Abstract
The heterotrimeric G proteins are known to have a variety of downstream effectors, but Gs was long thought to be specifically coupled to adenylyl cyclases. A new study indicates that activated Gs can also directly interact with a guanine nucleotide exchange factor for Rho family small GTPases, PDZ-RhoGEF. This novel interaction mediates activation of the small G protein Cdc42 by Gs-coupled GPCRs, inducing cytoskeletal rearrangements and formation of filopodia-like structures. Furthermore, overexpression of a minimal PDZ-RhoGEF fragment can down-regulate cAMP signaling, suggesting that this effector competes with canonical signaling. This first demonstration that the Gαs subfamily regulates activity of Rho GTPases extends our understanding of Gαs activity and establishes RhoGEF coupling as a universal Gα function.
The canonical G protein pathway consists of a cell surface receptor, a heterotrimeric G protein, and an effector protein that controls signaling within the cells. This fundamental paradigm, familiar to every biologist, is rooted in discoveries by the laboratories of Sutherland, Rodbell, and Gilman, which in the 1970s and 1980s dissected biochemical mechanisms of adenylyl cyclase activation by hormones. Their breakthrough came after experiments showing that the G protein Gs is essential to transfer agonist stimulation from the receptor to adenylyl cyclase (1). This G protein consists of the ∼42-kDa α subunit, which binds and hydrolyzes GTP, and the permanently associated dimer of 35-kDa β and ∼10-kDa γ subunits (Gβγ). Their findings helped establish a canonical model in which the agonist-bound receptor causes the G protein to release GDP, and the heterotrimer dissociates into Gα-GTP and free Gβγ; in this state, the G protein can activate its effector (i.e. Gαs will activate adenylyl cyclase until GTP is hydrolyzed). Although the rod photoreceptor G protein, transducin, was discovered by that time (2), the ubiquitously expressed Gs can be considered the founding member of the G protein family.
The subsequent cloning and identification of the other three families (Gi, Gq, and G12) completed the rough map of G protein–mediated transduction. These initial studies suggested that the α subunits were responsible for activation of one type of effector (e.g. Gαs for adenylyl cyclase and cAMP; Gαq for phospholipase C, phosphoinositides, and Ca2+; and Gαi for ion channels and inhibition of adenylyl cyclase), whereas the free Gβγ complexes interact with a remarkably large number of binding partners, including some effector enzymes and ion channels (3). Later, Gα12 and Gα13 were found to regulate a distinct type of effectors, the RhoGEFs (4, 5). These multidomain proteins contain pleckstrin homology (PH) domains, which facilitate their membrane localization, and Dbl homology (DH) domains, which catalyze GDP-for-GTP exchange (guanine nucleotide exchange factor; GEF) in the Rho family of small (∼20-kDa) G proteins. At the time, the G12-RhoGEF pathway seemed odd as it contained two G proteins: the receptor-activated “large” G12 class protein and the “small” Rho G protein, which is activated by RhoGEF. However, it was then discovered that Gαq could activate a RhoGEF called Trio (6), and that Gβγ complexes activate other RhoGEFs, indicating that this pathway, if unusual, is at least popular. Gαs, however, mostly appeared to be faithful to its originally determined role—to stimulate adenylyl cyclase(s)—possibly contributing to the enduring perception that regulation of a second messenger–generating enzyme is the “real” function of a heterotrimeric G protein.
In the current issue of JBC, Castillo-Kauil et al. (7) force a reexamination of the existing canon, presenting data that show Gαs can also interact with a specific RhoGEF, in this case PDZ-RhoGEF (PRG). The authors made this discovery as part of an examination of the regulation of cell shape by the Rho family. They began by expressing a series of short constructs of three RhoGEF proteins, p115RhoGEF, PRG, and LARG, all of which activated RhoA as expected, promoting cell contraction. However, they noticed that the DH/PH domain of PRG also activated Cdc42 and induced filopodia-like cell protrusions. To investigate which G protein is responsible for activation of this Cdc42-mediated pathway, they overexpressed constitutively active mutants of different Gα subunits. These mutants are stabilized in the active GTP-bound state due to substitution of the glutamine residue crucial for GTP hydrolysis. Surprisingly, the PRG-Cdc42 pathway was stimulated by GαsQ227L, the one Gα subtype not known for interaction with RhoGEFs. Furthermore, they showed that binding of PRG to Cdc42 was promoted only by Gs-coupled receptors, and not by Gq- or Gi-coupled GPCRs. The authors then investigated the PRG site responsible for the interaction with Gαs, narrowing it down to the isolated PRG DH and PH domains and their linker region. A construct encompassing these domains was able to inhibit (i) GPCR-mediated activation of Cdc42, (ii) the GαsQ227L-promoted interaction of PRG with Cdc42, and (iii) some protein phosphorylation events downstream of the canonical cAMP pathway. Taken together, their work identifies PRG as a novel effector for Gs; the Gαs-PRG interaction mediates activation of Rho family protein Cdc42, leading to cytoskeletal remodeling.
The unexpected results of Castillo-Kauil et al. open up new opportunities to explore this mechanism at different levels of biology. The experiments described in the paper were performed in vitro using cultured cells, imaging, and pulldown of protein complexes containing the overexpressed Gαs Q227L mutant. Considering the multitude of Gs-coupled receptors and RhoGEFs in the body (8, 9), it will be important to understand the physiological context where the new Gs-mediated pathway plays a significant role. This will require experimentation in vivo and possibly reevaluation of the phenotypes associated with known pathogenic mutations in Gαs (GNAS) and other relevant genes. At the molecular level, it would be important to delineate the biochemical mechanisms of Gαs interaction with PRG. For example, at what stage of the GTP/GDP cycle does Gαs bind to PRG: in the GTP-bound state, which also activates adenylate cyclase, or in the transition state (i.e. just before the terminal phosphate of GTP is removed)? Indeed, there is precedent for proteins that bind preferentially with the transition state—specifically RGS proteins, which accelerate the GTPase reaction. Another possibility is that, by analogy with p115RhoGEF, which stimulates GTPase activity of Gα12 and Gα13, PRG (and other RhoGEFs with similar DH-PH sequences) can influence interaction of Gαs with nucleotides, Gβγ, and other partners.
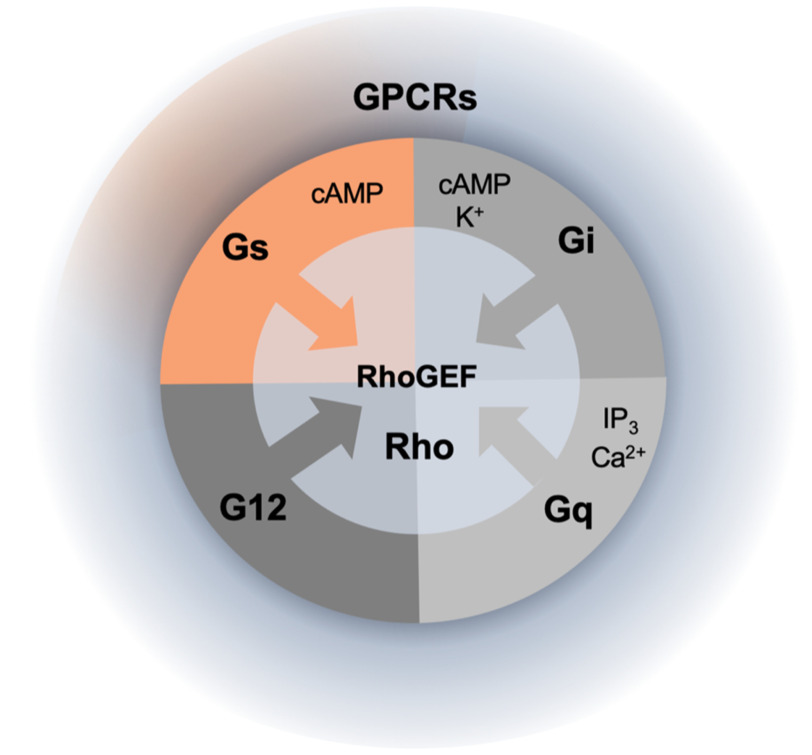
Since defining the receptor, G protein, and effector as the three essential members of the G protein pathway, researchers have discovered many additional proteins that regulate the amplitude and duration of the stimulus and/or participate in cross-talk with other signaling circuits. These “new” proteins include arrestins, receptor kinases, nonreceptor exchange factors, GTPase-activating proteins, special chaperones, etc. Thus, in a way, discovering a novel binding partner for a signaling molecule is not as surprising as it would have been 20 years ago. However, the new partner identified by Castillo-Kauil et al. makes the result of extra significance; until now, we knew that three of four G protein subfamilies could regulate Rho GTPases by activating RhoGEFs: G12 and Gq via their α subunits and Gi via the Gβγ subunits (10). The demonstration that the Gs subfamily is no exception shows that activation of RhoGEFs by heterotrimeric G proteins may be a truly universal mechanism (Fig. 1). The significance of this insight is that the multitude of biological processes regulated by Rho-GTPase networks can potentially respond to the entire repertoire of GPCR-mediated stimuli.
Figure 1.
Activation of the Rho family by heterotrimeric G proteins. The Rho family of small GTPases is activated by RhoGEF proteins, some of which can be stimulated by heterotrimeric G proteins. Of four families of heterotrimeric G proteins, three (G12, Gq, and Gi, shown in shades of gray) were known to activate certain RhoGEFs. The new results (highlighted in orange) (7) show that Gs, the G protein known to stimulate production of cAMP, can also stimulate a particular RhoGEF; this suggests that the Rho GTPases can potentially be stimulated by the multitude of signals from the entire class of GPCRs, including those coupled to Gs. IP3, inositol 1,4,5-trisphosphate.
Funding and additional information—This work was supported in part by National Institutes of Health Grant R56DK119262 (to V. Z. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- PH
- pleckstrin homology
- DH
- Dbl homology
- GEF
- guanine nucleotide exchange factor
- PRG
- PDZ-RhoGEF
- GPCR
- G protein–coupled receptor.
References
- 1. Northup, J. K., Smigel, M. D., Sternweis, P. C., and Gilman, A. G. (1983) The subunits of the stimulatory regulatory component of adenylate cyclase: resolution of the activated 45,000-dalton α subunit. J. Biol. Chem. 258, 11369–11376 [PubMed] [Google Scholar]
- 2. Fung, B. K., Hurley, J. B., and Stryer, L. (1981) Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc. Natl. Acad. Sci. U. S. A. 78, 152–156 10.1073/pnas.78.1.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smrcka, A. V. (2008) G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell Mol. Life Sci. 65, 2191–2214 10.1007/s00018-008-8006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hart, M. J., Jiang, X., Kozasa, T., Roscoe, W., Singer, W. D., Gilman, A. G., Sternweis, P. C., and Bollag, G. (1998) Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Gα13. Science 280, 2112–2114 10.1126/science.280.5372.2112 [DOI] [PubMed] [Google Scholar]
- 5. Fukuhara, S., Murga, C., Zohar, M., Igishi, T., and Gutkind, J. S. (1999) A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J. Biol. Chem. 274, 5868–5879 10.1074/jbc.274.9.5868 [DOI] [PubMed] [Google Scholar]
- 6. Rojas, R. J., Yohe, M. E., Gershburg, S., Kawano, T., Kozasa, T., and Sondek, J. (2007) Gαq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J. Biol. Chem. 282, 29201–29210 10.1074/jbc.M703458200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castillo-Kauil, A., García-Jiménez, I., Cervantes-Villagrana, R. D., Rafael Adame-García, S., Mabell Beltrán-Navarro, Y., Gutkind, J. S., Reyes-Cruz, G., and Vázquez-Prado, J. (2020) Gαs directly drives PDZ-RhoGEF signaling to Cdc42. J. Biol. Chem. 295, 16920–16928 10.1074/jbc.ac120.015204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cook, D. R., Rossman, K. L., and Der, C. J. (2014) Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene 33, 4021–4035 10.1038/onc.2013.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rossman, K. L., Der, C. J., and Sondek, J. (2005) GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6, 167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- 10. Aittaleb, M., Boguth, C. A., and Tesmer, J. J. (2010) Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 77, 111–125 10.1124/mol.109.061234 [DOI] [PMC free article] [PubMed] [Google Scholar]