Abstract
In nucleotide excision repair, bulky DNA lesions such as UV-induced cyclobutane pyrimidine dimers are removed from the genome by concerted dual incisions bracketing the lesion, followed by gap filling and ligation. So far, two dual-incision patterns have been discovered: the prokaryotic type, which removes the damage in 11–13-nucleotide-long oligomers, and the eukaryotic type, which removes the damage in 24–32-nucleotide-long oligomers. However, a recent study reported that the UvrC protein of Mycobacterium tuberculosis removes damage in a manner analogous to yeast and humans in a 25-mer oligonucleotide arising from incisions at 15 nt from the 3´ end and 9 nt from the 5´ end flanking the damage. To test this model, we used the in vivo excision assay and the excision repair sequencing genome-wide repair mapping method developed in our laboratory to determine the repair pattern and genome-wide repair map of Mycobacterium smegmatis. We find that M. smegmatis, which possesses homologs of the Escherichia coli uvrA, uvrB, and uvrC genes, removes cyclobutane pyrimidine dimers from the genome in a manner identical to the prokaryotic pattern by incising 7 nt 5´ and 3 or 4 nt 3´ to the photoproduct, and performs transcription-coupled repair in a manner similar to E. coli.
Keywords: nucleotide excision repair, excision repair sequencing (XR-Seq), transcription-coupled repair (TCR), tuberculosis, smegmatis, UvrC, UvrD, Mfd, Mycobacterium tuberculosis, Mycobacterium smegmatis, DNA damage, antibiotic resistance
Nucleotide excision repair (excision repair) is a nearly universal DNA repair mechanism (1–5) that removes bulky lesions from DNA, including UV-induced cyclobutane pyrimidine dimers (CPDs) and (4–6) photoproducts, by concerted dual incisions bracketing the lesion (6). Although the concerted dual-incision mechanism is conserved from bacteria to humans (7), the proteins necessary for the dual incision in prokaryotes and eukaryotes are not evolutionarily related, and similarly the dual-incision patterns in prokaryotes and eukaryotes are different. In prokaryotes, proteins encoded by three genes, uvrA, uvrB, and uvrC, acting in concert, incise the damaged DNA 7 nt 5´ and 3 or 4 nt 3´ to damage (such as a CPD), generating an excised fragment of 12 or 13 nt (1, 4, 6). In eukaryotes, 6 repair factors encompassing 16 proteins incise 20 nt 5´ and 5 nt 3´ to the damage to remove the damage in the form of a 27-nt-long oligomer (7). Although the prokaryotic incision pattern is relatively narrow, in the range of 11–13 nt, in eukaryotes, the incision sites exhibit some variability such that in humans the excision products range in size from 24 to 32 nt (median, 26–27 nt), and in yeast the median is 24 nt.
Although the dual-incision/excision pattern has been shown in numerous eukaryotes ranging from humans to Drosophila to Arabidopsis to yeast (5), the study of excision repair in prokaryotes has been largely confined to Escherichia coli and some thermophilic bacteria. Of the third kingdom of life, Archaea, only excision in Methanobacterium thermoautotrophicum has been analyzed, and it was found that this species performs prokaryotic-type dual incisions removing CPD in the form of an 11-mer (8). In fact, phylogenetic analysis of Archaea has shown that UvrA, -B, and -C homologs are found in all Euryarchaeota such as M. thermoautotrophicum and Halobacterium sp. NRC-1, but not in members of Crenoarcheota (9), which remains a major branch of the biological kingdom in which the presence (or the mechanism) of nucleotide excision repair is not known.
The mechanistic aspects of prokaryotic excision repair have been investigated in considerable detail in E. coli (3–6). The damage is recognized by (UvrA)2 or (UvrA)2UvrB, in which the ATPase activities of UvrA and UvrB provide the requisite energy for kinetic proofreading (5) for damage recognition and formation of a stable UvrB–DNA complex in which the DNA is kinked and wrapped around UvrB (10). This complex is recognized by UvrC, which contains endonuclease sites: the 5´ endonuclease at the C-terminal half (11) which exhibits RNaseH homology (9) and the 3´ endonuclease in the N-terminal domain (12) containing GIY-YIG homing endonuclease motifs (9, 13). UvrC makes the dual incisions in a concerted manner, but under certain experimental conditions and in 5´ endonuclease or 3´ endonuclease active-site mutants, uncoupled incisions do occur (1). Nevertheless, the major products generated by WT repair proteins in E. coli are 12- and 13-mers (1, 4–6). Following the concerted dual incisions, UvrB and UvrC are displaced by UvrD (helicase II) and DNA polymerase I (14, 15), and the gap is filled and ligated (16).
The same mechanism of damaged DNA incision and removal occurs in transcription-coupled excision repair (TCR). In TCR, template strand damage, which blocks RNA polymerase, is targeted for repair by the Mfd translocase. Mfd, widely distributed among prokaryotes, removes RNA polymerase and delivers UvrA and UvrB to the transcription-blocking damage to initiate repair as described above. However, characterization of TCR has largely been limited to E. coli, and to date, TCR has been assumed to occur similarly in all prokaryotes possessing full-length Mfd (17, 18).
Against this background, then, it was rather unexpected when it was recently reported that in Mycobacterium tuberculosis (which possess the UvrA, B, and C homologs), the UvrC protein alone was capable of making dual incisions bracketing a bulky adduct 15 nt 3´ and 9 nt 5´ flanking the damage to generate a 25-mer, and it was concluded that dual incisions made by UvrC in M. tuberculosis are analogous to those made by its orthologs in yeast and humans (19). In light of this report, we decided to investigate dual incisions in mycobacteria using Mycobacterium smegmatis as representative of the genus. We find that M. smegmatis makes dual incisions analogous to those made in E. coli and other prokaryotes. By extension, the entire Mycobacteria genus, including M. tuberculosis, performs dual incisions in the prokaryotic and not the eukaryotic mode. We also find direct evidence for transcription-coupled repair in M. smegmatis, consistent with the presence of the mfd gene in this organism.
Results and discussion
Phylogenetic analysis of UvrC of E. coli, M. smegmatis, and M. tuberculosis
Of the three proteins UvrA, -B, and -C, UvrC exhibits the most sequence variability among microbial species. Thus, in contrast to UvrA and UvrB, which reveal ∼50% sequence identity across the phylogenetic tree, including bacterial and archaeal species, the UvrC protein is the least conserved, with an overall sequence identity of ∼30% across species (9). In particular, the middle of the protein exhibits high variability. With this consideration, we decided to compare the sequence of E. coli UvrC (which contains the 5´ and 3´ incision active sites in the C- and N-terminal domains of the protein, respectively) with UvrC proteins of M. smegmatis and M. tuberculosis (Fig. 1). E. coli UvrC exhibits ∼35% sequence identity with both mycobacteria, which themselves demonstrate ∼75% sequence identity with each other. Importantly, the 3´ endonuclease active site is embedded in the GIY-YIG intron endonuclease catalytic domain at the N-terminal half, and the 5´ endonuclease active site is embedded in the RNaseH homology region in the C-terminal half of E. coli UvrC protein. Moreover, single amino acid mutations in these active sites have been reported to abrogate nuclease activity. As seen in the sequence alignment, the GVY–10aa–YVG motif is conserved in all three proteins, and most importantly, the Arg42 residue that is essential for the 3´ incision (12) and the Asp399 and Asp466 residues (E. coli numbering) that are essential for the 5´ incision (11) are similarly conserved in all three UvrC proteins, suggesting that these residues are also responsible for the 3´ and 5´ dual-incision activities of mycobacterial UvrC.
Figure 1.
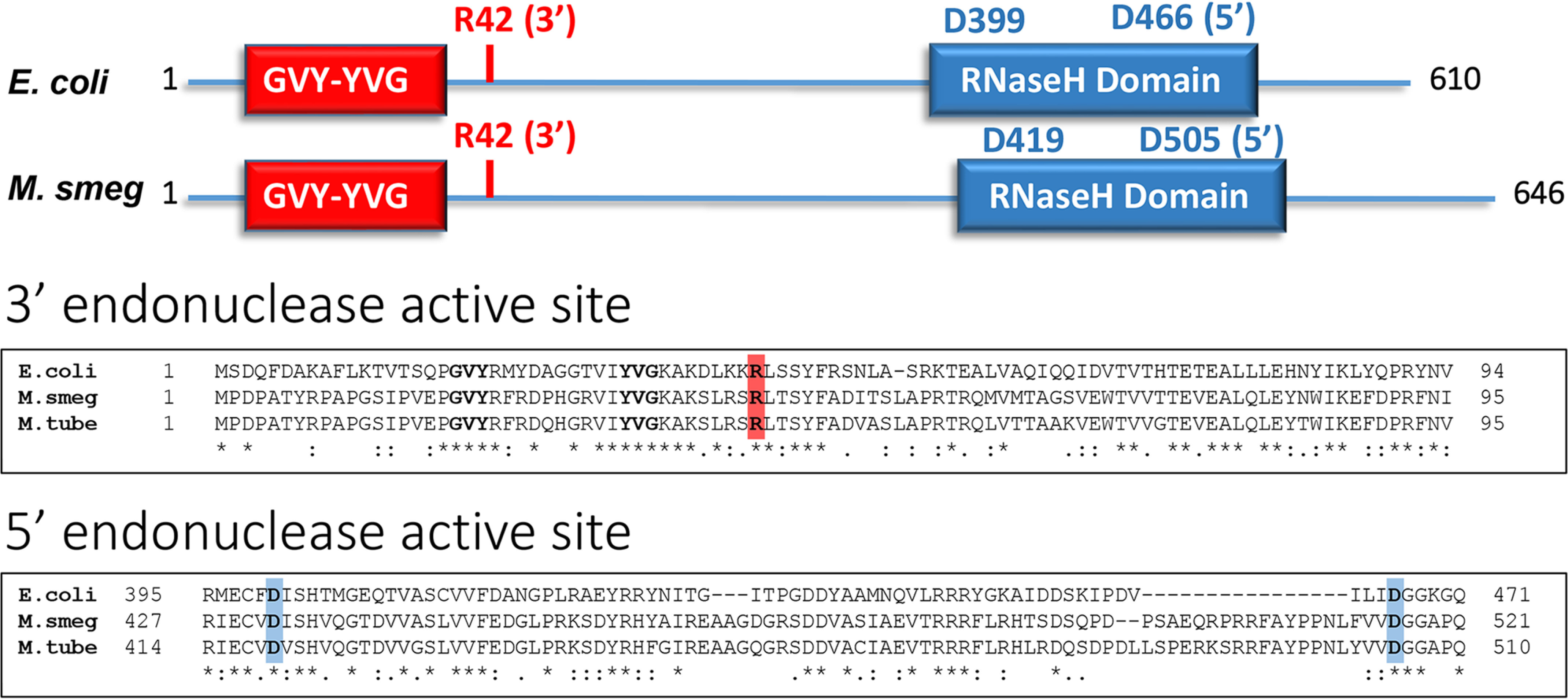
Top panel, the domain structure of UvrC showing the relative positions of the conserved endonuclease domains and active-site residues. Bottom panel, partial sequence alignment of UvrC from E. coli, M. smegmatis, and M. tuberculosis (NCBI reference sequences WP_123123263.1, WP_158166742.1, and WP_057159806.1, respectively.) The sequences were aligned using the CLUSTAL O (1.2.4) multiple sequence alignment program. An asterisk indicates identical residues, a colon indicates conservation of amino acid functional groups, and a period indicates conservation of functional groups of amino acids with weakly similar properties. The active-site arginine required for the 3´ incision is indicated in red, and the two aspartic acids required for the 5´ incision are indicated in blue.
M. smegmatis removes cyclobutane pyrimidine dimers from the genome by the prokaryotic dual-incision pattern
Because of biosafety restrictions, we conducted our mycobacterial experiments with WT (MC2155) and the uvrD1 mutant (MGM224) of M. smegmatis (20–23). The uvrD mutant was used because of the role of UvrD in excision product degradation, as discussed below.
To determine how M. smegmatis repairs CPD-damaged DNA, we first performed the in vivo excision assay recently developed in our laboratory. In this assay, following UV irradiation of cells and time for repair, the cells are lysed, and excision products containing CPDs are isolated, radiolabeled, and separated on a sequencing gel. The gel in Fig. 2 shows that CPDs are removed from the M. smegmatis genome principally in the form of 12- to 13-mers, with a bias to the shorter product in WT cell, as discussed further below. In contrast, the human cells release CPDs in the form of 26- to 29-mers.
Figure 2.

In vivo excision assay in M. smegmatis. WT or uvrD mutant cells were irradiated with 100 Jm−2 of 254 nm UV and incubated at room temperature for 10 min. Then the excised oligonucleotides were isolated, mixed with a 50-mer internal control, 3´-end labeled, and separated on a DNA sequencing gel. Note the higher level of 12-mer detected in the uvrD mutant because the “excised” oligomer remains bound to the duplex and is protected from nucleolytic degradation in the stable postincision complex containing UvrB–UvrC proteins as seen in E. coli (24). Rep1 and Rep2 refer to duplicate samples. A separate gel was run to resolve excision products from HeLa cells. This gel (lane 8) was aligned beside the M. smegmatis gel (lanes 1–7). An accidental tear in the M. smegmatis gel between the 25- and 32-nt markers occurred after running the gel, which slightly alters the orientation of the lanes above and below this point.
We then performed XR-seq analysis on excision products isolated from UV-irradiated M. smegmatis. In this procedure, DNA fragments isolated from treated cells are ligated to adapters, photoreactivated to repair the CPDs, amplified by PCR, and sequenced by next-generation sequencing, and reads are mapped to the genome. In Fig. 3 (A and B), the reads are plotted to show the read length as a function of frequency. The prevalent reads are 12- to 13-mers in uvrD− cells and 10- to 13-mers in WT cells. The same pattern of additional small fragments in uvrD+ versus uvrD− cells was seen in E. coli (24). The smaller excision products in WT cells are produced when UvrD releases excision products annealed to the genome, thereby exposing them to nucleases (24, 25). UvrD-dependent degradation of excision products in WT cells also leads to the appearance of reduced levels of incision as seen in Fig. 2. The appearance of reduced repair in WT cells is an artifact. In uvrD− cells, UvrB and UvrC remain trapped with the (nuclease-protected) excised oligochromosomal DNA postincision complex, unable to function catalytically (14, 15). In WT cells, the Uvr protein subunits do turn over, repairing more damage than in uvrD− cells, but the excised oligonucleotide is degraded to levels below the level protected in uvrD− cells.
Figure 3.
XR-seq analysis of excision sites in M. smegmatis. The frequency distribution histograms in A and B show that the predominant excision product length in uvrD− cells is 13 nt. In WT cells, the excision product is released from the genome and vulnerable to nuclease degradation (24), producing elevated levels of 10–12-nt sized products at the expense of the 13-mer. The histograms in C and D show the nucleotide frequency (y axis) at each position (x axis) of the 11–13-nt excision products. Reads were plotted by locating the 5´ residue at position 1. Notably, the position of the dipyrimidine peak (presumptive CPD damage site) in the 12- and 13-nt excision products is consistent with the formation of the 5´ incision 7 nt from the CPD and the 3´ incision 3- or 4-nt from the CPD. Interestingly, the progressive loss of nucleotides from the 3´ end (going from 13 nt to 12 nt excision products) is consistent with the degradation of excision products by an intracellular 3´ exonuclease activity.
Excision products were then analyzed by plotting the nucleotide distribution along the length of excision fragments (Fig. 3, C and D). As apparent, positions 8–9 nt from the 5´ end and 5–6 (or 4–5) nt from the 3´ end contain almost exclusively T–T (or more precisely dipyrimidine) dinucleotides, consistent with the excision assay in Fig. 2 and with the prokaryotic pattern of dual incisions 7 nt 5´ and 3–4 nt 3´ to the dipyrimidine. A different set of incision sites were described for M. tuberculosis repair, likely because of the use of indirect evidence for the 3´ incision site and the failure to use appropriate amounts of UvrA and UvrB in repair reactions (19).
Transcription-coupled repair in mycobacteria
Although TCR analysis has been performed in yeast, Arabidopsis, humans, and a few other eukaryotic species (26–29), it has not been extensively performed in prokaryotes with the exception of E. coli (17, 24, 25), even though the TCR factor Mfd has been found in all nonendosymbiotic bacteria including mycobacteria (9). Here we use our XR-seq data to analyze TCR in M. smegmatis as a sentinel for all other Mfd-containing bacteria. Fig. 4A shows a screenshot illustrating levels of repair in each strand of a representative region of the M. smegmatis genome. Genes in this region exhibit divergent transcription, such that the TS for the mfd and ABK72606 genes is the + strand (repair in blue) and the TS for glmU and ABK72893 is the – strand (repair in red). Visual inspection reveals TCR in the strongly transcribed mfd, ABK72606, and ABK72893 genes. Quantification of repair in each gene is illustrated in Fig. 4B, which confirms TCR in these three genes, and also shows that TCR is absent in the nontranscribed glmU gene. These results are consistent with prior findings in E. coli and other organisms that show an association between transcription and TCR levels (24, 25, 27, 28, 29). We extended our analysis to include the top 25% of M. smegmatis genes over 100 bp. Relative repair of the two strands of these genes is illustrated in Fig. 4C as a frequency distribution plot of log2-transformed TS/NTS ratios. The shift of the histograms to positive values is indicative of the influence of TCR on the cellular repair response to DNA damage. The influence of TCR appears essentially the same in WT and uvrD mutant cells, which was also observed in E. coli (24, 25). Overall, these results show that coupled repair is common among transcribed genes, which is a characteristic of TCR in E. coli, and likely occurs in all prokaryotes that possess full-length Mfd.
Figure 4.
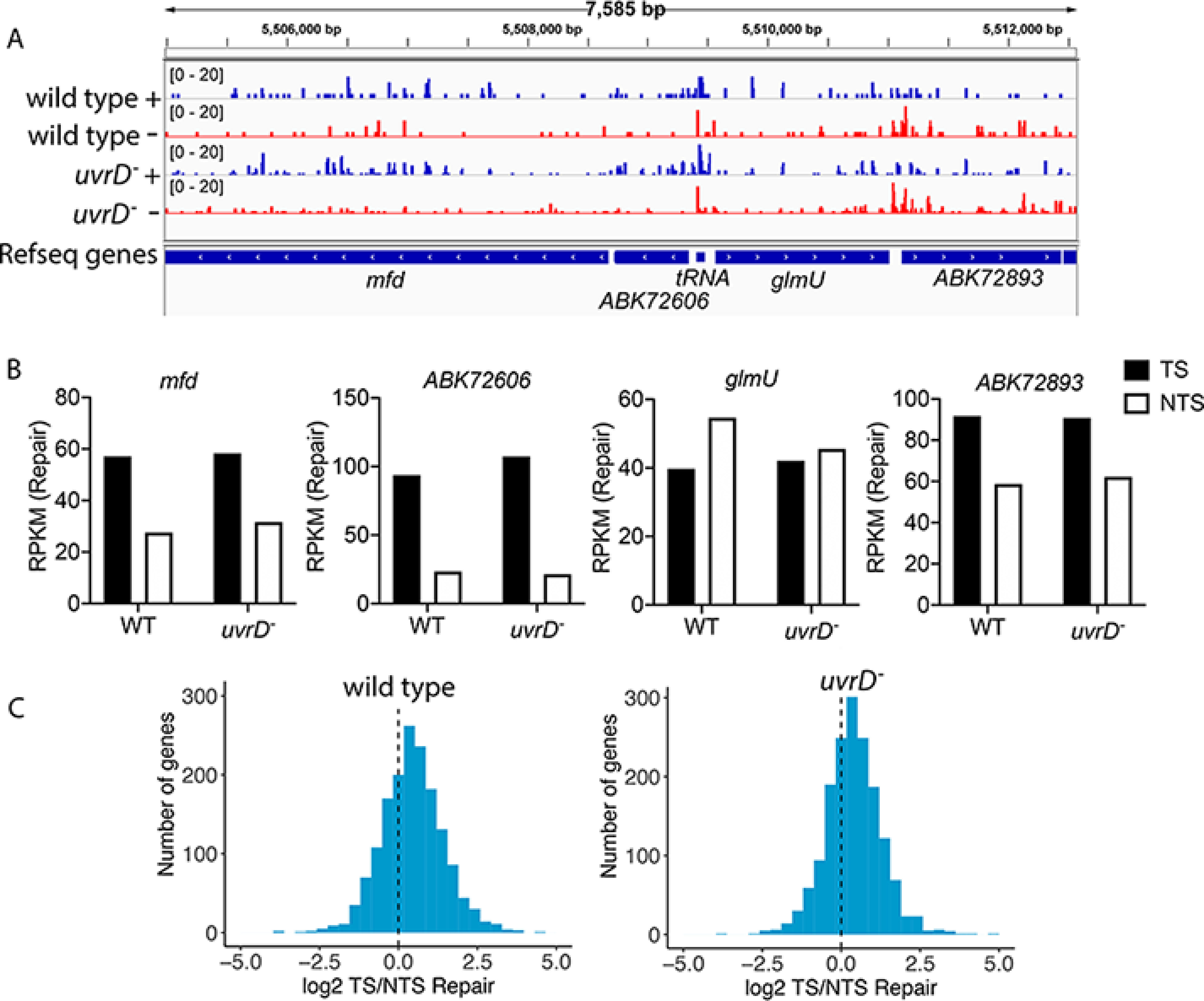
Transcription-coupled repair in M. smegmatis. The screenshot in A illustrates repair in the + and – strands of a representative 7585-bp region of the M. smegmatis genome. Repair is plotted as reads per million total reads (RPM) with the scale from 0 to 20. This region includes actively transcribed genes (mfd, ABK72606, and ABK72893) and a nontranscribed gene (glmU). For mfd and ABK72606, the + strand (repair in blue) is the TS, and for glmU and ABK72893 the – strand (repair in red) is the TS. TCR is evident in the actively transcribed genes by visual inspection. B illustrates quantitative values for each gene in A as RPKM. The absence of TCR in the nontranscribed glmU gene is evident. C shows frequency distribution of log2-transformed TS/NTS repair (as RPKM) in the 25% most highly transcribed M. smegmatis genes over 100 bp. The dotted line represents where repair in the two strands is equivalent. The shift toward positive values reflects TCR (24).
Conclusion
In conclusion, both phylogenetic and biochemical analyses indicate that M. smegmatis performs excision repair by the prokaryotic pattern. Even though for biosafety considerations we used M. smegmatis in our experiments, the phylogenetic data and the fact that M. tuberculosis UvrB and UvrD mutants are as UV-sensitive as the corresponding E. coli mutants (21) indicate that M. tuberculosis performs excision repair by the same mechanism: damage recognition by UvrA and kinetic proofreading by UvrB and dual incisions by UvrC in the UvrB–UvrC–DNA complex. A comparison of the eukaryotic and bacterial (including M. smegmatis) dual-incision patterns is illustrated in Fig. 5.
Figure 5.
Nucleotide excision repair patterns for prokaryotes, archaea, and eukaryotes. In the two prokaryotes, E. coli and M. smegmatis, dual incisions occur 7 nt 5´ and 3–4 nt 3´ to the UV photoproduct, resulting in excision products predominantly 12–13 nt in length. In the archaea, M. thermoautotrophicum, dual incisions occur 5–6 nt 5´ and 3–4 nt 3´ to the UV photoproduct, resulting in excision products predominantly 11 nt in length (8). In yeast cells, the photoproducts are excised by dual incisions 13–18 nt 5´ and 6–7 nt 3´ to the damage (28). In humans, plants, and insects, the dual-incision sites are at 19–22 nt 5´ and 5–6 nt 3´ to the UV damage (27, 29, 42). In eukaryotes there is considerable variability in incision sites, principally at the 5´ incision site, giving rise to excision products with a median length of 24 nt in S. cerevisiae and 26–27 nt in D. melanogaster, H. sapiens, and A. thaliana.
Finally, we wish to make some comments on the so-called noncanonical functions of Mfd (18), in particular as it relates to the development of resistance to antibacterial drugs by promoting mutations in various genes involved in cell wall biosynthesis, translation, and transcription by an unknown mechanism. This has led to Mfd being referred to as a “proevolutionary factor” (18) or “evolvability factor” (30). Although it is clear that Mfd plays a role in recombination (31) and repair of double-strand breaks, perhaps by aiding in resolution of R-loops and the consequent replication fork collapse (18, 30), the requirement of Mfd for development of resistance to antimicrobial drugs remains poorly explained. It should be noted that the antimicrobial drugs to which resistance develops in an Mfd-dependent manner are not DNA-damaging drugs, and the role of Mfd in this phenomenon and the related stationary phase mutagenesis remain to be mechanistically defined. What is clear about Mfd function is that it is an antimutator gene. In fact, even though MFD was a phenomenon discovered for mutation avoidance in tRNA suppressor genes, the Mfd protein performs a genome-wide antimutagenic function in protein-encoding genes as exemplified by the fact that Mfd reduces UV-induced mutation frequency in the E. coli lacI gene 3–5-fold and at one particular site by more than 300-fold (32).
Experimental procedures
Excision and XR-seq assays
M. smegmatis cultures (21) were grown with shaking at 37 °C in Luria Broth containing 0.5% (w/v) Tween 80. Exponentially growing cells were transferred to R150 tissue culture dishes in volumes of 15 ml when at an A600 of ∼0.6 and then irradiated at room temperature with 100 J/m2 UVC. For excision assay, the dishes were then incubated for 10 min at room temperature, and then the cells were chilled, harvested, and maintained on ice. Excision products were then isolated using a modified Hirt procedure that employs shearing to assist cell lysis. The cells were pelleted at 4 °C, resuspended in ice-cold TE (10 mm Tris, 1 mm EDTA, pH 8.0), transferred to ice-cold Eppendorf tubes, and pelleted at 4 °C, and the supernatants were removed. The pellets were then resuspended with 360 μl of ice-cold TE. Then 50 μl of room temperature 10% (w/v) SDS was added, and the cells were lysed by passage through a 26-gauge needle 40 times followed by incubation at room temperature for 20 min. A 120-μl volume of room temperature NaCl (5 m) was then added, tubes were gently mixed, and suspensions were incubated at 4 °C overnight. After centrifuging at high speed for 1 h at 4 °C, supernatants (∼480 μl each) were taken, and each was mixed with and incubated with 12 μl of RNaseA (R4642; Sigma) for 1 h at 37 °C and then with 12 μl of proteinase K (P8107S; NEB) for 1.5 h at 60 °C. Samples were then extracted twice with phenol/chloroform/isoamyl alcohol and precipitated with ethanol. The samples were immunoprecipitated with an anti-CPD antibody and washed as described (24, 26). Extraction, precipitation, and labeling of the 3′ ends with cordycepin were as described (24). One fmol of a 50-nt oligonucleotide was added to each sample before labeling to serve as a labeling and loading control. Samples were then extracted with phenol/chloroform/isoamyl alcohol and precipitated with ethanol and resolved with a 16% (w/v) polyacrylamide sequencing gel. We note that the Hirt procedure removes UvrB and UvrC that remain associated with DNA after dual incision and thus causes release of the excised oligonucleotide from genomic DNA. Excision assay of HeLa cells was as described previously (26).
XR-seq was by procedures previously described (24) using excision products isolated as described above. Approximately 20 plates of WT cells, which underwent 5-min repair were used, and ∼8 plates of uvrD mutant cells, which underwent 10 min repair, were used for XR-seq. XR-seq libraries were pooled and then sequenced on the HiSeq 2500 Rapid Run platform at the University of North Carolina–Chapel Hill High-Throughput Sequencing Facility.
Data analysis
At least 2 million uniquely mapped reads were obtained for each sample. Analyses of sequencing reads and data visualization were as described previously (24). Reads were trimmed to remove flanking adapter sequences by cutadapt (33), and then duplicate reads were removed by fastx_toolkit/0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Trimmed reads were aligned to the M. smegmatis WT strain MC2155 (GCA_000015005.1 was downloaded from EnsemblBacteria) by using bowtie2 with arguments −f −very-sensitive (34, 35). The output sam files were converted into bam files by using samtools (36) and then were converted into bed files using bedtools (37). For Fig. 3, excised oligonucleotides were aligned using bedtools with arguments getfasta −fi −s. Aligned reads in the 5–35-nt range totaled 2,309,284 (WT) and 2,060,476 (uvrD). Oligonucleotide lengths and nucleotide distributions were plotted by R. The analyses in Fig. 4 used only 13-nt reads with the TT dinucleotide at positions 8 and 9. With this filtering, there were 278,237 (WT) and 606,989 (uvrD−) reads. Bedtools was used to obtain reads for each gene's TS and NTS.
To determine the upper quartile of transcribed genes, we used a publicly available RNA-seq data set (data for cells in midexponential phase, 16 h) (38). SALMON (39) was used to obtain transcripts per million. The 25% most highly transcribed genes over 100 bp in length were selected for our analysis.
Data visualization utilized bedtools (37) with arguments genomecov –bg –strand −scale to generate bedgraph files. After sorting, ucsc tools (http://hgdownload.soe.ucsc.edu/admin/exe/) were used to convert bedgraph files int bigwig files. Bigwig files were visualized by IGV (RRID:SCR_007073, Broad Institute, and the Regents of the University of California) (40, 41). RPKM for each gene was plotted with Prism 8 (RRID:SCR_002798).
Data availability
The raw data and alignment data have been deposited in the Gene Expression Omnibus under accession number GSE159048. All codes used in this paper are available at https://github.com/yanyanyangunc/DNA-Damage-Repair-Circadian-Clock/tree/master/Mycobacterium_smegmatis_str_mc2_155.
Acknowledgments
We thank Michael Glickman (Sloan Kettering Institute) and Miriam Braunstein (University of North Carolina–Chapel Hill) for kindly providing M. smegmatis cultures.
Author contributions—C. P. S. and Y. Y. formal analysis; C. P. S., L. A. L.-B., and Y. Y. investigation; C. P. S., L. A. L.-B., and Y. Y. methodology; C. P. S., L. A. L.-B., Y. Y., and A. S. writing-review and editing; Y. Y. data curation; A. S. conceptualization; A. S. funding acquisition.
Funding and additional information—This work was supported by National Institutes of Health Grant GM118102 (to A. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- TCR
- transcription-coupled repair
- XR-seq
- excision repair sequencing
- TS
- transcribed strand or template strand
- NTS
- nontranscribed strand or coding strand
- RPKM
- reads per kbp per million total reads
- CPD
- cyclobutane pyrimidine dimer
- nt
- nucleotide(s).
References
- 1. Sancar, A. (1996) DNA excision repair. Annu. Rev. Biochem. 65, 43–81 10.1146/annurev.bi.65.070196.000355 [DOI] [PubMed] [Google Scholar]
- 2. Sancar, A., Lindsey-Boltz, L. A., Unsal-Kaçmaz, K., and Linn, S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- 3. Goosen, N., and Moolenaar, G. F. (2008) Repair of UV damage in bacteria. DNA Repair (Amst) 7, 353–379 10.1016/j.dnarep.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 4. Sancar, A. (2016) Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8502–8527 10.1002/anie.201601524 [DOI] [PubMed] [Google Scholar]
- 5. Hu, J., Selby, C. P., Adar, S., Adebali, O., and Sancar, A. (2017) Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J. Biol. Chem. 292, 15588–15597 10.1074/jbc.R117.807453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sancar, A., and Rupp, W. D. (1983) A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33, 249–260 10.1016/0092-8674(83)90354-9 [DOI] [PubMed] [Google Scholar]
- 7. Huang, J. C., Svoboda, D. L., Reardon, J. T., and Sancar, A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5´ and the 6th phosphodiester bond 3´ to the photodimer. Proc. Natl. Acad. Sci. U.S.A. 89, 3664–3668 10.1073/pnas.89.8.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ogrünc, M., Becker, D. F., Ragsdale, S. W., and Sancar, A. (1998) Nucleotide excision repair in the third kingdom. J. Bacteriol. 180, 5796–5798 10.1128/JB.180.21.5796-5798.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aravind, L., Walker, D. R., and Koonin, E. V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242 10.1093/nar/27.5.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi, Q., Thresher, R., Sancar, A., and Griffith, J. (1992) Electron microscopic study of (A)BC excinuclease: DNA is sharply bent in the UvrB–DNA complex. J. Mol. Biol. 226, 425–432 10.1016/0022-2836(92)90957-L [DOI] [PubMed] [Google Scholar]
- 11. Lin, J. J., and Sancar, A. (1992) Active site of (A)BC excinuclease: I. Evidence for 5´ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J. Biol. Chem. 267, 17688–17692 [PubMed] [Google Scholar]
- 12. Verhoeven, E. E., van Kesteren, M., Moolenaar, G. F., Visse, R., and Goosen, N. (2000) Catalytic sites for 3´ and 5´ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem. 275, 5120–5123 10.1074/jbc.275.7.5120 [DOI] [PubMed] [Google Scholar]
- 13. Kowalski, J. C., Belfort, M., Stapleton, M. A., Holpert, M., Dansereau, J. T., Pietrokovski, S., Baxter, S. M., and Derbyshire, V. (1999) Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 27, 2115–2125 10.1093/nar/27.10.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Husain, I., Van Houten, B., Thomas, D. C., Abdel-Monem, M., and Sancar, A. (1985) Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 82, 6774–6778 10.1073/pnas.82.20.6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orren, D. K., Selby, C. P., Hearst, J. E., and Sancar, A. (1992) Post-incision steps of nucleotide excision repair in Escherichia coli: disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J. Biol. Chem. 267, 780–788 [PubMed] [Google Scholar]
- 16. Sibghat-Ullah, Sancar, A., and Hearst, J. E. (1990) The repair patch of E. coli (A)BC excinuclease. Nucleic Acids Res. 18, 5051–5053 10.1093/nar/18.17.5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Selby, C. P. (2017) Mfd protein and transcription–repair coupling in Escherichia coli. Photochem. Photobiol. 93, 280–295 10.1111/php.12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strick, T. R., and Portman, J. R. (2019) Transcription-coupled repair: from cells to single molecules and back again. J. Mol. Biol. 431, 4093–4102 10.1016/j.jmb.2019.05.040 [DOI] [PubMed] [Google Scholar]
- 19. Thakur, M., Agarwal, A., and Muniyappa, K. (2020) The intrinsic ATPase activity of Mycobacterium tuberculosis UvrC is crucial for its damage-specific DNA incision function. FEBS J. 10.1111/febs.15465 [DOI] [PubMed] [Google Scholar]
- 20. Darwin, K. H., Lin, G., Chen, Z., Li, H., and Nathan, C. F. (2005) Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 55, 561–571 10.1111/j.1365-2958.2004.04403.x [DOI] [PubMed] [Google Scholar]
- 21. Sinha, K. M., Glickman, M. S., and Shuman, S. (2009) Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling. Biochemistry 48, 4019–4030 10.1021/bi900103d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sinha, K. M., Stephanou, N. C., Gao, F., Glickman, M. S., and Shuman, S. (2007) Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 282, 15114–15125 10.1074/jbc.M701167200 [DOI] [PubMed] [Google Scholar]
- 23. Güthlein, C., Wanner, R. M., Sander, P., Davis, E. O., Bosshard, M., Jiricny, J., Böttger, E. C., and Springer, B. (2009) Characterization of the mycobacterial NER system reveals novel functions of the uvrD1 helicase. J. Bacteriol. 191, 555–562 10.1128/JB.00216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adebali, O., Chiou, Y. Y., Hu, J., Sancar, A., and Selby, C. P. (2017) Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc. Natl. Acad. Sci. U.S.A. 114, E2116–E2125 10.1073/pnas.1700230114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adebali, O., Sancar, A., and Selby, C. P. (2017) Mfd translocase is necessary and sufficient for transcription-coupled repair in Escherichia coli. J. Biol. Chem. 292, 18386–18391 10.1074/jbc.C117.818807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu, J., Li, W., Adebali, O., Yang, Y., Oztas, O., Selby, C. P., and Sancar, A. (2019) Genome-wide mapping of nucleotide excision repair with XR-seq. Nat. Protoc. 14, 248–282 10.1038/s41596-018-0093-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu, J., Adar, S., Selby, C. P., Lieb, J. D., and Sancar, A. (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 10.1101/gad.261271.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li, W., Adebali, O., Yang, Y., Selby, C. P., and Sancar, A. (2018) Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U.S.A. 115, E3408–E3415 10.1073/pnas.1801687115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oztas, O., Selby, C. P., Sancar, A., and Adebali, O. (2018) Genome-wide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat. Commun. 9, 1503 10.1038/s41467-018-03922-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brugger, C., Zhang, C., Suhanovsky, M. M., Kim, D. D., Sinclair, A. N., Lyumkis, D., and Deaconescu, A. M. (2020) Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd. Nat. Commun. 11, 3740 10.1038/s41467-020-17457-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ayora, S., Rojo, F., Ogasawara, N., Nakai, S., and Alonso, J. C. (1996) The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 256, 301–318 10.1006/jmbi.1996.0087 [DOI] [PubMed] [Google Scholar]
- 32. Oller, A. R., Fijalkowska, I. J., Dunn, R. L., and Schaaper, R. M. (1992) Transcription-repair coupling determines the strandedness of ultraviolet mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 11036–11040 10.1073/pnas.89.22.11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin, M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 34. Langmead, B., and Salzberg, S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langmead, B., Wilks, C., Antonescu, V., and Charles, R. (2019) Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 35, 421–432 10.1093/bioinformatics/bty648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quinlan, A. R. (2014) BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li, X., Mei, H., Chen, F., Tang, Q., Yu, Z., Cao, X., Andongma, B. T., Chou, S. H., and He, J. (2017) Transcriptome landscape of Mycobacterium smegmatis. Front. Microbiol. 8, 2505 10.3389/fmicb.2017.02505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017) Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., and Mesirov, J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deger, N., Yang, Y., Lindsey-Boltz, L. A., Sancar, A., and Selby, C. P. (2019) Drosophila, which lacks canonical transcription-coupled repair proteins, performs transcription-coupled repair. J. Biol. Chem. 294, 18092–18098 10.1074/jbc.AC119.011448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data and alignment data have been deposited in the Gene Expression Omnibus under accession number GSE159048. All codes used in this paper are available at https://github.com/yanyanyangunc/DNA-Damage-Repair-Circadian-Clock/tree/master/Mycobacterium_smegmatis_str_mc2_155.




