Abstract
Factor-dependent transcription termination mechanisms are poorly understood. We determined a series of cryo-electron microscopy structures portraying the hexameric ATPase ρ on path to terminating NusA/NusG-modified elongation complexes. An open ρ ring contacts NusA, NusG, and multiple regions of RNA polymerase, trapping and locally unwinding proximal upstream DNA. NusA wedges into the ρ ring, initially sequestering RNA. Upon deflection of distal upstream DNA over the RNA polymerase Zinc-binding domain, NusA rotates underneath one capping ρ subunit, which subsequently captures RNA. Following detachment of NusG and clamp opening, RNA polymerase loses its grip on the RNA:DNA hybrid and is inactivated. Our structural and functional analyses suggest that ρ and other termination factors across life may utilize analogous strategies to allosterically trap transcription complexes in a moribund state.
One Sentence Summary:
Structure-function analyses reveal how termination factor ρ captures and inactivates a transcription elongation complex.
Pervasive transcription of cellular genomes is kept in check by surveillance mechanisms that ensure that synthesis of unwanted RNAs is terminated early. In bacteria, this function is performed by ρ, originally identified as a factor that terminates transcription in Escherichia coli bacteriophage λ (1). E. coli ρ defines boundaries of many transcription units (2), silences horizontally-acquired genes and antisense RNAs (2–4), removes stalled RNA polymerase (RNAP) from the path of the replisome to maintain chromosome integrity (5), and inhibits R-loop formation (6). Five decades of mechanistic studies of E. coli ρ led to a model in which its motor activity takes a center stage. ρ is a hexameric ring-shaped RecA-family RNA translocase that exists in open and closed states capable of loading onto RNA and translocation, respectively (7). A ρ monomer is composed of two domains. The N-terminal domain (NTD) contains a primary RNA-binding site (PBS) that engages unstructured C-rich ρ-utilization (rut) sites; the C-terminal domain (CTD) contains the secondary RNA-binding site (SBS) and ATPase/translocase determinants. Following rut recognition, the ring closes, trapping RNA at the SBSs in a central pore (7). The closed hexamer engages in ATP-powered 5’-to-3’ translocation along the RNA towards RNAP, maintaining contacts to the rut RNA, a race described as “kinetic coupling” (8). When RNAP pauses, ρ catches up and dissociates an otherwise very stable elongation complex (EC) by a still-debated mechanism (9).
Primed by a canonical rut site, ρ terminates transcription by phage and eukaryotic RNAPs (10, 11) and displaces streptavidin from a biotin anchor (12), arguing that ρ could dissociate any EC. However, in context of the physiological mechanism, glaring discrepancies have been noted. For example, ρR353A is severely defective in ring closure but terminates efficiently, whereas ρW381A closes readily but has termination defects (13, 14). A lack of perfect correlation among ATPase, helicase, and termination activities suggests that ρ motor and termination functions are separable and that ρ/RNAP interactions, first reported in 1984 (15), may control termination. Direct interactions with RNAP would also explain how ρ is targeted to actively-synthesized RNAs and excluded from completed transcripts. Furthermore, elongation factors NusA and NusG modulate termination. NusA stimulates ρ binding to RNAP (15), yet paradoxically delays termination in vitro (16). NusG promotes early termination (17); it allosterically stimulates ring closure (13, 18), enabling ρ to act at non-canonical sites (2). In support of ρ trafficking with the EC in vivo, ChIP-chip analysis showed that ρ and NusA bind to RNAP immediately after promoter escape, with NusG lagging behind (19). An allosteric model, in which ρ is recruited to RNAP rather than RNA and traps the EC in an inactive state prior to dissociation (20), explains how ρ is excluded from transcripts that have been released from RNAP. However, while RNAP substitutions that confer resistance to ρ are known (8), they are unlikely to alter RNAP binding to ρ. Instead, these mutant RNAPs are insensitive to pauses and are thought to simply outrun ρ.
To reveal ρ action in the context of complete E. coli ρ/NusA/NusG/rut ECs (ρ-ECs), we elucidated their atomic structures by single-particle cryo-electron microscopy (cryoEM) and conducted structure-guided functional analyses. Our data are consistent with a series of steps along a termination pathway, in which ρ allosterically inactivates the EC via interactions with RNAP, NusA, NusG, upstream DNA and rut RNA.
Results
NusA and NusG are the only general elongation factors that modulate ρ
Six general elongation factors are present in E. coli: NusA, NusG, cleavage factors GreA/B, recycling factor RapA, and transcription-repair coupling factor Mfd. We assessed their potential effects in vitro on a DNA template encoding bacteriophage λ tR1, an archetypical ρ-dependent terminator (Fig. S1A and S2A). In the absence of other proteins, RNAP generated predominantly readthrough (RT) transcripts. ρ alone promoted termination at several sites, NusG stimulated RNA release at promoter-proximal sites, whereas NusA shifted the termination window downstream (Fig. S1A). By contrast, Gre factors, RapA and Mfd did not alter the efficiency or pattern of ρ-dependent termination (Fig. S1A). We conclude that a minimal system to study termination comprises EC, NusA, NusG and ρ.
Assembly and structural analysis of ρ-ECs
While ECs are readily amenable to structural studies, RNAP dissociates rapidly once committed to termination. We assembled ECs on a DNA scaffold with a 15-base pair (bp) downstream DNA (dDNA), a 9-nucleotide (nt) bubble, and a 30-bp upstream DNA (uDNA). The 99-nt RNA contained the λ tR1 rut region (also used in all transcription assays; Fig. S1A), which is followed by a well-defined ρ release window on long templates (Fig. S2). However, to capture the metastable complex prior to dissociation, in this scaffold RNAP is poised at the upstream edge of the ρ termination region. ρ-ECs were assembled stepwise with Nus factors, incubated with the ATP analog, ADP-BeF3, that supports ρ ring closure (7) and subjected to single-particle cryoEM analysis without cross-linking (Fig. S3–S9). From ~10,000 micrographs, we picked ~2,100,000 particle images, ~390,000 of which contained ρ; multi-particle 3D refinement (21) led to nine cryoEM maps, corresponding to complexes I-V, IΔNusG, IIIΔNusG, IIIa and IVa (Fig. S4). The local resolution varied from below 3 Å in some core regions to 8–12 Å in some peripheral elements (Fig. S5–S7; Table S1). C-terminal regions of NusA were tentatively placed into weakly-defined cryoEM density in the region where they reside in other ECs (22–24). While backbones of all other described elements could be traced unequivocally, assignment of side chain conformations is tentative at the present resolutions. Therefore, in the following narrative we used individual residues mostly as landmarks of specific regions.
Our cryoEM structures can be sorted along a pathway in which ρ initially engages the EC, is then primed for rut RNA binding, subsequently captures rut RNA, and finally inactivates RNAP (Fig. S1B–J). In the following, we describe the main structures (complexes I-V) individually along this presumed sequence of events and then discuss how additional structures fit into the picture. We encourage the reader to view animated versions of the process (Movie S1; Movie S2) first.
EC engagement
Domain structures of NusA and NusG are shown in Fig. S2C, and Table S2 lists relevant regions of RNAP and factors. In complex I (Fig. 1A; Fig. S1B; Fig. S9; Movie S1), RNAP (α2ββ’ω subunit composition) assumes a conformation observed in an unmodified post-translocated EC (25) (root-mean-square deviation [RMSD] of 1.24 Å for 2,687 pairs of aligned Cα atoms; Fig. 1B). NusGNTD is bound at its canonical site (26) next to proximal uDNA (Fig. 2A). NusANTD is sandwiched between the β flap tip (FT) and α1CTD, as in a NusA-modified hairpin-paused EC (22) (Fig. 1A). The NusA S1-KH RNA-binding region and AR1 extend outwards across β’ Zinc binding domain (β’ZBD), whereas AR2 angles down towards ω. Additional contacts of AR2 to α2CTD observed in (22) are possible, and would explain how in our structures AR2 is displaced from an auto-inhibitory position on NusAS1-KH in isolated NusA (27), but are not clearly resolved in the map.
Fig. 1. Engagement.
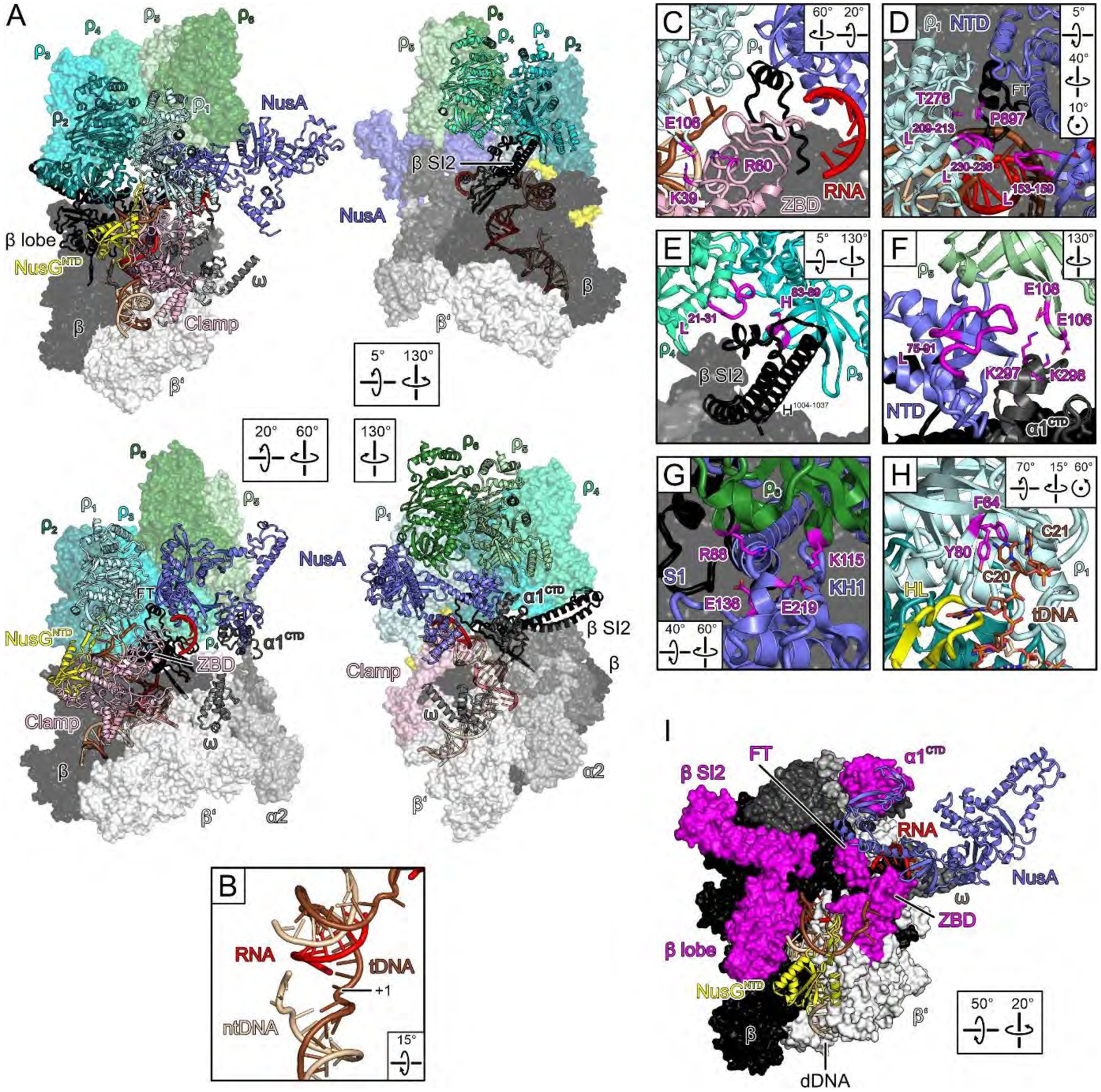
(A) Semi-transparent surface/cartoon representations of the engagement complex, highlighting contact sites of ρ subunits. Rotation symbols in this and the following figures indicate views relative to (A), upper left. (B) Post-translocated state of the nucleic acids at the active site; tDNA, template DNA; ntDNA, non-template DNA; +1, template nucleotide pairing with the next incoming NTP. (C-H) Close-up views of ρ/EC contacts. Elements discussed in the text, magenta. (I) ρ-cutaway view; ρ-contacting RNAP elements around the RNA exit, magenta.
Fig. 2. Effects of ρ PBS/SBS ligands and NusGNTD.
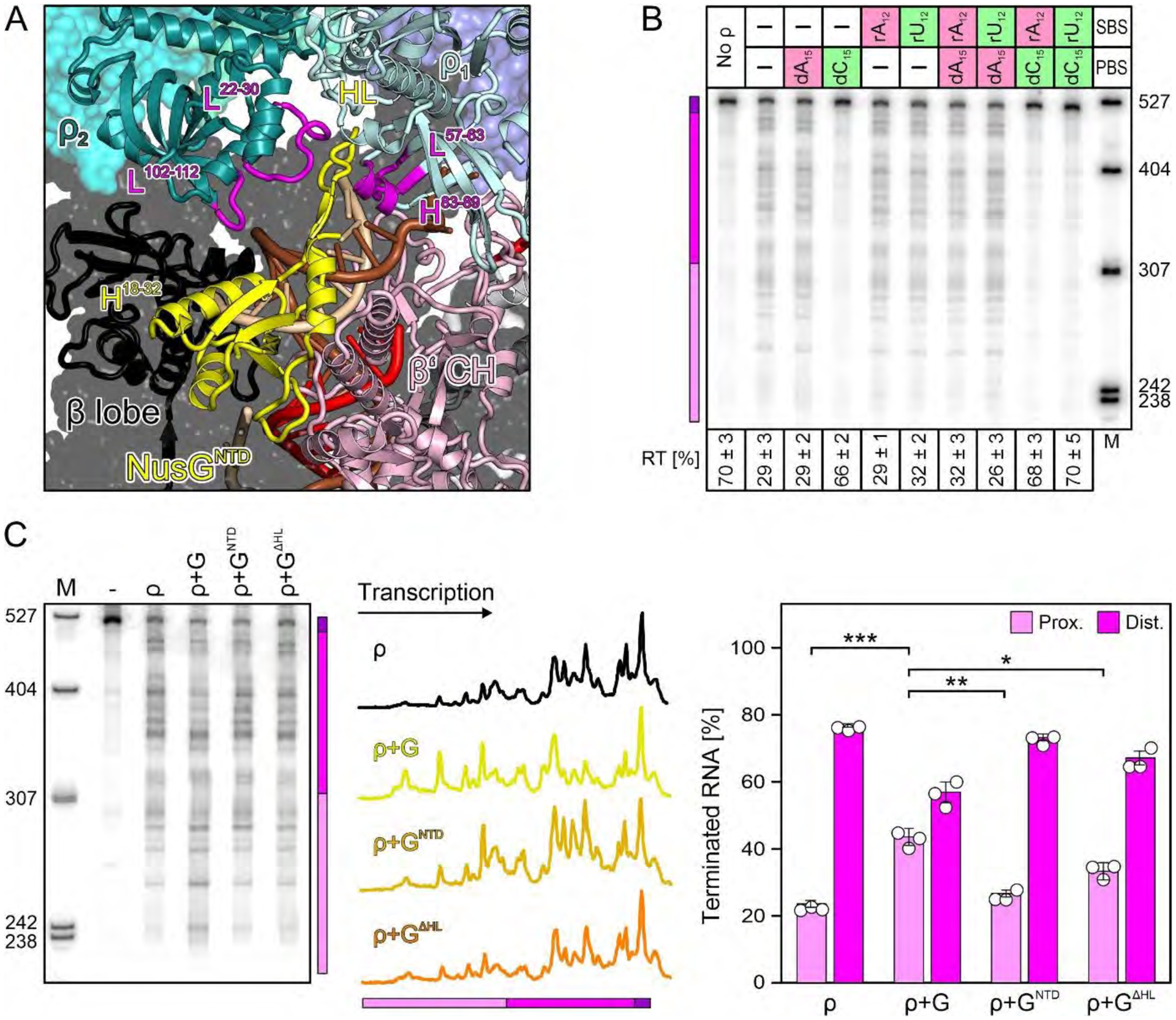
(A) Effects of optimal (green) and poor (red) PBS/SBS ligands on ρ termination; here and in other figures, positions of proximal (pink) and distal (magenta) terminated RNAs and the read-through transcript (RT; purple) are indicated with a colored bar. PBS (dN15) ligands were present at 5 μM, SBS (rN12) ligands at 500 nM. A fraction of RT versus the sum of all RNA products is shown at the bottom. Values represent means ± SD of three independent experiments. (B) Close-up view on NusGNTD in the engagement complex. Elements discussed in the text, magenta. (C) Modulation of ρ effects by the indicated NusG (“G”) variants. The center panel shows lane profiles from the gel on the left; the Y-axis signals were normalized based on the total signal in that lane. The right panel shows a distribution of ρ-terminated RNAs between the proximal and distal regions. Values represent means ± SD of three independent experiments. * P<0.01; ** P<0.001; *** P<0.0001 [unpaired Student’s t-test].
Despite the presence of ADP-BeF3 and NusG (13), ρ adopts an open-ring conformation and binds above the active site cleft around the β flap, with ρNTDs oriented towards RNAP (Fig. 1A). We modeled ADP-BeF3 at the five intact nucleotide binding sites in this and other complexes. Looking from CTD to NTD, we labeled the protomers clockwise ρ1-ρ6, starting at the ring opening (Fig. 1A), ρ1NTD lies next to β’ZBD, with β’ZBD-K39/R60 forming electrostatic contacts with ρ1E106 (Fig. 1C). One edge of ρ1CTD (T276) is positioned next to βFT-P897 opposite NusANTD (Fig. 1D). Loop209−213 and loop230−236 of ρ1CTD contact loop153−159 of NusAS1 (Fig. 1D). The hairpin loop (HL) of NusGNTD is bent over the proximal uDNA, sandwiched between loop57−63 and helix83−89 of ρ1NTD and loop22−30 of ρ2NTD (Fig. 2A). Loop102−112 of ρ2PBS lies on top of NusGNTD helix18−32, while the ρ2PBS cavity hovers above the β lobe/protrusion (Fig. 2A). ρ3PBS accommodates helix1004−1037 of the lineage-specific β SI2 insertion, while neighboring edges of the NTDs of ρ3 (helix83−89) and ρ4 (loop21−31) sandwich the globular tip of SI2 (Fig. 1E). ρ5PBS binds the protruding loop75−91 of NusANTD, and ρ5NTD-E106/E108 form an electrostatic network with α1CTD-K297/K298 (Fig. 1F). ρ6 does not directly contact RNAP; instead, ρ6PBS rests on NusAS1-KH1, opposite ρ1CTD, with direct ρ6R88-S1E136 and ρ6K115-KH1E219 contacts (Fig. 1G). Thus, ρ subunits engage multiple RNAP elements (FT, ZBD, lobe, SI2, α1CTD), NusGNTD and NusANTD-S1-KH1, which are circularly arranged around the RNA exit tunnel, matching the spiral pitch of the open ρ ring (Fig. 1I).
Multifaceted contacts with the EC may enable ρ to achieve a precisely tuned termination activity. For example, SI2 may be important for initial ρ recruitment, in which case its deletion should suppress termination, but SI2 blocks ρ3PBS (Fig. 1E) and helps stabilize ρ in an open conformation, such that its deletion should promote termination. We found that SI2 deletion clearly shifted ρ termination to more promoter-proximal sites in vitro (Fig. S10A). Interestingly, an opposite effect of ΔSI2 is observed in vivo (Fig. S10B), supporting the idea of fine tuning, e.g., by changes in the chemical environment. NusA is also expected to exert opposing effects. While observed ρ-NusA contacts and gel filtration data (Fig. S2D) are in line with a reported contribution of NusA to ρ recruitment (15), NusA also hinders ρ ring closure: the S1 and KH1 domains are wedged between ρ1 and ρ6, with the βFT/NusANTD/α1CTD array additionally stabilizing the ρ spiral (Fig. 1A). Furthermore, a clear but poorly contoured region of density above the RNA exit tunnel opening indicates flexible exiting RNA guided between NusAS1 and β’ZBD (Fig. 1C). Thus, NusA keeps the ρ ring open and, acting with β’ZBD, may sequester exiting RNA from ρ, as suggested previously (28). Both these effects could explain how NusA delays ρ termination observed by us (Fig. S1A) and others (16, 17).
A striking feature of complex I is continuous density, corresponding to single-stranded template DNA (tDNA) that extends from the proximal uDNA into ρ1PBS (Fig. 1H; Fig. 2A). The finding that ρ ATPase activity is stimulated by DNA ligands that can bind to PBS but not SBS (29) are commonly used to distinguish the PBS and SBS effects, and DNA-PBS interactions were observed in structures (30), yet presumed to be artifactual. We used dN15 and rN12 oligomers specific for the PBS and SBS, respectively, to assess the importance of ρ-DNA interactions. Our results show that dC15, the optimal PBS ligand (31), strongly inhibits termination (Fig. 2B) when present alone or with the SBS ligands. By contrast, dA15, which does not bind PBS, or rU12, a canonical SBS ligand (31), had no effect on ρ activity. These results support a model in which ρPBS interactions with tDNA are functionally important. However, it is also possible that dC15 oligomers could compete with the nascent RNA at a later step in the pathway. Capture of uDNA would be expected to hinder continuous DNA movement through RNAP, revealing a first mechanism by which ρ can inhibit RNAP.
NusGHL is pushed against and displaces the complementary non-template (nt) strand (Fig. 2A). To test if HL contributes to termination, we replaced NusG residues 47–63 with Gly2 and evaluated its effect in vitro. In the absence of NusG, ρ predominantly releases longer RNAs (distal region, magenta in Fig. 2C). Consistent with published reports (13, 17), the wild-type (WT) NusG shifted the termination window upstream: the fraction of proximal ρ-terminated RNAs increased from 24 to 43 % (violet in Fig. 2C). NusGΔHL was partially defective in stimulating early termination (33 %), whereas the isolated NTD was almost completely inactive (27 %), as shown previously (13, 32). Based on these findings, we interpret complex I as an engagement complex, from which ρ can trigger further steps towards termination.
Priming for RNA capture
In complex II, RNAP, NusGNTD, the hybrid, dDNA, proximal uDNA, and ρ1-ρ3 subunits are essentially unaltered. However, a drastic rotation of NusANTD/βFT towards αNTDs is observed (Fig. 3A–C), and NusANTD-βFT interactions change upon repositioning (Fig. 4A). The tip of NusANTD moves from ρ5PBS to ρ4PBS, with concomitant handover of NusANTD from α1CTD to α2CTD, which consolidates the NusANTD-ρ4PBS interaction (Fig. 3B,C). NusAS1 now resides underneath ρ6PBS (Fig. 3B), and loop213−221 of NusAKH1 is inserted between helix83−89 of ρ5 and loop22−30 of ρ6 (Fig. 4B). As NusA moves underneath, ρ4-ρ6 are slanted upwards (Fig. 3C).
Fig. 3. Priming.
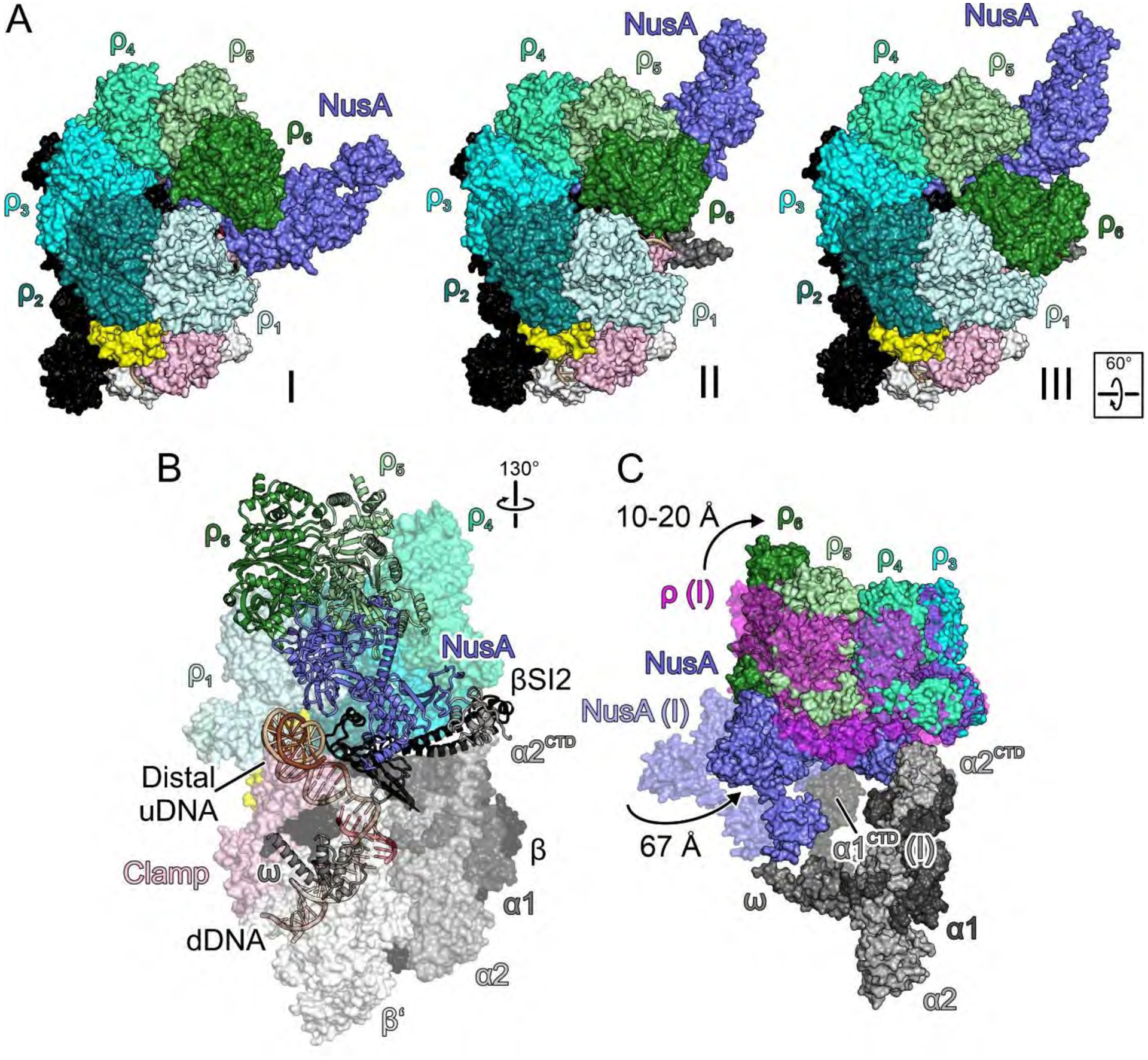
(A) Surface views of the engagement (I), primed (II) and RNA capture (III) complexes, illustrating rotation of NusA underneath ρ6 (I to II) and shift of ρ6 from ρ5 to ρ1 (II to III). (B) Semi-transparent surface/cartoon representations of the primed complex, highlighting contact sites of ρ subunits and distal uDNA on top of β’ZBD. (C) Overlay of selected elements of the primed complex (solid surfaces) and engagement complex (semi-transparent surfaces; ρ, magenta), highlighting movements of NusA and ρ, and handover of NusANTD from α1CTD to α2CTD.
Fig. 4. NusA interactions.
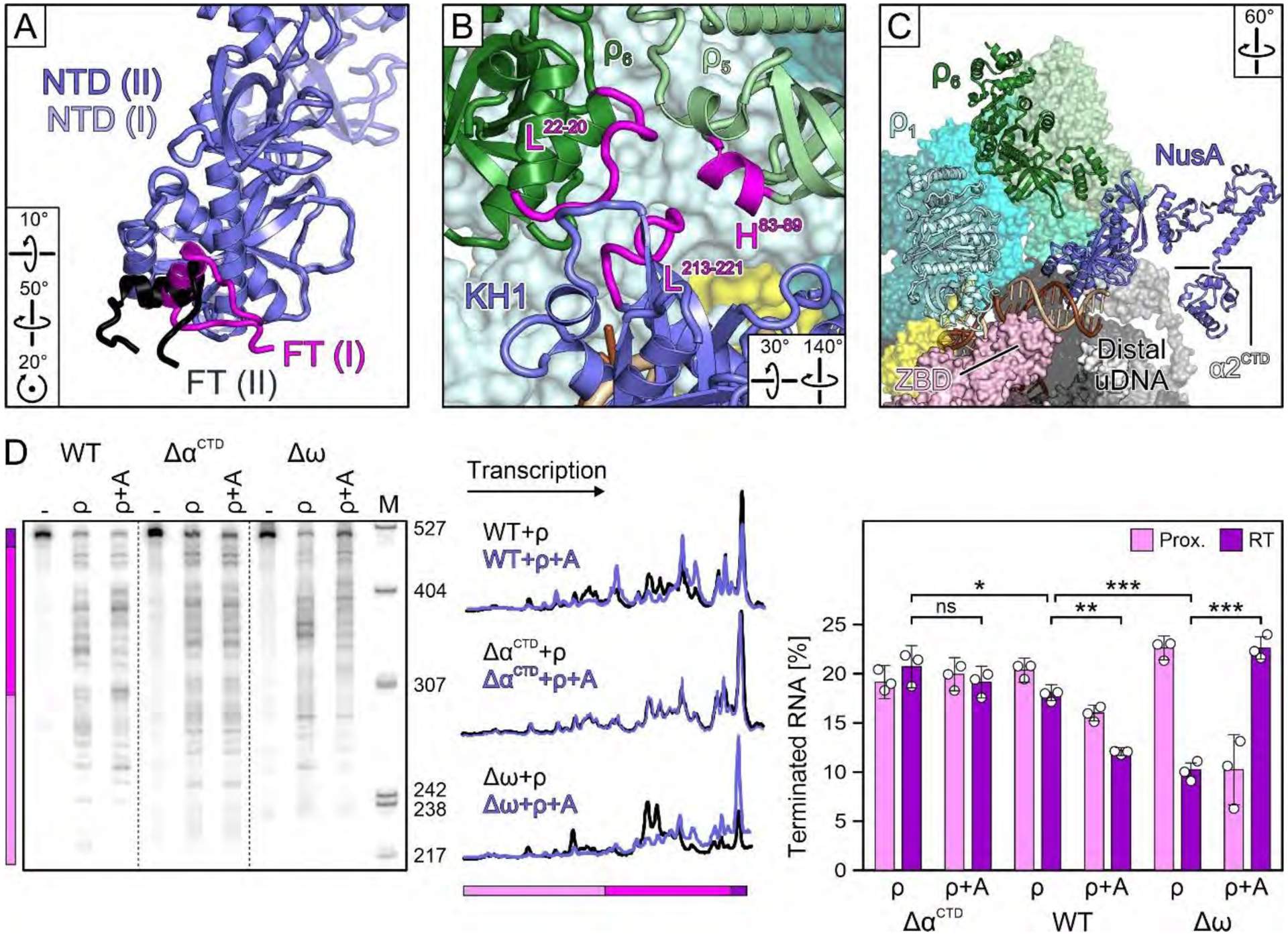
(A) Comparison of βFT-NusANTD interactions in the primed and engagement complexes, after superposition of NusANTDs. (B) ρ5/ρ6/NusAKH1 interaction network in the primed complex. (C) Correlation of accommodation of distal uDNA on the β’ZBD and NusA rotation underneath ρ6 in the primed complex. (D) NusA (“A”) effects on termination by WT RNAP, or RNAP variants lacking αCTDs or ω; dashed lines indicate spliced images. The RNA fractions are means ± SD of three independent experiments. ns, not significant; * P<0.1; ** P<0.001; *** P<0.0001.
While NusA has moved away from β’ZBD, the distal uDNA duplex is running across the ZBD (Fig. 4C). Thus, the transition to complex II might be fueled by competition of distal uDNA and NusA for β’ZBD, as well as by the interchangeability of the NusANTD/ρ5/α1CTD (complex I) and NusANTD/ρ4/α2CTD (complex II) interaction networks. Consistent with an earlier report (33), we found that deletion of αCTDs modestly inhibited termination while essentially eliminating the effect of NusA (Fig. 4D). In stark contrast, deletion of the ω subunit potentiated ρ termination and the NusA effect thereon (Fig. 4D). As NusAAR2 approaches ω in complex I (Fig. 1A) and as this interaction is broken in complex II, ω deletion may assist the transition to complex II.
ρ6PBS hovers some 45 Å above β’ZBD and is not bound to RNA (Fig. 4C), but a weak neighboring density (not modeled) might indicate an approaching RNA. Thus, we consider complex II to be primed for RNA capture by ρ.
RNA capture
Upon transition to complex III, RNAP, dDNA, the hybrid, proximal uDNA, NusGNTD, NusA and ρ subunits 1–5 remain unaltered. In contrast, ρ6 detaches from ρ5, steps down by about 45 Å from on top of NusAKH1 in the primed complex to β’ZBD, displacing distal uDNA, and links up with ρ1 (Fig. 3A; Fig. 5A). ρ6 now interacts laterally with NusAS1 as does ρ1 in the engagement complex (Fig. 5B). The ring opening thereby migrates from ρ1/ρ6 to ρ6/ρ5.
Fig. 5. RNA capture.
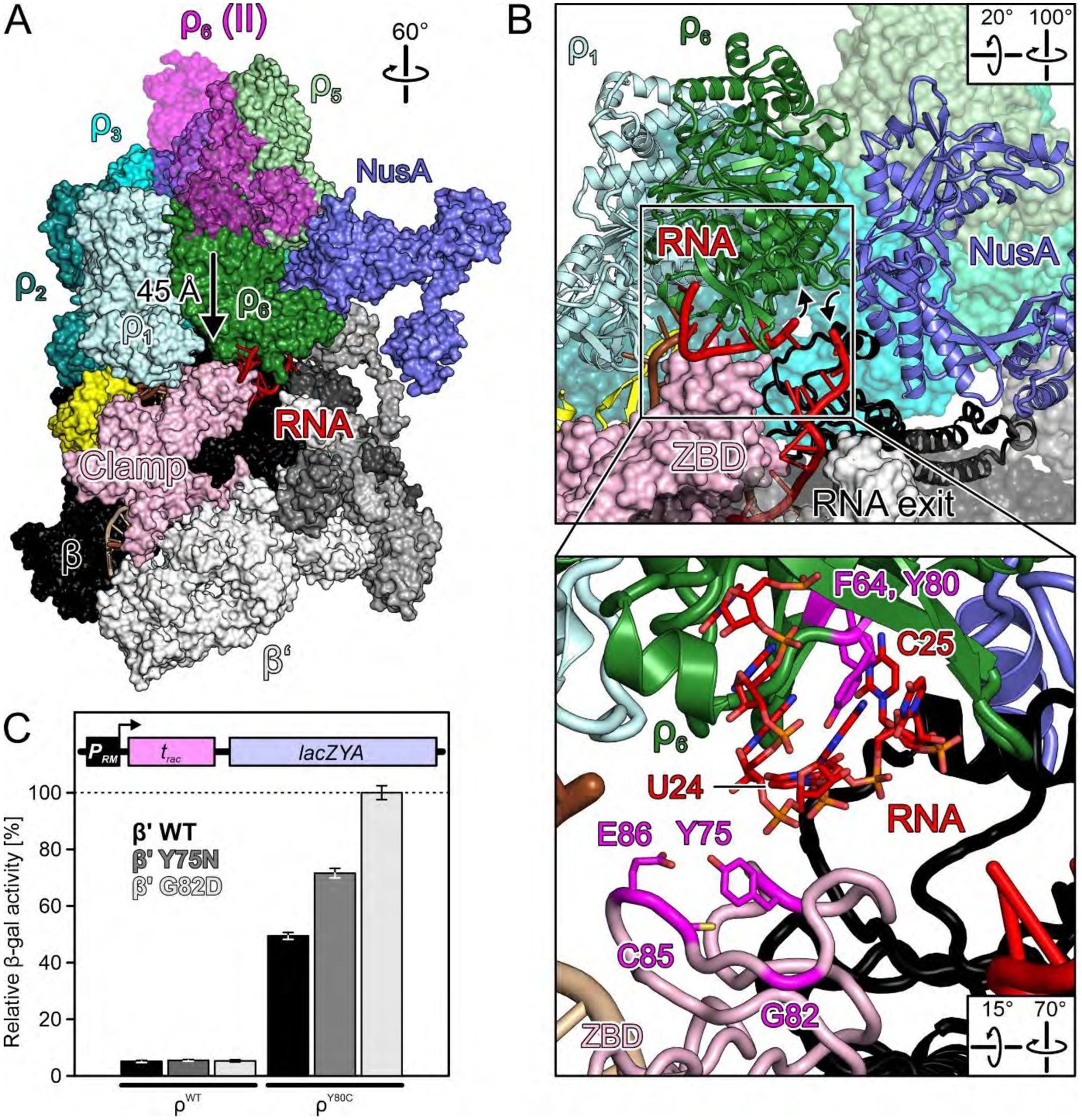
(A) Surface view of the RNA capture complex (nucleic acids as cartoon) with superimposed ρ6 from the primed complex. Arrow, movement of ρ6 during the transition from the primed to the RNA capture state. (B) Close-up views on ρ6PBS with bound RNA. Angled arrows, direction of intervening RNA region that might ascend 5’-to-3’ through the open ρ ring and return on the outside. Inset, details of RNA binding at ρ6PBS. 5’-portion of the RNA and selected ρ6PBS residues as sticks colored by atom type. In this and the following figures: Carbon RNA, red; carbon ρ residues, magenta; oxygen, light red, nitrogen blue; phosphorus, orange. (C) Quantification of β-gal activity derived from a reporter construct (scheme) in cells with ρWT or ρY80c, in the presence of the indicated plasmid-encoded β’ variants. Values represent means ± SEM of at least nine independent experiments.
ρ6PBS captures two nucleotides of rut RNA and sandwiches them with the underlying β’ZBD, while a rather featureless density next to β’ZBD above the RNA exit represents exiting RNA (Fig. 5B). It can be envisaged that, as ρ6 steps down onto β’ZBD, portions of RNA between exiting RNA and the captured rut nucleotides are funneled into the open ρ ring (Fig. 5B). With the known pyrimidine preference of ρPBS (7, 30), we, therefore, tentatively assigned U24 and C25 from the upstream rut site (Fig. S2B) as the ρ6PBS ligands. We term complex III the RNA capture complex, as ρ engages RNA for the first time.
The ZBD/RNA/ρ6PBS contacts observed in complex III suggest that ρ PBS variants could have synergistic defects with β’ZBD variants. We screened for synthetic termination defects of β’ variants in the presence of ρY80C that weakens rut affinity (34). We randomly mutagenized the rpoC gene on a plasmid and transformed the mutant library into E. coli ρWT or ρY80C strains containing a chromosomal PRM-racR-trac-lacZYA reporter fusion. trac is a NusG-dependent terminator at which ρY80C exhibits a milder defect (35). Screening yielded a β’G82D ZBD variant with a two-fold enhanced termination defect in combination with ρY80C (Fig. 5C). A previously reported β’Y75N substitution (36) had a similar effect (Fig. 5C). Many additional β’ZBD variants constructed by site-directed mutagenesis, particularly C72H, C85H and E86K, showed synthetic growth defects with ρY80C (Fig. S10C, Table S3). The affected residues reside on the upper ZBD surface that supports ρ6PBS-bound RNA (Fig. 5B), and substitutions of zinc-coordinating C72 and C85 likely disturb the ZBD structure. While we cannot exclude a possibility that ZBD substitutions may affect other steps of RNA synthesis or its coupling to translation (37, 38), our results support the notion of direct ρ/ZBD cooperation revealed by the RNA capture complex.
EC inhibition
Several major changes distinguish complex IV from the RNA capture complex. The density for NusGNTD is missing, and the bottom part of the uDNA duplex swings outwards to a position where it would sterically clash with NusGNTD (Fig. S11A), while the template strand is partly pulled back from ρ1PBS (Fig. S11B). The N-terminal part of the β’ clamp rotates away from dDNA, widening the primary channel by about 8 Å (Fig. 6A), β’lid rearranges (Fig. S11C), and β’SI3 and β’jaw pivot away from dDNA (Fig. S11D). Concurrently with rearrangements in nucleic acid-guiding elements, the tDNA acceptor nt is destabilized at the templating position (Fig. 6B), reminiscent of a paused bacterial EC (39) and an α-amanitin-stalled eukaryotic RNAPII (40).
Fig. 6. Inhibition.
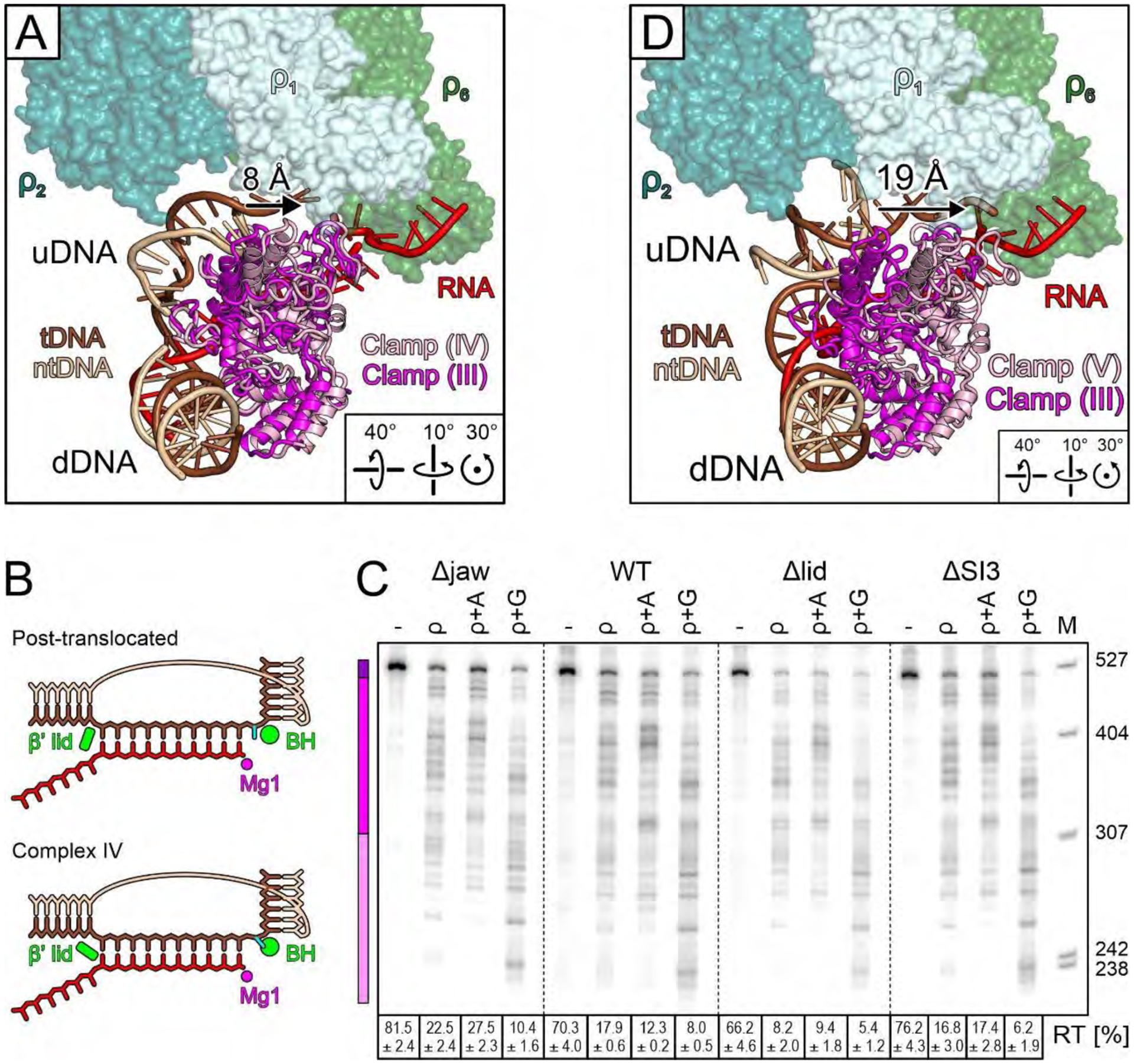
(A) Comparison of selected elements of the inhibited complex (regular colors) with the β’ clamp of the RNA capture complex (magenta), illustrating partial clamp opening (arrow). (B) tDNA is post-translocated in complexes I-III, but β’ lid moves and the +1 nucleotide is rotated out of the templating position in complex IV. Templating nt, cyan; BH, bridge helix; Mg1, catalytic magnesium ion. (C) Effects of deleting β’ jaw, lid, or SI3, alone or in the presence of NusA or NusG. Reactions were run on the same gel; dashed lines indicate positions where intervening lanes were removed. (D) Comparison of selected elements of the moribund complex (regular colors) with the β’ clamp of the RNA capture complex (magenta), illustrating dramatic clamp opening (arrow).
ρ-induced rearrangements of the lid, SI3, or jaw suggest that their removal may influence termination. To test this idea, we determined ρ effects on RNAPs lacking these elements. While the lid deletion increased termination more than twofold (P<0.001), as expected, deletions of the SI3 and jaw had minor effects (Fig. 6C), in apparent contradiction with our hypothesis. However, Δjaw and ΔSI3 enzymes are pause-insensitive and are thus expected to be strongly resistant to ρ. Our results show that decreased pausing (Fig. S10D) and increased susceptibility to ρ-induced allosteric changes (Fig. 6C) may cancel out, yielding near-WT termination. By comparison, the lid deletion does not alter elongation and its effects on ρ are direct. We stress that interpretation of these and other enzymes’ sensitivities to ρ necessitates evaluation of their responses to other signals that modulate elongation.
Complex IV, with a partially open clamp, lost NusGNTD and destabilized templating nt, represents a further step towards the ρ-induced RNAP inactivation. We thus termed it the inhibited complex.
EC inactivation
In complex V, RNAP is fully inactivated. The tip of the β’ clamp helices is displaced from the dDNA duplex by about 19 Å (Fig. 6D), while β’SI3 and β’jaw return to their positions in complex III, indicating that RNAP has lost its firm grip on dDNA. The rearrangements result in an opening of the primary channel (βgate loop E374 to β’clamp E162) from ~16 Å in complex III to ~30 Å in complex V. This opening is wide enough to allow escape of dDNA, which is further destabilized by a reorganization of β’rudder and β’switch 2 that guide nucleic acids near the active site in elongation-competent ECs, and by complete collapse of the lid (Fig. 7A). However, dDNA remains in place, held back by dramatic further rearrangements: the entire RNA:DNA hybrid swings into a pseudo-continuous helix with dDNA, displacing the RNA 3’-end about 35 Å from the active site (Fig. 7B), and shifting proximal uDNA back to its position in complex III. Complex V thus represents a trapped complex postulated by Nudler and colleagues (20). Remarkably, ρ achieves RNAP inactivation while remaining in an open state.
Fig. 7. Inactivation.
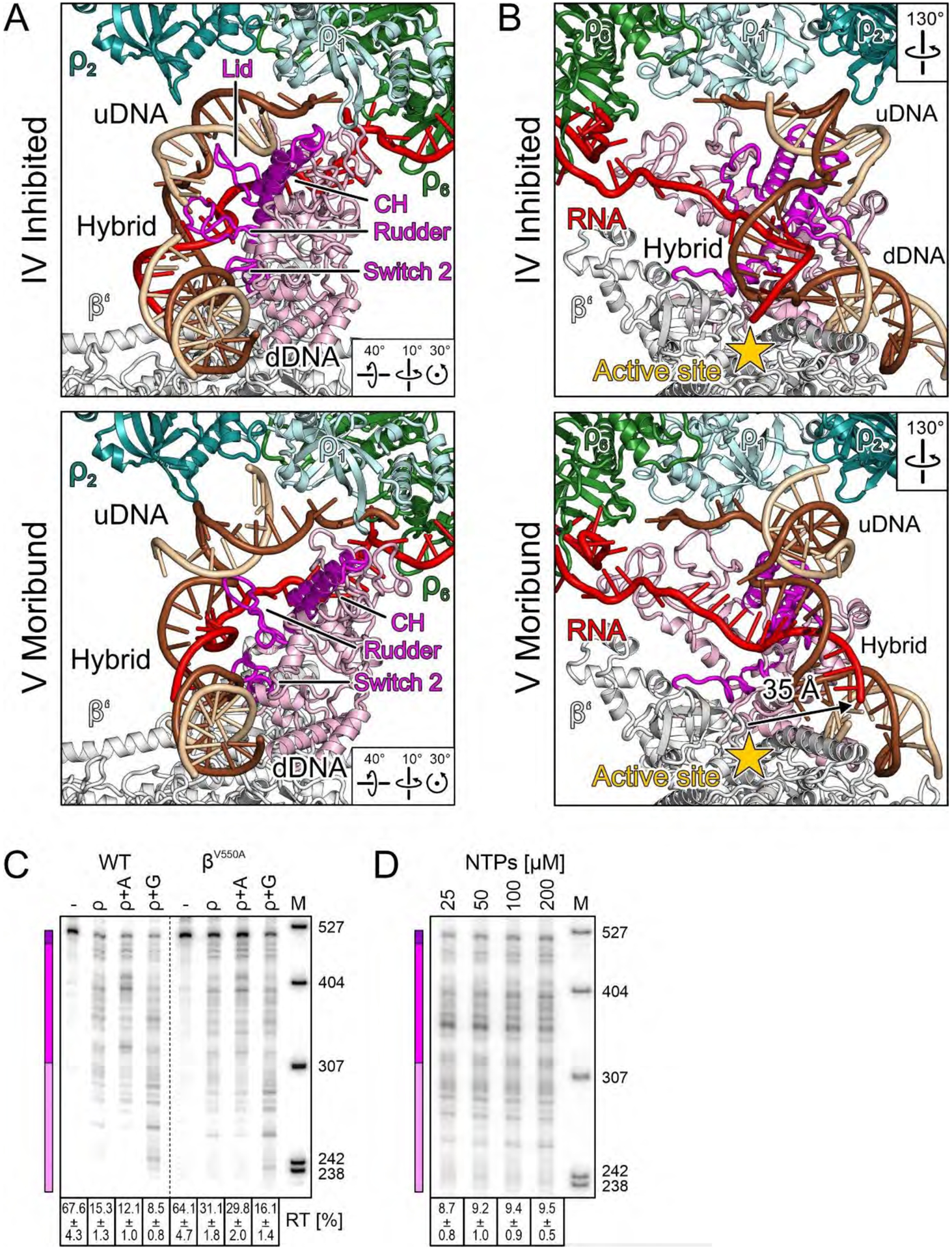
(A,B) Side-by-side comparison of selected elements in the inhibited complex (top) and in the moribund complex (bottom), highlighting movement of the β’ clamp helices (CH, magenta) and nucleic acid-guiding loops (lid/rudder/switch 2, magenta) (A), as well as repositioning of the hybrid and displacement of the RNA 3’-end from the active site (arrow) (B). (C) Pause-resistant βV550A substitution decreases ρ termination. Reactions were run on the same gel, and a dashed line indicates the splice position. (D) Effects of NTP concentration at λ tR1.
Our findings are at odds with the kinetic coupling model (8), which explains why ρ releases RNAP at pause sites and why fast RNAPs are resistant to termination. However, fast RNAPs do not display significantly increased pause-free rates (41), suggesting that their resistance to pausing, rather than faster rate of RNA synthesis, confers protection against ρ. In support of this idea, we found that a pause-resistant βV550A RNAP that is only marginally faster than the WT enzyme (46 vs. 36 nt/s) (41) was also resistant to ρ (31 % RT RNA as compared to 15 % for WT RNAP; P<0.0001; Fig. 7C). In startling contrast, altering the rate of elongation by titrating NTPs had little effect; when NTP concentrations were increased from 25 to 200 μM, a change that enhances the rate of elongation six-fold (42), termination by WT RNAP was decreased only ~1.1 times (Fig. 7D). We conclude that the RNAP propensity to undergo conformational changes associated with pausing determines its sensitivity to ρ.
Discussion
Our findings suggest a pathway for ρ-mediated EC disassembly in which RNAP and general transcription factors NusA and NusG play key roles (Fig. 8; Movie S2). We presume that ρ can passively traffic on an EC in an open configuration because the ring closure inhibitor bicyclomycin (31) does not alter early ρ occupancy (19). At a pause site, ρ engages the EC, contacting NusA, NusGNTD and several circularly arranged elements on RNAP, with NusA wedged between ρ1 and ρ6 (engagement complex). ρ1 locally melts uDNA with the help of NusGHL, and distal uDNA is directed towards β’ZBD, causing NusA to rotate underneath ρ6, preparing ρ6 for rut RNA binding (primed complex). The up-lifted ρ6PBS captures rut RNA and steps down onto β’ZBD, displacing distal uDNA (RNA capture complex); the nascent RNA that loops between ρ6PBS and RNAP may be guided into the open ring. By pressing on NusGNTD, the proximal uDNA duplex may facilitate NusGNTD detachment, initiating clamp opening and inhibiting tDNA translocation (inhibited complex). Upon further clamp opening, RNAP loses its grip on the nucleic acids, allowing the hybrid to dislodge from the active site (moribund complex).
Fig. 8. Model for an EC-dependent ρ-mediated termination pathway.
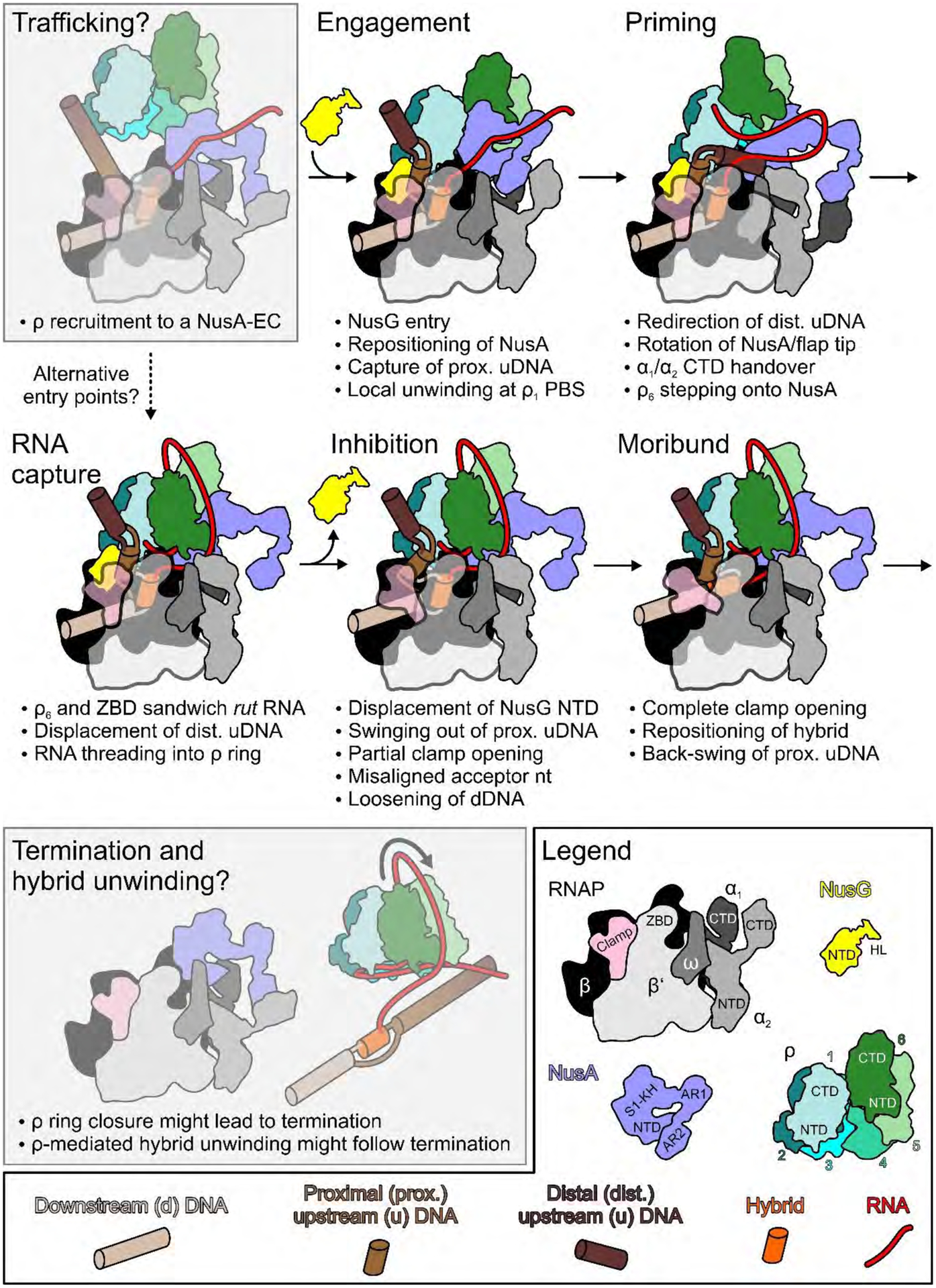
Trafficking and termination/hybrid unwinding correspond to hypothetical steps (behind semi-transparent gray boxes) preceding and following the stages resolved by cryoEM in this work. Legend on the lower right and bottom. Coloring as in structural figures except: DNA, upstream to downstream progressively lighter brown; hybrid, orange.
Notably, we also observe structures that represent intermediates between the RNA capture and inhibited complexes (IIIa; intermediate displacement of proximal uDNA and clamp opening; Fig. S1E) and between the inhibited and moribund complexes (IVa; intermediate clamp opening and hybrid displacement; Fig. S1I), which strongly support a continuous path from complex III to V. However, we recognize that some of our complexes may represent different modes by which ρ directly engages a paused EC (Fig. 8, dashed arrow). We note that other configurations, either representing extra steps in a continuous pathway or additional forms of ρ attack, likely exist.
Our ρ-EC preparation contained ADP-BeF3, rut RNA, and NusG, all of which support ring closure (13), yet ρ remains open throughout all stages imaged here. In fact, only the open ρ can realize all observed contacts to the EC and several ρPBSs are inaccessible to RNA. Thus, the ρ-EC conformation is incompatible with ring closure, preventing immediate termination upon ρ engagement. We envision that the moribund EC is only marginally stable, and will eventually allow ρ ring closure and subsequent ρ dissociation from the EC. Astonishingly, cryoEM has allowed us to capture the transient moribund state (Fig. S1J; Fig. 7A,B), possibly because we designed the nascent RNA to be just below the length sufficient to fill all ρPBSs. Loss of NusGNTD will facilitate ρ subunits linking up with the NusA-bound end during subsequent ring closure. With RNAP wide-open, upon ring closure ρ may detach with bound nucleic acids, followed by release of RNA from DNA (Fig. 8). Alternatively, ρ may translocate and release the stalled EC, either while remaining bound via a subset of contacts or after disengagement. Thus, our results do not exclude the possibility that ρ eventually closes and translocates the RNA.
Irrespective of its precise details, our model stands in stark contrast to the textbook model, in which ρ first engages the nascent RNA and uses its ATP-powered motor to translocate towards RNAP. Upon encounter, it was suggested that ρ might push RNAP forward (43) or pull RNA from the catalytic cleft (44). The latter mode of action is used by some spliceosomal RNA helicases to act from a distance (45). However, evidence for the direct role of translocase/helicase activity in EC dissociation by ρ is presently missing. Instead, observations that E. coli ρ can be replaced by phage T4 RNA:DNA helicase UvsW or RNaseH (6) argue that, although critical for cell viability, RNA:DNA unwinding can be uncoupled from transcription.
The textbook, RNA-dependent ρ recruitment could be utilized in some circumstances, but our results strongly argue against this mechanism representing the major physiological pathway of termination. Decades of in vitro experimentation have demonstrated that after ρ loads onto a perfect rut site, it can strip off any obstacle from RNA. However, in the cell, ρ has to terminate synthesis of all useless RNAs, whether or not they have rut sites (2), and appears to engage RNAP at the promoter (19). If ρ failed to bind to RNAP early on, it is certainly capable of binding to an exposed rut site, but this RNAP-independent targeting poses two major quandaries for ρ, which needs to (i) select RNAs that are still attached to RNAP and (ii) avoid being trapped on high-affinity RNAs. Our results show that ρ directly binds RNAP and captures RNA later, thereby selecting nascent transcripts from a vast pool of cellular RNAs.
Importantly, each step in our proposed pathway could serve as a potential checkpoint for regulation. As ρ in each of these states realizes similar types and extents of contacts to the EC, the pathway could be readily reversible, allowing ρ to probe the RNA sequence. If no rut site is available, the pathway may be halted prior to RNA capture. If ρ encounters a perfect rut, termination will ensue with a high probability whereas a sub-optimal rut may support termination with an intermediate likelihood. Likewise, if some of our structures represent independent attempts by ρ to terminate, rather than a continuous pathway, each state will have different probability to lead to termination. Both scenarios also provide an explanation for ρ terminating throughout a window rather than at a specific site, as the process may be interrupted and reversed in every case, necessitating several attempts of ρ at termination.
The proposed pathway also provides insights into regulation by RNAP-associated factors. For example, our hierarchical clustering analysis showed that while the engagement and the RNA capture complexes can form in the absence of NusG (complexes IΔNusG and IIIΔNusG), we do not find particles conforming to the primed complex lacking NusG (Fig. S4). Thus, NusGNTD may stabilize additional intermediate steps and influence the pathway reversibility. As NusA seems to initially prevent RNA capture by ρ (Fig. 1), there is a regulatory potential via a particular RNA region exhibiting differential affinities to NusA or ρ PBS. Structural comparisons reveal how transcription anti-termination complexes (23, 24) or a closely trailing ribosome (37, 38) can fend off ρ by erecting physical barriers (Fig. S12).
NusG also modulates ρ-mediated termination via its CTD, by promoting ring closure on suboptimal RNAs (13, 18) and mutations at the crystallographically-defined NusGCTD-ρCTD interface (13) lead to termination defects in vivo (34, 46); NusGCTD sequestration by NusE (S10) in anti-termination complexes (23, 24) and a coupled ribosome (37, 38) is thought to underpin their resistance to ρ. Surprisingly, none of our refined maps revealed density for NusGCTD. In the binary complex, NusGCTD appears to capture and stabilize the dynamic ρ ring in a closed state (13). In our structures, the ring is held open by multiple interactions with EC components, likely inhibiting stable NusGCTD binding. We thus can only speculate how NusGCTD could affect the suggested pathway. It is possible that, via transient contacts not captured here, NusGCTD (i) mediates transitions between ρ-EC states or (ii) serves to retain NusG in the complex following the clamp opening (Fig. 6A,D) and perhaps promotes subsequent ring closure.
Taken together, the available data clearly support a model in which ρ hitchhikes on RNAP and subsequently traps it in a moribund state (20). This contrasts with “torpedo” termination mechanisms (47, 48), in which exoribonucleases engage the upstream RNA after cleavage and must catch up with the EC for timely dissociation. Slowing RNAPII down upon entry into a polyadenylation site (47) promotes recruitment of cleavage factors (49) and subsequent EC capture by “torpedo” exonucleases (50). Yet, ample support for a hybrid model that incorporates allosteric effects also exists (47).
All transcription termination mechanisms must trigger dissociation of a stable EC. While the nucleic acid signals and protein factors that elicit termination differ across life, the structures of the ECs are remarkably similar, suggesting that termination signals may act upon analogous key elements, such as the clamp and the RNA:DNA hybrid. The exact sequence of events during EC dissociation remain to be determined, and may differ for different termination scenarios, but there is evidence that allosteric effects contribute to termination. In bacteria, termination of most genes is triggered by formation of an RNA hairpin. Among different models of hairpin-induced termination (9), one posits that the hairpin allosterically inactivates the EC (51), acting similarly to ρ in our structures. Furthermore, clamp opening for DNA release during intrinsic termination (52) seems to parallel the ρ-mediated mechanism detailed here. In eukaryotes, an RNA/DNA helicase Sen1, a functional analog of ρ, releases RNAPII from non-coding RNAs and must interact with RNAPII to elicit efficient termination (53) via a long-lived inactive EC intermediate (54). Thus, a sequential trap/release strategy emerges as a ubiquitous mechanism of termination.
Supplementary Material
Acknowledgements:
We thank Sonia Agarwal, Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India, for help with genetic screening. We acknowledge access to electron microscopic equipment at the core facility BioSupraMol of Freie Universität Berlin, supported through grants from the Deutsche Forschungsgemeinschaft (HA 2549/15-2), and from the Deutsche Forschungsgemeinschaft and the state of Berlin for large equipment according to Art. 91b GG (INST 335/588-1 FUGG, INST 335/589-1 FUGG, INST 335/590-1 FUGG), and at the core facility operated by the Microscopy & Cryo-Electron Microscopy service group at the Max Planck Institute for Molecular Genetics, Berlin. We are grateful for access to high-performance computing resources at the Zuse Institut Berlin.
Funding: This work was supported by grants from the Deutsche Forschungsgemeinschaft (RTG 2473-1 and WA 1126/11-1 to M.C.W.); the Bundesministerium für Bildung und Forschung (01DQ20006 to M.C.W.); the Indian Council of Medical Research (AMR/INDO/GER/219/2019-ECD-II to R.S.); Department of Biotechnology, Government of India (BT/PR27969/BRB/10/1662/2018 to R. S.); the National Institutes of Health (GM067153 to I.A.) and the Sigrid Jusélius Foundation to G.A.B.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: CryoEM data have been deposited in the Electron Microscopy Data Bank (http://www.emdatabank.org) with accession codes EMD-11087 (complex I), EMD-11088 (complex II), EMD-11089 (complex III), EMD-11090 (complex IV), EMD-11091 (complex V), EMD-11722 (complex IΔNusG), EMD-11723 (complex IIIΔNusG), EMD-11724 (complex IIIa) and EMD-11725 (complex IVa). Structure coordinates have been deposited in the RCSB Protein Data Bank (https://www.rcsb.org/) with accession codes 6Z9P (complex I), 6Z9Q (complex II), 6Z9R (complex III), 6Z9S (complex IV), and 6Z9T (complex V), 7ADB (complex IΔNusG), 7ADC (complex IIIΔNusG), 7ADD (complex IIIa) and 7ADE (complex IVa). CryoEM data and coordinates will be released upon publication. All other data are available in the main text or the supplementary materials.
Supplementary Materials:
Materials and Methods
Figures S1-S12
Tables S1-S5
Movies S1-S2
References and Notes:
- 1.Roberts JW, Termination factor for RNA synthesis. Nature 224, 1168–1174 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Peters JM et al. , Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26, 2621–2633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardinale CJ et al. , Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedlyarova N et al. , sRNA-Mediated Control of Transcription Termination in E. coli. Cell 167, 111–121 e113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E, Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leela JK, Syeda AH, Anupama K, Gowrishankar J, Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 110, 258–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomsen ND, Lawson MR, Witkowsky LB, Qu S, Berger JM, Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor. Proc Natl Acad Sci U S A 113, E7691–E7700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin DJ, Burgess RR, Richardson JP, Gross CA, Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A 89, 1453–1457 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JM, Vangeloff AD, Landick R, Bacterial transcription terminators: the RNA 3’-end chronicles. J Mol Biol 412, 793–813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang WH, Platt T, Reeder RH, Escherichia coli rho factor induces release of yeast RNA polymerase II but not polymerase I or III. Proc Natl Acad Sci U S A 95, 4900–4905 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasman Z, von Hippel PH, Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry 39, 5573–5585 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Schwartz A, Margeat E, Rahmouni AR, Boudvillain M, Transcription termination factor rho can displace streptavidin from biotinylated RNA. J Biol Chem 282, 31469–31476 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Lawson MR et al. , Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein. Mol Cell 71, 911–922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Kohn H, Widger WR, Mutations in the rho transcription termination factor that affect RNA tracking. J Biol Chem 277, 30023–30030 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Schmidt MC, Chamberlin MJ, Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J Biol Chem 259, 15000–15002 (1984). [PubMed] [Google Scholar]
- 16.Lau LF, Roberts JW, Rho-dependent transcription termination at lambda R1 requires upstream sequences. J Biol Chem 260, 574–584 (1985). [PubMed] [Google Scholar]
- 17.Burns CM, Richardson LV, Richardson JP, Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol 278, 307–316 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Valabhoju V, Agrawal S, Sen R, Molecular Basis of NusG-mediated Regulation of Rho-dependent Transcription Termination in Bacteria. J Biol Chem 291, 22386–22403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney RA et al. , Regulator trafficking on bacterial transcription units in vivo. Mol Cell 33, 97–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epshtein V, Dutta D, Wade J, Nudler E, An allosteric mechanism of Rho-dependent transcription termination. Nature 463, 245–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loerke J, Giesebrecht J, Spahn CM, Multiparticle cryo-EM of ribosomes. Methods Enzymol 483, 161–177 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Guo X et al. , Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell 69, 816–827 e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YH et al. , Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis. Mol Cell, (2020). [DOI] [PubMed] [Google Scholar]
- 24.Krupp F et al. , Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor. Mol Cell 74, 143–157 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Kang JY et al. , Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife 6, e25478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang JY et al. , Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell 173, 1650–1662 e1614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweimer K et al. , NusA interaction with the alpha subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure 19, 945–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qayyum MZ, Dey D, Sen R, Transcription Elongation Factor NusA Is a General Antagonist of Rho-dependent Termination in Escherichia coli. J Biol Chem 291, 8090–8108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson JP, Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem 257, 5760–5766 (1982). [PubMed] [Google Scholar]
- 30.Skordalakes E, Berger JM, Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Lawson MR, Dyer K, Berger JM, Ligand-induced and small-molecule control of substrate loading in a hexameric helicase. Proc Natl Acad Sci U S A 113, 13714–13719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R, Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391, 341–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kainz M, Gourse RL, The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J Mol Biol 284, 1379–1390 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Chalissery J, Banerjee S, Bandey I, Sen R, Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex. J Mol Biol 371, 855–872 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shashni R, Qayyum MZ, Vishalini V, Dey D, Sen R, Redundancy of primary RNA-binding functions of the bacterial transcription terminator Rho. Nucleic Acids Res 42, 9677–9690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King RA, Banik-Maiti S, Jin DJ, Weisberg RA, Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell 87, 893–903 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Wang C et al. , Structural basis of transcription-translation coupling. Science, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster MW et al. , Structural basis of transcription-translation coupling and collision in bacteria. Science, (2020). [DOI] [PubMed] [Google Scholar]
- 39.Weixlbaumer A, Leon K, Landick R, Darst SA, Structural basis of transcriptional pausing in bacteria. Cell 152, 431–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brueckner F, Cramer P, Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 15, 811–818 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Malinen AM et al. , CBR antimicrobials alter coupling between the bridge helix and the beta subunit in RNA polymerase. Nat Commun 5, 3408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I, Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res 35, 5694–5705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JS, Roberts JW, Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci U S A 103, 4870–4875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson JP, Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta 1577, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Semlow DR, Blanco MR, Walter NG, Staley JP, Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell 164, 985–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miwa Y, Horiguchi T, Shigesada K, Structural and functional dissections of transcription termination factor rho by random mutagenesis. J Mol Biol 254, 815–837 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Eaton JD, Francis L, Davidson L, West S, A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev 34, 132–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikova M et al. , The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes. EMBO J 39, e102500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parua PK et al. , A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature 558, 460–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cortazar MA et al. , Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism. Mol Cell 76, 896–908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E, An allosteric path to transcription termination. Mol Cell 28, 991–1001 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Bellecourt MJ, Ray-Soni A, Harwig A, Mooney RA, Landick R, RNA Polymerase Clamp Movement Aids Dissociation from DNA but Is Not Required for RNA Release at Intrinsic Terminators. J Mol Biol 431, 696–713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han Z et al. , Termination of non-coding transcription in yeast relies on both an RNA Pol II CTD interaction domain and a CTD-mimicking region in Sen1. EMBO J 39, e101548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Han Z, Libri D, Porrua O, Strick TR, Single-molecule characterization of extrinsic transcription termination by Sen1 helicase. Nat Commun 10, 1545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svetlov V, Artsimovitch I, Purification of bacterial RNA polymerase: tools and protocols. Methods Mol Biol 1276, 13–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baba T et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Said N et al. , Structural basis for lambdaN-dependent processive transcription antitermination. Nat Microbiol 2, 17062 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardone G, Heymann JB, Steven AC, One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol 184, 226–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asarnow D, Palovcak E, Cheng Y. (2019). UCSF pyem v0.5. Zenodo 10.5281/zenodo.3576630. [DOI]
- 63.Scheres SH, Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol 189, 114–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grant T, Rohou A, Grigorieff N, cisTEM, user-friendly software for single-particle image processing. Elife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muteeb G, Sen R, Random mutagenesis using a mutator strain. Methods Mol Biol 634, 411–419 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Cheeran A et al. , Escherichia coli RNA polymerase mutations located near the upstream edge of an RNA:DNA hybrid and the beginning of the RNA-exit channel are defective for transcription antitermination by the N protein from lambdoid phage H-19B. J Mol Biol 352, 28–43 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Artsimovitch I, Landick R, The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Gogol EP, Seifried SE, von Hippel PH, Structure and assembly of the Escherichia coli transcription termination factor rho and its interaction with RNA. I. Cryoelectron microscopic studies. J Mol Biol 221, 1127–1138 (1991). [DOI] [PubMed] [Google Scholar]
- 71.Hu K, Artsimovitch I, A Screen for rfaH Suppressors Reveals a Key Role for a Connector Region of Termination Factor Rho. mBio 8, e00753–00717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ederth J, Artsimovitch I, Isaksson LA, Landick R, The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem 277, 37456–37463 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Toulokhonov I, Landick R, The role of the lid element in transcription by E. coli RNA polymerase. J Mol Biol 361, 644–658 (2006). [DOI] [PubMed] [Google Scholar]
- 74.O’Reilly FJ et al. , In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richardson JP, Preventing the synthesis of unused transcripts by Rho factor. Cell 64, 1047–1049 (1991). [DOI] [PubMed] [Google Scholar]
- 76.Chen VB, Wedell JR, Wenger RK, Ulrich EL, Markley JL, MolProbity for the masses-of data. Journal of biomolecular NMR 63, 77–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strauss M et al. , Transcription is regulated by NusA:NusG interaction. Nucleic Acids Res 44, 5971–5982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vassylyeva MN et al. , The carboxy-terminal coiled-coil of the RNA polymerase beta’-subunit is the main binding site for Gre factors. EMBO Rep 8, 1038–1043 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N, Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci U S A 109, 3353–3358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Artsimovitch I, Svetlov V, Murakami KS, Landick R, Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem 278, 12344–12355 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Ruff EF et al. , E. coli RNA Polymerase Determinants of Open Complex Lifetime and Structure. J Mol Biol 427, 2435–2450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turtola M, Belogurov GA, NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife 5, e18096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheeran A, Kolli NR, Sen R, The site of action of the antiterminator protein N from the lambdoid phage H-19B. J Biol Chem 282, 30997–31007 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


