Abstract
Protein biosynthesis is fundamental to cellular life and requires the efficient functioning of the translational machinery. At the center of this machinery is the ribosome, a ribonucleoprotein complex that depends heavily on Mg2+ for structure. Recent work has indicated that other metal cations can substitute for Mg2+, raising questions about the role different metals may play in the maintenance of the ribosome under oxidative stress conditions. Here, we assess ribosomal integrity following oxidative stress both in vitro and in cells to elucidate details of the interactions between Fe2+ and the ribosome and identify Mn2+ as a factor capable of attenuating oxidant-induced Fe2+-mediated degradation of rRNA. We report that Fe2+ promotes degradation of all rRNA species of the yeast ribosome and that it is bound directly to RNA molecules. Furthermore, we demonstrate that Mn2+ competes with Fe2+ for rRNA-binding sites and that protection of ribosomes from Fe2+-mediated rRNA hydrolysis correlates with the restoration of cell viability. Our data, therefore, suggest a relationship between these two transition metals in controlling ribosome stability under oxidative stress.
Keywords: ribosomal ribonucleic acid (rRNA), RNA folding, ribosome, ribosome structure, degradation of ribosomes, oxidative stress, oxidants, iron, iron metabolism, metals, manganese, yeast, cell viability
All organisms require a number of metal elements in trace amounts, with manganese, iron, copper, zinc, selenium, cobalt, and molybdenum all considered essential for plants and animals, whereas larger amounts of magnesium, calcium, potassium, and sodium are also required (1). Divalent metal cations have a long-established involvement in biomolecules including stabilizing structures and participating in the active sites of enzymes operating across a vast catalytic range (2–4).
Enzymes coordinating divalent metal ions act in important processes such as DNA replication (5–7), cellular metabolism and respiration (8, 9), the phosphorylation underpinning much of cellular signaling (10), the oxygen-evolving function of photosystem II (11), and the oxygen-transporting function of hemoglobin (12). Additionally, metal cations have essential stabilizing roles in structures including phospholipid bilayers (13) and the ribosome (14). Although a given binding site may exhibit a preference for a given ion, competition between and substitution of ions occurs throughout metal-coordinating biomolecules with a variety of effects (15–18). Many questions remain unresolved regarding how a preferred metal cofactor is selected and whether promiscuous coordination of ions is advantageous (19).
Among biological molecules that utilize divalent metal cations, magnesium is the most common (20–22), because of a combination of its abundance and amenable chemical properties such as its small radius and lack of redox activity (7, 23). Although other divalent cations may be similar enough to be coordinated in place of Mg2+, the differing properties can impact the biomolecule to which they are bound. For example, kinases that usually utilize Mg2+ can associate with other trace metal ions but suffer a loss in efficiency (10). DNA polymerases require divalent metal cations, and most often employ Mg2+ in this role. Coordinating other metal cofactors including Mn2+ and Co2+ can increase activity, but negatively affect fidelity, and can be carcinogenic as a result (5–7).
The ribosome is a massive ribonucleoprotein complex that is particularly dependent on Mg2+ (14) to fold and maintain stability because of the negative charge of the RNA backbone, and as many as 200 Mg2+ ions can be associated with just the large subunit, coordinated in six distinct geometries (23–25). How other ions with different properties are able to promote RNA folding is not well-understood (26). Ribosomal Mg2+ can be substituted with other divalent cations, including Fe2+ or Mn2+ (27–30), and the ribosome can competently mediate translation in this state (30).
The intricately folded rRNA (rRNA) of the functional ribozyme core has remained largely the same since it evolved 3–4 billion years ago (31, 32). Dozens of ribosomal proteins (r-proteins) interact with rRNA residues to assemble compact, flexible, and stable subunits (33), which are abundant and long-lived in a cytoplasmic environment populated with RNases. However, in physiologically challenging environments such as nutrient scarcity or oxidative stress, the ribosome can lose stability and undergo degradation (34, 35), but the mechanisms initiating this process are still not well-understood.
Recent work has drawn on the interesting observation that although evolution has conserved the ribosomal core through billions of years, the earth environment has changed dramatically over this time and with it the bioavailability of the metals on which the ribosome structure depends. Metal levels in the aqueous environment prior to abiogenesis included high concentrations of Fe2+ (36), whereas Mg2+ was less abundant than today (30, 37). With the evolution of photosynthesis and the increase in molecular oxygen, much of the iron was lost from the aqueous environment through precipitation (38). Similarly, abundant Mn2+ was oxidized and precipitated out, resulting in vast amounts of both transition metals in sedimentary rocks (39). Therefore, the interaction of Fe2+ with rRNAs presents a potential stabilizing interaction used by ancient ribosomes in the differing conditions of the earth when the core structure evolved, a role that was then filled by Mg2+ as Fe2+ availability decreased. Although this establishes Fe2+ and Mn2+ interactions with the ribosome as evolutionarily relevant, little is known about these associations in the present-day oxidative environment and how ribosomal stability is impacted.
As well as the drop in available iron, the oxygenation of the atmosphere produced a further reason for the translation machinery to select another cofactor. The proclivity for Fe2+ to participate in the Fenton reaction with H2O2 (which produces hydroxyl radicals and Fe3+) is of consequence in cells under oxidative stress conditions because it can enhance oxidant-induced damage to biomolecules. In an increasingly oxygen-rich environment, continuous close associations with Fe2+ would have become a dangerous proposition (30). Conversely, the higher reduction potential of manganese prevents its participation in Fenton chemistry (40). Aside from this difference, the biochemistry of these transition metal ions is linked by several similar parameters (including oxidation states, atomic radius, and preferred coordination geometries), as well as their abundant bioavailability in the environment (16, 18, 30).
In our previous work, we have demonstrated that Fe2+ is a factor that contributes to destabilization of ribosomes and provided evidence that it promotes chemical hydrolysis of rRNA. Based on the structural predictions of the ES7L region of the 60S subunit (41, 42), we proposed that Fe2+ replaces Mg2+ at specific binding sites on a ribosome (41) and that these sites become primed to undergo rapid hydrolysis upon exposure to ROS. In agreement with this model, cellular systems that limit the amounts of either cellular oxidant species or ribosome-associated Fe2+ act to protect ribosomal integrity (35, 41, 43).
Here we add detail to the understanding of divalent cation interactions with the ribosome by demonstrating that Fe2+ interacts directly with rRNA and in the presence of oxidants is sufficient to degrade all yeast rRNA molecules in reproducible patterns. Moreover, we identify Mn2+ as a factor that can protect the rRNA from these cleavage events by substituting Fe2+ at the ribosomal binding sites, which correlates with improved cell viability under oxidative stress.
Results
ROS-induced degradation of rRNA is mediated by Fe2+ throughout the ribosome
We have demonstrated in a previous study (41) that Fe2+ has a direct role in cleaving the sugar-phosphate backbone of rRNA under oxidative stress conditions. Specifically, one prominent cleavage site was observed within the ES7L region of the 25S rRNA in the large (60S) ribosomal subunit. To further understand the roles of Fe2+ in rRNA degradation, we chose to investigate whether an iron-dependent oxidative mechanism is responsible for the fragmentation of the 5S and 5.8S rRNAs of the 60S subunit or the 18S rRNA of the 40S subunit. For these experiments, we employed a strain deleted for the gene encoding the mitochondrial glutaredoxin Grx5, loss of which results in increased endogenous levels of labile iron in the cytoplasm (44, 45) and has proved to be a useful system for modeling the effects of increased Fe2+ availability in cells. In our previous work, we observed that all rRNAs are unstable in a grx5Δ mutant (41), but it remained unclear whether the detected cleavages resulted primarily from enzymatic digestion by ribonucleases or Fe2+-dependent nonenzymatic reaction mechanisms.
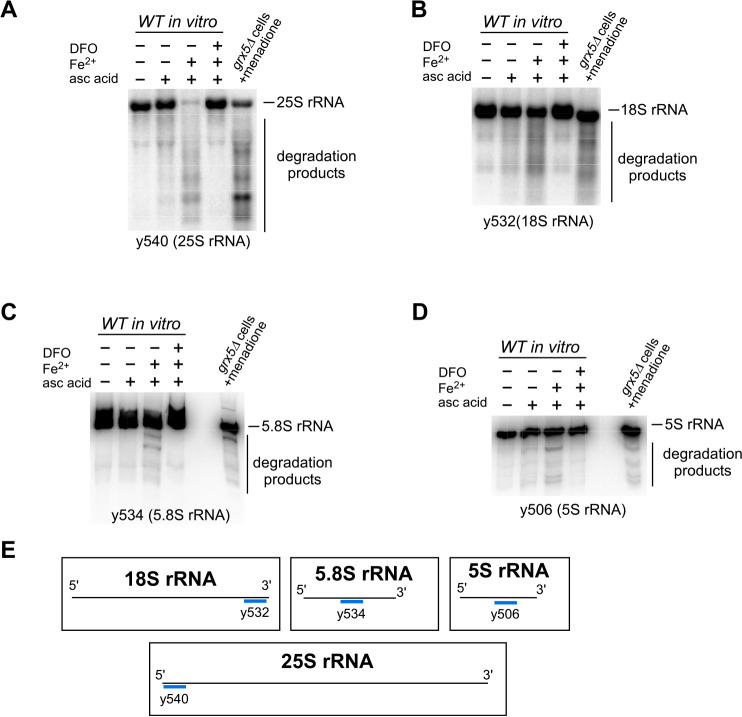
To investigate the extent of fragmentation of different rRNA species by a Fe2+-dependent mechanism, we utilized an in vitro Fe2+/ascorbic acid assay that was developed as part of our previous study (41), further validation of which is shown in Fig. S1. In this assay, we use purified ribosomes as a substrate, ammonium iron(II) sulfate hexahydrate (Fe(NH4)2(SO4)2) to supply extra Fe2+ ions, whereas ascorbic acid is used for its ability to generate H2O2 by inducing redox cycling at metal-binding sites (46, 47). Cellular lysates from exponentially growing WT cells were centrifuged through a sucrose cushion to obtain purified ribosomes, which were treated with ascorbic acid alone, ascorbic acid in combination with Fe(NH4)2(SO4)2, or ascorbic acid with both Fe(NH4)2(SO4)2 and the iron chelator deferoxamine (DFO). In a complementary experiment, yeast grx5Δ cells were subjected to oxidative stress by treatment with menadione before RNA extraction. RNA samples from the in vitro Fe2+/ascorbic acid assay and those from menadione-treated grx5Δ cells were resolved adjacent to one another on the same gel and analyzed by Northern hybridization with probes for each of the four rRNAs (Fig. 1, A–D and probe hybridization schematic in E).
Figure 1.
Ribosomal RNAs from 40S and 60S subunits are degraded in the presence of Fe2+ and ROS both in vitro and in cells. A–D, Northern blotting analysis of in vitro cleavage of rRNA purified from WT cells treated with 0.5 mm ascorbic acid in the presence or absence of 1 μm Fe(NH4)2(SO4)2, using probes for 25S (y540, A), 18S (y532, B), 5.8S (y534, C), and 5S (y506, D) rRNAs. Where indicated, the reactions were pretreated with iron chelator DFO (0.5 mm). In vitro reaction products were resolved in seven repeats on 1.2% agarose–formaldehyde gels (two repeats are shown in A and B; five repeats are shown on Fig. S2A) or in two repeats on 8% polyacrylamide, 8 m urea-containing gels (C and D). Adjacent to these is total RNA from grx5Δ cells that were treated with 50 μm menadione for 2 h at 30 °C. The sequences of all probes used in this study are listed in Table S1. Representative hybridizations are shown. E, schematic representation of the annealing sites of the probes used. See Table S1 for probe sequences.
Consistent with our previous findings (41), purified ribosomes treated with ascorbic acid alone produced distinct degradation fragments detected using a probe complementary to the 5′ end of the 25S rRNA (y540) (Fig. 1A). Addition of Fe2+ greatly increased the degradation, resulting in well-defined rRNA fragments and disappearance of full-length 25S rRNA. The inclusion of the iron chelator DFO in the reaction completely prevented degradation. Similarly, an increase in rRNA fragmentation was observed for the 18S rRNA (Fig. 1B), 5.8S rRNA (Fig. 1C), and 5S rRNA (Fig. 1D) when treated with ascorbic acid and Fe2+, and the degradation was again prevented by the inclusion of the chelator DFO. Interestingly, when rRNAs degradation products generated in vitro were compared side by side with those in menadione-treated grx5Δ cells, we noticed a clear resemblance between the rRNAs' fragmentation patterns, regardless of a probe used (Fig. 1, compare third and fifth lanes on each blot). The similarities were seen most clearly when using probes that detect well-resolved and defined rRNA fragments, such as probe y540 for 25S rRNA (Fig. 1A), probe y532 for 18S rRNA (Fig. 1B), probe y534 for 5.8S rRNA (Fig. 1C), and probe y506 for 5S rRNA (Fig. 1D). Further probes spanning the length of both the 25S and 18S rRNAs were also used to assess the fragmentation across the length of the molecules (Fig. S2C). In all cases, the addition of Fe2+ increased the degradation observed with ascorbic acid alone, and this degradation was prevented by inclusion of DFO. However, for both the 25S and 18S rRNAs, the degradation products appeared as a smear of low molecular weight fragments as probes moved away from the 5′ ends (Fig. S2).
Restoration of integrity of rRNAs by an iron chelator in the presence of Fe2+ and ascorbic acid (Fig. 1, A–D) supports the hypothesis that Fe2+ is responsible for inducing rRNA cleavage in vitro. Additionally, probes targeting regions throughout the 18S and 25S rRNAs were able to hybridize with degradation products, suggesting that Fe2+-mediated degradation occurs at multiple sites in these molecules (Fig. S2). The similarity of rRNA degradation patterns observed between menadione-treated grx5Δ cells and those from the in vitro Fe2+/ascorbic acid assay indicates that cellular RNases do not substantially contribute to these cleavage events (Fig. 1, A–D) and that a Fe2+-mediated mechanism is primarily responsible for cleavages initiated in the 5.8S, 5S, and at the 5′ end of the 25S and 18S rRNAs when cellular redox state is perturbed. Thus, iron plays a pivotal role in the degradation of ribosomes occurring during oxidative stress.
Fe2+-mediated rRNA degradation does not require protein components of the ribosome
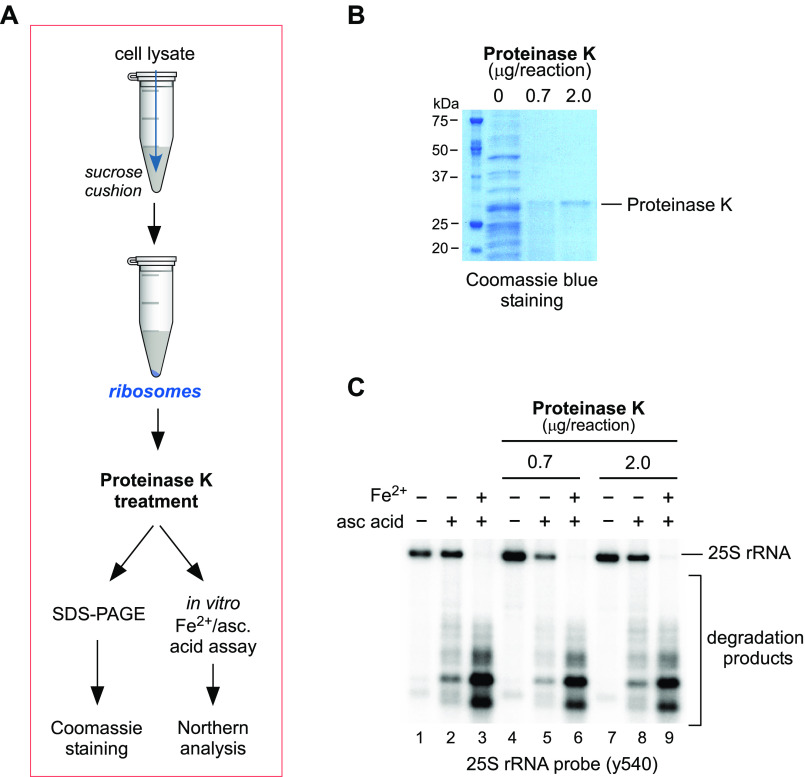
Having established the broad ability of Fe2+ to promote degradation of rRNAs, we next examined whether Fe2+ is coordinated by the rRNA itself or by a protein component of the ribosome. For instance, Fe2+ ions could be brought into proximity with rRNAs' sugar-phosphate backbone by one or more ribosome-associated proteins, which coordinate Fe2+ without inhibiting its redox activity. Alternatively, Fe2+ might bind rRNA directly, likely by substituting magnesium ions in rRNA structures, as was proposed previously (41, 48, 49). To distinguish between these two possibilities, we purified ribosomes from WT yeast cells and treated them with proteinase K under nondenaturing conditions to remove all accessible ribosomal and ribosome-associated proteins prior to assaying rRNA degradation in the presence of ascorbic acid and Fe2+ ions (scheme in Fig. 2A). To assess degradation of proteins within purified ribosomes following proteinase K treatment, the samples were analyzed by SDS-PAGE and stained with Coomassie Blue (Fig. 2B), demonstrating complete degradation of protein content at both enzyme concentrations. We detected no difference in the appearance of 25S rRNA degradation products between ribosomes treated with proteinase K and those that remained untreated (Fig. 2C). Consistent with previous data (Fig. 1), degradation was observed with ascorbic acid treatment only and was increased with the addition of Fe2+ (Fig. 2C). Finding that ascorbic acid alone was able to induce hydrolysis indicates that Fe2+ remained associated with the purified ribosomes after the degradation of proteins and therefore must associate directly with the rRNA. Furthermore, the observation that additional Fe2+ could enhance ribosomal degradation implies that protein-free rRNAs retain the ability to coordinate Fe2+ ions independently of any protein component.
Figure 2.
rRNA hydrolysis in vitro occurs independently of ribosome-associated proteins. A, schematic representation of the workflow to test the effect of removing the protein from ribosomes on rRNA cleavage in the presence of Fe2+ and ascorbic acid. Ribosomes were purified from WT cells by centrifugation through a 50% sucrose cushion, and ribosomal pellets containing 10 µg of RNA were treated with the indicated amounts of proteinase K for 5 min on ice or remained untreated. B, samples of purified ribosomes treated with proteinase K at the indicated concentrations were analyzed by SDS-PAGE and stained for total protein content with Coomassie Blue. C, samples of purified ribosomes treated with proteinase K at the indicated concentrations were treated with ascorbic acid and Fe(NH4)2(SO4)2, as in Fig. 1, and analyzed by Northern hybridization using the 25S rRNA probe y540. A representative hybridization is shown.
Ribosome-bound iron primes rRNA for cleavage
The generation of the similar patterns of rRNAs' degradation products both in vitro and in cells (Fig. 1) suggests that Fe2+-mediated rRNA cleavages are not random but site-specific, implying the existence of ribosomal iron-binding loci. We aimed to further establish that Fe2+ binds to ribosomes and that these bound ions contribute to the observed cleavage events.
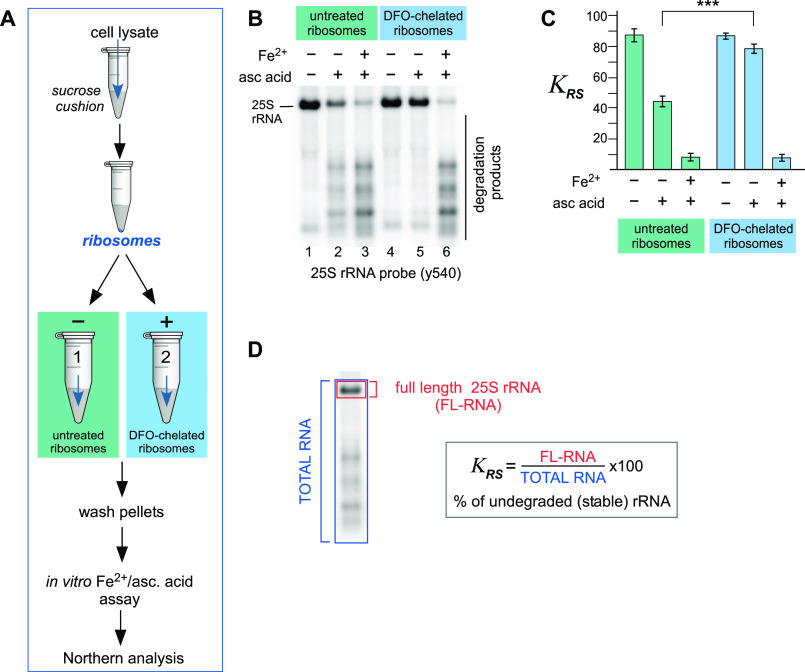
Having observed that ascorbic acid alone was able to induce degradation of rRNAs from purified ribosomes (Fig. 1) (41), we reasoned that Fe2+ ions present in the ribosome remain associated with it through the purification process and are active in the ascorbic acid–induced cleavage reaction. To test this hypothesis, purified ribosome pellets were either treated with DFO to chelate any bound iron or were left untreated (scheme in Fig. 3A). Subsequently, the pellets were purified through a second sucrose cushion and washed to remove excess reagent, before being subjected to the Fe2+/ascorbic acid assay to analyze the effect of iron chelation of purified ribosomes on the ascorbic acid–induced degradation of their rRNA (scheme in Fig. 3A). The y540 probe for the 5′ end of the 25S rRNA was used because of the distinct degradation fragment pattern detected by this probe, which was also sensitive enough to allow identification of rRNA fragments when treated with ascorbic acid only (Fig. 1) (41). Northern hybridization analysis indicated that chelation of ribosomes with DFO completely inhibited 25S rRNA hydrolysis with ascorbic acid (Fig. 3B, compare lanes 2 and 5), whereas addition of Fe2+ along with ascorbic acid restored 25S rRNA cleavages in both DFO-treated and untreated ribosomes (Fig. 3B, lanes 3 and 6).
Figure 3.
Treatment of purified ribosomes with the iron chelator DFO abolishes rRNA fragmentation. A, schematic representation of the double-cushion workflow for the chelation of ribosome-bound iron, followed by the assessment of this metal's presence within ribosomal structures via an in vitro ascorbate reaction. B, ribosomes purified from WT yeast cells were treated on ice with 0.5 mm DFO for 20 min or remained untreated and pelleted through a second 50% sucrose cushion to remove DFO excess. Equal ribosome amounts, equivalent to 3.5 µg of RNA, were incubated with 0.5 mm ascorbic acid and 1 μm Fe(NH4)2(SO4)2. The reaction products were analyzed by Northern hybridization with probe y540; a representative hybridization is shown. C, assessment of rRNA integrity by calculating the coefficient of rRNA stability (KRS) values. Three replicates of the reactions shown in B were used for the calculation. The error bars represent S.D. ***, P < 0.001 (two-tailed two-sample unequal variance t test). D, calculation of the coefficient of rRNA stability (KRS) exemplified here for 25S rRNA. Radioactive signal of the probe hybridized to undegraded, full-length 25S rRNA (FL-RNA, area shown in red) is divided by the total hybridization signal in the same lane of the blot (TOTAL RNA, blue) and multiplied by 100.
To better understand the extent of the observed rRNA degradation, we quantified the amounts of full-length 25S rRNA in the various reaction conditions (Fig. 3C). To do this, we converted probe hybridization signals for the 25S rRNA and its degradation products into phosphorimager units and calculated the ratio of full-length rRNA to the total rRNA present (Fig. 3D). The resulting number multiplied by 100 represents a percentage of undegraded full-length rRNA in a sample, and we termed this the “rRNA stability factor,” ΚRS. Using this approach, we determined that in ribosomes untreated with the chelator, approximately half of the 25S rRNA is degraded by addition of ascorbic acid, whereas DFO-chelated ribosomes are protected from hydrolysis because there is no significant loss of the full-length rRNA compared with the untreated sample. In both untreated and DFO-chelated ribosomes, the addition of Fe2+ (along with ascorbic acid) resulted in degradation of over 75% of the 25S rRNA (Fig. 3, B and C).
Taken together, these data provide evidence that Fe2+ is bound directly to yeast ribosomes and is retained through the sucrose cushion purification procedure. These ribosome-bound Fe2+ ions appear to be active participants in rRNA hydrolysis, because treatment with a chelator abolished the RNA cleavages, and are likely responsible for the degradation observed following treatment with ascorbic acid only. This agrees with the reported interaction of Fe2+ with ribosomes (48) and indicates that these ions in the cleavage events demonstrated in vitro and in cells in Fig. 1.
Genetic mutations that decrease cellular Fe2+ levels protect rRNA from degradation in yeast cells subjected to low-level oxidative stress
Having observed that chelating Fe2+ bound to purified ribosomes protects rRNA from degradation (Fig. 3), we next investigated whether a decrease in Fe2+ availability in cells would have the inverse effect on rRNA stability to that observed with the increased Fe2+ availability in grx5Δ cultures. To achieve this, we selected two genes responsible for the maintenance of cellular Fe2+ levels: AFT1, which encodes a transcription factor central to iron uptake when intracellular iron is depleted (50, 51), and FET3, an Aft1-regulated gene that encodes an accessory protein to the cell-surface Fe2+ transporter Ftr1 (51–53).
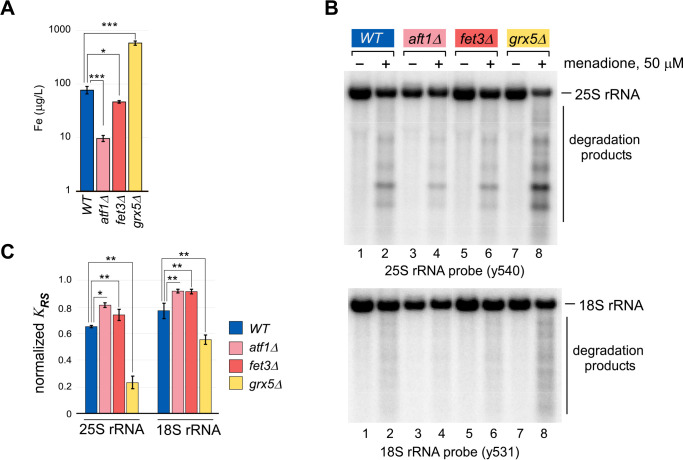
To confirm the decreased intracellular iron levels in null mutants for each of these genes, we measured iron in whole-cell extracts from equal cell numbers of WT and mutant cells using inductively coupled plasma (ICP)–MS (Fig. 4A). Cellular iron levels in the grx5Δ mutant were more than 8-fold greater than in the WT, whereas both aft1Δ and fet3Δ mutations resulted in significantly lower cellular iron levels, supporting the reported iron dysregulation in each of these mutant strains.
Figure 4.
Increased cellular iron content correlates with lowered 25S and 18S rRNA stability during oxidative stress. A, whole-cell lysates prepared from equal cell numbers of the indicated yeast strains were analyzed by ICP-MS for levels of iron. Three replicates of each sample from independent isolates were analyzed. ***, P < 0.001; *, P < 0.05 (two-tailed two-sample unequal variance t test). B, indicated yeast cultures were grown overnight in YPDA at 30 °C, diluted with fresh medium to A600 of ∼0.3, grown for an additional 4 h, and treated with 50 μm menadione for 2 h (+) or left untreated (−). RNA was isolated and analyzed by Northern hybridization with the indicated probes. A representative blot from three independent experimental repeats is shown. C, quantification of rRNAs stability in the experiment shown in B. KRS determined as described in Fig. 3D for menadione-treated samples was normalized by the corresponding values in untreated samples for each strain **, P < 0.01; *, P < 0.05, (two-tailed two-sample unequal variance t test).
The null strains were next assayed by Northern blotting for degradation of the 25S and 18S rRNAs under oxidative stress, alongside cultures of the increased intracellular iron mutant grx5Δ and the WT strain (Fig. 4B). Each lane of the Northern blotting was quantified and converted to KRS, and the values for the menadione-treated samples were normalized to the untreated sample from the same strain (Fig. 4C).
As previously observed, extensive degradation of the 25S and 18S rRNAs was detected in the grx5Δ mutant (Fig. 4, B, lanes 7 and 8, and C, yellow columns). In contrast, low levels of iron in aft1Δ and fet3Δ strains were accompanied by decreased menadione-induced fragmentation of both rRNAs tested (Fig. 4B, lanes 3–6). This manifested as an increase in KRS, indicating that a greater proportion of the total RNA remained full length following treatment (Fig. 4C, pink and red columns) when compared with WT cells (Fig. 4C, blue columns). The proportion of total 25S rRNA that remained full length was decreased by ∼35% by menadione treatment in WT cells, whereas in grx5Δ cells it was decreased by ∼75%. In aft1Δ- and fet3Δ-derived samples, this decrease was only 20–25% (Fig. 4C). The extent of degradation of 18S rRNA was less dramatic but showed the same pattern as 25S rRNA, and differences were statistically significant using probes against both, 18S and 25S rRNA (Fig. 4C). The levels of cellular iron in these strains (Fig. 4A) inversely correlates with ribosomal stability (Fig. 4C).
The data in Figs. 3 and 4 together demonstrate that iron associates with the ribosome and that there is a correlation between the level of available intracellular iron and rRNA stability. This suggests that available iron in the cell associates with binding sites on the ribosome and influences cleavage events in a manner that depends on redox activity.
Mn2+ protects rRNAs from Fe2+-mediated ROS-induced cleavage in vitro
Our data indicate that Fe2+ is bound directly to rRNA and in the presence of ROS induces hydrolysis at adjacent sites on the sugar-phosphate backbone. We have previously postulated (41) that the observed Fe2+-dependent rRNA cleavage events may be a result of the generation of reactive hydroxyl (•OH) radicals from H2O2 via the localized Fenton reaction (54).
Given the examples of Mn2+ and Fe2+ binding at the same sites, but with the crucial difference that Fe2+ participates in Fenton chemistry, whereas Mn2+ does not, we chose to investigate whether the Fe2+ bound to rRNA can be substituted by Mn2+, and if so, what effect this might have on the rRNA fragmentation in the presence of H2O2. We reasoned that if Fe2+ induces rRNA hydrolysis by generating localized hydroxyl radicals via the Fenton reaction, then rRNA in which the Fe2+ is substituted with Mn2+ would not generate radicals, and as such, the rRNA stability in the presence of ascorbic acid (as a source of H2O2) would be increased.
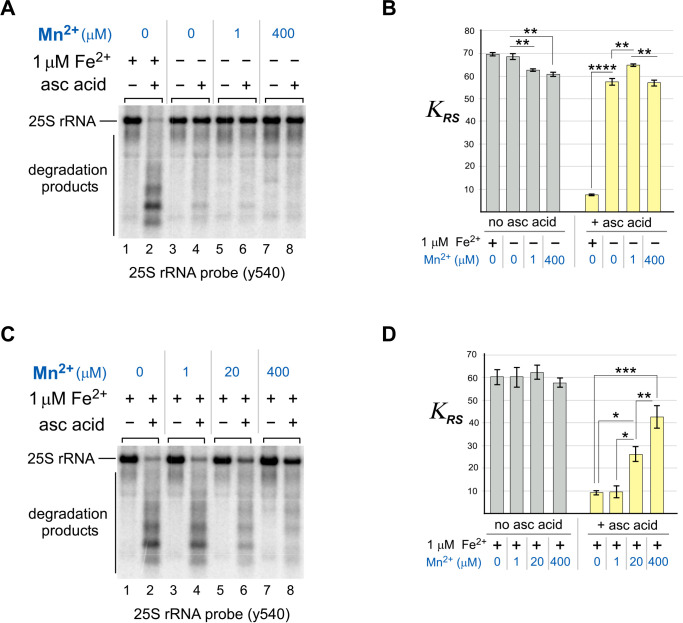
To address this hypothesis, ribosomes purified from WT cells were simultaneously treated with ascorbic acid and different concentrations of MnCl2 in vitro and analyzed for 25S rRNA degradation by Northern blotting using the y540 probe. As previously, the results of Northern blots were quantified and expressed as KRS (Fig. 3D) as a measure of rRNA stability. First, we tested the effect of treating purified ribosomes with Mn2+ instead of Fe2+ (Fig. 5A; quantification in Fig. 5B). In contrast to the strong rRNA-hydrolyzing activity of Fe2+, 25S rRNA from ribosomes were treated instead with Mn2+ (at 1 or 400 μm concentrations), and ascorbic acid largely remained intact and full-length (Fig. 5A). A small amount of 25S rRNA degradation was detected from the addition of ascorbic acid alone (Fig. 5A, compare lanes 3 and 4). It was also observed that in the absence of Fe2+, 1 μm Mn2+ improved stability in the presence of ascorbic acid, whereas at 400 μm Mn2+ there was a small increase in degradation compared with the 1 μm treatment.
Figure 5.
Mn2+ ions compete with Fe2+ in the in vitro cleavage of 25S rRNA. A and B, ribosomes isolated from WT cells were simultaneously treated with both 0.5 mm ascorbic acid (asc acid) and either 1 μm Fe(NH4)2(SO4)2 or the indicated concentrations of MnCl21. Reaction products were analyzed by Northern hybridization with the 25S rRNA probe y540. The KRS values obtained for each reaction represent the mean from three experimental repeats; the error bars represent S.D. The differences between MnCl2-free samples and samples that were treated with 0, 1, and 400 μm MnCl2 were significant. ****, P < 0.0001; **, P < 0.01 (two-tailed two-sample unequal variance t test). A representative blot is shown. C and D, same as in A and B, except that 1 μm Fe(NH4)2(SO4)2 was added into every reaction. Different concentrations of MnCl2 (0, 1, 20, and 400 μm) were also added as indicated. The differences between 25S rRNA stability were nonsignificant in the absence of ascorbic acid in the reaction (D, gray bars) and significant in the presence of ascorbic acid/Fe(NH4)2(SO4)2. ***, P < 0.001; **, P < 0.01; *, P < 0.05 (two-tailed two-sample unequal variance t test).
To further investigate the interplay between the two transition metal ions, Mn2+ and Fe2+, we examined whether Mn2+ can compete with Fe2+ for 25S rRNA fragmentation in the presence of ascorbic acid. We carried out the in vitro reaction in the presence of 1 μm Fe2+ and ascorbic acid alone or with a range of Mn2+ concentrations up to 400 μm (Fig. 5C; quantification in Fig. 5D). Consistent with our initial observation (Fig. 5, A and B), Mn2+ suppressed the Fe2+-mediated hydrolysis of 25S rRNA in a dose-dependent manner (Fig. 5, C and D). At the highest treatment level used, Mn2+ concentration was in excess of Fe2+ by 400-fold, resulting in the most prominent inhibition of Fe2+-mediated rRNA cleavage (Fig. 5C, compare lanes 2 and 8). A 20-fold excess of Mn2+ over Fe2+ had a moderate protective effect (Fig. 5C, compare lanes 2 and 6), whereas equimolar amounts of metals had only minor effects (Fig. 5C, compare lanes 2 and 4). Quantifications illustrated that Mn2+ is capable of restoring 25S rRNA in a dose-dependent manner (Fig. 5D), whereas the highest protection rate, at ∼30%, was detected with 400 μm MnCl2.
To corroborate our biochemical data, we analyzed the purified ribosomal fraction prepared from WT and grx5Δ cells by ICP-MS to assess the presence and amounts of iron and manganese on ribosomes. As a positive control, we also measured magnesium, because it binds and coordinates ribosomal structures promoting rRNA folding and ribosome assembly and therefore is present in large quantities (23). As expected, ICP-MS detected high concentrations of magnesium in purified yeast ribosomes from both strains (Fig. S3). We were also able to detect both iron and manganese in the WT and grx5Δ-derived ribosomal fractions, with ribosomes purified from grx5Δ cells containing higher Fe2+ levels than those purified from the WT cells. Interestingly, the manganese level was lower in grx5Δ cells, which may reflect the increased Fe2+ in this strain out-competing Mn2+ ions for ribosomal binding sites. These data support the possibility that the observed effect of Mn2+ on rRNA in vitro may have a physiological effect.
Collectively, these results show that Mn2+ does not induce 25S rRNA fragmentation in the presence of ascorbic acid in the manner that Fe2+ does and in fact inhibits Fe2+-mediated rRNAs hydrolysis when both metal ions are present. This would suggest that Mn2+ occupies and competes for the same sites as Fe2+ on the ribosome and prevents the Fe2+-dependent Fenton reaction mechanism for the hydrolysis of the sugar-phosphate backbone of rRNAs. We also uncovered a necessity for Mn2+ to be greatly in excess of Fe2+ to confer a significant protective effect on rRNA integrity.
Mn2+ protects ribosomal RNAs from the effects of dysregulated Fe2+ in cells
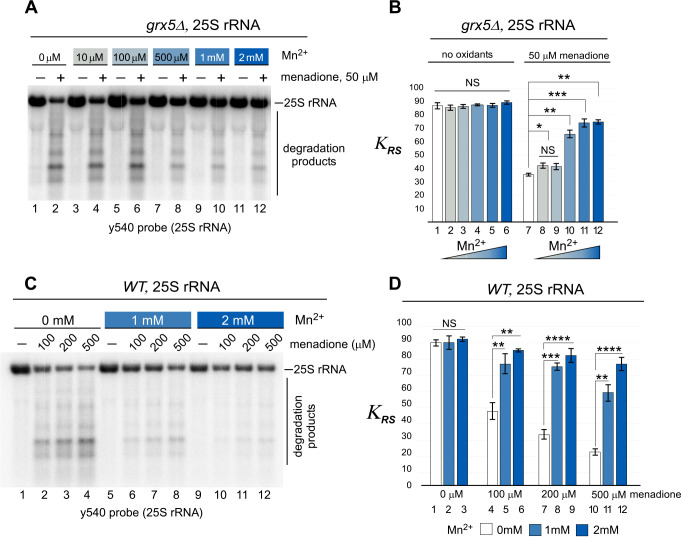
To examine whether the protection conferred to 25S rRNA by Mn2+ in vitro (Fig. 5) can occur in cells, we used the yeast grx5Δ strain (because it contains high levels of Fe2+) and supplied Mn2+ exogenously by supplementing the medium with MnCl2. As described previously, menadione was used to induce oxidative stress (Fig. 1) (41). Mid-log grx5Δ cultures grown in rich medium supplemented with a range of concentrations of Mn2+ (from 10 μm to 2 mm) were treated with 50 μm menadione or remained untreated, and total RNA was analyzed by Northern hybridization using probes against the 25S rRNA (Fig. 6A; quantification in Fig. 6B). None of the Mn2+ concentrations tested had an effect on the integrity of 25S rRNA in cells without menadione treatment (Fig. 6, A, odd lanes, and B, left columns). However, upon addition of menadione, the Fe2+-dependent 25S rRNA degradation was counteracted by Mn2+ in a dose-dependent manner (Fig. 6, A, even lanes, and B, right columns). This effect was most apparent at higher concentrations of Mn2+ (1 and 2 mm), in which KRS is as much as double that in those without Mn2+ (Fig. 6B). This stark difference is consistent with the in vitro data (Fig. 5D).
Figure 6.
Exogenously supplied manganese protects ribosomes from menadione-induced 25S rRNA degradation in cells. A, mid-log grx5Δ cultures grown in YPDA were shifted to medium supplemented with the indicated concentrations of MnCl2 and grown for 4 h at 30 °C. The cultures were adjusted to the same A600 of ∼0.6 and grown for an additional 2 h in the presence (+) or absence (−) of 50 μm menadione. RNA was isolated and analyzed by Northern hybridization with the 25S rRNA probe y540. A representative blot from three biological replicates is shown. B, quantification of the Northern hybridization data from A. The KRS values for 25S rRNA were determined for every culture treated or untreated with menadione and/or MnCl2. The data show mean values of three independent experiments; error bars represent S.D. The differences between KRS values determined for samples treated with menadione (right columns) grown in MnCl2-free medium and those grown in 1–10 μm, 0.5 mm, 1 mm, and 2 mm MnCl2-containing medium were significant. ***, P < 0.001; **, P <0.01; *, P < 0.05; NS, not significant. Two-tailed two-sample unequal variance t test was used for statistical analysis. C and D, the experiment was performed and the data were quantified as in A and B, except that WT cells were used, MnCl2 ranged from 0 to 2 mm, and menadione concentrations were 0, 100, 200, and 500 μm as indicated. Representative blots from three biological replicates are shown for all experiments. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, not significant (two-tailed two-sample unequal variance t test).
We next asked whether the ribosome-protective effect of manganese is a general effect or a peculiarity of excess-iron grx5Δ cells. To address this question, we examined 25S and 18S rRNAs stability and cell viability in WT cells subjected to oxidative stress. Unlike for oxidant-sensitive grx5Δ cells, a higher concentration of menadione was required for WT cells to induce oxidative stress. Therefore, a menadione concentration range of 100–500 μm was used to treat WT cultures for growth with or without MnCl2 in the medium. As in grx5Δ cells, Mn2+ did not affect the integrity of 25S rRNA in cells without menadione treatment (Fig. 6, C, lanes 1, 5, and 9, and D, columns 1–3). Again, upon addition of menadione, rRNA degradation was decreased by Mn2+ in a dose-dependent manner at all menadione concentrations tested (Fig. 6, C and D). At the highest menadione concentration (500 μm), at which the degradation of full-length rRNA is most extensive (Fig. 6C, lane 4), the presence of 2 mm Mn2+ was able to almost completely restore the integrity of rRNA, because we detected full-length 25S rRNAs in these samples (Fig. 6C, lane 10–12), which was consistent with increase of KRS by three times (Fig. 6D, compare columns 10 and 12).
Additionally, we carried out the Northern blotting analyses from Fig. 6 using probes for the 18S rRNA (Fig. S4). Although degradation of 18S rRNAs induced by menadione treatment occurs to a lesser extent compared with 25S rRNAs in both WT and grx5Δ cells (Fig. 6 and Fig. S4), the addition of Mn2+ was again able to rescue 18S rRNA from degradation (Fig. S4, A–C; quantifications are in Fig. S4, B–D). To test whether the observed RNA degradation in response to Fe2+ and ascorbic acid treatments was specific to ribosomal RNAs, we also analyzed degradation of the noncoding snoRNA U3 and found that stability was not affected (Fig. S5).
These data indicate that Mn2+ does not affect rRNA stability during normal growth without stress but has a protective role during oxidative stress in cells, similar to the observations from our in vitro experiments (Fig. 5). Additionally, the protective effect of Mn2+ in cells is shown here for both the 25S rRNA and 18S rRNA. Considering that 18S rRNA was found to be a target for Fe2+-dependent oxidant-induced degradation (Fig. 1B and Fig. S2B) (41), it is logical that Mn2+ also competes with Fe2+ in the small ribosomal subunit.
Mn2+ restores viability in yeast cells subjected to oxidative stress
To address the physiological relevance of the Mn2+-protective effect on yeast ribosomes, we tested whether manganese would rescue the menadione-induced decrease in viability of grx5Δ cells (Fig. 7A). In fact, previous studies have shown that treatment of grx5Δ cells with low-dose oxidants results in inhibition of cell growth (41, 55). We incubated grx5Δ cultures in medium supplemented with either 1 mm (Fig. 7A, left panel) or 2 mm (Fig. 7A, right panel) MnCl2 for 4 h prior to adding 50 or 100 μm menadione. As a control, we used grx5Δ cultures that were treated with menadione in the absence of Mn2+ (Fig. 7A, upper panels). Serial dilutions of the tested cultures were plated on the rich-medium agar plates, and cell growth was monitored over time. In the absence of Mn2+, grx5Δ cell viability was almost entirely lost with 50 μm menadione treatment, and no growth was observed with 100 μm menadione treatment. However, either concentration of Mn2+ tested allowed increased cell viability following menadione treatment. The rescue effect of manganese was comparable with that of the iron chelator 1,2-phenanthroline (PHL; Fig. S6), suggesting that redox activity of Fe2+ contributes to the decreased viability and that preventing this activity via chelation has the same outcome as substituting Fe2+ with Mn2+ at the ribosome.
Figure 7.
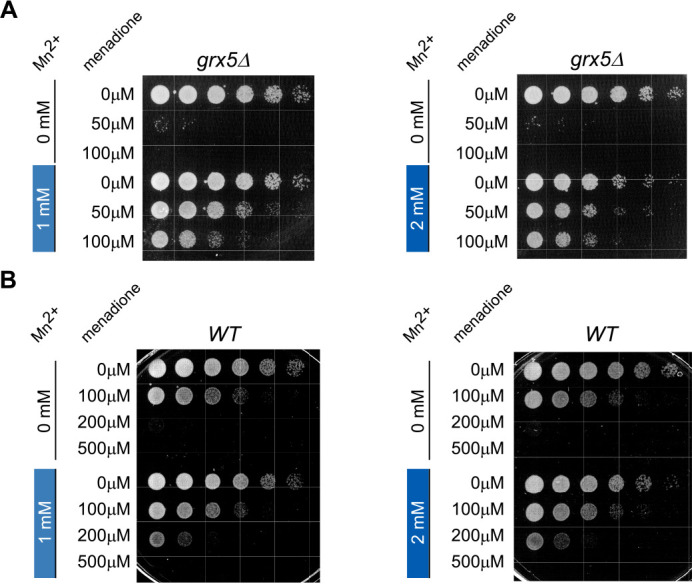
Exogenously supplied manganese restores viability of menadione-treated cells. A and B, overnight grx5Δ (A) and WT (B) cultures grown in YPDA were diluted with fresh YPDA medium to A600 of ∼0.2 and grown for 4 h in the presence of 1 or 2 mm MnCl2 or in the absence of MnCl2. The cultures were next treated with the indicated concentrations of menadione for 2 h at 30 °C with shaking. Thereafter, the cells were collected, washed twice, adjusted to the same cell counts (2 × 106 cells/ml), serially diluted (1:5), and plated on YPDA agar plates. The plates were incubated at 30 °C for ∼2–3 days (WT) or ∼3–5 days (grx5Δ). Viability assays were repeated three times, and representative images are shown.
The effect of Mn2+ on WT cell viability was also tested using the higher menadione concentrations (Fig. 7B), as in the rRNA degradation assay (Fig. 6, C and D). Growth of WT cells was completely abolished at 200 μm menadione treatment in the absence of Mn2+ in the growing culture, whereas 1 or 2 mm Mn2+ treatments resulted in partial restoration of the cell viability (Fig. 7B).
Although these data suggest that a decrease in redox-active Fe2+ at rRNAs is responsible for the protective effects of Mn2+, we cannot rule out other antioxidant functions of the additional Mn2+ in this assay. Nevertheless, our biochemical characterization of Fe2+/Mn2+ competition in the rRNA degradation process (Fig. 5, C and D) provides strong evidence that Mn2+ protects the entire ribosome from Fe2+-dependent H2O2-induced degradation and that this contributes to the restoration of cell growth in oxidative stress.
Discussion
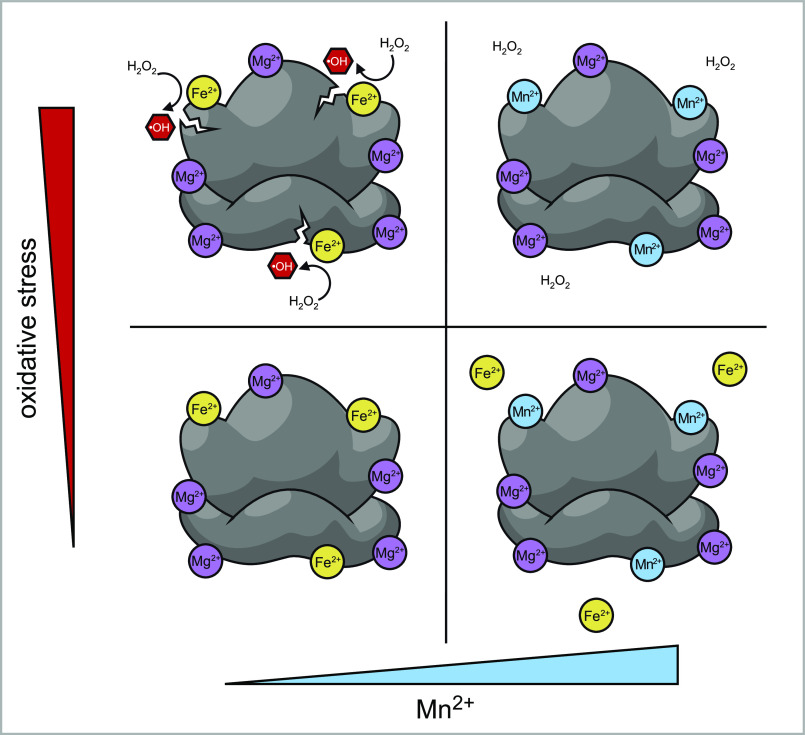
In this study, we have demonstrated that Fe2+ is coordinated directly by rRNA and from this position promotes fragmentation of the rRNA under oxidative conditions. These data support a model whereby Fe2+-dependent, Fenton reaction–mediated hydrolysis is attenuated by Mn2+, which competes with and excludes Fe2+ from binding sites on the rRNA. The working model for how the competition between these cations affects the integrity of ribosomes under oxidative stress is outlined in Fig. 8.
Figure 8.
Model for the roles of Fe2+ and Mn2+ in oxidant-induced ribosome damage. Under normal metal homeostasis conditions, rRNA in unstressed cells is bound by divalent metal cations, predominantly Mg2+, as well as some amount of Fe2+, with both cations occupying sites capable of coordinating other metals including Mn2+ (bottom left panel). An increase in the ratio of available Mn2+ to Fe2+ results in the displacement of Fe2+ from these rRNAs sites (bottom right panel). Under oxidative stress, ribosome-bound Fe2+ generates hydroxyl radicals in the Fenton reaction, causing rRNA strand breaks in the vicinity of bound Fe2+ ions (top left panel). Increased Mn2+ concentrations have the opposite effect: by displacing bound Fe2+, Mn2+ prevents rRNA strand breaks produced through Fenton chemistry and makes ribosomes more resistant to oxidative stress conditions (top right panel).
The polypeptide synthesis activity of ribosomes with Fe2+ and Mn2+ binding has been previously addressed in conditions that simulate the primordial earth environment (30), raising the question about the interplay between these two transition metal ions and the preferred divalent cation of the rRNA, Mg2+, in present-day ribosomes. In our previous study (41), which focused on a specific cleavage event mediated by bound Fe2+ in the ES7 region of the 25S rRNA, it was noted that three Mg2+-binding sites exist close to the cleavage site and that Fe2+ is likely to bind in place of Mg2+ in this context. Other work has shown that sites of RNA cleavage that occurs with Fe2+ treatment correlate with Mg2+ sites (56). We therefore consider it likely that the Fe2+-dependent cleavages we observed throughout multiple rRNAs correspond to what are normally Mg2+-binding sites. Given the large number of Mg2+ ions bound to rRNA, it is then perhaps surprising that any large fragments remain following Fe2+/oxidant damage. These fragments may simply represent larger sections of the rRNA that do not readily coordinate any metal ions. Alternatively, Fe2+ cannot readily replace Mg2+ at certain binding sites. We also observed a lower efficiency of Mn2+ in protecting small ribosomal subunits. This may reflect fewer metal-coordinating elements present in the smaller 18S rRNA molecule (42).
In several cases where we detected large distinct fragments following Fe2+-dependent rRNA cleavage, the rRNA degradation occurring inside cells was compared with that which occurred in the in vitro experiments (Fig. 1). That these same distinct fragment bands appear in both assays indicates the close similarity in cleavage mechanisms in the cell and those in the limited chemical environment of the in vitro assay. The results are presently less explicit, with several of the probes that did not allow resolution of individual fragments and produced a low-molecular-weight smear on the Northern blots. Overall, however, the degradation profile of yeast rRNAs produced with Fe2+ and ascorbic acid (Fig. 1 and Fig. S2) argues that not only is iron necessary for initiating ROS-induced rRNA degradation, but it is also sufficient for the multiple instances of fragmentation to happen, with little to no enzymatic hydrolysis involved.
The addition of Mn2+ clearly decreases Fe2+-mediated rRNA degradation (Fig. 5, C and D), but Mn2+ concentrations that are orders of magnitude higher than that of Fe2+ are required to have a major protective effect. An excess of 400 times results in an ∼30% increase in full-length rRNA, suggesting that Mn2+ has a lower affinity for these ribosomal binding sites than Fe2+. The stability of complexes with divalent transition metal cations tends to increase with atomic number (57), supporting Mn2+ having a weaker affinity than Fe2+. In fact, both ions are reported to have relatively low dissociation constants when bound to proteins compared with other divalent cations (19) and can be readily replaced by Mg2+ (58). The selection between Mn2+ and Fe2+ in other biomolecules that can comfortably coordinate either ion, including SOD enzymes (17, 59), appears to depend primarily on available ion concentration and may reflect shifts in metal homeostasis under stress conditions (19, 40). Magnesium-binding sites in the ribosome have been classified by distinct geometries (23), and it is possible that other cations have different affinities depending on the arrangement of side-chain ligands. Investigating the binding-site architecture and conformations of coordinated Fe2+ and Mn2+ in rRNA structures may clarify these differences.
A protective effect of Mn2+ being substituted for Fe2+ has been observed previously in nonredox enzymes in bacteria (60, 61). This phenomenon occurs following oxidation of the Fe2+ in a Fenton reaction, causing it to dissociate from the active site and reaction of the hydroxyl product with the adjacent residues. In this Escherichia coli system under oxidative stress conditions, the cells enact transcriptional adjustments that increase both uptake of manganese and the sequestration of iron, contributing to the substitution of Fe2+ with Mn2+ at active sites to protect against iron-dependent oxidative damage (15). Furthermore, the protective effect was also demonstrated with cobalt, which unlike manganese does not have any inherent antioxidant activity (60). Cobalt is not imported by E. coli but can enter nonspecifically when included in the medium at high concentrations. This supports the model that the protective effect of manganese is due to substitution of iron rather than other oxidant defensive activities. It may be interesting to investigate whether cobalt is able to replicate the manganese protection of ribosomes, because this would confirm that the effect is due to iron displacement rather than general antioxidant functions associated with manganese.
Our data from live-cell experiments (Fig. 7 and Fig. S6) do not exclude the possibility of Fe2+ or Mn2+ interactions also occurring away from the ribosome, which could influence the outcome of the oxidative stress experienced by the cell. The grx5Δ mutant exhibits a 6-fold increase in cellular iron (45), and although the oxidant sensitivity of this strain may in part be attributed to the increase in rRNA hydrolysis, the effects of loss of GRX5 also include impaired synthesis of the iron–sulfur clusters and lack of mitochondrial respiration (62). Also of note is the manganese-dependent Sod2 enzyme, which can incorporate Fe2+ instead of Mn2+ (63), resulting in a decreased oxidant defense. Conversely, Sod2 activity may increase with Mn2+ availability, thus enhancing the cell oxidant defense capability. However, SOD enzymes protect the cell from damaging superoxide anion but in doing so produce H2O2, the substrate of the Fenton reaction (64). Nonproteinaceous manganese antioxidant species are also up-regulated under oxidative stress (40). The in vitro data presented show that the RNA stability effects are so closely recreated in purified ribosomes as to suggest that additional antioxidants are not required for the protective capacity of manganese on ribosomes, but we cannot exclude that there are further protective influences in the complex cellular environment.
Although we were not able to demonstrate that Mn2+ could rescue the ability of oxidatively stressed ribosomes' to synthesize polypeptides immediately after the stress was removed, we suspect that this is due to the broad effects of oxidant insult throughout cells. Logically, protecting rRNA stability has only a limited capacity to maintain translational activity after such pervasive stress. In fact, the damaging effects of oxidative species extend beyond nucleic acids and affect lipids and proteins throughout the cell, thus damaging other components of the translational machinery (65–67).
Both iron and manganese are essential elements in humans while also being toxic at high doses, and thus their import and metabolism is of medical relevance. Metal ion imbalances in cells can occur as a result of either environmental exposure or genetic mutations influencing metabolism and cellular availability (68–70). A guidance value for toxic dose of manganese is only approximately five times the recommended nutritional intake for humans (8), which leaves a relatively small range of acceptable levels. The pathologies associated with dysregulation of manganese are interlinked with those of iron because of their overlapping interactions in biological systems (71). Chronic overexposure to manganese is associated with broad neurological effects including impaired motor control and cognition (72). Iron accumulation in the brain is associated with Alzheimer's and Parkinson's diseases (73), and neurodegenerative effects are also linked to manganese accumulation, although direct links to Alzheimer's and Parkinson's diseases are less clear (72, 74). Therefore interdependence between iron and manganese (as well as other metals (74)) creates a delicate situation to navigate in the maintenance of homeostasis.
In conclusion, it has been well-established that manganese can protect cells from oxidative stress, whereas iron can enhance oxidant-induced damage. That these two transition metal neighbors can substitute each other at the same sites in biomolecules presents a potent target for modulation of cellular machinery under oxidative stress. Here we report a novel observation of the protective role for Mn2+ at the ribosome, wherein it can defend against oxidative damage by displacing Fe2+ and preventing hydrolytic cleavages throughout the ribosomal RNAs. We propose that an ancient rivalry between divalent metal cations continues to influence the stability of the translation machinery. These observations highlight the multilayered competition that defines the relationship between iron and manganese, from the regulation of importers, competition for uptake, to further competition at binding targets inside the cell including the ribosome.
Experimental procedures
Yeast, media, and reagents
WT BY4742 (MATα his3-1 leu2-0 met15-0 ura3-0) strain and its derivative deletion strains aft1Δ and fet3Δ were obtained from Thermo Fisher. The grx5Δ strain was generated as described before (41). We used a standard recipe for YPDA medium (1% yeast extract, 2% peptone, 2% dextrose, and 10 mg/liter adenine). We used the following chemicals: menadione (Enzo), PHL (VWR Life Science), deferoxamine (DFO) (BioVision), Chelex 100 (Sigma), Fe(NH4)2(SO4)2 (Sigma), and ascorbic acid (Alfa Aesar). Proteinase K was from Roche, and 1 m MnCl2 solution was from Sigma.
Extraction of RNA from cells and preparation of ribosome-enriched fractions
To purify total RNA from cells for Northern blotting analysis, we used the published formamide extraction method (75). To purify ribosomes from cells for the in vitro ascorbic acid assay, the cells were grown to mid-log phase (A600 = ∼0.8), collected, and washed in buffer A (30 mm HEPES-KOH, pH 7.4, 100 mm KOAc, 3 mm MgOAc) that was pretreated with Chelex 100 for 15 min at room temperature with shaking to remove trace iron. The cells were then lysed in buffer A by glass bead shearing. Lysates were clarified by centrifugation at 21,000 × g for 10 min and layered on 0.5 ml of 50% (w/v) of sucrose cushion prepared in Chelex-treated buffer A. Ribosomes were precipitated by centrifugation for 90 min at 150,000 × g (55,000 rpm, Beckman tabletop Optima ultracentrifuge, rotor TLA-55). Ribosomal pellets were washed twice with buffer A and kept frozen at −80 °C. For an ascorbic acid assay, pellets were defrosted on ice, resuspended in buffer A supplemented with 0.15 unit/ml of RiboLock (Thermo Fisher), and the total RNA concentration was measured spectrophotometrically.
In vitro ascorbic acid assay and Fe2+/Mn2+ competition assay
The in vitro iron/ascorbic acid reactions were described in Ref. 41. In brief, we used a total of 3.5 µg RNA per in vitro reaction. Solutions of ascorbic acid, Fe(NH4)2(SO4)2, and/or MnCl2 were added to RNA as indicated in the figure legends. The reaction volume was adjusted to 100 µl with buffer A; reactions were incubated for 10 min on ice, followed by precipitation of RNA with 50% isopropanol. RNA was pelleted by centrifugation at 21,000 × g for 1 h at 4 °C, washed with 80% ethanol, and dissolved in 5–10 µl of FAE (98% formamide, 10 mm EDTA).
Iron chelation and proteinase K treatment of ribosomes
For iron chelation, ribosomal pellets prepared by ultracentrifugation through the sucrose cushion (as described above) were dissolved in buffer A, DFO was added to a final concentration of 0.5 mm, and the solution was incubated on ice for 20 min. DFO-treated ribosomes were collected by another round of ultracentrifugation (90 min, 150,000 × g) to remove the excess of DFO. Ribosomal pellets were washed and stored at −80 °C or analyzed further.
For treatments with proteinase K, ribosomal pellets prepared by ultracentrifugation through the sucrose cushion were resuspended in buffer A, and 3 µg of total RNA was treated with 0.7 or 2 units of proteinase K (Roche) for 5 min on ice. The reactions were divided into two aliquots each: one aliquot of the proteinase K–treated ribosomes was resolved by SDS-PAGE and analyzed by Coomassie staining, whereas the second aliquot was used for an ascorbic acid assay.
RNA analysis
We used 1.2% agarose gels containing 1.3% formaldehyde (76) to analyze large rRNA species (25S and 18S rRNAs), whereas for small rRNAs, we used 8% polyacrylamide gels containing 8 m urea as described previously (41). RNA was transferred to nylon membranes (Hybond N, GE Biosciences). Individual rRNA species and their degradation derivatives were hybridized with 32P-labeled oligonucleotide probes (Table S1) as described in Ref. 77. Hybridizations were visualized with the GE Amersham Biosciences Typhoon 5 imager and analyzed with ImageQuant software (GE Healthcare). For quantification, the volume of the hybridization signal corresponding to the area of interest was converted to phosphorimaging units. For quantification of rRNA stability, we calculated the ribosome stability factor (ΚRS) by dividing the number of phosphorimager units corresponding to nondegraded full-length rRNA by the number of phosphorimager units derived from the entire sample lane. Where indicated, the ΚRS values determined in cells treated with oxidants were normalized to ΚRS values determined in untreated cells. The P values were calculated using two-tailed, two-sample unequal variance t tests.
Cell viability assays
Overnight cultures were diluted with YPDA to A600 of ∼0.2, grown for 4 h at 30 °C in the presence or absence of 1 or 2 mm MnCl2, or in medium supplemented with 80 μm PHL. For oxidant treatment, the cells were incubated with various concentrations of menadione for 2 h at 30 °C with shaking. For treatment of WT cells, we used 100, 200, and 500 μm menadione; for grx5Δ cells, we used 50 and 100 μm menadione. Following the treatments, 5-fold serial dilutions of each culture were plated on YPD agar plates and incubated at 30 °C for 3–5 days.
ICP-MS analysis
The cell samples were extracted by addition of 200 µl of sub-boiled nitric acid (HNO3), vortexed for 30 s, and let sit for 30 min at room temperature. Subsequently, 10 ml of sub-boiled water was added, and the sample was centrifuged for 8 min at 1,000 × g before the supernatant was removed and amended with 50 µl of sub-boiled HCl. Purified ribosomes were resuspended in 120 µl of distilled H2O, extracted with concentrated nitric acid (32% HNO3 v/v in extraction) at room temperature for 3 h, and then centrifuged at 17,000 × g for 10 min; the supernatant was then diluted and amended with HCl to achieve final acid concentrations of 2% HNO3 and 0.5% HCl (v/v). Acidified aqueous samples were analyzed via ICP-MS (Agilent 7900) using a reaction-collision cell to minimize polyatomic interferences. A calibration curve was created using standards from Inorganic Ventures and was analyzed at the beginning of the batch and for determination of the limit of quantification. Batch quality assurance measured also included assessment of seven blanks (to determine the detection limit) and on-line internal standards for assessment of signal stability. A sample was considered quantitative only if all quality assurance/quality control standards were achieved, including: above the limit of quantification, reproducible element and internal standard values from triplicate measurements, and appropriate internal standard recovery. Sample dilutions were employed if the concentration was above the calibration curve or if the sample matrix interfered with analysis (i.e. altered internal standard recovery).
Data availability
All data are presented in the article.
Supplementary Material
Acknowledgments
We are thankful to Arnab Ghosh, Brandon Trainor, and Russell Sapio for the critical evaluation of this work and constructive comments on the manuscript.
This article contains supporting information.
Author contributions—D. G. J. S., N. K., E. R. M., D. G. P., and N. S. conceptualization; D. G. J. S. and N. S. resources; D. G. J. S., D. G. P., and N. S. data curation; D. G. J. S., N. K., E. R. M., D. G. P., and N. S. formal analysis; D. G. J. S. and N. S. supervision; D. G. J. S., N. K., E. R. M., D. G. P., and N. S. validation; D. G. J. S., N. K., E. R. M., D. G. P., and N. S. investigation; D. G. J. S., N. K., and E. R. M. visualization; D. G. J. S. and N. S. writing-original draft; D. G. J. S. and N. S. project administration; D. G. J. S., N. K., D. G. P., and N. S. writing-review and editing; N. K., E. R. M., D. G. P., and N. S. methodology; D. G. P. and N. S. software.
Funding and additional information—This work was supported by National Institutes of Health Grant R01GM114308 (to N. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- DFO
- deferoxamine
- PHL
- 1,2-phenanthroline
- ICP
- inductively coupled plasma
- ROS
- reactive oxygen species.
References
- 1. Zoroddu, M. A., Aaseth, J., Crisponi, G., Medici, S., Peana, M., and Nurchi, V. M. (2019) The essential metals for humans: a brief overview. J. Inorg. Biochem. 195, 120–129 10.1016/j.jinorgbio.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 2. Andreini, C., Bertini, I., Cavallaro, G., Holliday, G. L., and Thornton, J. M. (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218 10.1007/s00775-008-0404-5 [DOI] [PubMed] [Google Scholar]
- 3. McCall, K. A., Huang, C., and Fierke, C. A. (2000) Function and mechanism of zinc metalloenzymes. J. Nutr. 130, 1437S–1446S 10.1093/jn/130.5.1437S [DOI] [PubMed] [Google Scholar]
- 4. Weston, J. (2009) Biochemistry of magnesium. In PATAI'S Chemistry of Functional Groups (Rappoport, Z., ed) John Wiley & Sons, Chichester, UK [Google Scholar]
- 5. Vashishtha, A. K., Wang, J., and Konigsberg, W. H. (2016) Different divalent cations alter the kinetics and fidelity of DNA polymerases. J. Biol. Chem. 291, 20869–20875 10.1074/jbc.R116.742494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ricchetti, M., and Buc, H. (1993) E. coli DNA polymerase I as a reverse transcriptase. EMBO J. 12, 387–396 10.1002/j.1460-2075.1993.tb05670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cowan, J. A. (1998) Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev. 98, 1067–1088 10.1021/cr960436q [DOI] [PubMed] [Google Scholar]
- 8. Smith, M. R., Fernandes, J., Go, Y.-M., and Jones, D. P. (2017) Redox dynamics of manganese as a mitochondrial life-death switch. Biochem. Biophys. Res. Commun. 482, 388–398 10.1016/j.bbrc.2016.10.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poyner, R. R., Cleland, W. W., and Reed, G. H. (2001) Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme. Biochemistry 40, 8009–8017 10.1021/bi0103922 [DOI] [PubMed] [Google Scholar]
- 10. Knape, M. J., Ballez, M., Burghardt, N. C., Zimmermann, B., Bertinetti, D., Kornev, A. P., and Herberg, F. W. (2017) Divalent metal ions control activity and inhibition of protein kinases. Metallomics 9, 1576–1584 10.1039/c7mt00204a [DOI] [PubMed] [Google Scholar]
- 11. Shen, J.-R. (2015) The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 10.1146/annurev-arplant-050312-120129 [DOI] [PubMed] [Google Scholar]
- 12. Sánchez, M., Sabio, L., Gálvez, N., Capdevila, M., and Dominguez-Vera, J. M. (2017) Iron chemistry at the service of life: iron chemistry at the service of life. IUBMB Life 69, 382–388 10.1002/iub.1602 [DOI] [PubMed] [Google Scholar]
- 13. Martín-Molina, A., Rodríguez-Beas, C., and Faraudo, J. (2012) Effect of calcium and magnesium on phosphatidylserine membranes: experiments and all-atomic simulations. Biophys. J. 102, 2095–2103 10.1016/j.bpj.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sigel, R. K. O., and Pyle, A. M. (2007) Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 107, 97–113 10.1021/cr0502605 [DOI] [PubMed] [Google Scholar]
- 15. Anjem, A., Varghese, S., and Imlay, J. A. (2009) Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72, 844–858 10.1111/j.1365-2958.2009.06699.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dudev, T., and Lim, C. (2014) Competition among metal ions for protein binding sites: determinants of metal ion selectivity in proteins. Chem. Rev. 114, 538–556 10.1021/cr4004665 [DOI] [PubMed] [Google Scholar]
- 17. Beyer, W. F., and Fridovich, I. (1991) In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J. Biol. Chem. 266, 303–308 [PubMed] [Google Scholar]
- 18. Foster, A. W., Osman, D., and Robinson, N. J. (2014) Metal preferences and metallation. J. Biol. Chem. 289, 28095–28103 10.1074/jbc.R114.588145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cotruvo, J. A., Jr., and Stubbe, J. (2012) Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4, 1020–1036 10.1039/c2mt20142a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wacker, W. E. C. (1969) The biochemistry of magnesium. Ann. N.Y. Acad. Sci. 162, 717–726 10.1111/j.1749-6632.1969.tb13003.x [DOI] [PubMed] [Google Scholar]
- 21. Nierhaus, K. H. (2014) Mg2+, K+, and the ribosome. J. Bacteriol. 196, 3817–3819 10.1128/JB.02297-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sissi, C., and Palumbo, M. (2009) Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 37, 702–711 10.1093/nar/gkp024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein, D. J., Moore, P. B., and Steitz, T. A. (2004) The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10, 1366–1379 10.1261/rna.7390804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petrov, A. S., Bowman, J. C., Harvey, S. C., and Williams, L. D. (2011) Bidentate RNA–magnesium clamps: on the origin of the special role of magnesium in RNA folding. RNA 17, 291–297 10.1261/rna.2390311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guth-Metzler, R., Bray, M. S., Frenkel-Pinter, M., Suttapitugsakul, S., Montllor-Albalate, C., Bowman, J. C., Wu, R., Reddi, A. R., Okafor, C. D., Glass, J. B., and Williams, L. D. (2020) Cutting in-line with iron: ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res. 48, 8663–8674 10.1093/nar/gkaa586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nguyen, H. T., Hori, N., and Thirumalai, D. (2019) Theory and simulations for RNA folding in mixtures of monovalent and divalent cations. Proc. Natl. Acad. Sci. U.S.A. 116, 21022–21030 10.1073/pnas.1911632116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Athavale, S. S., Petrov, A. S., Hsiao, C., Watkins, D., Prickett, C. D., Gossett, J. J., Lie, L., Bowman, J. C., O'Neill, E., Bernier, C. R., Hud, N. V., Wartell, R. M., Harvey, S. C., and Williams, L. D. (2012) RNA folding and catalysis mediated by iron (II). PLoS One 7, e38024 10.1371/journal.pone.0038024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grosshans, C. A., and Cech, T. R. (1989) Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry 28, 6888–6894 10.1021/bi00443a017 [DOI] [PubMed] [Google Scholar]
- 29. Pyle, A. M. (2002) Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7, 679–690 10.1007/s00775-002-0387-6 [DOI] [PubMed] [Google Scholar]
- 30. Bray, M. S., Lenz, T. K., Haynes, J. W., Bowman, J. C., Petrov, A. S., Reddi, A. R., Hud, N. V., Williams, L. D., and Glass, J. B. (2018) Multiple prebiotic metals mediate translation. Proc. Natl. Acad. Sci. U.S.A. 115, 12164–12169 10.1073/pnas.1803636115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrov, A. S., Bernier, C. R., Hsiao, C., Norris, A. M., Kovacs, N. A., Waterbury, C. C., Stepanov, V. G., Harvey, S. C., Fox, G. E., Wartell, R. M., Hud, N. V., and Williams, L. D. (2014) Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 10251–10256 10.1073/pnas.1407205111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fox, G. E. (2010) Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2, a003483–a003483 10.1101/cshperspect.a003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melnikov, S., Ben-Shem, A., Garreau de Loubresse, N., Jenner, L., Yusupova, G., and Yusupov, M. (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 10.1038/nsmb.2313 [DOI] [PubMed] [Google Scholar]
- 34. Kraft, C., Deplazes, A., Sohrmann, M., and Peter, M. (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- 35. Mroczek, S., and Kufel, J. (2008) Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 36, 2874–2888 10.1093/nar/gkm1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bengtson, S. (ed.) (1994) Early Life on Earth: Nobel Symposium, No. 84, Columbia University Press, New York [Google Scholar]
- 37. Jones, C., Nomosatryo, S., Crowe, S. A., Bjerrum, C. J., and Canfield, D. E. (2015) Iron oxides, divalent cations, silica, and the early earth phosphorus crisis. Geology 43, 135–138 10.1130/G36044.1 [DOI] [Google Scholar]
- 38. Feig, A. L., and Uhlenbeck, O. C. (1999) The role of metal ions in RNA biochemistry. Cold Spring Harb. Monograph Arch. 37, 287–319 [Google Scholar]
- 39. Johnson, J. E., Webb, S. M., Ma, C., and Fischer, W. W. (2016) Manganese mineralogy and diagenesis in the sedimentary rock record. Geochim. Cosmochim. Acta 173, 210–231 10.1016/j.gca.2015.10.027 [DOI] [Google Scholar]
- 40. Aguirre, J. D., and Culotta, V. C. (2012) Battles with iron: manganese in oxidative stress protection. J. Biol. Chem. 287, 13541–13548 10.1074/jbc.R111.312181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zinskie, J. A., Ghosh, A., Trainor, B. M., Shedlovskiy, D., Pestov, D. G., and Shcherbik, N. (2018) Iron-dependent cleavage of ribosomal RNA during oxidative stress in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 293, 14237–14248 10.1074/jbc.RA118.004174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ben-Shem, A., Garreau de Loubresse, N., Melnikov, S., Jenner, L., Yusupova, G., and Yusupov, M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 43. Shedlovskiy, D., Zinskie, J. A., Gardner, E., Pestov, D. G., and Shcherbik, N. (2017) Endonucleolytic cleavage in the expansion segment 7 of 25S rRNA is an early marker of low-level oxidative stress in yeast. J. Biol. Chem. 292, 18469–18485 10.1074/jbc.M117.800003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gomez, M., Pérez-Gallardo, R. V., Sánchez, L. A., Díaz-Pérez, A. L., Cortés-Rojo, C., Meza Carmen, V., Saavedra-Molina, A., Lara-Romero, J., Jiménez-Sandoval, S., Rodríguez, F., Rodríguez-Zavala, J. S., and Campos-García, J. (2014) Malfunctioning of the iron–sulfur cluster assembly machinery in Saccharomyces cerevisiae produces oxidative stress via an iron-dependent mechanism, causing dysfunction in respiratory complexes. PLoS One 9, e111585 10.1371/journal.pone.0111585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodríguez-Manzaneque, M. T., Tamarit, J., Bellí, G., Ros, J., and Herrero, E. (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121 10.1091/mbc.01-10-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Samuni, A., Aronovitch, J., Godinger, D., Chevion, M., and Czapski, G. (1983) On the cytotoxicity of vitamin C and metal ions: a site-specific Fenton mechanism. Eur. J. Biochem. 137, 119–124 10.1111/j.1432-1033.1983.tb07804.x [DOI] [PubMed] [Google Scholar]
- 47. Khan, M. M., and Martell, A. E. (1967) Metal ion and metal chelate catalyzed oxidation of ascorbic acid by molecular oxygen: I. Cupric and ferric ion catalyzed oxidation. J. Am. Chem. Soc. 89, 4176–4185 10.1021/ja00992a036 [DOI] [PubMed] [Google Scholar]
- 48. Bray, M. S., Lenz, T. K., Haynes, J. W., Bowman, J. C., Petrov, A. S., Reddi, A. R., Hud, N. V., Williams, L. D., and Glass, J. B. (2018) Multiple prebiotic metals mediate translation. Proc. Natl. Acad. Sci. U.S.A. 115, 12164–12169 10.1073/pnas.1803636115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hsiao, C., Chou, I.-C., Okafor, C. D., Bowman, J. C., O'Neill, E. B., Athavale, S. S., Petrov, A. S., Hud, N. V., Wartell, R. M., Harvey, S. C., and Williams, L. D. (2013) RNA with iron(II) as a cofactor catalyses electron transfer. Nat. Chem. 5, 525–528 10.1038/nchem.1649 [DOI] [PubMed] [Google Scholar]
- 50. Martínez-Pastor, M. T., Perea-García, A., and Puig, S. (2017) Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 33, 75 10.1007/s11274-017-2215-8 [DOI] [PubMed] [Google Scholar]
- 51. Poor, C. B., Wegner, S. V., Li, H., Dlouhy, A. C., Schuermann, J. P., Sanishvili, R., Hinshaw, J. R., Riggs-Gelasco, P. J., Outten, C. E., and He, C. (2014) Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc. Natl. Acad. Sci. U.S.A. 111, 4043–4048 10.1073/pnas.1318869111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Askwith, C. C., and Kaplan, J. (1998) Site-directed mutagenesis of the yeast multicopper oxidase Fet3p. J. Biol. Chem. 273, 22415–22419 10.1074/jbc.273.35.22415 [DOI] [PubMed] [Google Scholar]
- 53. Shakoury-Elizeh, M., Tiedeman, J., Rashford, J., Ferea, T., Demeter, J., Garcia, E., Rolfes, R., Brown, P. O., Botstein, D., and Philpott, C. C. (2004) Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 1233–1243 10.1091/mbc.e03-09-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wardman, P., and Candeias, L. P. (1996) Fenton chemistry: an introduction. Radiat. Res. 145, 523–531 10.2307/3579270 [DOI] [PubMed] [Google Scholar]
- 55. Rodríguez-Manzaneque, M. T., Ros, J., Cabiscol, E., Sorribas, A., and Herrero, E. (1999) Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 8180–8190 10.1128/MCB.19.12.8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hanson, S., Bauer, G., Fink, B., and Suess, B. (2005) Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA 11, 503–511 10.1261/rna.7251305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Irving, H., and Williams, R. J. P. (1953) 637. The stability of transition–metal complexes. J. Chem. Soc. 3192 10.1039/jr9530003192 [DOI] [Google Scholar]
- 58. Zhu, W., and Richards, N. G. J. (2017) Biological functions controlled by manganese redox changes in mononuclear Mn-dependent enzymes. Essays Biochem. 61, 259–270 10.1042/EBC20160070 [DOI] [PubMed] [Google Scholar]
- 59. Yang, M., Cobine, P. A., Molik, S., Naranuntarat, A., Lill, R., Winge, D. R., and Culotta, V. C. (2006) The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 25, 1775–1783 10.1038/sj.emboj.7601064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sobota, J. M., and Imlay, J. A. (2011) Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U.S.A. 108, 5402–5407 10.1073/pnas.1100410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Anjem, A., and Imlay, J. A. (2012) Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J. Biol. Chem. 287, 15544–15556 10.1074/jbc.M111.330365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shakoury-Elizeh, M., Protchenko, O., Berger, A., Cox, J., Gable, K., Dunn, T. M., Prinz, W. A., Bard, M., and Philpott, C. C. (2010) Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 285, 14823–14833 10.1074/jbc.M109.091710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Naranuntarat, A., Jensen, L. T., Pazicni, S., Penner-Hahn, J. E., and Culotta, V. C. (2009) The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284, 22633–22640 10.1074/jbc.M109.026773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thorpe, G. W., Reodica, M., Davies, M. J., Heeren, G., Jarolim, S., Pillay, B., Breitenbach, M., Higgins, V. J., and Dawes, I. W. (2013) Superoxide radicals have a protective role during H2O2 stress. Mol. Biol. Cell 24, 2876–2884 10.1091/mbc.E13-01-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cabiscol, E., Tamarit, J., and Ros, J. (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8 [PubMed] [Google Scholar]
- 66. Castro, F. A. V., Mariani, D., Panek, A. D., Eleutherio, E. C. A., and Pereira, M. D. (2008) Cytotoxicity mechanism of two naphthoquinones (menadione and plumbagin) in Saccharomyces cerevisiae. PLoS One 3, e3999 10.1371/journal.pone.0003999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Costa, V., and Moradas-Ferreira, P. (2001) Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22, 217–246 10.1016/S0098-2997(01)00012-7 [DOI] [PubMed] [Google Scholar]
- 68. Fitsanakis, V. A., Zhang, N., Garcia, S., and Aschner, M. (2010) Manganese (Mn) and iron (Fe): interdependency of transport and regulation. Neurotox Res. 18, 124–131 10.1007/s12640-009-9130-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pantopoulos, K. (2018) Inherited disorders of iron overload. Front. Nutr. 5, 103 10.3389/fnut.2018.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Camaschella, C. (2005) DMT1 mutations: mice and humans are not alike. Blood 105, 916–917 10.1182/blood-2004-11-4260 [DOI] [Google Scholar]
- 71. Crossgrove, J., and Zheng, W. (2004) Manganese toxicity upon overexposure. NMR Biomed. 17, 544–553 10.1002/nbm.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Balachandran, R. C., Mukhopadhyay, S., McBride, D., Veevers, J., Harrison, F. E., Aschner, M., Haynes, E. N., and Bowman, A. B. (2020) Brain manganese and the balance between essential roles and neurotoxicity. J. Biol. Chem. 295, 6312–6329 10.1074/jbc.REV119.009453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Salvador, G. A., Uranga, R. M., and Giusto, N. M. (2010) Iron and mechanisms of neurotoxicity. Int. J. Alzheimers Dis. 2011, 720658 10.4061/2011/720658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dusek, P., Roos, P. M., Litwin, T., Schneider, S. A., Flaten, T. P., and Aaseth, J. (2015) The neurotoxicity of iron, copper and manganese in Parkinson's and Wilson's diseases. J. Trace Elements Med. Biol. 31, 193–203 10.1016/j.jtemb.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 75. Shedlovskiy, D., Shcherbik, N., and Pestov, D. G. (2017) One-step hot formamide extraction of RNA from Saccharomyces cerevisiae. RNA Biol. 14, 1722–1726 10.1080/15476286.2017.1345417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mansour, F. H., and Pestov, D. G. (2013) Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal. Biochem. 441, 18–20 10.1016/j.ab.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pestov, D. G., Lapik, Y. R., and Lau, L. F. (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol., Chapter 22, Unit 22.11 10.1002/0471143030.cb2211s39 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented in the article.