Abstract
The severe acute respiratory syndrome associated coronavirus-2 (SARS-CoV-2) poses a threat to human life worldwide. Since early March, 2020, coronavirus disease 2019 (COVID-19), characterized by an acute and often severe form of pneumonia, has been declared a pandemic. This has led to a boom in biomedical research studies at all stages of the pipeline, from the in vitro to the clinical phase. In line with this global effort, known drugs, currently used for the treatment of other pathologies, including antivirals, immunomodulating compounds and antibodies, are currently used off-label for the treatment of COVID-19, in association with the supportive standard care. Yet, no effective treatments have been identified. A new hope stems from medical oncology and relies on the use of immune-checkpoint inhibitors (ICIs). In particular, amongst the ICIs, antibodies able to block the programmed death-1 (PD-1)/PD ligand-1 (PD-L1) pathway have revealed a hidden potential. In fact, patients with severe and critical COVID-19, even prior to the appearance of acute respiratory distress syndrome, exhibit lymphocytopenia and suffer from T-cell exhaustion, which may lead to viral sepsis and an increased mortality rate. It has been observed that cancer patients, who usually are immunocompromised, may restore their anti-tumoral immune response when treated with ICIs. Moreover, viral-infected mice and humans, exhibit a T-cell exhaustion, which is also observed following SARS-CoV-2 infection. Importantly, when treated with anti-PD-1 and anti-PD-L1 antibodies, they restore their T-cell competence and efficiently counteract the viral infection. Based on these observations, four clinical trials are currently open, to examine the efficacy of anti-PD-1 antibody administration to both cancer and non-cancer individuals affected by COVID-19. The results may prove the hypothesis that restoring exhausted T-cells may be a winning strategy to beat SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, cancer, immune-checkpoint inhibitors, immunotherapy, anti-PD-1 monoclonal antibody
1. Introduction
In December, 2019, following an outbreak in a fish market in Wuhan (Hubei, China), an infection leading to severe pneumonia rapidly and uncontrollably spread worldwide. The World Health Organization (WHO) declared the status of a pandemic threat on March 11, 2020 (1), leading all countries to impose lockdowns with devastating socio-economic consequences.
The infection is caused by a highly pathogenic coronavirus, the severe acute respiratory syndrome associated corona-virus-2 (SARS-CoV-2), which is the identified etiological agent of the outbreak, named coronavirus disease 2019 (COVID-19) (2).
Despite global lockdown measures adopted by most countries, as of October 11, 2020 over 37 million COVID-19 cases and 1 million deaths have been reported globally, with half of these cases and deaths reported in the regions of the Americas (3).
SARS-CoV-2 transmission occurs through respiratory droplets, via direct person-to-person contact, with an incubation period of approximately 2 to 12 days following exposure (4). The symptoms of COVID-19 caused by viral infection of the lower respiratory tract include dry cough, fever, fatigue hyposmia and hypogeusia. The infection may rapidly degenerate to pneumonia, and it can be associated with mild complications, including dyspnea, lymphocytopenia, myalgia, or with more severe comorbidities, including significant hypoxia, associated with the appearance of the acute respiratory distress syndrome (ARDS) (5). Due to ARDS, a non-negligible proportion of COVID-19-affected patients may require oxygen therapy and admission to an intensive care unit (ICU), as the ARDS may be swiftly followed by septic shock and multi-organ dysfunction, thus resulting in a 1 to 4% case fatality rate (6).
Viral genome sequencing generated from samples of 5 patients hospitalized with pneumonia led to the identification of this novel SARS-CoV-2 as part of the β-coronaviridae genera, exhibiting an 88% identity to the sequence of 2 bat-derived CoVs. In fact, bats are considered reservoir hosts for CoVs and, it is considered that SARS-CoV-2 originated from bats, later crossing species before infecting humans (7). Additionally, SARS-CoV-2 exhibits an 80 and 50% identity, respectively, with the genome of 2 other clinically relevant human-β-CoV, the SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (8). SARS-CoV emerged in 2002 in China, and caused an outbreak in 26 countries, with a case fatality rate of 9.5%. MERS-CoV originated in 2012 in Saudi Arabia, spreading to 27 countries, with a case fatality rate of 34.4% (8). Therefore, SARS-CoV-2 is considered the third highly pathogenic human-adapted CoV of the 21st century, representing a threat for human health with a heavy global impact (9).
To preserve humanity from this noxious CoV, biomedical research is proceeding at a rapid pace, with the intention of developing a vaccine and/or a therapy to prevent and/or eradicate the virus (10).
The present review article reports up-to-date knowledge of SARS-CoV-2 and COVID-19, with particular attention being paid to novel research approaches aimed at identifying old and novel compounds for the treatment of COVID-19, as well as all therapeutic options currently tested. Among these, the importance of modulating the host immune system is widely discussed, with particular emphasis on the clinical testing of a drug currently used in oncology, the immune-checkpoint inhibitor (ICI) anti-programmed death-1 (PD-1) blocking antibody.
2. SARS-CoV-2 immunopathology
SARS-CoV-2 is a single-stranded positive-sense RNA virus, whose RNA-genome is approximately 30 Kb in length, with a 5′-CAP and a 3′-poly-A tail and containing 10 open reading frames (ORFs) (11,12). Genomic variations of SARS-CoV-2 have been observed; however, further studies are warranted in order to determine whether differently virulent strains may be associated with the differential infection severity range observed in patients with COVID-19 (13).
To enter the host cells, SARS-CoV-2, similar to SARS-CoV, binds to the extracellular enzymatic domain of the single pass transmembrane angiotensin-converting enzyme 2 (ACE2) receptor (14). Physiologically, ACE2 negatively modulates blood pressure by degrading the vasoconstrictor angiotensin II peptide (15). SARS-CoV-2 binding is mediated by the viral glycoprotein spike (S), which also determines the crown-shaped aspect of the virus (16). The difference between SARS-CoV-2 and SARS-CoV is that SARS-CoV-2 possesses a furin cleavage site within the S protein secondary sequence. This is likely to increase virus pathogenicity as cleavage of the S domain during protein synthesis highly increases the affinity for the ACE2 receptor (17), subsequently promoting SARS-CoV-2 endocytosis inside the target cell (18).
ACE2 receptor is expressed by alveolar epithelial cells, but also by lung endothelial cells, and, distally, by heart, lung, kidney, intestinal and neuronal cells. Whether SARS-CoV-2 is able to infect organs located far from the lungs (by directly binding ACE2 receptor in other cells distant from the primary site) is currently under evaluation (19). Within the alveoli, resident dendritic cells (DCs) and macrophages may also be invaded by the SARS-CoV-2, although it remains unclear as to whether this occurs via primary viral infection or through the phagocytosis of epithelial and endothelial pre-infected apoptotic cells (20).
After entering host cells, SARS-CoV-2 uses the host cellular replication machinery to synthesize the RNA negative strand and to produce up to 10 protein products, including the 4 structural proteins S, envelope (E), nucleocapsid (N) and membrane (M) proteins. The viral particles take shape inside the endoplasmic reticulum-Golgi intermediate cellular compartment. Once formed, the host vesicles containing the virus fuse with the plasma membrane, thereby releasing multiple copies of the replicated SARS-CoV-2 into the inter-cellular space, ready to spread to other cells (21).
As SARS-CoV-2 is a cytopathic virus, infected cells become deeply injured and may undergo cell death upon infection. For this reason, viral infection can trigger extensive lung damage (22). In particular, within infected alveolar cells, a peculiar type of inflammatory cell death termed pyroptosis is observed. Pyroptosis is associated with a concurrent endothelial leakage and triggers an acute pro-inflammatory host immune response, mediated by the intracellular activation of the NLRP3 inflammasome (23-25).
Following SARS-CoV-2 infection, a high rate of immune inflammatory cell infiltration within the alveolar space enhances the local synthesis and secretion of cytokines [interferon (IFN)-α, IFN-β, IFN-γ, tumor necrosis factor (TNF)-α, TGFβ, granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin (IL)-1β, IL-6, IL-8, IL-12, IL-18, IL-33, etc.] and chemokines (CCL2, CCL3, CCL5, CXCL8, etc.) by immune effector cells, thereby resulting in acute lung injury. If unresolved, this exacerbated production may lead to the so-called 'cytokine storm' (CS) (26). In the near future, it would be crucial to better understand the full dynamics of the cytokine storm kinetics (27).
It has been demonstrated that the severity of COVID-19 is proportional to the pro-inflammatory cytokine IL-6 levels (28). Moreover, the release of pro-IL-1β, subsequently cleaved to produce mature IL-1β, favors the onset of a pro-inflammatory milieu followed by pyroptosis in the lungs (29). Additionally, GM-CSF if produced in excess, can lead to local tissue damage (30). The CS, if uncontrolled, represents a lethal inflammatory response, which determines ARDS, respiratory defeat and multiple organ failure, finally leading to death (27).
The CS itself may determine a high and unrestrained immune cell infiltration rate within the lungs, which may be linked to thrombosis and pulmonary embolism, often observed in the severe forms of the COVID-19 (31). Moreover, the subsequent endothelial cellular damage and death promoted by direct viral infection and by the activity of the immune cells recalled over the alveolar surface, can increase lung permeability and promote systemic SARS-CoV-2 invasion (32). Such damage is similar to that induced by other noxae, including natural or glass fibers, other viruses and bacteria, etc. (33-35). A prospective autopsy cohort study on 21 deceased COVID-19-infected patients demonstrated the existence of an extensive systemic inflammatory response with diffused aggregates of neutrophils and platelets (36). Based on these results, the authors of that study suggested that COVID-19 was associated with a maladaptive immune response. Thus, immunomodula-tory molecules may be used as therapeutic approach in severe COVID-19 (36).
The rise of the CS is soon followed by the immune adaptive response which, in COVID-19-infected patients with unresolved infection, is particularly low and thus unable to effectively eradicate the virus (37). Specifically, SARS-CoV-2- infected cells may expose viral proteins (via mechanisms that are not yet fully characterized) to antigen-presenting cells (APCs), thus promoting B-cell activation and the subsequent production of IgM and then IgG (38). Importantly, it has been observed that immunoglobulins are detectable within 7 days from the beginning of infection and the increased IgG production is associated with the severity of COVID-19 (39,40).
T-cells also become activated, although the peripheral blood count of both CD4+ and CD8+ T-cells in patients with COVID-19 is extremely low (particularly CD8+ T-cells). T-cell activation is triggered during the acute phase of the disease (41). Notably, in patients with severe and critical COVID-19 infection, the T-cell status shifts from extreme activation at early stages to exhaustion. As a consequence, T-cells exhibit a high prominence of surface markers typical of both activation (including CD69, CD38 and CD44), as well as exhaustion (such as mucin-3, PD-1, NKG2A) (42,43). Additionally, natural killer (NK) cells decrease in number in peripheral blood and express the exhaustion marker, NKG2A (44).
Some studies have hypothesized that the exhaustion of T-cells may be a leading cause of the severe and critical forms of COVID-19. In particular, a recent preliminary report assessed the increased expression of NKG2A and PD-1 inhibitory receptors on T- and NK cell surfaces of patients with COVID-19, suggesting that anticancer immunotherapy could be repurposed as a first line of defense to trigger SARS-CoV-2 clearance (45). In general, SARS-CoV-2 infection may negatively affect the host antiviral immunity at an early stage of the infection process. Importantly, both dysregulated innate and adaptive immune responses largely contribute to the SARS-CoV-2 immunopathology (42). On the other hand, SARS-CoV-2 can evade the host immune system and use it to its own advantage. Therefore, the disruption of such immune evasion may play an important role in the development of novel therapeutic approaches to eradicate SARS-CoV-2 infections (46-48).
COVID-19 symptomatology is very heterogeneous and is strictly dependent on the hosts' response: i) Mild, with upper respiratory tract infection and digestive tract impairment; ii) moderate, with lower respiratory tract infection, pneumonia, no obvious hypoxemia with lung lesions; iii) severe, with pneumonia and elevated hypoxemia; and iv) critical, characterized by the appearance of ARDS, septic shock, myocardial and kidney injuries, coagulation dysfunctions (49).
COVID-19-infected patients who have recovered, even when moderately affected, can develop chronic lung damage at a diverse grade, as the injured tissue, once repaired, hardens. These fibrotic scars, if accumulated into the alveoli, determine blood vessel blockage and a low oxygen absorbance (50).
In patients with COVID-19 who have succumbed to the disease, the lung presents diffused bilateral alveolar damage and the formation of a hyaline membrane with the thickening of the alveolar wall (51). Moreover, in a number of cases, a strong accumulation of mononuclear cells and infiltrating macrophages is also present, which occludes the air-exposed surfaces. Following electron microscopy analyses, the SARS-CoV-2 virus has been detected inside bronchial and alveolar type 2 cells. Apart from the lungs, kidney hemorrhage, inflamed liver and neuron degeneration have often been recorded in a number of patients (51).
Additionally, in 70% of deaths, a disseminated intravascular coagulation has also been observed (52). Although vascular endothelial cells express ACE2 receptor, the exact mechanism of such a coagulopathy has not yet been clarified. Moreover, apart from being isolated from the lungs, SARS-CoV-2 viral particles have also been found in urine and stool samples (53). A total of 70% of deaths of patients with COVID-19 were caused by ARDS, whereas the other 30% by complications, including sepsis and multi-organ failure (possibly related to the CS) (54). Sepsis and septic shock have been reported in severe and critical cases (55). In almost 80% of patients with COVID-19 with sepsis, SARS-CoV-2 is the only etiological agent found in both blood and lower respiratory tract specimens, indicating that it is a viral sepsis. Viremia and sepsis are hypothesized to play a pivotal role in the dissemination of SARS-CoV-2 and the subsequent multi-organ damage (56,57).
The immune system of the host plays a fundamental role in the individual outcome of SARS-CoV-2. Based on available data from patients with COVID-19, a recently published medical hypothesis postulated that in mild COVID-19 cases, lung resident macrophages initiated the anti-viral inflammatory response. In turn, macrophages may contain the infection and can trigger the innate and adaptive immune responses that help to clear infection and induce recovery (58).
On the contrary, in severe and/or critical COVID-19 cases, due to the dysregulated immune system, the epithelial-endothelial lung blood barrier is severely damaged, allowing SARS-CoV-2 to attack alveolar and endothelial lung resident cells, thus attracting additional immune cells into the alveoli (59). This accumulation, with a positive feedback loop, exacerbates the local production of cytokines and chemokines, resulting in an uncontrollable pro-inflammatory state (the CS), therefore worsening the lung insult and damage (60). CS during SARS-CoV-2 infection has been ascribed mainly to the rapid viral replication during the first phase of the infection which triggers the pro-inflammatory response of the host (60). Additionally, CoV-derived molecules may induce the accumulation of pro-inflammatory monocytes in the lungs that, as a positive feedback loop, increase the local secretion of cytokines (61). Moreover, lymphocytes attracted to the lungs soon become overactivated and finally exhausted, thereby losing their ability to fight off the infection. Systemically, both the development of a CS and the concurrent lymphopenia, negatively disrupt organ function, resulting in viral sepsis, microcirculation issues and multiorgan dysfunction (62).
An ongoing clinical study, whose preliminary results were published recently by Laing et al, is currently aiming to assess the COVID-19 immune signature, which is a current challenge given the patient heterogeneity (in terms of underlying comorbidities, sex, age, ethnicity etc.). The preliminary findings of the project, termed COVID-IP (COVID-immunophenotyping), revealed distinct features of the immune system in patients with COVID-19, confirming the overproduction of IgG, the presence of a dysregulated cytokine response, disrupted monocyte and dendritic cell phenotype and selective cytopenia in particular T-cell subsets. All these parameters are significantly associated with a worse patient prognosis (63).
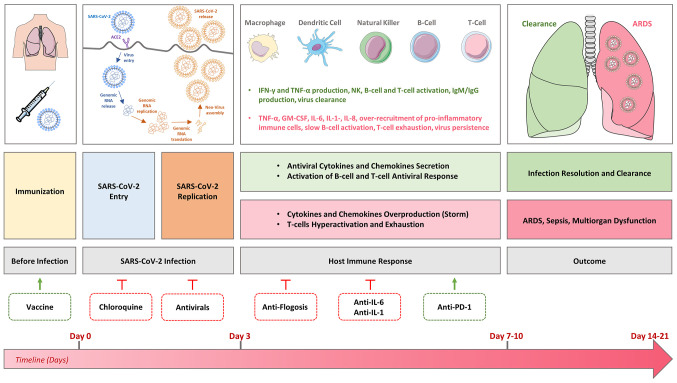
Given all these factors, a therapy able to modulate the innate immune response of the host and/or restore a positive and controlled anti-viral adaptive immune reactivity may improve the outcome of patients with COVID-19 (64-66). A summary of the SARS-CoV-2 immunopathology is represented in Fig. 1.
Figure 1.
Representation of the phases of SARS-CoV-2 infection and host immune response. Green boxes represent the infection dynamics in the case of a good immune response and successful infection clearance. Red boxes indicate the infection dynamics in case of severe or critical complication and infection persistence. All the preventive (vaccine) and therapeutic approaches currently tested clinically are reported (dotted lines) and placed in function of the infection timeline. Green arrows indicate an activating effect of the intervention, red inhibitory arrows indicate an inhibitory effect of the intervention. SARS-CoV-2, severe acute respiratory syndrome associated coronavirus-2; TNF, tumor necrosis factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IL, interleukin; ARDS, acute respiratory distress syndrome; PD-1, programmed death-1.
3. Management of COVID-19: Prevention, diagnosis and treatment
In order to prevent the spread of COVID-19, the WHO has elaborated standard guidelines, meant to reduce the overall risk of SARS-CoV-2 transmission, which include: i) Hand hygiene; ii) respiratory hygiene/cough etiquette; iii) the use of personal protection equipment based on the risk assessment; and iv) environmental cleaning maintenance rules (67).
Currently, there is no treatment approved by the European Medicines Agency (EMA) or the US Food and Drug Administration (FDA) available for the prevention and cure of COVID-19 (68,69). Since July, 2020, there has been an open debate regarding the approval for the treatment of COVID-19 using a specific inhibitor of viral RNA-dependent RNA polymerase, Remdesivir has exhibited promising preclinical efficacy (70). In spite of this, however, clinical trials have yielded controversial results. The American FDA approved the use of Remdesivir for the treatment of COVID-19, based on compelling clinical evidence (i.e., ACTT-1 trial, SIMPLE trials) (71). However, based on the results of the SOLIDARITY WHO-sponsored trial, which did not demonstrate any significant improvement in patients treated with such a molecule, the EMA disposed only a conditional authorization, while the WHO presently discouraged the use of Remdesivir in patients with COVID-19 (72-74).
The management of patients with COVID-19 is mainly based on the provision of supportive care, including oxygen therapy, which represents the main treatment intervention. Although an approved anti-SARS-CoV-2 drug is still lacking, therapy using anti-inflammatories and antivirals is encouraged (75-77). A complete interim guidance produced by the WHO, last updated on May 27, 2020, is currently available, with the main goals of drawing standard procedures to: i) Attenuate and stop transmission; ii) provide optimized care for all patients; and iii) minimize the impact of the pandemic on the health system and the socio-economical activities (78).
As regards the diagnosis of COVID-19, the WHO document recommends to proceed with the collection of upper respiratory tract specimens (nasopharyngeal and oropharyngeal) for testing by reverse transcription-polymerase chain reaction (RT-PCR). Additionally, where clinical suspicion remains, and tested specimens are negative, the advice is to collect specimens from the lower respiratory tract for further testing. More recently, novel high-sensitive molecular methods based on the usage of the high-sensitive droplet digital PCR have been further proposed for the effective diagnosis of COVID-19 patients with low viral load (79,80). On the contrary, SARS-CoV-2 antibody tests are not recommended by the WHO for the diagnosis of infection with COVID-19. Finally, chest imaging (radiograph, computed tomography scan, ultrasound) may assist in diagnosis and may identify or exclude pulmonary complications (78).
According to a report from the EMA on September 24, 2020, 39 potential COVID-19 vaccines and 163 potential COVID-19 treatments are currently under investigation for their use in human subjects (81). Regarding vaccination, 13 out of 34 candidate vaccines have been so far approved for clinical evaluation, based either on attenuated or inactivated viral particles or on viral-derived nucleic acids or protein subunits (82,83). A summary of the current candidate vaccines under clinical evaluation, adapted from the WHO relative document (updated on June 22, 2020), is reported in Table I (84). Importantly, as of October 11, 2020, preliminary data on two COVID-19 vaccines (one from AstraZeneca and another from BioNTech) are currently under evaluation by the EMA human medicines committee using a rapid rolling review procedure (81).
Table I.
Current COVID-19 vaccines under clinical investigation, adapted from a previous study (74).
| Vaccine Strategy | Developer | Associated Clinical Trials |
|---|---|---|
| ChAdOx1-S (non-replicating virus) | University of Oxford/AstraZeneca | ISRCTN89951424, 2020-001228-32, 2020-001072-15 |
| Adenovirus type 5 (non-replicating virus) | CanSino Biological/Beijing Institute of Biotechnology | ChiCTR2000031781, ChiCTR2000030906 |
| Nanoparticle-encapsulated viral RNA | Moderna/NIAID | NCT04405076, NCT04283461 |
| Inactivated virus | Wuhan Institute of Biological Products/Sinopharm | ChiCTR2000031809 |
| Inactivated virus | Beijing Institute of Biological Products/Sinopharm | ChiCTR2000032459 |
| Inactivated virus | Sinovac | NCT04383574, NCT04352608 |
| Nanoparticle-encapsulated viral glycoprotein | Novavax | NCT04368988 |
| Nanoparticle-encapsulated viral RNA | BioNTech/Fosun Pharma/Pfizer | 2020-001038-36, NCT04368728 |
| Inactivated virus | Institute of Medical Biology/Chinese Academy of Medical Sciences | NCT04412538 |
| DNA plasmid vaccine | Inovio Pharmaceuticals | NCT04336410 |
| Adenovirus (non-replicating virus) | Gamaleya Research Institute | NCT04436471, NCT04437875 |
| Nanoparticle-encapsulated viral RNA | Imperial College London | ISRCTN17072692 |
| Viral RNA | Curevac | n.a. |
COVID-19, coronavirus disease 2019; n.a., not available.
The heterogeneous development of COVID-19 is strictly dependent on the interaction between SARS-CoV-2 and the individual immune system. SARS-CoV-2 related factors are: The virus genetics, virulence and titer. Individual depending factors are: Genetics (e.g., HLA genes), sex, age, immune system health, as well as the presence of comorbidities (85).
With respect to the anti-COVID-19 potential treatments under investigation, the currently undergoing clinical trials include: i) Antiviral drugs; ii) antimalarial drugs; iii) anti-inflammatory molecules; and iv) monoclonal antibodies with activity against components of the immune system. A detailed description of all the available options (out of the scope of the present review) is carefully reviewed elsewhere (86-89). A summary of the main drugs currently under clinical investigation, together with their specific features is reported in Table II. A highlight of the leading preventive and therapeutic interventions currently tested with respect to the COVID-19 progression timeline, is reported in Fig. 1.
Table II.
Current COVID-19 repurposed drugs under clinical investigation.
| Drug | Primary pathology | Mechanism of action | Target |
|---|---|---|---|
| Hydroxychloroquine | Malaria | Cellular endocytosis inhibitor | SARS-CoV-2 |
| Chloroquine | Malaria | Cellular endocytosis inhibitor | SARS-CoV-2 |
| Remdesivir | Ebola virus | Viral RNA-dependent RNA polymerase inhibitor | SARS-CoV-2 |
| Favipiravir | Huma influenza virus; Ebola virus | Viral RNA-dependent RNA polymerase inhibitor | SARS-CoV-2 |
| Lopinavir/Ritonavir | Human immunodeficiency virus | Viral protease inhibitor | SARS-CoV-2 |
| Ribavirin | Hepatitis C virus | Viral RNA-dependent RNA polymerase inhibitor; RNA capping inhibitor | SARS-CoV-2 |
| Thalidomide | Cancer | Immunomodulatory | Host immune system |
| Human immunoglobulin | Primary immunodeficiency | Anti-inflammatory | Host immune system |
| Heparin | Coagulopathies | Anticoagulant, anti-inflammatory | Host immune system |
| Corticosteroids | Inflammatory diseases | Anti-inflammatory | Host immune system |
| Tocilizumab | Rheumatoid arthritis | Monoclonal antibody against IL-6 receptor | Host immune system |
| Sarilumab | Rheumatoid arthritis | Monoclonal antibody against IL-6 receptor | Host immune system |
| Canakinumab | Rheumatoid arthritis | Monoclonal antibody against IL-1β | Host immune system |
| Anakinra | Rheumatoid arthritis | Human IL-1 receptor antagonist protein | Host immune system |
| Gimsilumab | Inflammatory diseases; cancer | Monoclonal antibody against GM-CSF | Host immune system |
| Baricitinib | Rheumatoid arthritis | Janus kinase inhibitor; anti-inflammatory | Host immune system |
| Ruxolitinib | Myelofibrosis | Janus kinase inhibitor; anti-inflammatory | Host immune system |
| Nivolumab | Cancer | Monoclonal antibody against PD-1 | Host immune system |
COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome associated coronavirus-2; IL, interleukin; GM-CSF, granulocyte-macrophage colony-stimulating factor; PD-1, programmed death-1.
4. Role of computational biology in the response to COVID-19
The interdisciplinary collaboration from diverse scientific fields is truly relevant in these times. In general, not only is computer science helping to increase the rate of COVID-19 research, it also contributes to the management of severe consequences of the SARS-CoV-2 threat, for healthcare and socio-economical systems, worldwide. A recent review thoroughly described all the modern technologies which are currently helping to tackle the COVID-19 pandemic (90).
Computer science assists by efficiently monitoring and mapping the global spread of the disease, thereby contributing to a more rapid identification of newborn clusters, factors involved in the modulation of the infection rate, as well as to rapidly organize an effective public health response to novel outbreaks (91).
Importantly, bioinformatics represents a fundamental tool for researchers to speed up the process of the identification of drugs and vaccines against COVID-19. A number of web-based platforms have been implemented to promote the public availability of sequences and other complex data, as well as COVID-19-related literature, including the EBI COVID-19 Portal and the NIH SARS-CoV-2 Data Hub (92,93).
In the USA, the White House Office of Science and Technology Policy, the US Department of Energy and IBM founded a public-private partnership, termed the COVID-19 High Performance Computing (HPC) Consortium, aimed at providing researchers globally with massive computing resources, and counting already >100 active projects (94).
With the aid of computational modeling, a number of studies have recently been carried out to predict potential old and novel drug targets, but also candidate vaccines against COVID-19. A Japanese study, recently published in the Journal of Human Genetics - Nature, developed a bioinformatics-based predictive tool for the screening of potential T-cell epitopes for SARS-CoV-2, useful for the better understanding of the immunization dynamic (95). On the same line, a Chinese study developed a virtual screening tool for enzymatic inhibitory molecules, to predict their efficiency to block i) viral cellular entrance, ii) viral replication in the cells, and iii) viral-dependent immune evasion (96). Moreover, another Chinese study developed a drug repurposing platform to assess the efficiency of binding of known protease inhibitors to SARS-CoV-2 enzyme, in order to identify potential effective molecules (97). Another multicenter study set up a computational method to track and map single cell RNA-sequencing. The application to COVID-19 patient specimens proved that this tool may be applied for the identification of the molecular signatures involved in pathogenesis of SARS-CoV-2, thereby assisting in the identification of novel druggable targets (98). For example, using network proximity analyses of drug targets and CoV-host interactions, Zhou et al identified potential repurposable drugs against COVID-19 (99).
Predictive algorithms are also under rapid development, with the goal of identifying an effective SARS-CoV-2 vaccine. For example, an American study performed a bioinformatics analysis to screen potential S protein features which may be highly immunogenic (100). Computer simulation may also accelerate the search for an effective vaccine. Importantly, a study carried out at the University of Catania, in Italy, led to the development of a useful platform to predict in silico, the efficiency of selected anti-SARS-CoV-2 monoclonal antibodies in generating an adequate host immune response (101). The Universal Immune System Simulator (UISS) is able to simulate the dynamics of single entities of the immune system, following a stimulus or a therapeutic intervention, by using an agent-based methodology. This methodology already provided useful prediction for the development of SARS-CoV-2 vaccines. This platform may be applied in the future for the effective screening of novel and existing candidate vaccines against SARS-CoV-2 (101). On the same line, Kar et al from Bangalore University (India) used a computational approach to design a suitable candidate multi-epitope vaccine against SARS-CoV-2 (102). In conclusion, computational approaches and prediction platforms may be applied for the effective screening of potential vaccination and therapeutic strategies against SARS-CoV-2, with the aim of remodulating the impaired immune system to in SARS-CoV-2 infected individuals.
5. Immune-checkpoint inhibitors against COVID-19: A lesson learnt from cancer
The outcome of COVID-19 has been reported to be more severe in patients with co-existing pathologies, which are associated with an impaired immune system (6). For example, elderly subjects or individuals with comorbidities, such as diabetes, obesity, hypertension or cancer, possess an immune system that cannot efficiently contain and combat SARS-CoV-2 infection. In these cases, COVID-19, may rapidly degenerate towards a severe or critical status (6,103,104).
Of note, cancer is a multifactorial disease often associated with viral or bacterial infections. In particular, several studies have demonstrated a direct involvement of certain viruses (HBV, HCV, HPV, etc.) in the pathogenesis of tumors (105,106). On the other hand, cancer patients are highly vulnerable to infections, including SARS-CoV-2. They represent fragile subjects, as the cancer itself may be associated with an extensive immunosuppressive state (107) or as their immunosuppression may be exacerbated by myelosuppressive therapies, such as chemotherapy or radiotherapy (108).
Given their immune-compromised status, cancer patients infected by SARS-CoV-2 may be at a higher risk of developing ARDS, septic shock and acute myocardial infarction (109-111). An early nationwide study conducted in China demonstrated that cancer patients have a significantly higher risk of developing COVID-19 and of suffering severe complications (112). A larger and more recent clinical study performed on 928 cancer patients affected by COVID-19, confirmed that an increased 30-day mortality was associated with age, male sex, smoking and an active cancer status (113). Importantly, that clinical study, confirmed by several others, found that while the presence of comorbidities aggravated the mortality rate associated with COVID-19, the provision of chemotherapy, targeted therapy, or immunotherapy was not associated with an increased mortality in cancer patients (114-118). Possibly, the effects of a more advanced malignant disease may explain the overall severity of infection, as well as the COVID-19-associated death toll (119).
The question remains of how to take care of cancer patients to protect them from SARS-CoV-2. Recent recommendations, based on observations made on lung cancer patients, suggest to weigh the impact of interrupting any programmed cancer treatment, using a case-by-case approach, as there is no universal solution to oncological care during this pandemic. To note a warning from the authors was: 'primum nil nocere' (first do no harm) (120,121).
ICIs, including anti-PD-1, anti-PD ligand-1 (PD-L1) and anti-CTLA-4 antibodies, represent an innovative therapy for the treatment of numerous solid tumors, as well as some hematological malignancies (ie., certain lymphomas) (122) as they restore cellular-mediated immunocompetence (123-126).
As said, chemotherapies and radiotherapies may induce myelosuppression; therefore they lower the overall humoral, as well as the cellular mediated immune response (127). On the contrary, cancer patients treated with ICIs have been demonstrated to be able to restore their immunocompetence during HIV, hepatitis B, or hepatitis C viral infection, suggesting that these individuals may be highly immunocompetent compared to the other cancer patients undergoing chemotherapy or radiotherapy (128).
In rare cases, cancer patients who have undergone ICI therapy, may develop lung immune-related adverse events (irAEs), including pneumonia. This should be taken into consideration as a risk factor in cancer patients affected by COVID-19 and under ICI therapeutic regimens (129). In spite of this, a recent analysis of a cohort of patients affected by lung cancer (TERAVOLT), suggested that ICI-based therapy did not increase the overall mortality risk in cancer patients affected by COVID-19. Coherently, Gonzalez-Cao et al observed, through a retrospective analysis, of 50 cancer patients included in the Spanish registry, that anticancer immunotherapy did not significantly increase the risk of mortality by COVID-19 in melanoma patients (130). However, while the therapy itself may not affect the infection risk, the fragility of cancer patients represents an issue that needs to be assessed with a greater attention being paid in terms of protective care for these exposed individuals (110).
ICIs may restore individual cellular-mediated immuno-competence and this lesson from cancer may be transferred to non-cancer COVID-19 patients. ICIs have been already used beyond cancer for the treatment of, for example, sepsis-induced immunosuppression (131,132). Moreover, ICIs were safely administered to cancer patients vaccinated for influenza virus (133,134). Overall, ICIs are a promising tool to be studied against infective diseases, including SARS-CoV-2 (135,136).
The cytokine release syndrome, determining the CS, is an important complication in patients with severe SARS-CoV-2 infection, as it may lead to ARDS (27). As widely described in the section above, following a first hyper-inflammatory phase triggered by the CS, SARS-CoV-2 prolonged infection may induce T-cell hyperactivation and finally exhaustion, associated with concurrent lymphopenia in patient (137). High pro-inflammatory IL-6 levels, together with an elevated exhaustion and reduced functional diversity of T-cells in peripheral blood, may predict poorer outcomes of patients with COVID-19 (138). Although both CD4+ and CD8+ T-cells in patients with COVID-19 are produced, particularly against the SARS-CoV-2 S antigen, such cells are reduced in abundance and are less activated in the case of severe SARS-CoV-2 infection. Therefore, the viral clearance may be delayed. Additionally, the excessive exhaustion of CD8+ T-cells in patients with severe COVID-19 may reduce their cellular-mediated immune response to the virus (138,139). In detail, peripheral T-cells are impaired and express high levels of both the immunosuppressive markers, mucin-3 and PD-1, which deeply impairs their effector functionality (137). In association with this T-cell disruption, patients with severe and critical COVID-19 may develop viral sepsis (57). The identification of a therapy to restore the T-cell functionality in patients with COVID-19 may be used to prevent viral sepsis and therefore, the development of ARDS.
In a previous study, PD-1/PD-L1 blocking antibodies administered to mice chronically infected with lymphocytic choriomeningitis virus, enhanced the viral control and virus-specific CD8+ T-cell responses, demonstrating that the PD-1 inhibitory pathway is of particular importance in exhausted T-cell formation. Following the PD-1/PD-L1 blockage, T-cells CD8+ restarted to proliferate in lymphoid and non-lymphoid tissues. Importantly, the effect seemed to be specific for PD-1/PD-L1, as anti-CTLA-4 blocking antibody administration had no effect on either T-cell function or viral control (140). In line with these results, PD-1 inhibition was also found relevant in human viral disease. In fact, HIV-infected patients were shown to exhibit T-cell exhaustion with a high PD-1 expression. In such subjects, PD1-blocking antibodies restored both CD4+ and CD8+ T-cell abundance and functionality (141).
Based on results obtained with other models of viral infections, in both mice and humans, the use of ICIs (and in particular anti-PD-1 antibody) has been suggested by several authors for the treatment of SARS-CoV-2-infected subjects, alone or in combination with the concurrent blockage of the pro-inflammatory IL-6 pathway (142-145). The rationale behind this co-administration is to contemporarily block both the detrimental humoral CS with anti-IL-6 pathway-targeting antibodies, in association with restoring T-cell cellular-mediated immunity, by using the anti-PD-1 antibodies (146). These two treatments, if used in synergy, may re-educate the defeated host immune system to finally wipe off the SARS-Cov-2 infection. Importantly, it has also been suggested that an earlier anti-PD-1 intervention in patients with COVID-19 may block the development of ARDS, and thus minimize the need for further ICU support (143). Alternatively, anti-PD-1 administration may be associated with other modulators of the innate immunity, such as Toll-like receptor (TLR) agonists/antagonists, as recently suggested (64).
Based on the reported observations, it seems reasonable to suggest that ICIs may be used in both cancer and non-cancer patients affected by COVID-19 (147). Indeed, 5 clinical studies are currently registered at clinicaltrials.gov with the common goal of studying the potentialities of administering anti-PD-1 antibody to cure COVID-19.
The first one is being conducted on metastatic and advanced cancer patients, affected by COVID-19 and which are not eligible for a transfer to an ICU. It is a French, phase II, prospective, controlled, randomized study, which has already enrolled 384 patients since its opening. The study will assess the difference in the efficacy to eradicate SARS-CoV-2 infection between COVID-19 patients treated with anti-PD-1 antibody nivolumab in association with standard care protocol, versus standard care protocol offered alone (NCT04333914).
Notably, the other 4 registered studies will be enrolling and testing the safety and the efficacy of anti-PD-1 administration on NON-cancer COVID-19-infected patients. The first one is a Chinese, phase II, interventional randomized study, aimed at recruiting a total of 120 COVID-19-infected patients. Given that PD-1 is a key mediator of T-cell depletion and viral sepsis in patients with COVID-19, the investigators would like to assess the clinical efficacy of PD-1 blockade (administering a PD-1 blocking antibody), in association with standard of care treatments, in COVID-19-infected patients with pneumonia and lymphocytopenia (NCT04268537).
A second, Hong Kong, phase II, interventional, open-label, controlled pilot study on 15 adult patients with COVID-19 has been set up to evaluate: i) The efficacy of anti-PD1 anti-body (nivolumab) in clearing the SARS-CoV-2 infection; and ii) the safety of the anti-PD1 treatment in COVID-19-infected patients. In that study, nivolumab will be administered in association with the optimal standard supportive care for COVID-19 (NCT04356508).
A third French, phase II, interventional randomized study will recruit about 100 COVID-19-infected patients which will be treated with standard of care with or without nivolumab administration. As in previous studies, the primary goal is to assess the time required for clinical improvement and the overall efficacy of administering anti-PD-1 antibody to patients with COVID-19 (NCT04343144).
A fourth, French, phase II, randomized study, is enrolling 120 obese COVID-19-infected patients, to evaluate the efficacy of anti-PD-1 nivolumab in treating severe forms of COVID-19 in such high-risk class of infected subjects. Obesity markedly increases the risk of developing a severe, or even critical, form of COVID-19 (103). Obese individuals develop a chronic meta-inflammatory status associated with a dysregulated immune system. This obesity-related status is called inflammaging (148). That study will be divided into 2 arms. All participants will receive routine standard of care for COVID-19, while one arm will receive anti-PD-1 nivolumab in combination (NCT04413838).
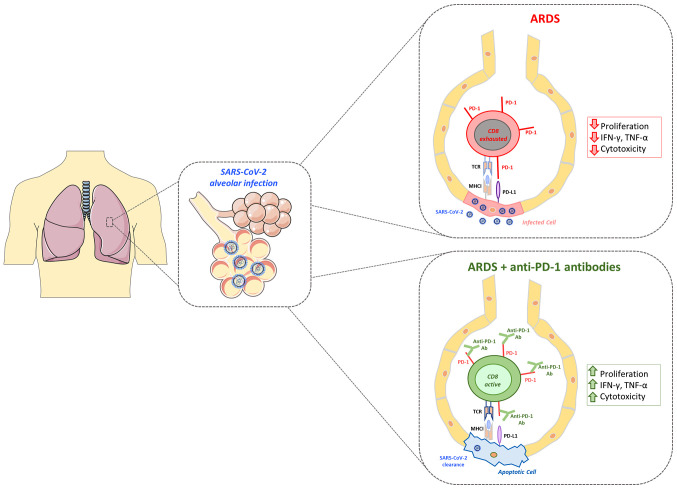
These clinical studies hold immense potential in that they may soon uncover the hidden capability of ICIs, that have revolutionized the field of oncology over the past 10 years, and may thus identify a cure for severe cases of COVID-19. A summary of the expected effects of anti-PD-1 administration is presented in Fig. 2.
Figure 2.
SARS-CoV-2 alveolar infection in severe cases may promote acute respiratory distress syndrome (ARDS; box on upper right, red color). Therapeutic intervention with anti-PD-1 antibody may restore T-cell cytotoxicity towards alveolar infected cells and help optimal viral clearance (box on lower right, green color). SARS-CoV-2, severe acute respiratory syndrome associated coronavirus-2; ARDS, acute respiratory distress syndrome; IFN, interferon; TNF, tumor necrosis factor; PD-1, programmed death-1; PD-L1, PD ligand-1.
6. Conclusions and future perspectives
Over the past 10 months, the threat of COVID-19 has led to a marked increase in both research and medical efforts internationally, with the collective aim of identifying a successful cure with which to eradicate SARS-CoV-2. Although standard care proto-cols and guidelines have been elaborated in order to ameliorate the outcomes of patients with COVID-19 and to contain the global spread of the disease, a cure has not yet been identified. Research has moved forward at a rapid pace and, regarding the vaccine, as of December 2, 2020 the UK gave emergency authorization to the Pfizer and BioNTech's vaccine. As of December 8, 2020, the UK began the vaccination campaign, which will be soon followed by Canada and other nations.
A number of drugs have been adapted from other pathologies to either block viral entry and intracellular replication or to redirect the impaired immune system of compromised severe and critical COVID-19-infected patients.
As regards the host immune system remodulation, two clinical approaches are currently tested in patients. On the one hand, tests are ongoing to identify molecules able to block the exacerbated CS that is responsible for extensive multi-organ damage and ultimately death. This category of molecules includes monoclonal antibodies and small molecules targeting both the IL-6 and IL-1 pro-inflammatory pathways, which may reduce unrestrained cytokine production. On the other hand, novel trials are exploring specific ICIs able to re-activate the exhausted T-cell immune response, and thus prevent important COVID-19-related complications, including viral septicemia. For example, it is hoped that in COVID-19-infected patients, anti-PD-1 monoclonal antibodies, which are already being used in cancer patients to re-activate the immune system against cancer cells may restore the cytotoxic activity of T-cells, particularly CD8+ cytotoxicity against the SARS-CoV-2 pathogen. It may be possible in the future to extend the study of ICIs by also blocking PD-L1 expressed by infected cells; however, additional translational studies will be required.
The vast number of clinical trials on COVID-19-infected patients that have begun at the onset of 2020 will undoubtedly uncover a huge quantity of novel results, hopefully leading to the identification of valid therapies and vaccines that may help humanity to overcome this testing and troublesome time in history.
Acknowledgments
The authors would like to acknowledge the Italian League Against Cancer (LILT) for its support. The authors would like to specially thank Dr Golnar Kolahgar, University of Cambridge, for her support with the English language revision and editing of this manuscript.
Funding
No funding was received.
Availability of data and materials
The original contributions presented in the study are publicly available. These data can be found at: www.pubmed.com; www.clinicaltrials.gov.
Authors' contributions
ML conceptualized and supervised the drafting of the manuscript and provided critical revisions. SV assisted in the conceptualization of the manuscript and wrote the first draft of the manuscript, including the drawing of the 2 figures and the preparation of the both tables. LF, FT, GS, RB, DAS and GRu contributed to the writing of the contents presented in Chapters 1, 2 and 5 and both tables of the manuscript. GRa and FP contributed to the writing of the contents presented in Chapters 3 and 4 of the manuscript. All authors contributed to manuscript revision, and read, and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article. The other authors declare that the review was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Chan JFW, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) Coronavirus disease (COVID-19): Situation reports. https://www.who.int/emer-gencies/diseases/novel-coronavirus-2019/situation-reports.
- 4.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Ma X. Acute respiratory failure in COVID-19: Is it 'typical' ARDS? Crit Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renu K, Prasanna PL, Valsala Gopalakrishnan A. Coronaviruses pathogenesis, comorbidities and multi-organ damage - A review. Life Sci. 2020;255:117839. doi: 10.1016/j.lfs.2020.117839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Costa VG, Moreli ML, Saivish MV. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arab-Zozani M, Hassanipour S. Features and limitations of LitCovid hub for quick access to literature about COVID-19. Balkan Med J. 2020;37:231–232. doi: 10.4274/balkanmedj.galenos.2020.2020.4.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J. New coronavirus threat galvanizes scientists. Science. 2020;367:492–493. doi: 10.1126/science.367.6477.492. [DOI] [PubMed] [Google Scholar]
- 13.Lokman SM, Rasheduzzaman M, Salauddin A, Barua R, Tanzina AY, Rumi MH, Hossain MI, Siddiki AMAMZ, Mannan A, Hasan MM. Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: A computational biology approach. Infect Genet Evol. 2020;84:104389. doi: 10.1016/j.meegid.2020.104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, et al. HCA Lung Biological Network Electronic address: lung-network@humancellatlas.org HCA Lung Biological Network: SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Kleine-Weber H, Pöhlmann S. A Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaux CA, Rolain J-M, Raoult D. ACE2 receptor poly-morphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L, Sakagami H, Miwa N. ACE2: The key molecule for understanding the pathophysiology of severe and critical conditions of COVID-19: Demon or Angel? Viruses. 2020;12:491. doi: 10.3390/v12050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A Structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye M, Druce J, Tran T, Kostecki R, Chibo D, Morris J, Catton M, Birch C. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Jiang L, Li X, Lin F, Wang Y, Li B, Jiang T, An W, Liu S, Liu H, et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5:e138070. doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah A. Novel coronavirus-induced NLRP3 Inflammasome activation: a potential drug target in the treatment of COVID-19. Front Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the 'Cytokine Storm' in COVID-19. J Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, Li B, Song X, Zhou X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong EZ, Chan YFZ, Leong WY, Lee NMY, Kalimuddin S, Haja Mohideen SM, Chan KS, Tan AT, Bertoletti A, Ooi EE, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–882.e2. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27:863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thachil J, Srivastava A. SARS-2 coronavirus-associated hemostatic lung abnormality in COVID-19: is it pulmonary thrombosis or pulmonary embolism? Semin Thromb Hemost. 2020;46:777–780. doi: 10.1055/s-0040-1712155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapisarda V, Loreto C, Ledda C, Musumeci G, Bracci M, Santarelli L, Renis M, Ferrante M, Cardile V. Cytotoxicity, oxidative stress and genotoxicity induced by glass fibers on human alveolar epithelial cell line A549. Toxicol In Vitro. 2015;29:551–557. doi: 10.1016/j.tiv.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong SM, Wang C, Tigdi J, Si X, Dumpit C, Charles S, Gamage A, Moraes TJ, Lee WL. Influenza infects lung microvascular endothelium leading to microvascular leak: Role of apoptosis and claudin-5. PLoS One. 2012;7:e47323. doi: 10.1371/journal.pone.0047323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohmura T, Tian Y, Sarich N, Ke Y, Meliton A, Shah AS, Andreasson K, Birukov KG, Birukova AA. Regulation of lung endothelial permeability and inflammatory responses by prostaglandin A2: Role of EP4 receptor. Mol Biol Cell. 2017;28:1622–1635. doi: 10.1091/mbc.e16-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schurink B, Roos E, Radonic T, Barbe E, Bouman CSC, de Boer HH, de Bree GJ, Bulle EB, Aronica EM, Florquin S, et al. Viral presence and immunopathology in patients with lethal COVID-19: A prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmadpoor P, Rostaing L. Why the immune system fails to mount an adaptive immune response to a COVID-19 infection. Transpl Int. 2020;33:824–825. doi: 10.1111/tri.13611. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Wang L, Kong X, Geng J, Xiao D, Ma C, Jiang XM, Wang PH. Long-term coexistence of SARS-CoV-2 with antibody response in COVID-19 patients. J Med Virol. 2020;92:1684–1689. doi: 10.1002/jmv.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Zhou X, Zhu C, Song Y, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B, Wang J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fathi N, Rezaei N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol Int. 2020;44:1792–1797. doi: 10.1002/cbin.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Liu Y, Wang G, et al. Reduction and functional exhaustion of T Cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Biasi S Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, Gozzi L, Iannone A, Lo Tartaro D, Mattioli M. et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with Covid-19 pneumonia. Nat Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaqinuddin A, Kashir J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med Hypotheses. 2020;140:109777. doi: 10.1016/j.mehy.2020.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvelli J, Demaria O, Vely F, Batista L, Benmansour NC, Fares J, Carpentier S, Thibult ML, Morel A, André P, et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cully M. Immune status could determine efficacy of COVID-19 therapies. Nat Rev Drug Discov. 2020;19:431–434. doi: 10.1038/d41573-020-00110-3. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Liu S, Liu J, Zhang Z, Wan X, Huang B, Chen Y, Zhang Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortaz E, Tabarsi P, Varahram M, Folkerts G, Adcock IM. The Immune Response and Immunopathology of COVID-19. Front Immunol. 2020;11:2037. doi: 10.3389/fimmu.2020.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuen KS, Ye ZW, Fung SY, Chan CP, Jin DY. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carsana L, Sonzogni A, Nasr A, Rossi RS, Pellegrinelli A, Zerbi P, Rech R, Colombo R, Antinori S, Corbellino M, et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infect Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lodigiani C, Iapichino G, Carenzo L, Cecconi M, Ferrazzi P, Sebastian T, Kucher N, Studt JD, Sacco C, Bertuzzi A, et al. Humanitas COVID-19 Task Force: Venous and arterial thromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb Res. 2020;191:9–14. doi: 10.1016/j.thromres.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 54.Zaim S, Chong JH, Sankaranarayanan V, Harky A. COVID-19 and multiorgan response. Curr Probl Cardiol. 2020;45:100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi Y, Lagniton PNP, Ye S, Li E, Xu RH. COVID-19: What has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, Su X, Cao B. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet. 2020;395:1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lescure F-X, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati F, Le Hingrat Q, et al. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Endeman H, van der Zee P, van Genderen ME, van den Akker JPC, Gommers D. Progressive respiratory failure in COVID-19: A hypothesis. Lancet Infect Dis. 2020;20:1365. doi: 10.1016/S1473-3099(20)30366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de la Rica R, Borges M, Gonzalez-Freire M. COVID-19: In the Eye of the Cytokine Storm. Front Immunol. 2020;11:558898. doi: 10.3389/fimmu.2020.558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, Muñoz-Ruiz M, McKenzie DR, Hayday TS, Francos-Quijorna I, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 64.Onofrio L, Caraglia M, Facchini G, Margherita V, Placido S, Buonerba C. Toll-like receptors and COVID-19: a two-faced story with an exciting ending. Future Sci OA. 2020;6:FSO605. doi: 10.2144/fsoa-2020-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Florindo HF, Kleiner R, Vaskovich-Koubi D, Acúrcio RC, Carreira B, Yeini E, Tiram G, Liubomirski Y, Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skalny AV, Rink L, Ajsuvakova OP, Aschner M, Gritsenko VA, Alekseenko SI, Svistunov AA, Petrakis D, Spandidos DA, Aaseth J, et al. Zinc and respiratory tract infections: Perspectives for COVID-19 (Review) Int J Mol Med. 2020;46:17–26. doi: 10.3892/ijmm.2020.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.World Health Organization (WHO) Standard precautions in health care. https://www.who.int/publications/i/item/standard-precautions-in-health-care. Accessed September 30, 2007.
- 68.Nitulescu GM, Paunescu H, Moschos SA, Petrakis D, Nitulescu G, Ion GND, Spandidos DA, Nikolouzakis TK, Drakoulis N, Tsatsakis A. Comprehensive analysis of drugs to treat SARS-CoV-2 infection: Mechanistic insights into current COVID-19 therapies (Review) Int J Mol Med. 2020;46:467–488. doi: 10.3892/ijmm.2020.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dehelean CA, Lazureanu V, Coricovac D, Mioc M, Oancea R, Marcovici I, Pinzaru I, Soica C, Tsatsakis AM, Cretu O. SARS-CoV-2: Repurposed drugs and novel therapeutic approaches-insights into chemical structure-biological activity and toxicological screening. J Clin Med. 2020;9:2084. doi: 10.3390/jcm9072084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, et al. ACTT-1 Study Group Members: Remdesivir for the treatment of Covid-19 - Final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.FDA FDA Remdesivir update, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-drug-combination-treatment-covid-19. Accessed December 1 2020.
- 72.World Health Organization (WHO) WHO recommends against the use of remdesivir in COVID-19 patients. https://www.who.int/news-room/feature-stories/detail/who-recommends-against-the-use-of-remdesivir-in-covid-19-patients. Accessed November 20 2020.
- 73.European Medicines Agency (EMA) Update on remdesivir -EMA will evaluate new data from Solidarity trial. https://www.ema.europa.eu/en/news/update-remdesivir-ema-will-evaluate-new-data-solidarity-trial. Accessed November 20 2020.
- 74.World Health Organization (WHO) Therapeutics and COVID-19: living guideline. https://www.who.int/publications/i/item/therapeutics-and-covid-19-living-guideline. Accessed November 20 2020. [PubMed]
- 75.Nicola M, O'Neill N, Sohrabi C, Khan M, Agha M, Agha R. Evidence based management guideline for the COVID-19 pandemic - Review article. Int J Surg. 2020;77:206–216. doi: 10.1016/j.ijsu.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kakodkar P, Kaka N, Baig MN. A comprehensive literature review on the clinical presentation, and management of the pandemic coronavirus disease 2019 (COVID-19) Cureus. 2020;12:e7560. doi: 10.7759/cureus.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bimonte S, Crispo A, Amore A, Celentano E, Cuomo A, Cascella M. Potential antiviral drugs for SARS-Cov-2 treatment: Preclinical findings and ongoing clinical research. In Vivo (Brooklyn) 2020;34:1597–1602. doi: 10.21873/invivo.11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.World Health Organization (WHO) Clinical management of COVID-19. https://www.who.int/publications/i/item/clinical-management-of-covid-19. Accessed May 20 2020. [PubMed]
- 79.Falzone L, Musso N, Gattuso G, Bongiorno D, Palermo CI, Scalia G, Libra M, Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int J Mol Med. 2020;46:957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suo T, Liu X, Feng J, Guo M, Hu W, Guo D, Ullah H, Yang Y, Zhang Q, Wang X, et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.European Medicines Agency (EMA) Treatments and vaccines for COVID-19. https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19. Last updated November 26 2020.
- 82.Kim YC, Dema B, Reyes-Sandoval A. COVID-19 vaccines: Breaking record times to first-in-human trials. NPJ Vaccines. 2020;5:34. doi: 10.1038/s41541-020-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calina D, Docea AO, Petra k is D, Egorov A M, Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F, Vinceti M, et al. Towards effective COVID-19 vaccines: Updates, perspectives and challenges (Review) Int J Mol Med. 2020;46:3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.World Health Organization (WHO) Draft landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Accessed December 8 2020.
- 85.Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu R, Wang L, Kuo HD, Shannar A, Peter R, Chou PJ, Li S, Hudlikar R, Liu X, Liu Z, et al. An Update on Current Therapeutic Drugs Treating COVID-19. Curr Pharmacol Rep. 2020;6:1–15. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Magro G. COVID-19: Review on latest available drugs and therapies against SARS-CoV-2. Coagulation and inflammation cross-talking. Virus Res. 2020;286:198070. doi: 10.1016/j.virusres.2020.198070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID-19 treatment. Rev Panam Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lythgoe MP, Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar A, Gupta PK, Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. Diabetes Metab Syndr. 2020;14:569–573. doi: 10.1016/j.dsx.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rossman H, Keshet A, Shilo S, Gavrieli A, Bauman T, Cohen O, Shelly E, Balicer R, Geiger B, Dor Y, et al. A framework for identifying regional outbreak and spread of COVID-19 from one-minute population-wide surveys. Nat Med. 2020;26:634–638. doi: 10.1038/s41591-020-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.European Bioinformatics Institute (EBI) European Molecular Biology Laboratory (EMBL): EMBL-EBI launches COVID-19 Data Portal. https://www.ebi.ac.uk/about/news/press-releases/embl-ebi-launches-covid-19-data-portal. Accessed April 18 2020.
- 93.National Institutes of Health (NIH) National Library Of Medicine, National Center for Biotechnology Information (NCBI):SARS-CoV-2 Data. https://www.ncbi.nlm.nih.gov/sars-cov-2/
- 94.White House Office, US Department of Energy, IBM The COVID-19 high performance computing consortium. https://covid19-hpc-consortium.org/
- 95.Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quimque MTJ, Notarte KIR, Fernandez RAT, Mendoza MAO, Liman RAD, Lim JAK, Pilapil LAE, Ong JKH, Pastrana AM, Khan A, et al. Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J Biomol Struct Dyn. 2020 Jun 16; doi: 10.1080/07391102.2020.1776639. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J Chem Inf Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, Blecher-Gonen R, Cohen M, Medaglia C, Li H, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Veljkovic V, Perovic V, Paessler S. Prediction of the effectiveness of COVID-19 vaccine candidates. F1000 Res. 2020;9:365. doi: 10.12688/f1000research.23865.1. [DOI] [Google Scholar]
- 101.Russo G, Pennisi M, Viceconti M, Pappalardo F. In silico trial to test COVID-19 candidate vaccines: a case study with UISS platform. doi: 10.1186/s12859-020-03872-0. arXiv:2005.02289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kar T, Narsaria U, Basak S, Deb D, Castiglione F, Mueller DM, Srivastava AP. A candidate multi-epitope vaccine against SARS-CoV-2. Sci Rep. 2020;10:10895. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petrakis D, Margină D, Tsarouhas K, Tekos F, Stan M, Nikitovic D, Kouretas D, Spandidos DA, Tsatsakis A. Obesity - a risk factor for increased COVID-19 prevalence, severity and lethality (Review) Mol Med Rep. 2020;22:9–19. doi: 10.3892/mmr.2020.11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goumenou M, Sarigiannis D, Tsatsakis A, Anesti O, Docea A, Petrakis D, Tsoukalas D, Kostoff R, Rakitskii V, Spandidos DA, et al. COVID-19 in Northern Italy: An integrative overview of factors possibly influencing the sharp increase of the outbreak (Review) Mol Med Rep. 2020;22:20–32. doi: 10.3892/mmr.2020.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ledda C, Loreto C, Zammit C, Marconi A, Fago L, Matera S, Costanzo V, Fuccio Sanzà G, Palmucci S, Ferrante M, et al. Non-infective occupational risk factors for hepatocellular carcinoma: A review (Review) Mol Med Rep. 2017;15:511–533. doi: 10.3892/mmr.2016.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Falzone L, Marconi A, Loreto C, Franco S, Spandidos DA, Libra M. Occupational exposure to carcinogens: Benzene, pesticides and fibers (Review) Mol Med Rep. 2016;14:4467–4474. doi: 10.3892/mmr.2016.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wargo JA, Reuben A, Cooper ZA, Oh KS, Sullivan RJ. Immune effects of chemotherapy, radiation, and targeted therapy and opportunities for combination with immunotherapy. Semin Oncol. 2015;42:601–616. doi: 10.1053/j.seminoncol.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan J, Yang C. Prevention and control strategies for the diagnosis and treatment of cancer patients during the COVID-19 pandemic. Br J Cancer. 2020;123:5–6. doi: 10.1038/s41416-020-0854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garassino MC, Whisenant JG, Huang L-C, Trama A, Torri V, Agustoni F, Baena J, Banna G, Berardi R, Bettini AC, et al. TERAVOLT investigators: COVID-19 in patients with thoracic malignancies (TERAVOLT): First results of an international, registry-based, cohort study. Lancet Oncol. 2020;21:914–922. doi: 10.1016/S1470-2045(20)30314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H, Zhang L. Risk of COVID-19 for patients with cancer. Lancet Oncol. 2020;21:e181. doi: 10.1016/S1470-2045(20)30149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liang W, Guan W, Chen R, Wang W, Li J, Xu K, Li C, Ai Q, Lu W, Liang H, et al. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuderer NM, Choueiri TK, Shah DP, Shyr Y, Rubinstein SM, Rivera DR, Shete S, Hsu CY, Desai A, de Lima Lopes G, Jr, et al. COVID-19 and Cancer Consortium: Clinical impact of COVID-19 on patients with cancer (CCC19): A cohort study. Lancet. 2020;395:1907–1918. doi: 10.1016/S0140-6736(20)31187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee LY, Cazier J-B, Angelis V, Arnold R, Bisht V, Campton NA, Chackathayil J, Cheng VW, Curley HM, Fittall MW, et al. UK Coronavirus Monitoring Project Team: COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: A prospective cohort study. Lancet. 2020;395:1919–1926. doi: 10.1016/S0140-6736(20)31173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee LYW, Cazier J-B, Starkey T, Briggs SEW, Arnold R, Bisht V, Booth S, Campton NA, Cheng VWT, Collins G, et al. UK Coronavirus Cancer Monitoring Project Team: COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demo-graphics: A prospective cohort study. Lancet Oncol. 2020;21:1309–1316. doi: 10.1016/S1470-2045(20)30442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, et al. Patients with cancer appear more vulnerable to SARS-COV-2: A multi-center study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robilotti EV, Babady NE, Mead PA, Rolling T, Perez-Johnston R, Bernardes M, Bogler Y, Caldararo M, Figueroa CJ, Glickman MS, et al. Determinants of COVID-19 disease severity in patients with cancer. Nat Med. 2020;26:1218–1223. doi: 10.1038/s41591-020-0979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Horn L, Whisenant JG, Torri V, Huang L-C, Trama A, Paz-Ares LG, Felip E, Pancaldi V, De Toma A, Tiseo M, et al. Thoracic Cancers International COVID-19 Collaboration (TERAVOLT): Impact of type of cancer therapy and COVID therapy on survival. J Clin Oncol. 2020;38(Suppl 18):LBA111. doi: 10.1200/JCO.2020.38.18_suppl.LBA111. [DOI] [Google Scholar]
- 119.Russell B, Moss C, Papa S, Irshad S, Ross P, Spicer J, Kordasti S, Crawley D, Wylie H, Cahill F, et al. Factors affecting COVID-19 outcomes in cancer patients: a first report from guy's cancer center in London. Front Oncol. 2020;10:1279. doi: 10.3389/fonc.2020.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Banna G, Curioni-Fontecedro A, Friedlaender A, Addeo A. How we treat patients with lung cancer during the SARS-CoV-2 pandemic: Primum non nocere. ESMO Open. 2020;5(Suppl 2):e000765. doi: 10.1136/esmoopen-2020-000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Derosa L, Melenotte C, Griscelli F, Gachot B, Marabelle A, Kroemer G, Zitvogel L. The immuno-oncological challenge of COVID-19. Nat Cancer. 2020;1:946–964. doi: 10.1038/s43018-020-00122-3. [DOI] [PubMed] [Google Scholar]
- 122.Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp Mol Med. 2018;50:1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.La-Beck NM, Nguyen DT, Le AD, Alzghari SK, Trinh ST. Optimizing Patient Outcomes with PD-1/PD-L1 Immune Checkpoint Inhibitors for the First-Line Treatment of Advanced Non-Small Cell Lung Cancer. Pharmacotherapy. 2020;40:239–255. doi: 10.1002/phar.2364. [DOI] [PubMed] [Google Scholar]
- 124.Leonardi GC, Candido S, Falzone L, Spandidos DA, Libra M. Cutaneous melanoma and the immunotherapy revolution (Review) Int J Oncol. 2020;57:609–618. doi: 10.3892/ijo.2020.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Falzone L, Salomone S, Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front Pharmacol. 2018;9:1300. doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Christofi T, Baritaki S, Falzone L, Libra M, Zaravinos A. Current perspectives in cancer immunotherapy. Cancers (Basel) 2019;11:1472. doi: 10.3390/cancers11101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.May JE, Donaldson C, Gynn L, Morse HR. Chemotherapy-induced genotoxic damage to bone marrow cells: Long-term implications. Mutagenesis. 2018;33:241–251. doi: 10.1093/mutage/gey014. [DOI] [PubMed] [Google Scholar]
- 128.Shah NJ, Al-Shbool G, Blackburn M, Cook M, Belouali A, Liu SV, Madhavan S, He AR, Atkins MB, Gibney GT, et al. Safety and efficacy of immune checkpoint inhibitors (ICIs) in cancer patients with HIV, hepatitis B, or hepatitis C viral infection. J Immunother Cancer. 2019;7:353. doi: 10.1186/s40425-019-0771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Das S, Johnson DB. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. J Immunother Cancer. 2019;7:306. doi: 10.1186/s40425-019-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gonzalez-Cao M, Antonazas-Basa M, Puertolas T, Munoz-Consuelo E, Manzano JL, Carrera C, Marquez-Rodas I, Lopez-Criado P, Rodriguez-Moreno JF, Garcia-Castano A, et al. Cancer immunotherapy does not increase the risk of death by COVID-19 in melanoma patients. MedRxiv. doi: 10.1101/2020.05.19.20106971. [DOI]
- 131.Hotchkiss RS, Colston E, Yende S, Crouser ED, Martin GS, Albertson T, Bartz RR, Brakenridge SC, Delano MJ, Park PK, et al. Immune checkpoint inhibition in sepsis: A Phase 1b randomized study to evaluate the safety, tolerability, pharmacokinetics, and pharmacodynamics of nivolumab. Intensive Care Med. 2019;45:1360–1371. doi: 10.1007/s00134-019-05704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hotchkiss RS, Colston E, Yende S, Angus DC, Moldawer LL, Crouser ED, Martin GS, Coopersmith CM, Brakenridge S, Mayr FB, et al. Immune checkpoint inhibition in sepsis: a phase 1b randomized, placebo-controlled, single ascending dose study of antiprogrammed cell death-ligand 1 antibody (BMS-936559) Crit Care Med. 2019;47:632–642. doi: 10.1097/CCM.0000000000003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chong CR, Park VJ, Cohen B, Postow MA, Wolchok JD, Kamboj M. Safety of inactivated influenza vaccine in cancer patients receiving immune checkpoint inhibitors. Clin Infect Dis. 2020;70:193–199. doi: 10.1093/cid/ciz202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bayle A, Khettab M, Lucibello F, Chamseddine AN, Goldschmidt V, Perret A, Ropert S, Scotté F, Loulergue P, Mir O. Immunogenicity and safety of influenza vaccination in cancer patients receiving checkpoint inhibitors targeting PD-1 or PD-L1. Ann Oncol. 2020;31:959–961. doi: 10.1016/j.annonc.2020.03.290. [DOI] [PubMed] [Google Scholar]
- 135.Gatto L, Franceschi E, Nunno VD, Brandes AA. Potential protective and therapeutic role of immune checkpoint inhibitors against viral infections and COVID-19. Immunotherapy. 2020;12:1111–1114. doi: 10.2217/imt-2020-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vivarelli S, Falzone L, Grillo CM, Scandurra G, Torino F, Libra M. Cancer management during COVID-19 pandemic: is immune checkpoint inhibitors-based immunotherapy harmful or beneficial? Cancers (Basel) 2020;12:2237. doi: 10.3390/cancers12082237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zheng H-Y, Zhang M, Yang C-X, Zhang N, Wang XC, Yang XP, Dong XQ, Zheng YT. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 141.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 142.Magro G. SARS-CoV-2 and COVID-19: Is interleukin-6 (IL-6) the 'culprit lesion' of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine X. 2020;2:100029. doi: 10.1016/j.cytox.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Di Cosimo S, Malfettone A, Pérez-García J M, Llombart-Cussac A, Miceli R, Curigliano G, Cortés J. Immune checkpoint inhibitors: A physiology-driven approach to the treatment of coronavirus disease 2019. Eur J Cancer. 2020;135:62–65. doi: 10.1016/j.ejca.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Riva G, Nasillo V, Tagliafico E, Trenti T, Luppi M. COVID-19: Room for treating T cell exhaustion? Crit Care. 2020;24:229. doi: 10.1186/s13054-020-02960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buonaguro FM, Puzanov I, Ascierto PA. Anti-IL6R role in treatment of COVID-19-related ARDS. COVID-19: Room for treating T cell exhaustion? J Transl Med. 2020;18:165. doi: 10.1186/s12967-020-02333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Arnaldez FI, O'Day SJ, Drake CG, Fox BA, Fu B, Urba WJ, Montesarchio V, Weber JS, Wei H, Wigginton JM, et al. The Society for Immunotherapy of Cancer perspective on regulation of interleukin-6 signaling in COVID-19-related systemic inflammatory response. J Immunother Cancer. 2020;8:e000930. doi: 10.1136/jitc-2020-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maio M, Hamid O, Larkin J, Covre A, Altomonte M, Calabrò L, Vardhana SA, Robert C, Ibrahim R, Anichini A, et al. Immune checkpoint inhibitors for cancer therapy in the COVID-19 era. Clin Cancer Res. 2020;26:4201–4205. doi: 10.1158/1078-0432.CCR-20-1657. [DOI] [PubMed] [Google Scholar]
- 148.Salvestrini V, Sell C, Lorenzini A. Obesity may accelerate the aging process. Front Endocrinol (Lausanne) 2019;10:266. doi: 10.3389/fendo.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found at: www.pubmed.com; www.clinicaltrials.gov.