Abstract
In addition to their well-known role in the control of cellular proliferation and cancer, cell cycle regulators are increasingly identified as important metabolic modulators. Several GWAS have identified SNPs near CDKN2A, the locus encoding for p16INK4a (p16), associated with elevated risk for cardiovascular diseases and type-2 diabetes development, two pathologies associated with impaired hepatic lipid metabolism. Although p16 was recently shown to control hepatic glucose homeostasis, it is unknown whether p16 also controls hepatic lipid metabolism. Using a combination of in vivo and in vitro approaches, we found that p16 modulates fasting-induced hepatic fatty acid oxidation (FAO) and lipid droplet accumulation. In primary hepatocytes, p16-deficiency was associated with elevated expression of genes involved in fatty acid catabolism. These transcriptional changes led to increased FAO and were associated with enhanced activation of PPARα through a mechanism requiring the catalytic AMPKα2 subunit and SIRT1, two known activators of PPARα. By contrast, p16 overexpression was associated with triglyceride accumulation and increased lipid droplet numbers in vitro, and decreased ketogenesis and hepatic mitochondrial activity in vivo. Finally, gene expression analysis of liver samples from obese patients revealed a negative correlation between CDKN2A expression and PPARA and its target genes. Our findings demonstrate that p16 represses hepatic lipid catabolism during fasting and may thus participate in the preservation of metabolic flexibility.
Keywords: CDKN2A, PPARα, liver, lipid metabolism, fatty acid oxidation, AMPKα, steatosis, cell cycle, peroxisome proliferator-activated receptor (PPAR), AMP-activated kinase (AMPK)
Cell cycle regulators have been extensively studied in the context of proliferation, cancer development, and aging (1). Progression through the cell cycle requires specific metabolic programs for synthesis of cellular building blocks or ATP production (2). Interestingly, recent work has shown that several cell cycle regulators also modulate metabolism in nonproliferative cells, suggesting new physiological functions of this large family of proteins (3).
P16INK4a (p16) is a cyclin-dependent kinase inhibitor that blocks activation of E2F transcription factors via inhibition of CDK4/6 (4). Interestingly, several GWAS have identified single nucleotide polymorphism near CDKN2A, the locus encoding for p16, as associated with elevated risk for cardiovascular disease and type-2 diabetes (T2D) development (5). In line, we recently reported that p16-deficient mice display elevated hepatic gluconeogenesis during fasting due to activation of a cascade involving CDK4, PKA, CREB, and PGC1α in hepatocytes, suggesting an important role for p16 in metabolic control (6). Interestingly, other cell cycle regulators in the CDK4/Cyclin D-E2F1 pathway, downstream of p16, have also been implicated in the control of hepatic lipid metabolism (7, 8). In addition, impaired hepatic lipid metabolism (e.g. as seen during aging or exposure to high fat diet feeding) is associated with increased hepatic expression of p16 (9, 10). However, whether p16 directly regulates hepatic lipid homeostasis remains unknown.
The AMP-activated protein kinase (AMPK) is an important regulator of hepatic lipid homeostasis. During prolonged fasting, AMPK senses cellular energetic deficit and activates fatty acid oxidation (FAO) to reestablish normal energy balance (11). AMPK is a heterotrimeric complex composed of one catalytic subunit α and two regulatory subunits β and γ, each of which have several different isoforms. There are two catalytic subunit isoforms, AMPKα1 (PRKAA1) and AMPKα2 (PRKAA2), and their phosphorylation at Thr-172 is critical for AMPK activation. The tissue expression of these two isoforms is different, however, whether there are specific roles for each isoform remains unresolved. Interestingly, CDK4 phosphorylates AMPKα2, and not AMPKα1, at several sites (other than Thr-172) thereby suppressing AMPK activity and FAO (12). Moreover, other cell cycle regulators such as Cyclin D1, a CDK4 partner (13), inhibit the activity of PPARα, a master regulator of lipid metabolism gene expression and downstream effector of AMPK activation. Overall, these studies highlight the close interplay between cell cycle and energy balance regulators.
In this study, we assessed the effects of hepatic p16 expression on fasting lipid metabolism, and found that modulation of p16 expression regulates hepatic FAO, mitochondrial function, and ketogenesis. p16-deficiency leads to activation of a cascade involving AMPKα2, SIRT1, and PPARα, which drives enhanced expression of lipid catabolism genes. Interestingly, we found these effects of p16-deficiency to be independent of changes in CDK4 activity. Moreover, adenovirus-mediated overexpression of p16 led to accumulation of LD in vitro, and decreased hepatic mitochondrial activity and ketogenesis in vivo. Our findings highlight a new role for p16 in the hepatic response to fasting and uncover a novel mechanism by which p16 may contribute to the development of metabolic diseases via modulation of hepatic mitochondrial function.
Results
p16-deficiency increases fatty acid catabolic gene expression in hepatocytes
To determine whether p16-deficiency affects hepatic lipid metabolism, we performed transcriptomic profiling of primary hepatocytes isolated from p16+/+ and p16−/− mice cultured under conditions mimicking fasting (as described under “Materials and methods”). There were 3289 differentially expressed genes between p16+/+ and p16−/− cells (false discovery rate < 0.2). Gene Ontology terms enrichment analysis revealed that several metabolic pathways are significantly affected by p16 deficiency, including oxidation reduction, α-amino acid metabolism, and xenobiotic metabolism (Fig. 1A), suggesting that p16 regulates mitochondrial and peroxisomal functions. As expected, principle component analysis of the 3289 differentially expressed genes identified a clear separation based on p16 genotype (Fig. 1B). Interestingly, the loadings for several genes involved in FAO and ketogenesis (e.g. Cpt1a, Ehhadh, Hmgcs2, Bdh1) were highly correlated with PC1 (Fig. 1B), indicating that their expression pattern contributes strongly to the difference between the p16+/+ and p16−/− transcriptomes. We then specifically investigated metabolic pathways associated with fatty acid (FA) mobilization, storage and utilization (Fig. 1C). Interestingly, p16-deficiency was associated with marked changes in these pathways, including activation of both mitochondrial and peroxisomal catabolic pathways (e.g. Ehhadh, Cpt1a, Hmgcs2, and Bdh1, Fig. S1A).
Figure 1.
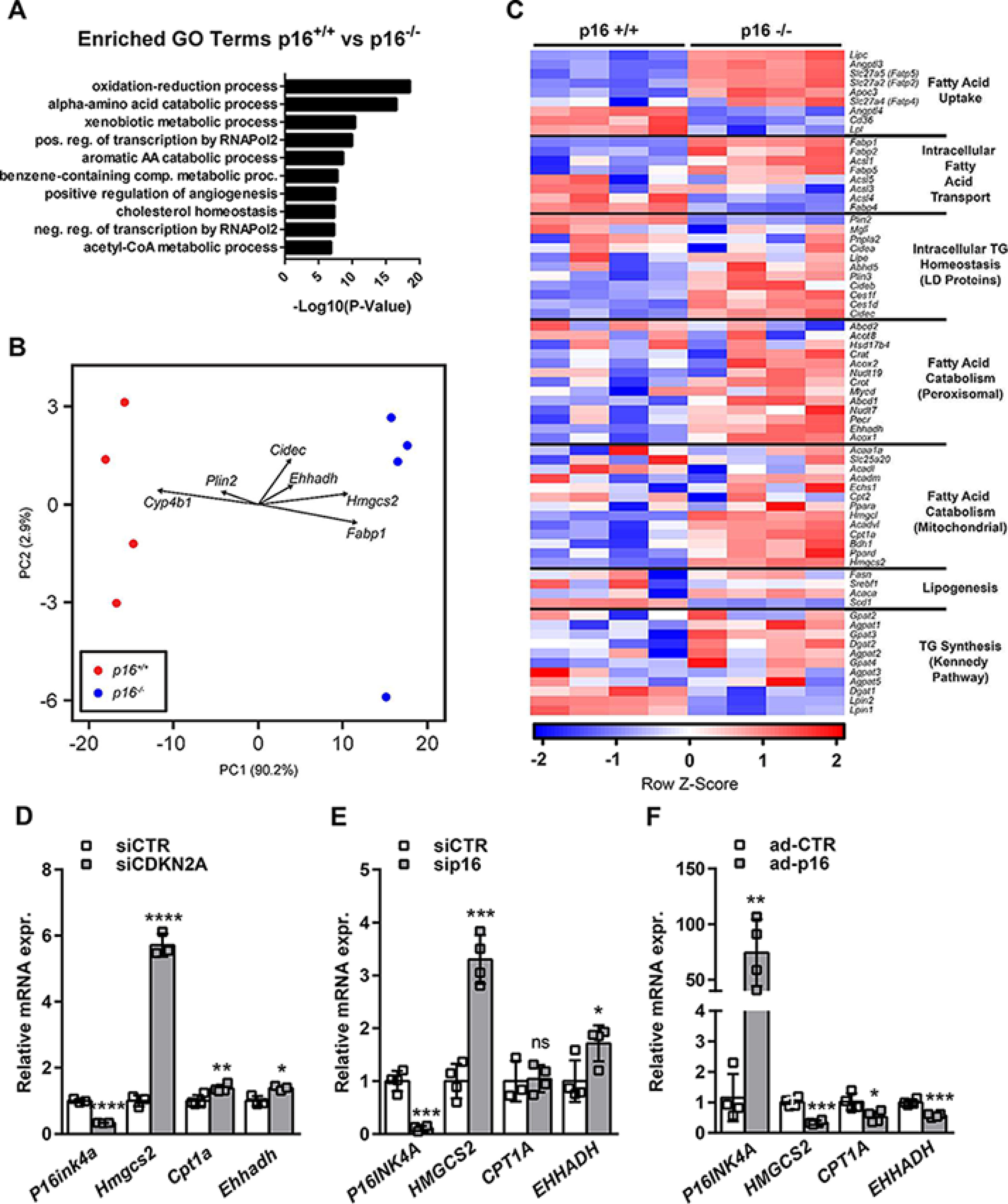
p16-deficiency increases fatty acid catabolism gene expression in hepatocytes. A–C, primary hepatocytes were isolated from p16+/+ and p16−/− mice and incubated for 8 h in 1 mm glucose DMEM supplemented with 10 μm FSK. A, Gene Ontology terms enrichment for differentially expressed genes between p16+/+ and p16−/− primary hepatocytes. B, principle component analysis of differentially expressed genes between p16+/+ and p16−/− primary hepatocytes with selected FAO and ketogenesis gene loadings plotted. C, a heatmap of genes in the major fatty acid metabolic pathways in p16+/+ and p16−/− primary hepatocytes. D–F, mRNA relative expression of P16INK4A, HMGCS2, CPT1A, and EHHADH were measured by qPCR in (D) the murine hepatocyte cell line AML12 transfected with siRNA-CDKN2A (siCDKN2A), (E) the human hepatocyte cell line IHH transfected with siRNA-p16 (sip16), and (F) the human hepatocyte cell line HepG2 infected with p16-adenovirus (ad-p16). All values are expressed as mean ± S.D. *, compared with siCTR or ad-CTR, unpaired t test: ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; and *, p < 0.05. See also Fig. S1.
Because primary hepatocytes from p16-deficient mice could be affected by an altered developmental program due to p16-deficiency, we next tested whether acute modulation of p16 expression was sufficient to induce similar changes in gene expression in vitro. In murine AML12 cells (Fig. 1D) and immortalized human hepatocytes (IHH, Fig. 1E), siRNA-mediated reduction of p16 led to a strong increase in several genes identified in the transcriptomic analysis, including HMGCS2, CPT1A, and EHHADH. Furthermore, adenovirus-mediated p16-overexpression in HepG2 cells resulted in reduced expression of these same genes (Fig. 1F). Taken together, these results indicate that p16-deficiency in hepatocytes leads to the induction of a transcriptional program indicative of increased fatty acid utilization.
p16 controls mitochondrial fatty acid oxidation and lipid droplet formation in hepatocytes
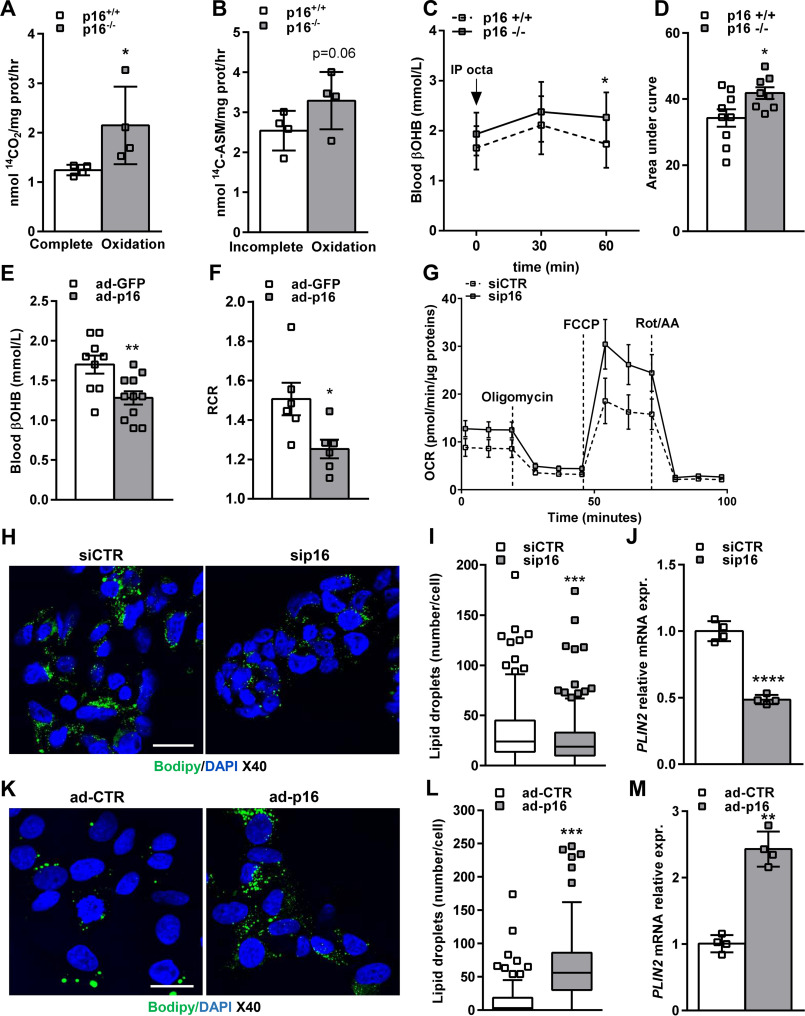
We next sought to investigate whether activation of lipid metabolism gene expression as a result of p16-deficiency is reflected by corresponding functional changes in hepatocyte lipid metabolism and mitochondrial function. Measurement of β-oxidation with 14C-labeled oleic acid in p16+/+ and p16−/− primary hepatocytes revealed that formation of 14CO2 (complete oxidation, Fig. 2A) was increased in p16−/− compared with p16+/+ cells, and associated with a tendency toward higher 14C-acid soluble metabolites, mostly ketone bodies in hepatocytes (incomplete oxidation, Fig. 2B). To test whether hepatic mitochondrial activity and ketone body production are also modulated by p16 in vivo, we first assessed fasting blood β-hydroxybutyrate (βOHB) levels, which account for ∼75% of circulating ketone bodies in mice (14). Unexpectedly, βOHB concentration did not differ between p16+/+ and p16−/− mice after overnight fasting (Fig. 2C). However, acute injection of overnight fasted mice with octanoate, a medium chain fatty acid shown to specifically activate hepatic ketogenesis (15), resulted in a statistically significant augmentation in blood βOHB levels in p16−/− mice within 60 min. Moreover, the area under the curve of βOHB was also higher in p16−/− mice compared with p16+/+ littermate controls (Fig. 2D). Finally, we tested whether liver-specific p16 overexpression by adenovirus is sufficient to decrease hepatic mitochondrial function in C57BL6/J mice. Hepatic p16 overexpression (Fig. S2A) resulted in a significant decrease in fasting blood βOHB levels (Fig. 2E) and hepatic mitochondrial respiratory control ratio (RCR) measured in the presence of palmitoylcarnitine (Fig. 2F) when compared with GFP-overexpressing controls. Importantly, these changes were independent of differences in body weight (Fig. S2B) and plasma free fatty acids (Fig. S2C), the main substrate for ketone body synthesis.
Figure 2.
p16 controls mitochondrial fatty acid oxidation and lipid droplet accumulation in hepatocytes. A, complete (14CO2), and B, incomplete fatty acid oxidation (14C-ASM) measured in primary hepatocytes isolated from p16+/+ and p16−/− mice and incubated with 14C-labeled oleic acid. C, ketone body (β-OHB) production in p16+/+ (n = 9) and p16−/− (n = 9) 12-week–old male mice fasted overnight and injected with a solution of sodium octanoate (250 mm). D, area under the curve (AUC) of sodium octanoate-stimulated β-OHB production. E and F, 12-week–old male mice were injected with adenovirus GFP (ad-GFP) (n = 10) or adenovirus p16 (ad-p16) (n = 11). E, circulating β-OHB was measured following an overnight fasting. F, RCR was measured on liver homogenates from ad-GFP (n = 5) and ad-p16 (n = 5) injected mice. G, OCR in IHH cells transfected with either siCTR or sip16 for 48 h and incubated for 1 h in Seahorse assay media. Oligomycin, ATP synthase inhibitor. FCCP, mitochondrial uncoupler. Rot/AA, rotenone and antimycin A, specific inhibitors for ETC complex I and III, respectively. H, representative images of BODIPY 493/503 staining of neutral lipids in IHH cell line transfected with siCTR or sip16; scale bar = 20 μm. I, quantification of lipid droplet number per cell with spot tracking plug-in of Icy software. J, PLIN2 relative mRNA expression in IHH measured by qPCR. K–M, the human hepatocyte cell line HepG2 was infected with an adenovirus expressing the human p16 (ad-p16) or a control adenovirus (ad-CTR) for 72 h. K, BODIPY staining of neutral lipids in HepG2 cells; scale bar = 20 μm. L, quantification of lipid droplets. M, PLIN2 relative mRNA expression in HepG2 measured by qPCR. All values are expressed as mean ± S.D. (A, B, G, I, J, L, ands M) or mean ± S.E. (C–F). t test: ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; and *, p < 0.05. See also Fig. S2 and S3.
To test which mitochondrial processes were most affected by p16 deficiency, oxygen consumption measurements using the Seahorse Extracellular Flux Analyzer were performed. Interestingly, p16 siRNA-transfected IHH cells displayed increased maximal respiration capacity compared with control siRNA-transfected cells (Fig. 2G and Fig. S3A). Finally, we assessed the effect of p16-knockdown on intracellular triglyceride and lipid droplet (LD) accumulation by BODIPY staining. P16-knockdown resulted in a significant decrease in LD number per cell (Fig. 2, H and I), which was associated with decreased expression of perilipin 2 (PLIN2, Fig. 2J), an LD-associated protein whose expression correlates with intracellular lipid storage. In line with the decreased mitochondrial activity observed in vivo (Fig. 2, E and F), p16 overexpression in HepG2 cells resulted in a strong increase (>20-fold) in LD per cell compared with control infected cells (Fig. 2, K and L), and associated with increased PLIN2 expression (Fig. 2M). These results indicate that p16 overexpression leads to accumulation of intracellular LD accompanied by reduced mitochondrial FAO.
p16 expression modulates HMGCS2 expression independently of CDK4
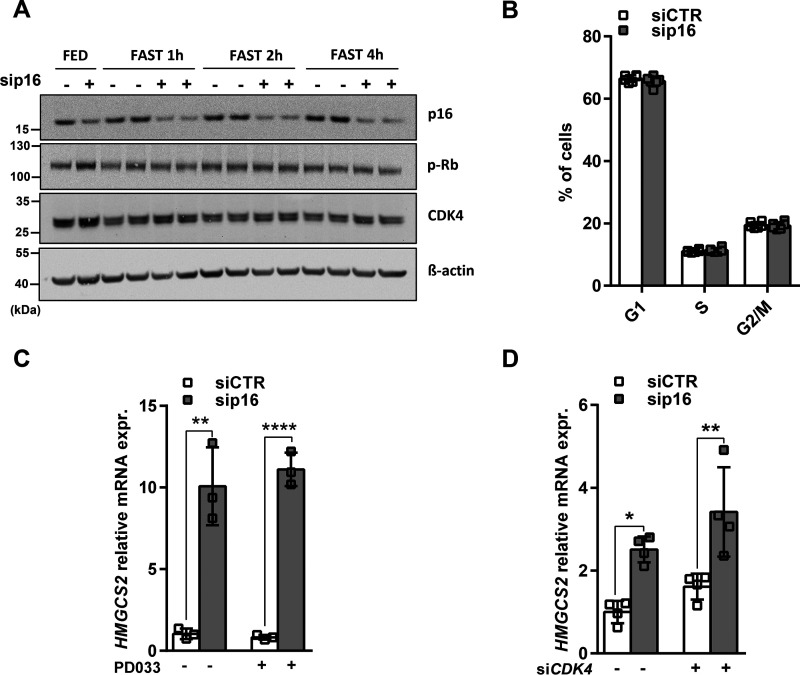
Thus far, we have demonstrated that modulation of p16 expression leads to changes in fatty acid metabolism in both nonproliferative primary hepatocytes and proliferating hepatocyte cell lines, indicating that p16's action on this pathway may be independent of its role in cell cycle regulation. The canonical function of p16 is to block the G0 to S phase transition via inhibition of CDK4-mediated phosphorylation of Rb protein (4). Consequently, we analyzed whether the p16-dependent changes in mitochondrial activity were associated with altered cell cycle progression. To do so, we first analyzed changes in CDK4 protein levels and Rb phosphorylation in p16-silenced IHH cells (Fig. 3A). As expected (16), both CDK4 expression and Rb phosphorylation were reduced within 1-4 h of shifting cells to conditions mimicking fasting (low glucose, no insulin, supplemented with forskolin). Interestingly, p16 silencing did not affect total CDK4 nor Rb phosphorylation under these conditions (Fig. 3A). Moreover, p16 silencing in these cells did not modify cell cycle progression as measured by propidium iodide staining (Fig. 3B). Finally, we assessed whether CDK4 activation was also required for the regulation of HMGCS2 gene expression upon p16 deletion. Pharmacological CDK4 inhibition by PD0332991 (Palbociclib, PD033) (Fig. 3C) or CDK4 silencing by siRNA (Fig. 3D and Fig. S4A) did not affect the siRNA-p16-mediated increase in HMGCS2 expression.
Figure 3.
p16 expression modulates HMGCS2 expression independent of CDK4-Rb cell cycle control in IHH cells. The human hepatocyte cell line IHH was transfected with either siCTR or sip16. A, Western blotting of p16, CDK4 and the phosphorylation of Rb in fed condition (FED) and after 1-, 2-, and 4-h incubations in 1 mm glucose DMEM supplemented with 10 ± μm FSK (FAST). β-Actin was used as a loading control. B, FACS analysis by propidium iodide staining of the percentage of IHH cells in each phase of the cell cycle after 4 h incubation in 1 mm glucose DMEM supplemented with 10 μm FSK. C and D, relative HMGCS2 mRNA expression in IHH cells transfected with siCTR or sip16 and incubated for 8 h in 1 mm glucose DMEM supplemented with 10 ± μm FSK and (C) treated or not with 1 μm PD0332991 (PD033), the pharmacological inhibitor of CDK4 or (D) cotransfected with siRNA-CDK4 (siCDK4) as described under “Materials and methods.” All values are expressed as mean ± S.D. *, compared with siCTR of the same condition, one-way ANOVA: ****, p < 0.0001; **, p < 0.01; and *, p < 0.05. See also Fig. S4.
Because p16 overexpression is associated with a senescent phenotype and senescence increases LD accumulation in hepatocytes (9), we assessed several parameters of senescence in ad-p16 overexpressing HepG2 cells (Fig. 2, K–M). A kinetic study of adenovirus-mediated p16 overexpression in HepG2 cells showed no differences in p21 mRNA expression, a marker of senescence, between ad-p16 and ad-Control cells at 24 and 48 h post-infection. However, an increase of p21 transcript levels was observed in ad-p16-infected HepG2 cells after 72 h (Fig. S4, B and C). Interestingly, the modest increase in p21 mRNA observed at the 72-h time point was not associated with modulation of cell cycle progression nor with an increase of SA-β-galactosidase activity, as measured by C12FDG staining, suggesting that p16 overexpression may not fully induce senescence in this time frame (Fig. S4, D and E). Conversely, BODIPY staining showed that ad-p16–infected HepG2 cells present significantly more lipid droplets per cell as early as 24 h post-infection and this is maintained up to 72 h (Fig. S4, F and G). This effect is associated with a gradual and significant increase of PLIN2 mRNA expression at 48 and 72 h and a significant decrease of HMGCS2 mRNA expression at 72 h post-infection (Fig. S4, H and I). In addition, adenovirus-mediated p16 overexpression in the liver was associated with decreased p21 mRNA expression and no changes in hepatic β-galactosidase activity (Fig. S4, J and K). Taken together, these data suggest that p16 overexpression in hepatocytes may impact lipid metabolism prior to a possible induction of cellular senescence. These results indicate that the effects of p16 on hepatocyte mitochondrial activity are unlikely to be mediated by modification of cell cycle progression and occur independently of CDK4 activity.
p16 protein expression is modulated by nutritional changes in IHH cells
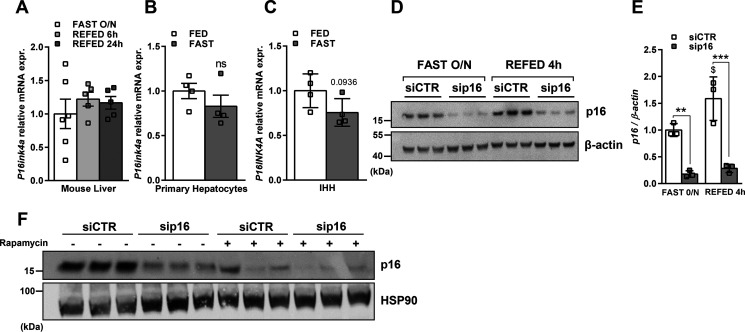
We then wondered if suppression of p16 expression was a physiological mechanism occurring during fasting. p16 mRNA expression was assessed in the liver of mice fasted overnight and refed for 6 or 24 h (Fig. 4A) or in mouse primary hepatocytes (Fig. 4B) and IHH cells (Fig. 4C) incubating in media mimicking fed or fasted conditions. None of these nutritional changes altered the expression of p16 at the mRNA level. Interestingly, p16 protein levels were increased in IHH cells in the refeeding experiment (Fig. 4, D and E), suggesting that p16 is regulated post-translationally. A recent report demonstrated that p16 is degraded by autophagy (17). Accordingly, rapamycin treatment for 8 h in IHH cells led to a strong decrease of p16 protein (Fig. 4F), suggesting that autophagy-mediated p16 degradation likely links nutritional status to p16 activity in hepatocytes.
Figure 4.
p16 protein expression is modulated by nutritional changes in IHH cells. p16 mRNA expression was measured by qPCR in A. Liver from mice fasted overnight and refed for either 6 or 24 h; B, primary hepatocytes incubated in media containing 10% serum, 11 mm glucose, and 100 nm insulin (FED) or without serum, 1 mm glucose and with glucagon for 8 h (FAST); C, IHH hepatocyte cell line incubating in complete media containing serum and insulin (FED) or in fasting media for 8 h as described under “Materials and methods.” D, p16 protein expression was measured by Western blotting in IHH cell line transfected with siRNA-CTR or siRNA-p16 and incubating overnight in fasting media and refed for 4 h and E, quantification of p16 protein level normalized to β-actin. F, IHH cells were transfected with siRNA-CTR or siRNA-p16 and treated with 100 nm rapamycin or DMSO for 8 h in a media containing 10% serum, 11 mm glucose, and 100 nm insulin. p16 protein expression was measured by Western blotting. All values are expressed as mean ± S.D. C and E, or mean ± S.E. (A and B). *, compared with siCTR of the same condition (FAST overnight or REFED 4 h), one-way ANOVA: ***, p < 0.001; **, p < 0.01; and *, p < 0.05; $, compared with siCTR or sip16 in two different conditions (FAST overnight versus REFED 4 h), $$$$, p < 0.0001; $$$, p < 0.001; $$, p < 0.01; $, p < 0.05.
p16 silencing activates the AMPKα2-SIRT1 pathway
To further dissect the mechanism of gene regulation by p16, we focused on HMGCS2 expression. We first assessed whether p16 may inhibit AMPK, an important regulator of hepatic FAO and mitochondrial function, to drive changes in gene expression. Interestingly, p16-silencing in IHH cells resulted in increased AMPK phosphorylation at Thr-172 (Fig. 5A), a marker of AMPK activation. Moreover, siRNA knockdown of the AMPKα2 catalytic subunit (siPRKAA2) (Fig. 5, B and C, and Fig. S5A) abrogated the p16-dependent increase in HMGCS2 expression, whereas knockdown of the AMPKα1 (siPRKAA1) catalytic subunit had no effect (Fig. S5, B–D). Interestingly, we found that knockdown of AMPKα1 increased HMGCS2 expression and decreased PLIN2 expression in IHH cells (Fig. S5H), whereas knockdown of the AMPKα2 increased PLIN2 expression (Fig. S5I), mimicking the effects observed after knockdown or overexpression of p16, respectively. p16 silencing in IHH cells also increased mRNA and protein expression/nuclear localization of AMPKα2, but not AMPKα1 (Fig. 5, B–D, and Fig. S5, C–G), suggesting that p16 silencing may also increase AMPK activity specifically through activation of the AMPKα2 catalytic subunit. In line, reduced total AMPK and AMPK phosphorylation at Thr-172 were observed in livers of overnight fasted mice overexpressing p16 (Fig. 5E). Finally, p16-knockdown in IHH cells was associated with increased mitochondrial ROS, an activating signal for AMPK (18), but not total intracellular ROS (Fig. 5, F and G).
Figure 5.
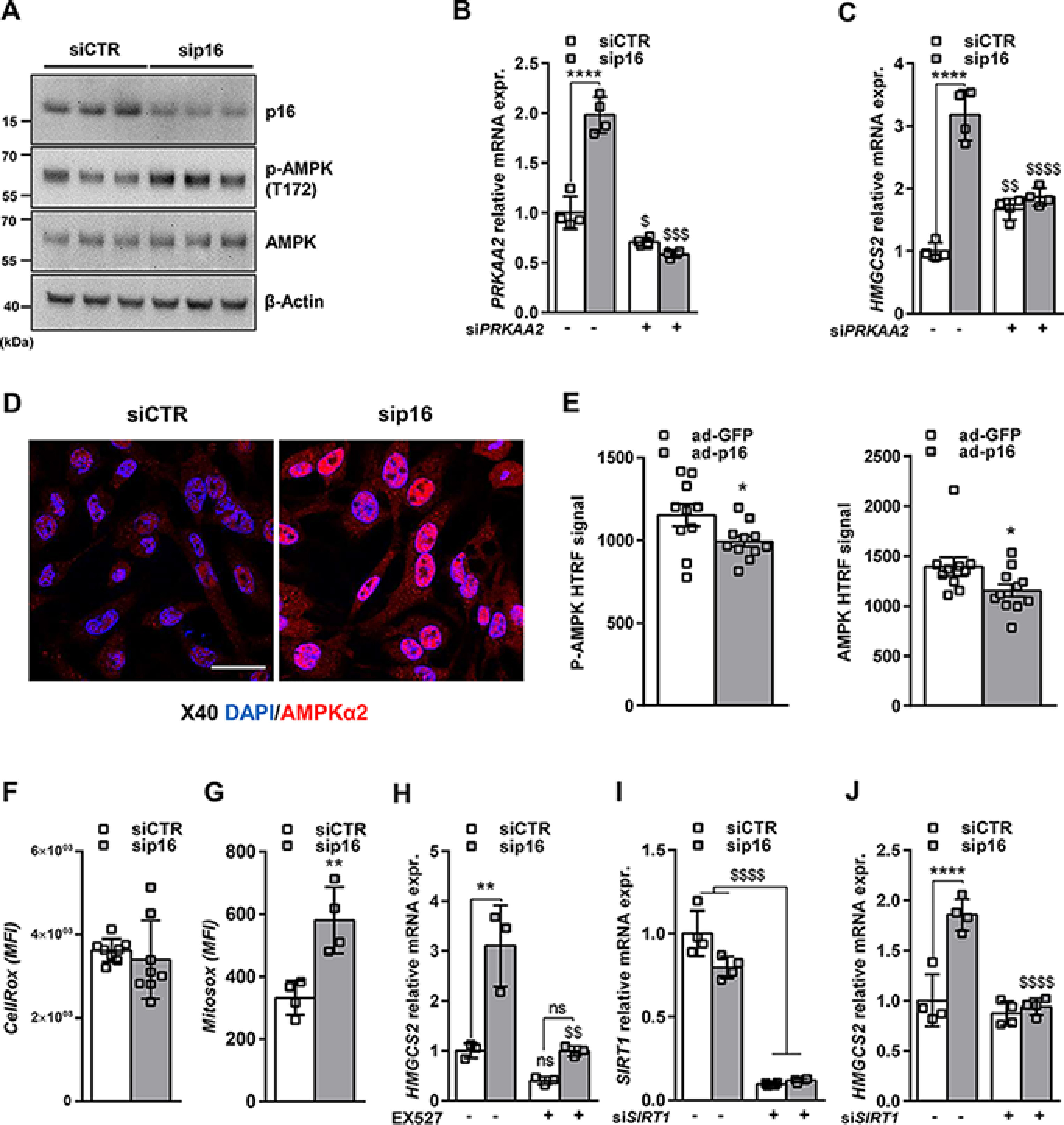
p16 silencing increases HMGCS2 expression by activating the AMPKα2-SIRT1 pathway. The human hepatocyte cell line IHH was transfected with either siCTR or sip16 and incubated for 8 h in 1 mm glucose DMEM supplemented with 10 μm FSK. A, Western blotting assay showing protein levels of p16, AMPK, and the phosphorylation of AMPK on Thr-172. β-Actin was used as a loading control. B, PRKAA2, and C, HMGCS2 relative mRNA expression in IHH cell line cotransfected with sip16 and siRNA-PRKAA2 (siPRKAA2). D, representative images of AMPKα2 immunofluorescent staining in IHH cell line transfected with siCTR or sip16. Scale bar = 20 μm. E, phospho-AMPK (p-AMPK) and total AMPK signals were analyzed by HTRF technology on liver homogenates from ad-GFP (n = 10) or ad-p16 (n = 11) infected mice. F, total and G, mitochondrial ROS measured, respectively, by CellROX and MitoROX fluorescent probes in IHH cell line as described under “Materials and methods.” H, HMGCS2 relative mRNA expression in IHH Cell line transfected with siCTR or sip16 and treated with 10 μm EX527. I, SIRT1 and J, HMGCS2 relative mRNA expression in IHH cell line cotransfected with sip16 and siRNA-SIRT1 (siSIRT1). All values are expressed as mean ± S.D. (B, C, F–J). *, compared with siCTR of the same condition (DMSO, treatment or siRNA), one-way ANOVA: ****, p < 0.0001; ***, p < 0.001; **, p < 0.01 and *, p < 0.05; $, compared with siCTR or sip16 in two different conditions (DMSO versus treatment or scramble versus siRNA); $$$$, p < 0.0001; $$$, p < 0.001; $$, p < 0.01; $, p < 0.05. All values are expressed as mean ± S.E. E and F, t test: *, p < 0.05. See also Fig. S5.
Because AMPK activation increases the NAD+/NADH ratio leading to SIRT1 activation (19), the role of SIRT1 in the p16-mediated regulation of HMGCS2 gene expression was also assessed. Treatment of IHH cells with EX527 (Fig. 5H and Fig. S5J), a pharmacological SIRT1 inhibitor, or siRNA-mediated silencing of SIRT1 (Fig. 5, I and J, Fig. S5K) abrogated the increase of HMGCS2 gene expression upon p16-silencing. Altogether, these data suggest that p16-silencing increases HMGCS2 expression and mitochondrial activity via an AMPKα2 and SIRT1-dependent signaling pathway.
p16 regulates fatty acid catabolism via activation of PPARα in hepatocytes
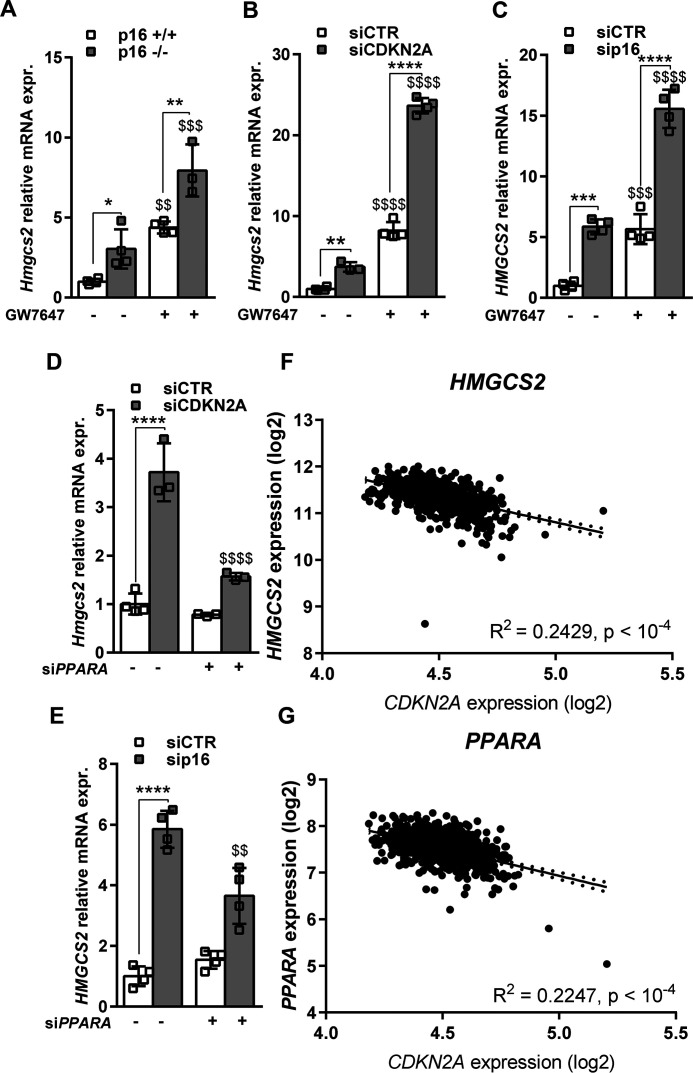
As PPARα is a master regulator of fasting-induced ketogenesis and a key effector of AMPK activation (20), we next assessed the effects of p16-silencing on PPARα activation in vitro. PPARα activation using the specific agonist GW7647 increased HMGCS2 mRNA expression in primary murine hepatocytes, AML12, IHH, and HepG2 cells and p16-silencing potentiated this induction (Fig. 6, A–C, Fig. S6, A–D) in all models. Conversely, PPARα knockdown partially blocked the induction of HMGCS2 mRNA upon p16-silencing in AML12 and IHH cells (Fig. 6, D and E, Fig. S6, E–H). To assess whether p16 may regulate FAO and mitochondrial function in human liver, expression of CDKN2A, PPARA, and HMGCS2 was measured in liver biopsies from a cohort of obese patients (n = 910 patients) (21). Interestingly, CDKN2A gene expression negatively correlated with HMGCS2 (Pearson r = −0.49; p < 1 × 10−16, Fig. 6F) and PPARA (r = −0.68; p < 1 × 10−16, Fig. 6G) gene expression, suggesting that p16 may also regulate hepatic ketogenesis in humans.
Figure 6.
p16-deficiency increases fatty acid catabolism gene expression via activation of PPARα in hepatocytes. A–C, HMGCS2 relative mRNA expression measured by qPCR in A, primary hepatocytes isolated from p16+/+ and p16−/− mice, B, murine AML12 cell line transfected with siRNA-CTR (siCTR) or siRNA-CDKN2A (siCDKN2A) for 48 h, C, human IHH cell line transfected with siCTR or siRNA-p16 (sip16) for 48 h and treated with 600 nm GW7647 for 8 h or DMSO. D and E, HMGCS2 relative mRNA expression measured by qPCR in D, murine AML12 and E, human IHH cell lines transfected, respectively, with siCDKN2A or sip16 and cotransfected with siRNA-PPARA (siPPARA) for 48 h. All values are expressed as mean ± S.D. *, compared with siCTR of the same condition (DMSO, treatment, siCTR or siPPARA), one-way ANOVA: ***, p < 0.001; **, p < 0.01; and *, p < 0.05; $, compared with siCTR or sip16 in two different conditions (DMSO versus GW7647 or siCTR versus siPPARA). $$$$, p < 0.0001; $$$, p < 0.001; $$, p < 0.01; $, p < 0.05. F and G, plots of hepatic CDKN2A expression versus (F) HMGCS2, (G) PPARA expression in human liver biopsies obtained during abdominal surgery (n = 910 patients) as described under “Materials and methods.” Values are reported for R2 as well as the linear regression best fit equation. The best fit line and 95% confidence intervals are plotted on the graphs. See also Fig. S6 and Table S1.
Discussion
In the present study, we describe a novel function of the cell cycle inhibitor p16 as a regulator of hepatic fatty acid metabolism. p16 deletion in both murine and human hepatocytes enhanced fatty acid oxidation and ketogenesis by a mechanism involving activation of AMPKα2, SIRT1, and consequently, PPARα. Moreover, our data indicate that regulation of these target genes by p16 is independent of its action on CDK4 and independent of alterations in cell cycle progression.
Cell cycle-independent regulation of metabolism is increasingly becoming an established function of many cell cycle regulators. The CDK4-E2F1 pathway was shown to increase hepatic lipogenesis and decrease FAO in nonproliferating cells under feeding conditions (7, 8), thus contributing to steatosis and NAFLD development. In contrast, Cyclin D1, a CDK4 partner, was demonstrated to inhibit hepatic lipogenesis through repression of ChREBP, HNF4 (22), and PPARγ (23). More recently, Cyclin D1 was found to inhibit PPARα transcriptional activity in a CDK4-independent manner (13), highlighting the complexity of metabolic regulation by cell cycle regulators. Our data indicate that p16 inhibits fasting-induced FAO through PPARα modulation independently of CDK4 inhibition, suggesting that p16 and CDK4 may exert opposing actions on FA metabolism, perhaps depending on the nutritional context. Further studies are necessary to better understand the interplay of p16 and CDK4 on hepatic metabolic functions.
Our results show that p16 acts via AMPK and SIRT1 to modulate PPARα transcriptional activity in hepatocytes. AMPK and SIRT1 are important nutrient sensors regulating FAO during fasting. SIRT1 deacetylates PGC1α, thereby increasing its association with PPARα and subsequent transcriptional activity (24). Previous studies suggest that the AMPKα2 catalytic isoform controls the balance between lipogenesis and FAO in the liver (25). More recently, AMPKα2 was also shown to be required for efficient FAO in mouse embryonic fibroblasts (12). Several studies have shown that the AMPKα2 subunit could be nuclear localized (26, 27), and previous work from our group has shown that AMPK is recruited to chromatin and participates in transcriptional regulation with glucocorticoid receptor and PPARα (28). In the present study, we show that p16-silencing is associated with increased AMPK Thr-172 phosphorylation and a specific increase in the mRNA and protein expression of the AMPKα2 (but not α1). Furthermore, only knockdown of AMPKα2 was sufficient to reverse the effects of p16-silencing on HMGCS2 gene expression. In line, adenovirus-mediated p16 overexpression in the liver in vivo decreases FAO and AMPK expression and phosphorylation. Together, these results suggest that p16 could specifically activate the AMPKα2 isoform to modulate PPARα transcriptional activity.
The mechanism by which AMPKα2 activity is regulated in hepatocytes is not fully understood. AMPK is an energetic sensor that is also highly sensitive to the cellular redox state. Indeed, AMPK can be activated by ROS-induced ATP depletion (18) and various mechanisms of direct ROS interaction (29, 30). Increased mtROS production, as observed upon p16-knockdown, could thus explain increased AMPK activity (Fig. 5G). However, further studies are required to better characterize the signaling cascade leading to a specific activation of AMPKα2 in p16-deficient hepatocytes.
We observed that the p16 protein level is increased in IHH cells during refeeding conditions, without modulation of p16 mRNA expression by fasting. In line, the p16 protein was recently demonstrated to be degraded by autophagy (17), a process induced by fasting and inhibited by refeeding (31, 32). Moreover, partial deletion of ATG5 in mice, resulting in inhibition of autophagy, increased p16 protein levels in the liver (33). We propose that p16 protein degradation during fasting, likely via an autophagy-dependent mechanism, could participate in the physiological activation of FAO by PPARα in hepatocytes.
In line with our previous study showing that p16-deficiency enhances gluconeogenesis through the CDK4-PKA-CREB pathway (6), the increased PPARα activity upon p16-deficiency could also contribute to this increase via regulation of the expression of several genes involved in the conversion of glycerol to glucose (34) or modulation of substrate utilization. Indeed, both pyruvate (via expression of Pdk4) (35) and glycerol (36) metabolism are sensitive to PPARα modulation.
Finally, we observed that whereas overexpression of p16 in HepG2 cells increases lipid accumulation, p16 impacts neither the cell cycle nor senescence in hepatocytes in the conditions studied. In line, adenovirus-mediated overexpression of p16 in the mice livers led to a decrease of circulating ketone bodies and mitochondrial activity in the presence of palmitoylcarnitine without induction of senescence. Aging and senescence are associated with decreased fatty acid oxidation resulting in increased hepatic fat accumulation (9). Although p16 expression is a widely used marker of senescence, recent evidence suggests that a senescence-associated increase in liver p16 expression may not be related to its expression in hepatocytes (37, 38). It is also possible that increased p16 may impact these metabolic pathways prior to the onset of outright senescence in a chronic setting.
Overall, our results provide further evidence that p16 plays an important role in the regulation of hepatic glucose and lipid metabolism and again support the genetic link between p16/CDKN2A and metabolic disorders such as T2D and cardiovascular disease. Lowering p16 expression may help to restore hepatic FAO and lipid homeostasis by activating the AMPKα2-SIRT1-PPARα signaling pathway.
Materials and methods
Genetic terminology: CDKN2A versus p16
The use of CDKN2A in the manuscript refers to the entire CDKN2A locus (P16 and P14ARF in human or P19arf in mouse), when analysis of P16 mRNA expression alone or its modulation was not possible (human liver transcriptomic data or siRNA-mediated knockdown of p16 in the murine AML12 cell line). Conversely, when using the terminology P16, this means that analysis of P16 mRNA expression alone or its modulation was technically possible (siRNA in human cell lines, adenovirus-mediated overexpression of P16 or measure of P16 mRNA expression by qPCR).
Mouse model and diets
p16−/− and littermate control (p16+/+) mice on a C57Bl6/J background (provided by P. Krimpenfort and backcrossed more than 8 generations) were housed under SPF conditions (22 ± 2 °C) in conventional cages with free access to water and food unless indicated otherwise. Experimental procedures were conducted with the approval of the ethics committee for animal experimentation of the Nord Pas-de-Calais region (APAFIS number 12317-2015121612289958 V15).
Ketogenesis assay in vivo
Following an overnight fast, 12-week–old p16+/+ (n = 9) and p16−/− (n = 9) male mice were injected with 250 mm sodium octanoate and β-hydroxybutyrate was measured with a ketometer and β-ketone test strips (Abbott Diabetes Care, Freestyle Optium Neo) before the injection and every 30 min following the injection.
Adenoviral-mediated overexpression of p16
16-Week–old male mice were injected via the tail vein with a solution containing 1 × 108 genome copies of ad-GFP (n = 10) or ad-p16 (n = 11). 4 days after adenovirus injections, mice were fasted overnight, plasma β-hydroxybutyrate was measured as described above, and livers were harvested for mitochondrial function analysis.
Primary hepatocyte isolation, culture, and treatments
Primary hepatocytes were isolated from 12-week–old male mice using the two-step collagenase perfusion method, essentially as described previously (6). Following isolation, cells were seeded on collagen-coated plates in Williams E media (Life Technologies) supplemented with 0.1% BSA, 2 mm glutamine, 100 nm bovine insulin, 100 nm dexamethasone, and penicillin and streptomycin. The following morning, cells were washed with PBS and incubated in DMEM, 1 mm glucose, supplemented with 0.1% BSA and antibiotics for 8 h, described as fasting conditions.
Microarray analysis
Mouse primary hepatocytes transcriptomics
Analysis was performed using Mouse Gene 2.0ST arrays (Affymetrix). Array data processing was performed using Bioconductor in the R-environment (r-project.org). Gene expression changes were calculated after signal normalization using robust multichip averaging in the oligo package (39). Differential gene expression was assessed using the limma package (40) with a threshold of 5% false discovery rate. Gene Ontology (GO) terms enrichment of selected clusters was performed using the clusterProfiler package (41, 42).
Human liver transcriptomics
Liver biopsies were obtained from morbidly obese patients during abdominal surgery. Patients underwent a full clinical workup to assess metabolic characteristics and surgical risks. All experimental procedures abide by the Declaration of Helsinki Principles and written informed consent was obtained from each participant as part of the ABOS protocol (ID: NCT01129297). The protocol was also reviewed and approved by the Lille University Hospital ethical committee. RNA quality was assessed using the Agilent Bioanalyzer and only samples with RIN > 5 were included for analysis. Transcriptomic analysis of human liver biopsies was performed using Affymetrix HTA 2.0 arrays (21). Array pre-processing and gene expression analysis were performed as described for murine arrays. Basic clinical characteristics of the patient population are provided in Table S1.
Cell culture, siRNA, and adenovirus experiments
Alpha mouse liver 12 (AML12), IHH, and the human hepatoma cell line HepG2 were maintained as described under in details, under a humidified incubator at 37 °C under 5% CO2.
Alpha mouse liver 12 (AML12)
AML12 (catalog number CRL2254; ATCC) cells were cultured in deprivation medium–Ham's F-12 (Gibco–ThermoFisher Scientific) supplemented with 10% FBS (Invitrogen), 5 g/ml of insulin (ThermoFisher Scientific), 5 g/ml of transferrin (Sigma-Aldrich), 5 ng/ml of selenium (Sigma-Aldrich), 1% glutamine (Gibco-Life Technologies), and 1% penicillin-streptomycin (Gibco-Life Technologies). The cells were plated in 12-well–plates at a density of 200,000 cells/ml. AML12 cells were transfected with siRNA (see supporting “Methods”) using Dharmafect-1 (Horizon) according to the manufacturer's instructions. 48 h after siRNA transfection, AML12 cells were incubated in DMEM (Gibco – ThermoFisher Scientific) containing 1 mm glucose, 0.1% fatty acid-free BSA (SKU 08810681, MPBio) and supplemented with 10 μm forskolin (FSK) (Sigma-Aldrich) for 8 h.
Immortalized human hepatocyte (IHH)
IHH (43) cells were cultured in Williams E medium (Invitrogen, CergyPontoise, France) containing 11 mm glucose and supplemented with 10% FBS (Invitrogen), 100 units/ml of penicillin, 100 μg/ml of streptomycin, 20 milliunits/ml of bovine insulin (Sigma-Aldrich), and 50 nm dexamethasone (Sigma-Aldrich) (these conditions were stated as “feeding” or “FED”). The cells were plated in 12-well–plates at a density of 250,000 cells/ml. IHH cells were transfected with siRNA (see supporting “Methods”) using the Dharmafect1 reagent according to the manufacturer's instructions. After 48 h of siRNA transfection, IHH cells were incubated in DMEM (11966025, Gibco) containing 1 mm glucose, 0,1% fatty acid-free BSA (SKU 08810681, MPBio), and supplemented with 10 μm FSK for 8 h, referenced as “fasting” or “FAST” media.
Human hepatoma cell line HepG2
HepG2 cells (catalog number HB-8065; ATCC) were seeded on 24-well–plates and starved during 24 h for cell cycle synchronization. Starvation medium was replaced by medium supplemented with 2% FCS containing p16-adenovirus (kind gift of Dr. A. Mazo, University of Barcelona, Spain) or empty adenovirus with a multiplicity of infection of 60. 24 h after infection, medium was replaced by fresh medium with 10% FCS. 72 h after infection, cells were collected for BODIPY staining or mRNA analysis.
Reverse transcription real time quantitative PCR analysis
Total RNA was isolated from cells using EXTRACT ALL® (Eurobio). cDNA was generated using the high capacity reverse transcription kit (Applied Biosystems, Life Technologies, USA). Real-time qPCR was performed on a Stratagene Mx3005P system (Agilent Technologies, Santa Clara, CA) using specific primers (see supporting “Methods”). mRNA levels were normalized to cyclophilin A mRNA and fold-induction was calculated using the ΔΔCt method.
Protein extraction and Western blotting analysis
Cells were scraped in protein lysis buffer (50 mm Tris-HCl, pH 8, 137 mm NaCl, 5 mm EDTA, 2 mm EGTA, 1% Triton X-100) on ice, transferred to 1.5-ml Eppendorf tubes and rotated for 20 min at 4 °C, followed by sonication for 5 min and centrifugation at 13,000 × g for 10 min at 4 °C. The resulting supernatants were stored in aliquots at −20 °C until analysis. Cell lysate protein concentrations were determined using a BCA protein assay kit (Pierce). The protein concentration of samples was equalized and cell lysates were mixed with 4× LDS nonreducing loading buffer (Life Technologies). Samples were heated at 95 °C for 5 min before loading and separated on precast 4–12% BisTris polyacrylamide gels (Life Technologies). Proteins were transferred to a nitrocellulose membrane using the iBlot2 Dry Blotting System (Life Technologies). Membranes were then incubated overnight at 4 °C with various primary antibodies in blocking buffer containing 5% nonfat milk at the dilution specified by the manufacturers. The following antibodies were used: p16 (BD Biosciences, number 554079), CDK4 (Santa Cruz Biotechnology, number sc-260), p-Rb (Abcam, ab10921), AMPK (Cell Signaling, number 2532), and p-AMPK T172 (Cell Signaling, number 2531).
Cell cycle analysis
Cells were washed with PBS, harvested, and fixed with cold 100% ethanol for 10 min at 4 °C. Cells were again washed with PBS, centrifuged, and incubated with a solution of 50 μg/ml of propidium iodide (PI) (Sigma-Aldrich) with 200 μg/ml of RNase A (Qiagen) and NucBlue® (Invitrogen) for 30 min at 37 °C. Cells were then washed, resuspended in PBS, and analyzed by flow cytometry with BD LSR Fortessa X-20 cell analyzer (BD Biosciences). PI fluorescence was analyzed with blue laser 488- and 575-nm emission filters. NucBlue® fluorescence was analyzed with UV laser 355- and 496-nm emission filters. Acquisition was performed with FACS DIVA software (BD Biosciences). PI fluorescence was analyzed on live single-cell population. The percentage of cells in each cell cycle phase was analyzed by using the FlowJo™ software algorithm (BD Biosciences).
Immunofluorescence analysis
IHH cells seeded on coverslips were washed with cold PBS and fixed in 4% paraformaldehyde for 10 min at 4 °C. Cells were permeabilized with 0.2% Triton X-100 (Euromedex) in PBS for 20 min at room temperature and blocked for 30 min using blocking solution (BSA 1%, 0.2% Triton X-100 in PBS). Cells were then incubated overnight at 4 °C with AMPKα1 (Abcam, ab32047) and AMPKα2 (Abcam, ab3760) antibodies in blocking solution before being washed and incubated 1 h at room temperature with secondary antibodies coupled to Alexa 594 (Molecular Probes by Life Technologies) and incubated for 10 min with Hoechst (Invitrogen). Cells were mounted in fluorescence mounting medium (GBl Laboratory). Acquisition was performed using an inverted confocal microscope (LSM 710, Zeiss) with a ×40 oil-immersion lens (NA 1.3 with an optical resolution of 176 nm). Alexa 594 and NucBlue were imaged using UV, argon 488 nm and 561 nm lasers. Images were processed with Zen software and analyzed with Icy software.
BODIPY 493/503 staining
Cells seeded on coverslips were fixed in 4% paraformaldehyde for 10 min at 4 °C and incubated with BODIPY 493/503 (Molecular Probes) at 200 ng/ml and NucBlue® for 20 min at room temperature. Cells were mounted in fluorescent mounting medium (GBl Laboratory). Acquisitions were performed using an inverted confocal microscope (LSM 710, Zeiss) with an airyscan detector and ×40 oil-immersion lens (NA 1.3 with an optical resolution of 176 nm) and 1.8× digital zoom. BODIPY and NucBlue fluorescence were imaged using argon 488 nm and UV lasers, respectively. Images were processed with Zen and analyzed with Icy programs respectively.
Mitochondrial activity measurement
Fatty acid oxidation test
Fatty acid oxidation was measured essentially as described previously (44). Freshly isolated primary hepatocytes were plated in DMEM-high glucose (25 mm) + 10% FBS for 4-6 h and changed to DMEM-low glucose (5 mm) overnight. 24 h after isolation, cells were incubated with 500 μm [14C]palmitic acid:BSA complex. After 3-4 h, media was transferred to a CO2 trap and 14CO2 was captured in 1 n NaOH in adjacent wells. Captured 14CO2 (complete oxidation) and 14C-acid soluble metabolites (ASMs, incomplete oxidation) were subjected to scintillation counting.
Seahorse analysis
Measurements of oxygen consumption rate (OCR) were performed using the XF24 apparatus (Seahorse Bioscience, North Billerica, MA). Briefly, cells were plated into XF24 (V7) polystyrene cell culture plates (Seahorse Bioscience, North Billerica). IHH cells were seeded at 70,000/well (XF24 plate). The cells were incubated for 24 h and transfected with siRNA for 48 h. Sensor cartridges were calibrated prior to each assay. IHH cells were incubated for 1 h in complete Seahorse assay medium (10 mm glucose, 2 mm glutamine) supplemented or not with 0.5 mm Etomoxir (Eto) in a 37 °C/non-CO2 incubator for 60 min prior to the start of an assay. All experiments were performed at 37 °C. Each measurement cycle consisted of a mixing time of 3 min, a waiting time of 2 min, and a data acquisition period of 3 min. OCR data points represent to the average rates during the measurement cycles. Oligomycin, FCCP, and Rotenone/Antimycin A were prepared at, respectively, 2 and 1 and 0.5 μm concentrations in the desired assay medium and adjusted to pH 7.4. A volume of 50 to 60 μl of compound was added to each injection port. In a typical experiment, 3 baseline measurements were taken prior to the addition of any compound, and 3 response measurements were taken after the addition of each compound. OCR were normalized to protein concentration (pmol/min/μg of proteins).
Oroboros analysis
A piece of liver (125 mg) was minced and homogenized using a Dounce homogenizer at 8-10 strokes in MIR05 respiratory buffer (20 mm HEPES, 10 mm KH2PO4, 110 mm sucrose, 20 mm taurine, 60 mm K-lactobionate, 0.5 mm EGTA, 3 mm MgCl2·6H2O, 1 g/liter of BSA (fatty acid free)). Liver homogenates containing mitochondria (50 μl) were introduced into O2K oxygraph chambers (Oroboros Instruments, Innsbruck, Austria) to assess oxygen consumption in the presence of the β-oxidation substrate palmitoylcarnitine (25 μm), malate (2 mm), followed by ADP (0.5 mm) injection. The RCR was calculated as the ratio of the state 3 to state 2. Finally, cytochrome c (10 μm) was added to the chamber to measure mitochondrial integrity. Citrate synthase (CS) activity was measured on liver homogenates after the Oroboros experiment.
ROS measurement
Total ROS and mitochondrial superoxide generation was determined using the CellROX Deep Red (Invitrogen) and MitoSOX red (Invitrogen), respectively. Briefly, IHH cells were seeded on a 12-well–plate and transfected with siRNA-CTR or siRNA-p16. 48 h after transfectection, IHH cells were harvested and resuspended in culture media containing 5 μm CellROX or MitoSOX and incubating for 20 min at 37 °C in the dark. Cells were then washed, resuspended in PBS, and analyzed by flow cytometry with BD LSR Fortessa X-20 cell analyzer (BD Biosciences). CellROX fluorescence was analyzed with extinction and emission at 640 and 665 nm. MitoSOX fluorescence was analyzed with extinction and emission at 510 and 580 nm.
HTRF® P-AMPK and total AMPK assays
A piece of liver (∼50 mg) was mechanically disrupted with a Polytron in ∼500 μl of lysis buffer. Homogenates were centrifuged and supernatant was collected for protein quantification. Samples containing 0.15 or 0.5 mg/ml of proteins were, respectively, added into 96-well HTRF plates for P-AMPK and total AMPK analysis, according to manufacturer's instructions (Cisbio).
Statistics
All data were expressed as mean ± S.D. or S.E. of measurement, as indicated in the figure legends. Student's unpaired t test was used for assessing statistical differences between two groups, whereas comparison more than two groups was performed using one-way ANOVA. Differences between two groups under two different factors were examined by a two-way ANOVA. In all analyses, a p values of p < 0.05 was considered significant.
Data availability
Raw datasets for mouse primary hepatocytes transcriptomics have been deposited in the GEO database through GEO Series accession number GSE134625.
Supplementary Material
Acknowledgments
We thank CPER-Région Hauts-de-France for the O2K Oroboros oxygraph.
This article contains supporting information.
Author contributions—Y. D., J. T. H., and R. P. conceptualization; Y. D., A. K. C., S. A. H., N. H., L. B., B. D., S. C., V. L., N. M., S. L., K. B., A. P., V. R., F. P., P. L., C. A., B. S., and R. P. investigation; Y. D., A. K. C., J. T. H., and R. P. writing-original draft; Y. D. and A. K. C. writing-review and editing; S. A. H., N. H., L. B., V. L., and J. T. H. formal analysis; B. D., S. C., E. V., E. D., N. M., S. L., J-S. A., K. B., and V. R. methodology; A. P., F. P., C. A., B. S., J. T. H., and R. P. funding acquisition; V. R. data curation; B. S., J. T. H., and R. P. supervision; J. T. H. project administration.
Funding and additional information—This work was supported in part by grants from ANR and European Union Grant EGID ANR-10-LABX-46 (to B. S.), PreciNASH ANR-16-RHUS-0006 (to F. P., J. T. H., and B. S.), the Société Francophone de Diabète-SFD (to J. T. H.), the Société Française d'Hépatologie-AFEF (to J. T. H.), and the Contrat Plan État-Région Hauts-de-France-Centre Transdisciplinaire de la Recherche sur la Longévité (CPER-CTRL) (to R. P. and A. C.). Y. D. was supported by fellowships from INSERM/Région Hauts-de-France, the Nouvelle Société Française d'Atherosclèrose, and EGID. J. T. H. was supported by an EMBO long-term fellowship ALTF-277. B. S. is a recipient of a European Research Council (ERC) Advanced Grant 694717.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- GWAS
- genome-wide association studies
- AMPKα
- AMP-activated protein kinase α
- CDK
- cyclin-dependent kinase
- CDKN2A
- cyclin-dependent kinase inhibitor 2A
- FAO
- fatty acid oxidation
- LD
- lipid droplets
- NAFLD
- non-alcoholic fatty liver disease
- p16
- p16INK4a
- PPARα
- peroxisome proliferator-activated receptor α
- PRKAA1/2
- 5'-AMP-activated protein kinase catalytic subunit 1/2
- Rb
- retinoblastoma protein
- SIRT1
- Sirtuin 1
- T2D
- type 2 diabetes
- ORC
- oxygen consumption rate
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- RCR
- respiratory control ratio
- FA
- fatty acid
- CREB
- cAMP-response element-binding protein
- IHH
- immortalized human hepatocytes
- ANOVA
- analysis of variance
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- qPCR
- quantitative PCR
- PI
- propidium iodide
- βOHB
- β-hydroxybutyrate
- FSK
- forskolin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. LaPak, K. M., and Burd, C. E. (2014) The molecular balancing act of p16INK4a in cancer and aging. Mol. Cancer Res. 12, 167–183 10.1158/1541-7786.MCR-13-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salazar-Roa, M., and Malumbres, M. (2017) Fueling the cell division cycle. Trends Cell Biol. 27, 69–81 10.1016/j.tcb.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 3. Aguilar, V., and Fajas, L. (2010) Cycling through metabolism. EMBO Mol. Med. 2, 338–348 10.1002/emmm.201000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharpless, N. E., Ramsey, M. R., Balasubramanian, P., Castrillon, D. H., and DePinho, R. A. (2004) The differential impact of p16 INK4a or p19 ARF deficiency on cell growth and tumorigenesis. Oncogene 23, 379–385 10.1038/sj.onc.1207074 [DOI] [PubMed] [Google Scholar]
- 5. Hannou, S. A., Wouters, K., Paumelle, R., and Staels, B. (2015) Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol. Metab. 26, 176–184 10.1016/j.tem.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 6. Bantubungi, K., Hannou, S.-A., Caron-Houde, S., Vallez, E., Baron, M., Lucas, A., Bouchaert, E., Paumelle, R., Tailleux, A., and Staels, B. (2014) Cdkn2a/p16Ink4a regulates fasting-induced hepatic gluconeogenesis through the PKA-CREB-PGC1α pathway. Diabetes 63, 3199–3209 10.2337/db13-1921 [DOI] [PubMed] [Google Scholar]
- 7. Denechaud, P.-D., Lopez-Mejia, I. C., Giralt, A., Lai, Q., Blanchet, E., Delacuisine, B., Nicolay, B. N., Dyson, N. J., Bonner, C., Pattou, F., Annicotte, J.-S., and Fajas, L. (2016) E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. J. Clin. Invest. 126, 137–150 10.1172/JCI81542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin, J., Valanejad, L., Nguyen, T. P., Lewis, K., Wright, M., Cast, A., Stock, L., Timchenko, L., and Timchenko, N. A. (2016) Activation of CDK4 triggers development of non-alcoholic fatty liver disease. Cell Rep. 16, 744–756 10.1016/j.celrep.2016.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ogrodnik, M., Miwa, S., Tchkonia, T., Tiniakos, D., Wilson, C. L., Lahat, A., Day, C. P., Burt, A., Palmer, A., Anstee, Q. M., Grellscheid, S. N., Hoeijmakers, J. H. J., Barnhoorn, S., Mann, D. A., Bird, T. G., et al. (2017) Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 8, 15691 10.1038/ncomms15691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang, X., Xu, G. B., Zhou, D., and Pan, Y.-X. (2018) High-fat diet modifies expression of hepatic cellular senescence gene p16(INK4a) through chromatin modifications in adult male rats. Genes Nutr. 13, 6 10.1186/s12263-018-0595-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foretz, M., Even, P. C., and Viollet, B. (2018) AMPK activation reduces hepatic lipid content by increasing fat oxidation in vivo. Int. J. Mol. Sci. 19, 2826 10.3390/ijms19092826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lopez-Mejia, I. C., Lagarrigue, S., Giralt, A., Martinez-Carreres, L., Zanou, N., Denechaud, P.-D., Castillo-Armengol, J., Chavey, C., Orpinell, M., Delacuisine, B., Nasrallah, A., Collodet, C., Zhang, L., Viollet, B., Hardie, D. G., et al. (2017) CDK4 phosphorylates AMPKα2 to inhibit its activity and repress fatty acid oxidation. Mol. Cell 68, 336–349.e6 10.1016/j.molcel.2017.09.034 [DOI] [PubMed] [Google Scholar]
- 13. Kamarajugadda, S., Becker, J. R., Hanse, E. A., Mashek, D. G., Mashek, M. T., Hendrickson, A. M., Mullany, L. K., and Albrecht, J. H. (2016) Cyclin D1 represses peroxisome proliferator-activated receptor alpha and inhibits fatty acid oxidation. Oncotarget 7, 47674–47686 10.18632/oncotarget.10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Puchalska, P., and Crawford, P. A. (2017) Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284 10.1016/j.cmet.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGarry, J. D., and Foster, D. W. (1971) The regulation of ketogenesis from octanoic acid. The role of the tricarboxylic acid cycle and fatty acid synthesis. J. Biol. Chem. 246, 1149–1159 [PubMed] [Google Scholar]
- 16. Lee, Y., Dominy, J. E., Choi, Y. J., Jurczak, M., Tolliday, N., Camporez, J. P., Chim, H., Lim, J.-H., Ruan, H.-B., Yang, X., Vazquez, F., Sicinski, P., Shulman, G. I., and Puigserver, P. (2014) Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510, 547–551 10.1038/nature13267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coryell, P. R., Goraya, S. K., Griffin, K. A., Redick, M. A., Sisk, S. R., and Purvis, J. E. (2020) Autophagy regulates the localization and degradation of p16INK4a. Aging Cell 19, e13171 10.1111/acel.13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hinchy, E. C., Gruszczyk, A. V., Willows, R., Navaratnam, N., Hall, A. R., Bates, G., et al. (2018) Mitochondria-derived ROS activate AMP-activated protein kinase (AMPK) indirectly. J. Biol. Chem. 293, 17208–17217 10.1074/jbc.RA118.002579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price, N. L., Gomes, A. P., Ling, A. J. Y., Duarte, F. V., Martin-Montalvo, A., North, B. J., Agarwal, B., Ye, L., Ramadori, G., Teodoro, J. S., Hubbard, B. P., Varela, A. T., Davis, J. G., Varamini, B., Hafner, A., et al. (2012) SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15, 675–690 10.1016/j.cmet.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grabacka, M., Pierzchalska, M., Dean, M., and Reiss, K. (2016) Regulation of ketone body metabolism and the role of PPARα. Int. J. Mol. Sci. 17, 2093 10.3390/ijms17122093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margerie, D., Lefebvre, P., Raverdy, V., Schwahn, U., Ruetten, H., Larsen, P., Duhamel, A., Labreuche, J., Thuillier, D., Derudas, B., Gheeraert, C., Dehondt, H., Dhalluin, Q., Alexandre, J., Caiazzo, R., et al. (2019) Hepatic transcriptomic signatures of statin treatment are associated with impaired glucose homeostasis in severely obese patients. BMC Med. Genomics 12, 80 10.1186/s12920-019-0536-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanse, E. A., Mashek, D. G., Becker, J. R., Solmonson, A. D., Mullany, L. K., Mashek, M. T., Towle, H. C., Chau, A. T., and Albrecht, J. H. (2012) Cyclin D1 inhibits hepatic lipogenesis via repression of carbohydrate response element binding protein and hepatocyte nuclear factor 4α. Cell Cycle 11, 2681–2690 10.4161/cc.21019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu, M., Rao, M., Bouras, T., Wang, C., Wu, K., Zhang, X., Li, Z., Yao, T.-P., and Pestell, R. G. (2005) Cyclin D1 inhibits peroxisome proliferator-activated receptor γ-mediated adipogenesis through histone deacetylase recruitment. J. Biol. Chem. 280, 16934–16941 10.1074/jbc.M500403200 [DOI] [PubMed] [Google Scholar]
- 24. Purushotham, A., Schug, T. T., Xu, Q., Surapureddi, S., Guo, X., and Li, X. (2009) Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327–338 10.1016/j.cmet.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foretz, M., Ancellin, N., Andreelli, F., Saintillan, Y., Grondin, P., Kahn, A., Thorens, B., Vaulont, S., and Viollet, B. (2005) Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes 54, 1331–1339 10.2337/diabetes.54.5.1331 [DOI] [PubMed] [Google Scholar]
- 26. Salt, I. P., Johnson, G., Ashcroft, S. J., and Hardie, D. G. (1998) AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic beta cells, and may regulate insulin release. Biochem. J. 335, 533–539 10.1042/bj3350533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGee, S. L., Howlett, K. F., Starkie, R. L., Cameron-Smith, D., Kemp, B. E., and Hargreaves, M. (2003) Exercise increases nuclear AMPK alpha2 in human skeletal muscle. Diabetes 52, 926–928 10.2337/diabetes.52.4.926 [DOI] [PubMed] [Google Scholar]
- 28. Ratman, D., Mylka, V., Bougarne, N., Pawlak, M., Caron, S., Hennuyer, N., Paumelle, R., De Cauwer, L., Thommis, J., Rider, M. H., Libert, C., Lievens, S., Tavernier, J., Staels, B., De Bosscher, K., et al. (2016) Chromatin recruitment of activated AMPK drives fasting response genes co-controlled by GR and PPARα. Nucleic Acids Res 44, 10539–10553 10.1093/nar/gkw742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zmijewski, J. W., Banerjee, S., Bae, H., Friggeri, A., Lazarowski, E. R., and Abraham, E. (2010) Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J. Biol. Chem. 285, 33154–33164 10.1074/jbc.M110.143685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao, D., Oka, S.-I., Liu, T., Zhai, P., Ago, T., Sciarretta, S., Li, H., and Sadoshima, J. (2014) A redox-dependent mechanism for regulation of AMPK activation by thioredoxin1 during energy starvation. Cell Metab. 19, 232–245 10.1016/j.cmet.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma, D., Panda, S., and Lin, J. D. (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 30, 4642–4651 10.1038/emboj.2011.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ezaki, J., Matsumoto, N., Takeda-Ezaki, M., Komatsu, M., Takahashi, K., Hiraoka, Y., Taka, H., Fujimura, T., Takehana, K., Yoshida, M., Iwata, J., Tanida, I., Furuya, N., Zheng, D.-M., Tada, N., et al. (2011) Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7, 727–736 10.4161/auto.7.7.15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cassidy, L. D., Young, A. R. J., Young, C. N. J., Soilleux, E. J., Fielder, E., Weigand, B. M., Lagnado, A., Brais, R., Ktistakis, N. T., Wiggins, K. A., Pyrillou, K., Clarke, M. C. H., Jurk, D., Passos, J. F., Narita, M., et al. (2020) Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat. Commun. 11, 307 10.1038/s41467-019-14187-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patsouris, D., Mandard, S., Voshol, P. J., Escher, P., Tan, N. S., Havekes, L. M., Koenig, W., März, W., Tafuri, S., Wahli, W., Müller, M., and Kersten, S. (2004) PPARα governs glycerol metabolism. J. Clin. Invest. 114, 94–103 10.1172/JCI200420468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang, B., Wu, P., Bowker-Kinley, M. M., and Harris, R. A. (2002) Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator–activated receptor-α ligands, glucocorticoids, and insulin. Diabetes 51, 276–283 10.2337/diabetes.51.2.276 [DOI] [PubMed] [Google Scholar]
- 36. Cotter, D. G., Ercal, B., d'Avignon, D. A., Dietzen, D. J., and Crawford, P. A. (2014) Impairments of hepatic gluconeogenesis and ketogenesis in PPARα-deficient neonatal mice. Am. J. Physiol. Endocrinol. Metab. 307, E176–E185 10.1152/ajpendo.00087.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grosse, L., Wagner, N., Emelyanov, A., Molina, C., Lacas-Gervais, S., Wagner, K.-D., and Bulavin, D. V. (2020) Defined p16 high senescent cell types are indispensable for mouse healthspan. Cell Metab. 32, 87–99 10.1016/j.cmet.2020.05.002 [DOI] [PubMed] [Google Scholar]
- 38. Omori, S., Wang, T.-W., Johmura, Y., Kanai, T., Nakano, Y., and Kido, T. (2020) Generation of a p16 reporter mouse and its use to characterize and target p16high cells in vivo. Cell Metab. 1–15 10.1016/j.cmet.2020.09.006 [DOI] [PubMed] [Google Scholar]
- 39. Carvalho, B. S., and Irizarry, R. A. (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Phipson, B., Lee, S., Majewski, I. J., Alexander, W. S., and Smyth, G. K. (2016) Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10, 946–963 10.1214/16-AOAS920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alexa, A., Rahnenführer, J., and Lengauer, T. (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 10.1093/bioinformatics/btl140 [DOI] [PubMed] [Google Scholar]
- 42. Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schippers, I. J., Moshage, H., Roelofsen, H., Müller, M., Heymans, H. S. A., Ruiters, M., and Kuipers, F. (1997) Immortalized human hepatocytes as a tool for the study of hepatocytic (de-)differentiation. Cell Biol. Toxicol. 13, 375–386 10.1023/A:1007404028681 [DOI] [PubMed] [Google Scholar]
- 44. Haas, J. T., Miao, J., Chanda, D., Wang, Y., Zhao, E., Haas, M. E., Hirschey, M., Vaitheesvaran, B., Farese, R. V., Kurland, I. J., Graham, M., Crooke, R., Foufelle, F., and Biddinger, S. B. (2012) Hepatic insulin signaling is required for obesity-dependent expression of SREBP-1c mRNA but not for feeding-dependent expression. Cell Metab. 15, 873–884 10.1016/j.cmet.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J., and Toussaint, O. (2019) Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 10.1038/nprot.2009.191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw datasets for mouse primary hepatocytes transcriptomics have been deposited in the GEO database through GEO Series accession number GSE134625.