Abstract
Melanoma is an aggressive, deadly skin cancer derived from melanocytes, a neural crest cell derivative. Melanoma cells mirror the developmental program of neural crest cells in that they exhibit the same gene expression patterns and utilize similar cellular mechanisms, including increased cell proliferation, epithelial-mesenchymal transition, and migration. Here we studied the role of neural crest regulator PRDM1 in melanoma onset and progression. In development, Prdm1a functions to promote neural crest progenitor fate, and in melanoma, we found that PRDM1 has reduced copy number and is recurrently deleted in both zebrafish and humans. When examining expression of neural crest and melanocyte development genes, we show that sox10 progenitor expression is high in prdm1a−/− mutants, while more differentiated melanocyte markers are reduced, suggesting that normally Prdm1a is required for differentiation. Data mining of human melanoma datasets indicates that high PRDM1 expression in human melanoma is correlated with better patient survival and decreased PRDM1 expression is common in metastatic tumors. When one copy of prdm1a is lost in the zebrafish melanoma model Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/−, melanoma onset occurs more quickly, and the tumors that form have a larger area with increased expression of sox10. These data demonstrate a novel role for PRDM1 as a tumor suppressor in melanoma.
Keywords: melanoma, neural crest cells, PRDM1, zebrafish
1 |. INTRODUCTION
Melanoma is an aggressive cancer that accounts for the majority of human skin cancer deaths. The 5-year survival rate of metastatic melanoma is only 23%. Early-stage melanoma can be cured by surgical excision; however, only a few treatment options exist once the melanoma has metastasized. More than 50% of melanoma patients present with a somatic BRAF mutation,2 and up to 90% of these are from a single amino acid change from valine (V) to glutamic acid (E) at position 600 (V600E).3 As such, the high frequency of BRAFV600E mutations among melanoma patients has prompted the development of targeted therapies such as BRAF and MEK inhibitors that dampen BRAF/MEK pathway signaling.4 Dabrafenib and trametinib were the first combined inhibitors to be approved by the Food and Drug Administration for patients with advanced BRAFV600E melanoma.5,6 The combined inhibitors improve patient survival rates and have fewer adverse side effects compared to monotherapy treatments. About one-third of melanoma patients experience disease recurrence or metastasis years after initial treatment.7,8 The acquirement of additional somatic mutations (ie, point mutations, chromosomal rearrangements, copy-number variations, etc) that bypass the targets of BRAF and MEK inhibitors may account for some cases of incomplete response to the primary line of targeted therapy. However, other mechanisms of resistance or melanoma progression exist that have yet to be studied. For example, it has been previously shown that gene amplification of SETDB1, in conjunction with a BRAFV600E mutation, accelerates melanoma tumor formation in a zebrafish model by causing disruptions in transcriptional regulation, including a decrease in expression of HOX genes.9 Identifying additional genes and pathways involved in melanoma initiation is critical, as it can lead to the development of future therapeutic and treatment options.
The cell origins of melanoma tumors are thought to arise from transformed melanocytes,10 which are derived from neural crest cells, a multipotent population of cells that forms between the neural and nonneural ectoderm. During embryonic development, neural crest cells undergo an epithelial-to-mesenchymal transition and migrate to different regions of the developing embryo before giving rise to a variety of cell types, including peripheral neurons and glia, chondrocytes, osteoblasts, and melanocytes. As neural crest cells are a highly migratory population of cells, it has been hypothesized that the same genes and pathways utilized by neural crest cells during development to aid in the trafficking of cells to their final destinations in the embryo can be later hijacked by cancer cells to enhance their proliferation and migration.11,12 Indeed, several publications have shown that there are distinct parallels in behaviors and gene expression patterns between melanoma cells and neural crest cells.13 For instance, Brn3a, a neural crest transcription factor in the neuronal lineage, is frequently expressed in melanoma tumors and has been shown to accelerate tumor growth by promoting survival.14 Interestingly, mouse melanoma cells can arise from a population of seemingly differentiated melanocytes that respond to environmental cues to undergo transcriptional reprogramming.15 One cue is likely a neural crest growth factor, as melanoma cells express CD271 receptors on the cell surface.16
Apart from these studies that have implicated single neural crest-related genes in melanoma progression, there is strong evidence that a neural crest transcriptional program plays a causative role in melanoma progression. Genes such as crestin, sox10, ednrb, tyr, and dct, which are characteristic markers of neural crest progenitors and melanocytes, are highly enriched in zebrafish and mouse melanoma tumors.17,18 In zebrafish, crestin is normally downregulated at later stages in development after neural crest cells have fully differentiated.19 Using a crestin:EGFP reporter in a zebrafish model, Kaufman et al20 used live-cell imaging of melanoma tumor formation and showed that a single melanocyte can initiate tumor growth by reactivating crestin. Moreover, crestin:EGFP-positive zebrafish scales, but not adjacent crestin-negative scales, exhibit gene signatures that were enriched with neural crest and melanoma genes, such as mitfa and sox10.20 This work suggests that reactivation of specific neural crest cell genes coincides with tumor initiation. It is evident that inappropriate activation of developmental genes can lead to cancer; however, it is not well-known whether expression of neural crest cell genes can also be protective.
Here, we identify neural crest regulator PRDM1 as a novel tumor suppressor in melanoma. PRDM1 (zebrafish ortholog Prdm1a) is a highly conserved transcription factor, whose structure consists of an N-terminal PR/SET domain, followed by a proline-rich domain, and five DNA-binding C2H2 zinc fingers. PRDM1 regulates transcription both by direct DNA binding21 and by forming complexes with chromatin-modifying proteins including histone methyltransferases Prmt522 and G9a23 and histone deacetylases HDAC1/2.24 PRDM1 is a critical factor in neural crest cell development. It is highly expressed at the neural plate border in zebrafish,25,26Xenopus laevis,27 and lamprey28 and is required for Rohon-Beard sensory neuron and neural crest cell formation.25,26 Homozygous loss of prdm1a in developing zebrafish embryos results in a significant reduction of neural crest cell derivatives, including pigment cells and craniofacial cartilage,25 suggesting that prdm1a is required for neural crest differentiation. Similarly, PRDM1 has also been shown to be required for the differentiation of both adaxial cells into slow-twitch muscle cells,29 as well as retinal progenitors to photoreceptor cells.30,31 In addition to being important for cell fate specification during development, PRDM1 and other members of the PRDM family function as tumor suppressors for many cancers, including B, T, and natural killer cell lymphomas, colorectal cancer, and lung cancer.32–35 In lymphoid malignancies, genomic deletions, missense mutations, and premature truncations of PRDM1 are frequently observed,34,36,37 causing inactivation of the protein and misregulation of lymphocyte differentiation and cell cycle progression. These studies provide strong evidence and support a role for PRDM1 as a tumor suppressor. In this study, we show that Prdm1a is required for melanocyte differentiation from neural crest cells during development and that decreased PRDM1 expression is correlated with worse melanoma patient survival. We also report that loss of PRDM1 in a zebrafish model leads to an acceleration of melanoma onset and progression.
2 |. MATERIALS AND METHODS
2.1 |. Zebrafish
Zebrafish were maintained as previously described.38 The Institutional Animal Care and Use Committee at the University of Colorado Denver Anschutz Medical Campus (UCD-AMC) reviewed and approved all animal procedures. Wildtype strains include AB and EKK lines. Additional lines include prdm1am805 (nrd),39 referred to as prdm1a−/−; tp53M214/M214, referred to as p53−/−; Tg(mitfa:BRAFV600E);p53−/−17,40; and Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/−.
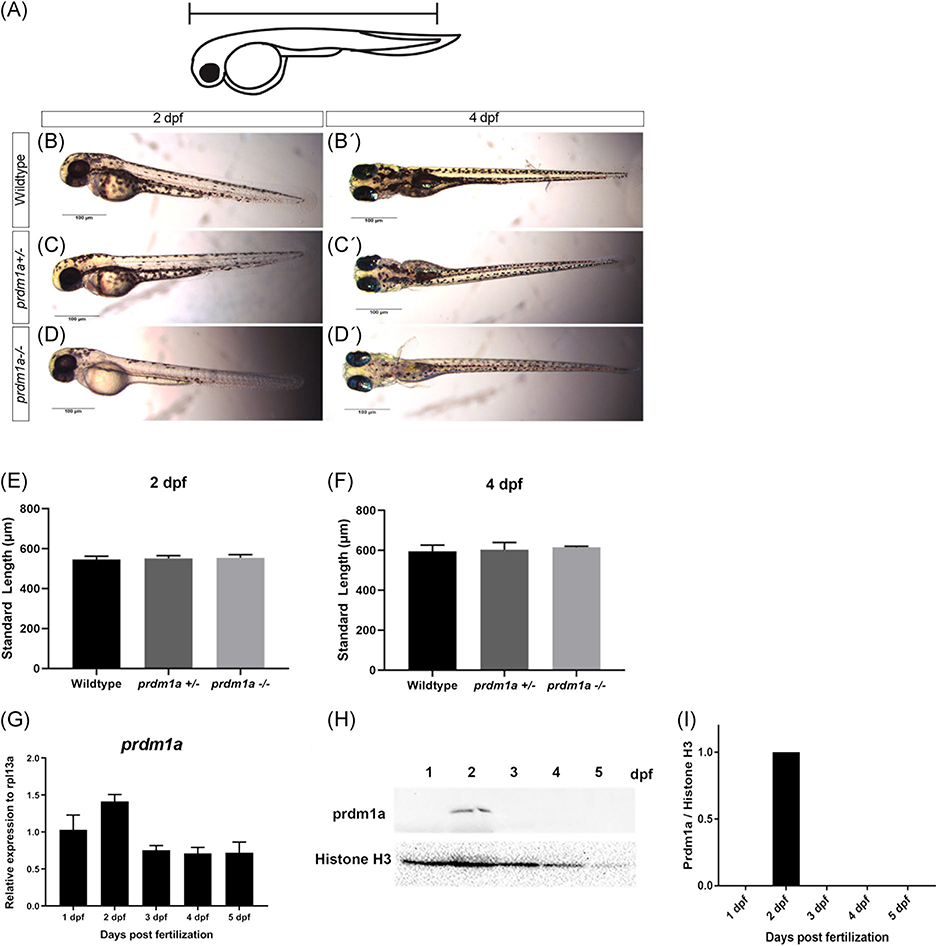
The standard length of the zebrafish was measured from the tip of the snout to the base of the tail (Figure 2A).
FIGURE 2.
prdm1aexpression during early development. A, Schematic showing how the standard length of the embryo was measured. Lateral images of wildtype (B), prdm1a+/− (C), and prdm1a−/− (D) at 2 dpf. Ventral images of wildtype (B’), prdm1a+/− (C’) and prdm1a−/− (D’) at 4 dpf. Quantification of standard length at 2 (E) and 4 (F) dpf. Statistical analysis was performed with a one-way ANOVA. G, RT-qPCR was performed on 20 pooled embryos at each time point (n = 3 replicate experiments) for prdm1a. H, Proteins were resolved by SDS-PAGE and Western blotting for prdm1a and total H3 was performed. I, Quantification of band intensity from the Western blot (H). Relative intensity was normalized to total histone H3 for each sample. ANOVA, analysis of variance; dpf, days postfertilization; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Microinjections were performed in p53−/− and prdm1a+/− zebrafish at the single-cell stage. The mitfa:BRAFV600E vector was a gift from Richard M. White.17 The vector was linearized with SalI and BssHI, purified, and resuspended in 100mM KCl for a final concentration of 100 ng/mL. A total of 500 pL were injected into each embryo.
Zebrafish were observed and scored weekly for hyperpigmentation, which indicated the presence of melanoma tumors. We compared tumor-free curves over time using a log-rank test where P < .05 was statistically significant. The tumor area was measured using ImageJ and normalized to body length.
2.2 |. aCGH, JISTIC, and comparative analysis
Array comparative genomic hybridization (aCGH) was performed and analyzed as previously described.41 Briefly, we custom designed an array with 398 426 unique probes covering 97% of the zebrafish genome (based on Zv9 assembly). Genomic DNA was extracted from melanoma tumors in Tg(mitfa:BRAFV600E);p53−/−17,40 zebrafish and from a normal region of the same fish using the Qiagen DNeasy Blood and Tissue kit (#69504; Qiagen, Germantown, MD). The DNA was fragmented and hybridized to the array. Raw data was generated from scanned images with the Agilent Feature Extraction software (v10.7) and normalized using the Agilent Genomic workbench to identify copy-number differences (Agilent, Santa Clara, CA). The JISTIC algorithm was used to calculate significantly altered regions, and peak calling was done using a q-value cutoff of 0.25. For human melanomas, copy-number data were downloaded from Tumorscape42,43 and JISTIC analysis was conducted as described previously.41 Genes from within peaks were pooled to define species-specific sets of recurrently deleted genes. Human orthologs of zebrafish genes were determined using Ensembl and BLAST.
2.3 |. Survival analysis of The Cancer Genome Atlas cutaneous melanoma data
Clinical and mutational data of 329 The Cancer Genome Atlas (TCGA) melanoma patients were obtained from the Supporting Information Tables of the TCGA publication (http://cancergenome.nih.gov/). Normalized RNA-seq gene expression profiles (level 3, RSEM value) for these patients were downloaded from the cBioPortal website (http://www.cbioportal.org). Patients were assigned to high (hi) and low (lo) groups based on above or below mean gene expression of PRDM1. In this study, we only focus on the 286 metastatic melanoma patients with outcome data. Kaplan-Meier survival analysis was performed comparing hi and lo groups. A log-rank test of P < .05 was considered statistically significant.
2.4 |. Mining public gene expression datasets
Gene expression profiles of the Riker dataset (GSE7553)44 were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds). This dataset contains 87 melanoma patients and was profiled by the Affymetrix Human Genome U133 Plus 2.0 microarray. Raw data were downloaded and normalized with the Robust Multiarray Average method using the Affymetrix Power Tools. From this dataset, there were 4 normal skin, 14 primary melanoma, and 40 metastatic melanoma patients. Gene expression for PRDM1 was compared across these patients, and a one-way analysis of variance (ANOVA) test was performed to determine the different expressions. Error bars represent mean ± standard deviation (SD).
2.5 |. Histology
Zebrafish samples were fixed in 4% paraformaldehyde and embedded in paraffin. The samples were then sectioned at a 5 μM thickness and fixed to glass slides. Hematoxylin and eosin stain (H&E) staining was performed by the Gates Center Regenerative Medicine Histology Core at UCD-AMC. Histology images were captured on an Olympus BX51 microscope equipped with a four megapixel Macro-fire digital camera (Optronics, Goleta, CA) using the Picture Frame Application 2.3 (Optronics).
2.6 |. RNA extraction and real-time PCR
Total RNA was isolated from dissected zebrafish tumors and zebrafish embryos (20 pooled embryos). The zebrafish samples were stored in TRIzol reagent (#15596018; Invitrogen Corp, Carlsbad, CA) and then purified with the Direct-zol RNA kit (Zymo Research, Irvine, CA). RNA (1 μg) was converted to complementary DNA (cDNA) using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). All procedures were conducted according to the manufacturers’ suggestions.
Quantitative reverse transcription polymerase chain reactions (RT-qPCR) were performed in the Step One Plus Real-time PCR System (Thermo Fisher Scientific). Taqman primers for PRDM1, prdm1a, sox10, dct, mitfa, mc1r, and tyr were purchased from Thermo Fisher Scientific. rpl13a Was used as the zebrafish internal control. (rpl13a: probe 5′-/56-FAM/ATGAAGTTGGCTGGAAGTACCA/3lABlk_FQ/−3′, forward 5′-CACACGCAAATTTGCCCTGCTTGG-3′, and reverse 5′-GCCTGCTTAGTCAGCTTCATC-3′). Reactions were performed in three biological and technical replicates. Transcript abundance and relative fold change were quantified using the 2−ΔΔCt method relative to control. Relative abundance was compared using an unpaired, independent t test. Error bars represent the mean ± SD. Results were statistically significant when P < .05.
2.7 |. Western blotting
Western blotting was performed as previously described.45 A total of 50 to 100 zebrafish embryos were pooled at each time point. Calcium-free Ginzberg Fish Ringer’s solution (NaCl, KCl, NaHCO3) was added to the embryos for deyolking. Samples were then washed in deyolking wash buffer (NaCl, Tris [pH 8.5], KCl, and CaCl2) and lysed in lysis buffer using a syringe (0.1% glycerol, 0.01% sodium dodecyl sulfate, 0.1M Tris-HCL [pH6.8]). Total protein concentration was determined with the Bio-Rad DC Protein assay (Bio-Rad, Hercules, CA). Proteins (20 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) and transferred to polyvinylidene difluoride membranes. Primary antibodies against PRDM1 (1:750 dilution; gift from Dr Philip W. Ingham)46 and total histone H3 (1:5000 dilution; #9715; Cell Signaling Technology, Danvers, MA) were used. The blots were incubated with secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5000 dilution; #A0545; Sigma-Aldrich, St Louis, MO) for 2 hours. Chemiluminescent detection was performed with Luminata Classico Western HRP substrate (Millipore Sigma, Burlington, MA). Bio-Rad Chemidoc multiplex imaging system was used to detect and quantify the protein levels. Band intensity was quantified in ImageJ and normalized to the total histone H3 loading control. The experiment was performed at least twice.
2.8 |. Tyrosinase assay
The tyrosinase assay was performed as previously described with a few modifications.47,48 About 100 embryos at 2 days postfertilization (dpf) were dechorionated and sonicated in Pro-Prep protein extraction solution (#17081; iNtRON Biotechnology, Burlington, MA). A total of 200 μg of total protein in 75 μL of lysis buffer was added to 75 μL of 1.0mM of L-3,4-dihydroxyphenylalanine (L-DOPA) in a 96-well plate. The control well contained 75 μL of lysis buffer and 75 μL of 1.0 mM L-DOPA. After incubation at 37°C for 60minutes, absorbance was read at 425 nm. The assay was performed twice with three technical replicates in each trial. Error bars represent the mean ± SD. The data were analyzed using a one-way ANOVA test followed by a multiple comparisons test where P < .05 was statistically significant.
3 |. RESULTS
3.1 |. PRDM1 is recurrently deleted in human and zebrafish melanoma and is required for melanocyte differentiation
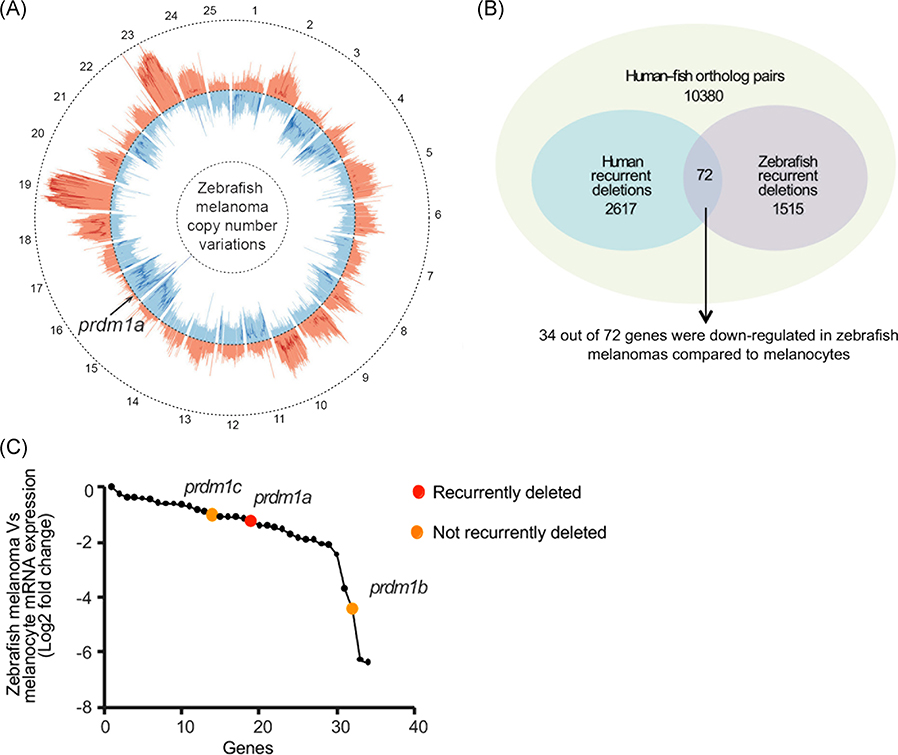
Somatic gene mutations, including gene copy-number variations, are frequently observed in cancer, including melanoma.9,43,49,50 We used a cross-species oncogenomics approach to identify recurrently deleted genes in both human and zebrafish melanomas. As previously described, we performed aCGH on Tg(mitfa:BRAFV600E);p53−/− zebrafish to generate copy-number variant profiles for zebrafish that develop melanomas (see below)17,40,41 (Figure 1A). Human data were obtained from Tumorscape, a dataset that contains copy-number analyses for over 3000 human cancer specimens.42,43 The aCGH values and human data were analyzed using the JISTIC algorithm,51 which then listed recurrently amplified and deleted genes. We identified 2617 genes recurrently deleted in human melanomas and 1515 genes in zebrafish melanomas. Of these recurrently deleted genes, 72 were deleted in both human and zebrafish melanomas (Figure 1B), and furthermore 34 of these genes were also identified as transcriptionally downregulated in a microarray dataset comparing expression in zebrafish melanoma tissue and normal melanocytes.41 Among the 34 downregulated genes in zebrafish melanoma was the family of prdm1 paralogs, prdm1a/b/c (Figure 1C). prdm1a was selected as a candidate gene because of its known role as a neural crest regulator.25 Paralogs prdm1b and prdm1c were also downregulated, but they were not recurrently deleted based on the JISTIC analysis (Figure 1C). Therefore, we did not explore these paralogs further.
FIGURE 1.
PRDM1 is recurrently deleted in human and zebrafish melanoma. A, Circos plot displaying gene copy-number gains and losses in zebrafish melanomas as previously described.41 JISTIC G-scores are displayed as pale red shading (amplifications [minimum = 0; maximum = 1550]) and blue shading (deletions [minimum = 0; maximum = 250]). −log10-transformed JISTIC Q-values with a cutoff of 0.6 (corresponding to an untransformed Q-value of 0.25) are shown as bold red lines (amplifications [minimum = 0; maximum = 11]) and bold blue (deletions [minimum = 0; maximum = 11]). Dotted circles represent the –log10-transformed Q-value of 0 (center) and 11 (outer: amplification; inner: deletion). B, Venn diagram of orthologous genes significantly deleted in human and zebrafish melanomas from 10 380 human-zebrafish gene pairs (hypergeometric test, P value: 2.0E-16). C, Genes downregulated in zebrafish melanomas as compared with melanocytes (analyzed from previously obtained microarray dataset41). prdm1a (red dot) and its paralogs prdm1b and prdm1c (orange dots) are indicated
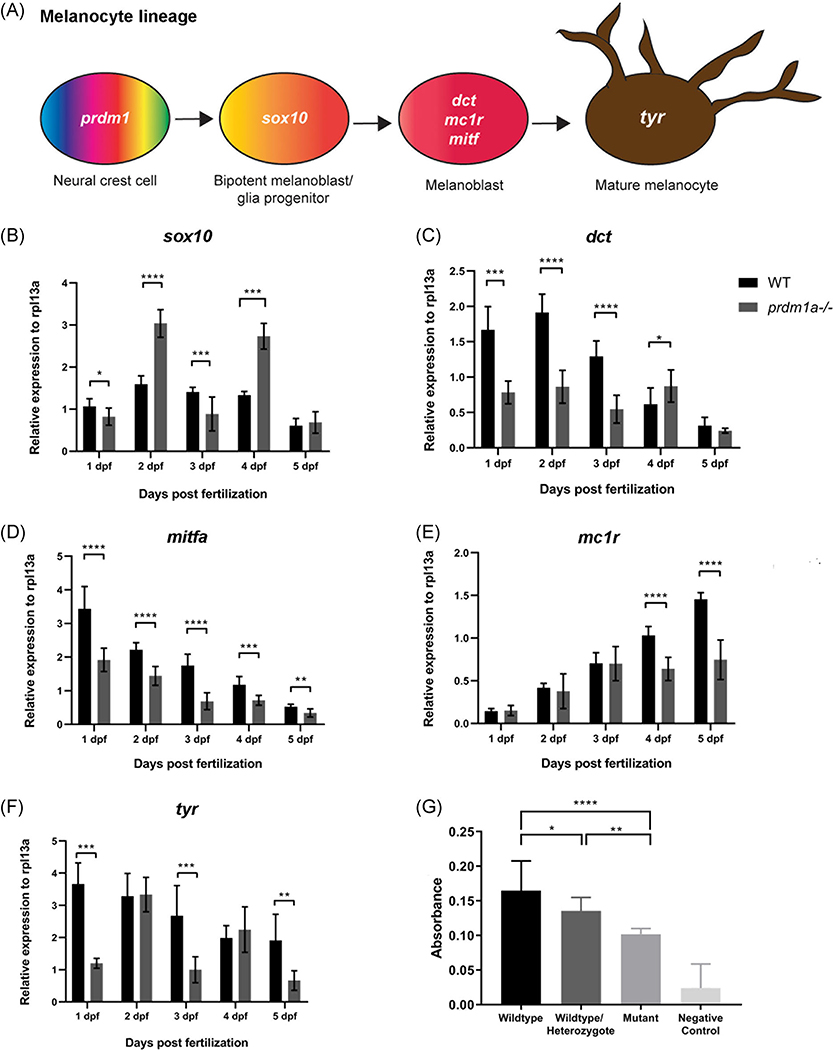
PRDM1 is a known tumor suppressor in multiple cancers, including B and T-cell lymphoma,36,52 colorectal cancer,53 lung cancer,54 and glioma.55 Moreover, Prdm1a has been shown to play a role in neural crest differentiation in zebrafish.25,39 Homozygous loss of prdm1a results in reduced neural crest cell derivatives, including peripheral neurons and mature pigment cells (Figure 2A–D).39 prdm1a−/− mutants die at 6 to 7 dpf, and wildtype and heterozygotes are indistinguishable by phenotype (Figure 2B–C’). The standard length of the zebrafish from the tip of the snout to the base of the tail is not affected when one or both copies of prdm1a are mutated (P > .05) (Figure 2E,F). prdm1a transcripts (Figure 2G) and protein levels (Figure 2H,I) are highest a 2 dpf, when pigment cells are differentiating along body of the zebrafish.56 To better understand the role of Prdm1a in melanocyte development, we performed RT-qPCR on whole wildtype and prdm1a−/− mutant embryos for a series of genes that mark early (sox10) and late melanocyte lineage (dct, mc1r, mitfa, tyr)10 during a time course of development from 1 to 5 dpf (schematic Figure 3A). When neural crest cells commit to the melanocyte lineage, they express different markers that indicate their stage of differentiation (Figure 3A). sox10 Is an early melanocyte stem cell marker and is required for melanocyte differentiation from neural crest cells in that it activates expression of mitf, dct, and tyr.10,57 Interestingly, in wildtype embryos, sox10 expression is steady over time, but prdm1a−/− mutants exhibit a significant increase in expression at 2 and 4 dpf, corresponding to the first and second waves of melanocyte migration during zebrafish development58 (P < .0001) (Figure 3B). Expression of dct, mitfa, and tyr, markers of the melanoblast precursor state, decrease steadily over developmental time, while mitfa increases in wildtype embryos (Figure 3C–F). In contrast, the prdm1a−/− mutants consistently exhibit an overall decrease in the later melanocyte markers compared to wildtype embryos (P < .05) (Figure 3C–F). The increase in sox10, along with the overall decrease in later melanocyte markers, suggests that the cells could be maintained as early melanocyte progenitors, unable to fully differentiate into mature, pigment-producing melanocytes.
FIGURE 3.
prdm1a is required for melanocyte differentiation and pigment production. A, Melanocyte lineage from neural crest to mature melanocytes and their characteristic markers. Whole wildtype embryos were pooled for RNA isolation and protein extraction over developmental time. B-F, Whole wildtype and prdm1a−/− mutant embryos were pooled for RNA isolation over developmental time. Twenty whole embryos were pooled for each genotype and at each time point (n = 3 replicate experiments). RT-qPCR was performed for sox10 (B), dct (C), mitfa (D), mc1r (E), and tyr (F) over developmental time. Expression was compared using an unpaired, independent t test. J, 100 Embryos were pooled and incubated with L-DOPA for a tyrosinase functional assay. Pigment production was measured by spectrometry in whole wildtype, wildtype/heterozygote mix, and prdm1a−/− mutant embryos. Statistical analysis was performed with a one-way ANOVA followed by a multiple comparisons test. Error bars represent mean ± SD. *P < .05, **P < .01, ***P < .001, ****P < .0001. ANOVA, analysis of variance; dpf, days postfertilization; L-DOPA, L-3,4-dihydroxyphenylalanine: SD, standard deviation
To further test whether loss of prdm1a affected proper melanogenesis, we tested for tyrosinase activity, the enzyme responsible for pigment production, in prdm1a−/− mutant, wildtype (EKK strain), and wildtype/heterozygote mixed embryos at 2 dpf. Pooled embryos were incubated with the tyrosinase substrate L-DOPA, and spectrophotometry was used to quantify pigment production. Compared to both the wildtype and wildtype/heterozygote mix, prdm1a−/− mutants had significantly decreased pigment production (P < .01) and this loss of tyrosinase activity aligns with the levels of prdm1a, as lysates from a combination of wildtype and prdm1a+/− embryos exhibited an intermediate loss of tyrosine activity that was significantly different from that of both the wildtype and prdm1a−/− lysates (Figure 3G). Together with our RT-qPCR results demonstrating that prdm1a mutants are deficient in the expression of late melanocyte markers, these data suggest that melanocyte progenitors require Prdm1a to terminally differentiate and produce pigment. Moreover, this suggests that even a partial loss of prdm1a affects melanogenesis, which is important to note given that our adult melanoma models are prdm1a+/− heterozygotes. Together, the data indicate that Prdm1a is important for melanocyte development from neural crest cells. Given that there are key similarities between melanoma and neural crest cells and that PRDM1 is already a known tumor suppressor, we determined whether it played a role in melanoma as well.
3.2 |. Low PRDM1 expression is correlated with worse patient outcomes and metastatic melanoma in publicly available datasets
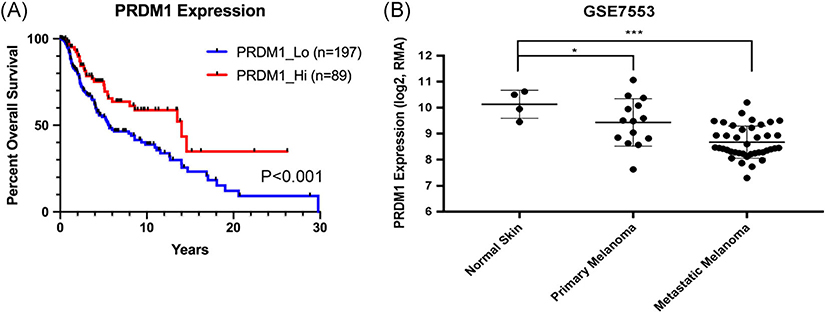
To determine whether PRDM1 expression was associated with human melanoma patient prognosis, we analyzed TCGA cutaneous melanoma dataset and plotted the overall survival of patients with metastatic melanoma based on their expression of PRDM1. Lower PRDM1 expression is correlated with a worse overall survival rate (P < .001) (Figure 4A). We also mined the Riker melanoma dataset (GSE7553 Moffitt data) and observed a significant decrease in PRDM1 expression in metastatic melanoma tumors compared to normal skin (P < .001) and primary melanoma tumors (P < .05) (Figure 4B)
FIGURE 4.
Low PRDM1 expression in humans is correlated with worse patient outcomes and metastatic melanoma. A, Overall survival of patients with metastatic melanoma compared to PRDM1 expression obtained from The Cancer Genome Atlas (TCGA). Statistical analysis was performed with a log-rank test. B, PRDM1 expression in normal skin, primary melanoma tumors, and metastatic melanoma tumors obtained from the Riker melanoma dataset. Statistical analysis was performed using a one-way ANOVA. Error bars represent mean ± SD. *P < .05, **P < .01, ***P < .001. ANOVA, analysis of variance; SD, standard deviation
3.3 |. Heterozygous loss of prdm1a in zebrafish increases melanoma onset and progression
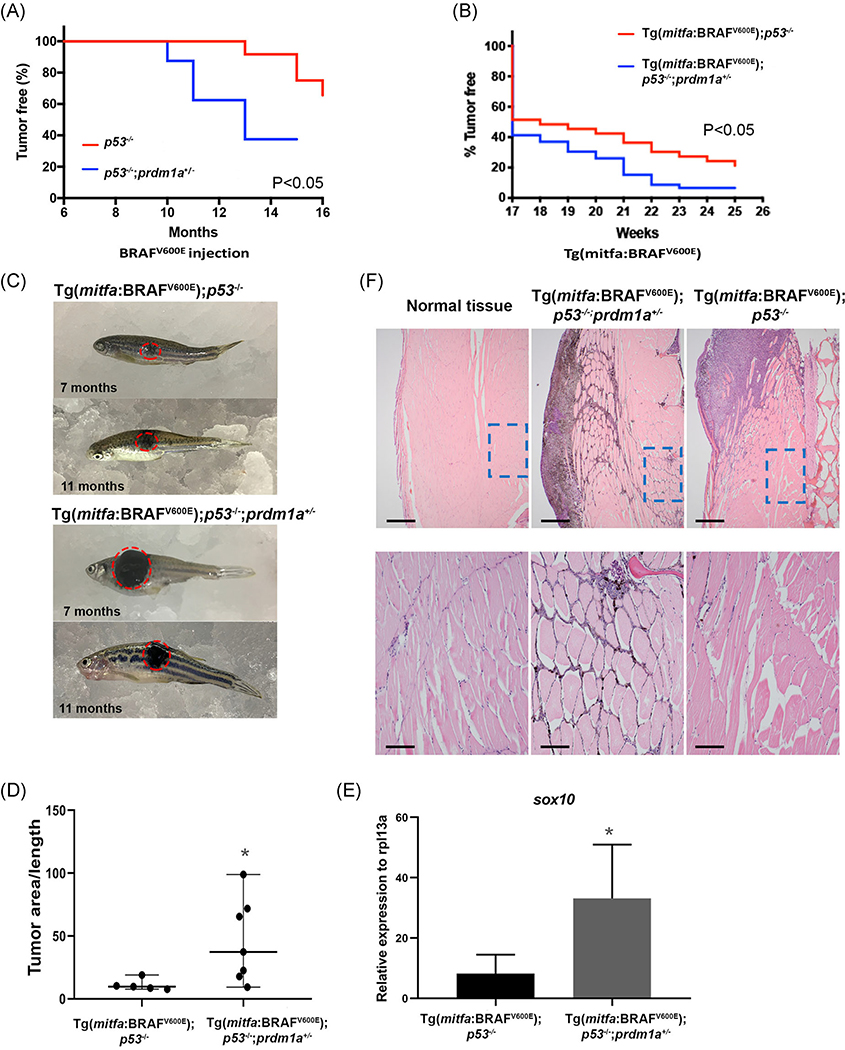
To determine whether loss of PRDM1 promotes melanoma in vivo, we used two zebrafish models, an injected and transgenic mitfa:BRAFV600E line. In both models, the BRAF mutation is driven by a mitfa promoter to drive melanoma in melanocytes specifically, with an additional p53−/− mutant background that further accelerates melanoma onset.17,40 For the injected model, the oncogenic mitfa:BRAFV600E construct was introduced into p53−/− and p53−/−; prdm1a+/− zebrafish embryos at the single-cell stage.17,40 Again, homozygous prdm1a−/− mutants die at 6 to 7 dpf. The construct is integrated randomly into the genome and is expressed transiently. We monitored the zebrafish and noted when hyperpigmentation occurred in concentrated areas of the body. This hyperpigmentation was indicative of the initiation of melanoma tumor formation. Due to transient expression of the mitfa:BRAFV600E mutation in the injected model, tumor formation was slower than previously reported.17,20,40 The p53−/− zebrafish formed tumors 12 months postinjection (n = 12), while the p53−/−;prdm1a+/− zebrafish developed tumors significantly more quickly at 10 months postinjection (n = 8) (P < .05) (Figure 4A). Unfortunately, due to the p53−/− mutant background, the zebrafish often developed malignant peripheral nerve sheath tumors (MPNST)59 at about 10 months postinjection before melanoma formation. Therefore, these fish were excluded from our melanoma study.
To circumvent this issue, we established two stable transgenic lines, Tg(mitfa:BRAFV600E)p53−/− and Tg(mitfa:BRAFV600E)p53−/−;prdm1a+/−, and monitored tumor formation. As expected, these tumors formed more quickly than in the injected model. By 17 weeks, about 50% of the zebrafish for both genotypes had formed tumors (Figure 5B). With one copy of prdm1a mutated, the Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish (n = 33) quickly surpassed their wildtype Tg(mitfa:BRAFV600E); p53−/− siblings (n = 18) and developed melanoma tumors (P < .05) (Figure 5B). By week 19, more than 60% of the Tg(mitfa:BRAFV600E); p53−/−;prdm1a+/− zebrafish had visible tumors. In contrast, approximately 50% of the wildtype siblings were still tumor-free, suggesting that loss of prdm1a accelerates tumor onset. Representative images of melanoma tumors are shown at 7 and 11 months for both genotypes (Figure 5C), and tumor area was measured and normalized to the body length (Figure 5D). The tumor area of Tg(mitfa:BRAFV600E); p53−/−;prdm1a+/− (n = 7) was significantly larger than wildtype Tg(mitfa:BRAFV600E);p53−/− tumors (n = 5) (P < .05) (Figure 5C,D). However, this may be because tumor onset occurred earlier in Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish (Figure 5B). Similar to the injected model, several transgenic fish developed MPNST tumors due to the p53−/− mutant background.59 Therefore, the melanoma tumors on these fish were not large enough to measure and were not included in the quantification.
FIGURE 5.
Loss of prdm1a in zebrafish accelerates tumor onset. A, Tumor-free curve from p53−/− (n = 12) and p53−/−;prdm1a+/− (n = 8) zebrafish injected with BRAFV600E construct to induce melanoma. B, Tumor-free curve from Tg(mitfa:BRAFV600E);p53−/− (n = 18) and Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− (n = 33) zebrafish. Statistical analysis for tumor-free curves was performed with a log-rank test. C, Representative images of melanoma tumors from Tg(mitfa:BRAFV600E);p53−/− and Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish at 7 and 11 months. D, Tumor area was measured in five Tg(mitfa:BRAFV600E);p53−/− tumors and seven Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish tumors. The area was then divided by the body length to normalize. Relative tumor area was compared using an unpaired, Mann-Whitney U test. Error bars represent median and range. E, RNA was isolated from dissected tumors and RT-qPCR was performed for sox10. Expression was compared using an unpaired, Mann-Whitney U test. Error bars represent mean ± SD. *P < .05. F, H&E staining of size-matched tumors for Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− (7 months) and Tg(mitfa:BRAFV600E);p53−/− (9 months) zebrafish compared to normal skin. Upper panels show ×40 magnification. The lower panels are zoomed into ×200 magnification. H&E, hematoxylin and eosin stain; SD, standard deviation
As previously described, Sox10 is a conserved marker of neural crest cells and potentially co-opted in melanoma progenitor cells in many vertebrate models,20,60 thus we compared its expression between dissected Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− and Tg(mitfa:BRAFV600E);p53−/− tumors. For each tumor, sox10 expression was compared to normal, adjacent muscle tissue of the same individual. sox10 expression varied but was elevated for both genotypes. On average, tumor samples from Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish had a threefold increase in sox10 compared to their Tg(mitfa:BRAFV600E);p53−/− siblings (P < .01) (Figure 5E). This suggests that reduction of prdm1a expression causes an upregulation of sox10 in both neural crest development and melanoma cells.21,61 In melanoma, we predict that the gain in sox10 expression supports the ability of melanoma to grow and spread.
To examine the zebrafish tumors in more detail, we paired zebrafish with similarly sized tumors and then sectioned the samples for H&E staining. In both tumors, we observed invasion into the neighboring muscle tissue with variable levels of melanin across the tumors (n = 3 for each genotype; Figure 5F). The melanoma in Tg(mitfa:BRAFV600E);p53−/−;prdm1a+/− zebrafish have an overall larger tumor area (Figure 5D), with melanin observed in the neighboring tissue (Figure 5F) whereas the Tg(mitfa:BRAFV600E);p53−/− tumor remains somewhat more localized at the skin surface with less invasion into the surrounding skeletal muscle.
4 |. DISCUSSION
Melanoma cells often acquire features of neural crest cells during tumor formation by reactivating genes and pathways utilized by neural crest cells.12,13 The work presented in this study suggest that Prdm1a, a known neural crest regulator, is required for melanocyte differentiation from neural crest cells and that loss of Prdm1a accelerates melanoma onset and progression. During development, we have shown that prdm1a−/− mutants have a reduction in neural crest derivatives, including mature pigment cells.25,26 Here, we demonstrate that loss of prdm1a leads to an increase in expression of sox10, an early melanocyte stem cell marker, and an overall decrease in later melanocyte markers. prdm1a−/− mutants also have significantly reduced pigment production by tyrosinase. Together, these data suggest that Prdm1a is required for the differentiation of neural crest cells to mature melanocytes. It is likely that in the absence of Prdm1a, the cells are maintained as early melanocyte progenitors and thus function in supporting melanoma formation and progression. The case for PRDM1 functioning as a tumor suppressor in melanoma is further supported by the fact that PRDM1/prdm1a copy number is significantly reduced in both human and zebrafish melanomas, and its expression is reduced in melanoma cells compared to normal melanocytes. Moreover, in human patients, low PRDM1 expression is correlated with worse human patient survival. Finally, we demonstrate that heterozygous loss of prdm1a promotes melanoma tumorigenesis in a zebrafish model and results in an increase in sox10 expression in the tumors. Thus, our data demonstrate a novel role for the neural crest regulator PRDM1 as a melanoma tumor suppressor.
Melanoma is known to have increased somatic mutation rates compared to other cancers due to mutagenesis by UV exposure,49,50 with BRAFV600E being the most common mutation.3 Other mutations, such as NRAS, are common in large congenital nevi, which are often melanogenic and can be precursors to melanoma.62 Interestingly when NRASQ61K is driven by a tyrosinase promoter, transgenic mice develop congenital naevi and melanoma and express the neural crest progenitor marker Sox10,18 again iterating that Sox10 expression is required for malignant melanoma formation. Reducing one copy of Sox10 in this background reduces the number of congenital naevi and melanoma tumors by regulating cell proliferation.18 This suggests that Sox10’s function in development, to maintain neural crest stem cells, may be dysregulated during disease progression.
In our previous work and this current body of work, we have demonstrated that the developmental regulator, Prdm1a, is required for neural crest differentiation to mature melanocytes, which is consistent with its function in other cell types during development.29–31 We show that prdm1a may promote differentiation through regulation of sox10,61 as suggested by the increase in its expression in prdm1a−/− mutants. However, because the prdm1a−/− mutants have visibly less pigmentation compared to wildtype embryos, this may have affected our interpretation of the melanocyte marker gene expression data. Moreover, just from their reduced pigmentation and reduced expression of late melanocyte markers, we cannot conclusively claim that prdm1a−/− larvae in fact have less melanoblasts or melanocytes, as prdm1a loss may disrupt neural crest cells’ ability to differentiate into these cell types or impair the proliferation of these neural crest derivatives. In future studies, we can address these issues by performing live-imaging experiments and/or lineage tracing experiments to parse out the exact effects of prdm1a loss on neural crest derivatives. However, it is interesting that reducing prdm1a in heterozygous embryos also reduces tyrosinase activity, even when there is not a discernable difference in pigmentation. Therefore, our results suggest that Prdm1a is an important regulator of neural crest cell differentiation into melanocytes. Our data contribute to the growing literature regarding the parallels between neural crest cell development and melanoma tumor formation.13
Interestingly, PRDM1 is known to act as a tumor suppressor in other cancers, particularly B, T, and natural killer cell lymphomas and lung cancer.34,36,37,54 We have now shown in a zebrafish model that heterozygous loss of prdm1a can accelerate melanoma onset, and it is likely acting as a tumor suppressor. We hypothesize that loss of PRDM1 coordinates with mutations in BRAF and/or TP53 to promote tumorigenesis. PRDM1 deletions have been reported to be associated with loss of TP53 in anaplastic large cell lymphoma patients,37 and this association likely contributed to the pathogenicity of the cancer. Our zebrafish models carried mutations in both BRAF and TP53. Additional mutations in PRDM1 led to accelerated melanoma tumor onset and suggested a trend toward metastasis (data not shown), similar to what has been shown with PRDM1 and lung cancer.54 Whether PRDM1 coordinates with these or additional tumor suppressor/oncogenic genes during tumorigenesis must be explored further.
In conclusion, this study demonstrates a novel role for PRDM1 as a tumor suppressor in p53 mutated melanoma. We showed that even partial loss of prdm1a in zebrafish accelerates melanoma onset, and this can potentially be sourced to the impairment of melanocyte differentiation from neural crest cells during embryonic development observed upon prdm1a loss. Broadly, these results compound on accumulating evidence that neural crest cells and melanomas operate on overlapping gene regulatory networks and suggest that factors that regulate neural crest development are a source of therapeutic value for melanomas.
ACKNOWLEDGMENTS
We thank the Artinger and Shellman labs for experimental feedback; Morgan Singleton and UCD-AMC CLAC for zebrafish care; Virginia Ware and Aaron Clark for technical help with experiments; and Richard White for sharing microarray data, fish lines, and advice. This work is supported by pilot grants from the Skin Disease Research Center at the University of Colorado Anschutz Medical Campus P30AR057212-10, The State of Colorado, Tobacco Settlement funds pilot grants, and P30CA046934 to the University of Colorado Cancer Center.
Funding information
Skin Disease Research Center, Grant/Award Number: P30AR057212; University of Colorado Cancer Center, Grant/Award Number: P30CA046934
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Cancer Facts & Figures. Atlanta, GA: American Cancer Society; 2019. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/cancer-facts-and-figures-2019.pdf [Google Scholar]
- 2.Ascierto PA, Kirkwood JM, Grob J-J, et al. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012;10(85):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradish JR, Cheng L. Molecular pathology of malignant melanoma: changing the clinical practice paradigm toward a personalized approach. Hum Pathol. 2014;45(7):1315–1326. [DOI] [PubMed] [Google Scholar]
- 4.Mackiewicz J, Mackiewicz A. BRAF and MEK inhibitors in the era of immunotherapy in melanoma patients. Contemp Oncol (Pozn). 2018; 22(1A):68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372(1):30–39. [DOI] [PubMed] [Google Scholar]
- 6.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tas F, Erturk K. Relapse patterns in patients with local and regional cutaneous melanoma. Clin Transl Oncol. 2019;21(4):412–419. [DOI] [PubMed] [Google Scholar]
- 8.Soong S, Harrison R, McCarthy W, Urist M, Balch C. Factors affecting survival following local, regional, or distant recurrence from localized melanoma. J Surg Oncol. 1998;67(4):228–233. [DOI] [PubMed] [Google Scholar]
- 9.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White RM, Zon LI. Melanocytes in development, regeneration, and cancer. Cell Stem Cell. 2008;3(3):242–252. [DOI] [PubMed] [Google Scholar]
- 11.Powell DR, Blasky AJ, Britt SG, Artinger KB. Riding the crest of the wave: parallels between the neural crest and cancer in epithelial-to-mesenchymal transition and migration. Wiley Interdiscip Rev: Syst Biol Med. 2013;5(4):511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maguire LH, Thomas AR, Goldstein AM. Tumors of the neural crest: common themes in development and cancer. Dev Dyn. 2015;244: 311–322. [DOI] [PubMed] [Google Scholar]
- 13.Shakhova O Neural crest stem cells in melanoma development. Curr Opin Oncol. 2014;26(2):215–221. [DOI] [PubMed] [Google Scholar]
- 14.Hohenauer T, Berking C, Schmidt A, et al. The neural crest transcription factor Brn3a is expressed in melanoma and required for cell cycle progression and survival. EMBO Mol Med. 2013;5(6):919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler C, Nittner D, Rambow F, et al. Mouse cutaneous melanoma induced by mutant BRaf arises from expansion and dedifferentiation of mature pigmented melanocytes. Cell Stem Cell. 2017;21(5): 679–693 e676. [DOI] [PubMed] [Google Scholar]
- 16.Boiko AD, Razorenova OV, van de Rijn M, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466(7302):133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RM, Cech J, Ratanasirintrawoot S, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shakhova O, Zingg D, Schaefer SM, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol. 2012;14(8):882–890. [DOI] [PubMed] [Google Scholar]
- 19.Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev Dyn. 2001;220(2):169–174. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman CK, Mosimann C, Fan ZP, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351(6272):aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell DR, Hernandez-Lagunas L, LaMonica K, Artinger KB. Prdm1a directly activates foxd3 and tfap2a during zebrafish neural crest specification. Development. 2013;140(16):3445–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ancelin K, Lange UC, Hajkova P, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8(6):623–630. [DOI] [PubMed] [Google Scholar]
- 23.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5(3):299–308. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20(7):2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Lagunas L, Choi IF, Kaji T, et al. Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev Biol. 2005;278(2):347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–1777. [DOI] [PubMed] [Google Scholar]
- 27.Rossi CC, Hernandez-Lagunas L, Zhang C, et al. Rohon-Beard sensory neurons are induced by BMP4 expressing non-neural ectoderm in Xenopus laevis. Dev Biol. 2008;314(2):351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikitina N, Tong L, Bronner ME. Ancestral network module regulating prdm1 expression in the lamprey neural plate border. Dev Dyn. 2011; 240(10):2265–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy S, Wolff C, Ingham PW. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes & Development. 2001;15:1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brzezinski JA, Lamba DA, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010;137(4):619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci. 2010;30(19):6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mzoughi S, Tan YX, Low D, Guccione E. The role of PRDMs in cancer: one family, two sides. Curr Opin Genet Dev. 2016;36:83–91. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438(7066):374–378. [DOI] [PubMed] [Google Scholar]
- 34.Küçük C, Iqbal J, Hu X, et al. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci USA. 2011;108(50): 20119–20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boi M, Zucca E, Inghirami G, Bertoni F. PRDM1/BLIMP1: a tumor suppressor gene in B and T cell lymphomas. Leuk Lymphoma. 2015; 56(5):1223–1228. [DOI] [PubMed] [Google Scholar]
- 36.Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18(6):568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boi M, Rinaldi A, Kwee I, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122(15): 2683–2693. [DOI] [PubMed] [Google Scholar]
- 38.Westerfield M The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio). 4th EditionEugene, OR: University of Oregon Press; 2000. https://zfin.org/zf_info/zfbook/zfbk.html [Google Scholar]
- 39.Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126(18):3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patton EE, Widlund HR, Kutok JL, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15(3):249–254. [DOI] [PubMed] [Google Scholar]
- 41.Venkatesan AM, Vyas R, Gramann AK, et al. Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma. J Clin Invest. 2018;128(1):294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008; 68(3):664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283): 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riker AI, Enkemann SA, Fodstad O, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1(13):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shull L, Sen R, Menzel J, Artinger KB. The conserved and divergent role of Prdm3 and Prdm16 in zebrafish and mouse craniofacial development. Dev Biol. 2020;461(2):132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008; 9(7):683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi TY, Kim JH, Ko DH, et al. Zebrafish as a new model for phenotype-based screening of melanogenic regulatory compounds. Pigment Cell Res. 2007;20(2):120–127. [DOI] [PubMed] [Google Scholar]
- 48.Cha SH, Ko SC, Kim D, Jeon YJ. Screening of marine algae for potential tyrosinase inhibitor: those inhibitors reduced tyrosinase activity and melanin synthesis in zebrafish. J Dermatol. 2011;38(4): 354–363. [DOI] [PubMed] [Google Scholar]
- 49.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Garcia F, Akavia UD, Mozes E, Pe’er D. JISTIC: identification of significant targets in cancer. BMC Bioinformatics. 2010; 11(189):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calado DP, Zhang B, Srinivasan L, et al. Constitutive canonical NFkappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18(6):580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Banister CE, Weige CC, et al. PRDM1 silences stem cell-related genes and inhibits proliferation of human colon tumor organoids. Proc Natl Acad Sci USA. 2018;115(22):E5066–E5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, Wang H, Wei Y, Meng F, Liu Z, Zhang Z. Downregulation of PRDM1 promotes cellular invasion and lung cancer metastasis. Tumour Biol. 2017;39(4):1010428317695929. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Wang K, Han L, et al. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/beta-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013; 331(2):211–219. [DOI] [PubMed] [Google Scholar]
- 56.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203: 253–310. [DOI] [PubMed] [Google Scholar]
- 57.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010; 23(4):496–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milos N, Dingle AD. Dynamics of pigment pattern formation in the zebrafish, Brachydanio rerio. I. Establishment and regulation of the lateral line melanophore stripe during the first eight days of development. J Exp Zool. 1978;205(2):205–216. [Google Scholar]
- 59.Berghmans S, Murphey RD, Wienholds E, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102(2):407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohamed A, Gonzalez RS, Lawson D, Wang J, Cohen C. SOX10 Expression in malignant melanoma, carcinoma, and normal tissues. Appl Immunohistochem Mol Morphol. 2013;21(6):506–510. [DOI] [PubMed] [Google Scholar]
- 61.Olesnicky E, Hernandez-Lagunas L, Artinger KB. prdm1a Regulates sox10 and islet1 in the development of neural crest and Rohon-Beard sensory neurons. Genesis. 2010;48(11):656–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127(1):179–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.