Abstract
The organic anion transporters (OATs) and organic anion–transporting polypeptides (OATPs) belong to the solute carrier (SLC) transporter superfamily and play important roles in handling various endogenous and exogenous compounds of anionic charge. The OATs and OATPs are often implicated in drug therapy by impacting the pharmacokinetics of clinically important drugs and, thereby, drug exposure in the target organs or cells. Various mechanisms (e.g. genetic, environmental, and disease-related factors, drug-drug interactions, and food-drug interactions) can lead to variations in the expression and activity of the anion drug-transporting proteins of OATs and OATPs, possibly impacting the therapeutic outcomes. Previous investigations mainly focused on the regulation at the transcriptional level and drug-drug interactions as competing substrates or inhibitors. Recently, evidence has accumulated that cellular trafficking, post-translational modification, and degradation mechanisms serve as another important layer for the mechanisms underlying the variations in the OATs and OATPs. This review will provide a brief overview of the major OATs and OATPs implicated in drug therapy and summarize recent progress in our understanding of the post-translational modifications, in particular ubiquitination and degradation pathways of the individual OATs and OATPs implicated in drug therapy.
Keywords: transporter, membrane trafficking, intracellular processing, protein degradation, glycosylation, phosphorylation, oligomerization, ubiquitylation (ubiquitination), protein phosphorylation, drug transporters, organic anion transporters, organic anion-transporting polypeptides
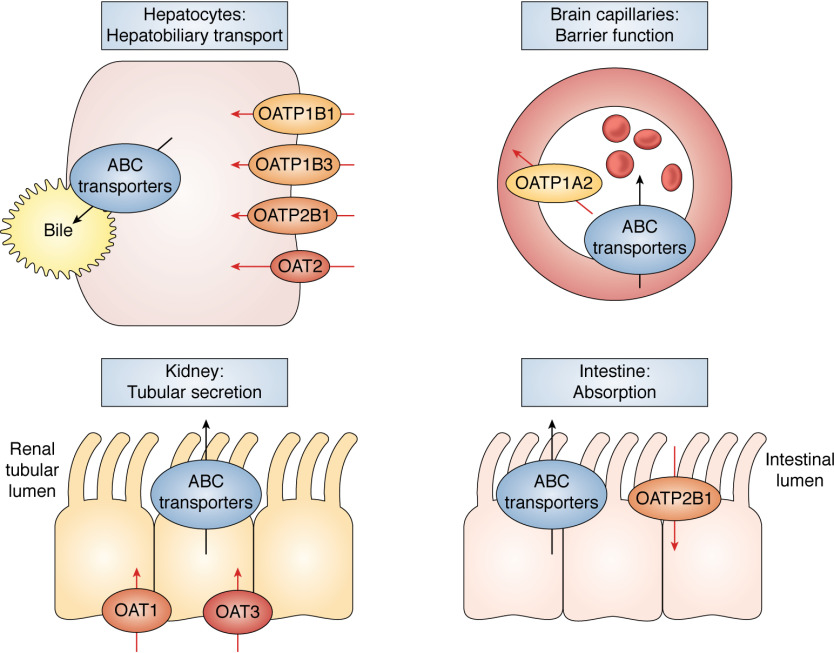
Membrane transporters are essential proteins that facilitate the directional movement of endogenous solutes and xenobiotics across cell and organelle membranes. Their functions are multi-fold, covering from homeostasis, cell communication, stress resistance, and cellular protection against toxins to drug sensitivity and resistance (1). Transporters are broadly classified into two classes, namely the solute carrier (SLC) and ATP-binding cassette (ABC) transporter superfamilies. The ABC transporters are organized into seven families (ABCA through ABCG) and include the well-known members that function as drug efflux pumps, contributing to chemotherapy resistance (e.g. ABCB1 (P-glycoprotein, Pgp) and ABCG2 (breast cancer–related protein, BCRP)). The ABC transporters have been most extensively investigated for their roles in drug therapy and also with regard to their regulatory and cellular processing mechanisms (2, 3). The SLC transporters are subdivided into 50+ families that can act as either influx or efflux transporters. Among them, the organic anion transporters (OATs) and the organic anion–transporting polypeptides (OATPs) belong to the SLC22A and SLCO superfamily, respectively, and display tissue-dependent expression profiles coordinated by genetic and epigenetic controls (4). By working jointly with the ABC transporters, the OATs and OATPs play important roles in the hepatobiliary transport, renal secretion, intestinal absorption, and brain penetration of various drug molecules (Fig. 1).
Figure 1.
Tissue-dependent expression of the major OATs and OATPs implicated in drug therapy. The OATs and OATPs facilitate the directional movement of various endogenous and xenobiotic substrates, including clinically used drugs. Genetic and epigenetic controls coordinate the tissue-dependent expression profiles of these transporters. The OATs and OATPs play important roles in the hepatobiliary transport, renal secretion, intestinal absorption, and blood-brain barrier function by working with the ABC transporters.
As anionic (hydrophilic) drug molecules tend to have a poor membrane permeability via passive diffusion, the transporters can play an important role in determining the cellular entry of anionic drugs and thereby influencing the pharmacokinetics (PKs; the profiles of the absorption, distribution, metabolism, and excretion processes in the body) and altering the drug exposure in the target organs and cells (possibly, the response and toxicity to drug therapies) (5). Accordingly, genetic, environmental, or disease-related factors and drug-drug interactions (DDIs) can influence the expression/activity of the transporters and consequently lead to the occurrence of adverse events in drug therapy and contribute to interindividual variability drug response (6). Severe and even life-threatening side effects of drug therapy may occur by the inhibition or impairment of these transporters (the tragic cases of the lipid-lowering statin drugs and fatalities caused by severe muscle toxicity of rhabdomyolysis (7–9) and many other cases of drug side effects and DDIs via transporter-mediated processes (10, 11)). Thus far, the cases of transporter-mediated DDIs have been mainly the combinations of the drugs that interact with transporters as substrates and/or inhibitors or as inducers and/or repressors (the modulators at the transcriptional level) (12). Changes in the transporters (due to genetic, environmental, or disease-related factors) can also occur via alternative modes that involve the post-translational regulation and processing of transporters (by impacting the localization, trafficking, post-translational modifications, or protein-protein interactions).
This review will focus on the major OATs and OATPs implicated in drug therapy and summarize the recent findings regarding their cellular processing, post-translational regulation, and degradation pathways. We start with a brief overview of the major OATs and OATPs implicated in drug therapy and the general cellular processing and trafficking of transporter proteins (more detailed and comprehensive reviews are available, but they do not focus on the anion drug–transporting proteins (13–15)). A previous review focusing on OATs and OATPs implicated in drug therapy was published in 2017 (16), and this review will cover some updates in the field.
Overview of the major anionic drug–transporting proteins in the families of OATs and OATPs
The SLC22 and SLCO superfamilies include transporters with broad substrate specificity, covering anionic, zwitterionic, and cationic molecules, and some members are more extensively studied for their pharmaceutical importance. The United States Food and Drug Administration, the European Medicines Agency, and other regulatory agencies recommend testing new drug candidates for possible interactions with selected members in the SLC22 and SLCO superfamilies. The SLC22 superfamily can be divided into at least six subfamilies based on the evolutionary analysis (17), and the SLC22A subfamily includes the major anion drug transporters, such as OAT1 and OAT3 (18). The SLCO superfamily includes 11 human OATPs, among which OATP1B1 and OATP1B3 are best-studied for their role in hepatic handling of the lipid-lowering statin drugs and anticancer drugs. More recently, OATP2B1 has attracted attention as another OATP member of pharmaceutical importance, regarding its role in impacting the intestinal drug absorption (19). Below is a brief overview focusing on the major OATs and OATPs implicated in drug therapy (OAT1, OAT2, OAT3, OAT4; OATP1B1, OATP1B3, OATP1A2, and OATP2B1). Table 1 lists their representative endogenous and exogenous substrates, including clinically important drugs and endogenous probes that are increasingly explored to assess the transporter functions in vivo and the potential for transporter-mediated DDIs (20, 21).
Table 1.
List of the major OATs and OATPs frequently implicated in drug therapy
This list includes the representative substrates; the endogenous probes used for the assessment of the transporter function in vivo and the risk for drug-drug interactions are underlined; comprehensive lists covering endogenous and exogenous substrates are available in previous reviews (1, 19, 21, 46, 151–154).
| Protein name (gene name) | Location | Example substrates |
|---|---|---|
| OAT1 (SLC22A6) | Kidney, brain, placenta, muscle; basolateral | Endogenous: α-ketoglutarate, urate, prostaglandin E2, taurine |
| Exogenous: adefovir, cidofovir, acyclovir, methotrexate, pravastatin, ciprofloxacin, chlorothiazide, ochratoxin A | ||
| OAT2 (SLC22A7) | Liver, kidney; basolateral | Endogenous: cGMP, creatinine, prostaglandin E2 |
| Exogenous: irinotecan, 5-fluorouracil, acyclovir, ganciclovir | ||
| OAT3 (SLC22A8) | Kidney, choroid plexus, testis; basolateral | Endogenous: estrone sulfate, bile acids, creatinine, prostaglandin E2, glycochenodeoxycholate-3-O-sulfate (GCDCA-S), 6β-hydroxyl cortisol |
| Exogenous: benzylpenicillin, cimetidine, ranitidine, famotidine, methotrexate, cidofovir, valacyclovir, sitagliptin, pravastatin | ||
| OAT4 (SLC22A11) | Kidney (apical), placenta (basolateral) | Endogenous: dehydroepiandrosterone sulfate, urate, estrone sulfate, prostaglandin E2 |
| Exogenous: indomethacin, tetracycline, ochratoxin A, methotrexate, olmesartan, levocetirizine | ||
| OATP1B1 (SLCO1B1) | Liver (basolateral) | Endogenous: estrone sulfate, estradiol 17β-glucuronide, dehydroepiandrosterone sulfate, coproporphyrin I and III (CPI and CPIII), GCDCA-S, unconjugated and conjugated bilirubin (UCB and CB) |
| Exogenous: bosentan, daunorubicin, fexofenadine, pitavastatin, atorvastatin, rosuvastatin, rifampicin, darunavir, mycrocystin | ||
| OATP1B3 (SLCO1B3) | Liver (basolateral) | Endogenous: cholecystokinin-8, CPI, CPIII, GCDCA-S, UCB, and CB |
| Exogenous: paclitaxel, pitavastatin, atorvastatin, rosuvastatin, rifampicin, telmisartan, mycophenolic acid, glucuronide | ||
| OATP1A2 (SLCO1A2) | Liver (cholangiocytes), brain capillary, kidney, retinal epithelium | Endogenous: estrone sulfate, retinoids, prostaglandin E2 |
| Exogenous: imatinib, fexofenadine, methotrexate, statins, microcystin | ||
| OATP2B1 (SLCO2B1) | Liver, intestine | Endogenous: estrone sulfate, dehydroepiandrosterone sulfate, prostaglandin E2 |
| Exogenous: glibenclamide, statins, fexofenadine, atenolol, montelukast |
OATs
By far, OAT1 and OAT3 are the most widely recognized drug transporters in the SLC22A subfamily. Both OAT1 (encoded by SLC22A6) and OAT3 (encoded by SLC22A8) are abundantly expressed in the kidney, moving anionic substrates across the basolateral membrane to the proximal tubular cells and enhancing the subsequent excretion into the urine. OAT1 and OAT3 display largely overlapping substrate specificity in handling drug molecules, but the recent analysis from a machine learning–based approach indicated that OAT3 may interact with drugs of a slightly cationic character, differing from OAT1 (22). When a similar approach was applied to the metabolomics data from mice lacking Oat1 or Oat3, a difference emerged in that Oat3 has a propensity for more complex substrates possessing more rings and chiral centers, compared with Oat1 (23).
OAT2 (encoded by SLC22A7) was the first cloned mammalian OAT and initially named as NLT (novel liver-specific transporter) (24). Later found to be expressed in the kidney as well as in the liver and closely related to OAT1, the NLT was renamed as OAT2 (25). A notable aspect of OAT2 is the presence of three splice variants: OAT2-546aa (NM_153320), OAT2-548aa (NM_006672), and OAT2-539aa (AF210455). The sequence difference between the two variants OAT2-546aa and OAT2-548aa is the insertion of 2 amino acids translated from 6 bp of nucleotides. The two variants display different substrate specificity and subcellular localization, but with some conflicting results from different laboratories (which is why OAT2 was called an enigmatic transporter in a recent review (26)). In recent years, OAT2 has emerged as an important transporter that handles clinically important drugs, including diclofenac (a nonsteroidal anti-inflammatory drug) and entecavir (an antiviral drug) (26, 27). Many of the OAT2 substrates are also substrates of OAT1 and/or OAT3, but OAT2 appears to use a transport mechanism distinct from OAT1 and OAT3 (28).
OAT4 (encoded by SLC22A11) is mainly located on the basolateral membrane of syncytiotrophoblasts in the placenta and on the apical membrane of renal tubular cells in the kidney (29–31). OAT4 can also operate as either uptake or efflux transporter (32), and the recently identified drug substrates of OAT4 include olmesartan (an antihypertensive drug blocking angiotensin receptor) and levocetirizine (an antihistamine drug) (33, 34).
Structurally, the members of the SLC22A/OAT subfamily share 12 transmembrane domains (TMDs) with a large extracellular loop between TMD1 and TMD2 (harboring glycosylation sites) and a large intracellular loop between TMD6 and TMD7 (harboring phosphorylation and ubiquitination sites).
OATPs
Among the 11 OATPs identified in humans, OATP1B1 and OATP1B3 are extensively studied for their roles in hepatic drug disposition and transporter-mediated DDIs. The genes encoding OATP1B1 (SLCO1B1) and OATP1B3 (SLCO1B3) are located in proximity to each other on the chromosome 12, and the two proteins are also highly homologous (80% identity at the amino acid level). OATP1B1 and OATP1B3 display nearly overlapping substrate specificity. The substrates commonly handled by OATP1B1 and OATP1B3 include many clinically important drugs (e.g. lipid-lowering statin drugs, anticancer and antidiabetic drugs), for which their entry to hepatocytes can be rate-determining in the overall hepatic drug elimination process (in which drug molecules are metabolized or excreted out of hepatocytes) (6). In such cases, impaired hepatic drug uptake due to either genetic variations or co-administered drugs can lead to the PK changes, impacting the drug efficacy and toxicity. For lipid-lowering statin drugs such as cerivastatin, impaired hepatic uptake was responsible for the occurrence of myotoxicity (muscle toxicity) ranging from mild cases to fatal rhabdomyolysis (the most severe form of muscle toxicity) (6, 35). To prevent similar tragic consequences, anionic drug candidates are screened for their possible interactions with OATP1B1 and OATP1B3. There is also a continuing effort to enhance our understanding of various mechanisms regulating the expression and function of these important hepatic uptake transporters.
Although OATP1B1 and OATP1B3 are present on the basolateral membrane of hepatocytes, they differ in terms of the zonal expression pattern in the liver: OATP1B1 is expressed throughout the liver parenchyma, but OATP1B3 is expressed in the region surrounding the central vein of hepatic lobules (36–38). Another notable difference was that unlike OATP1B1, OATP1B3 was detected in cancerous cells derived from various nonhepatic organs (38–40). It is now known that the OATP1B3 protein detected in the cancerous cells (called as cancer-type OATP1B3) arises from an alternative mRNA transcript and lacks the N-terminal 28 amino acids compared with the OATP1B3 protein expressed in nonmalignant hepatocytes (called liver-type OATP1B3) (40). Compared with the liver-type OATP1B3, the cancer-type OATP1B3 protein had an inferior efficiency of membrane trafficking, thus much reduced transport activity (40).
OATP1A2 (encoded by SLCO1A2) is another OATP that has been extensively studied for its interactions with a broad range of drugs (e.g. imatinib (an anticancer drug), methotrexate (an anticancer drug), and fexofenadine (an antihistamine drug)), cellular trafficking mechanisms, and differential regulation in disease conditions, including breast cancer (41, 42). OATP1A2 is expressed in the epithelium of various organs, such as the liver (cholangiocytes but not hepatocytes), kidney, brain capillaries, and eye (43, 44). An early study reported the positive immunohistochemical staining of OATP1A2 in the intestine (45), but subsequent studies did not detect the presence of OATP1A2 at either mRNA or protein level (see Ref. 46 and references therein). Those subsequent studies, however, verified the presence of OATP2B1 (encoded by SLCO2B1) in the human intestine. Based on its intestinal location and ability to transport clinically drug molecules, OATP2B1 is recognized as an important transporter that influences the intestinal drug absorption and serves as a target for food-drug interactions (19, 47). Interestingly, the two recent studies reported that the transport activity of OATP2B1 can be inhibited by common food and drug additives (small-molecule excipients added to food and drug products), such as azo dyes (48, 49). The food and drug additives had been presumed to be inactive and inert in the body, but these studies indicated that the food and drug additives can have unintended effects on the intestinal drug absorption and PK profiles by inhibiting OATP2B1 in the intestinal epithelium. Moreover, the OATP2B1-inhibitory additives were converted to metabolites that no longer inhibit OATP2B1 by the gut microbiota (48). These findings highlight the intriguing interplay of OATP2B1 and gut microbiota in influencing the intestinal drug absorption. In addition to the intestinal epithelium, OATP2B1 is also expressed in multiple organs, including the liver (hepatocytes), and its transport activity is pH-dependent with enhanced activity at acidic conditions (50, 51).
Structurally, the members of the SLCO/OATP family share 12 TMDs with an extracellular loop between TMD9 and TMD10 (harboring conserved cysteine residues and glycosylation sites). The highly conserved family signature sequence is located in the region that spans from extracellular loop 3 and TMD6 (52). For OATP1A2 and OATP2B1, their C-terminal regions harbor the PDZ-binding domain (53).
Overview of the general cellular processing of transporters in normal physiology and pathology
In normal physiology, it is generally believed that the translation of a transporter protein is tightly linked to its translocation (i.e. being co-translationally translocated) from ribosomes to the endoplasmic reticulum (ER), as illustrated in Fig. 2. Post-translational modifications play an important role in coordinating the folding and targeting of newly synthesized transporter proteins to the appropriate cellular organelles and the final destination of the plasma membrane (54–57). The processes that occur in the ER and Golgi apparatus are often referred to as “central quality control,” but the regulatory actions also take place near the plasma membrane. The mechanism is called “peripheral quality control,” and it regulates endosomal recycling and lysosomal degradation near the plasma membrane (54). The central and peripheral quality control pathways are interconnected and work together, shuffling the transporter proteins inside cells and to the plasma membrane, from the ER and Golgi to various intracellular degradation machinery (e.g. endosomes, lysosomes, proteasomes, aggresomes) (58, 59). Post-translational modifications and other protein-protein or protein-lipid interactions are involved in the internalization and recycling of the transporters located at the plasma membrane (15).
Figure 2.
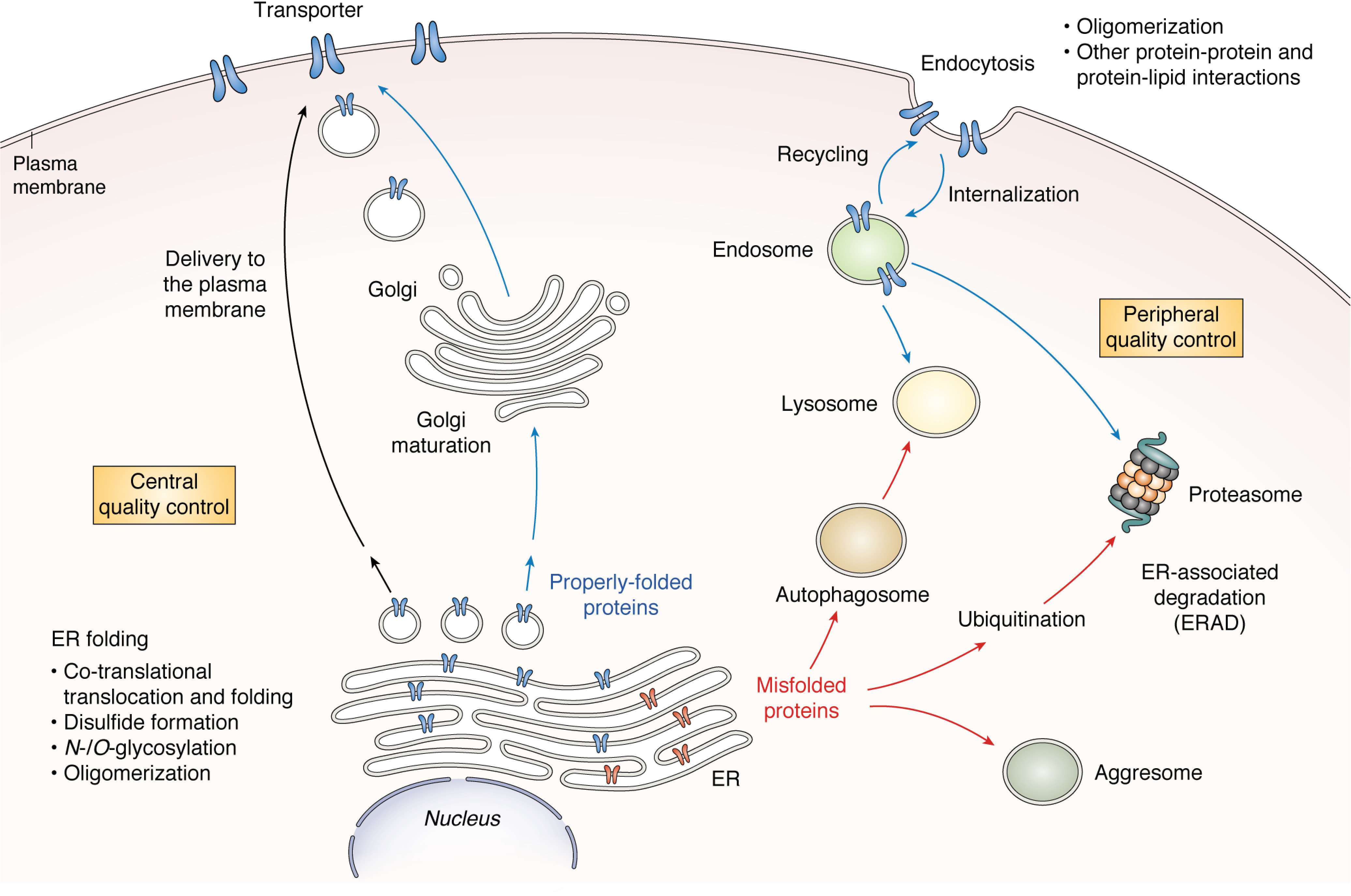
Overview of general cellular processing pathways for the membrane transporters. The central and peripheral quality control (QC) pathways are interconnected and work together, shuffling the transporter proteins inside cells and to the plasma membrane, from the ER and Golgi to various intracellular degradation machinery (e.g. endosomes, lysosomes, proteasomes, aggresomes).
In cellular processing of transporter proteins, it is increasingly recognized that the composition of membrane lipids can affect transporter conformation at the plasma membrane, leading to partial misfolding or modifications of cytoplasmic domains and altered susceptibility of the transporter to endocytic turnover processes (13). In particular, lipid rafts (microdomains enriched in cholesterol and glycosphingolipids at the external leaflet of the plasma membrane) have been shown to enhance oligomerization and other protein-protein interactions that impact the activity and trafficking for several transporters (60–63). This aspect is becoming of great interest and is another important layer of the post-translational regulatory mechanisms for the OATs and OATPs.
Defective processing and trafficking of transporters can cause various diseases. Cystic fibrosis is one of the well-known cases, and it is attributed to genetic variations causing misfolding and degradation of the cystic fibrosis transmembrane conductance regulator (CFTR, encoded by ABCC7) (64). With mechanistic understanding of the CFTR trafficking, there now exist therapeutic options for cystic fibrosis patients (e.g. lumacaftor-ivacaftor, tezacaftor-ivacaftor) (65, 66). Correction strategies using trafficking modifiers or chemical chaperones are being explored in preclinical and clinical settings for other transporters (e.g. ABCB11 encoding bile salt export pump (BSEP) and progressive familial intrahepatic cholestasis type 2 (67–69); ABCG2 encoding BCRP and gout (70, 71)). To date, a number of studies have reported alterations in the level and cellular localization of certain OATs and OATPs under disease conditions, such as cancer and liver diseases (72, 73). The OATs and OATPs have been rarely identified as single disease-causing factors, but the altered level/function of the OATs and OATPs may serve as disease-modifying factors and possibly impact the PK profiles of drug molecules in patients (74).
Three common types of post-translational modifications involved in cellular processing of the major anionic drug–transporting proteins in the families of OATs and OATPs
In response to various cellular stresses or stimuli, the transporter protein at the plasma membrane can receive multiple types of post-translational changes and protein-protein interactions that can alter the functional activity, protein internalization, and recycling. Below is a brief description of the three types of post-translational modifications (N-glycosylation, phosphorylation, and ubiquitination) that are commonly involved in the cellular processing and trafficking of the major anion drug transporters, OATs and OATPs.
N-glycosylation
In eukaryotic cells, N-glycosylation occurs at Asn residues within the consensus sequences of Asn-Xaa-Thr/Ser, where Xaa cannot be Pro or Asp. The types and linkages of N-glycosylation processed in the ER are diverse and complex, and the extent to which they exert discrete functions is not yet fully understood. Disruption of N-glycosylation can increase the accumulation of misfolded proteins in the ER, enhancing the ER-associated degradation.
Phosphorylation
Protein phosphorylation is a process of attaching a phosphate monoester group to a free hydroxyl group of Ser, Thr, or Tyr residues (or far less commonly to other amino acids, such as His, Lys, and Arg). For a single residue modifiable by phosphorylation, the modification can occur by different kinases and phosphatases, leading to different biological outcomes. For many proteins and signaling events, phosphorylation/dephosphorylation often serves as a functional on/off switch, interconnecting and cross-talking with other types of post-translational modifications. Prominent examples include the connection between phosphorylation and ubiquitination (75). Phosphorylation can either promote or inhibit ubiquitination, thereby affecting protein degradation or other molecular events. The activation of protein kinase C (PKC) has been associated with the internalization of the functional transporter from the cell surface, and the underlying mechanisms involve the protein phosphorylation promoting the ubiquitination of the transporter protein residing in a cholesterol-rich microdomain of the plasma membrane (lipid rafts) (76).
Ubiquitination
Ubiquitination is a process of attaching ubiquitin by forming an isopeptide bond between a Gly residue of ubiquitin and a specific Lys residue of a target protein. Ubiquitination is carried out in a multistep process involving a cascade of three enzymes: an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase enzyme (77). The E3 ubiquitin ligases confer substrate specificity and are classified into the two major families: HECT (homology to E6AP C terminus; ∼30 ligases) and RING (really interesting new gene; ∼600 ligases) (78). Different types (e.g. monoubiquitination (attachment of a single ubiquitin to a single Lys residue of the substrate), multiubiquitination (attachment of several ubiquitin molecules to multiple Lys residues), and polyubiquitination (attachment of polyubiquitin chain)) and linkages of ubiquitination can alter the biological outcomes. For example, Lys-48–linked polyubiquitination (Lys-48 residing in the ubiquitin molecules serves as a base for growing a polyubiquitin chain) commonly serves as the earmark for proteasome-mediated degradation (56). On the other hand, Lys-63–linked polyubiquitination (Lys-63 residing in the ubiquitin molecules also serves as a base for growing a polyubiquitin chain) is used for multiple purposes, including the regulation of the endosomal recycling and degradation events near the plasma membrane (79). Ubiquitination can be reversed by deubiquitinating enzymes (DUBs; >100 enzymes).
Cellular processing and post-translational modifications of the major anionic drug–transporting proteins in the families of OATs and OATPs
Below is a summary of current understanding of this topic, and major findings are also depicted in Fig. 3.
Figure 3.
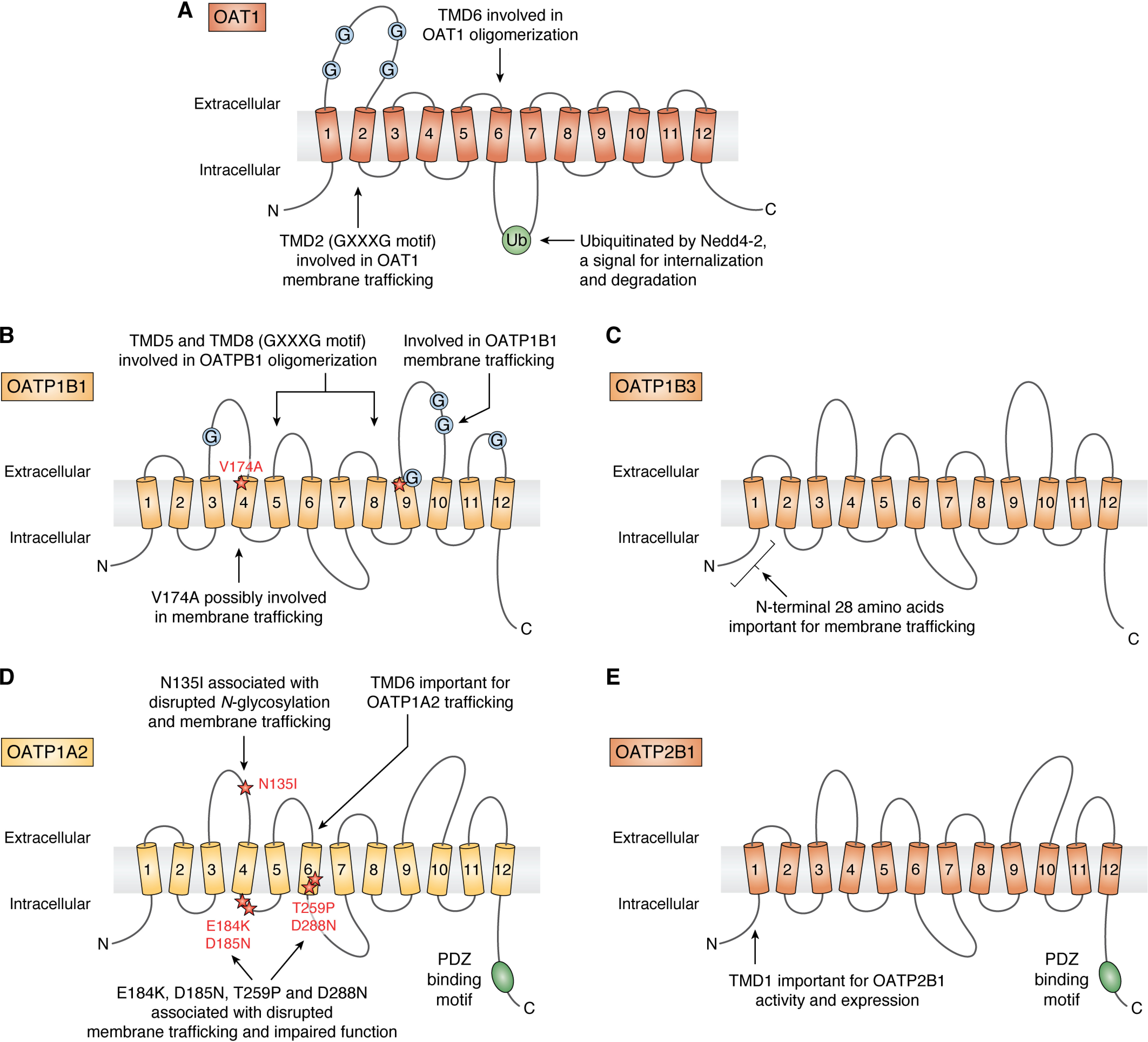
Schemes showing the published findings on the sites or regions involved in the post-translational regulation of OAT1 (A), OATP1B1 (B), OATP1B3 (C), OATP1A2 (D), and OATP2B1 (E).
OAT1
N-Glycosylation was initially identified to impact substrate binding or membrane trafficking of OAT1 (80). However, the mutation of multiple Asn residues was necessary for disruption of the OAT1 trafficking to the plasma membrane, and the responsible mechanisms or signaling pathways remain unknown.
The PKC activation triggered the internalization of OAT1 from the cell surface, as depicted in Fig. 4A (81). OAT1 was found to undergo the constitutive internalization and recycling at a rate of ∼10% of the initial OAT1 protein pool on the cell surface per 5 min, and the PKC activation accelerated the rate of OAT1 internalization without affecting OAT1 recycling (81). Subsequent studies reported that three Lys residues at positions of 297, 303, and 315 play a synergistic role in PKC-regulated OAT1 ubiquitination, trafficking, and transport activity (82) and that Lys-48–linked polyubiquitination is an important signal for internalization and degradation (83). These findings were further followed up by another observation on the connection between the PKC activation and polyubiquitination (84). The two E3 ubiquitin ligases belonging to the HECT family played an important role in the ubiquitination of OAT1, namely NEDD4-1 (neural precursor cell–expressed, developmentally down-regulated 4-1) and Nedd4-2 (84). In particular, NEDD4-2 was responsible for interconnecting the PKC activation and the internalization of OAT1: PKC activation leads to the phosphorylation and conformational change of NEDD4-2, which then leads to the ubiquitination and internalization of OAT1 (85). In contrast to the PKC activation linked to the OAT1 internalization (and thereby a decrease in the transport activity), the activation of SGK2 (serum- and glucocorticoid-inducible kinase 2) enhanced the surface level of OAT1 and increased the maximal transport activity (Vmax) without affecting the substrate-binding affinity (Km) (86). In either signaling event, OAT1 was not a substrate of direct phosphorylation (by PKC or SGK2 activation).
Figure 4.
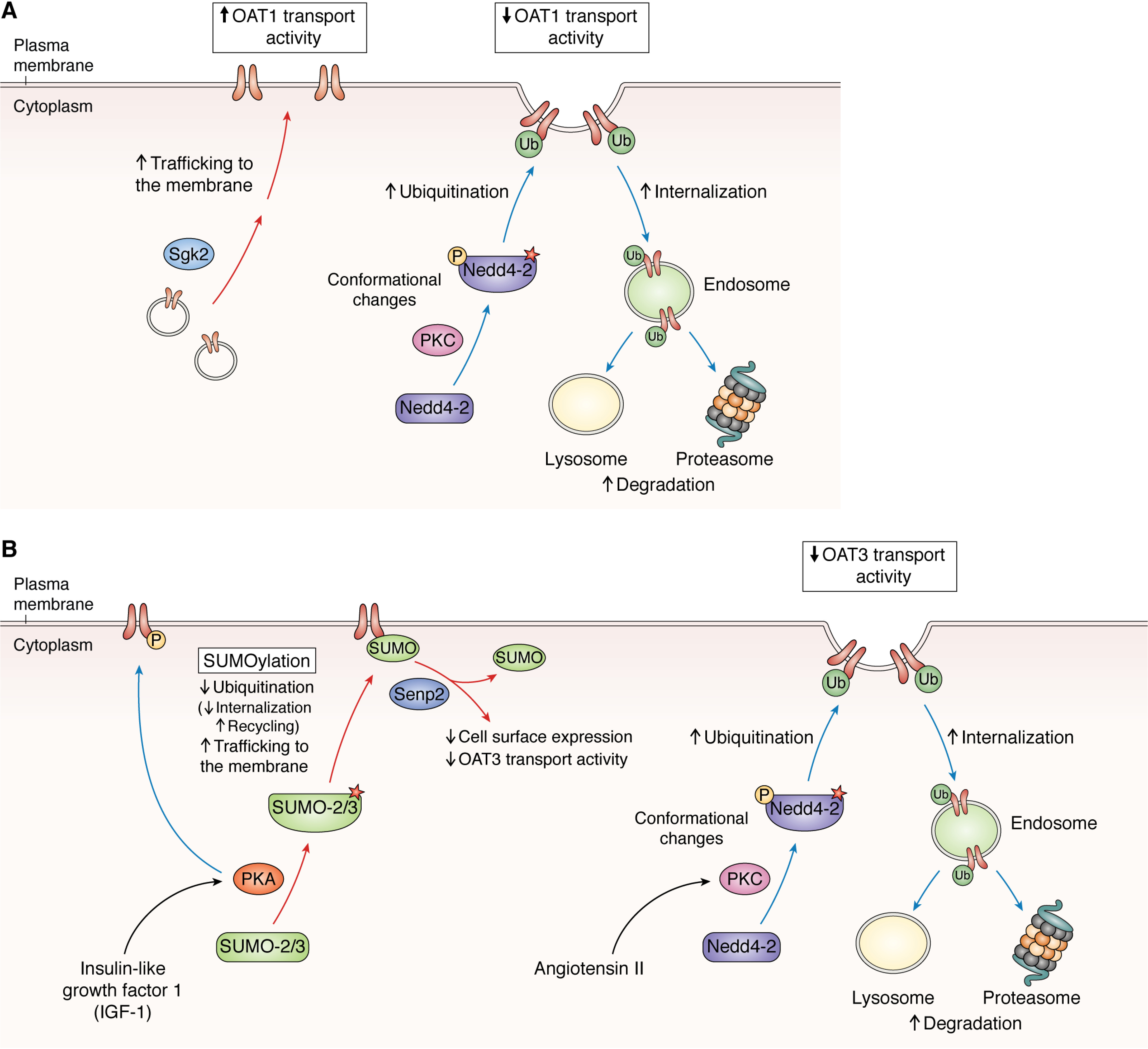
Proposed model based on the published findings of signaling pathways regulating the processing and post-translational regulation of OAT1 (A) and OAT3 (B).
In a recent study, the two clinically used proteasome inhibitor drugs (bortezomib and carfilzomib) increased the cellular levels of the ubiquitinated OAT1 protein and also enhanced the level of the functional OAT1 at the plasma membrane (87). Interestingly, the ubiquitinated OAT1 protein displayed a molecular size much higher than expected for a monomer (to an extent much higher than poly- or multiubiquitination). These results appear in accordance with the earlier finding that OAT1 can form homooligomers (88). The treatment with a chemical cross-linking agent led to the formation of immunoreactive OAT1 bands consistent with the formation of homooligomers. Immunoprecipitation experiments also revealed that the OAT1 proteins fused with different tags directly interact with each other. It remains to be investigated whether the homooligomerization is impacted by ubiquitination or treatment with proteasome inhibitors. The proteasome inhibitor therapy is often administered for an extended time in combination with other anticancer drugs to suppress the disease's progression. It is currently unknown whether OAT1 and other transporters' expression and activity may be impacted in cancer patients receiving the long-term proteasome inhibitor therapy.
Other findings on the post-translational regulation of OAT1 are also related to its oligomerization. The oligomerization of OAT1 was inhibited by co-expressing a short fusion peptide containing the TMD6 of OAT1 (89). Moreover, the membrane-trafficking and transport activity of OAT1 was abolished by the mutation of Gly residues in the TMD2 (within the motif of Gly-Xaa-Xaa-Xaa-Gly) (90). The mutated OAT1 proteins (G144A and G148A) were accumulated in the ER and subsequently degraded by the proteasome. The treatment with the proteasome inhibitor MG132 increased the protein level in total cell lysates but did not improve the trafficking of the mutated OAT1 protein to the cell surface (90). Together, these results suggest that the oligomerization and degradation of OAT1 are interconnected, but additional post-translational mechanisms may be involved.
OAT2
For OAT2, the alternatively spliced transcripts have been identified and investigated for their and function (91, 92). The variant OAT2-548aa contains two additional amino acids in the large extracellular loop (the addition of Ser and Gln between Glu-131 and Trp-132) and displays defective membrane trafficking (91). The previous study reported the presence of the putative sites (the consensus motifs) that could be phosphorylated by protein kinase A (PKA) or PKC (93), but further verification is unavailable yet.
OAT3
In a similar manner to OAT1, the PKC activation decreased the level of the functional OAT3 at the plasma membrane (83, 94) (Fig. 4B). The treatment with angiotensin II enhanced the internalization of OAT3 (resulting in a decrease in the Vmax value but no change in the Km value), connected to the PKC activation, accelerating the endocytosis of OAT3 (94). Short-term and long-term exposure to a PKC activator led to differential outcomes: the short-term exposure (<30 min) was associated with enhanced OAT3 internationalization without affecting the total expression level, but the long-term exposure (>2 h) led to OAT3 degradation by both lysosomes and proteasomes (83). A subsequent study showed a physical interaction between OAT3 and NEDD4-2, indicating that the PKC activation and internalization of OAT3 are interconnected via the ubiquitination by NEDD4-2, like OAT1 (95).
In contrast to the inhibitory effect of PKC on the OAT3 activity, PKA stimulated the OAT3 activity by enhancing its level at the plasma membrane (96, 97) (Fig. 4B). The PKA-mediated stimulation of OAT3 was mediated by SUMOylation (covalent attachment of SUMO (small ubiquitin-related modifier) by SUMO-2 and -3 but not by SUMO-1) (96). Further investigations revealed that SUMOylation could be reversed by the SUMO-specific protease Senp2, whose knockdown by siRNAs increased OAT3 SUMOylation and its transport activity and level. Insulin-like growth factor-1 (IGF-1) activated the PKA, subsequently increasing the SUMOylation of OAT3 (98), but a subsequent study reported that the effect of IGF-1–induced PKA activation was also linked to the phosphorylation of OAT3 (96). A more recent study verified direct phosphorylation of OAT3 by the treatment with Bt2-cAMP (a PKA activator) or IGF-1 (97). Together, the regulatory mechanisms for OAT3 showcase how the post-translational regulation and signaling pathways are interconnected.
In terms of the regulation via protein-protein interactions, OAT3 was shown to interact with lipid raft-associated proteins (β-actin, myosin, and caveolin-1) (99). When the authors exposed the rat renal cortical slices to methyl-β-cyclodextrin, which depletes cholesterol from the plasma membrane (thereby disrupting the lipid rafts), there was a dose-dependent reduction in the transport activity of the rat Oat3. In a more recent study, the membrane distribution of the rat Oat1 and Oat3 was assessed in animals that received bile duct ligation (leading to obstructive jaundice) or sham operation (100). The membrane distribution of Oat3 was not shifted in renal cortical cells isolated from the rats that received bile duct ligation. On the other hand, Oat1 displayed a significant shift in its membrane distribution (moving away from lipid raft domains). It remains to be investigated whether the level and function of human OAT1 and OAT3 may be impacted by the cholesterol content in the plasma membrane, which may well vary among individuals and depend on disease types and states.
OAT4
The PKC activation decreased the OAT4 activity by lowering its cell surface display (101). In the same study, progesterone, but not 17β-estradiol, decreased the OAT4 activity. The blockade of the PKC pathway did not reverse the progesterone's inhibitory effect on the OAT4 activity, suggesting that progesterone regulates the OAT4 activity via mechanisms independent of the PKC pathway (101). The PKC activation was later found to enhance the OAT4 internalization via clathrin-dependent pathways (102). Unlike the PKC activation, parathyroid hormone–related protein and a PKA activator stimulated the OAT4 activity by promoting the trafficking of OAT4 to the plasma membrane (103). The stimulatory effect of parathyroid hormone–related protein on the OAT4 activity was independent of the PKA pathway (103).
Similar to OAT1, the activity of OAT4 was reported to be regulated by the ubiquitin ligase NEDD4-2 (104). The enhancing effect of insulin on OAT4 level and activity was accompanied by the increased phosphorylation of Ser-327 on NEDD4-2, which in turn weakened the association of NEDD4-2 with OAT4 and reduced OAT4 internalization and degradation. The siRNA-based knockdown of NEDD4-2 abolished the enhancing effect of insulin on OAT4. The effect of insulin did not appear to be mediated by SGK2, another signaling event that enhanced the NEDD4-2 phosphorylation (86, 104). Together, these findings suggest that OAT4 processing and activity are dynamically regulated by the balance among multiple signaling pathways and cellular stimuli.
Another important aspect of OAT4 processing is its interaction with the scaffold proteins PDZK1 (PDZ domain–containing 1) and NHERF1 (Na+–H+ exchanger regulatory factor 1). Unlike OAT1, OAT2, and OAT3, OAT4 has a protein-protein interaction peptide sequence named the PDZ (PSD-95/Discs Large/ZO-1) motif at the C terminus. The co-immunoprecipitation results verified that OAT4 interacted with the two scaffold proteins PDZK1 and NHERF1 (105, 106). Interestingly, the interactions of OAT4 with PDZK1 and NHERF1 were observed in the LLC-PK cells (of kidney origin) but not in BeWo cells (of placenta origin) (106). These findings suggest that the interaction of OAT4 and the PDZ proteins may be cell- or tissue-specific. The oligomerization ability of scaffold proteins is known to be regulated by phosphorylation and other signaling pathways, via modifications of either scaffold proteins or their partnering proteins, including transporters (107, 108). Thus, it would also be important to consider whether the association of OAT4 with the scaffold proteins is impacted by any of the reported signaling pathways regulating OAT4 on the cell surface.
OATP1B1
The SLCO1B1 gene is highly polymorphic, and commonly occurring genetic variations (in particular, c.521T>C (dbSNP-ID of rs4149056; p.174Val>Ala)) have been firmly associated with PK changes and a high incidence of statin-induced myopathy (6). Most statins rely on the transporters to gain access to the hepatocytes, which becomes the rate-determining step in the overall hepatic elimination processes (6, 109, 110). The decreased hepatic uptake of statins can lower the hepatic elimination, increasing the drug exposure in plasma and drug distribution to the other organs, including the muscle, and the risk for side effects (6). The early investigations reported that the p.174Val>Ala variation was associated with a decreased membrane localization and a decreased Vmax value with no change in Km value (111, 112). A recent study, however, reported different findings that the V174A variation does not necessarily reduce the OATP1B1 level at the plasma membrane (113). Instead, the authors noted that V174A variation is associated with a modest increase in protein phosphorylation. Further investigations will be necessary to test whether the OATP1B1 phosphorylation is indeed causally linked to a decrease in the transport activity of OATP1B1 with V174A variation.
Other types of post-translational modifications have been investigated as the regulatory mechanisms for OATP1B1. The mutations at the three N-glycosylation sites disrupted the intracellular trafficking and accelerated the proteasomal degradation of OATP1B1 (114), yet the mutation of an individual glycosylation site did not disrupt the processing and transport activity of OATP1B1. By analyzing the human liver tissues from patients with nonalcoholic steatohepatitis, another study reported a possible disease-related impairment of N-glycosylation for OATP1B1 and OATP1B3 (115). It is currently unknown whether the variations in N-glycosylation can account for genetic and disease-related differences in OATP1B1 and OATP1B3 among individuals.
The activation of PKC triggered the internalization of OATP1B1 (116). In human primary hepatocytes and HepaRG cells, the treatment with a PKC activator reduced the levels of OATP1B1 protein (117). A large-scale phosphoproteomic analysis identified the phosphopeptides of OATP1B1 in human liver tissue samples (118). Another recent study reported further evidence regarding the phosphorylation of the OATP1B1 protein based on the phospholabeling experiments (119), but it remains unknown which signaling pathways and kinases are involved in the phosphorylation of OATP1B1.
The degradation mechanism of OATP1B1 was probed using the chemical inhibitors targeting the lysosomal or proteasomal pathways. The treatment with chloroquine (a lysosomal inhibitor; a drug used for the treatment of malaria and certain autoimmune diseases) increased the total OATP1B1 protein levels (based on the band intensities of OATP1B1 immunoblots) in HEK293 cells stably expressing OATP1B1 or human sandwich-cultured hepatocytes (120). Immunofluorescence imaging analysis, however, indicated that the OATP1B1 protein was located in the cytoplasm and associated with late endosome/lysosome in cells treated with chloroquine. In line with such findings, the chloroquine treatment was associated with a decrease in the Vmax value but no change in the Km value for OATP1B1-mediated transport of estradiol 17β-glucuronide (120). In the same study, the authors included the data from pharmacoepidemiological studies, in which female patients on co-medication of chloroquine and lipid-lowering statin drugs were associated with higher incidence of statin-associated myopathy. Further validation is however necessary, especially to probe the mechanisms for the observed gender differences in the pharmacoepidemiological studies. Treatment with bortezomib (a proteasome inhibitor) led to no changes in the total OATP1B1 protein levels (121), suggesting that the proteasome is likely to play a lesser role in the OATP1B1 degradation than the lysosome.
Regarding the protein-protein interactions, an earlier study reported that OATP1B1 displays immunoreactive bands around 190 kDa in nonreducing conditions and around 75 kDa in reducing conditions (122). The possibility of OATP1B1 homooligomers was further examined using a chemical cross-linking agent or by expressing the OATP1B1 protein fused with different tags in HEK293 cells, and the results verified the oligomer formation of OATP1B1, likely via disulfide bond formation (123). OATP1B1 harbors three Gly-Xaa-Xaa-Xaa-Gly motifs, among which the motif located in TMD5 was found to be important for oligomerization, based on the results obtained using the mutant G393A (a decreased oligomerization and a reduced uptake of estrone 3-sulfate) (123). Further investigations are however warranted, in particular, using other cellular systems with a more native membrane environment, such as human primary hepatocytes, and the question remains whether the oligomerization of OATP1B1 occurs primarily as a homooligomer or a heterooligomer with other OATP transporters and whether the oligomerization of OATP1B1 varies among individuals in healthy or diseased states.
OATP1B3
OATP1B3 is highly homologous to OATP1B1 and displays nearly overlapping substrate specificity with OATP1B1. Cholecystokinin-8 (a peptide gastrointestinal hormone) is an exception that is handled uniquely by OATP1B3 and not by OATP1B1. The SLCO1B3 gene is not as polymorphic as the SLCO1B1 gene, but a few nonsynonymous variations have been reported for OATP1B3 (124, 125). For the variants M233I, H520P, and V560A, the OATP1B3 protein levels at the membrane surface were decreased, accompanied by the reduction in the Vmax values for the transport of cholecystokinin-8 (125). In a study that assessed the systemic PK profiles of mycophenolic acid (an immunosuppressant drug), the subjects harboring the haplotype combination of p.233Met>Ile and p.112Ser>Ala (c.699G>A and c.334T>G present in linkage disequilibrium; dbSNP-IDs of rs7311358 and rs4149117) tended to have the elevated systemic levels of mycophenolic acid (126). More importantly, a recent retrospective study reported that the subjects homozygous for the variant haplotype have an improved clinical outcome (in terms of the risk for acute rejection and survival) following lung transplantation (127). Currently, a mechanistic understanding is lacking as to how the genetic variations lead to the reduction in the OATP1B3 level on the plasma membrane and the impaired transport activity.
Unlike OATP1B1 displaying nearly exclusive expression in hepatocytes, OATP1B3 was detected in cancerous cells derived from various nonhepatic organs also expressing OATP1B3, with the predominantly cytoplasmic pattern when probed using the OATP1B3 antibody detecting the C-terminal sequence) (38–40). It is now known that the positive cytoplasmic immunostaining was from the cancer-type OATP1B3 (674 aa, lacking the N-terminal 28 amino acids compared with the liver-type OATP1B3 protein of 702 aa) (40). Using the N-terminal truncation mutants, a follow-up study revealed that the N-terminal sequence of OATP1B3 (in particular, the amino acid positions between 12 and 18 within the region lacking in the cancer-type OATP1B3) is important for membrane trafficking of OATP1B3 (128). The structural motifs or individual amino acids responsible for the trafficking of OATP1B3 were not, however, identified in that region. But the importance of the N-terminal sequences of OATP1B3 as well as OATP1B1 was supported by the finding that the N-terminal peptides (50 amino acids) fused with a Myc tag were efficiently localized to the plasma membrane (128). On the other hand, the C-terminal sequences of both OATP1B3 and OATP1B1 were predicted to lack a PDZ-binding motif interacting with the scaffold proteins (53).
For the degradation of the liver-type OATP1B3 protein, the lysosome may play a more prominent role than the proteasome (121). When cells stably expressing the liver-type OATP1B3 were treated with chloroquine (a lysosomal inhibitor) or bortezomib (a proteasome inhibitor), the total OATP1B3 protein (assessed by OATP1B3 immunoblots) was increased only by chloroquine (121). Similar to the case of OATP1B1, the increased level of the total OATP1B3 protein by chloroquine treatment was associated with a decreased transport activity (121, 129). Despite having no impact on the OATP1B3 protein levels in total cell lysates or surface membrane fraction, the bortezomib treatment also led to a modest decrease in the Vmax value but no change in the Km value for OATP1B3-mediated transport of cholecystokinin-8 (121). The bortezomib treatment, however, had no impact on the OATP1B3-mediated transport of pitavastatin (a lipid-lowering statin drug) (121). The authors speculated that the bortezomib treatment may change the turnover rate of OATP1B3, but further experiments will be necessary to verify that possibility.
Like OATP1B1, OATP1B3 was found to homo- or heterooligomerize (130). In addition to homooligomers, OATP1B3 oligomerized with OATP1B1 and Na+-taurocholate–cotransporting polypeptide (NTCP) in HEK293 cells expressing those transporters. As the rat Ntcp was reported to be located in the lipid rafts of the plasma membrane (131), the question remains whether the heterooligomers of OATP1B3 and NTCP would be also located in the lipid rafts and whether the level and function of OATP1B3 could be affected by the cholesterol content in the membrane.
OATP1A2
The decreased OATP1A2 level at the plasma membrane was reported with naturally occurring genetic variations: variations disrupting N-glycosylation (43) and those replacing the negatively charged residues (Asp, Glu) in the intracellular loops or Thr residue in the putative TMD6 (132). A follow-up study focused on the putative TMD6 and narrowed down the amino acids that are important for the OATP1A2 trafficking and degradation via proteasomes or lysosomes (133).
Another important aspect of OATP1A2 processing is its interaction with the scaffold proteins PDZK1 and NHERF1, based on the results from the yeast two-hybrid library screening (53) and the co-immunoprecipitation experiments (134). In the case of PDZK1, no direct interaction was detected by co-immunoprecipitation experiments, but both PDZK1 and NHERF1 enhanced the OATP1A2 level at the membrane surface by reducing OATP1A2 internalization via the clathrin-dependent pathway (134).
A separate study indicated that the clathrin-dependent internalization of OATP1A2 from the cell surface can be accelerated by the PKC activation (135). In addition to PKC, casein kinase 2 (CK2) was involved in regulating the trafficking of OATP1A2 (136). Chemical or genetic inhibition of CK2 led to decreases in the OATP1A2 internationalization and recycling, causing a reduction in the Vmax value but no change in the Km value for the OATP1A2-mediated transport of estrone-3-sulfate (136). Interestingly, CK2 is reported to be dysregulated in many disease states, including breast cancer, in which the OATP1A2 level is elevated (42, 137). Another study showed that OATP1A2 trafficking is regulated by 5′-AMP–activated protein kinase signaling associated with an increased incidence of type II diabetes and nonalcoholic fatty liver disease (138). Further investigations will be necessary to assess how and to what extent these multiple kinases are connected and contribute to the processing of the functional OATP1A2 in healthy and disease states.
OATP2B1
The SLCO2B1 gene encoding OATP2B1 harbors polymorphic variations whose frequencies vary among different ethnic groups (139). Nonsynonymous genetic variations of OATP2B1 (c.935G>A and c.1457C>T; dbSNP-IDs of rs12422149 and rs230618) have been associated with the altered transport activity and PK changes in vivo, with some conflicting results (140–143). However, our understanding is limited as to whether and how these genetic variations differ at the post-translational level.
The OATP2B1 protein contains three consensus sequences for N-glycosylation, two of which are predicted to be intracellular (144). To date, no study has examined specifically whether those N-glycosylation sites play a role in the trafficking mechanism of OATP2B1. The study that examined the human liver tissues from patients with nonalcoholic steatohepatitis reported a possible impairment of N-glycosylation for OATP2B1, but to a lesser extent than OATP1B1 and OATP1B3 (115).
Similar to OATP1B1, the PKC activation accelerated the OATP2B1 internalization via the clathrin-dependent pathway and subsequent lysosomal degradation (145). No information is yet available regarding the residue(s) phosphorylated in OATP2B1 and other mediating signaling pathways that interconnect the PKC activation and OATP2B1 processing.
Another study identified that the TMD1 of OATP2B1 is important for its transporter function and stability (146). The replacement of a Phe residue at position 51 enhanced the OATP2B1 degradation, but the functional activity of OATP2B1 was recovered by neither lysosomal nor proteasomal inhibition. Like OATP1A2, OATP2B1 harbors a PDZ-binding motif at its C terminus (53). The localization and function of OATP2B1 was reported to be regulated by the interactions with PDZK1 (147). In HeLa cells stably expressing OATP2B1, the transient transfection of PDZK1 led to an increase in the functional OAT2B1. The N-terminal deletion mutant of OATP2B1 lacking the PDZ-binding motif, however, showed no enhancement by PDZK1 (147). Another intriguing finding was that the membrane localization of OATP2B1 appeared to switch from the apical to basolateral sides in the proximal and distal renal tubules, based on the immunohistochemical staining of the human kidney tissue sections (147). As the PDZK1 was found only on the apical side, the authors cautiously suggested that PDZK1 may be involved in the control of subcellular localization of OATP2B1. Further investigations will be necessary to determine whether similar interactions between OATP2B1 and PDZK1 occur in the intestine and whether variations in the PDZK1 expression may be a source of interindividual variability in the level of the functional OATP2B1.
Conclusions and future directions
This review summarized the recent progress in describing the cellular processing and trafficking of the major anion drug-transporting OATs and OATPs. Although some of the SLC transporters have been explored as therapeutic targets, the major anion drug–transporting proteins were not included (148). However, the anion drug–transporting proteins will continue to play a critical part in predicting the PK profiles of clinically important drugs and managing the risk of DDIs during drug development.
A better understanding of post-translational regulation and trafficking of the anion drug–transporting proteins can allow us to obtain an accurate prediction of the transporter-mediated handling of clinical used drugs and new drug candidates. Advances have been made in the quantitative proteomic analysis of transporters, providing increasingly precise measurements of the transporter level across various human tissues and experimental systems and relevant transporter-related parameters that can be scaled up for the prediction of PK profiles via in silico modeling–based approaches (149, 150). The more we understand and appreciate the contribution of post-translational mechanisms regulating the transporters, the more we recognize that the protein amounts detected either in cells or at the surface membrane may not necessarily correlate with the functional transporters. Another area that has made significant progress in recent years is the discovery and validation of endogenous probes that can serve as biomarkers for transporter function in vivo (20, 21). A number of endogenous probes for the major anion drug–transporting proteins have been identified among the endogenous metabolites or food-derived compounds (as listed in Table 1). The evaluation of the endogenous probes will continue in terms of their transporter selectivity and specificity and sensitivity to detect the intra- and interindividual variations in the transporter function in vivo. When discrepancies are found between the transporter level (e.g. the quantitative proteomics data) and the in vivo functional outcomes in healthy or diseased subjects (e.g. the assessment using drug probes or endogenous probes), a better understanding of post-translational regulation and trafficking may provide clues to resolving the disconnect. In coming years, more careful investigations will be necessary to examine and understand the changes in the processing and trafficking of transporters over time and in response to various cellular stresses and stimuli.
Protein-protein interactions (including oligomerization) and lipid-protein interactions (e.g. near the lipid rafts of the plasma membrane) are emerging as important players in regulating transporter functioning. In examining the interactions of transporters with drug molecules and the regulatory mechanisms for various transporters, in vitro cell line models expressing individual transporters in either a transient or stable manner have been widely employed. In such model systems, the cellular environment may not fully capture protein-protein and protein-lipid interactions that occur in the native environment in vivo, including possible variations in the cellular environment among individuals (e.g. gender, age, genetic, environmental, and disease-related). When the transport activity is assessed using commonly available cell line models or primary cells from human donors, there often exist substantial data variabilities within and among laboratories. The observed data variability may be attributable to variations in culturing conditions and possibly factors impacting the processing, trafficking, and degradation of the transporters at the post-translational level. Thus, careful comparative analysis and cross-validation of the results in different model systems, including primary cells, will be necessary to enhance our understanding of the regulatory mechanisms that have clinical significance.
As covered in this review, the processing and trafficking of the transporters are coordinated by different types of post-translational modification, including ubiquitination and phosphorylation. Ubiquitination and phosphorylation are highly dynamic processes controlled by the balance of the enzymes involved (kinases and phosphatases; E3 ubiquitin ligases and DUBs). Those enzymes are specifically targeted or affected by some approved drugs (e.g. various kinase inhibitors) or drug candidates in clinical and preclinical development. The approved drugs targeting the proteasomes (bortezomib, carfilzomib, and ixazomib) have brought a breakthrough in treating patients with multiple myeloma and other hematological malignancies and are used as long-term therapies. Currently, efforts are ongoing to develop next-generation drugs targeting the proteasomes and E3 ligases as well as drugs targeting DUBs. The lysosomal inhibitors chloroquine and hydroxychloroquine are currently used against some infectious and inflammatory diseases. When these drugs are used to treat patients on a long-term basis, they may have an impact on the cellular proteomic profiles, including transporters. The consequences of such changes may need to be carefully examined in relation to the PK and pharmacodynamic aspects in drug therapy.
Funding and additional information—This work was supported under the framework of international cooperation program managed by the National Research Foundation of Korea (NRF-2020K2A9A2A08000172) and the Creative-Pioneering Researchers Program through Seoul National University (to W. L.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SLC
- solute carrier
- ABC
- ATP-binding cassette
- OAT
- organic anion transporter
- OATP
- organic anion–transporting polypeptide
- ER
- endoplasmic reticulum
- CFTR
- cystic fibrosis transmembrane conductance regulator
- PK
- pharmacokinetic
- DDI
- drug-drug interaction
- TMD
- transmembrane domain
- BCRP
- breast cancer–related protein
- HECT
- homology to E6AP C terminus
- DUB
- deubiquitinating enzyme
- PKC
- protein kinase C
- PKA
- protein kinase A
- NEDD4-1
- neural precursor cell–expressed, developmentally down-regulated 4-1
- NEDD4-2
- neural precursor cell–expressed, developmentally down-regulated 4-2
- SGK2
- serum- and glucocorticoid-inducible kinase 2
- SUMO
- small ubiquitin-like modifier
- IGF-1
- insulin-like growth factor-1
- PDZ
- PSD-95/Discs Large/ZO-1
- PDZK1
- PDZ domain–containing 1
- NHERF1
- Na+/H+ exchange regulatory cofactor 1
- CK2
- casein kinase 2.
References
- 1. Nigam, S. K. (2015) What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 10.1038/nrd4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Durmus, S., Hendrikx, J. J., and Schinkel, A. H. (2015) Apical ABC transporters and cancer chemotherapeutic drug disposition. Adv. Cancer Res. 125, 1–41 10.1016/bs.acr.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 3. Crawford, R. R., Potukuchi, P. K., Schuetz, E. G., and Schuetz, J. D. (2018) Beyond competitive inhibition: regulation of ABC transporters by kinases and protein-protein interactions as potential mechanisms of drug-drug interactions. Drug Metab. Dispos. 46, 567–580 10.1124/dmd.118.080663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirota, T., Tanaka, T., Takesue, H., and Ieiri, I. (2017) Epigenetic regulation of drug transporter expression in human tissues. Expert Opin. Drug Metab. Toxicol. 13, 19–30 10.1080/17425255.2017.1230199 [DOI] [PubMed] [Google Scholar]
- 5. Giacomini, K. M., Galetin, A., and Huang, S. M. (2018) The international transporter consortium: summarizing advances in the role of transporters in drug development. Clin. Pharmacol. Ther. 104, 766–771 10.1002/cpt.1224 [DOI] [PubMed] [Google Scholar]
- 6. Shitara, Y., Maeda, K., Ikejiri, K., Yoshida, K., Horie, T., and Sugiyama, Y. (2013) Clinical significance of organic anion transporting polypeptides (oatps) in drug disposition: their roles in hepatic clearance and intestinal absorption. Biopharm. Drug Dispos. 34, 45–78 10.1002/bdd.1823 [DOI] [PubMed] [Google Scholar]
- 7. Yao, Y., Toshimoto, K., Kim, S. J., Yoshikado, T., and Sugiyama, Y. (2018) Quantitative analysis of complex drug-drug interactions between cerivastatin and metabolism/transport inhibitors using physiologically based pharmacokinetic modeling. Drug Metab. Dispos. 46, 924–933 10.1124/dmd.117.079210 [DOI] [PubMed] [Google Scholar]
- 8. Furberg, C. D., and Pitt, B. (2001) Withdrawal of cerivastatin from the world market. Curr. Control Trials Cardiovasc. Med. 2, 205–207 10.1186/cvm-2-5-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turner, R. M., and Pirmohamed, M. (2019) Statin-related myotoxicity: A comprehensive review of pharmacokinetic, pharmacogenomic and muscle components. J. Clin. Med. 9, 22 10.3390/jcm9010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feng, B., and Varma, M. V. (2016) Evaluation and quantitative prediction of renal transporter-mediated drug-drug interactions. J. Clin. Pharmacol. 56, S110–S121 10.1002/jcph.702 [DOI] [PubMed] [Google Scholar]
- 11. Alam, K., Crowe, A., Wang, X., Zhang, P., Ding, K., Li, L., and Yue, W. (2018) Regulation of organic anion transporting polypeptides (oatp) 1b1- and oatp1b3-mediated transport: an updated review in the context of oatp-mediated drug-drug interactions. Int. J. Mol. Sci. 19, 855 10.3390/ijms19030855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. König, J., Müller, F., and Fromm, M. F. (2013) Transporters and drug-drug interactions: important determinants of drug disposition and effects. Pharmacol. Rev. 65, 944–966 10.1124/pr.113.007518 [DOI] [PubMed] [Google Scholar]
- 13. Diallinas, G., and Martzoukou, O. (2019) Transporter membrane traffic and function: lessons from a mould. FEBS J. 286, 4861–4875 10.1111/febs.15078 [DOI] [PubMed] [Google Scholar]
- 14. Czuba, L. C., Hillgren, K. M., and Swaan, P. W. (2018) Post-translational modifications of transporters. Pharmacol. Ther. 192, 88–99 10.1016/j.pharmthera.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang, Y., and Hagenbuch, B. (2019) Protein-protein interactions of drug uptake transporters that are important for liver and kidney. Biochem. Pharmacol. 168, 384–391 10.1016/j.bcp.2019.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murray, M., and Zhou, F. (2017) Trafficking and other regulatory mechanisms for organic anion transporting polypeptides and organic anion transporters that modulate cellular drug and xenobiotic influx and that are dysregulated in disease. Br. J. Pharmacol. 174, 1908–1924 10.1111/bph.13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu, C., Nigam, K. B., Date, R. C., Bush, K. T., Springer, S. A., Saier, M. H., Jr., Wu, W., and Nigam, S. K. (2015) Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLoS ONE 10, e0140569 10.1371/journal.pone.0140569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burckhardt, G. (2012) Drug transport by organic anion transporters (OATs). Pharmacol. Ther. 136, 106–130 10.1016/j.pharmthera.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 19. Zamek-Gliszczynski, M. J., Taub, M. E., Chothe, P. P., Chu, X., Giacomini, K. M., Kim, R. B., Ray, A. S., Stocker, S. L., Unadkat, J. D., Wittwer, M. B., Xia, C., Yee, S. W., Zhang, L., and Zhang, Y., and International Transporter Consortium (2018) Transporters in drug development: 2018 ITC recommendations for transporters of emerging clinical importance. Clin. Pharmacol. Ther. 104, 890–899 10.1002/cpt.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodrigues, A. D., Taskar, K. S., Kusuhara, H., and Sugiyama, Y. (2018) Endogenous probes for drug transporters: balancing vision with reality. Clin. Pharmacol. Ther. 103, 434–448 10.1002/cpt.749 [DOI] [PubMed] [Google Scholar]
- 21. Chu, X., Liao, M., Shen, H., Yoshida, K., Zur, A. A., Arya, V., Galetin, A., Giacomini, K. M., Hanna, I., Kusuhara, H., Lai, Y., Rodrigues, D., Sugiyama, Y., Zamek-Gliszczynski, M. J., and Zhang, L., and International Transporter Consortium (2018) Clinical probes and endogenous biomarkers as substrates for transporter drug-drug interaction evaluation: Perspectives from the international transporter consortium. Clin. Pharmacol. Ther. 104, 836–864 10.1002/cpt.1216 [DOI] [PubMed] [Google Scholar]
- 22. Liu, H. C., Goldenberg, A., Chen, Y., Lun, C., Wu, W., Bush, K. T., Balac, N., Rodriguez, P., Abagyan, R., and Nigam, S. K. (2016) Molecular properties of drugs interacting with SLC22 transporters OAT1, OAT3, OCT1, and OCT2: a machine-learning approach. J. Pharmacol. Exp. Ther. 359, 215–229 10.1124/jpet.116.232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nigam, A. K., Li, J. G., Lall, K., Shi, D., Bush, K. T., Bhatnagar, V., Abagyan, R., and Nigam, S. K. (2020) Unique metabolite preferences of the drug transporters oat1 and oat3 analyzed by machine learning. J. Biol. Chem. 295, 1829–1842 10.1074/jbc.RA119.010729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simonson, G. D., Vincent, A. C., Roberg, K. J., Huang, Y., and Iwanij, V. (1994) Molecular cloning and characterization of a novel liver-specific transport protein. J. Cell Sci. 107, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 25. Sekine, T., Cha, S. H., Tsuda, M., Apiwattanakul, N., Nakajima, N., Kanai, Y., and Endou, H. (1998) Identification of multispecific organic anion transporter 2 expressed predominantly in the liver. FEBS Lett. 429, 179–182 10.1016/S0014-5793(98)00585-7 [DOI] [PubMed] [Google Scholar]
- 26. Shen, H., Lai, Y., and Rodrigues, A. D. (2017) Organic anion transporter 2: an enigmatic human solute carrier. Drug Metab. Dispos. 45, 228–236 10.1124/dmd.116.072264 [DOI] [PubMed] [Google Scholar]
- 27. Ma, Z., Lu, S., Sun, D., Bai, M., Jiang, T., Lin, N., Zhou, H., Zeng, S., and Jiang, H. (2019) Roles of organic anion transporter 2 and equilibrative nucleoside transporter 1 in hepatic disposition and antiviral activity of entecavir during non-pregnancy and pregnancy. Br. J. Pharmacol. 176, 3236–3249 10.1111/bph.14756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henjakovic, M., Hagos, Y., Krick, W., Burckhardt, G., and Burckhardt, B. C. (2015) Human organic anion transporter 2 is distinct from organic anion transporters 1 and 3 with respect to transport function. Am. J. Physiol. Renal Physiol. 309, F843–F851 10.1152/ajprenal.00140.2015 [DOI] [PubMed] [Google Scholar]
- 29. Cha, S. H., Sekine, T., Kusuhara, H., Yu, E., Kim, J. Y., Kim, D. K., Sugiyama, Y., Kanai, Y., and Endou, H. (2000) Molecular cloning and characterization of multispecific organic anion transporter 4 expressed in the placenta. J. Biol. Chem. 275, 4507–4512 10.1074/jbc.275.6.4507 [DOI] [PubMed] [Google Scholar]
- 30. Ekaratanawong, S., Anzai, N., Jutabha, P., Miyazaki, H., Noshiro, R., Takeda, M., Kanai, Y., Sophasan, S., and Endou, H. (2004) Human organic anion transporter 4 is a renal apical organic anion/dicarboxylate exchanger in the proximal tubules. J. Pharmacol. Sci. 94, 297–304 10.1254/jphs.94.297 [DOI] [PubMed] [Google Scholar]
- 31. Ugele, B., St-Pierre, M. V., Pihusch, M., Bahn, A., and Hantschmann, P. (2003) Characterization and identification of steroid sulfate transporters of human placenta. Am. J. Physiol. Endocrinol. Metab. 284, E390–E398 10.1152/ajpendo.00257.2002 [DOI] [PubMed] [Google Scholar]
- 32. Hagos, Y., Stein, D., Ugele, B., Burckhardt, G., and Bahn, A. (2007) Human renal organic anion transporter 4 operates as an asymmetric urate transporter. J. Am. Soc. Nephrol. 18, 430–439 10.1681/ASN.2006040415 [DOI] [PubMed] [Google Scholar]
- 33. Noguchi, S., Nishimura, T., Mukaida, S., Benet, L. Z., Nakashima, E., and Tomi, M. (2017) Cellular uptake of levocetirizine by organic anion transporter 4. J. Pharm. Sci. 106, 2895–2898 10.1016/j.xphs.2017.03.026 [DOI] [PubMed] [Google Scholar]
- 34. Noguchi, S., Nishimura, T., Fujibayashi, A., Maruyama, T., Tomi, M., and Nakashima, E. (2015) Organic anion transporter 4-mediated transport of olmesartan at basal plasma membrane of human placental barrier. J. Pharm. Sci. 104, 3128–3135 10.1002/jps.24434 [DOI] [PubMed] [Google Scholar]
- 35. Iwaki, Y., Lee, W., and Sugiyama, Y. (2019) Comparative and quantitative assessment on statin efficacy and safety: Insights into inter-statin and inter-individual variability via dose- and exposure-response relationships. Expert Opin. Drug Metab. Toxicol. 15, 897–911 10.1080/17425255.2019.1681399 [DOI] [PubMed] [Google Scholar]
- 36. König, J., Cui, Y., Nies, A. T., and Keppler, D. (2000) Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J. Biol. Chem. 275, 23161–23168 10.1074/jbc.M001448200 [DOI] [PubMed] [Google Scholar]
- 37. Ho, R. H., Tirona, R. G., Leake, B. F., Glaeser, H., Lee, W., Lemke, C. J., Wang, Y., and Kim, R. B. (2006) Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology 130, 1793–1806 10.1053/j.gastro.2006.02.034 [DOI] [PubMed] [Google Scholar]
- 38. Abe, T., Unno, M., Onogawa, T., Tokui, T., Kondo, T. N., Nakagomi, R., Adachi, H., Fujiwara, K., Okabe, M., Suzuki, T., Nunoki, K., Sato, E., Kakyo, M., Nishio, T., Sugita, J., et al. (2001) Lst-2, a human liver-specific organic anion transporter, determines methotrexate sensitivity in gastrointestinal cancers. Gastroenterology 120, 1689–1699 10.1053/gast.2001.24804 [DOI] [PubMed] [Google Scholar]
- 39. Lockhart, A. C., Harris, E., Lafleur, B. J., Merchant, N. B., Washington, M. K., Resnick, M. B., Yeatman, T. J., and Lee, W. (2008) Organic anion transporting polypeptide 1B3 (OATP1B3) is overexpressed in colorectal tumors and is a predictor of clinical outcome. Clin. Exp. Gastroenterol. 1, 1–7 10.2147/ceg.s3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thakkar, N., Kim, K., Jang, E. R., Han, S., Kim, K., Kim, D., Merchant, N., Lockhart, A. C., and Lee, W. (2013) A cancer-specific variant of the SLCO1B3 gene encodes a novel human organic anion transporting polypeptide 1B3 (OATP1B3) localized mainly in the cytoplasm of colon and pancreatic cancer cells. Mol. Pharm. 10, 406–416 10.1021/mp3005353 [DOI] [PubMed] [Google Scholar]
- 41. Miki, Y., Suzuki, T., Kitada, K., Yabuki, N., Shibuya, R., Moriya, T., Ishida, T., Ohuchi, N., Blumberg, B., and Sasano, H. (2006) Expression of the steroid and xenobiotic receptor and its possible target gene, organic anion transporting polypeptide-A, in human breast carcinoma. Cancer Res. 66, 535–542 10.1158/0008-5472.CAN-05-1070 [DOI] [PubMed] [Google Scholar]
- 42. Meyer zu Schwabedissen, H. E., Tirona, R. G., Yip, C. S., Ho, R. H., and Kim, R. B. (2008) Interplay between the nuclear receptor pregnane X receptor and the uptake transporter organic anion transporter polypeptide 1A2 selectively enhances estrogen effects in breast cancer. Cancer Res. 68, 9338–9347 10.1158/0008-5472.CAN-08-0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee, W., Glaeser, H., Smith, L. H., Roberts, R. L., Moeckel, G. W., Gervasini, G., Leake, B. F., and Kim, R. B. (2005) Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J. Biol. Chem. 280, 9610–9617 10.1074/jbc.M411092200 [DOI] [PubMed] [Google Scholar]
- 44. Chan, T., Zhu, L., Madigan, M. C., Wang, K., Shen, W., Gillies, M. C., and Zhou, F. (2015) Human organic anion transporting polypeptide 1A2 (OATP1A2) mediates cellular uptake of all-trans-retinol in human retinal pigmented epithelial cells. Br. J. Pharmacol. 172, 2343–2353 10.1111/bph.13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Glaeser, H., Bailey, D. G., Dresser, G. K., Gregor, J. C., Schwarz, U. I., McGrath, J. S., Jolicoeur, E., Lee, W., Leake, B. F., Tirona, R. G., and Kim, R. B. (2007) Intestinal drug transporter expression and the impact of grapefruit juice in humans. Clin. Pharmacol. Ther. 81, 362–370 10.1038/sj.clpt.6100056 [DOI] [PubMed] [Google Scholar]
- 46. Oswald, S. (2019) Organic anion transporting polypeptide (oatp) transporter expression, localization and function in the human intestine. Pharmacol. Ther. 195, 39–53 10.1016/j.pharmthera.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 47. Tamai, I. (2012) Oral drug delivery utilizing intestinal OATP transporters. Adv. Drug Deliv. Rev. 64, 508–514 10.1016/j.addr.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 48. Zou, L., Spanogiannopoulos, P., Pieper, L. M., Chien, H. C., Cai, W., Khuri, N., Pottel, J., Vora, B., Ni, Z., Tsakalozou, E., Zhang, W., Shoichet, B. K., Giacomini, K. M., and Turnbaugh, P. J. (2020) Bacterial metabolism rescues the inhibition of intestinal drug absorption by food and drug additives. Proc. Natl. Acad. Sci. U. S. A. 117, 16009–16018 10.1073/pnas.1920483117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pottel, J., Armstrong, D., Zou, L., Fekete, A., Huang, X. P., Torosyan, H., Bednarczyk, D., Whitebread, S., Bhhatarai, B., Liang, G., Jin, H., Ghaemi, S. N., Slocum, S., Lukacs, K. V., Irwin, J. J., et al. (2020) The activities of drug inactive ingredients on biological targets. Science 369, 403–413 10.1126/science.aaz9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varma, M. V., Rotter, C. J., Chupka, J., Whalen, K. M., Duignan, D. B., Feng, B., Litchfield, J., Goosen, T. C., and El-Kattan, A. F. (2011) pH-sensitive interaction of HMG-CoA reductase inhibitors (statins) with organic anion transporting polypeptide 2b1. Mol. Pharm. 8, 1303–1313 10.1021/mp200103h [DOI] [PubMed] [Google Scholar]
- 51. Nozawa, T., Imai, K., Nezu, J., Tsuji, A., and Tamai, I. (2004) Functional characterization of pH-sensitive organic anion transporting polypeptide OATP-B in human. J. Pharmacol. Exp. Ther. 308, 438–445 10.1124/jpet.103.060194 [DOI] [PubMed] [Google Scholar]
- 52. Taylor-Wells, J., and Meredith, D. (2014) The signature sequence region of the human drug transporter organic anion transporting polypeptide 1B1 is important for protein surface expression. J. Drug Deliv. 2014, 129849 10.1155/2014/129849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kato, Y., Yoshida, K., Watanabe, C., Sai, Y., and Tsuji, A. (2004) Screening of the interaction between xenobiotic transporters and PDZ proteins. Pharm. Res. 21, 1886–1894 10.1023/b:pham.0000045244.83999.43 [DOI] [PubMed] [Google Scholar]
- 54. Mózner, O., Bartos, Z., Zámbó, B., Homolya, L., Hegedűs, T., and Sarkadi, B. (2019) Cellular processing of the abcg2 transporter-potential effects on gout and drug metabolism. Cells 8, 1215 10.3390/cells8101215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosnoblet, C., Peanne, R., Legrand, D., and Foulquier, F. (2013) Glycosylation disorders of membrane trafficking. Glycoconj. J. 30, 23–31 10.1007/s10719-012-9389-y [DOI] [PubMed] [Google Scholar]
- 56. Yau, R., and Rape, M. (2016) The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 10.1038/ncb3358 [DOI] [PubMed] [Google Scholar]
- 57. Preston, G. M., and Brodsky, J. L. (2017) The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem. J. 474, 445–469 10.1042/BCJ20160582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hebert, D. N., and Molinari, M. (2007) In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87, 1377–1408 10.1152/physrev.00050.2006 [DOI] [PubMed] [Google Scholar]
- 59. Markossian, K. A., and Kurganov, B. I. (2004) Protein folding, misfolding, and aggregation. Formation of inclusion bodies and aggresomes. Biochemistry (Mosc.) 69, 971–984 10.1023/B:BIRY.0000043539.07961.4c [DOI] [PubMed] [Google Scholar]
- 60. Cecchetti, C., Pyle, E., and Byrne, B. (2019) Transporter oligomerisation: roles in structure and function. Biochem. Soc. Trans. 47, 433–440 10.1042/BST20180316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Alguel, Y., Cameron, A. D., Diallinas, G., and Byrne, B. (2016) Transporter oligomerization: form and function. Biochem. Soc. Trans. 44, 1737–1744 10.1042/BST20160217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Storch, C. H., Ehehalt, R., Haefeli, W. E., and Weiss, J. (2007) Localization of the human breast cancer resistance protein (BCRP/ABCG2) in lipid rafts/caveolae and modulation of its activity by cholesterol in vitro. J. Pharmacol. Exp. Ther. 323, 257–264 10.1124/jpet.107.122994 [DOI] [PubMed] [Google Scholar]
- 63. Szilagyi, J. T., Vetrano, A. M., Laskin, J. D., and Aleksunes, L. M. (2017) Localization of the placental bcrp/abcg2 transporter to lipid rafts: Role for cholesterol in mediating efflux activity. Placenta 55, 29–36 10.1016/j.placenta.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Veit, G., Avramescu, R. G., Chiang, A. N., Houck, S. A., Cai, Z., Peters, K. W., Hong, J. S., Pollard, H. B., Guggino, W. B., Balch, W. E., Skach, W. R., Cutting, G. R., Frizzell, R. A., Sheppard, D. N., Cyr, D. M., et al. (2016) From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol. Biol. Cell 27, 424–433 10.1091/mbc.E14-04-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Connett, G. J. (2019) Lumacaftor-ivacaftor in the treatment of cystic fibrosis: design, development and place in therapy. Drug Des. Devel. Ther. 13, 2405–2412 10.2147/DDDT.S153719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Clancy, J. P., Cotton, C. U., Donaldson, S. H., Solomon, G. M., VanDevanter, D. R., Boyle, M. P., Gentzsch, M., Nick, J. A., Illek, B., Wallenburg, J. C., Sorscher, E. J., Amaral, M. D., Beekman, J. M., Naren, A. P., Bridges, R. J., et al. (2019) CFTR modulator theratyping: current status, gaps and future directions. J. Cyst. Fibros. 18, 22–34 10.1016/j.jcf.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gonzales, E., Grosse, B., Cassio, D., Davit-Spraul, A., Fabre, M., and Jacquemin, E. (2012) Successful mutation-specific chaperone therapy with 4-phenylbutyrate in a child with progressive familial intrahepatic cholestasis type 2. J. Hepatol. 57, 695–698 10.1016/j.jhep.2012.04.017 [DOI] [PubMed] [Google Scholar]
- 68. Hayashi, H., and Sugiyama, Y. (2007) 4-Phenylbutyrate enhances the cell surface expression and the transport capacity of wild-type and mutated bile salt export pumps. Hepatology 45, 1506–1516 10.1002/hep.21630 [DOI] [PubMed] [Google Scholar]
- 69. Hayashi, H., Takada, T., Suzuki, H., Akita, H., and Sugiyama, Y. (2005) Two common PFIC2 mutations are associated with the impaired membrane trafficking of BSEP/ABCB11. Hepatology 41, 916–924 10.1002/hep.20627 [DOI] [PubMed] [Google Scholar]
- 70. Cleophas, M. C., Joosten, L. A., Stamp, L. K., Dalbeth, N., Woodward, O. M., and Merriman, T. R. (2017) ABCG2 polymorphisms in gout: insights into disease susceptibility and treatment approaches. Pharmgenomics Pers. Med. 10, 129–142 10.2147/PGPM.S105854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoque, K. M., Dixon, E. E., Lewis, R. M., Allan, J., Gamble, G. D., Phipps-Green, A. J., Halperin Kuhns, V. L., Horne, A. M., Stamp, L. K., Merriman, T. R., Dalbeth, N., and Woodward, O. M. (2020) The ABCG2 Q141K hyperuricemia and gout associated variant illuminates the physiology of human urate excretion. Nat. Commun. 11, 2767 10.1038/s41467-020-16525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thakkar, N., Lockhart, A. C., and Lee, W. (2015) Role of organic anion-transporting polypeptides (OATPs) in cancer therapy. AAPS J. 17, 535–545 10.1208/s12248-015-9740-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li, T. T., An, J. X., Xu, J. Y., and Tuo, B. G. (2019) Overview of organic anion transporters and organic anion transporter polypeptides and their roles in the liver. World J. Clin. Cases 7, 3915–3933 10.12998/wjcc.v7.i23.3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Evers, R., Piquette-Miller, M., Polli, J. W., Russel, F. G. M., Sprowl, J. A., Tohyama, K., Ware, J. A., de Wildt, S. N., Xie, W., Brouwer, K. L. R., and International Transporter Consortium (2018) Disease-associated changes in drug transporters may impact the pharmacokinetics and/or toxicity of drugs: a white paper from the International Transporter Consortium. Clin. Pharmacol. Ther. 104, 900–915 10.1002/cpt.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hunter, T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 76. Mayati, A., Moreau, A., Le Vee, M., Stieger, B., Denizot, C., Parmentier, Y., and Fardel, O. (2017) Protein kinases C-mediated regulations of drug transporter activity, localization and expression. Int. J. Mol. Sci. 18, 764 10.3390/ijms18040764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ciechanover, A. (2005) Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87 10.1038/nrm1552 [DOI] [PubMed] [Google Scholar]
- 78. Zheng, N., and Shabek, N. (2017) Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- 79. MacGurn, J. A., Hsu, P. C., and Emr, S. D. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259 10.1146/annurev-biochem-060210-093619 [DOI] [PubMed] [Google Scholar]
- 80. Tanaka, K., Xu, W., Zhou, F., and You, G. (2004) Role of glycosylation in the organic anion transporter OAT1. J. Biol. Chem. 279, 14961–14966 10.1074/jbc.M400197200 [DOI] [PubMed] [Google Scholar]
- 81. Zhang, Q., Hong, M., Duan, P., Pan, Z., Ma, J., and You, G. (2008) Organic anion transporter OAT1 undergoes constitutive and protein kinase c-regulated trafficking through a dynamin- and clathrin-dependent pathway. J. Biol. Chem. 283, 32570–32579 10.1074/jbc.M800298200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li, S., Zhang, Q., and You, G. (2013) Three ubiquitination sites of organic anion transporter-1 synergistically mediate protein kinase c-dependent endocytosis of the transporter. Mol. Pharmacol. 84, 139–146 10.1124/mol.113.086769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang, Q., Li, S., Patterson, C., and You, G. (2013) Lysine 48-linked polyubiquitination of organic anion transporter-1 is essential for its protein kinase C-regulated endocytosis. Mol. Pharmacol. 83, 217–224 10.1124/mol.112.082065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu, D., Wang, H., Zhang, Q., and You, G. (2016) Nedd4-2 but not Nedd4-1 is critical for protein kinase C-regulated ubiquitination, expression, and transport activity of human organic anion transporter 1. Am. J. Physiol. Renal Physiol. 310, F821–F831 10.1152/ajprenal.00522.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xu, D., Zhang, J., Zhang, Q., Fan, Y., Liu, C., and You, G. (2017) PKC/Nedd4-2 signaling pathway regulates the cell surface expression of drug transporter hOAT1. Drug Metab. Dispos. 45, 887–895 10.1124/dmd.117.075861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang, H., Xu, D., Toh, M. F., Pao, A. C., and You, G. (2016) Serum- and glucocorticoid-inducible kinase SGK2 regulates human organic anion transporters 4 via ubiquitin ligase Nedd4-2. Biochem. Pharmacol. 102, 120–129 10.1016/j.bcp.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fan, Y., and You, G. (2020) Proteasome inhibitors bortezomib and carfilzomib stimulate the transport activity of human organic anion transporter 1. Mol. Pharmacol. 97, 384–391 10.1124/mol.119.118653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hong, M., Xu, W., Yoshida, T., Tanaka, K., Wolff, D. J., Zhou, F., Inouye, M., and You, G. (2005) Human organic anion transporter hOAT1 forms homooligomers. J. Biol. Chem. 280, 32285–32290 10.1074/jbc.M501447200 [DOI] [PubMed] [Google Scholar]
- 89. Duan, P., Li, S., and You, G. (2011) Transmembrane peptide as potent inhibitor of oligomerization and function of human organic anion transporter 1. Mol. Pharmacol. 79, 569–574 10.1124/mol.110.070185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Duan, P., Wu, J., and You, G. (2011) Mutational analysis of the role of GXXXG motif in the function of human organic anion transporter 1 (hOAT1). Int. J. Biochem. Mol. Biol. 2, 1–7 [PMC free article] [PubMed] [Google Scholar]
- 91. Cropp, C. D., Komori, T., Shima, J. E., Urban, T. J., Yee, S. W., More, S. S., and Giacomini, K. M. (2008) Organic anion transporter 2 (SLC22A7) is a facilitative transporter of cGMP. Mol. Pharmacol. 73, 1151–1158 10.1124/mol.107.043117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hotchkiss, A. G., Berrigan, L., and Pelis, R. M. (2015) Organic anion transporter 2 transcript variant 1 shows broad ligand selectivity when expressed in multiple cell lines. Front. Pharmacol. 6, 216 10.3389/fphar.2015.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Srimaroeng, C., Perry, J. L., and Pritchard, J. B. (2008) Physiology, structure, and regulation of the cloned organic anion transporters. Xenobiotica 38, 889–935 10.1080/00498250801927435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Duan, P., Li, S., and You, G. (2010) Angiotensin ii inhibits activity of human organic anion transporter 3 through activation of protein kinase Cα: accelerating endocytosis of the transporter. Eur. J. Pharmacol. 627, 49–55 10.1016/j.ejphar.2009.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu, D., Wang, H., and You, G. (2016) An essential role of Nedd4-2 in the ubiquitination, expression, and function of organic anion transporter-3. Mol. Pharm. 13, 621–630 10.1021/acs.molpharmaceut.5b00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang, H., Zhang, J., and You, G. (2019) Activation of protein kinase a stimulates sumoylation, expression, and transport activity of organic anion transporter 3. AAPS J. 21, 30 10.1208/s12248-019-0303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang, J., Yu, Z., and You, G. (2020) Insulin-like growth factor 1 modulates the phosphorylation, expression, and activity of organic anion transporter 3 through protein kinase a signaling pathway. Acta Pharm. Sin. B 10, 186–194 10.1016/j.apsb.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang, H., and You, G. (2019) The SUMO-specific protease Senp2 regulates SUMOylation, expression and function of human organic anion transporter 3. Biochim. Biophys. Acta Biomembr. 1861, 1293–1301 10.1016/j.bbamem.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Srimaroeng, C., Cecile, J. P., Walden, R., and Pritchard, J. B. (2013) Regulation of renal organic anion transporter 3 (SLC22A8) expression and function by the integrity of lipid raft domains and their associated cytoskeleton. Cell. Physiol. Biochem. 31, 565–578 10.1159/000350077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nosetto, E. C., Campagno, R. V., Torres, A. M., and Brandoni, A. (2020) Distribution of the organic anion transporters Oat1 and Oat3 between renal membrane microdomains in obstructive jaundice. Pflugers Arch. 472, 711–719 10.1007/s00424-020-02402-4 [DOI] [PubMed] [Google Scholar]
- 101. Zhou, F., Hong, M., and You, G. (2007) Regulation of human organic anion transporter 4 by progesterone and protein kinase C in human placental BeWo cells. Am. J. Physiol. Endocrinol. Metab. 293, E57–E61 10.1152/ajpendo.00696.2006 [DOI] [PubMed] [Google Scholar]
- 102. Zhang, Q., Pan, Z., and You, G. (2010) Regulation of human organic anion transporter 4 by protein kinase C and NHERF-1: altering the endocytosis of the transporter. Pharm. Res. 27, 589–596 10.1007/s11095-009-9983-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Duan, P., Li, S., and You, G. (2012) Regulation of human organic anion transporter 4 by parathyroid hormone-related protein and protein kinase a. Int. J. Biochem. Mol. Biol. 3, 322–327 [PMC free article] [PubMed] [Google Scholar]
- 104. Wang, H., Zhang, J., and You, G. (2019) The mechanistic links between insulin and human organic anion transporter 4. Int. J. Pharm. 555, 165–174 10.1016/j.ijpharm.2018.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Miyazaki, H., Anzai, N., Ekaratanawong, S., Sakata, T., Shin, H. J., Jutabha, P., Hirata, T., He, X., Nonoguchi, H., Tomita, K., Kanai, Y., and Endou, H. (2005) Modulation of renal apical organic anion transporter 4 function by two PDZ domain-containing proteins. J. Am. Soc. Nephrol. 16, 3498–3506 10.1681/ASN.2005030306 [DOI] [PubMed] [Google Scholar]
- 106. Zhou, F., Xu, W., Tanaka, K., and You, G. (2008) Comparison of the interaction of human organic anion transporter hOAT4 with PDZ proteins between kidney cells and placental cells. Pharm. Res. 25, 475–480 10.1007/s11095-007-9359-4 [DOI] [PubMed] [Google Scholar]
- 107. Choi, J. H., Murray, J. W., and Wolkoff, A. W. (2011) PDZK1 binding and serine phosphorylation regulate subcellular trafficking of organic anion transport protein 1a1. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G384–G393 10.1152/ajpgi.00500.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Centonze, M., Saponaro, C., and Mangia, A. (2018) Nherf1 between promises and hopes: overview on cancer and prospective openings. Transl. Oncol. 11, 374–390 10.1016/j.tranon.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gillette, J. R., and Pang, K. S. (1977) Theoretic aspects of pharmacokinetic drug interactions. Clin. Pharmacol. Ther. 22, 623–639 10.1002/cpt1977225part2623 [DOI] [PubMed] [Google Scholar]
- 110. Roberts, M. S., and Rowland, M. (1986) A dispersion model of hepatic elimination: 2. Steady-state considerations–influence of hepatic blood flow, binding within blood, and hepatocellular enzyme activity. J. Pharmacokinet. Biopharm. 14, 261–288 10.1007/BF01106707 [DOI] [PubMed] [Google Scholar]
- 111. Tirona, R. G., Leake, B. F., Merino, G., and Kim, R. B. (2001) Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 276, 35669–35675 10.1074/jbc.M103792200 [DOI] [PubMed] [Google Scholar]
- 112. Kameyama, Y., Yamashita, K., Kobayashi, K., Hosokawa, M., and Chiba, K. (2005) Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet. Genomics 15, 513–522 10.1097/01.fpc.0000170913.73780.5f [DOI] [PubMed] [Google Scholar]
- 113. Crowe, A., Zheng, W., Miller, J., Pahwa, S., Alam, K., Fung, K. M., Rubin, E., Yin, F., Ding, K., and Yue, W. (2019) Characterization of plasma membrane localization and phosphorylation status of organic anion transporting polypeptide (OATP) 1B1 c.521 T>C nonsynonymous single-nucleotide polymorphism. Pharm. Res. 36, 101 10.1007/s11095-019-2634-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yao, J., Hong, W., Huang, J., Zhan, K., Huang, H., and Hong, M. (2012) N-Glycosylation dictates proper processing of organic anion transporting polypeptide 1B1. PLoS ONE 7, e52563 10.1371/journal.pone.0052563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Clarke, J. D., Novak, P., Lake, A. D., Hardwick, R. N., and Cherrington, N. J. (2017) Impaired N-linked glycosylation of uptake and efflux transporters in human non-alcoholic fatty liver disease. Liver Int. 37, 1074–1081 10.1111/liv.13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hong, M., Hong, W., Ni, C., Huang, J., and Zhou, C. (2015) Protein kinase C affects the internalization and recycling of organic anion transporting polypeptide 1B1. Biochim. Biophys. Acta 1848, 2022–2030 10.1016/j.bbamem.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 117. Mayati, A., Le Vee, M., Moreau, A., Jouan, E., Bucher, S., Stieger, B., Denizot, C., Parmentier, Y., and Fardel, O. (2015) Protein kinase C-dependent regulation of human hepatic drug transporter expression. Biochem. Pharmacol. 98, 703–717 10.1016/j.bcp.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 118. Bian, Y., Song, C., Cheng, K., Dong, M., Wang, F., Huang, J., Sun, D., Wang, L., Ye, M., and Zou, H. (2014) An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics 96, 253–262 10.1016/j.jprot.2013.11.014 [DOI] [PubMed] [Google Scholar]
- 119. Farasyn, T., Crowe, A., Hatley, O., Neuhoff, S., Alam, K., Kanyo, J., Lam, T. T., Ding, K., and Yue, W. (2019) Preincubation with everolimus and sirolimus reduces organic anion-transporting polypeptide (OATP)1B1- and 1B3-mediated transport independently of mTOR kinase inhibition: Implication in assessing OATP1B1- and OATP1B3-mediated drug-drug interactions. J. Pharm. Sci. 108, 3443–3456 10.1016/j.xphs.2019.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Alam, K., Pahwa, S., Wang, X., Zhang, P., Ding, K., Abuznait, A. H., Li, L., and Yue, W. (2016) Downregulation of organic anion transporting polypeptide (OATP) 1B1 transport function by lysosomotropic drug chloroquine: implication in OATP-mediated drug-drug interactions. Mol. Pharm. 13, 839–851 10.1021/acs.molpharmaceut.5b00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Alam, K., Farasyn, T., Crowe, A., Ding, K., and Yue, W. (2017) Treatment with proteasome inhibitor bortezomib decreases organic anion transporting polypeptide (OATP) 1B3-mediated transport in a substrate-dependent manner. PLoS ONE 12, e0186924 10.1371/journal.pone.0186924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang, P., Kim, R. B., Chowdhury, J. R., and Wolkoff, A. W. (2003) The human organic anion transport protein SLC21A6 is not sufficient for bilirubin transport. J. Biol. Chem. 278, 20695–20699 10.1074/jbc.M301100200 [DOI] [PubMed] [Google Scholar]
- 123. Ni, C., Yu, X., Fang, Z., Huang, J., and Hong, M. (2017) Oligomerization study of human organic anion transporting polypeptide 1B1. Mol. Pharm. 14, 359–367 10.1021/acs.molpharmaceut.6b00649 [DOI] [PubMed] [Google Scholar]
- 124. Letschert, K., Keppler, D., and Konig, J. (2004) Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8). Pharmacogenetics 14, 441–452 10.1097/01.fpc.0000114744.08559.92 [DOI] [PubMed] [Google Scholar]
- 125. Schwarz, U. I., Meyer zu Schwabedissen, H. E., Tirona, R. G., Suzuki, A., Leake, B. F., Mokrab, Y., Mizuguchi, K., Ho, R. H., and Kim, R. B. (2011) Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenet. Genomics 21, 103–114 10.1097/FPC.0b013e328342f5b1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Miura, M., Satoh, S., Inoue, K., Kagaya, H., Saito, M., Inoue, T., Suzuki, T., and Habuchi, T. (2007) Influence of SLCO1B1, 1B3, 2B1 and ABCC2 genetic polymorphisms on mycophenolic acid pharmacokinetics in Japanese renal transplant recipients. Eur. J. Clin. Pharmacol. 63, 1161–1169 10.1007/s00228-007-0380-7 [DOI] [PubMed] [Google Scholar]
- 127. Tague, L. K., Byers, D. E., Hachem, R., Kreisel, D., Krupnick, A. S., Kulkarni, H. S., Chen, C., Huang, H. J., and Gelman, A. (2020) Impact of SLCO1B3 polymorphisms on clinical outcomes in lung allograft recipients receiving mycophenolic acid. Pharmacogenomics J. 20, 69–79 10.1038/s41397-019-0086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chun, S. E., Thakkar, N., Oh, Y., Park, J. E., Han, S., Ryoo, G., Hahn, H., Maeng, S. H., Lim, Y. R., Han, B. W., and Lee, W. (2017) The N-terminal region of organic anion transporting polypeptide 1B3 (OATP1B3) plays an essential role in regulating its plasma membrane trafficking. Biochem. Pharmacol. 131, 98–105 10.1016/j.bcp.2017.02.013 [DOI] [PubMed] [Google Scholar]
- 129. Xu, C., Zhu, L., Chan, T., Lu, X., Shen, W., Madigan, M. C., Gillies, M. C., and Zhou, F. (2016) Chloroquine and hydroxychloroquine are novel inhibitors of human organic anion transporting polypeptide 1A2. J. Pharm. Sci. 105, 884–890 10.1002/jps.24663 [DOI] [PubMed] [Google Scholar]
- 130. Zhang, Y., Boxberger, K. H., and Hagenbuch, B. (2017) Organic anion transporting polypeptide 1B3 can form homo- and hetero-oligomers. PLoS ONE 12, e0180257 10.1371/journal.pone.0180257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Molina, H., Azocar, L., Ananthanarayanan, M., Arrese, M., and Miquel, J. F. (2008) Localization of the sodium-taurocholate cotransporting polypeptide in membrane rafts and modulation of its activity by cholesterol in vitro. Biochim. Biophys. Acta 1778, 1283–1291 10.1016/j.bbamem.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 132. Zhou, F., Zheng, J., Zhu, L., Jodal, A., Cui, P. H., Wong, M., Gurney, H., Church, W. B., and Murray, M. (2013) Functional analysis of novel polymorphisms in the human SLCO1A2 gene that encodes the transporter OATP1A2. AAPS J. 15, 1099–1108 10.1208/s12248-013-9515-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chan, T., Zheng, J., Zhu, L., Grewal, T., Murray, M., and Zhou, F. (2015) Putative transmembrane domain 6 of the human organic anion transporting polypeptide 1A2 (OATP1A2) influences transporter substrate binding, protein trafficking, and quality control. Mol. Pharm. 12, 111–119 10.1021/mp500459b [DOI] [PubMed] [Google Scholar]
- 134. Zheng, J., Chan, T., Cheung, F. S., Zhu, L., Murray, M., and Zhou, F. (2014) PDZK1 and NHERF1 regulate the function of human organic anion transporting polypeptide 1A2 (OATP1A2) by modulating its subcellular trafficking and stability. PLoS ONE 9, e94712 10.1371/journal.pone.0094712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhou, F., Lee, A. C., Krafczyk, K., Zhu, L., and Murray, M. (2011) Protein kinase C regulates the internalization and function of the human organic anion transporting polypeptide 1A2. Br. J. Pharmacol. 162, 1380–1388 10.1111/j.1476-5381.2010.01144.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chan, T., Cheung, F. S., Zheng, J., Lu, X., Zhu, L., Grewal, T., Murray, M., and Zhou, F. (2016) Casein kinase 2 is a novel regulator of the human organic anion transporting polypeptide 1A2 (OATP1A2) trafficking. Mol. Pharm. 13, 144–154 10.1021/acs.molpharmaceut.5b00576 [DOI] [PubMed] [Google Scholar]
- 137. Münstermann, U., Fritz, G., Seitz, G., Lu, Y. P., Schneider, H. R., and Issinger, O. G. (1990) Casein kinase II is elevated in solid human tumours and rapidly proliferating non-neoplastic tissue. Eur. J. Biochem. 189, 251–257 10.1111/j.1432-1033.1990.tb15484.x [DOI] [PubMed] [Google Scholar]
- 138. Lu, X., Chan, T., Cheng, Z., Shams, T., Zhu, L., Murray, M., and Zhou, F. (2018) The 5′-AMP-activated protein kinase regulates the function and expression of human organic anion transporting polypeptide 1A2. Mol. Pharmacol. 94, 1412–1420 10.1124/mol.118.113423 [DOI] [PubMed] [Google Scholar]
- 139. Namgoong, S., Cheong, H. S., Kim, J. O., Kim, L. H., Na, H. S., Koh, I. S., Chung, M. W., and Shin, H. D. (2015) Comparison of genetic variations of the SLCO1B1, SLCO1B3, and SLCO2B1 genes among five ethnic groups. Environ. Toxicol. Pharmacol. 40, 692–697 10.1016/j.etap.2015.08.033 [DOI] [PubMed] [Google Scholar]
- 140. Nozawa, T., Nakajima, M., Tamai, I., Noda, K., Nezu, J., Sai, Y., Tsuji, A., and Yokoi, T. (2002) Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J. Pharmacol. Exp. Ther. 302, 804–813 10.1124/jpet.302.2.804 [DOI] [PubMed] [Google Scholar]
- 141. Imanaga, J., Kotegawa, T., Imai, H., Tsutsumi, K., Yoshizato, T., Ohyama, T., Shirasaka, Y., Tamai, I., Tateishi, T., and Ohashi, K. (2011) The effects of the SLCO2B1 c.1457C > T polymorphism and apple juice on the pharmacokinetics of fexofenadine and midazolam in humans. Pharmacogenet. Genomics 21, 84–93 [DOI] [PubMed] [Google Scholar]
- 142. Mougey, E. B., Lang, J. E., Wen, X., and Lima, J. J. (2011) Effect of citrus juice and SLCO2B1 genotype on the pharmacokinetics of montelukast. J. Clin. Pharmacol. 51, 751–760 10.1177/0091270010374472 [DOI] [PubMed] [Google Scholar]
- 143. Ieiri, I., Doi, Y., Maeda, K., Sasaki, T., Kimura, M., Hirota, T., Chiyoda, T., Miyagawa, M., Irie, S., Iwasaki, K., and Sugiyama, Y. (2012) Microdosing clinical study: pharmacokinetic, pharmacogenomic (SLCO2B1), and interaction (grapefruit juice) profiles of celiprolol following the oral microdose and therapeutic dose. J. Clin. Pharmacol. 52, 1078–1089 10.1177/0091270011408612 [DOI] [PubMed] [Google Scholar]
- 144. Hänggi, E., Grundschober, A. F., Leuthold, S., Meier, P. J., and St-Pierre, M. V. (2006) Functional analysis of the extracellular cysteine residues in the human organic anion transporting polypeptide, OATP2B1. Mol. Pharmacol. 70, 806–817 10.1124/mol.105.019547 [DOI] [PubMed] [Google Scholar]
- 145. Köck, K., Koenen, A., Giese, B., Fraunholz, M., May, K., Siegmund, W., Hammer, E., Völker, U., Jedlitschky, G., Kroemer, H. K., and Grube, M. (2010) Rapid modulation of the organic anion transporting polypeptide 2B1 (OATP2B1, SLCO2B1) function by protein kinase C-mediated internalization. J. Biol. Chem. 285, 11336–11347 10.1074/jbc.M109.056457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fang, Z., Huang, J., Chen, J., Xu, S., Xiang, Z., and Hong, M. (2018) Transmembrane domain 1 of human organic anion transporting polypeptide 2b1 is essential for transporter function and stability. Mol. Pharmacol. 94, 842–849 10.1124/mol.118.111914 [DOI] [PubMed] [Google Scholar]
- 147. Ferreira, C., Hagen, P., Stern, M., Hussner, J., Zimmermann, U., Grube, M., and Meyer Zu Schwabedissen, H. E. (2018) The scaffold protein PDZK1 modulates expression and function of the organic anion transporting polypeptide 2B1. Eur. J. Pharm. Sci. 120, 181–190 10.1016/j.ejps.2018.05.006 [DOI] [PubMed] [Google Scholar]
- 148. Garibsingh, R. A., and Schlessinger, A. (2019) Advances and challenges in rational drug design for SLCS. Trends Pharmacol. Sci. 40, 790–800 10.1016/j.tips.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kumar, V., Prasad, B., Patilea, G., Gupta, A., Salphati, L., Evers, R., Hop, C. E., and Unadkat, J. D. (2015) Quantitative transporter proteomics by liquid chromatography with tandem mass spectrometry: addressing methodologic issues of plasma membrane isolation and expression-activity relationship. Drug Metab. Dispos. 43, 284–288 10.1124/dmd.114.061614 [DOI] [PubMed] [Google Scholar]
- 150. Ohtsuki, S., Hirayama, M., Ito, S., Uchida, Y., Tachikawa, M., and Terasaki, T. (2014) Quantitative targeted proteomics for understanding the blood-brain barrier: towards pharmacoproteomics. Expert Rev. Proteomics 11, 303–313 10.1586/14789450.2014.893830 [DOI] [PubMed] [Google Scholar]
- 151. Zhou, F., Zhu, L., Wang, K., and Murray, M. (2017) Recent advance in the pharmacogenomics of human solute carrier transporters (SLCS) in drug disposition. Adv. Drug Deliv. Rev. 116, 21–36 10.1016/j.addr.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 152. Walker, N., Filis, P., Soffientini, U., Bellingham, M., O'Shaughnessy, P. J., and Fowler, P. A. (2017) Placental transporter localization and expression in the human: the importance of species, sex, and gestational age differences. Biol. Reprod. 96, 733–742 10.1093/biolre/iox012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hagenbuch, B., and Stieger, B. (2013) The SLCO (former SLC21) superfamily of transporters. Mol. Aspects Med. 34, 396–412 10.1016/j.mam.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. International Transporter Consortium, Giacomini, K. M., Huang, S. M., Tweedie, D. J., Benet, L. Z., Brouwer, K. L., Chu, X., Dahlin, A., Evers, R., Fischer, V., Hillgren, K. M., Hoffmaster, K. A., Ishikawa, T., Keppler, D., Kim, R. B., et al. (2010) Membrane transporters in drug development. Nat. Rev. Drug Discov. 9, 215–236 10.1038/nrd3028 [DOI] [PMC free article] [PubMed] [Google Scholar]