Abstract
Zika virus (ZIKV) is a neurotropic flavivirus that causes several diseases including birth defects such as microcephaly. Intrinsic immunity is known to be a frontline defense against viruses through host anti-viral restriction factors. Limited knowledge is available on intrinsic immunity against ZIKV in brains. Amyloid precursor protein (APP) is predominantly expressed in brains and implicated in the pathogenesis of Alzheimer's diseases. We have found that ZIKV interacts with APP, and viral infection increases APP expression via enhancing protein stability. Moreover, we identified the viral peptide, HGSQHSGMIVNDTGHETDENRAKVEITPNSPRAEATLGGFGSLGL, which is capable of en-hancing APP expression. We observed that aging brain tissues with APP had protective effects on ZIKV infection by reducing the availability of the viruses. Also, knockdown of APP expression or blocking ZIKV-APP interactions enhanced ZIKV replication in human neural progenitor/stem cells. Finally, intracranial infection of ZIKV in APP-null neonatal mice resulted in higher mortality and viral yields. Taken together, these findings suggest that APP is a restriction factor that protects against ZIKV by serving as a decoy receptor, and plays a protective role in ZIKV-mediated brain injuries.
Keywords: Amyloid precursor protein, Zika virus, restriction factor, intrinsic immunity, decoy receptor, flavivirus, amyloid precursor protein (APP), innate immunity, immunology, interferon, brain specific
Zika virus (ZIKV) belongs to Flaviviridae and the genus Flavivirus, and is thus related to the dengue, West Nile, and other flaviviruses. ZIKV is an enveloped virus with icosahedral symmetry and a nonsegmented, single-stranded, positive-sense RNA genome (1, 2). The RNA genome, which is ∼11 kb in length, has a single ORF that encodes three structural proteins: the capsid (C), membrane (M), and envelope (E) glycoproteins, and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).
Infections of ZIKV in humans are typically transmitted by Aedes mosquito, but ZIKV may spread through sexual contact, blood transfusion, and from mother-to-fetus (3–9). Only about 20% of ZIKV-infected individuals develop mild symptoms, however, ZIKV can transport across the placenta and infect fetal brain cells (10–12), and thereby being one of the causative agents of microcephaly in fetuses (13–16). Microcephaly is a neurodevelopmental disorder, which is characterized by a marked reduction in brain size and intellectual disability (17). Neural progenitor/stem cells (NPC/NSC) in developing brains are particularly vulnerable to ZIKV infection. In both human brain organoid culture as well as mouse models, ZIKV infection can be detected in NPCs/NSCs and lead to microcephaly (12, 15, 16, 18–23). Although there has been rapid progress in our understanding of ZIKV infection, no antiviral treatment or vaccine for ZIKV is clinically approved yet.
Intrinsic immunity is a form of innate immunity against viruses in eukaryotic cells, whereby pre-existing restriction factors limit specific virus infections (24, 25). Individual organs, such as the brains, have their own unique intrinsic immunity (26), and host cells have intrinsic immunity against flaviviruses (27). However, knowledge about restriction factors or intrinsic immunity against ZIKV, especially in the brains, is very limited (28–30).
Amyloid precursor protein (APP) is a membrane protein expressed predominantly in the brains and metabolized in a rapid and highly complex fashion by a series of sequential proteases (31). Alternate splicing of the APP transcript generates several forms, of which three are most common: the 695-amino acid form, which is expressed predominantly in the brains, and the 751- and 770-amino acid forms, which are more ubiquitously expressed. The precise physiological function of APP is not very clear, but it is well established that APP is a gene involved in the pathogenesis of Alzheimer's diseases (31).
We show in this work that APP interacts with ZIKV, is stabilized by ZIKV infection, and inhibits ZIKV replication in both in vitro in human NPCs/NSCs and in vivo in neonatal mouse brains. Aging brain cells with APP expression have a protective effect on other cells by reducing ZIKV availability. Therefore, we have established that APP is a restriction factor for ZIKV by serving as a host decoy receptor in the brains. Understanding intrinsic immunity against ZIKV is likely to be crucial for the treatment and prevention of ZIKV-mediated diseases.
Results
ZIKV interacts with APP protein
Using information about the ZIKV virion structure (32, 33), we applied protein structure alignment methods, such as SSM (34) and TM-alignment (35), to screen structurally homologous proteins to ZIKV virion and E protein. The binding partners of the homologous proteins were considered as potential ZIKV binding candidates. Dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN, also known as the cluster of differentiation 209) is one of the cellular receptors for ZIKV (36). This method predicted that ZIKV E protein interacted with members of the C-type lectin receptor family 4, including DC-SIGN (data not shown).
Beta-secretase 1 (BACE1) was a hit of the screening, and there were some structural similarities between BACE1 and ZIKV virion, particularly within the ZIKV E protein. BACE1 is a transmembrane protein that binds to and cleaves APP proteins (37). PDB IDs for structures of ZIKV E and BACE1, obtained from the Protein Data Bank (www.rcsb.org), are 5IRE chain A and 3HW1 chain A, respectively (38). Alignments of the ZIKV E and BACE1 showed only limited structural similarities between the two proteins: both proteins have a similar β−hairpin structure, a 2-stranded β-sheet (Fig. 1, a and b). Interestingly, the corresponding part in BACE1 was located in the binding region to APP (Fig. 1a). Amino acid residues, GSS, form a very short loop region in the β−hairpin of BACE1. In contrast, ZIKAV E has a longer corresponding part (Fig. 1c). Because APP is a target of BACE1, the similarity in the structures suggested that APP protein might be a potential candidate for ZIKV binding.
Figure 1.
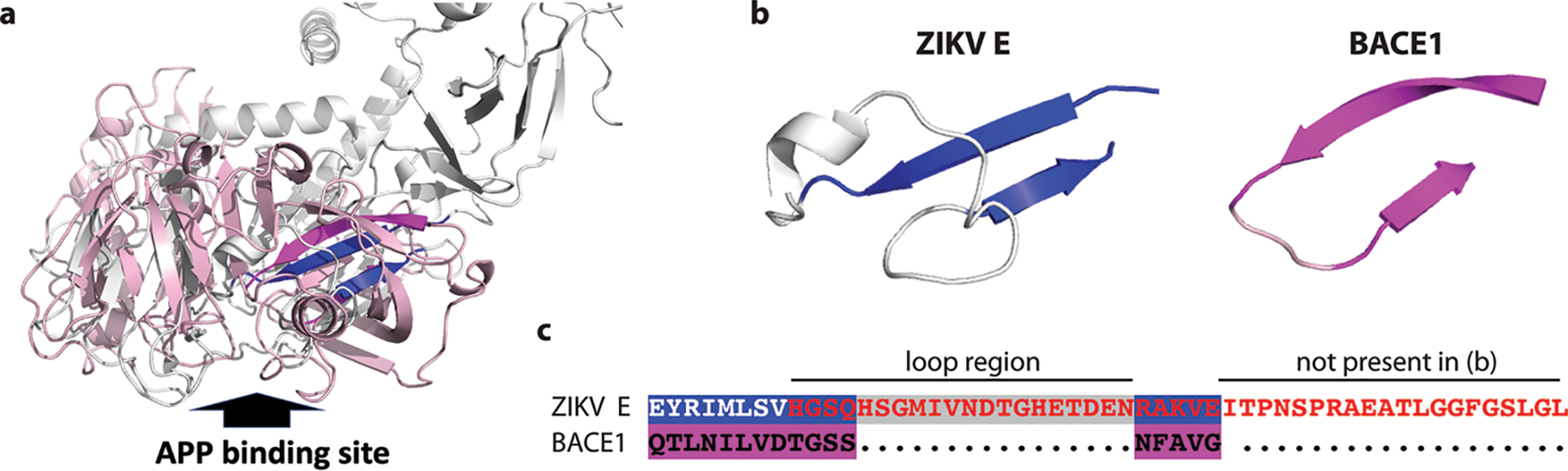
Structural similarity between ZIKV virions and APP protein. a, the structural alignment of ZIKV E and BACE1 proteins is as shown. The structure of ZIKV E protein is shown in gray and blue, whereas the structure of BACE1 is shown in light pink and magenta. The blue part in the ZIKV E protein and magenta part in BACE1 are the best aligned parts, which are two β strands with a loop. The aligned structures are around the APP-binding site. The APP-binding site of BACE1 is indicated by a big arrow. b, the best aligned structural parts on ZIKV E protein (blue) and on BACE1 (magenta) are displayed separately. c, the corresponding sequence alignment between the strand sheets on ZIKV E protein and BACE1 are displayed with corresponding color background. Red font: the amino acid sequence used for generating P2 peptide. Loop region and sequence in P2 that is not present in the panel b are also indicated.
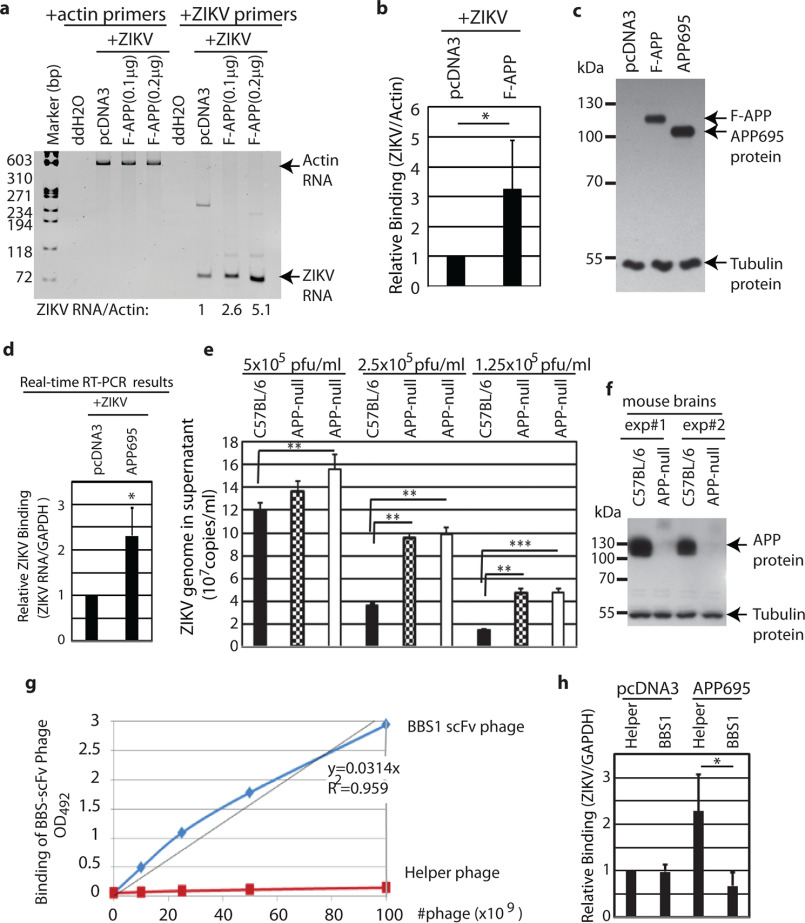
To examine whether APP was related to ZIKV infection, human embryonic kidney fibroblast 293 cells (HEK293) were used for two reasons: 1) the cells are not susceptible for ZIKV infection (12); and 2) the cells have very low levels of endogenous APP. First, we tested whether APP was a factor for ZIKV attachment to cells. N-terminal FLAG-tagged full-length APP expression plasmids (39) were transfected into the HEK293 cells, and 1 day later, the transfected cells were mixed with ZIKV for 1 h. The cells were washed extensively to remove the unbound viruses. The attachments of ZIKV to transfected cells were examined by semi-quantitative RT-PCR (RT-PCR). ZIKV bound to APP-expressing cells more strongly than the control vector-transfected HEK293 cells (Fig. 2, a and b). The expression of APP was confirmed by various methods and appeared to be located in the cytoplasm and possibly cell membranes (Figs. 2c and 3b). Of note, incubating viruses and cells for a short period of time and measuring viral components are a common approach for assessing viral attachment to cells (40–42). To avoid the potential interference from N-terminal inserted FLAG tag, an APP695 expression plasmid without any tags, was used for ZIKV-binding assays with quantitative real time RT-PCR (qRT-PCR) assays. APP695 is the predominant form of APP in the brains and more ZIKV could attach to APP695-expressing cells (Fig. 2d, Fig. S1a).
Figure 2.
APP inhibits ZIKV attachment to cells. a, HEK293 cells were transfected with vector (pcDNA3) or APP expression plasmids (pCAX FLAG APP or F-APP). One day later, cells were incubated with ZIKV (PRVABC59) at 0.5 m.o.i. in 37 °C for 1 h. Cells were vigorously washed to remove unadsorbed virus. RNA was isolated from the cells. Same amounts of cDNAs were used for semiquantitative PCR analyses. Primers for ZIKV and actin were added in separate tubes as shown on the top and amplified with different cycles based on previous experiments for linear ranges of amplifications. An 8% polyacrylamide gel was used to separate the PCR products. Standard dsDNA fragment markers are shown on the left in base pairs (bp). Relative ZIKV RNA level (ZIKV/Actin) is as shown on the bottom. b, three independent experiments were done as in panel a, and relative ZIKV RNA levels (ZIKV/Actin) in vector and APP-transfected cells were quantified and compared. Student's t tests were done. *, p < 0.05. c, pCAX FLAG APP or pCAX-APP695 were transfected into HEK293 cells and expression of human APP was determined by Western blotting. pcDNA3 was used as a vector control. Molecular weight markers are shown on the left in kDa. d, pCAX-APP695 or pcDNA3 plasmids were transfected into HEK293 cells and ZIKV attachment assays were done as in panel a. Real-time quantitative RT-PCR assays (qRT-PCR) were used for measurement of ZIKV RNA, with GAPDH RNA as internal control. The 2−ΔΔCt method was used for calculation for relative RNA levels. Relative ZIKV attachments are as shown. e, cortical cortexes from aging mouse brains were dissected, and passed through 70- and 40-μm cell strainers. Cells were washed and 5 × 106 cell clumps/ml were seeded and incubated with various amounts of ZIKV (pfu/ml) at 37 °C for 1 h. Supernatants were collected and used for qRT-PCR assays. Total three mice (> 1-year–old) were used for the experiments. C57BL/6J(wt), solid bar; two APP-null mice, dashed and open bars, respectively. Student's t tests were performed, *, p < 0.05; **, p < 0.01; ***, p < 0.001. f, equal volumes of explant cultures were collected and protein expression in cultured cells was examined by Western blotting analyses. The identity of proteins is as shown. Results represent two independent experiments from four mice. g, binding of Phage BBS1 scFv to BACE1 cleavage site peptide. A 96-well–plate was coated with MAP-[ISEVKMDA]8 (peptide representing the BACE1 cleavage site on APP). Different phage concentrations in equal volumes were added to the plate and allowed to bind for 1 h at 37 °C. The assay was developed using OPD substrate. h, HEK293 cells were transfected with target plasmids. Cells were incubated with ZIKV and BBS1 scFv expressing or corresponding helper phages simultaneously at 37 °C for 1 h. The cells were washed and RNA were isolated for qRT-PCR assays. Three biological replicates were done and the relative ZIKV/GAPDH amounts were calculated using 2−ΔΔCt method.
Figure 3.
ZIKV E protein and APP may interact with each other. a, the co-immunoprecipitation assays were used to detect the interaction between ZIKV E and APP in HEK cells. The expression plasmids for APP and ZIKV PrM-E were co-transfected into HEK293T cells (left panels), or the cells were transfected with vector (pcDNA3) or APP expression plasmid and infected with recombinant adenovirus expressing ZIKV-PrM-E, or GFP protein (right panels). Each experiment was repeated at least three times. Cell lysates were immunoprecipitated with ZIKV E antibody followed by Western blotting with various antibodies. The direct Western blots with cell lysates are also shown (bottom panel). b, the HEK293T cells were transfected with vector (pcDNA3) or APP expression plasmid and infected with ZIKV (1 m.o.i.) for 1 h. Cell lysates were immunoprecipitated with mixture of APP polyclonal antibodies or normal rabbit serum (NRS). The immunoprecipitates were used for RT-PCR analyses with ZIKV-specific primers (top panel) or Western blotting analyses (bottom panel). Each experiment was repeated at least three times. The direct Western blots with cell lysates are also shown (right panel). The experiment was repeated four times. c, HEK293 cells were transfected with PrM-E and APP695 expression plasmids. One day later, the cells were stained with rabbit ZIKV E and mouse APP antibodies followed by secondary antibodies conjugated with Alexa Fluor 488 and 647, respectively. The cells were examined under a confocal microscope. ZIKV E (green), APP (red), overlay of ZIKV E, APP and nuclei (blue) are shown. Amplified regions and scale bars are also shown.
In addition, there are some reports that HEK293 cells were infectable by ZIKV (43, 44). Although the ZIKV strain (PRVABC59) was not able to replicate in the HEK293 cells in our settings (data not shown), it was necessary to test a different line and/or attachment method to verify the absorption results. Adult brains seem to be more resistant to ZIKV infection (12, 45–47). One possible explanation is that adult brains have very few or no NPCs/NSCs (48–50). We suspected that adult brain cells, with the expression of APP, might trap ZIKV and thus protected NPCs/NSCs or other infectable cells from ZIKV infection. To test this hypothesis, cortical cortexes from aging (>1-year–old) APP-null (B6.129S7-Apptm1Dbo/J) and age-matched control WT mice (WT, C57BL/6J) were isolated and passed through cell strainers (see “Experimental procedures” for details). The same amounts of cell clumps were dispensed and immediately incubated with ZIKV for 1 h, and the amounts of free viruses remaining in the media were determined as a measure of viral absorptions to brain cortexes. Culture media from explant APP-null brain cortexes had higher levels of ZIKV viral genomes than those in WT mice, suggesting that ZIKV was being trapped by APP protein (Fig. 2e, Fig. S1b). Other than similar cell clumps, similar levels of tubulin were detected in the two batches of explant cortex cultures, and the genotype status of APP–null strain was also verified at the same time (Fig. 2f). Those results suggested that mature brain cells with APP could bind to and trap ZIKV.
Potential ZIKV E protein-binding site on APP was tested. Previous studies have shown that BBS1 antibody targets the BACE1-cleavage site of APP (51). Because ZIKV may bind to a similar site based on molecular structural analyses (Fig. 1a), we tested if BBS1 antibody could interfere with ZIKV attachments to cells. The single-chain variable fragment (scFv) of the BBS1 antibody was cloned and expressed in a phage, and the BBS1 scFv phage was able to bind to the target peptide (Fig. 2g). The BBS1 scFv expressing phages and its control helper phages were incubated with ZIKV for attachment assays to APP695 expressing HEK293 cells. The BBS1 scFv phages inhibited ZIKV attachments to APP695 expressing cells (Fig. 2h). The results suggested that the ZIKV specifically bound to APP and the region of the binding was located around the BACE1 cleavage site (Fig. 1a).
ZIKV E protein is considered as the major virion protein for attachment to host cells. Interestingly, PrM-E complex has been shown to be a good candidate for ZIKV vaccine (52), which suggested that PrM may help E protein to generate the correct conformation for interaction with host proteins. To further test the potential interaction between ZIKV-APP further, we employed co-immunoprecipitation assay (co-IP) for potential ZIKV and APP interactions. By co-transfection of PrM-E and APP expression plasmids in HEK293 cells, a ZIKV-E antibody immunoprecipitated APP proteins in transfected cell lysates (Fig. 3a). In addition, similar results were obtained by transfection of APP695 and infection with a recombinant adenovirus expressing ZIKV PrM-E proteins (53). Unfortunately, with multiple APP antibodies and extensive efforts, we could not use APP antibody to precipitate ZIKV E protein in the same or similar co-IP conditions (data not shown). We reasoned the sensitivity of the detection might be a problem and switched to viral RNA as a target. ZIKV were incubated with APP-expressing HEK293 cells for 1 h and cell lysates were used for immunoprecipitation with APP antibodies. The resulting immune precipitates were used for RNA isolation and RT-PCR analyses. As shown in Fig. 3b (left, top panel), the viral RNAs were immunoprecipitated with APP. The results suggest that APP interacts with ZIKV virions.
To further confirm the physical interactions between ZIKV E and APP proteins, whether ZIKV E and APP proteins were co-localized in the same cells was examined. ZIKV-E and APP were partially co-localized in the same transfected cells (Fig. 3c). In the majority (>90%) of APP and ZIKV E co-expressing cells, partial co-localization patterns of the two proteins were observed. Those data from computer modeling, virus attachments, as well as co-localization collectively indicate that ZIKV has a physical interaction with APP protein.
ZIKV enhances APP protein expression
APP is quickly metabolized through an orderly process by multiple proteases. Whether ZIKV binding could affect APP expression was examined. HEK293 cells were transfected with APP expressing plasmids and the cells were incubated with ZIKV overnight. APP-expressing cells exposed to ZIKV exhibited elevated levels of the APP protein than those nonexposed cells (Fig. 4, a and b). The higher expression of APP at time 0 had been consistently observed, and we reasoned it might be due to processing the samples required 5-10 min. It is interesting to note that the overexpression of transfected APP diminished the induction effects, suggesting that the expression of APP in a cell might have an upper limit (data not shown). Moreover, the induction by ZIKV was specific for APP protein because Epstein-Barr virus latent membrane protein 1 (LMP1) could not be enhanced by ZIKV infection (Fig. 4c). LMP1 is a membrane protein, partially located in endosome (54), and considered as a constitutively active receptor (55). To examine the mechanistic bases for the increase in APP protein in these cells with ZIKV, a protein synthesis inhibitor, cycloheximide (CHX), was added to the culture medium. The t1/2 of APP protein in the presence of ZIKV was increased significantly, suggesting that ZIKV attachments increased the stability of APP protein in these cells (Fig. 4, d and e). As NPCs/NSCs are major targets for ZIKV replication, we chose mature neurons to test if ZIKV attachments could enhance APP expression to avoid the complications from viral entry/replication. A mixture of human cortical neurons derived from human embryonic stem cells (56) was infected with ZIKV for 6 h, and higher expression of APP was observed (Fig. 4, f and g). In addition, the increases might be due to the increase of the protein t1/2 (Fig. 4h). Mature neurons are less susceptible to ZIKV and 1 h incubation is unlikely to synthesize any new viral proteins (57). Therefore, the observed effects were most likely due to the virion attachments. Whether in vivo infection of ZIKV would cause the accumulation of APP in mice was also examined. However, due to the variability of APP in different individual mice (see Fig. 2f for a reference) no conclusive results could be obtained with whole brain protein analyses (data not shown). An explant infection system was established whereby the mixtures of whole mouse brain cells were isolated and incubated with ZIKV. Mouse brain cells had a higher APP protein expression after 6 h of incubation with ZIKV (Fig. 4i). APP levels also increased after 24 h ZIKV infection (data not shown). Of note the endogenous APP levels in the mouse brains might determine the folds of inductions: the lower endogenous APP levels would lead to higher induction by ZIKV.
Figure 4.

ZIKV stabilizes APP proteins. a, HEK293 cells were transfected with human APP695 expression plasmid, pCAX-APP695, or vector control. Cells were infected with or without ZIKV at 1 m.o.i. overnight. Western blots were used for detection of APP and tubulin simultaneously. b, relative levels of APP proteins (APP/tubulin) in HEK293 cells are shown from three independent experiments. Student's t tests were performed, *, p < 0.05. c, HEK293 cells were transfected with EBV LMP1 expression plasmid or vector control (pcDNA3). One day later, the cells were infected with or without ZIKV overnight as in panel a. Western blots were used for detection of LMP1 and tubulin simultaneously. The images in the same box indicate they were derived from the same gels. The identity of the proteins is as shown. d, HEK293 cells were transfected with APP695, split into several smaller wells 4-6 h later. After overnight culture, the cells were treated with CHX, or CHX plus ZIKV simultaneously for various times. Cells were collected at indicated times (hours post treatment). Expression of APP and tubulin, determined by Western blots, is as shown. e, relative levels of APP proteins (APP/Tubulin) in HEK293 cells with or without ZIKV infection are shown. The data are the combinations of two independent experiments (one is shown in panel d). Solid line, ZIKV infected; dashed line, no ZIKV infection. f, the mixture of human mature cortical neurons was infected with ZIKV (10 m.o.i.) for 6 h, and expression of APP and tubulin was determined by Western blots. One representative result is as shown. g, relative levels of APP proteins (APP/Tubulin) in human mature neurons are shown from three independent experiments in two batches of human neurons as in panel e. Student's t tests were performed, *, p < 0.05. h, the mixture of human mature cortical neurons was infected with or without ZIKV (approximate 10 m.o.i.) for 1 h, and then cells were treated with CHX for the indicated times (minutes). The expression of APP proteins from two independent experiments is as shown. The relative APP expression levels (APP/Tubulin) are indicated in the bottom. i, mouse brains (WT, C57BL/6J) were dissected from euthanized mice. Mixture of cells were isolated and infected with ZIKV for 6 h. The approximate m.o.i. is shown in parentheses. The relative APP expression levels are indicated in the bottom.
We suspected that the loop in the specific structure of ZIKV E protein was responsible for the potential interaction with APP (Fig. 1, a and b). A peptide (P2) derived from the potential binding site, the β-hairpin region, was synthesized with sequence optimization (Fig. 1c). Located in the core region of the ZIKV E protein and being hydrophobic, the 8 amino acid stretch, EYRIMLSV, in the β-hairpin region of E protein was not included in the P2 sequence. Furthermore, the P2 peptide could potentially form a similar structure as in the native protein by including extra amino acids in the C terminus (Fig. 1, b and c) (58, 59). The P2 peptide is a water-soluble molecule but could not enhance APP expression when treating cells directly (data not shown). It is known that the interactions between BACE1 and APP protein happen in both biosynthetic and endocytic compartments inside cells (60, 61), and ZIKV E protein was predicted to bind targets in the endosomal compartments (62, 63). Therefore, we transfected P2 peptide into cells to ensure the peptide could meet APP in endocytic compartments physically. The P2 peptide could increase the expression of APP comparing with the BSA control (Fig. 5, a and b). At the same time, the expression of vial LMP1 could not be enhanced by the peptide co-transfection (Fig. 5c). Furthermore, a similar mechanism to ZIKV was established for the peptide-mediated enhancement (Fig. 5d). All these data suggest that ZIKV enhances APP expression at least partially through the modulation of the APP protein stability.
Figure 5.
A viral peptide could enhance APP expression. a, HEK293 cells were co-transfected with APP695 expression plasmid (0.5 µg) and P2 peptide or BSA protein (1 µg each) with PULSin reagents. One day later, the expression of APP and tubulin were examined by Western blotting analyses. One representative result is as shown. b, relative APP protein expression levels were calculated from three independent experiments and are as shown. Student's t tests were performed, *, p < 0.05. c, HEK293 cells were co-transfected with different amounts of LMP1 expression plasmid as shown on the top and P2 peptide, or BSA protein (1 µg each) with PULSin reagents. One day later, the expression of APP and tubulin were examined simultaneously by Western blotting analyses. The identity of the proteins is as shown. d, HEK293 cells were co-transfected with APP expression plasmid (0.5 µg) plus BSA (1 µg) or APP (0.2 µg) plus P2 peptide (1 µg) with PULSin reagents. Four h later, the cells were split into two wells. One day later, the cells were treated with CHX for 1 h and lysates were collected. The expression of APP and tubulin were examined. The relative expression of APP proteins is as shown at the bottom.
APP is a negative regulator of ZIKV replication
Although ZIKV could bind to APP-expressing cells (Figs. 1–3), APP did not facilitate viral replication in HEK293 cells as measured by either plaque or qRT-PCR assays 2 days after infection (data not shown). Therefore, APP was clearly not a receptor for the virus, although located on the cell surface. Next, the role of APP as a regulator for ZIKV was examined in NPCs/NSCs, a major target of the virus in the brains with APP expression. The human NPCs/NSCs used in the studies were derived from the NIH approved H9 human embryonic stem cells and characterized using standard protocols (12, 64). siRNA was used to specifically knockdown gene expressions. The recombinant lentiviruses expressing APP siRNA were used to infect NPCs/NSCs. The APP-knockdown cells were then infected by ZIKV. The viral replications were monitored by the qRT-PCR 2 days later. The culture supernatants from APP-knockdown cells had higher viral genomic RNA copies than those from control cells (Fig. 6a). The effectiveness of siRNA expressing lentivirus on APP expression was confirmed by Western blotting analyses (Fig. 6b).
Figure 6.

APP inhibits ZIKV replication. A, lentivirus expressing siRNA for APP (siAPP) and its scramble lentivirus (siScramble) were used to infect human NPCs/NSCs overnight at 1 m.o.i. Cells were washed and infected with ZIKV (1 MOI) and 1 h later, the media were replaced with fresh media. Two days later, cell media were collected and subjected to real time qRT-PCR analyses. Four independent experiments were done and the relative ZIKV genomic RNA contents in the media were calculated with 2−ΔΔCt method. Student's t tests were performed, **, p < 0.01. b, human NPCs/NSCs were treated with the siRNA expressing lentivirus and 1 day later, the cells were collected for Western blotting analyses. The identity of target proteins is as shown. c, human NPCs were incubated with ZIKV (1 m.o.i.) in the presence with either BBS1Ab scFv expressing or helper phages (1 transforming unit per cell) for 1 h at 37 °C. Uninfected viruses were removed and cells were washed with fresh medium. Two days later, cell media were subjected to qRT-PCR analysis. Four independent experiments were done and relative ZIKV RNAs in medium were calculated with 2−ΔΔCt method. Student's t tests were performed, **, p < 0.01. d, C57BL/6 (WT) and APP-null P4 baby mice were used for i.c. injections of ZIKV (PRVABC59; 1.5 × 104 pfu) or the same volumes of 1× PBS. Pink, ZIKV-infected neonates; blue, PBS control mice; solid line, APP-null neonates; dashed line, WT mice. The survival of the baby mice was monitored for 4 days. To compare the survival curves for ZIKV-infected cases of APP-null and WT, the Log-Rank test was applied and the p value is as shown. e, two strains of mice were i.c. injected with ZIKV. P4 mice: 1 × 103 pfu, n = 10 WT, n = 11 app−/−; P7 mice: 1.5 × 104 pfu, n = 9 WT, n = 9 app−/−; P28 mice: 5 × 104 pfu, n = 10 WT, n = 11 app−/−. The mice, both WT and APP-null, with i.c. injected with 1× PBS, were controls. Multiple ages and strains were clustered (n = 7). Relative levels of ZIKV plus S.E. are as shown. Student's t tests were performed, *, p < 0.05; ***, p < 0.001.
The scFv of BBS1 antibody expressing phages could inhibit ZIKV interactions with APP (Fig. 2h). Whether this interference affected ZIKV replication was examined. The BBS1 expression phages were mixed with ZIKV during viral infection of human NPCs/NSCs. The scFv of BBS1Ab treatment led to enhancement of ZIKV replication in human NPCs/NSCs (Fig. 6c).
The functional significance of APP in ZIKV infection in vivo was examined in APP-null mice. Because ZIKV cannot replicate in an immunocompetent mouse, and the efficiency of the viruses crossing the blood-brain barrier may complicate the interpretations, the intracranial injection of ZIKV into testing mice was chosen. Brains have different NPSs/NSCs proportions in different developmental stages, both neonatal and adult mice were examined for their susceptibility to ZIKV infections. The postnatal day 4 (P4) neonates were injected intracranially (i.c.) with ZIKV (1.5 × 104 pfu), and the differences in survival rates between APP-null and WT mice were compared (Fig. 6d). Clearly, APP-null neonates were more sensitive to ZIKV infection than their WT counterparts. Because of the higher mortality in P4 APP-null neonates (Fig. 6d), lower dosages of ZIKV (1 × 103 pfu) were chosen for another set of i.c. injections. Although all WT mice (10/10) survived, some APP-null neonates (3/14) died 2 days after ZIKV injection (data not shown). Whole brains were isolated after euthanasia from surviving neonates and RNAs were isolated. Relative levels of ZIKV RNA in the brains were determined by qRT-PCR. APP-null mice had higher viral loads in the brains than WT P4 ones (Fig. 6e).
In P7 neonates, however, the dosage of ZIKV (1.5 × 104 pfu) that caused serious neonate death in P4 (Fig. 3e) did not generate much effects on mortality in both APP-null as well as WT mice: all mice survived for 2 days. In the following viral load detections, APP-null mouse brains had 3-fold higher viruses than WT counterparts (Fig. 6e). Interestingly, when we tested the viral replication and mortality in P28 mice, there was no mortality in 4 days after i.c. injections with the higher ZIKV load, and no differences for viral replication were observed between the two strains of mice (Fig. 6e).
All those data suggest that APP is a negative regulator of ZIKV replication in vitro in human NPCs/NSCs, and in vivo in mouse brains. The inhibitory effects of APP on ZIKV replication seems to be associated with the age of mice.
Discussion
It is known that ZIKV may interact with many host proteins (65–67). APP is apparently another target of ZIKV from several lines of evidences including protein structural analyses, ZIKV attachment assay, and co-localization of APP and ZIKV-E in transfection experiments (Figs. 1–3). Because ZIKV has a limited effect on adult mouse brains, aging mouse brain tissues may have a protective effect on other susceptible cells by binding and trapping the virus (Fig. 2e). The interacting domain in APP is probably located around the BACE1-binding site based on findings that the scFv of antibody BSC1 could affect both ZIKV binding as well as viral replication in cells (Figs. 2h and 5). ZIKV at least uses its E protein to interact with APP, and the domain for the potential interaction may be partially around the similar structural element to BACE1: the ZIKV E protein-derived P2 peptide enhances APP expression (Figs. 1c and 5). It is interesting to note that the transfection of the viral P2 peptide is required for the enhancement of APP expression, suggesting that subcellular location of the APP-ZIKV interaction is inside the cells (Fig. 5). Furthermore, the results also suggest that ZIKV might attach to an insusceptible cell and even reach to the endosome (Figs. 2 and 5). Given the fact that we could only co-IP the APP from E antibody, but not in the other direction (Fig. 3a), there are some unknown factors involved in the APP-ZIKV interactions. However, from all the data available, it is clear that ZIKV and APP have a physical interaction, at least transiently.
One possible outcome associated with APP-ZIKV interaction might be the enhancement of APP expressions (Fig. 4). The increase of the APP expressions is at least partially through the stabilization of APP protein (Figs. 4 and 5). Because APP is quickly processed through a complicated and well-ordered proteolytic process, ZIKV might block BACE1 for APP digestion, stop the whole proteolytic processing, and thus increase the t1/2 of the APP protein. Of note, ZIKV replication is apparently not required for the increase of APP stability because of the time required for the enhancement and the insusceptible cells used for our experiments.
APP is a negative regulator for ZIKV replication in both human NPCs/NSCs and neonatal mouse brains (Fig. 6). It seems that age plays a role in APP-mediated inhibition of ZIKV replications in mouse brains in vivo. P4 mice are more susceptible to ZIKV infection than older mice (P7, P28) (Fig. 6, d and e). It is known that a higher proportion of NPCs/NSCs present in neonatal mouse brains than in mature ones (48–50). Therefore, there is an apparent correlation between the degree of NPCs/NSCs content and APP inhibition (Fig. 6e). APP is expressed in the embryos (68), and fetal brains are readily infectable by ZIKV (15). Considering that fetal brains may have similar or even higher proportions of NPCs/NSCs than those in P4/P7 neonatal brains, it is tempting to speculate that APP may exert similar anti-ZIKV effects in fetuses, and therefore be a factor in ZIKV-mediated microcephaly.
Unlike interferons, viral restriction factors tend to inhibit target viral replication in a modest fashion (27, 69, 70). Based on the facts that APP is predominantly expressed in the brains and our results, APP fits perfectly the definition as a viral restriction factor in the brains (24, 25). Our data provide a reasonable mechanism for APP as a restriction factor for ZIKV: viruses attach on the surface of the cells in a nonspecific manner first, concentrating the virus and facilitating binding to its specific receptor (71, 72). APP is expressed both on the cell surface and inside cells, and the extracellular ZIKV has the potential to interact with APP (Figs. 2 and 3b). APP traps ZIKV as a decoy receptor and prevents its interactions with receptors in the brains. The expression of APP can be enhanced by ZIKV attachments, which further exacerbates APP's negative effect. For a susceptible cell with no APP expression, the cell will fully support ZIKV replication (Fig. 7, situation A). This situation may account for the early ZIKV transmission in blood, because APP is predominantly expressed in the brains. For a susceptible cell with APP expression, APP may intercept ZIKV during viral infection as decoy receptor, and with the binding to ZIKV, the APP protein is stabilized by preventing its degradation (Fig. 4). As the additional APP may trap more viruses, ZIKV replication will be impaired (Fig. 7, situation B). This situation is present in NPCs/NSCs, the major targets of ZIKV in the brains. Another extreme situation is the cells that are not susceptible for ZIKV infection by lacking ZIKV receptors or some other essential factors. However, if APP is expressed in these cells, APP can still prevent ZIKV virus infection to other infectable cells by trapping the viruses (Fig. 7, situation C). This happens in aging brain cells with APP expression (Fig. 2e, Fig. S1b). This model may partly explain why adults are more resistant to ZIKV infections, not only because of reduced NPCs/NSCs numbers, but also the efficient trapping of biologically active viruses by APP. However, additional mechanisms are likely present in APP expressing cells for anti-ZIKV effects. Because other members of APP family (APLP-1 and -2) apparently have lower homologies to APP around the BACE1-binding site, whether APLP1 and -2 have a similar inhibitory function of ZIKV replication has not addressed yet.
Figure 7.
Model for APP as a restriction factor for ZIKV. APP could inhibit ZIKV replication in both susceptible (indicated with a receptor presence) and insusceptible cells (indicated by the lack of receptor). Situation A, APP would play no role in a susceptible cell without APP expression; situation B, in a susceptible cell with APP expression, APP would intercept ZIKV during viral infection by serving a decoy receptor. Furthermore, APP protein expression will be increased with the interactions, which in turn may bind to more viruses. Only a fraction of the viruses would enter the target cell for replication. Situation C, in an insusceptible cell with APP expression, the attachments to APP would reduce the overall viral burdens and availability in the host, and would lead to the inhibition of the viral replication as a whole.
Although APP is extensively studied, its biological function remains not very clear beyond being the parent protein of β-amyloid peptides. There is a consensus in the fields that APP is a factor involved in Alzheimer's disease. With this antiviral effect described herein, and known anti-bacteria property (73), APP might be a general factor against microbial mediated inflammations. Interestingly, some viruses, especially herpesviruses, are associated with Alzheimer's diseases (67). In addition, because APP proteins are undergoing constant processing, it is possible that a processed product may behave similarly to the intact APP. Moreover, other flaviviruses, such as dengue and West Nile virus (74, 75), infect human brains. Whether APP is a general restriction factor for flaviviruses, and whether ZIKV infection might be associated with long-term brain damages or Alzheimer's disease need further investigations.
Experimental procedures
Mice
C57BL/6J and APP-null mice (app−/−; B6.129S7-Apptm1Dbo/J) mice were purchased from Jackson Laboratories. All mice were bred and housed at the American Association for Accreditation of Laboratory Animal Care accredited facility under specific pathogen-free conditions. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Nebraska–Lincoln and followed federal guidelines.
Brain explant culture and ZIKV infection assays
For cortical cortex culture for viral attachment assay in Fig. 1, aged mice (>1-year–old) were euthanized with CO2. The cerebrum was removed after cutting the cranium from the neck to the nose. A midline incision between the hemispheres was performed, and the cortex from the brain was peeled, cut into small pieces (1-2 mm3), and passed through 70-μm cell strainers first, and a 40-μm strainer subsequently. The cell clumps were washed twice with Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS), counted, and dispensed at desired concentrations. The cells clumps were immediately incubated with ZIKV at 37 °C for 1 h for viral absorption study. For whole brain explant cultures in Fig. 2, 4-6–week–old mice were euthanized, whole brains were isolated, cut into smaller pieces, and passed through 70- and 40-μm cell strainers. The cell clumps were used directly for ZIKV infections, and collected for further analyses.
ZIKV infection study in mice
Four-day–old (P4), 1-week–old (P7), and 4-week–old (P28) mice were i.c. injected with 10-µl solutions with 31-gauge syringes for P4 and P7 mice or 50-µl solutions with 29-gauge syringes for P28 mice. The dosages per injection were: 103 pfu or 5 × 104 pfu for P4, 5 × 104 pfu for P7, and 5 × 105 pfu for P28 mice. Hypodermic anesthesia (P4, P7 mice) or isoflurane anesthesia (P28 mice) were used following the standard protocol (76). Animals were observed daily for clinical illness. The maintenance of the neonates was reported (77). The brains were collected at 2 day post-inoculation for P4 and P7 mice, and 4 days post-inoculation for P28 mice. RNA were isolated and used for measurement of ZIKV RNA. Mortality was observed for a period of 4 days.
Cells, virus, and plasmids
Human embryonic kidney fibroblast 293 (HEK293, CRL-1573) and its derivative 293T cells, human neuroblastoma cells SH-SY5Y were obtained from the ATCC. African green monkey kidney cell line Vero, and human hepatoma cell lines Huh7.5 were gifts from Dr. Padmanabhan. The cells were grown and maintained in DMEM containing 10% FBS and 1× penicillin-streptomycin (PS) in a humidified chamber with 5% CO2 at 37 °C.
Zika virus strain PRVABC59 was obtained from the Centers for Disease Control and Prevention, Fort Collins, CO, USA. The viruses were prepared in Vero cells or Huh7.5 cells. The stock virus was stored in small aliquots at −80 °C. Virus titers were determined by plaque assay on Vero cells. Basically, duplicates of serial 10-fold dilutions of virus were applied to Vero cell monolayers in 24-well–plates. The inoculum was removed after 1-h incubation, and the cell monolayers were overlaid with medium containing 1% low-gelling temperature agarose. The cells were fixed in 10% formaldehyde in PBS for 30 min, after 5 days incubation. The agarose plugs were removed, and the cells were stained with 0.1% crystal violet in 30% methanol. Plaques were counted and virus titers were calculated.
The pCAX FLAG APP (Addgene plasmid number 30154) is FLAG-tagged APP expression plasmid APP and pCAX APP 695 (Addgene plasmid number 30137), an expression plasmid for the predominant form of APP in the brains, were gifts from Dennis Selkoe and Tracy Young-Pearse (39). pCI-neo-ZIKV-prME is an expression plasmid for PrM-E protein and was a gift from Dr. Asit Patnaik. pcDNA3 (Invitrogen) is a cloning vector used as controls for transfections.
Human pluripotent stem cells (hPSCs) differentiation into neural progenitor cells (hNPCs) and mature cortical neurons and ZIKV infection
The hPSCs (H9 hESCs) were purchased from WiCell Research Institute (catalog number WA09, WiCell). The hPSCs were maintained in 6-well– plate coated with Matrigel (catalog number 354277, BD Biosciences) in Essential 8TM medium (E8, catalog number A1517001, Invitrogen)(78). Cells were passaged every 4 days with 0.5 mm EDTA (catalog number AM9260G, Invitrogen). Medium was changed daily. Cells were routinely checked for the expression of pluripotency markers, OCT4 (catalog number 962649, R&D System) and NANOG (catalog number 963488, R&D System), their capability to form teratomas in immunodeficient mice, their karyotypes, and bacterial and mycoplasma contaminations. The hPSCs were dissociated with accutase (catalog number A1110501, Life Technologies) and plated in Matrigel-coated 6-well–plates (2 × 106 cells/well) and cultured in E8 medium overnight to reach >90% confluence. E8 medium was removed and replaced with neural induction medium consisting of Essential 6TM medium (E6, catalog number A1516401, Invitrogen) supplied with 100 nm LDN193189 (catalog number S2618, Selleckchem) and 10 μm SB431542 (catalog number S1067, Selleckchem) for 11 days. The resulting cells were considered as hNPCs, and their identities were confirmed (56).
To differentiate hNPCs into the cortical neurons, hNPCs were harvested on day 11 and re-plated to Matrigel-coated 6-well–plates, and cultured in neural differentiation medium consisting of Neurobasal® Media (catalog number 21103049, Life Technologies), B27 (50×, catalog number 17504044, Life Technologies), brain-derived neurotrophic factor (20 ng/ml, catalog number 450-02, PeproTech), glial cell-derived neurotrophic factor (10 ng/ml, catalog number 450-10, PeproTech), l-ascorbic acid (200 μm, catalog number NC0602549, Life Technologies), and DAPT (2.5 μm, catalog number S2215, Selleckchem), Dibutyryl-cAMP (0.5 mm, catalog number sc-201567A, Santa Cruz Biotechnology) for another 19 days. Half medium was changed every 2 days. The identities of the mature neurons were Tbr1+ (catalog number ab31940, Abcam) and Tuj1+ (catalog number T8578, Sigma) dual positive (79). The cells were infected with ZIKV and treated with CHX as described in the text and figure legends.
Generation of BBS1 antibody ScFv expressing phages
Total RNA was extracted from BBS2 hybridoma cells using Tri-reagent (catalog number 93289, Sigma-Aldrich). The cDNA was synthesized from the total RNA by Moloney murine leukemia virus reverse transcriptase (11062603001, Sigma-Aldrich) using oligo(dT)23 primer (O4387, Sigma-Aldrich). The variable domains of the murine light and heavy chains were amplified and sequenced. Based on this, sequence-specific primers for BBS2 Vh and Vl were designed harboring the appropriate restriction sites. Next BBS Vh and Vl were cloned into PCC 16 phagemid. First the Vh was cloned using NcoI (number 10835315001, Sigma-Aldrich) and BclI restriction (number 10693952001, Sigma-Aldrich) sites, followed by Vl cloning based on EcoRI (number 10200310001, Sigma-Aldrich) and NotI (number 11014714001, Sigma-Aldrich) restriction sites, yielding PCC16-BBS ScFv phagemid. For production, TG1 bacteria (number LUC60502-2, Sigma-Aldrich) containing the PCC16-BBS ScFv phagemid were grown at 37 °C in 10 ml of 2YT medium (number Y2377, Sigma-Aldrich) with 100 μg/ml of Amp (number 40345717748211, Sigma-Aldrich) up to OD600 ∼ 0.5, infected with 1:100 M13K07 helper phage (number N0315S, New England Biolabs), and incubated for 30 min without shaking followed by 30 min with 100 rpm shaking at 37 °C. The infected cells were then grown in 1 liter of 2YT with 100 μg/ml of Amp and 70 μg/ml of Kan (number 60615, Sigma-Aldrich) overnight at 30 °C with 250 rpm shaking. The next day the growing media (containing the phages) was collected and phages were purified by PEG/NaCl (number 1546605, S7653 Sigma-Aldrich) precipitation twice followed by Cs gradient purification.
The binding capacity of the phages was evaluated using ELISA by binding of phage BBS1 scFv to BACE1 cleavage site peptide. A 96-well–plate was coated with MAP-[ISEVKMDA]8 (peptide representing the BACE1 cleavage site on APP), washed twice with PBST (0.05% Tween20) (PBS number 02-023-5A, BI; Tween-20 number P1379, Sigma-Aldrich) and blocked with 5% slim milk overnight at 4 °C. Different phage concentrations in equal volumes were added to the plate and allowed to bind for 1 h at 37 °C followed by 3 washes with PBST. Next, rabbit anti-phage polyclonal antibodies (number PA1-26758, Invitrogen) were added at 1:5,000 and allowed to bind for 1 h at 37 °C followed by 3 washes with PBST. For detection, goat anti-rabbit horseradish peroxidase-conjugated (number AP307P, Sigma-Aldrich) at 1:10,000 and allowed to bind for 1h at 37 °C followed by 3 washes with PBST. The assay was developed using OPD substrate.
For BBS1scFv phage treatments, hNPCs were incubated with ZIKV (1 m.o.i.) in the presence of either scFv expressing or helper phages (1 transforming unit per cell) for 1 h at 37 °C. Cells were washed with fresh medium and the uninfected viruses were removed. Two days later, cell media were subjected to qRT-PCR analysis.
ZIKV attachment assay
Attractene Transfection Reagent (catalog number 301007; Qiagen) was used for transfection of 293 or 293T cells. One day later, ZIKV were incubated with the cells for 1 h at 37 °C. The cells were washed three times with 1× PBS and used for RNA isolation for RT-PCR analyses. For explant cultures, different amounts of viruses were incubated with cortical cortex explant cultures at 37 °C for 1 h. We also preformed the experiments at 4 °C for 1 h, however, the background attachments were much higher than those at 37 °C (data not shown). The culture media were centrifuged in an Eppendorf centrifuge at the maximum speed for 1 min and the supernatants were used to isolate RNA using QIAamp Viral RNA Mini Kit (catalog number 52904, Qiagen). The equal amounts of RNA solutions were used for qRT-PCR analyses.
Immunofluorescence staining of cells
HEK293 cells were transfected with PrM-E and APP695 expression plasmids at 1:1 ratio. The next day the transfected cells were washed once with 1× PBS and fixed with 4% paraformaldehyde (Sigma) for 15 min at room temperature. Cells were permeabilized and blocked with 0.1% Triton X-100 (Sigma) and 0.4% BSA (A7906; Sigma) in PBS for 20 min. Samples were then incubated with APP C-terminal Ab (catalog number 802803, Biolegend; 1:200 dilutions) and ZIKV E antibody (catalog number GTX133314, GeneTex; 1:800 dilutions) for 1 h, followed by incubation with secondary antibodies (donkey anti-rabbit IgG secondary antibody Alexa Fluor 488 conjugate (catalog number A-21206, ThermoFisher Scientific) and donkey anti-mouse IgG secondary antibody Alexa Fluor 647 conjugate (catalog number A-31571, ThermoFisher Scientific) for 1 h more at room temperature. Both secondary antibodies were used with 1:1000 dilutions and the dilution buffer for primary and secondary antibodies was 0.1% Tween 20 (Sigma) and 0.4% BSA (A7906; Sigma) in 1× PBS. The samples were then stained with 4′,6-diamidino-2-phenylindole, washed three times with PBS, and mounded. All samples were examined a Nikon-Ti2 fluorescence microscope and images were obtained using a Nikon A1r-Ti2 confocal system at the Microscopy Core Facility at the UNL.
Western blotting analysis with enhanced chemiluminescence (ECL)
The antibodies for β-amyloid (B-4) (sc-28365), glyceraldehyde-3-phosphate dehydrogenase (0411) (sc-47724), and goat anti-mouse IgG-horseradish peroxidase (sc-2005) were from Santa Cruz. APP C-terminal Ab (Clone C1/6.1; catalog number 802803) was from Biolegend. Tubulin antibody (catalog number T6557) was purchased from Sigma. Separation of proteins on SDS-PAGE was carried out following the standard protocol. After the proteins were transferred to a nitrocellulose or Immobilon membrane, the membrane was blocked with 5% nonfat dry milk in TBST (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 0.05% Tween-20) at room temperature for 30 min. It was washed briefly with TBST, and incubated with the primary antibody in 1% milk in TBST for 1 h at room temperature, or overnight at 4 °C. After washing the membrane with TBST three times (10 min each), it was incubated with the secondary antibody at room temperature for 1 h. The membrane was then washed three times with TBST, treated with ECL detection reagents, and exposed to BlueBlotTM HS film from Life Science Products (XR-0810-100). The intensities of the target signals were measured by a Bio-Rad ChemiDoc MP Imaging system or using NIH ImageJ software.
RNAi
The hNPCs were infected with a pooled human APP siRNA/shRNA/RNAi Lentivirus (siAPP; catalog number iV001195) and its control, Scrambled siRNA GFP Lentivirus (siScramble; catalog number LVP015-G; both from Applied Biological Materials Inc.) at 1 m.o.i. overnight, and on the second day, cells were washed and infected with ZIKV (1 m.o.i.), and 1 h later the medium was replaced with fresh medium. Two days later, cell media were collected and subjected to real time qRT-PCR assays.
Transfection of plasmids and peptides
The P2 peptide (HGSQHSGMIVNDTGHETDENRAKVEITPNSPRAEATLGGFGSLGL, also shown in Fig. 1c) was synthesized by NovoPep Inc. at 98% purity and dissolve in water. BSA was purchased from New England BioLabs Inc. (B9000S). For peptide and plasmid co-transfections, HEK293 cells were cultured at 50–70% confluence in 24 wells and washed with DMEM without FBS twice. One μg of peptide or other proteins were mixed with the desired amounts of plasmids with the use of PULSin Reagent (Polyplus transfection) for 15 min at room temperature, and added to cells for 4 h at 37 °C. After removing the media, cells were incubated in DMEM plus FBS (10%) overnight. For transfections with plasmid only, HEK293 cells were seeded at 50–70% confluence in 12- or 24-well–plates. Plasmids were transfected using Attractene Transfection Reagent (catalog number 301007; Qiagen) following manufacturers' recommendations.
qRT-PCR
For RNA from brains or cells, TRIzol™ Reagents (catalog number 15596026; Invitrogen) were used for RNA isolation with standard protocol. For viral RNA in the media, QIAamp Viral RNA Mini Kits (catalog number 52904; Qiagen) were used following manufacturer's recommendations. SuperScript™ II Reverse Transcriptase (catalog number 18064014; Invitrogen) were used for first cDNA strand synthesis. Routine methods for semi-quantitative RT-PCR were performed as described. The ZIKV primers were: ZIK-F, 5′-CCGCTGCCCAACACAAG-3′; ZIK-R, 5′-CCACTAACGTTCTTTTGCAGACAT-3′. The human actin primers were: Actin 1, 5′-TTCTACAATGAGCTGCGTGT-3′, and Actin 2, 5′-GCCAGACAGCACTGTGTTGG-3′. For qRT-PCR, abundance of target RNAs was quantified by CFX96 Real-Time System (Bio-Rad) or Applied Biosystems Step One plus Real Time PCR system following the manufacturers' recommendations. The ZIKV primers were ZIK-F and -R. Primers for mouse GAPDH: 5′-GAAGGTGAAGGTCGGAGTA-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. The probes for ZIKV and GAPDH were 5′-AGCCT-ACCTTGACAAGCAATCAGACACTCAA-3′ and 5′-CAA-GCTTC-CCGTTCTCAGCC-3′, respectively. The probes were labeled with 6-carboxyfluorescein phosphoramidite (FAM) reporter dye at the 5' end and 6-carboxytetramethylrhodamine (TAMRA) at the 3' end. The 2−ΔΔCt method was used for calculation of the relative ZIKV expression (ZIKV/GAPDH) (80). The ZIKV primers and probe were designed based on sequence in E protein region of PRVABC59 (GenBank: MH158237.1), but the primers could be used for detection of many ZIKV isolates.
Protein stability assays
For transfection of 293 cells, Attractene Transfection Reagent (catalog number 301007; Qiagen) was used following the manufacturer's recommendations. Protein biosynthesis inhibitor, CHX, was purchased from Sigma (C7698). The transfected cells were treated with CHX (100 µg/ml) with ZIKV simultaneously and cell lysates were made at the indicated times and used for Western blotting analysis. For mature neurons, the cells were incubated with ZIKV for 1 h, and then CHX was added, and cell lysates were collected at the indicated times.
Co-immunoprecipitation of APP and ZIKV
HEK293T cells in 100-mm culture plates (CLS43016, Corning) were transfected with various plasmids with the use of Attractene Transfection Reagent (catalog number 301007; Qiagen). For transfection only experiments, APP695 and PrM-E expression plasmid (2 µg each) were be used, and the pcDNA3 vector was used to normalized the total DNA to 4 µg. For transfection and infection experiments, pcDNA3 and APP695-expressing plasmid (2 µg each) were used to transfect 2 plates each. A day later, the cells were infected with recombinant adenovirus expressing ZIKV PrM-E protein or GFP at 500 particles/cell for 24 h (53). One day later, cells were washed once with PBS and collected by centrifugations. The cells were resuspended in 1.5 ml of lysis buffer (25 mm Tris-HCl, pH 7.6, 75 mm NaCl, 0.5% Nonidet P-40) plus protease inhibitor mixture (11836170001, Roche Applied Science) and phenylmethylsulfonyl fluoride (PMSF) (1 mm), and rotated at 4 °C for 20 min. Cell lysates were obtained by centrifugation at maximum speed in an Eppendorf microcentrifuge at 4 °C for 15 min. The 0.7 ml of the cell lysates were first cleared with 4 µl of normal rabbit serum plus 20 µl of Pierce™ Protein G-agarose beads (20398, ThermoFisher Scientific Inc.) for 30 min. Following removal of Protein G-agarose beads, lysates were incubated with 4 μl of the Anti-Zika Virus Envelope (E) Protein Antibody (EFS001, Kerafast, Inc.) and 20 μl of Protein G-agarose beads overnight at 4 °C. The beads were subsequently washed 3 times at 10 min each with washing buffer TBST (20 mm Tris-HCl, pH 7.6, 200 mm NaCl, 0.1% Tween 20) plus 1 mm PMSF, resuspended in 20 μl of SDS sample buffer prior to analysis by SDS-PAGE. Mouse anti-pan flavivirus envelope E protein mAb 4G2 (EMD Millipore; catalog number MAB10216) was used in immunoblot with nonreducing conditions for detection of ZIKV E protein, and APP C-terminal Ab (clone C1/6.1; catalog number 802803; Biolegend) was used for detection of APP.
For the immunoprecipitation of ZIKV virions, HEK293T cells were transfected with APP695 expression plasmid or vector control. One day later, cells were extensively washed with 1× PBS and lysed in the buffer containing 50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 0.5% (v/v) Nonidet P-40, and 2 mm EDTA for 30 min at 4 °C. The lysates were cleared with 30 min centrifugation at 4 °C. The normal rabbit sera were added with protein G beads for 30 min. The cell lysates were then divided with different antibodies plus protein G beads and rotated overnight at 4 °C. The mixture of three antibodies (CloudClone, catalog number PAB020Mu01; Boster, catalog number P89091; Aviva, catalog number ARP3401; and all 1:500 dilutions) were used to pull down APP proteins. The beads were washed three times with TBST plus PMSF three times and processed for RNA isolation with TRIzol method with addition of 20 µg of glycogen (ThermoFisher, catalog number R0551 RNA grade). The RNA were dissolved in 20 µl of water and 8 µl of the RNA were used for cDNA synthesis with the SuperScript® III first-strand synthesis system (Invitrogen, catalog number 18080051). PCR amplification with ZikaF and ZikaR primers was followed and the resulting products were separated on 8% DNA polyacrylamide gels.
Statistical analysis
General statistical analyses were performed by functions implemented in Microsoft Excel and R. Two group comparisons were done with Student's two-tailed unpaired t test. The R package of “Survival” was used to fit and plot the survival curve and the Log-Rank test was used to calculate p values.
Data availability
All the data are in the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Asit Patnaik for providing the ZIKV PrM-E expression plasmid, Dr. Padmanabhan for Huh7.5 and Vero cells, and Drs. Dennis Selkoe and Tracy Young-Pearse for APP expression plasmids.
This article contains supporting information.
Author contributions—A. L. and C. Z. validation; A. L., H. L., Y. G., P. P., V. L., Y. L., T. M. P., and C. Z. investigation; Y. G., E. W., Y. L., and B. S. resources; C. Z. and L. Z. conceptualization; C. Z. and L. Z. formal analysis; C. Z. and L. Z. writing-review and editing; L. Z. data curation; L. Z. supervision; L. Z. funding acquisition; L. Z. methodology; L. Z. writing-original draft.
Funding and additional information—This work was supported by the Laymen Award and Revision Award from UNL (to L. Z.). P. P. was a Fogarty fellow supported in part by the National Institutes of Health Fogarty International Center Grant D43TW010354. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ZIKV
- Zika virus
- NPC
- neural progenitor cell
- NSC
- neural stem cells
- APP
- amyloid precursor protein
- DC-SIGN
- dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin
- BACE1
- beta-secretase 1
- qRT
- quantitative real time
- scFv
- single-chain variable fragment
- co-IP
- co-immunoprecipitation assay
- LMP1
- latent membrane protein 1
- CHX
- cycloheximide
- i.c
- intracranially
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- hPSC
- human pluripotent stem cells
- hNPC
- human neural progenitor cell
- PMSF
- phenylmethylsulfonyl fluoride
- m.o.i
- multiplicity of infection.
References
- 1. Faye, O., Freire, C. C., Iamarino, A., Faye, O., de Oliveira, J. V., Diallo, M., Zanotto, P. M., and Sall, A. A. (2014) Molecular evolution of Zika virus during its emergence in the 20th century. PLoS Negl. Trop. Dis. 8, e2636 10.1371/journal.pntd.0002636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berthet, N., Nakouné, E., Kamgang, B., Selekon, B., Descorps-Declere, S., Gessain, A., Manuguerra, J. C., and Kazanji, M. (2014) Molecular characterization of three Zika flaviviruses obtained from sylvatic mosquitoes in the Central African Republic. Vector Borne Zoonotic Dis. 14, 862–865 10.1089/vbz.2014.1607 [DOI] [PubMed] [Google Scholar]
- 3. Foy, B. D., Kobylinski, K. C., Chilson Foy, J. L., Blitvich, B. J., Travassos da Rosa, A., Haddow, A. D., Lanciotti, R. S., and Tesh, R. B. (2011) Probable non-vector-borne transmission of Zika virus, Colorado, U.S.A. Emerg. Infect. Dis. 17, 880–882 10.3201/eid1705.101939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Musso, D., Roche, C., Robin, E., Nhan, T., Teissier, A., and Cao-Lormeau, V. M. (2015) Potential sexual transmission of Zika virus. Emerg. Infect. Dis. 21, 359–361 10.3201/eid2102.141363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zamani, M., and Zamani, V. (2017) Sexual transmission of Zika virus: an assessment of the evidence. Iran J. Public Health 46, 1305–1306 [PMC free article] [PubMed] [Google Scholar]
- 6. D'Ortenzio, E., Matheron, S., Yazdanpanah, Y., de Lamballerie, X., Hubert, B., Piorkowski, G., Maquart, M., Descamps, D., Damond, F., and Leparc-Goffart, I. (2016) Evidence of sexual transmission of Zika virus. N. Engl. J. Med. 374, 2195–2198 10.1056/NEJMc1604449 [DOI] [PubMed] [Google Scholar]
- 7. Brasil, P., Pereira, J. P., Jr., Moreira, M. E., Ribeiro Nogueira, R. M., Damasceno, L., Wakimoto, M., Rabello, R. S., Valderramos, S. G., Halai, U. A., Salles, T. S., Zin, A. A., Horovitz, D., Daltro, P., Boechat, M., Raja Gabaglia, C., et al. (2016) Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 375, 2321–2334 10.1056/NEJMoa1602412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Driggers, R. W., Ho, C. Y., Korhonen, E. M., Kuivanen, S., Jääskeläinen, A. J., Smura, T., Rosenberg, A., Hill, D. A., DeBiasi, R. L., Vezina, G., Timofeev, J., Rodriguez, F. J., Levanov, L., Razak, J., Iyengar, P., et al. (2016) Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N. Engl. J. Med. 374, 2142–2151 10.1056/NEJMoa1601824 [DOI] [PubMed] [Google Scholar]
- 9. Petersen, E., Wilson, M. E., Touch, S., McCloskey, B., Mwaba, P., Bates, M., Dar, O., Mattes, F., Kidd, M., Ippolito, G., Azhar, E. I., and Zumla, A. (2016) Rapid spread of Zika virus in the Americas: implications for public health preparedness for mass gatherings at the 2016 Brazil Olympic games. Int. J. Infect. Dis. 44, 11–15 10.1016/j.ijid.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 10. Calvet, G., Aguiar, R. S., Melo, A. S., Sampaio, S. A., de Filippis, I., Fabri, A., Araujo, E. S., de Sequeira, P. C., de Mendonca, M. C., de Oliveira, L., Tschoeke, D. A., Schrago, C. G., Thompson, F. L., Brasil, P., Dos Santos, F. B., et al. (2016) Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 16, 653–660 10.1016/S1473-3099(16)00095-5 [DOI] [PubMed] [Google Scholar]
- 11. Besnard, M., Lastere, S., Teissier, A., Cao-Lormeau, V., and Musso, D. (2014) Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February. Euro. Surveill. 19, 20751 10.2807/1560-7917.ES2014.19.13.20751 [DOI] [PubMed] [Google Scholar]
- 12. Tang, H., Hammack, C., Ogden, S. C., Wen, Z., Qian, X., Li, Y., Yao, B., Shin, J., Zhang, F., Lee, E. M., Christian, K. M., Didier, R. A., Jin, P., Song, H., and Ming, G. L. (2016) Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18, 587–590 10.1016/j.stem.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rasmussen, S. A., Jamieson, D. J., Honein, M. A., and Petersen, L. R. (2016) Zika virus and birth defects: reviewing the evidence for causality. N. Engl. J. Med. 374, 1981–1987 10.1056/NEJMsr1604338 [DOI] [PubMed] [Google Scholar]
- 14. Miner, J. J., Cao, B., Govero, J., Smith, A. M., Fernandez, E., Cabrera, O. H., Garber, C., Noll, M., Klein, R. S., Noguchi, K. K., Mysorekar, I. U., and Diamond, M. S. (2016) Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165, 1081–1091 10.1016/j.cell.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li, C., Xu, D., Ye, Q., Hong, S., Jiang, Y., Liu, X., Zhang, N., Shi, L., Qin, C. F., and Xu, Z. (2016) Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19, 120–126 10.1016/j.stem.2016.04.017 [DOI] [PubMed] [Google Scholar]
- 16. Cugola, F. R., Fernandes, I. R., Russo, F. B., Freitas, B. C., Dias, J. L., Guimaraes, K. P., Benazzato, C., Almeida, N., Pignatari, G. C., Romero, S., Polonio, C. M., Cunha, I., Freitas, C. L., Brandão, W. N., Rossato, C., et al. (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271 10.1038/nature18296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ashwal, S., Michelson, D., Plawner, L., Dobyns, W. B., Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society (2009) Practice parameter: Evaluation of the child with microcephaly (an evidence-based review): report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 73, 887–897 10.1212/WNL.0b013e3181b783f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcez, P. P., Loiola, E. C., Madeiro da Costa, R., Higa, L. M., Trindade, P., Delvecchio, R., Nascimento, J. M., Brindeiro, R., Tanuri, A., and Rehen, S. K. (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- 19. Dang, J., Tiwari, S. K., Lichinchi, G., Qin, Y., Patil, V. S., Eroshkin, A. M., and Rana, T. M. (2016) Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258–265 10.1016/j.stem.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., Yao, B., Hamersky, G. R., Jacob, F., Zhong, C., Yoon, K. J., Jeang, W., Lin, L., Li, Y., Thakor, J., et al. (2016) Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Onorati, M., Li, Z., Liu, F., Sousa, A. M. M., Nakagawa, N., Li, M., Dell'Anno, M. T., Gulden, F. O., Pochareddy, S., Tebbenkamp, A. T. N., Han, W., Pletikos, M., Gao, T., Zhu, Y., Bichsel, C., et al. (2016) Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell. Rep. 16, 2576–2592 10.1016/j.celrep.2016.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao, Q., Herrlinger, S., Yang, S. L., Lai, F., Moore, J. M., Brindley, M. A., and Chen, J. F. (2016) Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development 143, 4127–4136 10.1242/dev.143768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu, K. Y., Zuo, G. L., Li, X. F., Ye, Q., Deng, Y. Q., Huang, X. Y., Cao, W. C., Qin, C. F., and Luo, Z. G. (2016) Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res 26, 645–654 10.1038/cr.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen, Z., Zhang, L., and Ying, S. (2012) SAMHD1: a novel antiviral factor in intrinsic immunity. Future Microbiol. 7, 1117–1126 10.2217/fmb.12.81 [DOI] [PubMed] [Google Scholar]
- 25. Yan, N., and Chen, Z. J. (2012) Intrinsic antiviral immunity. Nat. Immunol. 13, 214–222 10.1038/ni.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lafaille, F. G., Pessach, I. M., Zhang, S. Y., Ciancanelli, M. J., Herman, M., Abhyankar, A., Ying, S. W., Keros, S., Goldstein, P. A., Mostoslavsky, G., Ordovas-Montanes, J., Jouanguy, E., Plancoulaine, S., Tu, E., Elkabetz, Y., et al. (2012) Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491, 769–773 10.1038/nature11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brass, A. L., Huang, I. C., Benita, Y., John, S. P., Krishnan, M. N., Feeley, E. M., Ryan, B. J., Weyer, J. L., van der Weyden, L., Fikrig, E., Adams, D. J., Xavier, R. J., Farzan, M., and Elledge, S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zani, A., Zhang, L., McMichael, T. M., Kenney, A. D., Chemudupati, M., Kwiek, J. J., Liu, S. L., and Yount, J. S. (2019) Interferon-induced transmembrane proteins inhibit cell fusion mediated by trophoblast syncytins. J. Biol. Chem. 294, 19844–19851 10.1074/jbc.AC119.010611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soto-Acosta, R., Xie, X., Shan, C., Baker, C. K., Shi, P. Y., Rossi, S. L., Garcia-Blanco, M. A., and Bradrick, S. (2018) Fragile X mental retardation protein is a Zika virus restriction factor that is antagonized by subgenomic flaviviral RNA. Elife 7, 10.7554/eLife.39023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiramel, A. I., Meyerson, N. R., McNally, K. L., Broeckel, R. M., Montoya, V. R., Mendez-Solis, O., Robertson, S. J., Sturdevant, G. L., Lubick, K. J., Nair, V., Youseff, B. H., Ireland, R. M., Bosio, C. M., Kim, K., Luban, J., et al. (2019) TRIM5α restricts flavivirus replication by targeting the viral protease for proteasomal degradation. Cell Rep. 27, 3269–3283e6 10.1016/j.celrep.2019.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Brien, R. J., and Wong, P. C. (2011) Amyloid precursor protein processing and Alzheimer's disease. Annu. Rev. Neurosci. 34, 185–204 10.1146/annurev-neuro-061010-113613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai, L., Song, J., Lu, X., Deng, Y. Q., Musyoki, A. M., Cheng, H., Zhang, Y., Yuan, Y., Song, H., Haywood, J., Xiao, H., Yan, J., Shi, Y., Qin, C. F., Qi, J., et al. (2016) Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 10.1016/j.chom.2016.04.013 [DOI] [PubMed] [Google Scholar]
- 33. Sirohi, D., Chen, Z., Sun, L., Klose, T., Pierson, T. C., Rossmann, M. G., and Kuhn, R. J. (2016) The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 10.1126/science.aaf5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel, E., and Henrick, K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 35. Zhang, Y., and Skolnick, J. (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33, 2302–2309 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamel, R., Dejarnac, O., Wichit, S., Ekchariyawat, P., Neyret, A., Luplertlop, N., Perera-Lecoin, M., Surasombatpattana, P., Talignani, L., Thomas, F., Cao-Lormeau, V. M., Choumet, V., Briant, L., Despres, P., Amara, A., et al. (2015) Biology of Zika virus infection in human skin cells. J. Virol. 89, 8880–8896 10.1128/JVI.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cole, S. L., and Vassar, R. (2008) The role of amyloid precursor protein processing by BACE1, the beta-secretase, in Alzheimer disease pathophysiology. J. Biol. Chem. 283, 29621–29625 10.1074/jbc.R800015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Young-Pearse, T. L., Bai, J., Chang, R., Zheng, J. B., LoTurco, J. J., and Selkoe, D. J. (2007) A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J. Neurosci. 27, 14459–14469 10.1523/JNEUROSCI.4701-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Platt, E. J., Kozak, S. L., Durnin, J. P., Hope, T. J., and Kabat, D. (2010) Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J. Virol. 84, 3106–3110 10.1128/JVI.01958-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong, D., Zhang, T. H., Zhao, D., Du, Y., Chapa, T. J., Shi, Y., Wang, L., Contreras, D., Zeng, G., Shi, P. Y., Wu, T. T., Arumugaswami, V., and Sun, R. (2018) High-throughput fitness profiling of Zika virus E protein reveals different roles for glycosylation during infection of mammalian and mosquito cells. iScience 1, 97–111 10.1016/j.isci.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davis, C. W., Nguyen, H. Y., Hanna, S. L., Sánchez, M. D., Doms, R. W., and Pierson, T. C. (2006) West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 80, 1290–1301 10.1128/JVI.80.3.1290-1301.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith, D. R., Sprague, T. R., Hollidge, B. S., Valdez, S. M., Padilla, S. L., Bellanca, S. A., Golden, J. W., Coyne, S. R., Kulesh, D. A., Miller, L. J., Haddow, A. D., Koehler, J. W., Gromowski, G. D., Jarman, R. G., Alera, M. T. P., et al. (2018) African and Asian Zika virus isolates display phenotypic differences both in vitro and in vivo. Am. J. Trop. Med. Hyg. 98, 432–444 10.4269/ajtmh.17-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu, H., Liao, H. M., Li, B., Tsai, S., Hung, G. C., and Lo, S. C. (2019) Comparative genomics, infectivity and cytopathogenicity of American isolates of Zika virus that developed persistent infections in human embryonic kidney (HEK293) cells. Int. J. Mol. Sci. 20, 3035 10.3390/ijms20123035] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li, H., Saucedo-Cuevas, L., Regla-Nava, J. A., Chai, G., Sheets, N., Tang, W., Terskikh, A. V., Shresta, S., and Gleeson, J. G. (2016) Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 19, 593–598 10.1016/j.stem.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hughes, B. W., Addanki, K. C., Sriskanda, A. N., McLean, E., and Bagasra, O. (2016) Infectivity of immature neurons to Zika virus: a link to congenital Zika syndrome. EBioMedicine 10, 65–70 10.1016/j.ebiom.2016.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muñoz, L. S., Barreras, P., and Pardo, C. A. (2016) Zika virus-associated neurological disease in the adult: Guillain-Barre syndrome, encephalitis, and myelitis. Semin. Reprod. Med. 34, 273–279 10.1055/s-0036-1592066 [DOI] [PubMed] [Google Scholar]
- 48. Gage, F. H., and Temple, S. (2013) Neural stem cells: generating and regenerating the brain. Neuron 80, 588–601 10.1016/j.neuron.2013.10.037 [DOI] [PubMed] [Google Scholar]
- 49. Feliciano, D. M., and Bordey, A. (2013) Newborn cortical neurons: only for neonates? Trends Neurosci. 36, 51–61 10.1016/j.tins.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Semple, B. D., Blomgren, K., Gimlin, K., Ferriero, D. M., and Noble-Haeusslein, L. J. (2013) Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 106-107, 1–16 10.1016/j.pneurobio.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arbel, M., Yacoby, I., and Solomon, B. (2005) Inhibition of amyloid precursor protein processing by beta-secretase through site-directed antibodies. Proc. Natl. Acad. Sci. U.S.A. 102, 7718–7723 10.1073/pnas.0502427102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Larocca, R. A., Abbink, P., Peron, J. P., Zanotto, P. M., Iampietro, M. J., Badamchi-Zadeh, A., Boyd, M., Ng'ang'a, D., Kirilova, M., Nityanandam, R., Mercado, N. B., Li, Z., Moseley, E. T., Bricault, C. A., Borducchi, E. N., et al. (2016) Vaccine protection against Zika virus from Brazil. Nature 536, 474–478 10.1038/nature18952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bullard, B. L., Corder, B. N., Gorman, M. J., Diamond, M. S., and Weaver, E. A. (2018) Efficacy of a T cell-biased adenovirus vector as a Zika virus vaccine. Sci. Rep. 8, 18017 10.1038/s41598-018-35755-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Verweij, F. J., van Eijndhoven, M. A., Hopmans, E. S., Vendrig, T., Wurdinger, T., Cahir-McFarland, E., Kieff, E., Geerts, D., van der Kant, R., Neefjes, J., Middeldorp, J. M., and Pegtel, D. M. (2011) LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 30, 2115–2129 10.1038/emboj.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu, D., Brumm, K., and Zhang, L. (2006) The latent membrane protein 1 of Epstein-Barr virus (EBV) primes EBV latency cells for type I interferon production. J. Biol. Chem. 281, 9163–9169 10.1074/jbc.M511884200 [DOI] [PubMed] [Google Scholar]
- 56. Lin, H., Li, Q., and Lei, Y. (2017) An integrated miniature bioprocessing for personalized human induced pluripotent stem cell expansion and differentiation into neural stem cells. Sci. Rep. 7, 40191 10.1038/srep40191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ramos da Silva, S., Cheng, F., Huang, I. C., Jung, J. U., and Gao, S. J. (2019) Efficiencies and kinetics of infection in different cell types/lines by African and Asian strains of Zika virus. J. Med. Virol. 91, 179–189 10.1002/jmv.25306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lloyd-Williams, P., Albericio, F., and Giralt, E. (1997) Chemical Approaches to the Synthesis of Peptides and Proteins, CRC Press, Boca Raton, FL [Google Scholar]
- 59. Grant, G. A. (2002) Synthetic Peptides: A User's Guide, 2nd Ed., Oxford University Press, Oxford; New York [Google Scholar]
- 60. Das, U., Wang, L., Ganguly, A., Saikia, J. M., Wagner, S. L., Koo, E. H., and Roy, S. (2016) Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat. Neurosci. 19, 55–64 10.1038/nn.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang, X., and Song, W. (2013) The role of APP and BACE1 trafficking in APP processing and amyloid-β generation. Alzheimers Res. Ther. 5, 46 10.1186/alzrt211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Munjal, A., Khandia, R., Dhama, K., Sachan, S., Karthik, K., Tiwari, R., Malik, Y. S., Kumar, D., Singh, R. K., Iqbal, H. M. N., and Joshi, S. K. (2017) Advances in developing therapies to combat Zika virus: current knowledge and future perspectives. Front. Microbiol. 8, 1469 10.3389/fmicb.2017.01469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Owczarek, K., Chykunova, Y., Jassoy, C., Maksym, B., Rajfur, Z., and Pyrc, K. (2019) Zika virus: mapping and reprogramming the entry. Cell Commun. Signal. 17, 41 10.1186/s12964-019-0349-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., and Studer, L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Esteves, E., Rosa, N., Correia, M. J., Arrais, J. P., and Barros, M. (2017) New targets for Zika virus determined by human-viral interactomic: a bioinformatics approach. Biomed. Res. Int. 2017, 1734151 10.1155/2017/1734151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coyaud, E., Ranadheera, C., Cheng, D. T., Goncalves, J., Dyakov, B., Laurent, E., St-Germain, J. R., Pelletier, L., Gingras, A. C., Brumell, J. H., Kim, P. K., Safronetz, D., and Raught, B. (2018) Global interactomics uncovers extensive organellar targeting by Zika virus. Mol. Cell. Proteomics 17, 2242–2255 10.1074/mcp.TIR118.000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Scaturro, P., Stukalov, A., Haas, D. A., Cortese, M., Draganova, K., Płaszczyca, A., Bartenschlager, R., Götz, M., and Pichlmair, A. (2018) An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 561, 253–257 10.1038/s41586-018-0484-5 [DOI] [PubMed] [Google Scholar]
- 68. Fisher, S., Gearhart, J. D., and Oster-Granite, M. L. (1991) Expression of the amyloid precursor protein gene in mouse oocytes and embryos. Proc. Natl. Acad. Sci. U.S.A. 88, 1779–1782 10.1073/pnas.88.5.1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H., and Landau, N. R. (2003) Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- 70. Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P., and Sodroski, J. (2004) The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 10.1038/nature02343 [DOI] [PubMed] [Google Scholar]
- 71. Cruz-Oliveira, C., Freire, J. M., Conceição, T. M., Higa, L. M., Castanho, M. A., and Da Poian, A. T. (2015) Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol. Rev. 39, 155–170 10.1093/femsre/fuu004 [DOI] [PubMed] [Google Scholar]
- 72. Grove, J., and Marsh, M. (2011) The cell biology of receptor-mediated virus entry. J. Cell Biol. 195, 1071–1082 10.1083/jcb.201108131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kumar, D. K., Choi, S. H., Washicosky, K. J., Eimer, W. A., Tucker, S., Ghofrani, J., Lefkowitz, A., McColl, G., Goldstein, L. E., Tanzi, R. E., and Moir, R. D. (2016) Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci. Transl. Med. 8, 340ra72 10.1126/scitranslmed.aaf1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li, G. H., Ning, Z. J., Liu, Y. M., and Li, X. H. (2017) Neurological manifestations of Dengue infection. Front. Cell Infect. Microbiol. 7, 449 10.3389/fcimb.2017.00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Debiasi, R. L., and Tyler, K. L. (2006) West Nile virus meningoencephalitis. Nat. Clin. Pract. Neurol. 2, 264–275 10.1038/ncpneuro0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim, J. Y., Grunke, S. D., Levites, Y., Golde, T. E., and Jankowsky, J. L. (2014) Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J. Vis. Exp. 91, 51863 10.3791/51863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kreikemeier-Bower, C., Polepole, P., Pinkerton, K., and Zhang, L. (2020) A simple method for short-term maintenance of neonatal mice without foster mothers. J. Biol. Methods 7, e126 10.14440/jbm.2020.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen, G., Gulbranson, D. R., Hou, Z., Bolin, J. M., Ruotti, V., Probasco, M. D., Smuga-Otto, K., Howden, S. E., Diol, N. R., Propson, N. E., Wagner, R., Lee, G. O., Antosiewicz-Bourget, J., Teng, J. M., and Thomson, J. A. (2011) Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 8, 424–429 10.1038/nmeth.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin, H., Du, Q., Li, Q., Wang, O., Wang, Z., Liu, K., Elowsky, C., Zhang, C., and Lei, Y. (2018) Hydrogel-based bioprocess for scalable manufacturing of human pluripotent stem cell-derived neural stem cells. ACS Appl. Mater. Interfaces 10, 29238–29250 10.1021/acsami.8b05780 [DOI] [PubMed] [Google Scholar]
- 80. Livak, K. J., and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-DeltaDeltaC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are in the manuscript.