Abstract
Cholangiocarcinoma is the most common biliary duct malignancy and the second most common primary liver cancer, accounting for 10-20% of hepatic malignancies. With high mortality and poor prognosis, the 5-year survival rate of cholangiocarcinoma is only 10%. A previous study demonstrated a significant association between aspirin use and a decreased risk of cholangiocarcinoma. However, the effect of aspirin on cholangiocarcinoma remains unknown. Therefore, the aim of the present study was to investigate the effects of aspirin on cholangiocarcinoma in vitro and in vivo. Three cholangiocarcinoma cell lines were used to analyze the effect of aspirin on cell proliferation, cell cycle progression, apoptosis, and the regulation of microRNAs. MicroRNAs are known to regulate the development and progression of various types of cancer. An HuCCT-1 xenograft model was used for the in vivo study. It was determined that aspirin inhibited the proliferation of human cholangiocarcinoma cells (except TKKK cells). Aspirin induced cell cycle arrest in the G0/G1 phase and regulated cell-cycle related proteins in cholangiocarcinoma cells (HuCCT-1 cells) but did not induce apoptosis. The expression of miR-340-5p was significantly upregulated after treatment, and overexpression of miR-340-5p inhibited the proliferation of HuCCT-1 cells and decreased the levels of cyclin D1. TKKK cells had low miR-340-5p expression, which may explain why aspirin had no effect on their proliferation. In vivo, aspirin reduced the growth of xenografted tumors. In conclusion, the present study indicated that aspirin partially inhibited cholangiocarcinoma cell proliferation and tumor growth by inducing G0/G1 phase cell cycle arrest, potentially through the miR-340-5p/cyclin D1 axis.
Keywords: aspirin, cell cycle, cholangiocarcinoma, in vivo, microRNAs
Introduction
Cholangiocarcinoma (CCA) is the most common biliary duct malignancy and the second most common primary liver cancer, accounting for 10-20% of all primary hepatic malignancies (1). CCA arises from bile duct epithelial cells, and there are three subtypes based on anatomic location: Intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA). The etiology of CCA includes lithiasis (2), primary sclerosing cholangitis (3), parasitic infection (4), congenital abnormalities (5), chronic liver disease (6), cirrhosis (6), metabolic abnormalities (7), and lifestyle (7). The incidence of iCCA appears to be increasing (8); the rates of CCA in North America, Japan, and Australia have been rising over the past two decades (9). Although surgery is the preferred treatment, the 5-year postoperative survival rate is markedly low. Chemotherapy can be used for inoperable cases; however, the highly desmoplastic nature, rich tumor microenvironment, and profound genetic heterogeneity all contribute to iCCA therapeutic resistance (9).
The nonsteroidal drug aspirin is an anti-inflammatory and anticoagulation agent used to prevent and reduce the risk of cardiovascular events. According to some clinical analyses, long-term use is also associated with a reduction in cancer risk, including colon (10), breast (11), and hepatocellular carcinoma (12). Aspirin use also has a significant inverse association with the development of all three CCA subtypes and an approximately 3-fold reduction in CCA risk (13). Numerous anticancer mechanisms for aspirin have been identified, such as inhibition of cyclooxygenase (14), activating key molecular targets in the AMPK, mTOR, STAT3, and NF-κB pathways (15), decreasing the levels of reactive oxygen species and glucose consumption (16), inducing autophagy via JNK/p-Bcl2/Beclin-1, AMPK/mTOR, and GSK-3 signaling (17), inducing apoptosis and mitochondrial dysfunction by increasing oxidative stress (18), and changing the tumor microenvironment by affecting platelets (19,20). However, the effect of aspirin on CCA remains unknown. Elucidation of the mechanism of aspirin in CCA could contribute to the development of new therapeutic agents in the future.
The purpose of the present study was to determine the antitumor effects of aspirin in CCA and identify the key molecular targets and microRNAs (miRNAs) associated with this effect.
Materials and methods
Chemicals
Aspirin was obtained from Wako Pure Chemical Industries, Ltd. The prepared solution was diluted in cell culture medium as per the requirement of cells and fresh pH 7.2 to 7.5, within the range suitable for cell growth was used.
Cell lines and cell culture
Human CCA cell lines (HuCCT-1, RBE, and TKKK) were obtained from the Japanese Research Resources Bank. HuCCT-1 and RBE cells were grown in RPMI-1640 media (Gibco-Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS) (product no. 557-30355; FUJIFILM Wako Pure Chemical Industries, Ltd.) and penicillin/streptomycin (100 mg/l; Invitrogen; Thermo Fisher Scientific, Inc.). TKKK cells were maintained in DMEM (Gibco-Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and penicillin/streptomycin. All cell lines were grown in a humidified incubator at 5% CO2 and 37°C.
Cell viability assay
Cell viability assays were performed using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.) according to the manufacturer's instructions. Briefly, cells (5,000 cells in 100 µl/well) were seeded into 96-well plates and allowed to adhere, followed by treatment with 0, 2.5, 5, or 10 mmol/l aspirin for 48 h. The medium was replaced with 100 µl of fresh medium containing 10% of CCK-8 reagent, and cells were incubated at 37°C for 3 h. The absorbance was measured at 450 nm using a multi-grating microplate reader SH-9000Lab (CORONA Electric Co., Ltd.). Experiments were carried out three times. Compared with RBE, HuCCT-1 cells were more sensitive to aspirin treatment in the cell viability assay. Moreover, considering the xenograft model, HuCCT-1 cells can be easily transplanted. Thus, HuCCT-1 cells were selected for further study in vitro and in vivo.
Flow cytometric analysis of the cell cycle
Flow cytometric analyses were performed using the Cycle Phase Determination kit (Cayman Chemical Company). HuCCT-1 and TKKK cells (1.0×106 cells/100-mm dish) were treated with 2.5 mmol/l aspirin or without for 24 to 48 h. Cells were trypsinized and resuspended in phosphate-buffered saline (PBS) at a density of 1×106 cells/ml. Approximately 1×106 cells were stained in 100 µl of PBS with 10 µl RNase A (250 µg/ml) and 10 µl propidium iodide (PI) stain (100 µg/ml) and incubated at room temperature in the dark for 30 min. Flow cytometry (FCM) was performed to compare the proportion of aspirin-treated and control cells in each phase of the cell cycle. FCM was performed using a Cytomics FC 500 flow cytometer (Beckman Coulter, Inc.) with an argon laser (488 nm), and the percentages of cells were analyzed using the Kaluza software version v2.1 (Beckman Coulter, Inc.). The experiments were repeated thrice.
Apoptosis analysis
Aspirin-mediated apoptosis was analyzed using FCM and Annexin V-FITC Early Apoptosis Detection kit (Cell Signaling Technology, Inc.). HuCCT-1 cells (1.0×106 cells/100-mm dish) were treated with 2.5 mmol/l aspirin or without for 48 h. Apoptotic and necrotic cells were analyzed by double staining with FITC-conjugated Annexin V and PI per the manufacturer's instructions. FCM was conducted using a Cytomics FC 500 flow cytometer with an argon laser (488 nm) to compare the proportion of apoptotic cells in the aspirin-treated and control groups, and data were analyzed using the Kaluza software version v2.1. The experiments were repeated thrice.
Apoptosis analysis by ELISA
ELISA was performed to analyze the levels of caspase-cleaved cytokeratin 18 (cCK18) using the M30 Apoptosense ELISA kit (cat. no. 10011; Peviva; Diapharma) according to the manufacturer's instructions. HuCCT-1 cells (5,000 cells/well) were seeded into 96-well plates and treated with 2.5 mmol/l aspirin for 48 h. Subsequently, the cells were lysed in polyoxyethylene octyl phenyl ether (Pure Chemical Industries, Ltd.) and further analyzed according to the manufacturer's instructions. The experiments were repeated thrice.
Western blot analysis
HuCCT-1 and TKKK cells were seeded (1.0×106 cells/100-mm dish) and treated with 2.5 mmol/l aspirin for 24 or 48 h. The cells were lysed with PRO-PREP complete protease inhibitor mixture (iNtRON Biotechnology, Korea). Supernatants were collected, and the protein concentrations were measured using a NanoDrop 2000 spectrofluorometer (Thermo Fisher Scientific, Inc.). Protein aliquots (10 µg) were resuspended in sample buffer and separated on 12% Tris-glycine gradient gels via sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The resolved proteins were transferred to a nitrocellulose membrane and blocked with a blocking buffer containing 2% skimmed milk (GE Healthcare) in TBST with 0.1% Tween 20 (cat. no. T9142; Takara Bio, Inc.) for 30 min at room temperature. Subsequently, the membranes were incubated overnight at 4°C with primary antibodies in 5% serum (cat. no. 9048-46-8; FUJIFILM Wako Pure Chemical Industries, Ltd.) followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies in 2% skimmed milk. The following primary antibodies were used: Cyclin D1 (SP4) (1:2,000 dilution; cat. no. MA5-14512), retinoblastoma (Rb) (1:1,000 dilution; cat. no. MA1-34070), and cyclin E (HE-12) (1:1,000 dilution; MS-870-P1) were obtained from Thermo Fisher Scientific, Inc.; phosphorylated Rb (pS780) (1:1,000 dilution; cat. no. 558554) was obtained from BD Biosciences; Cdk2 (1: 5,000 dilution; cat. no. sc-163) was obtained from Santa Cruz Biotechnology, Inc.; and anti-β-actin (1:5,000 dilution; product no. A5441) was purchased from Sigma-Aldrich; Merck KGaA. The membranes were washed again with TBST and incubated with HRP-conjugated anti-mouse (dilution 1:2,000; product no. 7076) and anti-rabbit (dilution 1:2,000; product no. 7074) IgG secondary antibodies obtained from Cell Signaling Technology, Inc. for 1 h at room temperature. Finally, the signals were visualized using a typically enhanced chemiluminescent (ECL) kit (cat. no. 45-000-999; Cytiva), and blots were imaged using ImageQuant LAS 4010 (GE Healthcare).
miRNA microarray
Cells were treated with 2.5 mmol/l aspirin for 48 h, and total RNA was extracted using the miRNeasy Mini kit (Qiagen GmbH) according to the manufacturer's instructions. After confirming the purity and quantity of each sample using an Agilent 2100 Bioanalyzer and an RNA 6000 Nano kit (both from Agilent Technologies), respectively, the samples were labeled using a miRCURY Hy3 Power Labeling kit (Exiqon A/S) and hybridized to a human miRNA Oligo Chip (v.21; Toray Industries, Inc.). Chips were scanned using the 3D-Gene Scanner 3000 (Toray Industries, Inc.). The 3D-Gene extraction software version 1.2 (Toray Industries, Inc.) was used to calculate the raw signal intensity of the images. The raw data were analyzed using the GeneSpring GX 10.0 software (Agilent Technologies, Inc.) to assess the differences in miRNA expression between the aspirin-treated and control samples. Global normalization was performed on raw data obtained above the background level. Differentially expressed miRNAs were determined using Welch's t-test.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of miRNAs
qPCR was used to validate the expression levels obtained from the miRNA assay. Total RNA was extracted as previously described above and diluted to 2.0 ng/µl. TaqMan microRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) were adopted to determine the expression of miRNAs using U6 small nuclear RNA (RNU6B) as an internal control. miRNAs were reverse transcribed using the TaqMan microRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Reverse transcription was performed in 20-µl reaction volumes consisting of 5 µl of RNA, 3 µl of 5X RT primer, and 12 µl of reverse transcription Master Mix. qPCR was performed in the MicroAmp Fast Optical 96-Well Reaction Plate (Applied Biosystems; Thermo Fisher Scientific, Inc.). Each well contained a 20-µl reaction consisting of 2 µl of cDNA, 1 µl of 20X qPCR assay, 7 µl of nuclease-free water, and 10 µl of TaqMan Fast Advanced Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR was performed using the ViiA7 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following reaction steps: Hold at 50°C for 2 min, denaturation at 95°C for 20 sec followed by 40 cycles of 1 sec at 95°C and 20 sec at 60°C. The relative expression of miR-340-5p was calculated using the comparative Cq method according to the following formula: 2−ΔΔCq (ΔCq=miRCq-U6Cq, ΔΔCq=ΔCq-average control ΔCq) (21). The primer sequences are as follows: miR-340-5p forward, 5′-GCG GTT ATA AAG CAA TGA GA-3′ and reverse, 5′-GTG CGT GTC GTG GAG TCG-3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′.
Bioinformatics analysis used for prediction of the target genes of miR-340-5p and functional and network analyses
For bioinformatics analysis, miRDB database (www.mirdb.org) was used to predict the target genes of miR-340-5p and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses was used for upregulated miRNAs depending on the Database for Annotation, Visualization, and Integrated Discovery (DAVID database) (22). miRNA regulatory network used mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr) (23); miRGator v3.0 (http://mirgator.kobic.re.kr) (24) and miTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php) (25) online software to reveal the expression of miR-340-5p in related cancer and normal tissue and the relationship with the underlying target gene. The cut-off criterion was set as P<0.05.
Cell transfection
For transfection, 5×105 HuCCT-1cells were seeded in 6-well plates with antibiotic-free medium 1 day before transfection to reach a confluence of 90% at the time of transfection. Cells were transfected with miR-340-5p mimic (50 nM) (sense, 5′-UUA UAA AGC AAU GAG ACU GAU U-3′ and antisense, 5′-UCA GUC UCA UUG CUU UAU AAT T-3′), inhibitor (50 nM) (5′-AAU CAG UCU CAU UGC UUU AUA A-3′), or negative control miRNA (50 nM) (mimic negative control 5′-UUG UAC UAC ACA AAA GUA CUG-3′; inhibitor negative control 5′-CAG UAC UUU UGU GUA GUA CAA-3′) (Life Technologies; Thermo Fisher Scientific, Inc.) using Lipofectamine RNAiMAX Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The transfection complex was prepared according to the manufacturer's instructions and added to the cells, and the plates were incubated in a humidified atmosphere with 5% CO2 at 37°C. Cells cultured with transfection reagent as untransfected control. The medium was replaced 6 h post-transfection. Cell samples were collected at 0 or 48 h after transfection for further analysis.
Colony formation assay
Cells were trypsinized for 5 min and resuspended at a density of 1×103/ml. Five hundred microliters were seeded into 6-well plates, and 1.5 ml RPMI-1640 medium containing 10% FBS was added to each well. The plates were incubated at 37°C at 5% CO2, and the medium was changed every 3 days until conspicuous colonies were observed. Colonies were stained with 0.1% crystal violet for 5 min at room temperature and positive colony formation (>50 cells/colony) was evaluated by counting the number of colonies.
Xenograft model analysis
The animal study was conducted in accordance with the guidelines set by the Committee on Experimental Animals of the Kagawa University. All experimental protocols were approved (approval no. 18674) by the Institutional Review Board of the Department of Laboratory Animal Science of Kagawa University (Kawaga, Japan). Thirty-five female athymic mice (BALB/c-nu/nu; 6 weeks old; 19-21 g) were purchased from Japan SLC (Shizuoka, Japan). The mice were maintained at 20-25°C with 30-60% humidity under a 12:12 h light/dark cycle, using a laminar airflow rack and had continuous free access to sterilized (γ-irradiated) food (CL-2; CLEA Japan, Inc.) and autoclaved water. Mice were subcutaneously inoculated with 1.5×106 HuCCT-1 cells in the right flank. When the xenografts were palpable with an approximate diameter of 3 mm, we randomly assigned the animals to three groups of 10 animals each. These groups were treated with 60 mg/kg aspirin, 100 mg/kg aspirin, or vehicle (10% ethanol in PBS) by intraperitoneal injection every day. The tumor volume (mm3) was calculated as tumor length (mm) x tumor width (mm)2/2. For humane endpoints, if difficulty in feeding and/or intake of water; apparent poor physical condition; a rapid and non-recoverable weight loss (over 20% body weight); and/or a significant increase in tumor size was observed (tumor size more than 10% of body weight and/or tumor diameter more than 20 mm), the experiments were discontinued. Animals were euthanized through CO2 euthanasia with 20% displacement of cage volume/min. Sacrifice was confirmed by observation of unconsciousness, absence of heartbeat and absence of breathing. All animals were sacrificed on day 28 of treatment. The experiments were performed from February 20, 2019 to March 28, 2019.
Immunohistochemistry
Immunohistochemistry was performed on tumor tissues obtained from the xeno-grafted mice. We prepared 5-µm-thick sections from formalin-fixed (10% formalin at room temperature for 24 h), paraffin-embedded tissue blocks which were deparaffinized, rehydrated, and subjected to immunohistochemistry studies. Following a blocking step at room temperature for 30 min using a Vectastain Elite ABC kit (Vector Laboratories, Inc.), tissue sections were incubated with primary anti-bodies [Cyclin D1 (EP12); 1:200 dilution; cat. no. 241R-4, Sigma-Aldrich; Merck KGaA]. Sections were washed and incubated with anti-rabbit IgG Antibody (1:50 dilution; cat. no. BP-9100-50; Vector Laboratories) at room temperature for 1 h and then with a streptavidin-peroxidase solution. Color reactions involved the use of 3,3′-diaminobenzidine (DAB) with Mayer's hematoxylin counterstaining at room temperature for 10 sec. The specificity of immunostaining was evaluated using non-immune mouse IgG (1:50 dilution; cat. no. BP-9200-50; Vector Laboratories) at room tempera-ture for 1 h as a negative control for the primary antibody. Sections were examined microscopically for specific staining, and nuclei with a brown color regardless of staining intensity were regarded as positive. Cyclin D1 positivity was calculated at a magnification of ×40 by dividing the number of positive cells by the total number of cells counted in five random fields and expressed as a percentage. Images were captured using an Olympus BX51 microscope and Olympus DP72 camera (magnification, ×40; Olympus Corporation).
Statistical analysis
GraphPad Prism software version 6.0 (GraphPad Software, Inc.) was used for all analyses. A two-tailed unpaired Student's t-test was used to determine statistical significance between different groups. Two-way analysis of variance (ANOVA) or mixed ANOVA was performed to test the comparisons and corrected by Tukey's post hoc test. One-way analysis of variance before the Tukey's post hoc test was performed to test the comparisons. A P-value of <0.05 was considered to indicate a statistically significant difference.
Results
Aspirin inhibits the proliferation of most human CCA cells
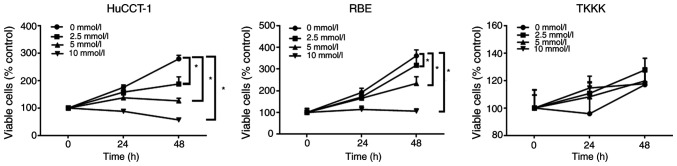
The anti-proliferative effects of aspirin on human CCA cells were determined using the HuCCT-1, RBE, and TKKK CCA cell lines. Cells were treated with 2.5, 5, or 10 mmol/l aspirin for 48 h, and the anti-proliferative effect of aspirin was assessed using the cell viability assay. Untreated cells were used as controls. Aspirin inhibited cell proliferation in CCA cells in a dose and time-dependent manner, except in TKKK cells (Fig. 1).
Figure 1.
Aspirin inhibits the proliferation of some CCA cells. All cell lines were treated with 0, 2.5, 5, or 10 mmol/l aspirin for 24 or 48 h. The data points represent the mean cell number from three independent cultures, and the error bars represent standard deviations. The antiproliferative effect was significantly higher in cells treated with aspirin treatment for 48 h than in control cells (0 mmol/l) as determined by two-way analysis of variance, except for TKKK cells. *P<0.01, vs. the control. CCA, cholangiocarcinoma.
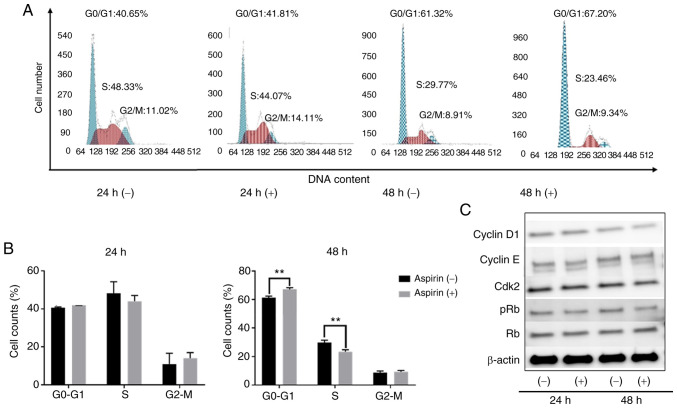
Aspirin induces cell cycle arrest in the G0/G1 phase and regulates cell-cycle related proteins in HuCCT-1 cells
To determine whether aspirin affected the cell cycle in CCA cells, HuCCT-1 cells were treated with aspirin and FCM was performed to examine cell cycle progression. Western blotting was also performed to evaluate the expression of cell-cycle related proteins. Cells were treated with 2.5 mmol/l aspirin for 24 or 48 h, and untreated cells were used as the control. Following aspirin treatment, the population in the G0/G1 phase significantly increased, whereas cells in the S phase decreased (Fig. 2A and B). Western blot results indicated that treatment with aspirin for 48 h significantly modulated cyclin D1, a key protein expressed in the early G1 phase. Cyclin D1 is a key regulator of the cell cycle and is involved in the transition from the G1 phase to the S phase (26). The levels of phosphorylated Rb decreased with aspirin treatment, suggesting that the treated cells were in G1 arrest (Fig. 2C).
Figure 2.
Aspirin causes cell cycle arrest in HuCCT-1 cells. (A) HuCCT-1 cells treated with or without 2.5 mmol/l aspirin were analyzed by flow cytometry to estimate the proportion of cells in each phase of the cell cycle. (B) Graphical representation of the proportion of cells in each phase of the cell cycle. Aspirin blocked the cell cycle at the G/G ** 0 1 phase. P<0.01, vs. the control. (C) Expression of cyclin D1, cyclin E, Cdk2, Rb, and pRb in HuCCT-1 cells 24 and 48 h after aspirin treatment. Rb, retinoblastoma; pRb, phosphorylated Rb.
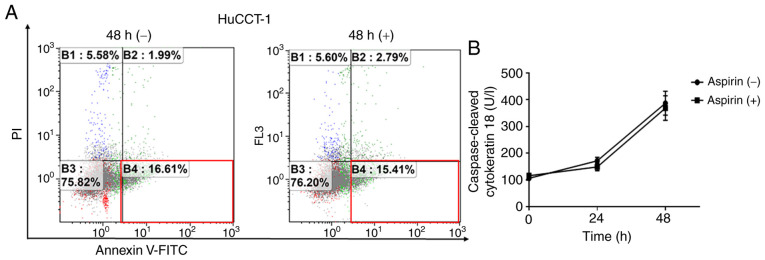
Aspirin does not induce cell apoptosis in HuCCT-1 cells
To determine whether aspirin induced apoptosis in HuCCT-1 cells, FCM was used to detect apoptotic cells after aspirin treatment. The different quadrants represent living cells (lower left quadrant), early apoptotic cells (lower right quadrant), and late apoptotic cells (upper right quadrant). The proportion of early apoptotic cells with aspirin treatment and without treatment were similar after 48 h (Fig. 3A). Additionally, there was no obvious difference in cCK-18 levels between treated and untreated cells after 24- or 48-h treatments (Fig. 3B).
Figure 3.
Aspirin does not induce apoptosis in HuCCT-1 cells. (A) Aspirin treatment (2.5 mmol/l for 48 h) did not alter the proportion of early apoptotic HuCCT-1 cells as determined by flow cytometry. (B) The expression of caspase-cleaved cytokeratin 18 as determined using ELISA after 24 or 48 h of 2.5 mmol/l aspirin treatment.
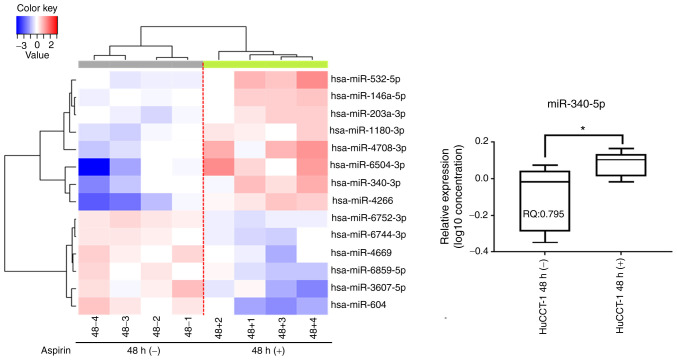
Aspirin affects miRNA expression in HuCCT-1 cells
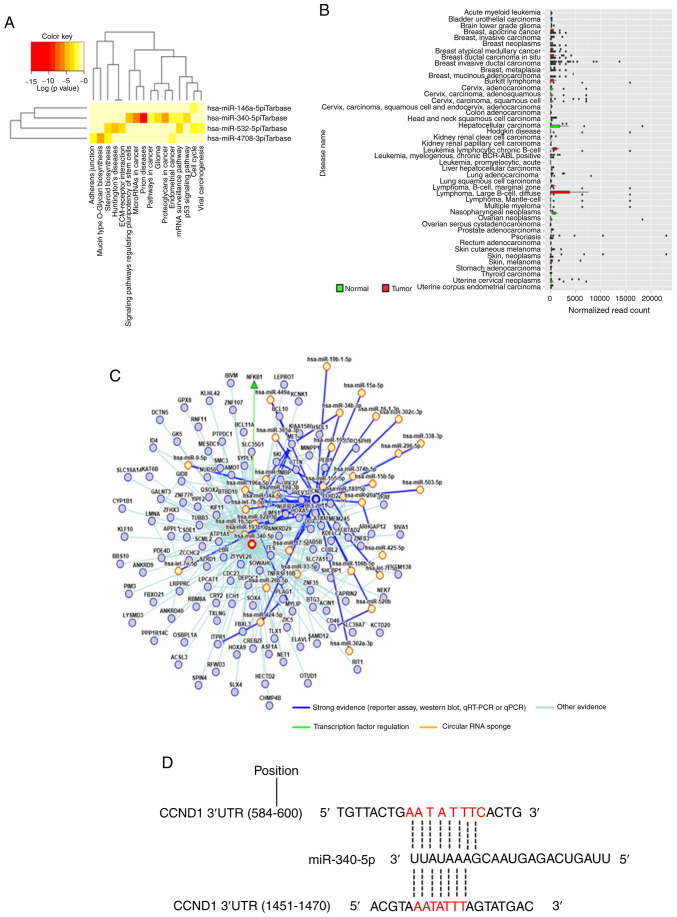
A customized microarray platform was used to analyze the expression of 2,555 miRNAs in aspirin-treated and control HuCCT-1 cells. Treatment with 2.5 mmol/l aspirin for 48 h induced the overexpression of eight miRNAs, whereas the expression of six miRNAs was decreased (Table I; Fig. 4). Unsupervised hierarchical clustering analysis was conducted by calculating Pearson's centered correlation coefficient, and the results indicated that differentially expressed miRNAs in aspirin-treated HuCCT-1 cells clustered together. Previous studies have indicated that miR-340-5p has tumor-suppressive properties (27,28), and miR-340-5p exhibited functions in different parts in the KEGG pathway analyses of upregulated miRNAs, which included 'prion diseases', 'microRNA in cancer', 'proteoglycans in cancer', and 'signaling pathways regulating pluripotency of stem cells' as well as others (Fig. 5A). Although the expression of miR-340-5p was not clearly revealed in CCA, it was decreased in most types of cancer (Fig. 5B). The miRNA regulatory network and miRDB database predicted that CCND1 may be a target of miR-340-5p (Fig. 5C and D). Thus, it was selected for further study, and RT-qPCR indicated that miR-340-5p levels were significantly upregulated in the aspirin-treated cells compared to the untreated cells (Fig. 4). Numerous other studies have revealed the effects of miRNAs on CCA in recent years (Table II) (29-47).
Table I.
Statistical results and chromosomal locations of microRNAs in HuCCT-1 cells.
| miRNA | Fold change (Treated/Untreated) | P-value | Chromosomal location |
|---|---|---|---|
| A, Upregulated
| |||
| hsa-miR-6504-3p | 3.092 | 0.021 | 16 |
| hsa-miR-4266 | 2.701 | 0.005 | 2q13 |
| hsa-miR-4708-3p | 2.158 | 0.028 | 14 |
| hsa-miR-532-5p | 2.055 | 0.039 | X |
| hsa-miR-340-5p | 2.029 | 0.039 | 5 |
| hsa-miR-203a-3p | 1.552 | 0.018 | 14 |
| hsa-miR-1180-3p | 1.547 | 0.009 | 17 |
| hsa-miR-146a-5p | 1.522 | 0.022 | 5 |
|
| |||
| B, Downregulated
| |||
| hsa-miR-6744-3p | 0.654 | 0.003 | 11 |
| hsa-miR-6752-3p | 0.652 | 0.000 | 11 |
| hsa-miR-6859-5p | 0.640 | 0.041 | |
| hsa-miR-4669 | 0.636 | 0.030 | 9q34.2 |
| hsa-miR-3607-5p | 0.526 | 0.045 | |
| hsa-miR-604 | 0.445 | 0.011 | 10p11.23 |
Cells treated with aspirin that exhibited a fold change (FC)>1.5, FC<0.67, or a P-value <0.05 compared with untreated cells are presented.
Figure 4.
Aspirin affects microRNA expression in HuCCT-1 cells. Hierarchical clustering of the expression profiles of numerous differentially expressed miRNAs from HuCCT-1 cells cultured with or without aspirin. Relative quantification of miRNA following aspirin treatment. miR-340-5p expression was significantly upregulated after aspirin treatment. *P<0.05, vs. the control. The log 2−ΔΔCq 10 values for microRNAs were used to create the image, and the lines represent averages with interquartile ranges.
Figure 5.
Bioinformatics analysis and the predicted target gene of miR-340-5p. (A) The Kyoto Encyclopedia of Genes and Genomes pathway analyses of upregulated miRNAs. (B) Expression levels of miR-340-5p in related cancer and normal tissue. Compared with normal tissue, the expression of miR-340-5p was revealed to be decreased in most types of cancer. (C) miRNA regulatory network of miR-340-5p. The intensity of evidence between miR-340-5p with predicted target genes and relationships with associated miRNAs are presented in this network. (D) The seed sequence of human miR-340-5p was complementary to the 3′UTR of CCND1.
Table II.
Previous studies examining microRNAs in cholangiocarcinoma.
| MicroRNA (promoter) | References | MicroRNA (inhibitor) | References |
|---|---|---|---|
| miR-191 | Li et al (2017) (29) | miR-195 | Li et al (2017) (30) |
| miR-205-5p | Kitdumrongthum et al (2018) (31) | miR-410 | Palumbo et al (2016) (32) |
| miR-21 | Lampis et al (2018) (33) | miR-203 | Li et al (2015) (34) |
| miR-181c | Wang et al (2016) (35) | miR-15a | Utaijaratrasmi et al (2018) (36) |
| miR-193-3p | Han et al (2018) (37) | miR-34a | Han et al (2016) (38) |
| miR-383 | Wan et al (2018) (27) | miR-433 | Mansini et al (2018) (28) |
| miR-22 | |||
| miR-199a-3p | Li et al (2017) (39) | ||
| miR-144 | Yang et al (2014) (40) | ||
| miR-590-3p | Zu et al (2017) (41) | ||
| miR-101 | Deng et al (2015) (42) | ||
| miR-26b-5p | Fan et al (2018) (43) | ||
| miR-24 | Ehrlich et al (2017) (44) | ||
| miR-122 | Liu et al (2015) (45) | ||
| miR-26a | Wang and Lv (2016) (46) | ||
| miR-551b-3p | Chang et al (2019) (47) |
miR-340-5p inhibits the proliferation of HuCCT-1 cells and decreases the expression levels of cyclin D1
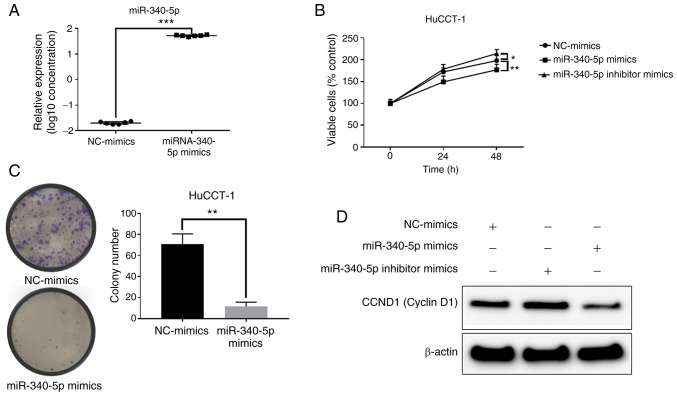
After transfection of miR-340-5p mimics, miR-340-5p expression was significantly increased in the HuCCT-1 cells (Fig. 6A). Transfection of miR-340-5p mimics decreased proliferation in HuCCT-1 cells (Fig. 6B). Moreover, overexpression of miR-340-5p decreased the levels of cyclin D1, whereas inhibition induced increased cyclin D1 levels (Fig. 6D). The colony formation assay indicated that overexpression of miR-340-5p decreased the cell proliferation ability of HuCCT-1 cells (Fig. 6C). Therefore, increasing the levels of miR-340-5p inhibited cyclin D1 expression and decreased the cell proliferation ability.
Figure 6.
Role of miR-340-5p in HuCCT-1 cells. (A) Relative quantification of miR-340-5p following transfection of miR-340-5p mimics or NC. (B) Cell proliferation in HuCCT-1 cells after transfection of NC-mimics, miR-340-5p mimics, or an miR-340-5p inhibitor. (C) miR-340-5p overexpression inhibited colony formation. (D) The levels of cyclin D1 after transfection of NC mimics, miR-340-5p mimics, or an miR-340-5p inhibitor. *P<0.05, **P<0.01 and ***P<0.001, vs. the control. NC, negative control.
Aspirin-nonresponsive TKKK cells express low levels of miR-340-5p
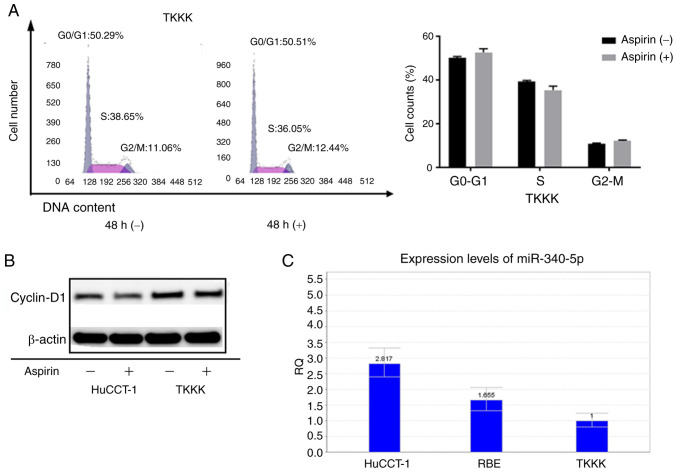
As the CCA cell line TKKK did not exhibit response to aspirin, the cell cycle progression in TKKK cells as compared to HuCCT-1 cells was assessed. Forty-eight hours of aspirin treatment in TKKK cells revealed no obvious difference in the proportion of cells in each phase of the cell cycle (Fig. 7A). In addition, the levels of cyclin D1 were not significantly altered (Fig. 7B). Relative quantification of miR-340-5p was assessed in all cell lines (Fig. 7C). Expression was lowest in TKKK cells, which may indicate that cell lines with high expression of miR-340-5p are more sensitive to cell cycle arrest with aspirin treatment.
Figure 7.
Comparison of the cell cycle and miR-340-5p in aspirin-nonresponsive TKKK cells and aspirin-responsive HuCCT1 cells. (A) Left image: TKKK cells treated with or without 2.5 mmol/l aspirin and analyzed by flow cytometry to estimate the proportion of cells in each phase of the cell cycle. Right image: Graphical representation of the proportion of cells in each phase of the cell cycle. (B) Expression of cyclin D1 in HuCCT-1 cells and TKKK cells after 48 h of aspirin treatment. (C) Relative quantification of miR-340-5p in cholangiocarcinoma cell lines.
Aspirin inhibits tumor proliferation in vivo
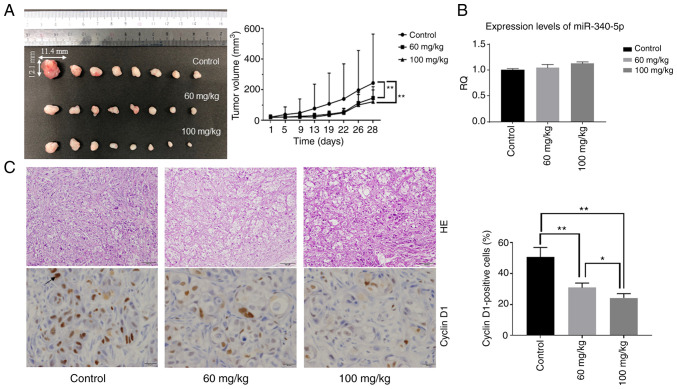
Based on the results obtained from in vitro studies, the effect of aspirin in an in vivo model of CCA was assessed. Nude mice were injected subcutaneously with HuCCT-1 cells followed by intraperitoneal injection of aspirin. The present results revealed that tumor growth was significantly inhibited in mice treated with aspirin compared to untreated mice (P<0.05) (Fig. 8A). No mice succumbed during the observation period. Expression levels of miR-340-5p in tumor tissue were not significantly different, although the RQ was slightly increased in the two aspirin-treated groups (Fig. 8B). H&E-stained images of the xenografted tumor tissues revealed no significant histo-pathological differences between the aspirin-treated and control mice. Immunohistochemical staining of cyclin D1 indicated that cyclin D1-positive cells in the aspirin-treated groups were reduced in number compared with the control group (Fig. 8C).
Figure 8.
Aspirin inhibits the growth of HuCCT-1 cell xenografts in nude mice. (A) The tumors were significantly smaller in aspirin-treated mice than in vehicle-treated mice. Each point represents the mean ± standard deviation of 10 animals. **P<0.01 by mixed analysis of variance. (B) Relative quantification of miR-340-5p in tumor tissue of the three groups of xenografted mice. (C) Left image: H&E-stained images of the xenografted tumor tissues between the aspirin-treated and control mice (scale bar, 100 µm). Immunohistochemical staining of cyclin D1 between the aspirin-treated and control mice (scale bar, 20 µm). Right image: Cyclin D1-positive cells in the aspirin-treated groups were reduced in number compared with the control group as determined by one-way analysis of variance. *P<0.05 and **P<0.01. H&E, hematoxylin and eosin.
Discussion
CCA is an aggressive cancer with high mortality and poor prognosis that accounts for approximately 3% of gastrointestinal malignancies (8). The 5-year survival rate of CCA is only 10%, and the median survival is 24 months (48). Initial analysis revealed that aspirin use was associated with a reduced iCCA risk in men (HR=0.64, 95% CI=0.42-0.98) (49). Subsequent analysis has revealed that aspirin use has a significant inverse association with the development of all three CCA subtypes and leads to an approximately 3-fold reduction in CCA risk (13). This is thought to be due to the anti-inflammatory effect of aspirin. In the present study, we further elucidated the mechanism of the effect of aspirin on CCA. To the best of our knowledge, the present study is the first study revealing that aspirin inhibits the proliferation of CCA cells in vivo.
The anti-inflammatory dose of aspirin varies from 0.5-2.5 mM (50); therefore, relying on data from the anti-proliferation assay, 2.5 mM was selected as the concentration of aspirin, which does not have off-target cytotoxicity. It was observed that aspirin inhibited the proliferation of CCA cells (HuCCT-1 and RBE) and induced cell cycle arrest (HuCCT-1) at the G0/G1 phase, which was correlated with a marked decrease in the expression of cyclin D1. Aspirin also decreased the phosphorylation of Rb. The expression of cell cycle-related molecules is related to cancer progression and prognosis (51), and aspirin has been revealed to inhibit the expression of cyclin D1 in other types of cancer, such as oral squamous cell carcinoma (52) and glioblastoma multiforme (53). To determine whether aspirin induced apoptosis, HuCCT-1 cells were treated with 2.5 mM aspirin and analyzed using FCM; the levels of cCK18 were also measured. However, there was no evidence that aspirin induced apoptosis in HuCCT-1 cells. These data indicated that aspirin inhibited CCA cell proliferation mainly through cell cycle arrest. However, aspirin has also been revealed to induce apoptosis in numerous types of cancers (52-55); this discrepancy could be due to differences in the properties of different types of cancers.
miRNAs are short, noncoding, endogenous, single-stranded RNA molecules 19-25 nucleotides in length that regulate target gene expression (56). They are known to regulate the development and progression of various cancers (57). A miRNA expression array was used to identify the miRNAs associated with the antitumor effects of aspirin. miR-340-5p that was significantly upregulated in response to aspirin treatment in cells, was not significantly different in tumor tissue, although the relative quantification (RQ) was slightly increased in the two aspirin-treated groups. Recent studies have indicated that miR-340-5p inhibited non-small cell lung cancer cell growth and metastasis by targeting ZNF503 (58) and suppressed osteo-sarcoma development via targeting STAT3 (59). In the present study, overexpression of miR-340-5p inhibited proliferation in HuCCT-1 cells. In addition, overexpression of miR-340-5p decreased the levels of cyclin D1. The predicted sequence from the miRDB database and bioinformatics analysis indicated that cyclin D1 may be an miR-340-5p target, and miR-340-5p overexpression decreased cyclin D1 levels in cells; immunohistochemical staining indicated that cyclin D1-positive cells in aspirin-treated mice were reduced compared with control mice. Therefore, it is theorized that aspirin works partially through the miR-340-5p/cyclin D1 axis to inhibit HuCCT-1 cell proliferation.
In the present study, aspirin treatment had no effect on TKKK cells. To investigate the difference between TKKK cells and HuCCT-1 cells, FCM was used to analyze changes in the cell cycle with aspirin treatment. Aspirin did not induce G0/G1 arrest in TKKK cells, and there was no difference in the levels of cyclin D1 after 48 h of treatment. In addition, the relative levels of miR-340-5p were assessed in all cell lines, and TKKK cells had the lowest expression. Therefore, aspirin may not suppress proliferation in TKKK cells because it cannot utilize the miR-340-5p/cyclin D1 axis.
In the present in vivo model, aspirin inhibited the growth of subcutaneous CCA tumors in athymic nude mice. In accordance with the in vitro results and previous studies (23,60), in the in vivo experiment, the tumor volumes in both treatment groups (low-dosage and high-dosage) were significantly decreased compared with the tumor volume in the control group. However, there was no obvious difference in tumor volume between the low-dosage and high-dosage groups; long-term treatment may be required to see a difference based on dosage.
In conclusion, the present study indicated that aspirin inhibited cell proliferation and tumor growth in some CCA cell lines by inducing G0/G1 phase cell cycle arrest, and the underlying mechanism may partially be through the miR-340-5p/cyclin D1 axis to induce cell cycle arrest.
Acknowledgments
We thank Ms. Kayo Hirose, Ms. Keiko Fujikawa, Ms. Miwako Watanabe, Ms. Megumi Okamura, and Ms. Fuyuko Kokado at the Department of Gastroenterology and Neurology of Kagawa University (Kagawa, Japan) for their assistance.
Abbreviations
- CCA
cholangiocarcinoma
- NSAID
non-steroidal anti-inflammatory drug
- FCM
flow cytometer
- AMPK
AMP-activated protein kinase
- miRNA
microRNA
- NF-κB
nuclear factor κ-light-chain-enhancer of activated B cells
- JNK
c-Jun-N-terminal kinase
- GSK-3
glycogen synthase kinase-3
- mTOR
mammalian target of rapamycin
- STAT3
signal transducers and activators of transcription 3
- p-Bcl-2
phosphorylated B-cell lymphoma-2
- ELISA
enzyme-linked immunosorbent assay
- Cdk
cyclin-dependent kinase
Funding
No funding was received.
Availability of data and materials
All data supporting the conclusions of the present study have been documented in this study.
Authors' contributions
TS, AM, HK and TM conceived and designed the experiments. TS, HI, JG, NN, SL, MN, HY, TN, KO, YS and KF analyzed and interpreted the data. TS performed the experiments. TS wrote, reviewed and edited the manuscript. TM reviewed and edited the manuscript for important intellectual content. All authors reviewed, read and approved the final manuscript.
Ethics approval and consent to participate
All experimental protocols were approved (approval no. 18674) by the Institutional Review Board of the Department of Laboratory Animal Science of Kagawa University (Kawaga, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Patel T. Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology. 2001;33:1353–1357. doi: 10.1053/jhep.2001.25087. [DOI] [PubMed] [Google Scholar]
- 2.Xiao J, Zhu J, Liu Z, Wan R, Li Y, Xiao W. Role of surgical treatment for hepatolithiasis-associated intrahepatic cholangio-carcinoma: A retrospective study in a single institution. J Cancer Res Ther. 2017;13:756–760. doi: 10.4103/jcrt.JCRT_356_17. [DOI] [PubMed] [Google Scholar]
- 3.Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, Erice O, Gonzalez E, Jimenez-Agüero R, Lacasta A, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocar-cinoma. Hepatology. 2017;66:1125–1143. doi: 10.1002/hep.29291. [DOI] [PubMed] [Google Scholar]
- 4.Kim TS, Pak JH, Kim JB, Bahk YY. Clonorchis sinensis, an oriental liver fluke, as a human biological agent of cholangiocar-cinoma: A brief review. BMB Rep. 2016;49:590–597. doi: 10.5483/BMBRep.2016.49.11.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang MH, Lee YJ, Kim H. Intrahepatic cholangiocarcinoma arising in Caroli's disease. Clin Mol Hepatol. 2014;20:402–405. doi: 10.3350/cmh.2014.20.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Jiang Z, Yang Y, Zheng P, Wei H, Lin Y, Lv G, Yang Q. Clonorchis sinensis infection and co-infection with the hepatitis B virus are important factors associated with cholan-giocarcinoma and hepatocellular carcinoma. Parasitol Res. 2017;116:2645–2649. doi: 10.1007/s00436-017-5572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blechacz B. Cholangiocarcinoma: Current knowledge and new developments. Gut Liver. 2017;11:13–26. doi: 10.5009/gnl15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Yang T, Wu M, Shen F. Intrahepatic cholangio-carcinoma: Epidemiology, risk factors, diagnosis and surgical management. Cancer Lett. 2016;379:198–205. doi: 10.1016/j.canlet.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Ng K, Meyerhardt JA, Chan AT, Sato K, Chan JA, Niedzwiecki D, Saltz LB, Mayer RJ, Benson AB, III, Schaefer PL, et al. Aspirin and COX-2 inhibitor use in patients with stage III colon cancer. J Natl Cancer Inst. 2014;107:345. doi: 10.1093/jnci/dju345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barron TI, Flahavan EM, Sharp L, Bennett K, Visvanathan K. Recent prediagnostic aspirin use, lymph node involvement, and 5-year mortality in women with stage I-III breast cancer: A nationwide population-based cohort study. Cancer Res. 2014;74:4065–4077. doi: 10.1158/0008-5472.CAN-13-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon TG, Ma Y, Ludvigsson JF, Chong DQ, Giovannucci EL, Fuchs CS, Meyerhardt JA, Corey KE, Chung RT, Zhang X, Chan AT. Association between aspirin use and risk of hepatocellular carcinoma. JAMA Oncol. 2018;4:1683–1690. doi: 10.1001/jamaoncol.2018.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J, Ghoz HM, Peeraphatdit T, Baichoo E, Addissie BD, Harmsen WS, Therneau TM, Olson JE, Chaiteerakij R, Roberts LR. Aspirin use and the risk of cholangiocarcinoma. Hepatology. 2016;64:785–796. doi: 10.1002/hep.28529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guenzle J, Garrelfs NWC, Goeldner JM, Weyerbrock A. Cyclooxygenase (COX) inhibition by acetyl salicylic acid (ASA) enhances antitumor effects of nitric oxide in glioblastoma in vitro. Mol Neurobiol. 2019;56:6046–6055. doi: 10.1007/s12035-019-1513-6. [DOI] [PubMed] [Google Scholar]
- 15.Henry WS, Laszewski T, Tsang T, Beca F, Beck AH, McAllister SS, Toker A. Aspirin suppresses growth in PI3K-mutant breast cancer by activating AMPK and inhibiting mTORC1 signaling. Cancer Res. 2017;77:790–801. doi: 10.1158/0008-5472.CAN-16-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YX, Feng JY, Sun MM, Liu BW, Yang G, Bu YN, Zhao M, Wang TJ, Zhang WY, Yuan HF, Zhang XD. Aspirin inhibits the proliferation of hepatoma cells through controlling GLUT1-mediated glucose metabolism. Acta Pharmacol Sin. 2019;40:122–132. doi: 10.1038/s41401-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z, Fang W, Liu W, Wang L, Liu B, Liu S, Liu S. Aspirin induces Beclin-1-dependent autophagy of human hepatocellular carcinoma cell. Eur J Pharmacol. 2018;823:58–64. doi: 10.1016/j.ejphar.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 18.Raza H, John A, Benedict S. Acetylsalicylic acid-induced oxidative stress, cell cycle arrest, apoptosis and mitochondrial dysfunction in human hepatoma HepG2 cells. Eur J Pharmacol. 2011;668:15–24. doi: 10.1016/j.ejphar.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Pavlovic N, Rani B, Gerwins P, Heindryckx F. Platelets as key factors in hepatocellular carcinoma. Cancers (Basel) 2019;11:1022. doi: 10.3390/cancers11071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malehmir M, Pfister D, Gallage S, Szydlowska M, Inverso D, Kotsiliti E, Leone V, Peiseler M, Surewaard BGJ, Rath D, et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat Med. 2019;25:641–655. doi: 10.1038/s41591-019-0379-5. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41(Database issue):D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target inter-action database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology. 2002;181-182:475–481. doi: 10.1016/S0300-483X(02)00460-2. [DOI] [PubMed] [Google Scholar]
- 27.Wan P, Chi X, Du Q, Luo J, Cui X, Dong K, Bing Y, Heres C, Geller DA. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF1. J Cell Biochem. 2018;119:9720–9729. doi: 10.1002/jcb.27286. [DOI] [PubMed] [Google Scholar]
- 28.Mansini AP, Lorenzo Pisarello MJ, Thelen KM, Cruz-Reyes M, Peixoto E, Jin S, Howard BN, Trussoni CE, Gajdos GB, LaRusso NF, et al. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology. 2018;68:561–573. doi: 10.1002/hep.29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Zhou ZQ, Yang ZR, Tong DN, Guan J, Shi BJ, Nie J, Ding XT, Li B, Zhou GW, Zhang ZY. MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology. 2017;66:136–151. doi: 10.1002/hep.29116. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Piontek K, Ishida M, Fausther M, Dranoff JA, Fu R, Mezey E, Gould SJ, Fordjour FK, Meltzer SJ, et al. Extracellular vesicles carry microRNA-195 to intrahepatic cholangiocarcinoma and improve survival in a rat model. Hepatology. 2017;65:501–514. doi: 10.1002/hep.28735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitdumrongthum S, Metheetrairut C, Charoensawan V, Ounjai P, Janpipatkul K, Panvongsa W, Weerachayaphorn J, Piyachaturawat P, Chairoungdua A. Dysregulated microRNA expression profiles in cholangiocarcinoma cell-derived exosomes. Life Sci. 2018;210:65–75. doi: 10.1016/j.lfs.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 32.Palumbo T, Poultsides GA, Kouraklis G, Liakakos T, Drakaki A, Peros G, Hatziapostolou M, Iliopoulos D. A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer. 2016;16:353. doi: 10.1186/s12885-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lampis A, Carotenuto P, Vlachogiannis G, Cascione L, Hedayat S, Burke R, Clarke P, Bosma E, Simbolo M, Scarpa A, et al. MIR21 drives resistance to heat shock protein 90 inhibition in cholangio-carcinoma. Gastroenterology. 2018;154:1066–1079.e5. doi: 10.1053/j.gastro.2017.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Gao B, Huang Z, Duan T, Li D, Zhang S, Zhao Y, Liu L, Wang Q, Chen Z, Cheng K. Prognostic significance of microRNA-203 in cholangiocarcinoma. Int J Clin Exp Pathol. 2015;8:9512–9516. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Xie C, Pan S, Liang Y, Han J, Lan Y, Sun J, Li K, Sun B, Yang G, et al. N-myc downstream-regulated gene 2 inhibits human cholangiocarcinoma progression and is regulated by leukemia inhibitory factor/MicroRNA-181c negative feedback pathway. Hepatology. 2016;64:1606–1622. doi: 10.1002/hep.28781. [DOI] [PubMed] [Google Scholar]
- 36.Utaijaratrasmi P, Vaeteewoottacharn K, Tsunematsu T, Jamjantra P, Wongkham S, Pairojkul C, Khuntikeo N, Ishimaru N, Sirivatanauksorn Y, Pongpaibul A, et al. The microRNA-15a-PAI-2 axis in cholangiocarcinoma-associated fibroblasts promotes migration of cancer cells. Mol Cancer. 2018;17:10. doi: 10.1186/s12943-018-0760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han YL, Yin JJ, Cong JJ. Downregulation of microRNA-193-3p inhibits the progression of intrahepatic cholangiocarcinoma cells by upregulating TGFBR3. Exp Ther Med. 2018;15:4508–4514. doi: 10.3892/etm.2018.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y, Meng F, Venter J, Wu N, Wan Y, Standeford H, Francis H, Meininger C, Greene J, Jr, Trzeciakowski JP, et al. miR-34a-dependent overexpression of Per1 decreases cholangiocarcinoma growth. J Hepatol. 2016;64:1295–1304. doi: 10.1016/j.jhep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Xia X, Ji J, Ma J, Tao L, Mo L, Chen W. MiR-199a-3p enhances cisplatin sensitivity of cholangiocarcinoma cells by inhibiting mTOR signaling pathway and expression of MDR1. Oncotarget. 2017;8:33621–33630. doi: 10.18632/oncotarget.16834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang R, Chen Y, Tang C, Li H, Wang B, Yan Q, Hu J, Zou S. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b. BMC Cancer. 2014;14:917. doi: 10.1186/1471-2407-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zu C, Liu S, Cao W, Liu Z, Qiang H, Li Y, Cheng C, Ji L, Li J, Li J. MiR-590-3p suppresses epithelial-mesenchymal tran-sition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget. 2017;8:34698–34708. doi: 10.18632/oncotarget.16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng G, Teng Y, Huang F, Nie W, Zhu L, Huang W, Xu H. MicroRNA-101 inhibits the migration and invasion of intrahe-patic cholangiocarcinoma cells via direct suppression of vascular endothelial growth factor-C. Mol Med Rep. 2015;12:7079–7085. doi: 10.3892/mmr.2015.4239. [DOI] [PubMed] [Google Scholar]
- 43.Fan F, Lu J, Yu W, Zhang Y, Xu S, Pang L, Zhu B. MicroRNA-26b-5p regulates cell proliferation, invasion and metastasis in human intrahepatic cholangiocarcinoma by targeting S100A7. Oncol Lett. 2018;15:386–392. doi: 10.3892/ol.2017.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrlich L, Hall C, Venter J, Dostal D, Bernuzzi F, Invernizzi P, Meng F, Trzeciakowski JP, Zhou T, Standeford H, et al. miR-24 inhibition increases menin expression and decreases cholangio-carcinoma proliferation. Am J Pathol. 2017;187:570–580. doi: 10.1016/j.ajpath.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu N, Jiang F, He TL, Zhang JK, Zhao J, Wang C, Jiang GX, Cao LP, Kang PC, Zhong XY, et al. The roles of MicroRNA-122 overexpression in inhibiting proliferation and invasion and stimulating apoptosis of human cholangiocarcinoma cells. miR-24 inhibition increases menin expression and decreases cholangio-carcinoma proliferation. Sci Rep. 2015;5:16566. doi: 10.1038/srep16566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P, Lv L. miR-26a induced the suppression of tumor growth of cholangiocarcinoma via KRT19 approach. Oncotarget. 2016;7:81367–81376. doi: 10.18632/oncotarget.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang W, Wang Y, Li W, Shi L, Geng Z. MicroRNA-551b-3p inhibits tumour growth of human cholangiocarcinoma by targeting Cyclin D1. J Cell Mol Med. 2019;23:4945–4954. doi: 10.1111/jcmm.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrick JL, Sahasrabuddhe VV, Chan AT, Alavanja MC, Beane-Freeman LE, Buring JE, Chen J, Chong DQ, Freedman ND, Fuchs CS, et al. NSAID use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The liver cancer pooling project. Cancer Prev Res (Phila) 2015;8:1156–1162. doi: 10.1158/1940-6207.CAPR-15-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dovizio M, Bruno A, Tacconelli S, Patrignani P. Mode of action of aspirin as a chemopreventive agent. Recent Results Cancer Res. 2013;191:39–65. doi: 10.1007/978-3-642-30331-9_3. [DOI] [PubMed] [Google Scholar]
- 51.Masaki T, Shiratori Y, Rengifo W, Igarashi K, Yamagata M, Kurokohchi K, Uchida N, Miyauchi Y, Yoshiji H, Watanabe S, et al. Cyclins and cyclin-dependent kinases: Comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology. 2003;37:534–543. doi: 10.1053/jhep.2003.50112. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Feng H, Li Z, Guo J, Li M. Aspirin is involved in the cell cycle arrest, apoptosis, cell migration, and invasion of oral squamous cell carcinoma. Int J Mol Sci. 2018;19:2029. doi: 10.3390/ijms19072029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pozzoli G, Marei HE, Althani A, Boninsegna A, Casalbore P, Marlier LNJL, Lanzilli G, Zonfrillo M, Petrucci G, Rocca B, et al. Aspirin inhibits cancer stem cells properties and growth of glio-blastoma multiforme through Rb1 pathway modulation. J Cell Physiol. 2019 Jan 30; doi: 10.1002/jcp.28194. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 54.Hossain MA, Kim DH, Jang JY, Kang YJ, Yoon JH, Moon JO, Chung HY, Kim GY, Choi YH, Copple BL, Kim ND. Aspirin induces apoptosis in vitro and inhibits tumor growth of human hepatocellular carcinoma cells in a nude mouse xenograft model. Int J Oncol. 2012;40:1298–1304. doi: 10.3892/ijo.2011.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi BH, Chakraborty G, Baek K, Yoon HS. Aspirin-induced Bcl-2 translocation and its phosphorylation in the nucleus trigger apoptosis in breast cancer cells. Exp Mol Med. 2013;45:e47. doi: 10.1038/emm.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paliouras AR, Monteverde T, Garofalo M. Oncogene-induced regulation of microRNA expression: Implications for cancer initiation, progression and therapy. Cancer Lett. 2018;421:152–160. doi: 10.1016/j.canlet.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 58.Lu G, Zhang Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell Mol Biol Lett. 2019;24:34. doi: 10.1186/s11658-019-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rongxin S, Pengfei L, Li S, Xiaochen J, Yihe H. MicroRNA-340-5p suppresses osteosarcoma development by down-regulating the Wnt/β-catenin signaling pathway via targeting the STAT3 gene. Eur Rev Med Pharmacol Sci. 2019;23:982–991. doi: 10.26355/eurrev_201902_16985. [DOI] [PubMed] [Google Scholar]
- 60.Yue W, Zheng X, Lin Y, Yang CS, Xu Q, Carpizo D, Huang H, DiPaola RS, Tan XL. Metformin combined with aspirin significantly inhibit pancreatic cancer cell growth in vitro and in vivo by suppressing anti-apoptotic proteins Mcl-1 and Bcl-2. Oncotarget. 2015;6:21208–21224. doi: 10.18632/oncotarget.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the conclusions of the present study have been documented in this study.