helix (Fig. 1A) (25). The three-peaked ellipticity displayed by the M8R tropomyosin indicates that, despite the amino acid substitution, it still forms an α-helical structure. Chemical denaturation, determined by monitoring tropomyosin CD ellipticity at 220 nm when the protein is exposed to increasing concentrations of urea (0–4 m), was used to determine the effects of the M8R mutation on the stability of the mutant's secondary and tertiary structure (25) (Fig. 1B). The M8R tropomyosin unfolding following urea treatment demonstrated a decreased overall stability (CUrea-unfold at 1.52 ± 0.09 m versus WT 1.98 ± 0.05 m) and less resistance to chemical denaturation (nHUrea-unfold 0.77 ± 0.15 versus WT 1.23 ± 0.17), suggesting that the M8R tropomyosin dimer was folded less compactly compared with WT. Because tropomyosin dimers form intermittent bonds to one another in solution, it is not apparent from these data alone whether the four-helix bundle in particular is affected or whether it is the N terminus alone that has been changed.
Figure 1.
Tropomyosin chemical unfolding in the presence of urea. A, uncorrected ellipticity of 2 μm M8R (dashed line) and WT (solid line) tropomyosin at 0 m urea shows typical peaks for α-helical coil (190, 200, and 220 nm). B, unfolding of tropomyosin, measured in ellipticity changes at 220 nm (see arrow in A), as chemical denaturant urea is added to buffer shows a difference in M8R tropomyosin and WT tropomyosin sensitivity. M8R (filled circles with dotted line) and WT (open circles with solid line) data are fit to a three-parameter Hill equation (CUrea-unfold at 1.52 ± 0.09 m versus WT 1.98 ± 0.05 m; nHUrea-unfold 0.77 ± 0.15 versus WT 1.23 ± 0.17).
Model of the M8R tropomyosin–tropomyosin overlap structure
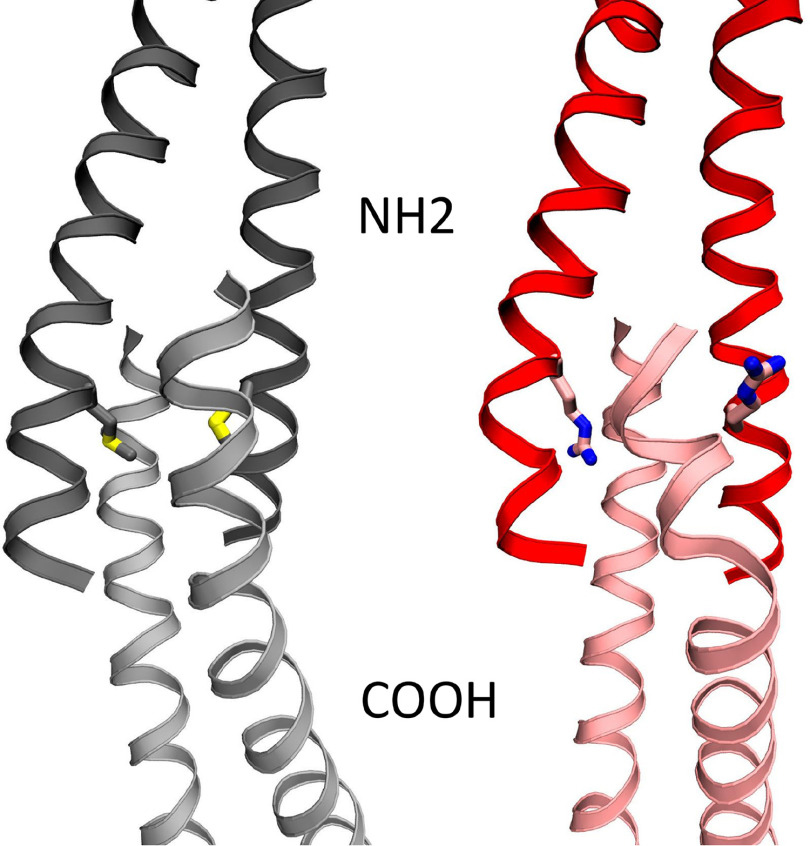
Previous computational work by our group on the head-to-tail bond between two tropomyosin dimers showed a compact N-terminal coiled-coil penetrating slightly spread coils of the C terminus to form a four-helix bundle, which is stabilized by hydrophobic interactions (26). Substitution of a charged arginine side chain at the methionine 8 position may disrupt the hydrophobic core of the overlap and expand the overlap domain, thus altering its structural and mechanical properties. We therefore examined the local effects of the M8R mutation on the formation and stability of the end-to-end bond four-helix bundle via MD simulation. Met8 of WT tropomyosin projects into the core of the tropomyosin end-to-end bundle; however, the four-helix bundle containing the M8R substitution transitioned to three helixes with a strand pushed out of the hydrophobic core, allowing for the arginine to project outward into solution (Fig. 2), resulting in an increased bending angle across the overlap (12 ± 2.7 versus WT 9.3 ± 4.8). This altered structure decreased the helix's buried solvent-accessible surface area (M8R in isolation 2212 ± 73 Å versus WT 2374 ± 65 Å2), consistent with the observed increased susceptibility to chemical denaturants (Fig. 1).
Figure 2.
Final image from molecular dynamics simulation of the end-to-end overlap structure of four tropomyosin molecules, following 30 ns of molecular dynamics simulation time. The 8R mutation (right, red with Arg8 in stick structure) disrupts the highly ordered end-to-end domain and sticks out into solution, compared with the WT tropomyosin (left, gray with Met8 in stick structure), leading to decrease in buried solvent-accessible surface area (M8R in isolation 2212 ± 73 Å versus WT 2374 ± 65 Å).
Disruption of the overlap junction reduces M8R tropomyosin affinity for actin
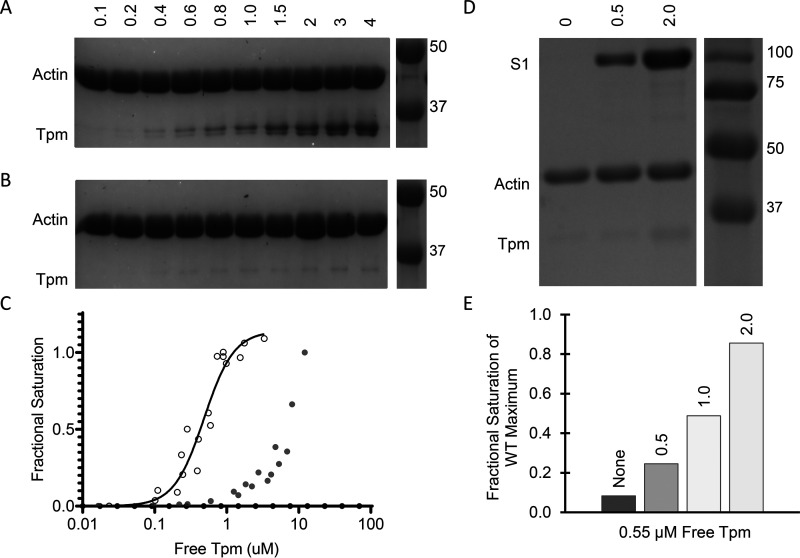
To perform its regulatory function, each tropomyosin molecule must interact weakly with actin to assure the ability to shift between regulatory positions. With such low-affinity interactions with actin, saturation of actin with tropomyosin relies on the tropomyosin–tropomyosin end-to-end linkage where tropomyosin dimers bind to adjacent dimers forming a chain along the thin filament. Given the M8R-induced structural disruptions observed in the molecular dynamics simulations, we therefore determined the effect of the M8R mutation on the binding of tropomyosin to actin. Consistent with previous studies, M8R tropomyosin exhibits a dramatically lowered actin affinity reaching the limit of sensitivity for the assay (27) (Fig. 3, A–C). By incubating 0.55 μm M8R tropomyosin with actin in the presence of increasing concentrations of S1 (up to 2 μm), we see that the M8R tropomyosin approaches the maximum, fully saturated WT tropomyosin/actin-binding ratio (Fig. 3, D and E). Therefore, consistent with previous studies (27), M8R tropomyosin was capable of binding actin in the presence of myosin subunit S1, which is known to increase tropomyosin–actin affinity and favor the M-state position of tropomyosin on actin (27).
Figure 3.
Raw binding images of pellet for co-sedimentation of tropomyosin on actin. Experiments were performed with 5 μm actin. A and B show quantity of tropomyosin in µm along top edge and molecular mass values in kDa in the spliced image from the same gel along the right edge, C shows the analysis of WT and M8R band densities, and D shows quantity of S1 in µm along top edge and molecular mass values in kDa in the spliced image from the same gel along the right edge. E shows the analysis of M8R band density compared with WT. A, WT tropomyosin shows typical binding pattern, with a KD of ∼0.48 μm. B, M8R tropomyosin demonstrates little to no binding to the actin filament at concentrations in which WT is fully saturated. C, M8R (filled circles) shows reduced affinity for actin compared with WT (open circles). D, in the presence of 2 μm S1, M8R tropomyosin (0.8 μm) binds the actin filament (5 μm). E, the binding of M8R in the presence of S1 scales with S1 concentration (values of S1 in µm listed on top of each bar).
M8R tropomyosin structure on actin shows long-range effects of N-terminal mutation
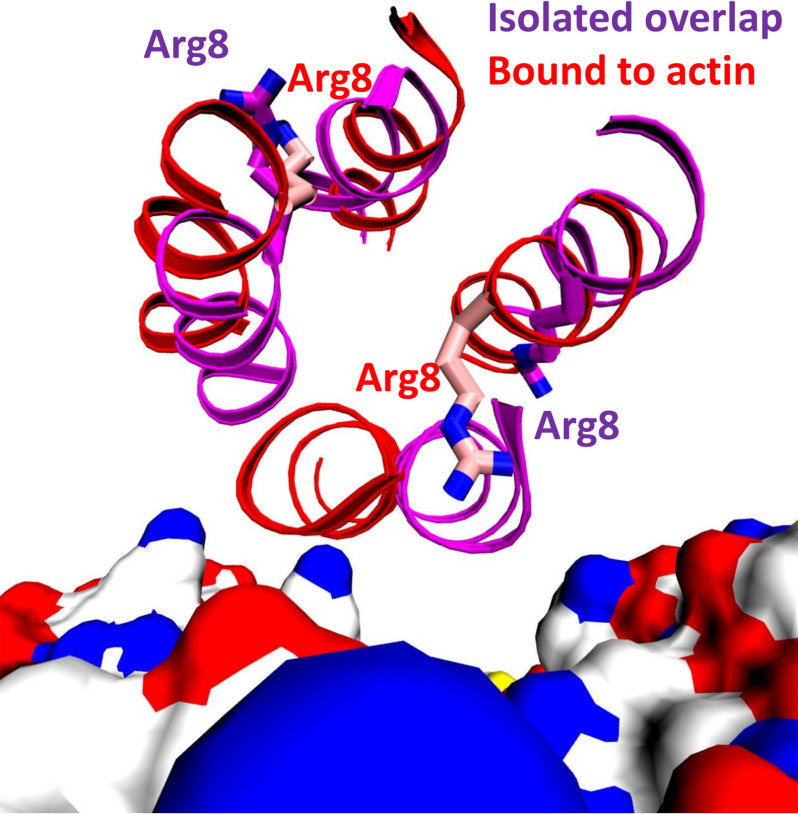
To examine the impact of the M8R mutation on the structure of, and interactions between, tropomyosin and actin, we performed molecular dynamics simulations of WT and mutant tropomyosin on actin. To study mutations in the tropomyosin end-to-end overlap, we exploited our recently developed model of actin–tropomyosin (8) derived from cryo-EM of the thin filament (9) that utilizes periodic boundary conditions to create an essentially infinite filament, as described previously (8). Comparison of the WT and M8R simulations show a similar result to the isolated overlap simulations, where the mutant arginine side chain moves from the inside of the four-helix bundle to a solvent-exposed position, with concurrent displacement of the helices in the bundle; a comparison of the isolated overlap and tropomyosin-on-actin structures shows relatively little change in the M8R tropomyosin helices, with the exception of the helix-contacting actin (Fig. 4). However, the geometry of the overlap is relatively intact, because there is little change between WT and M8R in the curvature (7.1 ± 2.0° versus 7.9 ± 3.1°, respectively), the junction twist angle (84.7 ± 1.9° versus 85.4 ± 2.1, respectively), or the buried solvent-exposed surface area in the overlap domain (2173 ± 61 Å2 versus 2135 ± 84 Å2, respectively). This result could be a consequence of restricting the tropomyosin cable to the actin filament, which may limit the accessible conformations in this simulation.
Figure 4.
Ribbon diagram of representative structures from the molecular dynamics simulations of actin with the M8R mutant (red) and the isolated M8R tropomyosin overlap (purple), viewed looking down the tropomyosin axis in the overlap domain. Actin is rendered as a surface-colored by element with carbon in white, oxygen in red, nitrogen in blue, and sulfur in yellow.
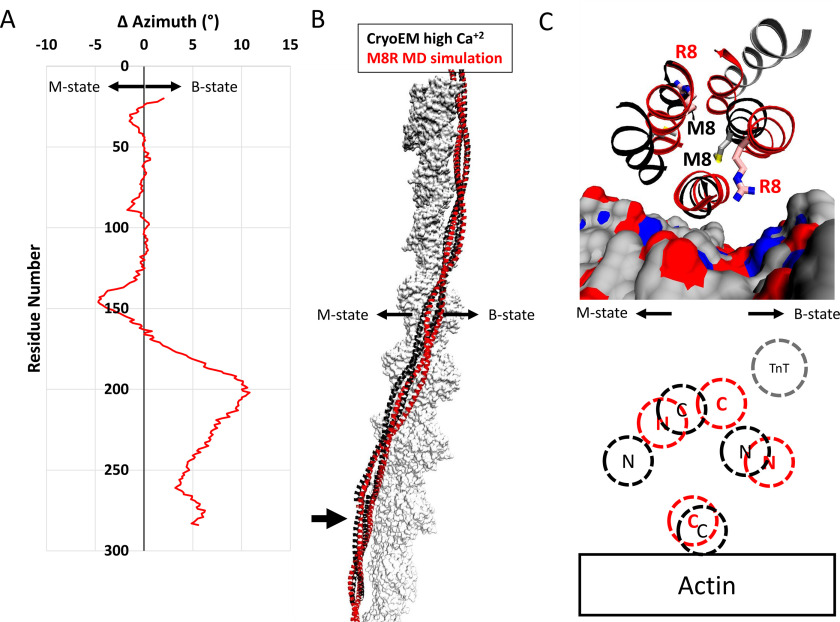
However, further inspection reveals some critical differences between the WT and M8R structures. In the WT, the dynamic persistence length of the overlap domain, which is associated with cooperativity of the thin filament, increases from 341 nm in isolation to 1969 nm when part of an infinite cable, an ∼6-fold increase in stiffness consistent with topologically constraining the overlap to the filament. However, the overlap dynamic persistence length of the M8R mutant is relatively unchanged when comparing the simulation in isolation (1080 nm) and in a cable (819 nm). Notably, the overlap dynamic persistence length is reduced in the cable by a factor of 2, suggesting that the mutation has created a less-stiff overlap.
The local distortions observed in the simulations are also predicted to alter tropomyosin's regulatory properties. The distortions to the overlap domain affect the troponin T–binding site. Although Met8 does not make direct contacts to troponin T in the recently published model (8), flanking residues Lys6, Lys7, Gln9, and Met10 interact with the troponin T N terminus. Thus, alterations to this region of tropomyosin are predicted to alter troponin T binding and regulatory function. Second, comparison of representative frames from the simulations on actin show that the mutation has resulted in a significant shift away from its starting, high-calcium C-state position toward the low-calcium position (Fig. 5A). The mutation-induced alterations of the overlap structure appear to change the twist of the overlap domain to a more B-state–like structure (Fig. 5C), and this twist is then propagated throughout the tropomyosin cable, stopping at the middle part of tropomyosin. These results suggest that the M8R cable adopts a conformation that would block myosin binding more effectively than the WT cable. Thus, the model suggests that the mutant filament would require a greater calcium concentration to achieve full activity.
Figure 5.
The alterations to the overlap domain caused by the M8R mutation result in a shift away from the closed state compared with the WT structure. A, change in azimuthal position by single residue pairs calculated by subtracting the azimuthal angle of the center of mass of the WT backbone atoms from the position of the M8R atoms. On average, the displacement of the tropomyosin dimer is ∼1° away from the closed/open states for the mutant. In the orientation used here, positive azimthual values represent motions toward the structural B-state, and negative azimuthal values represent motions toward the structural M-state. B, comparison of the WT refined high Ca2+ cryo-EM structure (black ribbon) and a representative structure of M8R mutant (red ribbon) on actin (gray surface) shows the differences in azimuthal positions calculated in A. C, close view of the structural changes of R8 on the end-to-end bond region of tropomyosin-on-actin colored as in B. Actin is rendered as surface-colored as in Fig. 3, and troponin T from the refined high Ca2+ cryo-EM structure is shown in gray ribbon. C, schematic diagram illustrating the relative positions of WT (black) and M8R (red) N and C termini on actin, and the position of troponin T N terminus binding (gray).
The M8R tropomyosin mutation alters interaction with the Troponin T N terminus
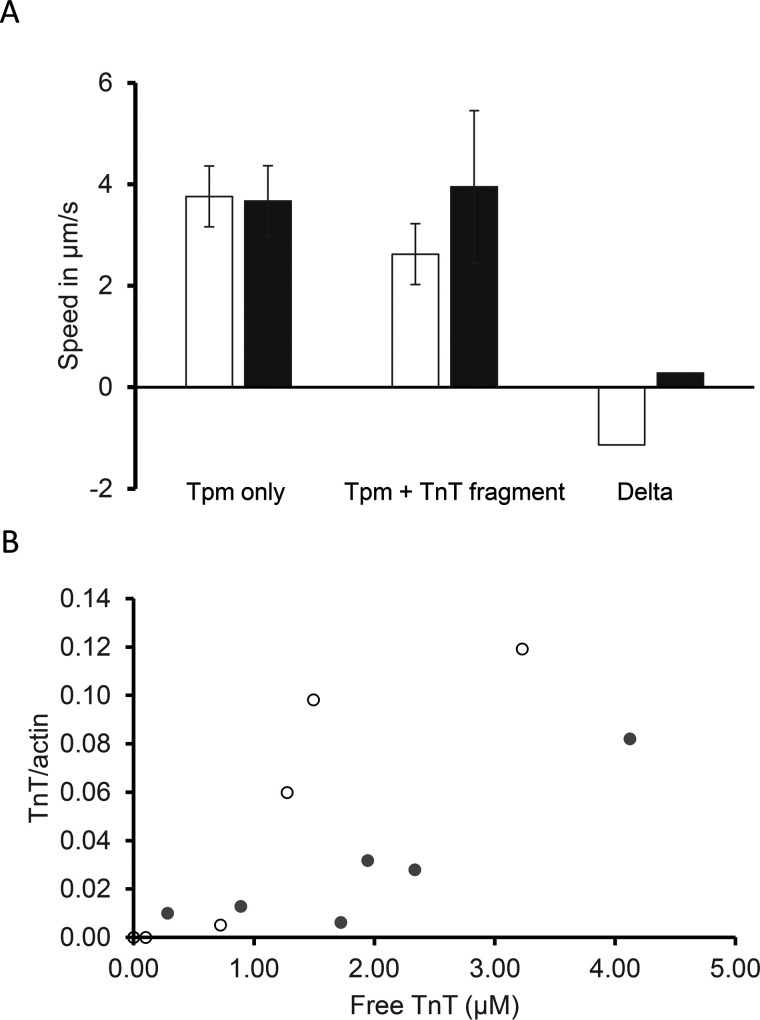
The N-terminal segment of troponin T binds across the end-to-end junction of adjacent tropomyosin molecules on the actin filament (diagram of Fig. 5D) and is known to depress the speed of tropomyosin-bound actin filaments at low myosin concentrations (12). We determined the speed of tropomyosin-decorated actin filaments in the presence of the troponin T fragment to determine whether structural and biochemical alterations caused by the M8R tropomyosin mutation affect troponin T function in tropomyosin–actin filaments. M8R-tropomyosin–decorated filaments showed no alteration in filament speed, although the troponin T-fragment did reduce WT tropomyosin speed, suggesting either that troponin T fragment binding to M8R tropomyosin is compromised or that the effect of the troponin T N terminus on filament sliding is lost in the presence of the M8R mutation (Fig. 6A).
Figure 6.
Troponin T N-terminal fragment does not affect the M8R tropomyosin. A, the diminished effect of troponin T N-terminal fragment (TnT) on the speed of actin–tropomyosin (Tpm) filaments bearing the M8R mutation (filled bars) versus WT (open bars). These values are from the in vitro motility assay, where tropomyosin and troponin T fragment were added in excess of 0.6 μm. B, analysis of multiple concentrations of troponin T binding to excess tropomyosin (10 μm for M8R, filled circles; 5 μm for WT, open circles) shows that the troponin T fragment binds M8R tropomyosin decorated actin filaments with greatly reduced affinity.
To further elucidate why the troponin T N terminus fragment failed to affect the M8R-tropomyosin–regulated actin in the in vitro motility assay, the ability of troponin T1 to actin filaments with bound mutant M8R- and WT-tropomyosin was determined via co-sedimentation. At saturating concentration for troponin T1 fragment binding to WT tropomyosin (5, 12), the presence of M8R tropomyosin dramatically diminished this interaction (Fig. 6B). This further supports the notion that contacts between the tropomyosin dimers' termini are structurally altered, suggesting that the M8R overlap complex binding to the troponin T N terminus fragment is compromised, consistent with the structural alterations observed (Figs. 1, 2, 4, and 5C, bottom panel).
M8R affects calcium regulation of actomyosin interactions
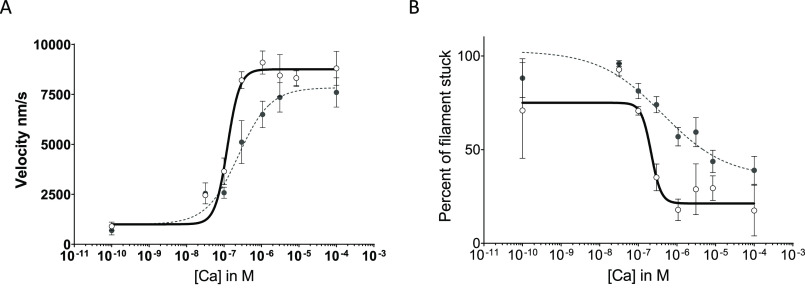
To determine whether the M8R-induced alterations in tropomyosin structure and binding interactions observed effect the Ca2+-dependent regulation and cooperative activation of actomyosin interactions, we measured the ability of calcium to regulate reconstituted cardiac thin filament movement using in vitro motility assays (Fig. 7). Both WT and M8R-mutant tropomyosin inhibited actin filament movement at low calcium concentrations, whereas actin filament velocity could be increased cooperatively with increasing calcium concentrations. Therefore, despite the mutation-induced alterations in actin and troponin T affinity (Fig. 6), M8R tropomyosin is still capable of binding troponin and regulating actomyosin interactions. However, cooperativity of thin filament activation, which is linked to tropomyosin's overall stiffness and ability to communicate over the end-to-end bond (28), is significantly lowered with the M8R mutation (nH: WT 2.64 ± 0.02 versus M8R 1.02 ± 0.01). The calcium sensitivity of the thin filament, a reflection calcium concentration required for 50% activation, is substantially decreased (pCa50: WT 6.91 ± 0.01 versus M8R 6.62 ± 0.01). The decreased cooperativity is consistent with a disruption of the M8R tropomyosin–tropomyosin junction (Figs. 1 and 2). The reduced calcium sensitivity observed with M8R tropomyosin is consistent with the notion that M8R-induced structural alterations shift the tropomyosin equilibrium position toward the B-state position (Fig. 5).
Figure 7.
In vitro motility of regulated filament actin on myosin. M8R is shown with filled circles and dotted lines. WT is shown with open circles and solid lines. A and B, velocity of top 5% of filaments (A) and percentage stuck of all filaments (B), each shown with four-parameter Hill fit equations. Each curve represents the average results of three separate experiments. Both cooperativity (nH: WT 2.64 ± 0.02 versus M8R 1.02 ± 0.01) and calcium sensitivity decreased (pCa50: WT 6.91 ± 0.01 versus M8R 6.62 ± 0.01).
Discussion
In this study we examined the effects of a disease-linked mutation on the end-to-end bond of tropomyosin, which is linked to both DCM and nemaline myopathy. Using a combination of biochemistry, computational modeling, and in vitro motility, we measured the structural effects of altered mutant tropomyosin end-to-end bonds on actin–tropomyosin and tropomyosin-troponin interactions and then correlated these with functional studies and phenotypic impact. First, we assessed the mutated protein's structure and stability, using computational and biochemical methods. We found that tropomyosin bearing the M8R mutation forms the canonical coiled-coil structure characteristic of WT tropomyosin, but MD simulations suggest that the helices of the end-to-end bonds between adjacent tropomyosins are splayed open. Thus, the M8R mutation, located near the end of the molecule, has both immediate, local effects and long-range distortions to the tropomyosin molecule. The local structural alterations reduced the ability for mutant tropomyosin to bind to actin or the N terminus of troponin. However, mutant tropomyosin is capable of binding to filamentous actin at concentrations similar to WT tropomyosin in the presence of myosin binding, thus allowing us to demonstrate the altered structure and biochemical properties of the mutant tropomyosin, which ultimately lead to dysregulation of cross-bridge formation.
The tropomyosin end-to-end bond contains a highly conserved sequence across mammals at both the C and N terminus for striated muscle α-tropomyosins (29). Within the tropomyosin end-to-end bond, the C-terminal α-helices of the tropomyosin coiled-coil partially splay open to accommodate the more tightly packed N-terminal coiled-coil of the adjacent tropomyosin (7, 26). The tight packing of the tropomyosin N terminus, and thus end-to-end bond formation and stability, is promoted by hydrophobic residues including the methionine at position eight in the sequence, which projects into the hydrophobic core of the end-to-end bond four-helix bundle (Fig. 5C).
Substitution of the hydrophobic amino acid side chain of methionine with the positively charged amino acid side chain found on arginine is predicted to distort the structure of the tropomyosin N terminus (27, 30), in agreement with the increased susceptibility to chemical denaturation observed here (Fig. 1). Our modeling also shows changes in the mutant tropomyosin N-terminal structure as a consequence of the mutant arginine side chain projecting outward away from the hydrophobic core and into solution. These structural alterations may reduce buried solvent-accessible surface, consistent with reduced tropomyosin end-to-end bond affinity.
Proper end-to-end association between tropomyosin molecules is critical for tropomyosin high affinity interactions with actin filaments. When the end-to-end bond is disrupted through such N-terminal modifications as lack of acetylation or truncation (14), tropomyosin is less likely to bind the thin filament. Similarly, the disruptions in N-terminal packing of M8R tropomyosin (Figs. 1 and 2) likely contribute to the mutant tropomyosin's lower affinity for actin (Fig. 3A). It is known that myosin increases the affinity of tropomyosin for actin and that despite the lower affinity for actin, M8R tropomyosin is capable of binding actin filaments in the presence of strongly bound myosin cross-bridges (Fig. 3, D and E) (27).
Depressed affinity alone is not necessarily a determinant of the lower cooperativity and calcium sensitivity observed with in vitro motility assays (Fig. 7), which are more complex processes that directly involve troponin proteins (31, 32), as well as tropomyosin's ability to smoothly transition between B-, C-, and M-state positions on actin. Changes to tropomyosin chain flexibility, to actin–tropomyosin contacts, and to tropomyosin twist will contribute to altered calcium sensitivity of the system by affecting the tropomyosin transitions (Figs. 5 and 7).
Strong head-to-tail association between tropomyosin molecules is considered a critical component for tropomyosin azimuthal position changes to be propagated along the cardiac filament during muscle activation and relaxation (reviewed in Ref. 33). The decreased cooperativity shown here (Fig. 7) is consistent with decreased strength of the tropomyosin end-to-end bond (30). It is important to note, however, that although often considered a direct hallmark of cooperativity, tropomyosin–tropomyosin affinity does not necessarily correlate with cooperativity (5). Rather, it appears that end-to-end bond mechanical rigidity (5), not affinity, indicates the ability for tropomyosin position to be transmitted between cooperative units.
Any stabilization of the end-to-end bond region by the troponin T N terminus (11, 34) would also presumably be compromised for M8R tropomyosin given the diminished troponin T N terminus binding observed here (Fig. 6B). With diminished troponin T N terminus binding, the transition of tropomyosin to the open state via cooperative activation would be reduced, as suggested by others (12, 35). Rat cardiac muscle fiber bundles lacking the troponin T N terminus showed depressed ATPase activity at the maximum activating calcium concentration (35). Consistent with this, we see that the percentage of motility in maximally activated in vitro motility is depressed with M8R tropomyosin (Fig. 7B).
Our results extend those previously described (27, 30, 36) and demonstrate that disruption of specific contacts within the tropomyosin end-to-end bond can not only have local effects at the overlap complex but can also affect its binding partners. Furthermore, we also show subtle molecular rearrangements that result in long-range effects that extend toward the middle of the tropomyosin molecule. In turn, these may affect the position of tropomyosin on actin and ultimately alter the inhibitory or activating states of the thin filament. MD simulations of M8R tropomyosin bound to actin show that these alterations and the subtle rearrangement of neighboring residues appear to deform the overall tropomyosin coiled-coil, resulting in global alterations that shift the mutant tropomyosin toward B-state position stabilization (Fig. 5), consistent with the reduced calcium sensitivity observed in Fig. 7.
The data presented here show that the M8R mutation has both local and long-range effects that reveal M8R-induced alterations in cardiac thin filament calcium regulation. On the one hand, the M8R substitution disrupts the end-to-end bond structure altering transmission between tropomyosin molecules along the thin filament strand, as well as disrupting binding of the troponin T N terminus to the end-to-end bond. Each of these mutation-induced perturbations likely drive the observed reduced cooperativity. On the other hand, stabilization of M8R tropomyosin in the B-state position via a stronger tropomyosin–actin interactions result in decreased calcium sensitivity. The approach applied here, involving a combination of biochemistry, mechanics, and molecular dynamics simulation, offer residue-specific strategies for understanding the effect of mutation-linked diseases at a molecular level. This information provides specific information to promote exploratory studies on potential therapeutics, as well as a framework for testing their complex effects.
Experimental procedures
Protein purification and acquisition
A M8R mutant of WT human striated α-tropomyosin (Tpm1.1), containing the N-terminal extension Met-Ala-Ser, was obtained from Genewiz (Cambridge, MA, USA) using the Met-Ala-Ser WT Tpm1.1 template in pET-3D (37). Vectors containing either WT or M8R tropomyosin were transformed into Rosetta cells for expression and then purified following standard procedures (38). A human cardiac troponin T, isoform 2 construct, matching from AAK92231.1, was truncated to residues 1–156, with a cysteine added at 157, and was isolated following previously described procedures (38).
Porcine cardiac troponin and actin were prepared from muscle ether powder as described previously (39, 40). Troponin aliquots were stored at −80 °C until use; actin, supplemented with 1 mm NaN3 and stored at 4 °C, was used for up to 6 months. Actin quality was confirmed by assaying regularly for degradation, polymerization, depolymerization, and tropomyosin co-sedimentation. The fast isoform of chicken skeletal muscle myosin was prepared from pectoralis major muscles from freshly slaughtered chickens following well-established methods (41) and was stored in 50% glycerol at −20 °C. Rabbit skeletal muscle myosin S1 fragments (Cytoskeleton, Inc.) were reconstituted following the manufacturer's instructions; then aliquots were flash-frozen in liquid N2 and stored at −80 °C until required.
CD
CD measurements were carried out as previously described (42), with the concentration of tropomyosin set at 2 μm in 5 mm KCl, 1.5 mm BES, and 0.45 mm MgCl2 at a pH of 7.4. Note that low-salt concentration is known to increase tropomyosin solution viscosity (43), presumably by enhancing tropomyosin end-to-end bonds. Protein unfolding as a function of urea concentration was measured across a wide spectrum range of wavelengths (180–500 nm), using a JASCO J-810 CD spectrometer and a Jasco J/1 quartz cell and averaging three scans from each solution. Ellipticity at 220 nm was extracted from the data, normalized to the maximum value for each protein, and fitted to a Hill equation (Y = Bottom + (Top − Bottom)/(1 + 10((LogIC50 − X) × HillSlope))) using Prism 8.0.2 to determine the unfolding cooperativity and the sensitivity of the protein to denaturant (GraphPad, La Jolla, CA, USA).
Actin co-sedimentation assay
All co-sedimentations were performed in triplicate and in sample buffer containing 15 mm BES, 5 mm MgCl2, 55 mm KCl, at pH 7.0, and carried out as described previously (5). Briefly, the samples were incubated at room temperature for 20 min, followed by centrifugation at 130,000 × g for 20 min at 4 °C. The supernatants were removed from the pellets immediately, and the pellets were rinsed with chilled buffer and then resuspended with sample buffer. Actin concentration was held at 5 μm for all experiments. S1 fragments were incubated with the tropomyosin and actin for 20 min in the standard procedure; however, for experiments containing the troponin T fragment, tropomyosin was incubated with actin for 20 min at room temperature, prior to adding the troponin T fragment and allowing an additional 20 min incubation at room temperature prior to sedimenting actin and actin bound proteins.
Analysis
Samples of supernatant and pellet were run on separate 12% SDS-PAGE mini-gels and then stained with Coomassie Brilliant Blue R-250. Gel profiles were acquired (ImageJ, National Institutes of Health, Bethesda, MD, USA); free tropomyosin concentration was estimated from the intensity of supernatant bands and plotted against the pelleted tropomyosin band intensity, normalized to the maximum intensity for bound tropomyosin. The Kd was estimated based on the fit of “specific binding with Hill slope least squares fit” using Prism 8.0.2 (GraphPad). For S1-enhanced M8R tropomyosin–actin binding, the ratio of M8R bound to actin was normalized to the ratio of WT tropomyosin–actin binding at saturating tropomyosin.
In vitro gliding filament assay
Solutions
Experimental solutions were formulated as previously described (37), and the compositions of the solutions are listed in Table 1. The experiments were performed in triplicate as described previously (4) with some modifications. Flow cells of ∼20 μl of volume were made by attaching a nitrocellulose-coated coverslip to a standard glass slide with 100-μm-thick 3M double stick tape, creating 2 channels. BSA concentration was increased from 0.5 to 1 mg/ml. Regulatory proteins, tropomyosin with and without troponin at equimolar concentrations were added but in large excess to that of actin (>600 nm). Solutions containing the following principal components were added to the flow cell channel in the following order: myosin, BSA, nonlabeled “shredded” actin, ATP, labeled actin, regulatory filaments, and finally calcium “movement” buffer. Actin filament movement was initiated by the addition of Ca2+-containing buffers with 1 mm ATP, containing regulatory proteins and oxygen scavengers (Table 1).
Table 1.
ATP- and calcium-containing solutions for the in vitro motility assay
| Component | pCa 4.0 | pCa 10.0 |
|---|---|---|
| BES | 25 mm | 25 mm |
| EGTA | 5 mm | 5 mm |
| MgCl2 | 4 mm | 4 mm |
| KOH | 40 mm | 31.6 mm |
| KCl | 142.8 mm | 158 mm |
| CaCl2 | 5.095 mm | 0 mm |
| ATP | 1 mm | 1 mm |
| DTT | 1 mm | 1 mm |
| Glucose | 2 mm | 2 mm |
| Glucose oxidase | 160 units | 160 units |
| Catalase | 2 μm | 2 μm |
| Methyl cellulose | 0.5% | 0.5% |
| pH 7.4 | High ionic strength: (∼0.21) | |
Data acquisition and analysis
Movies of actin filament motion were captured at up to five frames/s on an electron multiplied charge-coupled device camera (Andor iXon) using Solis (Andor by Oxford Instruments), with a pixel size of 190 nm/pixel. These movies were analyzed using a freeware program designed for the analysis of in vitro motility movies, FAST v1.1 (44). This program is a centroid-tracking program that automatically locates and tracks objects within a movie and reports the velocity of the object. Here, all filaments from a single movie that have been determined to be moving are then averaged to give a single value for that movie. The program will select the fastest 5% of filaments and provide the average velocity of those fastest filaments (44). These results were pooled and fitted with a four-parameter Hill fit using Prism 8 (GraphPad).
Statistics
The equation fitting referred to in this paper was performed using Prism 8. Student's t test was used to compare the actin sliding velocities between WT tropomyosin and M8R mutant tropomyosin, and nonoverlapping confidence intervals were used to determine statistical significance. These confidence intervals, as reported by Prism, were reported in the accompanying statistics. Significance was determined if the returned p values were <0.05 or if the confidence intervals did not overlap.
Molecular dynamics simulations
Molecular dynamics and analysis of tropomyosin end-to-end overlap domain
Molecular dynamics simulations were performed on the overlap domain of tropomyosin as described previously (5, 15). All simulations were performed in nanoscale molecular dynamics (45) in explicit solvent at 300 K and 1 atm. Frames were sampled every 20 ps of simulation time. The model simulated for intervals of 30–40 ns consisted of residues 1–80 at the N terminus joined to residues 205–284 at the C terminus of an adjacent tropomyosin, and analysis was calculated from the last 10 ns of the simulation. The coiled-coil radius of each frame was determined using the program Twister (46). The Twister output was also used to calculate the curvature (ω), defined as the angle between the vectors formed by the superhelical axis coordinates of residues 5 and 15 for the N terminus and 270 and 280 for the C terminus, the junction twist angle (θ), defined as the angle between the planes formed by the two helical axis coordinates at residue 5 and the superhelical axis coordinates at residue 15 for the N terminus and residue 270 and 280 for the C terminus, and the dynamic persistence length of the tropomyosin overlap domain, calculated from the standard deviation of the ω angle (26). An average structure was calculated from the last 5 ns of simulation.
Molecular dynamics of tropomyosin on actin
The coordinates for actin–tropomyosin based on Pavadai et al. (8) were used as the starting point for simulation of the M8R mutant in a polymeric cable on actin. This structure is derived from the cryo-EM structure of the thin filament in high calcium (9) in which the tropomyosin has been remodeled to the known α-helical rise. The M8R mutation was incorporated using the Mutator plugin in visual molecular dynamics (47). Molecular dynamics simulations were then performed in explicit solvent at 300 K with an initial box size of 160 Å × 160 Å × 772.76 Å using periodic boundary conditions to create an infinite thin filament as described (8). The model was simulated for 30 ns with frames sampled every 20 ps, and analysis of the overlap geometry is presented from the average of the last 10 ns. An average structure was also calculated over the last 3 ns of the simulation.
Data availability
The MD simulation data are available upon request to Jeffrey R. Moore (Jeffrey_Moore@uml.edu). All remaining data are contained within the article.
Acknowledgments
We thank Dr. Larry Tobacman (University of Illinois at Chicago) for providing us with the plasmid containing the troponin T fragment.
Author contributions—A. W. R., M. J. R., N. L., A. G., W. L., and J. R. M. conceptualization; A. W. R., M. J. R., N. L., A. G., W. L., and J. R. M. data curation; A. W. R., M. J. R., N. L., W. L., and J. R. M. formal analysis; W. L. and J. R. M. funding acquisition; A. W. R., M. J. R., N. L., W. L., and J. R. M. investigation; A. W. R. and A. G. methodology; A. W. R., M. J. R., W. L., and J. R. M. writing-original draft; A. W. R., M. J. R., N. L., A. G., W. L., and J. R. M. writing-review and editing; W. L. and J. R. M. supervision; W. L. and J. R. M. project administration.
Funding and additional information—This work was funded by the NHLBI, National Institutes of Health Grants HL123774 (to J. R. M. and W. L.) and HL036153 (to W. L.) and American Heart Association Grant 19POST34460005 (to A. W. R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- DCM
- dilated cardiomyopathy
- MD
- molecular dynamics
- BES
- N,N-Bis(2-hydroxyethyl)-2-aminoethanesulfonic acid
- pCa
- negative logarithm of the calcium concentration.
References
- 1. Chang, A. N., and Potter, J. D. (2005) Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail. Rev. 10, 225–235 10.1007/s10741-005-5252-6 [DOI] [PubMed] [Google Scholar]
- 2. Redwood, C., and Robinson, P. (2013) α-Tropomyosin mutations in inherited cardiomyopathies. J. Muscle Res. Cell Motil. 34, 285–294 10.1007/s10974-013-9358-5 [DOI] [PubMed] [Google Scholar]
- 3. Tardiff, J. C., Carrier, L., Bers, D. M., Poggesi, C., Ferrantini, C., Coppini, R., Maier, L. S., Ashrafian, H., Huke, S., and van der Velden, J. (2015) Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc. Res. 105, 457–470 10.1093/cvr/cvv023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farman, G. P., Rynkiewicz, M. J., Orzechowski, M., Lehman, W., and Moore, J. R. (2018) HCM and DCM cardiomyopathy-linked α-tropomyosin mutations influence off-state stability and crossbridge interaction on thin filaments. Arch. Biochem. Biophys. 647, 84–92 10.1016/j.abb.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sundar, S., Rynkiewicz, M. J., Ghosh, A., Lehman, W., and Moore, J. R. (2020) Cardiomyopathy mutation alters end-to-end junction of tropomyosin and reduces calcium sensitivity. Biophys. J. 118, 303–312 10.1016/j.bpj.2019.11.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li, X. E., Suphamungmee, W., Janco, M., Geeves, M. A., Marston, S. B., Fischer, S., and Lehman, W. (2012) The flexibility of two tropomyosin mutants, D175N and E180G, that cause hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 424, 493–496 10.1016/j.bbrc.2012.06.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenfield, N. J., Huang, Y. J., Swapna, G. V., Bhattacharya, A., Rapp, B., Singh, A., Montelione, G. T., and Hitchcock-DeGregori, S. E. (2006) Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J. Mol. Biol. 364, 80–96 10.1016/j.jmb.2006.08.033 [DOI] [PubMed] [Google Scholar]
- 8. Pavadai, E., Rynkiewicz, M. J., Ghosh, A., and Lehman, W. (2020) Docking troponin T onto the tropomyosin overlapping domain of thin filaments. Biophys. J. 118, 325–336 10.1016/j.bpj.2019.11.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada, Y., Namba, K., and Fujii, T. (2020) Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 11, 153 10.1038/s41467-019-14008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinto, J. R., Gomes, A. V., Jones, M. A., Liang, J., Nguyen, S., Miller, T., Parvatiyar, M. S., and Potter, J. D. (2012) The functional properties of human slow skeletal troponin T isoforms in cardiac muscle regulation. J. Biol. Chem. 287, 37362–37370 10.1074/jbc.M112.364927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sousa, D., Cammarato, A., Jang, K., Graceffa, P., Tobacman, L. S., Li, X. E., and Lehman, W. (2010) Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys. J. 99, 862–868 10.1016/j.bpj.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tobacman, L. S., Nihli, M., Butters, C., Heller, M., Hatch, V., Craig, R., Lehman, W., and Homsher, E. (2002) The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J. Biol. Chem. 277, 27636–27642 10.1074/jbc.M201768200 [DOI] [PubMed] [Google Scholar]
- 13. Jin, J.-P., and Chong, S. M. (2010) Localization of the two tropomyosin-binding sites of troponin T. Arch. Biochem. Biophys. 500, 144–150 10.1016/j.abb.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palm, T., Greenfield, N. J., and Hitchcock-DeGregori, S. E. (2003) Tropomyosin ends determine the stability and functionality of overlap and troponin T complexes. Biophys. J. 84, 3181–3189 10.1016/S0006-3495(03)70042-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li, X. E., Tobacman, L. S., Mun, J. Y., Craig, R., Fischer, S., and Lehman, W. (2011) Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 100, 1005–1013 10.1016/j.bpj.2010.12.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lorenz, M., Poole, K. J., Popp, D., Rosenbaum, G., and Holmes, K. C. (1995) An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin–tropomyosin gels. J. Mol. Biol. 246, 108–119 10.1006/jmbi.1994.0070 [DOI] [PubMed] [Google Scholar]
- 17. Rynkiewicz, M. J., Prum, T., Hollenberg, S., Kiani, F. A., Fagnant, P. M., Marston, S. B., Trybus, K. M., Fischer, S., Moore, J. R., and Lehman, W. (2017) Tropomyosin must interact weakly with actin to effectively regulate thin filament function. Biophys. J. 113, 2444–2451 10.1016/j.bpj.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lehman, W. (2017) Switching muscles on and off in steps: the McKillop–Geeves three-state model of muscle regulation. Biophys. J. 112, 2459–2466 10.1016/j.bpj.2017.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKillop, D. F., and Geeves, M. A. (1993) Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 65, 693–701 10.1016/S0006-3495(93)81110-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hershberger, R. E., Norton, N., Morales, A., Li, D., Siegfried, J. D., and Gonzalez-Quintana, J. (2010) Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1 and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ. Cardiovasc. Genet. 3, 155–161 10.1161/CIRCGENETICS.109.912345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lakdawala, N. K., Funke, B. H., Baxter, S., Cirino, A. L., Roberts, A. E., Judge, D. P., Johnson, N., Mendelsohn, N. J., Morel, C., Care, M., Chung, W. K., Jones, C., Psychogios, A., Duffy, E., Rehm, H. L., et al. (2012) Genetic testing for dilated cardiomyopathy in clinical practice. J. Card. Fail. 18, 296–303 10.1016/j.cardfail.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Driest, S. L., Ellsworth, E. G., Ommen, S. R., Will, M. L., Tajik, A. J., Gersh, B. J., and Ackerman, M. J. (2003) Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation 108, 445–451 10.1161/01.CIR.0000080896.52003.DF [DOI] [PubMed] [Google Scholar]
- 23. Rao, J. N., Rivera-Santiago, R., Li, X. E., Lehman, W., and Dominguez, R. (2012) Structural analysis of smooth muscle tropomyosin α and β isoforms. J. Biol. Chem. 287, 3165–3174 10.1074/jbc.M111.307330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ilkovski, B., Mokbel, N., Lewis, R. A., Walker, K., Nowak, K. J., Domazetovska, A., Laing, N. G., Fowler, V. M., North, K. N., and Cooper, S. T. (2008) Disease severity and thin filament regulation in M9R TPM3 nemaline myopathy. J. Neuropathol. Exp. Neurol. 67, 867–877 10.1097/NEN.0b013e318183a44f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennion, B. J., and Daggett, V. (2003) The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. U.S.A. 100, 5142–5147 10.1073/pnas.0930122100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li, X. E., Orzechowski, M., Lehman, W., and Fischer, S. (2014) Structure and flexibility of the tropomyosin overlap junction. Biochem. Biophys. Res. Commun. 446, 304–308 10.1016/j.bbrc.2014.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moraczewska, J., Greenfield, N. J., Liu, Y., and Hitchcock-DeGregori, S. E. (2000) Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophys. J. 79, 3217–3225 10.1016/S0006-3495(00)76554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore, J. R., Campbell, S. G., and Lehman, W. (2016) Structural determinants of muscle thin filament cooperativity. Arch. Biochem. Biophys. 594, 8–17 10.1016/j.abb.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geeves, M. A., Hitchcock-DeGregori, S. E., and Gunning, P. W. (2015) A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 36, 147–153 10.1007/s10974-014-9389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matyushenko, A. M., Shchepkin, D. V., Kopylova, G. V., Popruga, K. E., Artemova, N. V., Pivovarova, A. V., Bershitsky, S. Y., and Levitsky, D. I. (2017) Structural and functional effects of cardiomyopathy-causing mutations in the troponin T–binding region of cardiac tropomyosin. Biochemistry 56, 250–259 10.1021/acs.biochem.6b00994 [DOI] [PubMed] [Google Scholar]
- 31. Kremneva, E., Boussouf, S., Nikolaeva, O., Maytum, R., Geeves, M. A., and Levitsky, D. I. (2004) Effects of two familial hypertrophic cardiomyopathy mutations in α-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin-bound tropomyosin. Biophys. J. 87, 3922–3933 10.1529/biophysj.104.048793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tobacman, L. S. (2008) Cooperative binding of tropomyosin to actin. Adv. Exp. Med. Biol. 644, 85–94 10.1007/978-0-387-85766-4_7 [DOI] [PubMed] [Google Scholar]
- 33. Gordon, A. M., Homsher, E., and Regnier, M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 10.1152/physrev.2000.80.2.853 [DOI] [PubMed] [Google Scholar]
- 34. Jackson, P., Amphlett, G. W., and Perry, S. V. (1975) The primary structure of troponin T and the interaction with tropomyosin. Biochem. J. 151, 85–97 10.1042/bj1510085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chandra, M., Montgomery, D. E., Kim, J. J., and Solaro, R. J. (1999) The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J. Mol. Cell. Cardiol. 31, 867–880 10.1006/jmcc.1999.0928 [DOI] [PubMed] [Google Scholar]
- 36. Matyushenko, A. M., Koubassova, N. A., Shchepkin, D. V., Kopylova, G. V., Nabiev, S. R., Nikitina, L. V., Bershitsky, S. Y., Levitsky, D. I., and Tsaturyan, A. K. (2019) The effects of cardiomyopathy-associated mutations in the head-to-tail overlap junction of α-tropomyosin on its properties and interaction with actin. Int. J. Biol. Macromol. 125, 1266–1274 10.1016/j.ijbiomac.2018.09.105 [DOI] [PubMed] [Google Scholar]
- 37. Orzechowski, M., Fischer, S., Moore, J. R., Lehman, W., and Farman, G. P. (2014) Energy landscapes reveal the myopathic effects of tropomyosin mutations. Arch. Biochem. Biophys. 564, 89–99 10.1016/j.abb.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coulton, A. T., Koka, K., Lehrer, S. S., and Geeves, M. A. (2008) Role of the head-to-tail overlap region in smooth and skeletal muscle β-tropomyosin. Biochemistry 47, 388–397 10.1021/bi701144g [DOI] [PubMed] [Google Scholar]
- 39. Murray, J. M. (1982) Hybridization and reconstitution of the thin filament. Methods Enzymol. 85, 15–17 [DOI] [PubMed] [Google Scholar]
- 40. Potter, J. D. (1982) Preparation of troponin and its subnits. Methods Enzymol. 85, 241–263 10.1016/0076-6879(82)85024-6 [DOI] [PubMed] [Google Scholar]
- 41. Margossian, S. S., and Lowey, S. (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 85, 55–71 10.1016/0076-6879(82)85009-X [DOI] [PubMed] [Google Scholar]
- 42. Paulucci, A. A., Hicks, L., Machado, A., Miranda, M. T., Kay, C. M., and Farah, C. S. (2002) Specific sequences determine the stability and cooperativity of folding of the C-terminal half of tropomyosin. J. Biol. Chem. 277, 39574–39584 10.1074/jbc.M204749200 [DOI] [PubMed] [Google Scholar]
- 43. Bailey, K. (1948) Tropomyosin: a new asymmetric protein component of the muscle fibril. Biochem. J. 43, 271–279 10.1042/bj0430271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spudich, J. A., Aksel, T., Bartholomew, S. R., Nag, S., Kawana, M., Yu, E. C., Sarkar, S. S., Sung, J., Sommese, R. F., Sutton, S., Cho, C., Adhikari, A. S., Taylor, R., Liu, C., Trivedi, D., et al. (2016) Effects of hypertrophic and dilated cardiomyopathy mutations on power output by human β-cardiac myosin. J. Exp. Biol. 219, 161–167 10.1242/jeb.125930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kalé, L., and Schulten, K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strelkov, S. V., and Burkhard, P. (2002) Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137, 54–64 10.1006/jsbi.2002.4454 [DOI] [PubMed] [Google Scholar]
- 47. Humphrey, W., Dalke, A., and Schulten, K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MD simulation data are available upon request to Jeffrey R. Moore (Jeffrey_Moore@uml.edu). All remaining data are contained within the article.