Directionality switching of the mitotic kinesin-5 Cin8 motor is caused by an asymmetric response of its active movement to drag.
Abstract
Directed active motion of motor proteins is a vital process in virtually all eukaryotic cells. Nearly a decade ago, the discovery of directionality switching of mitotic kinesin-5 motors challenged the long-standing paradigm that individual kinesin motors are characterized by an intrinsic directionality. The underlying mechanism, however, remains unexplained. Here, we studied clustering-induced directionality switching of the bidirectional kinesin-5 Cin8. Based on the characterization of single-molecule and cluster motility, we developed a model that predicts that directionality switching of Cin8 is caused by an asymmetric response of its active motion to opposing forces, referred to as drag. The model shows excellent quantitative agreement with experimental data obtained under high and low ionic strength conditions. Our analysis identifies a robust and general mechanism that explains why bidirectional motor proteins reverse direction in response to seemingly unrelated experimental factors including changes in motor density and molecular crowding, and in multimotor motility assays.
INTRODUCTION
The bipolar and tetrameric kinesin-5 motors perform essential functions in mitotic spindle dynamics by cross-linking and sliding antiparallel spindle microtubules (MTs) apart (1–3). In kinesin-5 motors, the catalytic domain is located at the N terminus. Because they share this attribute with all known plus-end–directed kinesins, they were previously believed to be exclusively plus-end directed. Recent studies showed that several kinesin-5 motor proteins, which are minus-end directed at the single-molecule level, can switch directionality under various experimental conditions. Thus far, three bidirectional kinesin-5 motors have been identified (4–7): Cin8 and Kip1 in Saccharomyces cerevisiae and Cut7 in Schizosaccharomyces pombe. Their in vitro behavior is puzzling: Single Cin8 and Kip1 motors are minus-end directed in high ionic strength but switch directionality under low-ionic-strength conditions (5, 7, 8), as well as in multimotor gliding assays (5–7). Cin8 was also shown to switch directionality when engaged in sliding antiparallel MTs apart (6, 7), as a function of the density of surface-bound motors that interact with MTs in multimotor gliding assays (9), and when it forms clusters on single MTs (10). Cut7 is minus-end directed in single-molecule experiments and multimotor MT gliding assays (4, 11) but switches directionality as a function of crowding of either motor or nonmotor proteins on MTs (12). Notably, two kinesin-14 motors were recently demonstrated to be bidirectional (13, 14), indicating that switchable directionality is more common in the kinesin superfamily than was previously appreciated. The plethora of observations related to directionality switching may indicate that bidirectionality is of physiological importance, a notion that is also supported by a recent theoretical study (15).
Presently, however, there is no mechanistic explanation for all these intriguing observations, and the remarkable ability of the kinesin-5 motors to switch directionality remains unexplained. Here, we study directionality switching due to clustering of motor proteins (10), using a combined experimental and theoretical approach. We developed a single-molecule fluorescence-based method of analysis that enabled us to characterize the relationship between Cin8 cluster size and directionality. We found that while single Cin8 motors show minus-end–directed motion, clusters of Cin8 containing two and more Cin8 motors are more likely to display plus-end–directed motion. The motion of single Cin8 motors exhibits a large diffusive component, which is markedly reduced for Cin8 pairs and higher-order clusters. To explain these experimental results, we propose a mechanism based on the interplay between an asymmetric response of plus-end– and minus-end–directed active motion to forces that oppose this motion (referred to as “drag”) and weak attractive forces between motors. The mathematical analysis of a corresponding theoretical model shows that this mechanism leads to clustering-induced directionality switching of Cin8 from minus-end– to plus-end–directed motility for realistic model parameters. The model shows excellent quantitative agreement with experimental data obtained under high- and low-ionic-strength conditions. Moreover, the very same mechanism not only explains directionality switching due to motor clustering but also provides a rationale for directionality switching under diverse, previously reported conditions, such as changes in the ionic strength or the motor density in the motility assay, and molecular crowding with other motor and nonmotor proteins. Thus, our combined experimental and theoretical approach provides general insight into the possible mechanism and origin of bidirectional motion of motor proteins.
RESULTS
Size-based classification of motile Cin8 motors
We have previously reported that clustering of Cin8 motors on MTs is one of the conditions that can switch its directionality from fast minus-end– to slow plus-end–directed movement (10). However, this analysis did not provide quantitative information on the relationship between directionality and the number of motors in a cluster, which is essential for unraveling the underlying mechanism. Hence, to establish a basis for the analysis of the link between Cin8 clustering and the preferred direction of motility, we first developed an experimental strategy to measure the cluster size, i.e., the number of Cin8 molecules in a cluster. By following the fluorescence intensity of Cin8–GFP (green fluorescent protein) as a function of time, we observed single photobleaching events of ~50 arbitrary units (a.u.), most likely stemming from bleaching of a single GFP molecule (Fig. 1A). Because single Cin8 motors are tetramers containing four identical subunits, the maximal fluorescence intensity of a single Cin8 motor containing four GFP molecules is likely to be ~200 a.u. On the basis of this method of quantification, the population of Cin8 motors or clusters was divided into three categories according to the number of Cin8 molecules in each: (i) individual or “single” tetrameric Cin8 molecules (intensities <200 a.u.); (ii) “pairs” of Cin8 molecules, i.e., dimers of single tetrameric Cin8 molecules (intensities 200 to 400 a.u.); and (iii) higher “oligomers” of Cin8 (intensities >400 a.u.) (Fig. 1B). Pairs and higher oligomers of Cin8 are also referred to as “clusters” henceforth. “Cin8 motors” refers to moving Cin8 particles of unspecified cluster size. Further critical assessment of the experimental approach used to determine the Cin8 cluster size is given in Materials and Methods.
Fig. 1. Motility of Cin8 motors of different size.
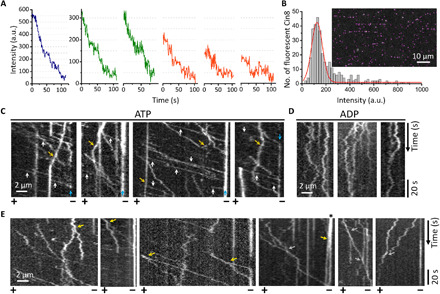
(A) Photobleaching profile of fluorescent Cin8-GFP motors. Single photobleaching steps, each likely representing the photobleaching of one GFP, lead to a drop in fluorescence intensity of ~50 a.u. The red, green, and blue traces represent single motors, pairs, and higher oligomers of Cin8, respectively. (B) Intensity distribution of Cin8-GFP motors in the first frame of a time-lapse sequence (inset). The Gaussian peak (red) represents single Cin8-GFP molecules constituting ~70% of the total Cin8 population. This peak is centered at ~120 a.u., which is the average intensity of a single Cin8 tetramer containing either one, two, three, or four fluorescent GFP molecules, with each GFP molecule contributing ~50 a.u. to the total intensity. Accordingly, the maximal fluorescence intensity of single Cin8 tetramers is 200 a.u. (C) Representative kymographs of Cin8-GFP motility along single MTs in the presence of 1 mM ATP. White arrows, fast-moving Cin8 motors; orange arrows, plus-end–directed Cin8 motors; blue arrows, clustering of Cin8 at the minus end of the MTs (10). Polarity of the MTs is indicated at the bottom. (D) Representative kymographs of Cin8-GFP motility along single MTs in the presence of 1 mM ADP. (E) Representative kymographs depicting dynamic clusters of Cin8-GFP, which split (yellow arrows) into smaller clusters or individual Cin8 molecules or merge (gray arrows) to form larger clusters while moving on single MTs. Asterisk indicates the splitting and subsequently plus-end–directed movement of a Cin8 cluster initially accumulated at the minus end.
To study the relation between Cin8 cluster size and motility, we performed experiments at saturating ATP (adenosine 5′-triphosphate) concentration and high ionic strength (Fig. 1C), where Cin8 motors were previously shown to be mostly minus-end directed and fast moving relative to kinesin-5 motors in higher eukaryotes (6, 7, 10, 16). In accordance with previous reports (6, 7, 10, 16), we observed that ~80% of the total motile Cin8 population was fast moving and minus-end directed and that almost all these trajectories fell into the category of single tetrameric Cin8 motors (Fig. 1, A to C; fig. SA1a, left panel; and Table 1, i). The remaining 20% comprise clusters of Cin8, which exhibit increased incidences of plus-end–directed and bidirectional trajectories (Fig. 1C; figs. SA1a, middle and right panels, and SA2a; and Table 1, i). We also observed that MT-bound clusters of Cin8 move along MTs and undergo splitting and merging events (Fig. 1E), indicating that these are active motors rather than nonspecific Cin8 aggregates. We found that ~19% of motile pairs and clusters of Cin8 (n = 96) exhibited splitting events, of which ~2% split into smaller clusters, ~5% split into a smaller cluster and a single Cin8 molecule, and ~12% split into two single Cin8 molecules. In addition, ~15% of clusters that accumulated at the minus end of the MTs (n = 46) exhibited directionality reversal and moved in the plus-end direction during the observation time. Moreover, clustering occurred regardless of the nucleotide state, both under high and low ATP concentrations and in the presence of ADP (adenosine 5′-diphosphate) [Fig. 1, C and D, and figs. SA1 and SA2 (b and c)], indicating that it is an intrinsic property of Cin8. Experiments carried out in the presence of ATP clearly showed that the cluster size correlated with the propensity for plus-end–directed motion. While only 3% of single-molecule trajectories were, on average, plus-end directed, 40 and 70% of the trajectories of pairs and oligomers of Cin8, respectively, exhibited net movement toward the plus end (fig. SA1a). Notably, in each cluster size category, the velocity of minus-end–directed motion was considerably faster than that of plus-end–directed motility (fig. SA1a).
Table 1. Velocities and diffusion coefficients obtained from MD/MSD analysis.
Values are presented for experimental data (i) and simulated using the fully quantified model for the collective motion of Cin8 (ii). na, not analyzed due to the paucity of this size of clusters.
| (i) Experimental | VMD (nm/s)* | VMSD (nm/s)* | D × 103 (nm2/s)† | ||||
| Nucleotide‡ | ATP | ATP | ATP | ADP | |||
| [KCl] (mM) | 165 | 110 | 165 | 110 | 165 | 110 | 165 |
| Single molecules | −237 ± 8 (71) | −18.1 ± 0.2 (39) | 229 ± 66 (71) | 31 ± 14 (39) | 102 ± 11 (71) | 22.3 ± 0.8 (39) | 10.4 ± 0.1 (28) |
| Pairs of molecules | 23.2 ± 0.7 (25) | −11.9 ± 0.1 (38) | 21 ± 5 (25) | 31 ± 6 (38) | 5.0 ± 0.8 (25) | 12.3 ± 0.2 (38) | 7.0 ± 0.3 (21) |
| Higher oligomers | 10.7 ± 0.2 (16) | 1.12 ± 0.02 (9) | 14 ± 2 (16) | 26 ± 3 (9) | 1.1 ± 0.1 (16) | 1.6 ± 0.1 (9) | na |
| (ii) Simulation§ | VMD (nm/s) | VMSD (nm/s) | D × 103 (nm2/s) | ||||
| Single molecules | −229 | 227 | 109 | ||||
| Pairs of molecules | 23.0 | 22.0 | 5.30 | ||||
| Higher oligomers║ | 16.8 | 15.7 | 2.26 | ||||
| Clusters with three molecules |
17.8 | 16.5 | 2.38 | ||||
| Clusters with four molecules |
11.5 | 9.58 | 1.79 | ||||
| Clusters with five molecules |
9.86 | 8.25 | 1.32 | ||||
*VMD and VMSD represent velocities evaluated from MD and MSD analyses, respectively. Negative values of velocity represent motility in the minus-end direction. Averages ± SEM are presented. Values in parentheses represent the numbers of Cin8 motors analyzed.
†Diffusion coefficient evaluated from the MSD analysis.
‡Nucleotide concentrations are 1 mM.
§Values were obtained by fitting the linear function 〈x(t)〉 = vMDt to the temporal evolution of the MD, and the quadratic function to the temporal evolution of the MSD, where x(t) denotes the position of the respective particles at time t on the MT, and angle brackets refer to an ensemble average over different trajectories. An attachment rate of was used. A lattice size of L = 5000 was used. Other model parameters are listed in table SB1. For further description on the evaluation and tracking method, see sections SB3 and SB4. Values correspond to experiments under high ionic strength conditions (165 mM KCl) in the presence of saturating ATP concentration.
║Parameters obtained from MD and MSD analyses of clusters containing three, four, and five Cin8 molecules, listed in the three lower rows of this table.
In summary, as a basis for further analyses, we developed an experimental approach that enabled us to assess the number of Cin8 molecules in a moving cluster. This, in turn, allowed us to quantitatively show that the preference for slow, plus-end–directed motion correlates with increasing size of Cin8 clusters and that pairs of Cin8 motors can switch directionality.
Individual Cin8 motors exhibit a large diffusive component
To better understand the implications of Cin8 clustering, we next sought to analyze and compare the motion of single Cin8 molecules and Cin8 clusters. As central quantities for the characterization of the motility, we used the average displacement velocity and the diffusion coefficient, obtained from an analysis of the mean displacement (MD) and mean-squared displacement (MSD) (Fig. 2, A and B, and Table 1, i). This quantitative analysis further supported the observation that single Cin8 motors are fast and minus-end directed (v = −237 nm/s), while pairs and clusters of Cin8, on average, are slow and plus-end directed with velocities of v = 23 nm/s and v = 11 nm/s, respectively (Fig. 2, A and B, and Table 1, i). In the presence of ATP, the motor dynamics deviates from diffusive behavior (Fig. 2B), confirming that single Cin8 molecules and Cin8 clusters exhibit active motion (8, 17). In contrast and as expected, we observed only diffusive motion, with no net displacement in the presence of ADP (Fig. 1D) (8, 17).
Fig. 2. MD and MSD of Cin8 motors of different size.
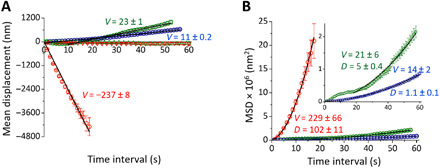
MD (A) and MSD (B) analyses are shown for single Cin8 molecules (red), pairs of Cin8 (green), and higher oligomers (blue) in the presence of 1 mM ATP (circles) or ADP (triangles). In the presence of ADP, every third point is indicated for better visibility of the two plots (single molecules and pairs). The black solid lines are linear (MD = vt) (MD) or statistically weighted, second-order polynomial fits (MSD = v2t2 + 2Dt) (MSD). Values of the mean velocity (V, nanometers per second) and the diffusion coefficient [D, square nanometers per second (×103)] are indicated. Error bars represent SEM. Negative values of velocity represent motility in the minus-end direction. In (B), the inset shows a zoomed-in plot depicting the MSD curves for Cin8 pairs (green) and oligomers (blue).
Next, we focused on the motility of single Cin8 molecules to establish a basis for a mechanistic model of Cin8 motion. In addition to the large velocity reported above, single Cin8 molecules also exhibited an exceptionally high effective diffusion coefficient of D ≈ 102 ∙103 nm2/s in the presence of ATP (Fig. 2B and Table 1, i). This value may relate to a large bidirectional component in the motion of Cin8, which could arise from active bidirectional stepping, Brownian motion, or a combination of both. In the presence of ADP, the diffusion coefficient was substantially lower with a value of D ≈10 ∙ 103 nm2/s (Table 1, i). The considerable decrease in the diffusion coefficient in the presence of ADP relative to ATP suggests at first sight that single Cin8 motors may indeed exhibit active bidirectional stepping that leads to additional effective diffusive behavior, which vanishes in the absence of ATP. There is, however, an additional counteracting effect. The average dwell times of ADP-bound Cin8 are longer, indicating an increased affinity of Cin8 for MTs under this condition (see section SA2.3), which, in turn, is expected to reduce the diffusion coefficient. To discriminate between the different possible origins of the large diffusion coefficient, we estimated the potential ATP turnover rate required to obtain the observed diffusion coefficient by active bidirectional stepping alone and obtained a value of approximately 3000 ATPs/s (see section SB2.1 for a detailed computation). Such a high turnover rate exceeds known values for other kinesins by at least one order of magnitude (18–20). Moreover, the diffusion coefficient measured for Cin8 in the presence of ATP shows a value that is typical for molecules that exhibit an ATP-independent diffusive component of motion along MTs (21–23). Thus, it is likely that the largest contribution to the diffusion coefficient of individual Cin8 motors originates from Brownian motion. Here, this argument does not exclude the presence of an additional, but notably smaller, contribution of ATP-dependent (active) bidirectional motion to the diffusion coefficient.
Overall, we conclude from the statistical analysis of the ensemble of stochastic motor trajectories that single Cin8 molecules show an active, ATP-dependent component, leading to minus-end–directed motion of single motors and plus-end–directed motion of clusters. Single Cin8 motors exhibit a strong diffusive component, which is largely attributable to Brownian motion.
Cin8 motors are subject to weak attractive interactions that reduce cluster diffusion
We next asked whether clusters of Cin8 display markedly different behavior. By performing an MSD analysis, as detailed in the previous section, we found that clusters of Cin8 motors indeed exhibited a substantially decreased diffusion coefficient as compared with single Cin8 motors (Fig. 2B and Table 1, i). For example, the diffusion coefficient for pairs of Cin8 was approximately 20 times smaller than that of single motors (Table 1, i). On the basis of our observations, we hypothesized that weak attractive interactions between Cin8 tetramers cause clustering of motors and might also be the underlying cause of the reduced diffusion coefficients of clusters.
To test these hypotheses, we formulated a detailed computational model that integrates molecular features and enabled us to predict the consequences of such weak interactions for the motion and clustering of Cin8 motors and, ultimately, for their directionality. This model comprises two main modules, one for the diffusive and one for the active components of the motions of the motors. Thus, we first quantified the model parameters related to Cin8 interactions by establishing a functional relation between cluster diffusion and attractive interactions (while neglecting active motion) (Fig. 3A and see Materials and Methods). This approach is justified as—based on the above considerations—the diffusive component of motion mainly reflects the impact of Brownian motion. In a next step (see the following paragraph), we added a module that accounts for active motion and its response to motor interactions, thereby establishing a mechanistic link between clustering and direction switching. Note that based on previous reports that indicate that the bipolar assembly domain of kinesin-5 motors is a rigid filament (24–26), we present in illustration a scenario where one pair of catalytic domains is bound to a single MT. On the basis of our computational analysis, increasing Cin8-Cin8 interaction strength decreases the diffusive motion of a Cin8 pair or cluster (Fig. 3B). From the relationship between the attractive force Fint and the diffusion coefficient D, we inferred that the experimentally measured diffusion coefficient of D = 5 ∙ 103 nm2/s for a pair of Cin8 corresponds to an attractive force of Fint = 1.4 pN (here, the sign convention is such that positive signs denote attractive forces) (Fig. 3B, dashed line). This value is close to the stall force for Cin8 (≈1.5 pN) observed in motility assay experiments (9). Thus, on the basis of the difference in the diffusion coefficient of single Cin8 molecules and pairs of Cin8, we conclude that one of the likely mechanisms of Cin8 clustering is the existence of attractive forces between MT-bound Cin8 molecules. With the inferred parameter, there is quantitative agreement between the computational model and experimental observations (see the section ‘Asymmetric response of active motion to drag explains directionality switching of Cin8 clusters’ and section SB5).
Fig. 3. Computational model for collective Cin8 motion.
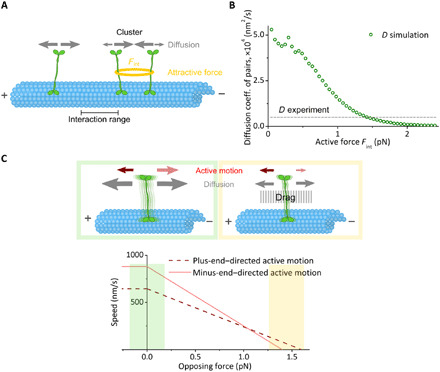
(A) The diffusive module simulates Cin8 motors as random walkers with symmetric dynamics (gray arrows) due to Brownian motion. Motors attract each other if they are within the interaction range R. The attractive force Fint (yellow ring) is assumed to be constant for distances <R and zero otherwise. Attractive interactions affect motor dynamics (detailed balance) (see the main text and section SB1). (B) The diffusion coefficient of pairs of Cin8 in simulations focusing on the diffusive module (circles) decreased with increasing attractive forces F between motors. Attractive forces of 1.4 pN result in a diffusion coefficient for Cin8 pairs that corresponds to that identified in vitro (dashed line; see also Table 1, i). (C) Top: Cin8 dynamics in the full computational model is composed of bidirectional active motion (red/pink arrows) and a diffusive component (gray arrows; illustrated by motor blurring). In the absence of an opposing force, the minus-end–directed active component is larger than the plus-end–directed component. Both the active and diffusive components are affected by opposing forces, referred to as drag (gray area in the top right). Drag suppresses Brownian motion exponentially, and minus-end–directed active motion (pink) is more strongly impeded by drag than is plus-end–directed active motion (red), such that the overall directionality switches under large drag. Bottom: The assumed force-velocity relation used in simulations, with green shading corresponding to low drag (top left) and yellow shading corresponding to high drag (top right). The sizes of the arrows are chosen for illustrative purposes only.
Asymmetric response of active motion to drag explains directionality switching of Cin8 clusters
After establishing the role of attractive interactions in the diffusive motion of Cin8 and cluster formation, we next investigated whether attractive interactions could cause clustering-induced directionality switching. To address this question, we extended the computational model to account for active motion. We assumed that Cin8 performs active stepping in both plus- and minus-end directions and that stepping in the two directions responds differently to opposing forces (Fig. 3C and see Materials and Methods). Note that this response is independent of the origin of the opposing force (drag) such that the model should be universally applicable. The observation that attractive forces between motors are in the range of the stall forces of plus-end– and minus-end–directed motions of Cin8, as estimated above from our data, suggests that an asymmetric drag response is likely to play an important role in collective Cin8 dynamics and could potentially play an essential role for switching of directionality in Cin8 clusters.
To test these ideas, we performed extensive stochastic simulations of the full model while accounting for all of the above features (see sections SB1 and SB2 for a detailed description of the computational model and the choice of parameters). Notably, we find that our model for Cin8 motion indeed exhibits directionality switching of Cin8 clusters. As explained further below, this is not built into the model per se but an emergent function due to a highly nontrivial stochastic effect (caterpillar mechanism; see below). The kymographs obtained from our stochastic simulations clearly show a tendency for clusters to move toward the plus end of MTs (Fig. 4), in remarkable agreement with experimental observations (Fig. 1C). To quantify this visual impression, we analyzed the stochastic trajectories obtained from the numerical simulation to calculate the MD velocity and the diffusion coefficient (Table 1, ii, and sections SB3 and SB4). In agreement with experiments, single Cin8 molecules show a minus-end–directed MD velocity, opposite to that of pairs and clusters. The model quantitatively reproduces the measured displacement velocities, as well as diffusion coefficients, of single Cin8 motors, pairs, and higher oligomers of motors to a very high degree of accuracy (Table 1, ii), with parameters independently determined from experimental data and general stability arguments but no further fitting parameters (see section SB2).
Fig. 4. Simulated kymographs exhibit directionality switching due to motor clustering.
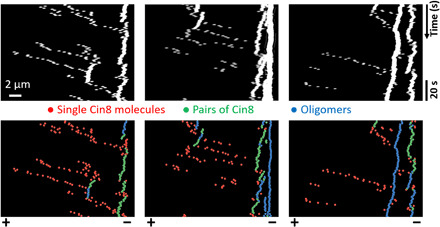
Cin8 clusters were biased toward the plus end, while single motors were biased toward the minus end. In the top row, simulation data were convoluted with a point-spread function to obtain images comparable to the experimental data, where bright traces represent clusters (see section SB9 for details of the convolution method). The bottom row shows the same kymographs, but here, single Cin8 motors (red), pairs of Cin8 (green), and larger clusters with more than two Cin8 molecules (blue) are depicted in different colors. Note that clusters with more than two motors (higher oligomers; blue traces) were not further distinguished in this image as motivated by the experimental analysis. Polarity of the MTs is indicated at the bottom. Identical parameters were used in the three simulations. To allow for initial motor binding and interactions, the first 1000 s of the simulation are not shown in the kymographs. See section SB2 and table SB1 for a detailed description and a complete list of the parameters used.
Because our computational model faithfully captured directionality switching as characterized by the displacement analysis, we performed a more detailed analysis of the distribution of cluster sizes and motion. To this end, we determined the full statistics of cluster size–dependent motility in simulations and experiments by computing and measuring the average velocities and degree of cluster intensity for each trajectory of single Cin8 motors or clusters. Figure 5 shows that the resulting distribution obtained from simulations is in excellent agreement with the data obtained from experiments. In addition, we found good agreement between experimental data and simulation results for the clustering behavior at different Cin8 concentrations. Increasing the Cin8 concentration in silico as well as in vitro led to an increased number of clusters (bright traces in kymographs) that slowly moved either in an undirected or plus-end–directed manner (Fig. 6, A and B, top). This agreement is supported by the statistics of simulated and experimentally determined cluster intensity (Fig. 6, A and B, bottom).
Fig. 5. Relationship between motor intensity and velocity, comparison between simulation results, and experimental data.
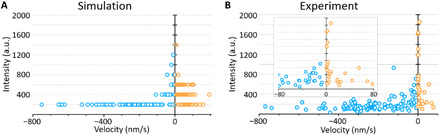
(A) Relationship between velocity and intensity of Cin8-GFP motors obtained by simulations based on the computational model. Intensity is determined by multiplying the number of single Cin8 molecules in a cluster by 200 a.u., the maximal intensity of a single Cin8 tetramer, based on the photobleaching curves (Fig. 1A). A detailed description and a complete list of the parameters used in (A) are found in section SB2 and table SB1. (B) Same as (A) but measured experimentally. Total n = 134; minus-end–directed n = 107, plus-end–directed n = 27. Of the total motors, ~20% are plus-end directed, and 70% of the total minus-end–directed motors are monomers. The majority (~88%) of plus-end–directed motors are clusters comprising >2 Cin8 molecules. Inset: Closeup distribution of slow-moving Cin8 motors. In (A) and (B), blue and yellow circles represent (−)- and (+)-end–directed Cin8 molecules, respectively.
Fig. 6. Dependence of motility and clustering on concentration of Cin8.
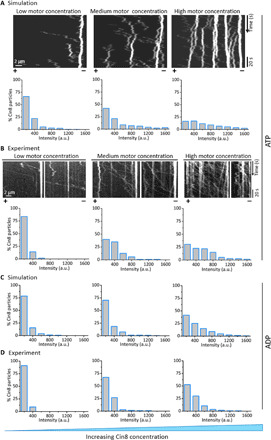
(A) Concentration-dependent clustering in simulations, in the presence of ATP. Top: Kymographs for different concentrations of Cin8, generated by simulations of the theoretical model. Bottom: Corresponding model-generated histograms of intensities of Cin8 motors bound to MTs at increasing concentrations of Cin8. (B) Experimental data for concentration-dependent clustering of Cin8 on single MTs. Top: Representative kymographs obtained from time-lapse measurements of Cin8 motility at different Cin8 concentrations (see Materials and Methods). Bottom: Corresponding histograms of intensities of Cin8 motors bound to MTs at increasing concentrations of Cin8. (C and D) Comparison of the distribution of cluster sizes determined in experiments in the presence of ADP with those obtained in simulations of the model without active motion at different concentrations of Cin8. The nucleotide condition is indicated on the right. In (A) and (C), the attachment rates were computed on the basis of the measured attachment rate at medium concentrations (1 to 2 pM): kon = {3.62 ∙ 10−5, 4.82 ∙ 10−5, 9.04 ∙ 10−5} s−1 from the left to the right. Lattice size in the simulations was L = 2000; other parameters were as in Fig. 5A. The percentage of the respective cluster size observed in the simulations was computed by relating the summed existence times of all single particles and clusters for each of the different size categories. Concentrations of Cin8 in simulations were c = {0.75 − 1.5,1 − 2, 1.875 − 3.75} pM from the left to the right.
To further understand the mechanism underlying the observed directionality switching of Cin8 clusters relative to the behavior of single Cin8 motors, we performed additional simulations in which we suppressed Brownian motion of Cin8. We found that directionality switching of clusters was absent when motors exhibited negligible Brownian motion (fig. SB6), which points to a decisive role of Brownian motion in the directionality switching of Cin8 clusters.
Our model for collective Cin8 motion, which accounts for active as well as Brownian motion, produces concentration-dependent cluster intensity distributions that agree with experimental data (Fig. 6, A and B, bottom). The similarity of these results to those obtained with the model that accounts for Brownian motion only (cf. Fig. 6, A to D) further supports the inference that clustering is only affected to a minor degree by active motion, as argued previously. Second, in our simulations, we found a sharp transition between a regime with unstable clusters and one in which cluster formation is strongly favored with increasing Cin8 concentration (fig. SB7). The concentration at which this sudden transition occurs agreed excellently with that which marks the onset of cluster formation in our experiments (see section SB8 for additional data and analysis).
Asymmetric response to drag is supported by Cin8 motility experiments at low ionic strength
The analysis based on the theoretical model suggests that the asymmetric response of Cin8 to drag is the mechanism underlying directionality switching from minus-end– to plus-end–directed motility. Therefore, we asked whether such an asymmetric response could also be observed more directly in experiments. Single Cin8 motors are likely to experience increased drag while moving along an MT if their affinity for the MTs is increased. Such a change of affinity can be achieved by reducing the ionic strength of the buffer, since electrostatic interactions are less shielded under such conditions. Accordingly, we performed experiments at a salt concentration of 110 mM KCl. We observed that decreasing the salt concentration drastically increased the number of MT-bound Cin8 motors compared with high-salt conditions at similar concentrations of Cin8 (Fig. 7A) and increased the landing rate of Cin8 (see section SA2.2). This indicates that lowering the salt concentration indeed increases the affinity of Cin8 for MTs and, hence, results in higher drag on moving Cin8 motors. To compensate for the increased affinity and still achieve a similar number of motors on the MTs, experiments under low-salt conditions were performed with ⁓5-fold lower Cin8 concentration (Fig. 7, B to E).
Fig. 7. Motility of Cin8 under low-salt conditions.
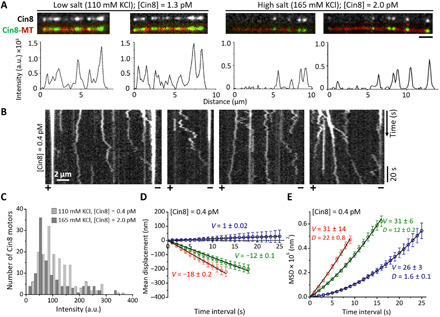
(A) MT-bound Cin8 in the presence of low or high salt concentration. Top: Representative images of MT-bound Cin8 at salt concentrations indicated at the top. Bottom: Corresponding Cin8-GFP intensity profile of MT-bound Cin8 motors. (B) Representative kymographs of Cin8-GFP motility along single polarity–marked MTs (polarity is indicated at the bottom) in the presence of 110 mM KCl and 1 mM ATP. (C) Intensity distribution histogram of MT-bound Cin8 at high (165 mM KCl, dark gray) and low salt (110 mM KCl, light gray). (D and E) MD (D) and MSD (E) for single Cin8 molecules (red), pairs of Cin8 (green), and higher oligomers (blue). The black solid lines are linear (MD = vt) (MD) or statistically weighted second-order polynomial fits (MSD = v2t2 + 2Dt) (MSD). Values of the mean velocity (V, nanometers per second) and the diffusion coefficient [D, square nanometers per second (×103)] are indicated. Error bars represent SEM. (A to E) Cin8 concentrations are indicated for each panel.
In agreement with previous reports (7, 8, 27, 28), we found that decreasing the salt concentration increases the fraction of plus-end–directed Cin8 trajectories and considerably diminishes the average displacement velocities (Fig. 7, B and D; fig. SA1(b); and Table 1, i). However, it had previously not been established whether this effect is caused by stronger Cin8-MT interaction or Cin8 clustering induced by low ionic strength. Our ability to analyze the motility of single molecules, pairs, and oligomers of Cin8 separately enabled us to differentiate between these distinct effects on Cin8 motility. In agreement with our hypothesis of an anisotropic response of active motion to drag, our analysis revealed that it is not clustering per se but an increased drag that switches the directionality of Cin8. Under high-salt conditions, only 3% of the trajectories of single Cin8 molecules were plus-end directed (fig. SA1a), while 33% of single Cin8 trajectories showed switched directionality at low ionic strengths (fig. SA1b). The observation that additional drag by lowering the salt concentration gives rise to directionality reversal is also in accordance with our theoretical model. To show this, we estimated the additional drag experienced by a single Cin8 motor moving on the MT in low-salt buffer. According to the Arrhenius law, drag suppresses diffusive motion by an exponential factor of e−∆E/kBT, where ∆E is the additional energy barrier a motor has to overcome when moving from one binding site on the MT to the next. We estimated that the energy barrier must be surmounted approximately halfway between two tubulin dimers, i.e., ⁓4 nm. Then, following the Arrhenius law, the fivefold decrease in the diffusion coefficient observed for single Cin8 molecules in low salt relative to high salt (Fig. 4E and Table 1, i) implies an additional drag of approximately 1.6 pN. This value agrees with our modeling hypothesis that forces of this magnitude which antagonize motion switch the directionality of Cin8 (see Fig. 3C, bottom).
Together, we found that consistent with our computational model, trajectories of single Cin8 motors become increasingly biased toward the plus-end direction along with experiencing a larger drag at low ionic strengths. Therefore, we conclude that not motor clustering per se but drag or any external force that opposes motion can switch the directionality of Cin8.
A caterpillar mechanism for directionality switching of motion in Cin8 clusters
How the combined effects of diffusive motion and an asymmetric response of active motion to drag led to clustering-induced directionality switching can be understood by the following mechanism, illustrated in Fig. 8. Note that although we do not assume a specific stepping mechanism, in the model presentation in Fig. 8, we show one pair of catalytic domains (and not both pairs of the tetrameter) interacting with a single MT. This presentation relies on previous reports indicating that the central mini filament of the bipolar assembly domain is rigid (24–26) and is likely to prevent extreme bending of the motor, required for simultaneous binding of the two pairs of catalytic domains to the same MT. We start from a configuration in which two interacting Cin8 molecules are adjacent to each other on the MT [Fig. 8, part (1)]. The center of mass of this cluster can change if either moves away from the other, referred to as stretching (Fig. 8, right). Therefore, stretching can occur by either plus-end–directed motion of the left motor or minus-end–directed motion of the right motor. Both of these (stochastic hopping) processes are counteracted by the attractive interaction (yellow rings in Fig. 8) between the motors. Because of the assumed asymmetry in the response of active motor motion to drag (Fig. 3C), the attractive interaction between the motors suppresses minus-end–directed active motion (pink arrows in Fig. 8) more strongly than plus-end–directed active motion. In other words, the left motor is more likely to move toward the plus end than the right motor is to shift in the direction of the minus end, thereby creating an overall bias for cluster motion toward the plus end, leading to a configuration in which the two motors have moved further apart [Fig. 8, part (2)]. Now, they can not only diverge further from each other but also move closer together, causing “cluster contraction” (Fig. 8, left). For cluster contraction, the direction of motion of the motors corresponds to the direction of the forces at work, i.e., a pulling force instead of a drag then acts on each of the two motors. In that case, the active component of motion has the same bias as for zero load, i.e., the left motor is more likely to move toward the minus end than the motor on the right is to step toward the plus end (Fig. 3C, bottom). Hence, this type of contractile motion creates a bias toward the minus end. Overall, the force-induced bias in the active motion acts in opposite directions during cluster stretching and cluster contraction, respectively.
Fig. 8. Model for plus-end directional preference of Cin8 clusters.
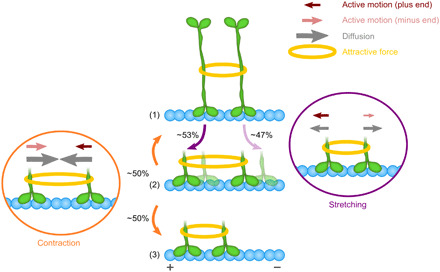
A fully contracted cluster of two Cin8 motors (1) may expand by movement of either motor away from the other (cluster stretching, right). Stretching is constrained by the attractive interaction (yellow ring), which opposes both diffusion (gray arrows) and active motion (red/pink arrows). Conversely, motor clustering implies that diffusion is suppressed exponentially, and active motion is biased toward the plus end owing to the assumed asymmetric response of the active motion of Cin8 to opposing forces (Fig. 3C). Consequently, stretching is slightly more likely (53%) to occur due to plus-end–directed motion of the left motor [opaque configuration in (2)] than by minus-end–directed motion of the right motor [transparent configuration in (2)]. Once the motors have moved further apart (2), the pair most likely contracts again [process (2) → (1) or (2) → (3)]. In the case of contraction (left), the attractive force supports both active motion and diffusion. Hence, active motion now shows a minus-end–directed bias (see Fig. 3C for supporting illustration). However, diffusive motion dominates cluster stretching, as it is supported (and not suppressed) by the attractive interaction. Because diffusive motion is inherently undirected, cluster contraction occurs almost equally often by motion of either motor (50.5% left motor; 49.5% right motor). As cluster stretching shows a weak bias toward the plus end while cluster contraction is almost unbiased, a pair of motors is more likely to move in a caterpillar-like fashion toward the plus end [sequence of processes (1) → (2) → (3)] than the minus end. The sizes of the arrows differ for illustrative purposes only; they are not proportional to the model parameters (see section SB6 for a detailed description of stochastic process and a computation of the conditional probabilities).
These two opposing directional biases, however, do no cancel out, owing to the key role played by diffusive motion. According to the Arrhenius law, thermal Brownian motion is exponentially reduced when movement ensues against a force as compared with moving along the direction of a force with the same magnitude. As a consequence, this thermal component of motor motion is much more pronounced for cluster contraction than for cluster stretching (compare gray arrows in the left and right panels of Fig. 8). Adding up the active and thermal components of motion, this implies that Brownian motion has very different effects on the relative bias for cluster motion toward the plus and minus ends. For cluster contraction, thermal motion is the dominating mode of motion and therefore strongly reduces the relative bias in cluster motion toward the minus end caused by active motion. Consequently, contracting clusters are almost equally likely to move toward either end (50.5% motion of the left motor, 49.5% motion of the right motor). In contrast, during cluster stretching, thermal motion is exponentially suppressed, such that a clear bias for cluster motion toward the plus end ensues (53% motion of the left motor, 47% motion of the right motor; see section SB6 for the computation of the conditional probabilities). Thus, plus-end–directed motion of Cin8 clusters occurs because they preferentially stretch toward the plus end, while there is no pronounced directional bias during cluster contraction. Whenever plus-end–directed cluster stretching is followed by plus-end–directed cluster contraction, a wave-like motion of the motors toward the plus end occurs, reminiscent of caterpillar motion [sequence of processes (1) → (2) → (3) of Fig. 8]. Note that although originating from completely different processes, the proposed caterpillar mechanism for cluster motion is reminiscent of the inchworm mechanism that was speculated for the motion of the two heads of an individual kinesin: In both models, motion is the result of a wave-like motion of the different building blocks (a single kinesin head for the inchworm mechanism and an individual tetrameric motor for the caterpillar mechanism, respectively), with always the same building block leading. Last, the same arguments as made here for a pair of motors can be made for the two outermost motors of a cluster with more than two motors, such that these motors bias cluster motion toward the plus end. However, unlike the case for a pair of Cin8, motility is further reduced by the motion of the motor(s) in the center of the cluster. Because these motors experience forces from both directions that cancel each other out, they behave like single motors with a large diffusive component in their motion and a minus-end–directed bias. This behavior leads to almost unbiased “steric clashes” of the center motors with the outermost ones. Hence, the plus-end–directed motion of the outermost motors will be slowed down but keep their plus-end–directed bias. Thus, in agreement with experimental data (Fig. 2, A and B, and Table 1, i), the motion of large clusters in the plus-end direction is slower than that of pairs of Cin8 (Table 1, ii).
DISCUSSION
Although bidirectional motility of kinesin motors was found nearly a decade ago (6, 7), the underlying mechanisms had remained obscure. Our analysis strongly suggests that clustering-induced switching of motor directionality arises from an interplay between active motion in the minus- and plus-end direction, a substantial diffusive component, and an asymmetric response of active motion to forces that oppose this motion.
How could the mechanistic features of the computational model, which agrees so well with a broad range of experimental observations, be related to known molecular structural-functional features of kinesin motors in general and Cin8 in particular? First, in the model, we assume that Cin8 has the ability to actively step in two directions. The kinesin neck-linker has been shown to be the major structural determinant that differentiates between the directionality of the N-terminal (plus-end–directed) and the C-terminal (minus-end–directed) kinesins (29–33) and modulates the stepping mechanism and processivity of plus-end–directed kinesin motors (34, 35). Hence, we speculate that to enable stepping in two directions, the neck-linker of Cin8 exhibits a higher degree of flexibility than those of unidirectional kinesin motors. Second, the model predicts that directionality reversal of Cin8 clusters can be facilitated by attractive forces between Cin8 molecules in a cluster. Regarding this model feature, it has recently been demonstrated that the large insert in loop 8 within the catalytic domain of Cin8, which is important for directionality switching (7), is required for the noncanonical binding of three to four motor domains of Cin8 per tubulin dimer, probably via an attractive motor-motor interaction (36). In light of this finding, we hypothesize that loop 8 of Cin8 may be one of the factors responsible for the weak attractive forces between Cin8 tetramers that enable them to form clusters. Last, the large loop 8 of Cin8 can interact with MTs, which may well affect the level of drag experienced by motors as they move along an MT and could therefore contribute directly to directionality switching. Future studies will likely shed light on how these and additional molecular domains affect their bidirectional motility of kinesin motors.
The asymmetric force-velocity relation of bidirectional motion of Cin8 proposed in this study explains why directionality switching can be triggered by a multitude of different factors, as observed previously. The asymmetric response to drag explains directionality switching due to clustering of motors as well as changes in motor concentration and/or ionic strength. It readily also explains several seemingly unrelated observations previously reported for directionality switching (5–7, 9, 10, 12). First, drag acting on motors can originate not only from interactions between Cin8 motors on MTs and their interactions with MTs but also in multimotor gliding and antiparallel sliding assays, in which a large ensemble of motors interact with the same MT. In such assays, changing the surface density of molecular motors alters the binding strength of the moving MT with the motor-covered surface. This, in turn, is expected to affect the drag underlying the gliding motion. Hence, an asymmetric response of active Cin8 motion to drag also explains the directionality switching in surface gliding and antiparallel sliding assays reported for the two kinesin-5 motors Cin8 and Kip1 (6, 7, 10). Note that from our modeling perspective, drag can be generated in a multimotor MT gliding assay for full-length tetrameric and truncated dimeric proteins, as long as a sufficient number of pairs of catalytic domains are interacting with the same MT at any given time. These conclusions are further supported by recent simulations of a gliding assay (37) in which a directionality switch in the motion of gliding MTs was observed when the displacement velocity of Cin8 motors depended asymmetrically on opposing forces—which is equivalent to our basic assumption of an asymmetric response of active motion to drag. Moreover, drag acting on a motor can also result from interactions with other proteins (with or without motor activity) that are bound to MTs. Our analysis suggests that such interactions lead to directionality switching, in agreement with recent findings indicating that crowding on MTs with nonmotor proteins switches the directionality of the bidirectional S. pombe kinesin-5 Cut7 (12). Last, directionality switching has also been observed for mitotic kinesin-14 motors (13, 14), and an asymmetric response to force has been directly demonstrated for the S. cerevisiae kinesin-14 Kar3 (14). Thus, the suggested mechanism of an asymmetric response of motor motion to drag may also explain directionality switching of other kinesins, therefore providing a unified view of directionality switching of kinesin motors.
MATERIALS AND METHODS
Experimental procedures
Full-length Cin8 purification from S. cerevisiae cells as well as MT polymerization and in vitro single-molecule motility experiments were performed as previously described (10) (see sections SA1.1 and SA1.2).
Data analysis
Directionality of the MTs was assigned on the basis of plus-end labeling and/or the direction of fast-moving minus-end–directed Cin8 molecules (10). Kymographs were created using ImageJ-Fiji software. MD and MSD analyses were performed as previously described (8, 17, 27). The coordinates of motile Cin8-GFP molecules and clusters were determined with the TrackMate plugin of the ImageJ-Fiji software or by manual tracking of the intensity center over time. Only those Cin8-GFP motors (Cin8 motors refers to moving Cin8 particles of unspecified cluster size) that moved more than three pixels were considered motile, and only those with motility times >4 s were tracked. Movements of the same molecule that included >6-s stall were considered as two separate movements. The number of Cin8-GFP motors traced in each size category is indicated in Table 1 (i). The coordinates obtained were used to calculate the Cin8-GFP displacement for each time interval. The MD and the MSD values were obtained by averaging the displacements and squared displacements, respectively, calculated for all motility recordings of Cin8-GFP on the MTs. Mean velocities (v) and diffusion constants (D) were derived by fitting the average MSD values to a weighted second-order polynomial equation MSD = v2t2 + 2Dt for facilitated directional motility conditions and to the linear equation MSD = 2Dt for purely diffusive motility conditions (17). Mean velocities (v) were also derived by fitting the MD functions to the linear equation MD = vt. For each time point, the average values and the SEM are indicated (Fig. 2, A and B). MSD data were plotted with statistically weighted second-order polynomial MSD curves, by using the Origin (OriginLab) software, in such a way as to give higher weight to the initial time points, as these points represent more data. The MSD plot fitting for the motility of pairs of Cin8 molecules at 165 mM KCl and 1 mM ATP was performed after 15 s so as to exclude the initial nonlinear part of the curve. The v and D values for single Cin8 molecules, pairs of Cin8 molecules, and oligomers of Cin8 obtained from the MSD and MD analyses are summarized in Table 1 (i). Each motor was categorized as MT plus-end directed if its net displacement was in the plus-end direction and if it remained continuously plus-end-directed for at least three quarters of the length of its overall run. All other moving motors were classified as minus-end directed.
Cin8 cluster size determination
To determine the size of Cin8 clusters, we performed photobleaching experiments to determine the contribution of single GFP molecules to the total intensity of these clusters (Fig. 1A). We followed the fluorescence intensity of Cin8-GFP as a function of time within a circle of radius of four pixels, using the TrackMate plugin of the ImageJ-Fiji software (38), following correction for uneven illumination and background subtraction. We observed that single photobleaching steps, probably representing photobleaching of one GFP, caused a reduction of ~50 a.u. in fluorescence (Fig. 1A). Because each Cin8 tetramer contains four GFP molecules, all the Cin8 motors having an intensity ≤200 a.u. are likely to be single tetrameric Cin8 molecules.
The intensity distribution analysis of Cin8-GFP motors was performed on a rectangular section (75 × 42 μm) in the middle of the full field of view (79 × 66 μm), avoiding the edges of the full field of view. This analysis was consistent with the determination of the cluster size of Cin8-GFP from the photobleaching experiments. The intensities of all the fluorescent Cin8-GFP motors in the first frame of a time-lapse sequence were measured as described above. The major peak of the histogram of Cin8-GFP motor intensity distribution was fitted to a Gaussian curve (Fig. 1B). The center of the Gaussian peak lays at ~120 a.u., which was consistent with the average intensity of single Cin8 molecules containing one, two, three, or four fluorescent (nonbleached) GFP molecules, with each fluorescent GFP molecule contributing ~50 a.u. to the total intensity. Thus, the Cin8 population within this Gaussian peak, which constituted ~70% of the total Cin8 population, probably represents single Cin8 molecules. In addition, measures were taken to minimize the effect of GFP photobleaching on the determination of the Cin8 cluster size. First, we determined the lifetime of a GFP molecule before photobleaching under our experimental conditions as 26 ± 4 (SEM) s (n = 22). Consequently, on the basis of this estimation, all our motility–cluster size measurements were performed only on those Cin8 motors that moved within the first 30 s of each measurement. Last, for motile Cin8 molecules and clusters, we measured the fluorescence intensity only in the first frame of their appearance, thereby substantially reducing the likelihood of photobleaching. By this method, we assigned intensity ranges of Cin8 motor fluorescence as <200, 200 to 400, and >400 for single Cin8 molecules, pairs of Cin8 molecules, and Cin8 oligomers, respectively.
For cluster size versus velocity analysis (Fig. 5B), the velocity of each Cin8-GFP particle was determined by measuring the slope of each end-to-end motility event on the kymograph. The intensities of MT-bound Cin8-GFP motors were measured in the same manner as for the intensity distribution analysis described above, but only for the motile MT-attached Cin8 motors. The intensity profile of MT-bound Cin8-GFP motors was determined using line-scan analysis in ImageJ-Fiji software and plotted using the Origin (Originlab) software (Fig. 7A).
Theoretical modeling and simulations
Modeling of the diffusive component of Cin8 motion
In the module that captures the diffusive behavior, we considered each single, noninteracting Cin8 molecule as a random walker moving bidirectionally on the MT lattice with equal hopping rates to the plus and minus ends; the hopping rate was obtained from the measured diffusion coefficient for single Cin8 molecules (Fig. 2B and Table 1, i). We assumed that Cin8 molecules interact via a weak and constant force Fint if they are within a given interaction range R of each other (Fig. 3A; see also section SB1.3 for a detailed description of this choice). As dictated by equilibrium statistical physics (detailed balance), this force must exert an effect on the hopping rates of interacting Cin8 motors through a Boltzmann factor e−ΔE/kBT, where ΔE = Fint a is the energy required to move two motors a distance of a = 8.4 nm (the length of a tubulin dimer) apart [see section SB1 for a technical description of the stochastic model and the numerical implementation of the stochastic particle dynamics using a Gillespie algorithm (39)]. An in silico parameter scan over different values for the magnitude of the attractive force and the interaction range yielded two crucial insights. First, our simulations strongly suggested an interaction range of approximately R = 25 nm, because smaller or larger values led to unstable clusters or very large clusters, respectively, which is at variance with experimental observations (see fig. SB3 and section SB2 for the corresponding simulation results with different values of R). Notably, this interaction range is in accordance with the length of kinesin-5 motors [approximately 80 nm (24–26, 40)]. Second, we observed that increasing the attractive force Fint between Cin8 molecules indeed causes a strong decrease in the diffusion coefficient of Cin8 pairs (as shown in Fig. 3B). This monotonic decrease originates from the fact that the interaction between two motors effectively acts as a load or drag when the motors move away from each other. As a consequence, increasing Cin8 interaction strength decreases the diffusive motion of a Cin8 pair or cluster.
Modeling of the active component of Cin8 motion
To account for both the diffusive and active components of Cin8 motility, we expand the theoretical model based on the following assumptions (Fig. 3C). First, motivated by the difference in diffusion coefficients of single Cin8 motors in the presence of ATP and ADP observed in vitro (Table 1, i), we hypothesized that single Cin8 motors show bidirectional active motion toward both ends (red/pink arrows in Fig. 3C). As single Cin8 molecules were found to be biased toward the minus end in the experiments, such active bidirectional motion must also be biased in this direction in the absence of external forces. Second, as a basic mechanism that could potentially switch the directionality of clusters (Fig. 2A and fig. SA1a), we hypothesized that motors that are moving toward the plus and minus ends, respectively, respond differently to forces that oppose this motion (collectively termed drag). Specifically, we assumed that drag suppresses active motion toward the minus end more effectively than movement toward the plus end; see the illustration in Fig. 3C and the corresponding force-velocity relations (Fig. 3C, bottom). Note that an asymmetric impact of drag on active forward and backward stepping very similar to that suggested here was recently observed experimentally for the kinesin-14 Kar3 of S. cerevisiae (14). Drag has different effects on the Brownian and the active components of the model. While it symmetrically reduces Brownian motion without introducing any bias toward either end of the MT, the asymmetric drag response of active motion leads, for large enough values, to a reversal of the existing bias at zero force (toward the minus end).
Acknowledgments
Funding: This research was supported in part by the Israel Science Foundation grant (ISF-386/18) awarded to L.G.; the NSF (NSF-1615991) and United States–Israel Binational Science Foundation grant (BSF-2015851), awarded to L.G. and J.A.-B.; the NIH (grant NIH-R01-GM11283), awarded to J.A.-B.; and the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) via project B02 awarded to E.F. within the Collaborative Research Center “Forces in Biomolecular Systems” (SFB 863, project ID 111166240). Author contributions: H.P., E.R., A.G.-L., J.A.-B., E.F., and L.G. designed the research. H.P., E.R., A.G.-L., E.F., and L.G. performed the research. H.P., E.R., J.A.-B., E.F., and L.G. analyzed the data. H.P., A.G.-L., and L.G. designed and carried out the experiments. H.P., J.A.-B., and L.G. analyzed the experimental data. E.R. and E.F. designed the theoretical models and performed the corresponding mathematical analyses. H.P., E.R., E.F., and L.G. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabc1687/DC1
REFERENCES AND NOTES
- 1.Mann B. J., Wadsworth P., Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 29, 66–79 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Goulet A., Moores C., New insights into the mechanism of force generation by kinesin-5 molecular motors. Int. Rev. Cell Mol. Biol. 304, 419–466 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Singh S. K., Pandey H., Al-Bassam J., Gheber L., Bidirectional motility of kinesin-5 motor proteins: Structural determinants, cumulative functions and physiological roles. Cell. Mol. Life Sci. 75, 1757–1771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edamatsu M., Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem. Biophys. Res. Commun. 446, 231–234 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Fridman V., Gerson-Gurwitz A., Shapira O., Movshovich N., Lakämper S., Schmidt C. F., Gheber L., Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 126, 4147–4159 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Roostalu J., Hentrich C., Bieling P., Telley I. A., Schiebel E., Surrey T., Directional switching of the Kinesin cin8 through motor coupling. Science 332, 94–99 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D. R., Schmidt C. F., Gheber L., Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30, 4942–4954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapira O., Gheber L., Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci. Rep. 6, 25597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallesen T., Roostalu J., Duellberg C., Pruessner G., Surrey T., Ensembles of bidirectional kinesin Cin8 produce additive forces in both directions of movement. Biophys. J. 113, 2055–2067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapira O., Goldstein A., Al-Bassam J., Gheber L., A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J. Cell Sci. 130, 725–734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edamatsu M., Molecular properties of the N-terminal extension of the fission yeast kinesin-5, Cut7. Genet. Mol. Res. 15, 15017799 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Britto M., Goulet A., Rizvi S., von Loeffelholz O., Moores C. A., Cross R. A., Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, E7483–E7489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popchock A. R., Tseng K.-F., Wang P., Karplus P. A., Xiang X., Qiu W., The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat. Commun. 8, 13999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molodtsov M. I., Mieck C., Dobbelaere J., Dammermann A., Westermann S., Vaziri A., A force-induced directional switch of a molecular motor enables parallel microtubule bundle formation. Cell 167, 539–552.e14 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Blackwell R., Edelmaier C., Sweezy-Schindler O., Lamson A., Gergely Z. R., O’Toole E., Crapo A., Hough L. E., McIntosh J. R., Glaser M. A., Betterton M. D., Physical determinants of bipolar mitotic spindle assembly and stability in fission yeast. Sci. Adv. 3, e1601603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapira O., Goldstein A., Al-Bassam J., Gheber L., Regulation and possible physiological role of BI-directional motility of the kinesin-5 Cin8. Biophys. J. 110, 460a (2016). [Google Scholar]
- 17.Kapitein L. C., Kwok B. H., Weinger J. S., Schmidt C. F., Kapoor T. M., Peterman E. J. G., Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 182, 421–428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbrugge S., van den Wildenberg S. M., Peterman E. J., Novel ways to determine kinesin-1’s run length and randomness using fluorescence microscopy. Biophys. J. 97, 2287–2294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yajima J., Alonso M. C., Cross R. A., Toyoshima Y. Y., Direct long-term observation of kinesin processivity at low load. Curr. Biol. 12, 301–306 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Schnitzer M. J., Block S. M., Kinesin hydrolyses one ATP per 8-nm step. Nature 388, 386–390 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Cooper J. R., Wordeman L., The diffusive interaction of microtubule binding proteins. Curr. Opin. Cell Biol. 21, 68–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., Hyman A. A., XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J., The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441, 115–119 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Scholey J. E., Nithianantham S., Scholey J. M., Al-Bassam J., Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. eLife 3, e02217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acar S., Carlson D. B., Budamagunta M. S., Yarov-Yarovoy V., Correia J. J., Niñonuevo M. R., Jia W., Tao L., Leary J. A., Voss J. C., Evans J. E., Scholey J. M., The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 4, 1343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashina A. S., Baskin R. J., Cole D. G., Wedaman K. P., Saxton W. M., Scholey J. M., A bipolar kinesin. Nature 379, 270–272 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duselder A., Fridman V., Thiede C., Wiesbaum A., Goldstein A., Klopfenstein D. R., Zaitseva O., Janson M. E., Gheber L., Schmidt C. F., Deletion of the Tail Domain of the Kinesin-5 Cin8 affects its directionality. J. Biol. Chem. 290, 16841–16850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiede C., Fridman V., Gerson-Gurwitz A., Gheber L., Schmidt C. F., Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. Bioarchitecture 2, 70–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endow S. A., Higuchi H., A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature 406, 913–916 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Case R. B., Pierce D. W., Hom-Booher N., Hart C. L., Vale R. D., The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90, 959–966 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Endow S. A., Waligora K. W., Determinants of kinesin motor polarity. Science 281, 1200–1202 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Henningsen U., Schliwa M., Reversal in the direction of movement of a molecular motor. Nature 389, 93–96 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D., A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Shastry S., Hancock W. O., Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253–16258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muretta J. M., Jun Y., Gross S. P., Major J., Thomas D. D., Rosenfeld S. S., The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc. Natl. Acad. Sci. U.S.A. 112, E6606–E6613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell K. M., Cha H. K., Sindelar C. V., Cochran J. C., The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner. J. Biol. Chem. 292, 14680–14694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito N., Kaneko K., Embedding dual function into molecular motors through collective motion. Sci. Rep. 7, 44288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tinevez J.-Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L., Eliceiri K. W., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Gillespie D. T., Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 58, 35–55 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Hildebrandt E. R., Gheber L., Kingsbury T., Hoyt M. A., Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 281, 26004–26013 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Edelstein A. D., Tsuchida M. A., Amodaj N., Pinkard H., Vale R. D., Stuurman N., Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou T., Mallick K., Zia R. K. P., Non-equilibrium statistical mechanics: From a paradigmatic model to biological transport. Rep. Prog. Phys. 74, 116601 (2011). [Google Scholar]
- 44.Hyman A. A., Chrétien D., Arnal I., Wade R. H., Structural changes accompanying GTP Hydrolysis in microtubules: Information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J. Cell Biol. 128, 117–125 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Block S. M., Asbury C. L., Shaevitz J. W., Lang M. J., Probing the kinesin reaction cycle with a 2D optical force clamp. Proc. Natl. Acad. Sci. U.S.A. 100, 2351–2356 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreasson J. O. L., Milic B., Chen G.-Y., Guydosh N. R., Hancock W. O., Block S. M., Examining kinesin processivity within a general gating framework. eLife 4, e07403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teimouri H., Kolomeisky A. B., Mehrabiani K., Theoretical analysis of dynamic processes for interacting molecular motors. J. Phys. A Math. Theor. 48, 065001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter N. J., Cross R. A., Mechanics of the kinesin step. Nature 435, 308–312 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Schnitzer M. J., Visscher K., Block S. M., Force production by single kinesin motors. Nat. Cell Biol. 2, 718–723 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Svoboda K., Block S. M., Force and velocity measured for single kinesin molecules. Cell 77, 773–784 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Visscher K., Schnitzer M. J., Block S. M., Single kinesin molecules studied with a molecular force clamp. Nature 400, 184–189 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Hua W., Young E. C., Fleming M. L., Gelles J., Coupling of kinesin steps to ATP hydrolysis. Nature 388, 390–393 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/6/eabc1687/DC1


